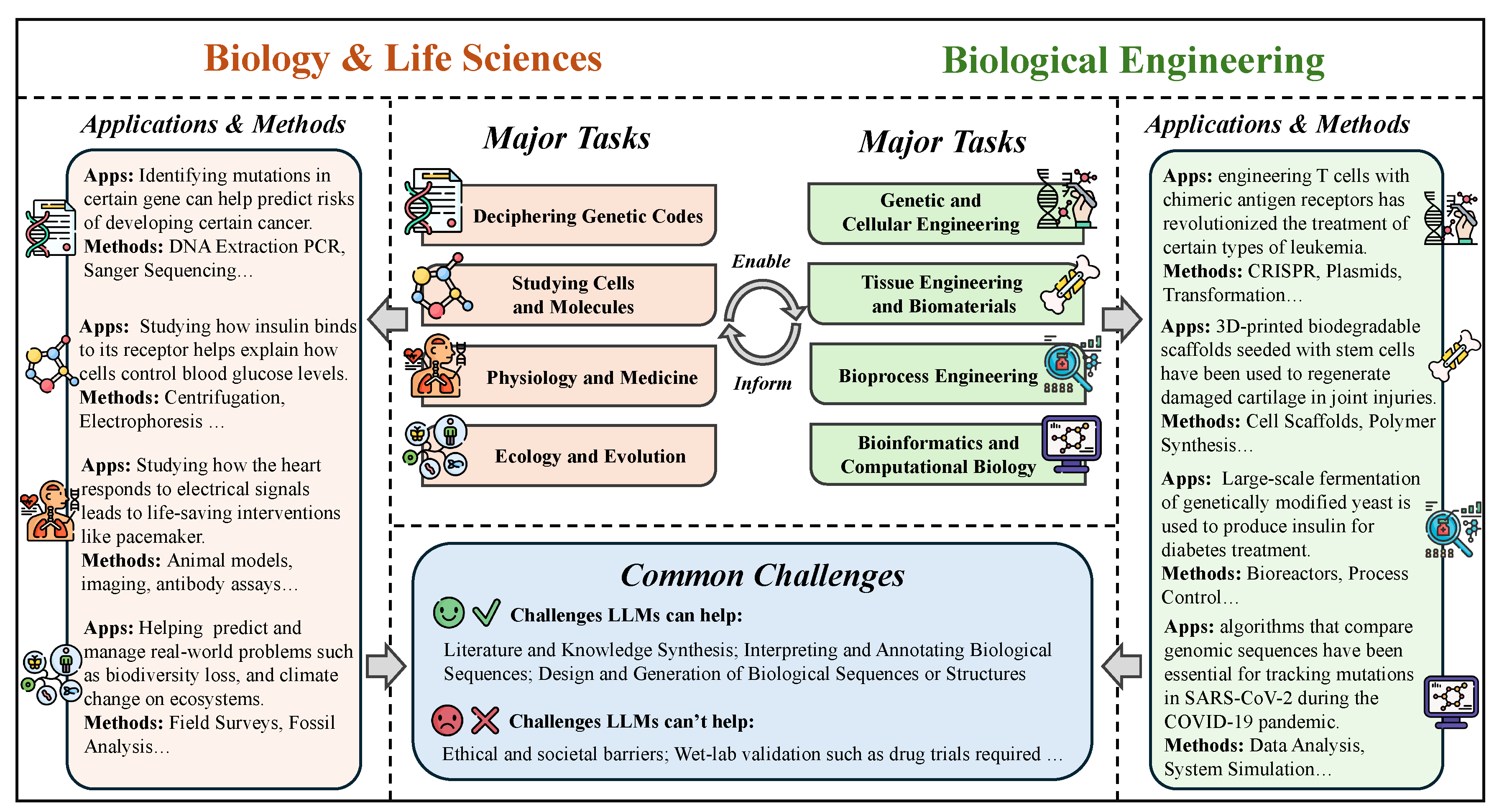
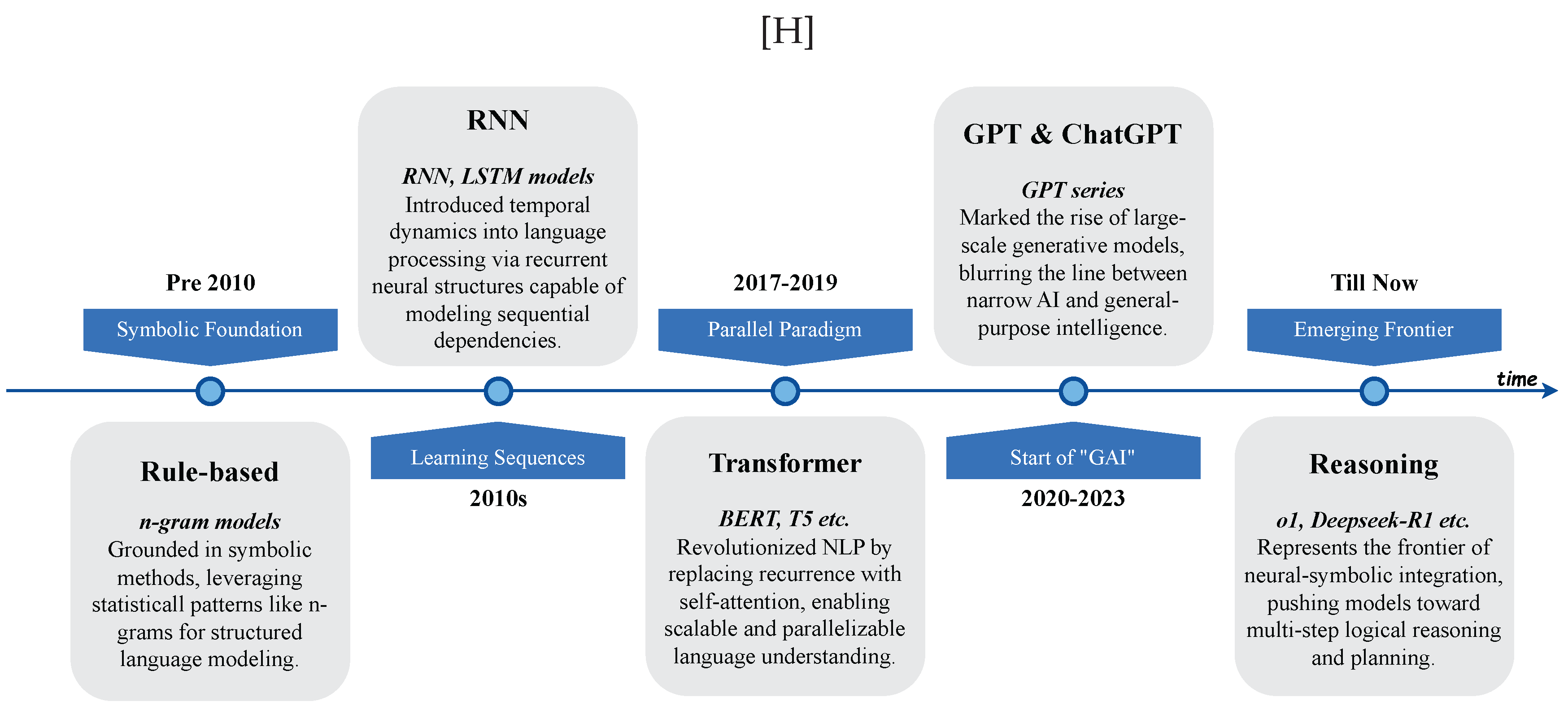
In this chapter, we chart how LLMs are utilized across science and engineering. The chapter spans mathematics; physics and mechanical engineering; chemistry and chemical engineering; life sciences and bioengineering; earth sciences and civil engineering; and computer science and electrical engineering. We open with mathematics—proof support, theoretical exploration and pattern discovery, math education, and targeted benchmarks. In physics and mechanical engineering, we cover documentation-centric tasks, design ideation and parametric drafting, simulation-aware and modeling interfaces, multimodal lab and experiment interpretation, and interactive reasoning, followed by domain-specific evaluations and a look at opportunities and limits. In chemistry and chemical engineering, we examine molecular structure and reaction reasoning, property prediction, materials optimization, test/assay mapping, property-oriented molecular design, and reaction-data knowledge organization, then compare benchmark suites. In life sciences and bioengineering section, we include genomic sequence analysis, clinical structured-data integration, biomedical reasoning and understanding, and hybrid outcome prediction, with emphasis on validation standards. In earth sciences and civil engineering section, we review geospatial and environmental data tasks, simulation and physical modeling, document workflows, monitoring and predictive maintenance, plus design/planning tasks, again with benchmarks. We close with computer science and electrical engineering: code generation and debugging, large-codebase analysis, hardware description language code generation, functional verification, and high-level synthesis, capped by purpose-built benchmarks and a final discussion of impacts and open challenges.
5.2. Physics and Mechanical Engineering
5.2.1. Overview
5.2.1.1 Introduction to Physics
Physics is a natural science that investigates the fundamental principles governing matter, energy, and their interactions through experimental observation and theoretical modeling [
702]. It spans from the smallest subatomic particles to the largest cosmic structures, aiming to establish predictive, explanatory, and unifying frameworks for natural phenomena [
703]. As the most fundamental natural science, physics provides conceptual foundations and methodological tools that support other sciences and engineering disciplines [
704].
Simply put, physics is the science of understanding how the world works. It seeks to explain everyday phenomena, such as why apples fall, why lights turn on, or why we can see stars [
702]. It not only provides explanations but also empowers us to harness these laws to develop new technologies [
705].
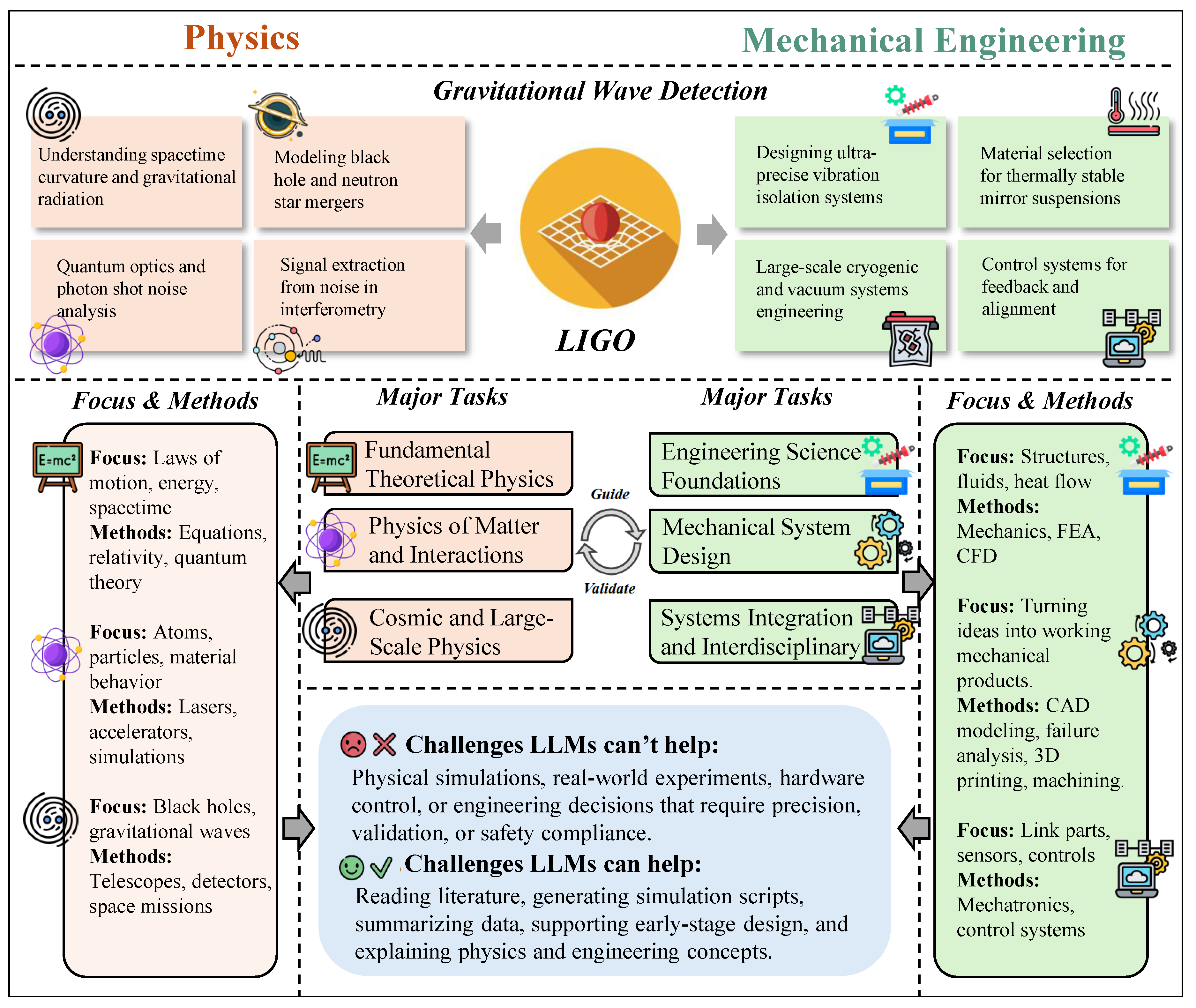
For example, the detection of gravitational waves marked one of the most significant breakthroughs in 21st-century physics [
706]. First predicted by Einstein’s theory of general relativity, gravitational waves had eluded direct observation for a century due to their extremely weak nature [
707]. In 2015, the LIGO interferometer in the United States successfully captured signals produced by the collision of two black holes [
706]. This discovery not only confirmed theoretical predictions but also launched a new field—gravitational wave astronomy [
708]—with far-reaching impacts on astrophysics, cosmology, and quantum gravity [
709,
710].
The discipline of physics is vast and typically organized into three core domains, each addressing a class of natural phenomena and associated methodologies [
711]:
Fundamental Theoretical Physics. This domain focuses on uncovering the basic laws of nature and forms the theoretical foundation of the entire field of physics [
712]. It encompasses classical mechanics, electromagnetism, thermodynamics, statistical mechanics, quantum mechanics, and relativity [
712]. The scope ranges from macroscopic motion (e.g., acceleration, vibration, and waves) to the behavior of subatomic particles (e.g., electron transitions, spin, self-consistent fields), and includes the structure of spacetime under extreme conditions such as near black holes [
713]. Researchers in this domain employ abstract mathematical tools such as differential equations, Lagrangian and Hamiltonian mechanics, and group theory to construct theoretical models and derive predictions [
714]. These models not only guide experimental physics but also provide essential frameworks for engineering applications [
715].
Physics of Matter and Interactions. This domain explores the microscopic structure of matter, the interaction mechanisms across different scales, and how these determine macroscopic material properties [
716]. Major subfields include condensed matter physics, atomic physics, molecular physics, nuclear physics, and particle physics [
717,
718]. Key research topics cover crystal structure, electronic band theory, spin and magnetism, superconductivity, quantum phase transitions, and the classification and interaction of elementary particles [
716,
718]. Common methodologies include first-principles calculations, quantum statistical modeling, and large-scale experiments involving synchrotron radiation, laser spectroscopy, or high-energy accelerators [
719]. The findings from this domain have led to breakthroughs in semiconductor devices, materials development, quantum computing, and energy systems, driving significant technological innovation [
720,
721].
Cosmic and Large-Scale Physics. This domain addresses phenomena at scales far beyond laboratory conditions and includes astrophysics, cosmology, and plasma physics [
722]. It investigates topics such as stellar evolution, galactic dynamics, gravitational wave propagation, early-universe inflation, and the nature of dark matter and dark energy [
723]. In parallel, plasma physics explores phenomena like solar wind, magnetospheric dynamics, and the behavior of ionized matter in space environments [
724]. Research in this domain typically relies on astronomical observations (e.g., telescopes, gravitational wave detectors, space missions), theoretical models, and large-scale simulations, forming a triad of “observation + simulation + theory” [
725].
Together, these three domains constitute the intellectual architecture of modern physics [
726]. From particle collisions in ground-based accelerators to observations of distant galaxies, physics consistently strives to understand the most fundamental laws of nature [
727]. With the emergence of artificial intelligence, high-performance computing, and advanced instrumentation, the research landscape of physics continues to expand—becoming increasingly intelligent, interdisciplinary, and precise [
728].
Figure 15.
The relationships between major research tasks between physics and mechanical engineering.
Figure 15.
The relationships between major research tasks between physics and mechanical engineering.
5.2.1.2 Introduction to Mechanical Engineering
Mechanical engineering is an applied discipline that focuses on the design, analysis, manufacturing, and control of mechanical systems driven by the principles of force, energy, and motion [
729]. It integrates engineering mechanics, thermofluid sciences, materials engineering, control theory, and computational tools to solve problems across a wide range of industries. As a foundational engineering field, mechanical engineering provides the backbone for technological advancement in transportation, energy, robotics, aerospace, and biomedical systems [
730].
In simple terms, mechanical engineering is the science and craft of making things move and work reliably. From engines and turbines to robots and surgical devices, it turns physical principles into functional products through design and manufacturing [
730].
For example, the construction of the LIGO gravitational wave observatory represents not only a milestone in fundamental physics but also a triumph of mechanical engineering. LIGO’s ultrahigh-vacuum interferometers required vibration isolation at nanometer precision, thermally stable mirror suspensions, and large-scale structural systems integrated with active control. These engineering feats enabled the detection of gravitational waves, a task demanding extreme precision in structure, thermal regulation, and dynamic stability [
706].
The field of mechanical engineering is vast and is typically categorized into three core domains, each covering a range of methodologies and applications:
Engineering Science Foundations. This domain forms the analytical and physical core of mechanical engineering. It encompasses:
Mechanics: including statics, dynamics, solid mechanics, and continuum mechanics, used to model the deformation, motion, and failure of structures [
731].
Thermal and fluid sciences: covering heat conduction, convection, fluid dynamics, thermodynamics, and phase-change phenomena [
732].
Systems and control: involving system dynamics, feedback control theory, and mechatronic integration [
733]. These fundamentals are implemented via tools such as finite element analysis (FEA), computational fluid dynamics (CFD), and system modeling platforms like Simulink and Modelica [
734,
735].
Mechanical System Design and Manufacturing. This domain addresses how ideas become real-world engineered products. It includes:
Mechanical design: CAD modeling, mechanism design, stress analysis, failure prediction, and design optimization [
736].
Manufacturing: traditional subtractive methods (e.g., milling, turning), additive manufacturing (3D printing), surface finishing, and process planning [
737].
Smart manufacturing and Industry 4.0: integration of sensors, data analytics, automation, and cyber-physical systems to create responsive and intelligent production environments [
738]. These technologies bridge the gap between virtual design and physical realization.
Systems Integration and Interdisciplinary Applications. Modern mechanical systems are often multi-functional and cross-disciplinary. This domain focuses on:
Robotics and mechatronics: combining mechanics, electronics, and computing to build intelligent machines [
739].
Energy and thermal systems: engines, fuel cells, solar collectors, HVAC systems, and sustainable energy technologies [
740].
Biomedical and bioinspired systems: development of prosthetics, surgical tools, and biomechanical simulations [
741].
Multiphysics modeling and digital twins: simulation of systems involving coupled fields (thermal, fluidic, mechanical, electrical) and virtual prototyping [
742]. This integration-driven domain reflects mechanical engineering’s evolution toward intelligent, efficient, and adaptive systems. Together, these three domains define the scope of modern mechanical engineering. From high-speed trains and wind turbines to nanomechanical actuators and wearable exoskeletons, mechanical engineers shape the physical world with ever-growing precision and complexity [
729,
730].
5.2.1.3 Current Challenges
Physics and mechanical engineering are closely interwoven disciplines that form the foundation for understanding and shaping the material and technological world. Physics seeks to uncover the fundamental laws of nature that govern matter, motion, energy, and forces, while mechanical engineering applies these principles to design, optimize, and control systems that power modern life. Together, they enable critical innovations across transportation, energy, manufacturing, healthcare, and space exploration. These disciplines are indispensable for solving complex, cross-scale challenges such as energy efficiency, automation, sustainable mobility, and precision instrumentation. Despite rapid progress in theoretical modeling, simulation, and intelligent design tools, both fields continue to grapple with the intricacies of nonlinear dynamics, multiphysics coupling, and real-world uncertainties in physical systems.
Still Hard with LLMs: The Tough Problems.
Complexity of Multiphysics Coupling and Governing Equations. Physical and mechanical systems are often governed by a series of highly coupled partial differential equations (PDEs), involving nonlinear dynamics, continuum mechanics, thermodynamics, electromagnetism, and quantum interactions [
734,
743]. Solving such systems requires professional numerical solvers, high-fidelity discretization techniques, and physics-informed modeling assumptions. Although LLMs can retrieve relevant equations or suggest approximate forms, they are incapable of deriving physical laws, ensuring conservation principles, or performing accurate numerical simulations.
Simulation Accuracy and Model Calibration. Accurate mechanical design and physical predictions typically rely on high-fidelity simulations such as finite element analysis (FEA), computational fluid dynamics (CFD), or multiphysics modeling [
744,
745]. These simulations demand precise geometry input, boundary conditions, material models, and experimental validation. LLMs may assist in interpreting simulation reports or proposing modeling strategies, but they lack the resolution, numerical rigor, and feedback integration necessary to execute or validate such models.
Experimental Prototyping and Hardware Integration. Engineering innovations ultimately require validation through physical experiments—building prototypes, tuning actuators, installing sensors, and measuring performance under dynamic conditions [
746,
747]. These tasks depend on laboratory facilities, fabrication tools, and hands-on experimentation, all of which are beyond the operational scope of LLMs. While LLMs can help generate test plans or documentation, they cannot replace real-world testing or iterative hardware development.
Materials and Manufacturing Constraints. Real-world engineering designs must account for constraints such as thermal stress, fatigue life, manufacturability, and cost-efficiency [
748]. Addressing these challenges often relies on materials testing, manufacturing standards, and domain experience in processes like welding, casting, and additive manufacturing. LLMs lack access to real-time physical data and material behavior, and thus cannot support tradeoff decisions in design or production.
Ethical, Safety, and Regulatory Considerations. From biomedical devices to autonomous systems, mechanical engineers must weigh ethical impacts, user safety, and legal compliance [
749]. Although LLMs can summarize policies or regulatory codes, they are not equipped to make decisions involving responsibility, risk evaluation, or normative judgment—elements essential for deploying certified, real-world systems.
Easier with LLMs: The Parts That Move.
Although current LLMs remain limited in core tasks such as physical modeling and experimental validation, they have shown growing potential in assisting a variety of supporting tasks in physics and mechanical engineering—particularly in knowledge integration, document drafting, design ideation, and educational support:
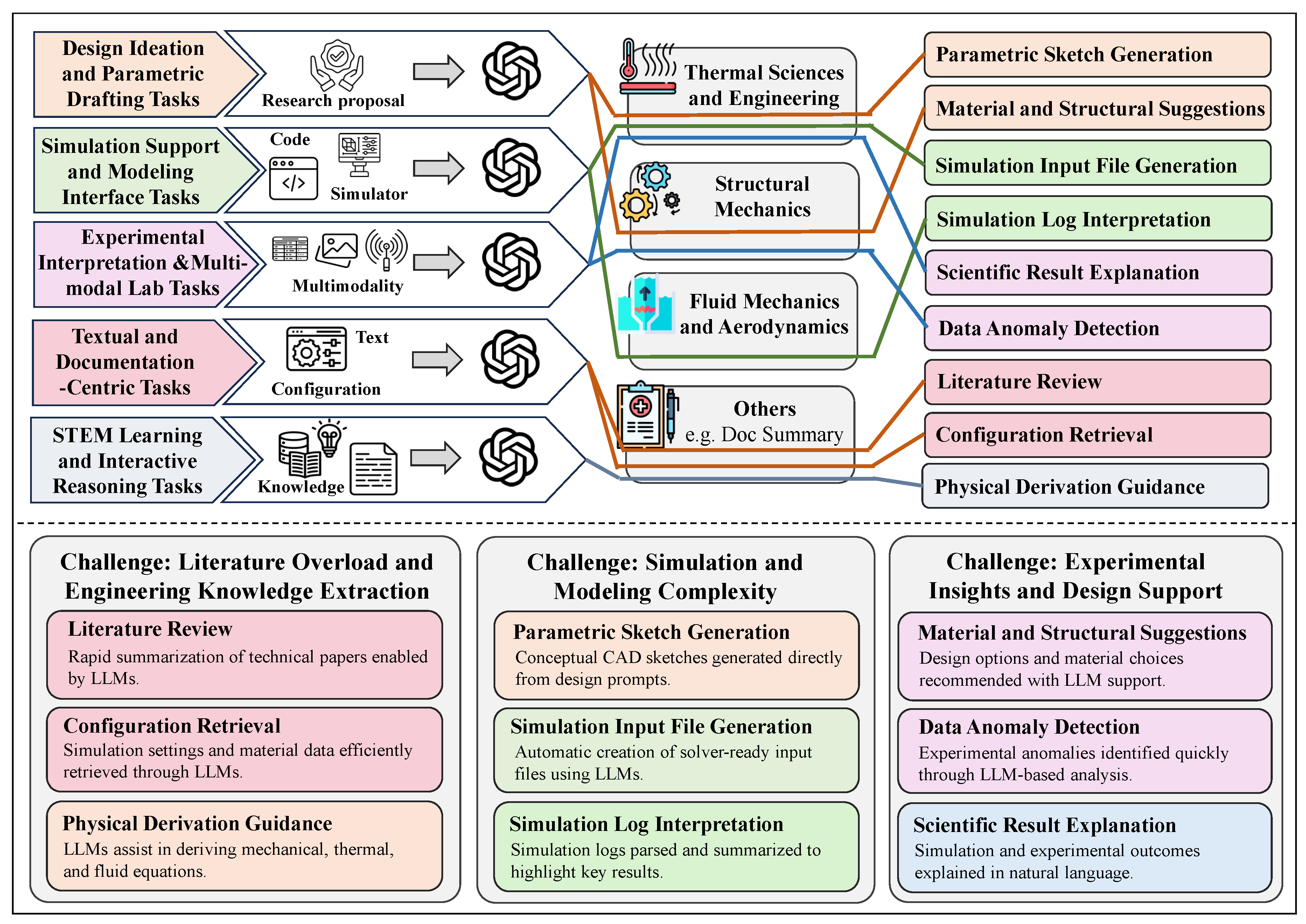
Literature Review and Standards Lookup. Both disciplines rely heavily on technical documents such as material handbooks, design standards, experimental protocols, and scientific publications. LLMs can significantly accelerate the literature review process by extracting key information about theoretical models, experimental conditions, or engineering parameters. For instance, an engineer could use an LLM to compare different welding codes, retrieve thermal fatigue limits of materials, or summarize applications of a specific mechanical model [
750,
751].
Assisting with Simulation and Test Report Interpretation. In simulations such as finite element analysis (FEA), computational fluid dynamics (CFD), or structural testing, LLMs can help parse simulation logs, identify setup issues, or generate summaries of experimental findings. When integrated with domain-specific tools, LLMs may even assist in generating simulation input files, interpreting outliers in results, or recommending appropriate post-processing techniques [
752,
753].
Supporting Conceptual Design and Parametric Exploration. During early-stage mechanical design or material selection, LLMs can suggest structural concepts, propose parameter combinations, or retrieve examples of similar engineering cases. For instance, given a prompt like “design a spring for high-temperature fatigue conditions,” the model might generate candidate materials, geometric options, and common failure modes [
754,
755].
Engineering Education and Learning Support. Education in physics and mechanical engineering involves both theoretical understanding and hands-on application. LLMs can generate step-by-step derivations, support simulation-based exercises, or simulate simple lab setups (e.g., free fall, heat conduction, beam deflection). They can also assist with terminology explanation or provide example problems to enhance interactive and self-guided learning [
756,
757].
In summary, while physical modeling, engineering intuition, and experimental testing remain essential in physics and mechanical engineering, LLMs are emerging as effective tools for information synthesis, design reasoning, documentation, and education. The future of these disciplines may be shaped by deep integration between LLMs, simulation platforms, engineering software, and laboratory systems—paving the way from textual reasoning to intelligent system collaboration.
Figure 16.
The pipelines of physics and mechanical engineering.
Figure 16.
The pipelines of physics and mechanical engineering.
5.2.1.4 Taxonomy
Research in physics and mechanical engineering spans a broad spectrum of problems, from modeling fundamental laws of nature to designing and validating engineered systems. With the rapid development of LLMs, many of these tasks are being redefined through human-AI collaboration, automation, and intelligent assistance. Traditionally, physics and mechanical engineering are divided along disciplinary lines—e.g., thermodynamics, solid mechanics, control systems—but from the perspective of LLM-based systems, it is more productive to reorganize tasks based on their computational characteristics and data modalities.
This functional, task-driven taxonomy helps distinguish where LLMs can take on primary responsibilities, where they act in a supporting role, and where traditional numerical methods and expert reasoning remain indispensable. Based on this perspective, we propose five major categories that capture the current landscape of LLM-integrated research in physics and mechanical engineering:
Textual and Documentation-Centric Tasks. LLMs are particularly effective in processing technical documents, engineering standards, lab reports, and scientific literature. For instance, Polverini and Gregorcic demonstrated how LLMs can support physics education by extracting and explaining key information from conceptual texts [
758], while Harle et al. highlighted their use in organizing and generating instructional materials for engineering curricula [
759].
Design Ideation and Parametric Drafting Tasks. In early-stage design and manufacturing workflows, LLMs can transform natural language prompts into CAD sketches, material recommendations, and parameter ranges. The MIT GenAI group systematically evaluated the capabilities of LLMs across the entire design-manufacture pipeline [
760], and Wu et al. introduced CadVLM, a multimodal model that translates linguistic input into parametric CAD sketches [
755].
Simulation-Support and Modeling Interface Tasks. Although LLMs cannot replace high-fidelity physical simulation, they can assist in generating model input files, translating specifications into solver-ready formats, and summarizing results. Ali-Dib and Menou explored the reasoning capacity of LLMs in physics modeling tasks [
761], while Raissi et al.’s PINN framework demonstrated how language-driven architectures can help solve nonlinear partial differential equations by encoding physics into neural representations [
762].
Experimental Interpretation and Multimodal Lab Tasks. In experimental workflows, LLMs can support data summarization, anomaly detection, and textual explanation of multimodal results. Latif et al. proposed PhysicsAssistant, an LLM-powered robotic learning system capable of interpreting physics lab scenarios and offering real-time feedback to students and instructors [
763].
STEM Learning and Interactive Reasoning Tasks. LLMs are increasingly integrated into educational settings to guide derivations, answer conceptual questions, and simulate physical systems. Jiang and Jiang introduced a tutoring system that enhanced high school students’ understanding of complex physics concepts using LLMs [
756], while Polverini’s work further confirmed the model’s utility in supporting structured, interactive learning [
758].
5.2.2. Textual and Documentation-Centric Tasks
In physics and mechanical engineering, researchers and engineers routinely interact with large volumes of unstructured text: scientific papers, technical manuals, design specifications, test reports, and equipment logs. These documents are often dense, domain-specific, and heterogeneous in format. LLMs provide a promising tool for automating the extraction, summarization, and interpretation of this information.
One of the primary use cases is literature review and standards extraction. LLMs can parse multiple engineering reports or scientific articles to extract key findings, quantitative parameters, or references to specific standards, thereby reducing time-consuming manual review. For example, Khan et al. (2024) showed that LLMs can effectively assist in requirements engineering by identifying constraints and design goals from complex textual documents [
764].
Another growing application is in log interpretation and structured report analysis. In mechanical systems testing and diagnostics, engineers often work with detailed experiment logs and operational narratives. Tian et al. (2024) demonstrated that LLMs can identify conditions, setup parameters, and key outcomes from such semi-structured text logs, making them useful in experiment-driven engineering workflows [
765].
Furthermore, LLMs have been applied in sensor data documentation and matching. Berenguer et al. (2024) proposed an LLM-based system that interprets natural language descriptions to retrieve relevant sensor configurations and data, effectively bridging the gap between textual requirements and structured data sources [
766].
These applications point to a broader role for LLMs as interfaces between human engineers and machine-readable engineering assets, enabling a smoother flow of information across documentation, modeling, and decision-making. While challenges remain—particularly in domain-specific precision and context disambiguation—the utility of LLMs in handling technical documentation is becoming increasingly evident.
5.2.3. Design Ideation and Parametric Drafting Tasks
In physics and mechanical engineering, the early stages of design—where ideas are generated and formalized into parameter-driven models—play a critical role in shaping the final product. These processes traditionally require both deep domain knowledge and creativity, often relying on iterative exploration using CAD tools, handwritten specifications, and physical prototyping. With the emergence of LLMs, this early design workflow is being significantly transformed. LLMs can help engineers rapidly generate, interpret, and modify design concepts using natural language, thus improving both accessibility and productivity in the drafting process.
Recent studies have shown that LLMs are capable of generating design concepts from textual prompts that describe functional requirements or contextual constraints. For instance, Makatura et al. (2023) introduced a benchmark for evaluating LLMs on design-related tasks, showing that these models can generate reasonable design plans and material suggestions purely based on natural language input [
767]. This capability supports brainstorming and variant generation, especially in multidisciplinary systems where engineers must evaluate many trade-offs quickly.
Beyond concept generation, LLMs are increasingly used to support parametric drafting. This involves translating natural language into design specifications, such as dimensioned geometry, material choices, and assembly constraints. Wu et al. (2024) proposed CadVLM, a model capable of generating parametric CAD sketches from language-vision input, bridging LLMs with traditional CAD workflows [
755]. Such models allow engineers to iterate on design through language-driven instructions (e.g., “Make the slot wider by 2 mm” or “Add a fillet at the bottom edge”), greatly simplifying the communication between design intent and digital geometry.
Some systems have also incorporated LLMs directly into CAD environments, allowing interactive, prompt-based drafting and editing. Tools like SketchAssistant and AutoSketch use LLMs to assist with geometry creation and layout proposals. These interfaces reduce the learning curve for non-expert users and open up early-stage design to a broader range of collaborators. However, challenges remain in aligning generated outputs with engineering standards, ensuring the manufacturability of outputs, and maintaining traceability between design versions and decision logic.
Overall, LLMs are becoming valuable collaborators in the ideation-to-drafting pipeline of physics and mechanical engineering design. While they are not yet substitutes for domain expertise or formal simulation, they significantly accelerate exploration, reduce iteration costs, and expand accessibility to design tools.
5.2.4. Simulation-support and Modeling Interface Tasks
In physics and mechanical engineering, simulations play a critical role in modeling complex systems, validating designs, and predicting behavior. Traditionally, configuring and running simulations requires significant domain expertise, specialized tools, and manual scripting. The integration of LLMs into simulation workflows is introducing new levels of efficiency and accessibility.
LLMs can translate natural language descriptions of physical setups into structured simulation code or configuration files. For example,
FEABench evaluates the ability of LLMs to solve finite element analysis (FEA) tasks from text-based prompts and generate executable multiphysics simulations, showing encouraging performance across benchmark problems [
752]. Similarly,
MechAgents demonstrates how LLMs acting as collaborative agents can solve classical mechanics problems (e.g., elasticity, deformation) through iterative planning, coding, and error correction [
753].
Beyond code generation, LLMs are being deployed as intelligent simulation interfaces.
LangSim, developed by the Max Planck Institute, connects LLMs to atomistic simulation software, enabling users to query and configure simulations via natural language [
768]. Such systems lower the barrier for non-experts to engage in simulation workflows, automate routine tasks, and reduce friction in setting up complex models.
Moreover, LLMs can help interpret simulation results, summarize outcome trends, and generate human-readable reports that connect raw numerical output with engineering reasoning. This interpretability is especially valuable in multi-physics scenarios where simulation logs and visualizations are often overwhelming.
While these advances are promising, there remain limitations in LLMs’ ability to ensure physical correctness, handle multiphysics coupling, and reason over temporal or boundary conditions. Nonetheless, their role as modeling assistants is becoming increasingly practical in early prototyping and parametric studies.
5.2.5. Experimental Interpretation and Multimodal Lab Tasks
In physics and mechanical engineering, laboratory experiments often generate complex datasets comprising textual logs, numerical measurements, images, and sensor outputs. Interpreting these multimodal datasets requires significant expertise and time. The advent of LLMs offers promising avenues to streamline this process by enabling automated analysis and interpretation of diverse data types.
LLMs can assist in translating experimental procedures and observations into structured formats, facilitating easier analysis and replication. For instance, integrating LLMs with graph neural networks has been shown to enhance the prediction accuracy of material properties by effectively combining textual and structural data [
769]. This multimodal approach allows for a more comprehensive understanding of experimental outcomes.
Moreover, LLMs have demonstrated capabilities in interpreting complex scientific data, such as decoding the meanings of eigenvalues, eigenstates, or wavefunctions in quantum mechanics, providing plain-language explanations that bridge the gap between complex mathematical concepts and intuitive understanding [
770]. Such applications highlight the potential of LLMs in making intricate experimental data more accessible.
Additionally, frameworks like GenSim2 utilize multimodal and reasoning LLMs to generate extensive training data for robotic tasks by processing and producing text, images, and other media, thereby enhancing the training efficiency for robots in performing complex tasks [
771].
Despite these advancements, challenges remain in ensuring the accuracy and reliability of LLM-generated interpretations, especially when dealing with noisy or incomplete data. Ongoing research focuses on improving the robustness of LLMs in handling diverse and complex experimental datasets.
5.2.6. STEM Learning and Interactive Reasoning Tasks
LLMs are increasingly being integrated into STEM education to enhance learning experiences and support interactive reasoning tasks. Their ability to process and generate human-like text allows for more engaging and personalized educational tools.
LLMs can simulate teacher-student interactions, providing real-time feedback and explanations that adapt to individual learning needs. This capability has been utilized to improve teaching plans and foster deeper understanding in subjects like mathematics and physics [
771]. Additionally, LLMs have been employed to interpret and grade student responses, offering partial credit and constructive feedback, which aids in the learning process [
772].
Interactive learning environments powered by LLMs, such as AI-driven tutoring systems, have shown promise in facilitating inquiry-based learning and promoting critical thinking skills. These systems can guide students through problem-solving processes, encouraging them to explore concepts and develop reasoning abilities [
770].
Despite these advancements, challenges remain in ensuring the accuracy and reliability of LLM-generated content. Ongoing research focuses on improving the alignment of LLM outputs with educational objectives and integrating multimodal data to support diverse learning styles.
Table 31.
Physics and Mechanical Engineering Tasks and Benchmarks
Table 31.
Physics and Mechanical Engineering Tasks and Benchmarks
| Type of Task |
Benchmarks |
Introduction |
| CAD and Geometric Modeling |
ABC Dataset [773] DeepCAD [774] Fusion 360 Gallery [775] CADBench [776] |
The ABC Dataset, DeepCAD, and Fusion 360 Gallery together provide a comprehensive foundation for studying geometry-aware language and generative models. While ABC emphasizes clean, B-Rep-based CAD structures suitable for geometric deep learning, DeepCAD introduces parameterized sketches tailored for inverse modeling tasks. Fusion 360 Gallery complements these with real-world user-generated modeling histories, enabling research on sequential CAD reasoning and practical design workflows. CADBench further supports instruction-to-script evaluation by providing synthetic and real-world prompts paired with CAD programs. It serves as a high-resolution benchmark for measuring attribute accuracy, spatial correctness, and syntactic validity in code-based CAD generation. |
| Finite Element Analysis (FEA) |
FEABench [? ] |
FEABench is a purpose-built benchmark that targets the simulation domain, offering structured prompts and tasks for evaluating LLM performance in generating and understanding FEA input files. It serves as a critical testbed for bridging the gap between symbolic physical language and numerical simulation. |
| CFD and Fluid Simulation |
OpenFOAM Cases [777] |
The OpenFOAM example case library provides a curated set of fluid dynamics simulation setups, widely used for training models to understand solver configuration, mesh generation, and boundary condition specifications in CFD contexts. |
| Material Property Retrieval |
MatWeb [778] |
MatWeb is a widely-used material database containing thermomechanical and electrical properties of thousands of substances. It plays an essential role in supporting downstream simulation tasks such as material selection, constitutive modeling, and multi-physics simulation setup. |
| Physics Modeling and PDE Learning |
PDEBench [779] PHYBench [780] |
PDEBench and PHYBench collectively advance the evaluation of LLMs in physical reasoning and numerical modeling. PDEBench focuses on classical PDEs like heat transfer, diffusion, and fluid flow in the context of scientific machine learning, while PHYBench introduces a broader spectrum of perception and reasoning tasks grounded in physical principles. Together, they support benchmarking across symbolic reasoning, equation prediction, and simulation-aware generation. |
| Fault Diagnosis and Health Monitoring |
NASA C-MAPSS [781] |
NASA C-MAPSS provides real-world time-series degradation data from turbofan engines, serving as a benchmark for predictive maintenance, anomaly detection, and reliability modeling in aerospace and mechanical systems. |
5.2.7. Benchmarks
In physics and mechanical engineering, tasks such as computer-aided design (CAD), finite element analysis (FEA), and computational fluid dynamics (CFD) are characterized by strong physical constraints, structured representations, and deep reliance on geometry or numerical solvers. The development of benchmarks to support LLMs in these domains is still in its infancy. Although recent datasets have enabled initial exploration of LLMs in these fields, they present multiple challenges with respect to scale, accessibility, and alignment with language-based modeling.
In the CAD domain, several large-scale datasets have been developed to support geometric learning and generative modeling. For example, the ABC Dataset [
773] provides over one million clean B-Rep (Boundary Representation) models, DeepCAD [
774] offers parameterized sketches for inverse modeling, and the Fusion 360 Gallery [
775] includes real-world design sequences from professional and amateur CAD users. However, most of these datasets represent geometry using numeric or parametric formats that lack symbolic or linguistic structure. Specifically, B-Rep trees and STEP files are low-level and require domain-specific parsers, making them difficult for LLMs to interpret or generate in a meaningful way.
While some recent efforts have attempted to represent CAD workflows through code-based formats such as FreeCAD Python scripts or Onshape feature code, these datasets are often small, sparse in supervision, and highly sensitive to syntactic or logical errors. Moreover, generating coherent and executable CAD programs remains a significant challenge due to the limited spatial reasoning capacity of current LLMs.
Recent advances, however, demonstrate that specialized instruction-to-code datasets and self-improving training pipelines can significantly improve LLM performance in CAD settings. For instance, BlenderLLM [
776] is trained on a curated dataset of instruction–Blender script pairs and further refined through self-augmentation. As shown in
Table 32, it achieves state-of-the-art results on the CADBench benchmark, outperforming models like GPT-4-Turbo and Claude-3.5-Sonnet across spatial, attribute, and instruction metrics, while maintaining a low syntax error rate. This indicates that domain-adapted LLMs, when paired with well-structured code-generation benchmarks, can overcome many of the geometric and syntactic limitations faced by general-purpose models.
To address these issues, several strategies can be explored. One direction involves decomposing full modeling workflows into modular sub-tasks, such as sketch creation, constraint placement, extrusion operations, and feature sequencing. This allows the LLM to focus on smaller, interpretable segments of the modeling pipeline. Another direction is to reframe CAD problems into geometric reasoning tasks—for instance, by translating design problems into 2D or 3D visual logic similar to those found in geometry exams. Prior studies have shown that LLMs such as GPT-4 perform surprisingly well on geometric puzzles when the problem is represented symbolically or visually. Furthermore, retrieval-augmented generation (RAG) can be employed to provide contextual examples from past designs or sketches, thus improving generation quality through analogy-based learning. Overall, bridging the gap between high-dimensional geometric information and language representation remains a central challenge in CAD-focused LLM research.
Similarly, simulation-based tasks in FEA and CFD also require structured input generation, including mesh topology, material properties, solver settings, and boundary conditions. These tasks often involve producing complete simulation decks compatible with engines such as CalculiX or OpenFOAM, followed by interpreting complex field outputs such as stress distributions or velocity gradients.
Benchmarks such as FEABench [
? ] and curated OpenFOAM case libraries [
777] provide valuable baselines for evaluating the simulation-awareness of LLMs. However, the availability of large-scale paired datasets—comprising natural language descriptions, simulation input files, and corresponding numerical results—remains limited, posing a bottleneck for supervised fine-tuning and instruction-based evaluation.
To address this gap, FEABench introduces structured tasks that assess LLMs’ ability to extract simulation-relevant information.
Table 33 presents the performance of various LLMs across multiple physics-aware metrics, including interface factuality, feature recall, and dimensional consistency. Models like Claude 3.5 Sonnet and GPT-4o demonstrate strong results in retrieving factual and geometric descriptors, particularly in interface and feature extraction. However, all models show relatively low performance in recalling physical properties and structured feature attributes, reflecting ongoing challenges in capturing physical relationships from text. These results suggest that while LLMs can reliably recover high-level simulation inputs, deeper understanding of numerical structure and physical laws remains an open research problem.
A promising solution is to integrate LLMs with external numerical solvers in a simulator-in-the-loop framework. In this approach, an LLM is tasked with generating a complete simulation setup given a natural language prompt or design goal. The generated setup is then executed by a physics-based solver to produce ground-truth outputs. The input-output pairs, along with the original language prompt, can be stored as a triplet dataset and reused for supervised training. This method enables semi-automated dataset construction at scale, facilitates error correction via feedback from the simulator, and promotes the development of LLM agents that can reason across symbolic and physical domains. Additionally, by iterating through prompt refinement and result validation, such frameworks could enable reinforcement learning with human or physical feedback for high-fidelity simulation tasks.
Together, these benchmarks and emerging methodologies form the foundation of an evolving research area at the intersection of language modeling, geometry, and physics. As more domain-specific tools and datasets are adapted for LLM-compatible formats, we expect substantial progress in generative reasoning, simulation co-pilots, and data-driven modeling for engineering systems.
5.2.8. Discussion
Opportunities and Impact. LLMs are beginning to reshape workflows in physics and mechanical engineering, particularly in tasks such as CAD modeling, finite element analysis (FEA), material selection, simulation setup, and result interpretation. As demonstrated by models like CadVLM [
755] which translates textual input into parametric sketches, FEABench [
? ] which evaluates LLMs on FEA input generation, and LangSim [
768] which enables natural language interaction with atomistic simulation tools, LLMs are emerging as intelligent intermediaries between domain experts and computational tools.
By converting natural language into structured engineering commands, LLMs greatly simplify early-stage design, parameter exploration, technical documentation, and preliminary simulation configuration. Through code generation, auto-completion, document retrieval, and example-based prompting, LLMs are becoming integral assistants in modern engineering workflows. As multimodal and multi-agent systems (e.g., MechAgents [
753]) become more common, LLMs are poised to play a key role in the next generation of closed-loop “design–simulate–validate” engineering pipelines.
Challenges and Limitations. Despite these promising applications, multiple challenges persist. First, physical modeling tasks such as FEA and CFD involve highly coupled, nonlinear partial differential equations (PDEs) that require domain-specific inductive biases, numerical stability, and conservation principles—capabilities that current LLMs fundamentally lack.
Second, existing datasets in engineering domains present significant structural barriers. Most CAD datasets (e.g., B-Rep, STEP) are stored in numeric or parametric formats with minimal symbolic representation, making them difficult for LLMs to understand or generate. Code-based CAD datasets are more interpretable by LLMs, but are often limited in size, brittle in syntax, and sensitive to logical correctness.
Moreover, LLMs struggle with tasks requiring unit consistency, physical constraint enforcement, and boundary condition reasoning. In real-world engineering, even small errors in design parameters or simulation configurations can lead to system failure, safety risks, or structural inefficiencies. This makes it difficult to rely on LLMs for mission-critical design tasks without rigorous validation.
Research Directions. To further improve the effectiveness of LLMs in physics and mechanical engineering, several research directions are particularly promising:
Simulation-Augmented Dataset Generation. Integrating LLMs with numerical solvers in a simulator-in-the-loop framework allows the generation of language-input–simulation-output triplets at scale. This enables supervised training, fine-tuning, and RLHF strategies grounded in physically valid feedback.
Task Decomposition and Geometric Reformulation. Decomposing CAD workflows into modular sub-tasks (e.g., sketching, constraints, extrusion) and reformulating modeling problems as geometric reasoning tasks can align better with LLM capabilities and improve interpretability.
Multimodal and Multi-agent Integration. Developing LLM systems that can call CAD tools, solvers, and databases autonomously—as seen in MechAgents or LangSim—will allow LLMs to reason, plan, and act across tools in complex design and simulation pipelines.
Standardized Benchmarks and Evaluation. Creating large-scale, task-diverse, and format-unified benchmark datasets (e.g., combining natural language prompts, simulation files, and result summaries) will accelerate model evaluation and fair comparison in this field.
Physics Validation and Safety Assurance. Embedding physical rule checkers and verification mechanisms into generation loops can help enforce unit consistency, structural validity, and simulation compatibility, ensuring that outputs are not just syntactically correct but physically plausible.
Conclusion. LLMs are becoming increasingly valuable assistants in physics and mechanical engineering, especially in peripheral tasks such as documentation, concept generation, parametric modeling, and simulation support. However, to deploy them in core workflows, future systems must integrate LLMs with symbolic reasoning, geometric logic, physics-based solvers, and expert feedback. This synergy will enable the transition from language-based assistance to trustworthy, intelligent co-creation in complex engineering design and modeling workflows.
5.3. Chemistry and Chemical Engineering
5.3.1. Overview
5.3.1.1 Introduction to Chemistry
Chemistry is the scientific discipline dedicated to understanding the properties, composition, structure, and behavior of matter. As a branch of the physical sciences, it focuses on the study of chemical elements and the compounds formed from atoms, molecules, and ions—their interactions, transformations, and the principles governing these processes [
782,
783].
Put simply, chemistry seeks to explain how matter behaves and how it changes [
784]. We must acknowledge that the field of chemistry is vast and encompasses a variety of branches. Given the particularly rich application scenarios in areas such as organic chemistry, life sciences—especially in relation to LLM-related work—we will discuss these branches in the following chapter to provide a detailed introduction to works closely related to biology and life sciences.
In the field of chemistry, there are numerous sub-tasks, and many scientists have made significant contributions and achieved groundbreaking results over the past few hundred years.
Before diving into LLM-related topics, we would like to provide an overview of major tasks and traditional methods in chemistry research. By integrating information from official websites [
785] and literature across various branches of the field [
786,
787,
788,
789,
790,
791,
792,
793,
794,
795], we have summarized the research tasks in the domain of chemistry as follows:
Analysis and Characterization. This task involves identifying the substances present in a sample (qualitative analysis) and determining the quantity of each substance (quantitative analysis) [
796]. In this section we emphasize experimental measurement and detection methods aimed at identifying which substances are present, as well as determining their composition, structure, and morphology; we do not here focus on how their properties change under varying conditions nor on prediction or modeling of those properties. It also includes elucidating the structure and properties of these substances at a molecular level [
797]. Traditional methods for analysis and characterization include techniques such as observing physical properties (color, odor, melting point), performing specific chemical tests to identify certain substances (like the iodine test for starch or flame tests for metals), and classical quantitative analysis using precipitation, extraction, and distillation [
798]. Modern research in this area heavily relies on sophisticated instruments. Spectroscopy, which studies how matter interacts with light, can provide significant insights into a molecule’s structure and composition [
797]. Chromatography is employed to separate complex mixtures into their individual components for analysis [
797]. Mass spectrometry (MS) is a powerful technique that can identify and quantify substances by measuring their mass-to-charge ratio with very high sensitivity and specificity [
797,
799].
Research on Properties. Research on properties in chemistry refers to the systematic exploration and analysis of the physical and chemical characteristics of substances, with the main objective being to reveal the behavior and reaction characteristics of substances under different conditions [
800,
801]. We take “Research on Properties” to include both experimental determination and prediction or modeling of physical and chemical properties, with a primary interest in how those properties behave or change under different conditions. Traditionally, researchers have employed experimental methods to determine these properties. For thermodynamic properties, calorimetry is a key technique used to measure heat flow during physical and chemical processes [
802,
803]. Equilibrium methods, such as measuring vapor pressure, can assist in determining energy changes during phase transitions [
802]. For kinetic properties, traditional methods involve monitoring the changes in concentration of reactants or products over time [
804].
Reaction Mechanisms. The primary objective of studying reaction mechanisms in chemistry is to reveal the specific processes and steps involved in chemical reactions, including the microscopic mechanisms by which reactants are converted into products. This research field focuses on the formation of various intermediates during the reaction, the reaction pathways, rate-determining steps, and their corresponding energy changes [
800,
805]. Traditional methods for investigating reaction mechanisms include kinetic studies, where the rate of a reaction is measured under different conditions to understand its progression [
806,
807,
808]. Isotopic labeling involves using reactants with specific isotopes to trace the movement of atoms during the reaction [
807]. Stereochemical analysis examines the spatial arrangement of atoms in reactants and products, providing insights into the reaction pathway [
807]. Identifying the intermediate products formed during the reaction is also a crucial aspect of this research.
Chemical Synthesis. Chemical Synthesis refers to actually producing molecules in the laboratory or pilot-scale. The synthesis of natural products is an important task in chemistry, aimed at using chemical methods to synthesize complex organic molecules found in nature [
809]. The realization of such synthesis in practice relies on several traditional experimental methods. Plant extraction separates compounds from plants using techniques like solvent extraction, cold pressing, or distillation, yielding various active ingredients. Fermentation technology utilizes microorganisms to produce natural products, commonly for antibiotics and bioactive substances [
810]. Organic synthesis constructs chemical structures through multi-step synthesis and the introduction of functional groups [
811]. Lastly, semi-synthetic methods modify simple precursors to create more complex natural compounds or their derivatives [
812].
Molecule Generation. Molecule Generation involves computational chemistry and molecular modeling techniques to predict, optimize, or generate new molecular structures with desired functions or properties [
813,
814]. It includes computer-aided design, virtual screening, property prediction, structure optimization, and theoretical modelling of molecules, etc. [
813,
814]. Molecular synthesis and design encompass both experimental synthesis [
814] and computer-aided design [
815].
Applied Chemistry. Applied Chemistry refers to the branch of chemistry that focuses on practical applications in various fields such as industry, medicine, and environmental science. It involves using chemical principles to solve real-world problems and improve processes, including material chemistry and drug chemistry [
816,
817,
818,
819]. Traditionally, several key methods are relied upon, including structure-activity relationship (SAR) studies, computer-aided drug design [
820], high-throughput screening [
821], and synthetic chemistry [
822].
5.3.1.2 Introduction to Chemical Engineering
Chemical engineering is an engineering field that deals with the study of the operation and design of chemical plants, as well as methods of improving production. Chemical engineers develop economical commercial processes to convert raw materials into useful products. Chemical engineering utilizes principles of chemistry, physics, mathematics, biology, and economics to efficiently use, produce, design, transport, and transform energy and materials [
823]. According to the Oxford Dictionary,
chemical engineering is a branch of engineering concerned with the application of chemistry to industrial processes, particularly involving the design, operation, and maintenance of equipment used to carry out chemical processes on an industrial scale [
824]. In summary, it serves as the bridge that applies chemical achievements to industry.
Similar to chemistry, chemical engineering encompasses multiple fields, including not only chemistry, but also mathematics, physics, and economics. Through a comprehensive review of previous research [
825,
826,
827,
828,
829], we have categorized the tasks in chemical engineering into the following types.
Chemical Process Engineering. Chemical process engineering includes chemical process design, improvement, control, and automation. Chemical process design focuses on the design of reactors, separation units, and heat exchange equipment to achieve efficient material conversion and energy utilization [
827,
830], typically employing computer-aided design software and process simulation tools [
831,
832]. Chemical process improvement involves the systematic analysis and optimization of existing chemical processes to enhance production efficiency, reduce resource consumption, and minimize environmental impact [
830]. It primarily relies on quality management tools [
833,
834] and process simulation software [
835]. Process control and automation aim to monitor and regulate chemical processes through control systems to ensure stable operation under set conditions, typically based on proportional–integral–derivative (PID) control systems [
836], combined with advanced control technologies such as Model Predictive Control [
837] to optimize processes. Distributed control systems and programmable logic controllers are also commonly used automation systems that can monitor and adjust process variables in real-time [
838,
839].
Equipment Design and Engineering. Equipment design and engineering focus on the design, selection, and maintenance of chemical engineering equipment to ensure its efficient and safe operation within specific processes. The reliability and functionality of the equipment directly impact overall efficiency and safety [
840]. Equipment design is typically carried out in accordance with industry standards and regulations, such as American Society of Mechanical Engineers (ASME) and American Petroleum Institute (API). Engineers use computer-aided design (CAD) software for detailed design and simulation [
841,
842]. Additionally, strength analysis and fluid dynamics simulation are critical components, generally relying on computational fluid dynamics software to ensure the safety and efficiency of equipment under various operating conditions [
840].
Sustainability and Environmental Engineering. Sustainability and environmental engineering focus on the impact of chemical processes on the environment and are dedicated to developing green chemical technologies to reduce pollution and resource consumption. This field emphasizes the importance of life cycle assessment and environmental impact assessment in achieving sustainability goals [
843].
Scale-up and Technology Transfer. The task of translating chemical achievements into practical applications in chemical engineering involves bridging the gap between laboratory discoveries and industrial-scale implementation, ensuring that innovative chemical processes and materials are effectively integrated into real-world production systems to meet societal and industrial demands [
844]. Traditionally, the application of chemical achievements employs methods such as pilot scale testing to validate the feasibility and stability of the technology [
844], and process simulation and optimization (e.g., using tools like Aspen Plus and CHEMCAD) to model and optimize process flows, thereby reducing costs and improving efficiency. Simultaneously, factors such as economic viability, supply chain and market dynamics, and safety and environmental compliance are also evaluated and optimized [
825,
827].
From the definition, we can see that there is a strong logical relationship between the fields of chemical engineering and chemistry at the macroscopic level. The main battleground of chemical science is in the laboratory, while the main battleground of chemical engineering is in the factory. Chemical engineering aims to translate processes developed in the lab into practical applications for the commercial production of products, and then work to maintain and improve those processes [
782,
783,
823,
845].
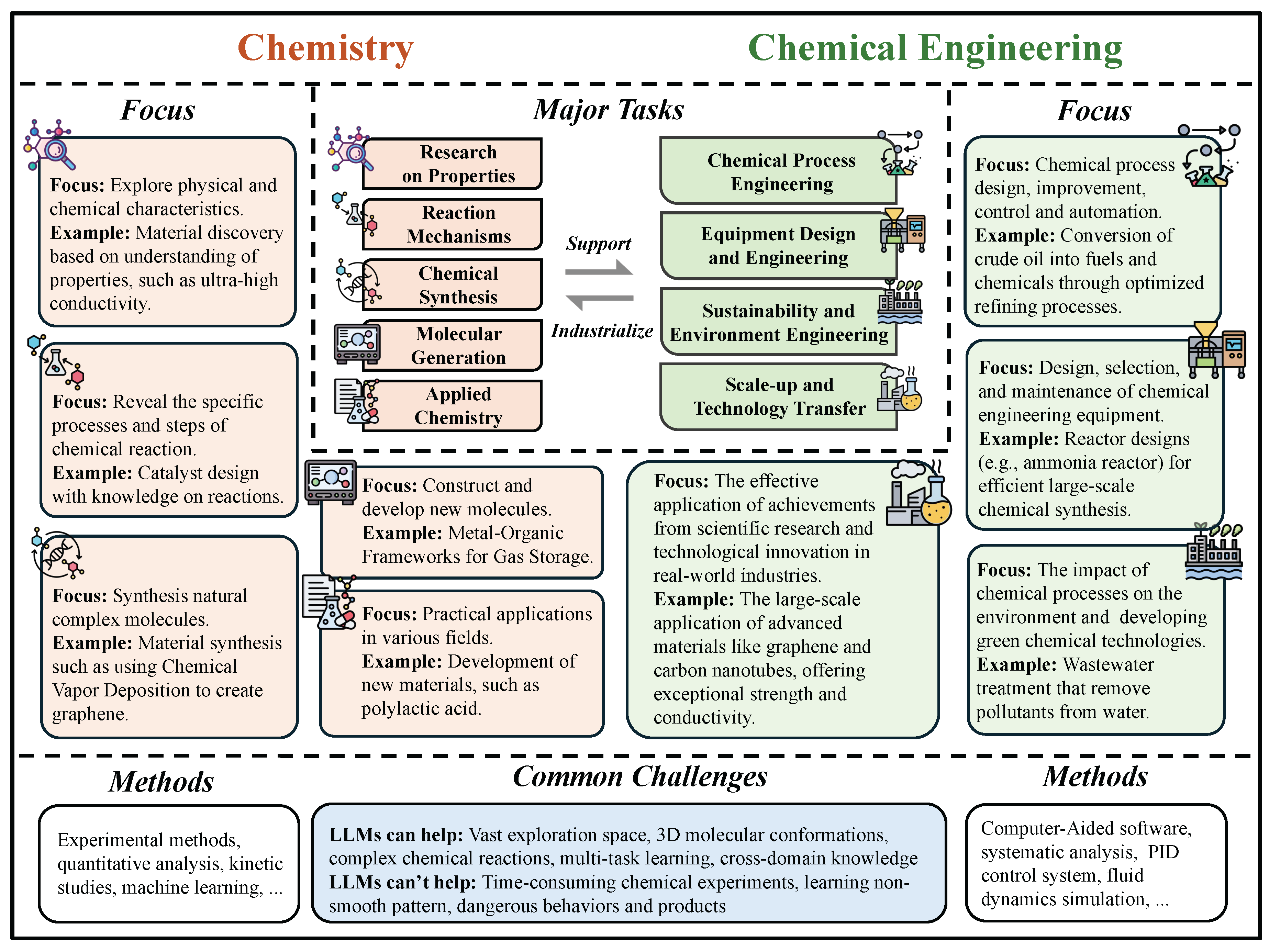
At the microscopic level, chemistry and chemical engineering share many common technologies, such as CAD and computational simulation. Moreover, there are varying degrees of connections between the different sub-tasks within these two fields. We have summarized the relationships among them in the form of a diagram in
Figure 17.
5.3.1.3 Contribution of Chemistry and Chemical Engineering
It is not difficult to imagine that chemistry, as a fundamental science, has profoundly impacted various aspects of human society, with its contributions evident in public health, materials innovation, environmental protection, and energy transition. Firstly, the contributions of chemistry to public health are significant. Through the synthesis and development of new pharmaceuticals, chemists have greatly improved human health [
846,
847]. For instance, the discovery of penicillin not only marked the beginning of the antibiotic era but also reduced the mortality risk associated with bacterial infections [
846,
848]. In recent years, the development of targeted therapies [
847,
849], such as drugs aimed at specific cancers, relies on a chemical understanding of the internal mechanisms of tumor cells, thereby significantly enhancing patient survival rates. Secondly, chemistry has a revolutionary impact on materials innovation. Through the development of polymers [
850], alloys [
851,
852], and nanomaterials [
853,
854], chemists have not only enhanced material properties but also advanced technological progress. For example, the application of modern lightweight and high-strength composite materials has enabled greater energy efficiency and safety in the aerospace and automotive industries [
850]. Moreover, the emergence of graphene and other nanomaterials has opened new possibilities for the development of electronic products [
853,
854].
In the realm of environmental protection, the contributions of chemistry cannot be overlooked. By developing efficient catalysts and clean technologies, chemists play a crucial role in reducing industrial emissions and tackling water pollution [
855]. For example, selective catalytic reduction reactions effectively convert harmful gases emitted by vehicles, significantly improving urban air quality [
856,
857]. Furthermore, the role of chemistry in energy transition is becoming increasingly important [
858,
859]. The development of renewable energy storage and conversion is fundamentally supported by chemical technologies [
860]. For instance, the research and development of lithium-ion batteries [
861] and hydrogen fuel cells [
862] depend on the optimization of chemical reactions and material innovations, making the use of clean energy feasible.
5.3.1.4 Challenges in the Era of LLMs
Despite the significant achievements in the fields of chemical science and chemical engineering, there remain unresolved challenges in these areas. The emergence of LLMs presents an opportunity to address these issues. We must acknowledge that, unfortunately, LLMs are not omnipotent; they cannot solve all the challenges within this field. However, for certain tasks, LLMs hold promise in assisting chemists in overcoming these challenges. We have listed the following difficulties that LLMs cannot solve:
The Irreplaceability of Time-consuming Chemical Experiments. LLMs-generated outcomes in chemical research still require experimental validation. Assessing the true utility of these generated molecules, such as evaluating their novelty in real-world applications, can be a time-consuming undertaking [
863]. While LLMs have their advantages in data processing and information retrieval, solely relying on the results generated by the model may not accurately reflect the actual experimental conditions. Moreover, LLMs are trained on existing data and literature; if a specific field lacks sufficient data support, the outputs of the model may be inaccurate or unreliable [
864].
Limitations in Learning Non-smooth Patterns. Traditional deep learning struggles to learn non-smooth target functions that map molecular data to labels, as these target functions are frequently non-smooth in molecular property prediction. This implies that minor alterations in the chemical structure of a molecule can lead to substantial changes in its properties [
865]. Additionally, LLMs also find it difficult to solve this problem under the limited size of molecular datasets.
Dangerous Behaviors and Products. The field of chemistry carries certain inherent risks, as some products or reactions can be hazardous (e.g., flammable, explosive, toxic gases, etc.). LLMs may generate scientifically incorrect or unsafe responses, and in some cases, they may encourage users to engage in dangerous behavior [
866]. Furthermore, LLMs can also be misused to create toxic or illegal substances [
863]. At the current stage of development, LLMs still cannot be fully trusted to ensure complete reliability.
On the other hand, despite the aforementioned limitations, the potential of LLMs in the fields of chemistry and chemical engineering is undeniable, as they hold promise in addressing many challenges:
Decrease the Vast Chemical Exploration Space. Inverse design enables the creation of synthesizable molecules that meet predefined criteria, accelerating the development of viable therapeutic agents and expanding opportunities beyond natural derivatives [
867]. However, this quest faces a combinatorial explosion in the number of potential drug candidates—the chemical space made up of all small drug-like molecules—which is unimaginably large (estimated at
) [
867]. Testing any significant fraction of these molecules, either computationally or experimentally, is simply impossible [
868]. This field has been revolutionized by machine learning methods, particularly generative models, which narrow down the search space and enhance computational efficiency, making it possible to delve deeply into the seemingly infinite chemical space of drug-like compounds [
869]. LLMs, such as MolGPT [
870], which employs an autoregressive pre-training approach, have proven to be instrumental in generating valid, unique, and novel molecular structures. The emergence of multi-modal molecular pre-training techniques has further expanded the possibilities of molecular generation by enabling the transformation of descriptive text into molecular structures [
871].
Generation of 3D Molecular Conformations. Generating three-dimensional molecular conformations is another significant challenge in the field of molecular design, as the three-dimensional spatial structure of a molecule profoundly impacts its chemical properties and biological activity. Traditional computational methods are often resource-intensive and time-consuming, making it difficult for researchers to design and screen new drugs effectively. Unlike conventional approaches based on molecular dynamics or Markov chain Monte Carlo, which are often hindered by computational limitations (especially for larger molecules), LLMs based on 3D geometry exhibit remarkable superiority in conformation generation tasks, as they can capture inherent relationships between 2D molecules and 3D conformations during the pre-training process [
871].
Automate Chemical Agents. Autonomous chemical agents combine LLM “brains” with planning, tool use, and execution modules to carry out experiments end-to-end. In the coscientist system, for example, GPT-4 first decomposes a high-level goal (“optimize a palladium-catalyzed coupling”) into sub-tasks (reagent selection, condition screening), retrieves relevant literature via a search tool, generates executable Python code for liquid-handling robots, and then interprets sensor feedback to iteratively refine the protocol—closing the loop between design and execution [
872]. Similarly, Boiko et al. built an agent that plans ultraviolet–visible spectroscopy (UV–Vis) experiments by writing code to control plate readers and analyzers, automatically processing spectral data to identify optimal wavelengths, and even adapting to novel hardware modules introduced after the model’s training cutoff [
873]. By leveraging LLMs for hierarchical task decomposition, self-reflection, tool invocation (e.g., search APIs, code execution, robotics control), and memory management, these systems drastically accelerate repetitive experimentation and free researchers to focus on hypothesis generation rather than manual protocol execution [
873].
Enhance Understanding of Complex Chemical Reactions. The field of reaction prediction faces several key challenges that affect the accuracy of forecasting chemical reactions. A significant issue is reaction complexity, stemming from multi-step pathways and dynamic intermediates, which complicates product predictions, especially with varying conditions like different catalysts. Traditional models often struggle with these complexities, leading to biased outcomes. Utilizing advanced transformer architectures, LLMs can model complex relationships in chemical reactions and adjust predictions based on varying conditions. They excel in learning from unlabeled data through self-supervised pretraining, helping identify patterns in chemical reactions, particularly useful for rare reactions.
Multi-task Learning and Cross-domain Knowledge. The complexity of multi-task learning makes the simultaneous optimization of diverse prediction tasks difficult, while LLMs effectively handle this via shared representations and multi-task fine-tuning [
874]. Traditional methods also struggle to integrate cross-domain knowledge from chemistry, biology, and physics, yet LLMs address this seamlessly through pre-training and knowledge graph enhancement.
5.3.1.5 Taxonomy
As summarized in
Table 34, in efforts to integrate chemistry research with artificial intelligence, particularly LLMs, many chemists primarily focus on tasks such as property prediction, property-directed inverse design, and synthesis prediction [
867,
874,
875]. However, other chemists highlight additional significant tasks, including data mining and predicting synthesis conditions [
876,
877]. By synthesizing insights from these studies along with other seminal works [
878,
879,
880], we propose a more comprehensive classification method. This method accounts for both the rationality of chemical task classification and the characteristics of computer science.
From the chemistry perspective, our taxonomy echoes the field’s established research divisions—such as molecular property prediction, property-directed inverse design, reaction type and yield prediction, synthesis condition optimization, and chemical text mining—ensuring that each category corresponds directly to a recognized experimental or theoretical task in chemical science. Concurrently, from the computer science perspective, by mapping every task onto a unified input–output modality framework, we add a computationally consistent structure that facilitates model development, benchmarking, and comparative analysis across diverse tasks within a single formal paradigm. Together, these dual alignments guarantee that our classification remains both chemically and algorithmically meaningful.
Figure 18.
A taxonomy of chemical tasks enabled by LLMs, categorized by input-output types and downstream objectives.
Figure 18.
A taxonomy of chemical tasks enabled by LLMs, categorized by input-output types and downstream objectives.
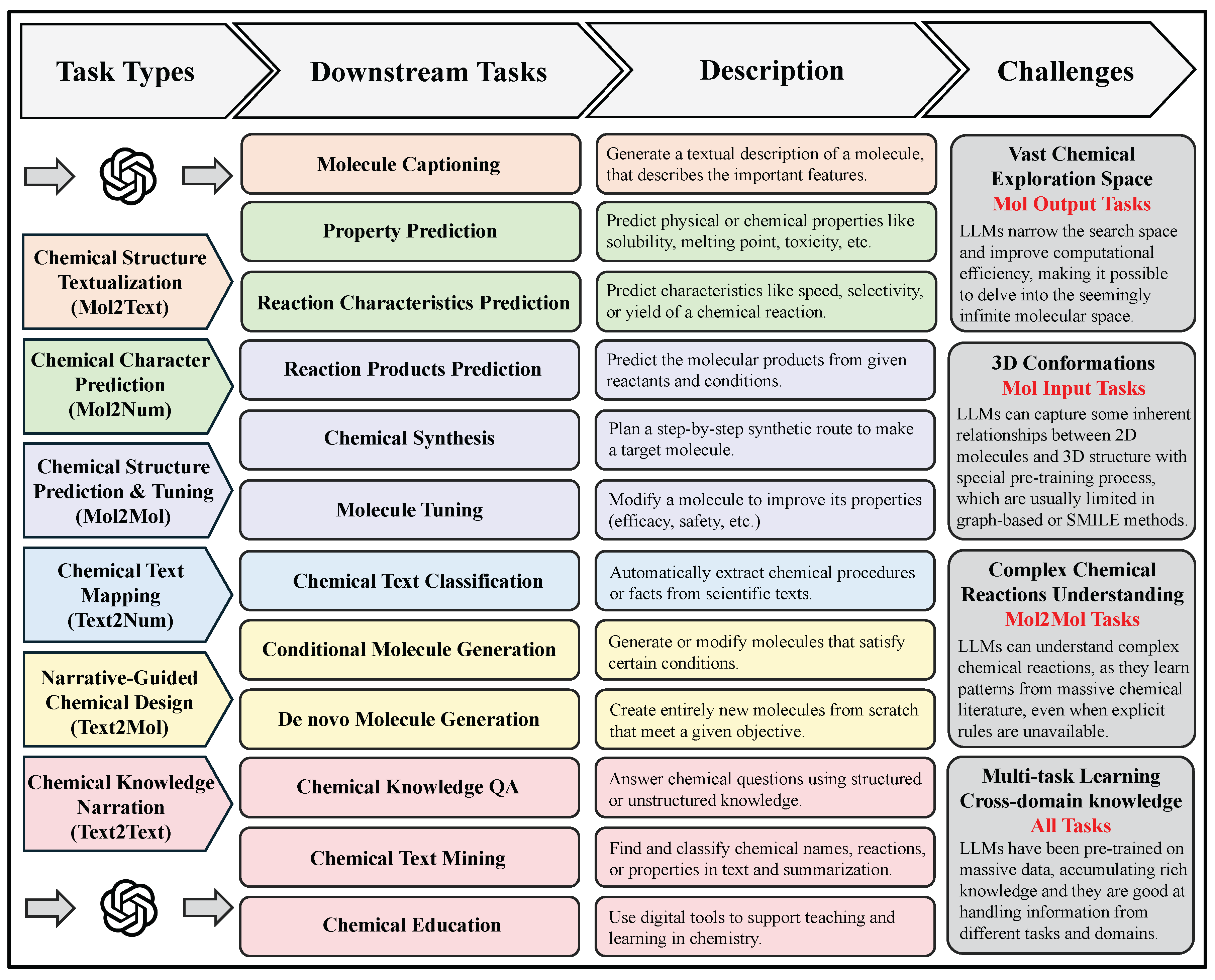
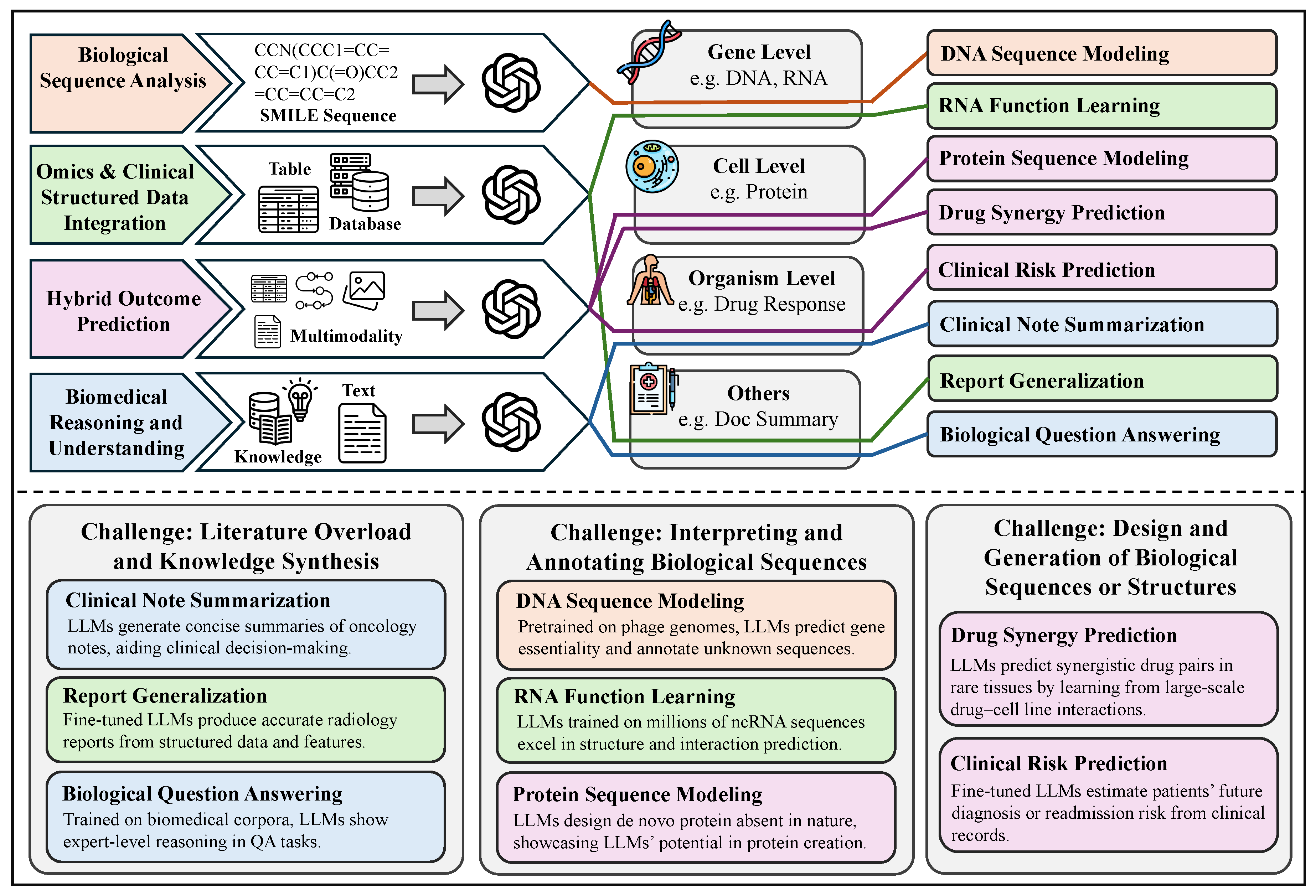
Chemical Structure Textualization. Chemical structure textualization is the process of taking a molecule’s SMILES sequence as input and producing a detailed textual depiction that highlights its structural features, physicochemical properties, biological activities, and potential applications. Here, SMILES (Simplified Molecular Input Line Entry System) encodes a molecule’s atomic composition and connectivity as a concise, linear notation—for example, “CCO” denotes ethanol (each “C” represents a carbon atom and “O” an oxygen atom), while “C1=CC=CC=C1” represents benzene (the digits mark ring closure and “=” indicates double bonds)—enabling computational models to capture meaningful structural patterns and relationships for downstream text generation. Subtasks include molecule captioning, which exemplifies the goal of generating rich, human-readable descriptions of molecules to give chemists and biologists rapid, accessible insights for experimental design and decision-making [
881].
Chemical Characteristics Prediction. Nowadays, SMILES provides a standardized method for encoding molecular structures into strings [
882]. This string-based representation enables efficient parsing and manipulation by computational models and underpins a variety of tasks in cheminformatics, drug discovery, and reaction prediction. Notably, many machine learning models, including large-scale language models like GPT, are pre-trained or fine-tuned using corpora of SMILES sequences. Among the tasks leveraging SMILES input are property prediction and reaction characteristics prediction, where the model takes a SMILES sequence as input and outputs numerical values, categorical labels, or multi-dimensional vectors representing chemical properties, reactivity, bioactivity, and other experimentally relevant quantities.
Chemical Structure Prediction & Tuning. Chemical structure prediction & tuning tasks represent a classical form of sequence-to-sequence modeling [
883], where the goal is to transform an input molecular sequence into an output sequence. In chemistry, this formulation is particularly intuitive because molecules are often represented as SMILES strings, which encode structural information in a linear textual format. Given an input SMILES sequence, the model learns to generate another SMILES string corresponding to a chemically meaningful transformation. This input–output structure underlies a variety of chemical modeling tasks, including reaction product prediction, chemical synthesis planning, and molecule tuning. For instance, the input may describe reactants or precursors, and the output may represent reaction products or structurally modified molecules, making these tasks central to computational reaction modeling and automated molecular design.
Chemical Text Mapping. Chemical text mapping tasks refer to the process of transforming unstructured textual input into numerical outputs such as labels, scores, or categories. At their core, these tasks involve analyzing chemical text—ranging from scientific articles to experimental protocols—and mapping the extracted information to structured numerical values for downstream applications like classification, relevance scoring, or trend prediction [
867,
884]. A typical example is document classification, where the input is natural language text and the output is a discrete or continuous number representing, for example, a document’s category or relevance score. These tasks enable scalable analysis of chemical literature and facilitate integration of textual knowledge into data-driven modeling workflows.
Narrative-Guided Chemical Design. Narrative-Guided chemical design is a generative modeling paradigm extensively applied in chemistry and materials science, with the core objective of deriving molecular structures or material candidates that fulfill specific target properties or functional requirements [
885,
886]. Unlike conventional forward design—which predicts properties from a given structure—inverse design begins with the desired outcome and works backward to propose compatible structures. In this context, the input is a description of the target properties, which may take the form of numerical constraints, categorical labels, or free-text descriptions, and the output is a molecular structure—typically represented as a SMILES string—that satisfies those specified criteria. This framework encompasses tasks such as de novo molecule generation and conditional molecule generation, enabling applications like targeted drug design, property-driven material discovery, and personalized molecular synthesis.
Chemical Knowledge Narration. Chemical knowledge narration tasks in chemistry refer to the transformation of one form of textual input into another, with both input and output grounded in chemical knowledge and language use [
867]. These tasks leverage Natural Language Processing (NLP) techniques to process, convert, or generate chemistry-related textual data, thereby facilitating a range of downstream applications in research, education, and industry. For instance, given a textual input such as a paragraph from a research paper, the model may extract key information, translate it into another language, generate a summary, or answer domain-specific questions. Such tasks—encompassing chemical text mining, chemical knowledge question answering, and educational content generation—typically operate on natural language input and produce human-readable textual output, making them essential tools for improving access to and understanding of chemical information.
Table 34.
Chemistry Tasks, Subtasks, Insights and References
Table 34.
Chemistry Tasks, Subtasks, Insights and References
| Type of Task |
Subtasks |
Insights and Contributions |
Key Models |
Citations |
| Chemical Structure Textualization |
Molecular Captioning |
LLMs, by learning structure–property patterns from data, generate meaningful molecular captions, thus improving interpretability and aiding chemical understanding. |
MolT5: generates concise captions by mapping substructures to descriptive phrases; MolFM: uses fusion of molecular graphs and text for richer narrative summaries. |
[887,888,889,890,891,892,893,894,895,896,897,898,899,900,901,902,892] |
| Chemical characteristics prediction |
Property Prediction |
LLMs, by capturing complex structure–property relationships from molecular representations, enable accurate property prediction, thereby providing mechanistic insights and guiding the rational design of molecules with desired functions. |
SMILES-BERT: self-supervised SMILES pretraining for robust property inference; ChemBERTa: masked SMILES modeling boosting solubility and toxicity predictions. |
[903,904,905,906,907,908,909,910,911,912,913,914] |
| |
Reaction Characteristics Classification |
LLMs, by modeling the relationships between reactants, conditions, and outcomes from large reaction datasets, can accurately predict reaction types, yields, and rates, thereby uncovering hidden patterns in chemical reactivity and enabling chemists to optimize reaction conditions and select efficient synthetic routes with greater confidence. |
RXNFP: fingerprint-transformer accurately classifies reaction types; YieldBERT: fine-tuned on yield data to predict experimental yields within 10% error. |
[863,915,916,917,918,919,920,921,922,923,924,925,926,927] |
| Chemical Structure Prediction & Tuning |
Reaction Products Prediction |
LLMs, by learning underlying chemical transformations from reaction data, can accurately predict reaction products, thus uncovering implicit reaction rules and supporting more efficient and informed synthetic planning. |
Molecular Transformer: state-of-the-art SMILES-to-product translation; |
[921,923,924,925,926,928,929,930,931,932,933] |
| |
Chemical Synthesis |
LLMs, by capturing patterns in reaction sequences and chemical logic from large datasets, can suggest plausible synthesis routes and rationales, thereby enhancing human understanding of synthetic strategies and accelerating discovery. |
Coscientist: GPT-4-driven planning and robotic execution. |
[872,932,934,935,936,937,938,939,940,941,942,943,944,945,946,947,948,949,950,951] |
| |
Molecule Tuning |
LLMs, by modeling structure–property relationships across diverse molecular spaces, enable targeted molecule tuning to optimize desired properties, thereby providing insights into molecular design and accelerating the development of functional compounds. |
DrugAssist: uses LLM prompts for ADMET property optimization; ControllableGPT: enables constraint-based molecular modifications. |
[952,953,954,955,956,957,958,959,960,961,962,963,964,964] |
| Chemical Text Mapping |
Chemical Text Mining |
LLMs, by capturing semantic and contextual nuances in chemical literature, enable accurate classification and regression in text mining tasks, thereby uncovering trends, predicting research outcomes, and transforming unstructured texts into actionable scientific insights. |
Fine-tuned GPT: specialized for chemical classification and regression; ChatGPT: adapts zero-shot classification of chemical text. |
[965,966,967,968,969,970,971,972,973,974,975,976,977,978] |
| Narrative-Guided Chemical Design |
De Novo Molecule Generation |
LLMs, by learning chemical syntax and patterns from large molecular corpora, enable de novo molecule generation with realistic and diverse structures, thus offering insights into unexplored chemical space and accelerating early-stage drug and material discovery. |
ChemGPT: unbiased SMILES sampling for novel molecules; MolecuGen: scaffold-guided generative modeling for improved novelty. |
[887,891,979,980,981,982,983,984,985,986,987,988,989,990,991,992] |
| |
Conditional Molecule Generation |
LLMs, by conditioning molecular generation on desired properties or scaffolds, enable the design of compounds that meet specific criteria, thereby offering insights into structure–function relationships and streamlining the discovery of tailored molecules. |
GenMol: multi-constraint text-driven fragment remasking. |
[887,889,931,957,983,984,986,987,988,993,994,995,996] |
| Chemical Knowledge Narration |
Chemical Knowledge QA |
LLMs, by integrating extensive chemical literature and diverse databases, can accurately address complex chemical knowledge questions, thereby uncovering valuable insights and enabling more informed, accelerated research and decision-making. |
ChemGPT: conditional SMILES generation for property-specific tuning; ScholarChemQA: domain-specific QA fine-tuned on scholarly chemistry data. |
[863,924,997,998,999,1000,1001,1002,1003,1004,1005,1006,1007,1008] |
| |
Chemical Text Mining |
LLMs, by understanding and extracting structured information from unstructured chemical texts, enable efficient chemical text mining, thereby revealing hidden knowledge, facilitating data-driven research, and accelerating the discovery of relationships across literature. |
ChemBERTa: BERT-based model fine-tuned for chemical text classification. SciBERT: pretrained on scientific text including chemical literature for robust retrieval. |
[965,966,973,976,1009,1010,1011,1012,1013,1014] |
| |
Chemical Education |
LLMs, by generating intuitive explanations and answering complex queries in natural language, support chemical education by making abstract concepts more accessible, thereby enhancing student understanding and promoting more interactive, personalized learning experiences. |
MetaTutor: LLM-based metacognitive tutor for chemistry learners. |
[997,998,1015,1016,1017,1018,1019,1020,1021,1022] |
5.3.2. Chemical Structure Textualization
Chemical tasks that map molecular structures to text serve as a bridge between the structural world of chemistry and human language. Chemical structure textualization is essentially “describing molecules in words,” akin to how one might recognize a complex object (like a new gadget) and then explain it in a common language [
863]. In everyday life, this is like looking at a detailed blueprint and verbally summarizing what it represents. In chemistry, such tasks are crucial: chemists often need to communicate structures verbally or in writing––for instance, by naming compounds or summarizing their features––so that others can understand them without seeing a drawing. Converting molecules to text makes chemistry more accessible and searchable (one can text-search a compound name in literature, but not a structure diagram). For example, a medicinal chemist might draw a novel molecule and want the International Union of Pure and Applied Chemistry (IUPAC) name or a simple description to include in a report. An environmental chemist might identify an unknown substance and need an AI to generate a description like “a chlorinated hydrocarbon solvent.” These translations from structure to language are essential for reporting, education, and integrating chemical information into broader databases.
Chemical Structure Textualization is fundamentally a Mol2Text task, mapping chemical structure representations (e.g., SMILES) as input to corresponding natural language descriptions as output. Mol2Text encompasses any scenario of “structure to language.” Key subtasks include chemical nomenclature generation (e.g. converting a molecular structure into its IUPAC name or common name), molecule captioning (generating a sentence describing the molecule’s class or properties), and even property or hazard annotations (predicting textual labels like “flammable” from a structure). For instance, nomenclature generation might take a SMILES string of a molecule and output “2-(4-Isobutylphenyl)propanoic acid” (the IUPAC name of ibuprofen). Molecule captioning could produce a phrase like “an aromatic benzene derivative with two nitro substituents” given a structure. They are LLM-friendly because large databases of molecules with names or descriptions exist (providing plenty of textual training data), and the output is structured text that follows rules (ideal for sequence generation models). We therefore highlight the area of molecule captioning in the following paragraph. These illustrate how LLMs evolved from simply reading text to “reading” chemistry and writing text.
Molecular Captioning. Beyond formal nomenclature, a growing chemical structure textualization application is molecular captioning––generating a natural-language description of a molecule’s structure or function. That is given a molecule’s representation (e.g. SMILES or graph), generating a coherent natural-language description that accurately captures its structural features, physicochemical properties, and potential biological activities. This is analogous to image captioning in computer vision (where an AI might look at a photo and say “a cat sitting on a sofa”). Here, the “image” is a chemical structure (which an LLM might ingest as a string like SMILES or another representation), and the output is a concise text description. For example, given the structure of caffeine, an ideal caption might be: “a bitter, white alkaloid of the purine class, commonly found in coffee and tea.” Or more straightforwardly: “Caffeine – a stimulant compound with a purine (xanthine) scaffold and multiple methyl groups.” This task is in its infancy, but its significance is clear: it would allow chemists to quickly get a textual summary of a molecule’s key features or known uses. In everyday life, this is akin to seeing a new plant and describing it as “a tall tree with waxy leaves and fragrant white flowers”––it communicates key identifying features in an intuitive way. MolT5 [
887] first achieved end-to-end text-SMILES translation by jointly training on vast amount of SMILES and textual descriptions, but relying solely on sequence information led to captions lacking intuitive structural understanding. To address this, MolFM [
891] introduced multimodal pretraining that combines SMILES, InChI, and text descriptions, significantly improving annotation accuracy and richness, yet it did not leverage molecular images for visual comprehension . Next, GIT-Mol [
890] integrated molecular graphs, images, and text through cross-modal fusion, achieving higher fidelity captions but at the cost of large model size and high deployment and inference overhead . To improve efficiency and deployment, MolCA [
899] designed a cross-modal projector and uni-modal adapters, greatly reducing fine-tuning and deployment costs while retaining multimodal capabilities, though its pretraining data coverage still needs expansion . Most recently, GraphT5 [
897] employs multimodal cross-token attention to tightly integrate molecular graph structures with a language model, balancing caption quality and model scale, and providing an efficient and scalable foundation for molecule captioning
Problems Solved by LLMs. LLMs have dramatically improved both chemical nomenclature generation and molecular captioning by learning from vast corpora of paired data. In the case of nomenclature, Transformer-based models such as Struct2IUPAC have achieved accuracies of 98–99% in converting SMILES strings to formal IUPAC names—performance that rivals rule-based systems like Open Parser for Systematic IUPAC Nomenclature (OPSIN), which itself set the benchmark for open-source name-to-structure parsing over a decade ago [
1023]. Simultaneously, proof-of-concept captioning studies have shown that LLMs can associate common substructures with descriptive terms (e.g., “–NO
2” → “nitro compound”), enabling models like MolT5 to generate concise textual summaries of molecular features [
887]. Together, these successes illustrate that LLMs can both “read” and “write” chemistry, transforming structural representations into human-readable language with high fidelity.
Remain Challenges. Despite these advances, challenges remain in both domains. Chemical naming models, while highly accurate, operate as black boxes; when they err on novel or highly complex molecules, it is difficult to trace the decision pathway or understand which structural elements led to a misnaming. Moreover, evolving IUPAC standards—such as recent organometallic nomenclature updates—require continual model retraining or fine-tuning to maintain correctness. In molecular captioning, the absence of a single “ground truth” means that errors are subtler and often manifest as partial truths or outright hallucinations (e.g., asserting a molecule is “used in perfumes” without evidence), and models struggle to calibrate the appropriate level of detail versus generalization [
1024]. This fuzziness poses risks in scientific communication, as speculative or incorrect descriptors can mislead users.
Future Work. Looking ahead, integrating LLMs with external knowledge sources and multimodal inputs promises to address many of these limitations. Hybrid pipelines that combine neural proposals with rule-based validation could ensure 100% naming accuracy while preserving flexibility for new nomenclature conventions. Likewise, coupling captioning models with structured databases—so that, upon recognizing “glucose,” an LLM retrieves and incorporates the formula C6H12O6—would enhance factual correctness. Finally, multimodal architectures capable of ingesting both SMILES and 2D structural images, or embedding property predictors to append numerical descriptors (e.g., molecular weight, logP), will yield richer, more reliable textual outputs and usher in a new era of AI-driven chemical communication.
5.3.3. Chemical Characteristics Prediction
Chemical characteristics prediction task focuses on predictive modeling of molecular data, aiming to predict specific outcomes related to molecular structures using machine learning techniques. The primary input for this task is a detailed representation of molecules, typically in the form of SMILES strings, these representations allow models to learn meaningful patterns and relationships that inform predictions. Chemical Characteristics Prediction is fundamentally a Mol2Number task, taking molecular representations as input and producing quantitative property values as output.
In regression tasks, the goal is to predict continuous numerical values based on molecular features. For example, given the SMILES string "CCO" representing ethanol, a model might predict its boiling point as approximately 78.37°C. Similarly, for the SMILES "CC(=O)O" representing acetic acid, the model could predict a solubility of about 1000 g/L in water [
1025]. In another case, the SMILES "C1=CC=CC=C1" for benzene might be used to predict its logP value as around 2.13. Similarly, in classification tasks, the objective is to determine discrete outcomes. For instance, given the SMILES "CC(=O)OC1=CC=CC=C1C(=O)O" for aspirin, the model might classify it as a non-steroidal anti-inflammatory drug (NSAID). Similarly, an example of reaction classification involves the reaction with the SMILES notation Brc1ccccc1.B(O)O >> c1ccccc1B(O)O, where the task is to classify the reaction type. The model classifies this reaction as a "Suzuki coupling" with a confidence of 98.2%.
The value of chemical characteristics prediction tasks in chemistry lies in their capacity to translate complex molecular and reaction information into quantitative predictions—enabling both experts and non-specialists to anticipate how molecules will behave under given conditions, including chemical properties, reaction types, yields, and reaction rates. For example, in property prediction, the SMILES string “CCO” succinctly encodes the structure of ethanol (“C” for carbon, “O” for oxygen), allowing a model to infer its physical and chemical properties without recourse to time-consuming quantum calculations. When we talk about reaction types, imagine mixing baking soda (NaHCO3) and vinegar (CH3COOH). A chemical characteristics prediction model would read the SMILES for each ingredient, see “NaHCOO” + “CC(=O)O,” and label it as an acid–base neutralization. That label helps chemists know that the reaction will produce CO2 gas. Predicting yields is like estimating how much carbon dioxide you’ll collect in a balloon when you mix your soda-and-yeast “volcano” experiment. A yield-prediction model might tell you, “Under these conditions, you’ll get about 80% of the maximum CO2 possible,” so you can plan ahead and avoid wasting ingredients. When discussing reaction-rate prediction, think of Alka-Seltzer fizzing in water. A model could predict how fast the tablet dissolves—does it take 30 seconds or three minutes?—based on temperature or how finely the tablet is crushed. In short, chemical characteristics prediction tasks let scientists—and even curious students—see, ahead of time, which “kitchen chemistry” will work best, how much product to expect, and how fast it will happen. This not only cuts down on costly trial-and-error in drug discovery or materials design, but also deepens our understanding of how molecules behave and reactions proceed.
Chemical characteristics prediction tasks encompass several categories: molecular property prediction (e.g., solubility — the maximum amount of a substance that can dissolve in a solvent, typically expressed in mol/L); bioactivity and binding-affinity prediction (e.g., IC50 — the concentration at which a compound inhibits 50% of a target’s biological activity); reaction characters prediction (e.g., reaction-type classification — categorizing a reaction by its mechanism, such as nucleophilic substitution or acid–base neutralization; and yield prediction — estimating the percentage of product obtained relative to the theoretical maximum under specified conditions); retrosynthesis-route scoring (e.g., feasibility scoring — assessing whether a proposed synthetic pathway can be practically executed in the laboratory; and cost scoring — predicting the total economic expense of reagents and operations); force-field and energy prediction (e.g., potential-energy surfaces — energy landscapes that map molecular geometry to potential energy; and interatomic forces — forces between atoms calculated as gradients on that energy surface); and molecular descriptor or embedding generation (e.g., low-dimensional embeddings — concise numerical vectors that capture key molecular characteristics in a reduced feature space).
Due to the intrinsic similarities among these tasks, we select two of the most representative ones—Property Prediction and Reaction Characters Prediction—for discussion in this paper. These two tasks benefit most from LLMs’ strengths: (1) they have abundant, well-structured textual and experimental data (e.g., MoleculeNet benchmarks, USPTO reaction corpora) that LLMs can readily learn from; (2) LLMs can provide both numerical predictions and human-readable rationales, enhancing interpretability over more opaque methods; and (3) improvements in these areas directly accelerate molecule prioritization and reaction planning in research and industry.
Property Prediction. The universal value of chemistry lies in accurately predicting compound properties to guide their practical use. Property Prediction is the task of predicting a molecule’s physicochemical or biological properties (e.g., solubility, binding affinity, toxicity) given its representation (such as a SMILES string, molecular graph, or descriptor vector). In pharmaceuticals, understanding how molecular structure influences bioactivity and toxicity enables the design of safer, more effective drugs; in materials science, predicting properties such as solubility, thermal stability, or mechanical strength from chemical structure accelerates the development of advanced polymers and functional materials. Traditional computational methods like quantum calculations and molecular dynamics offer high accuracy but demand extensive resources, whereas machine learning models can predict properties more efficiently. Recently, LLMs have demonstrated strong performance in molecular property and reaction-outcome prediction by leveraging vast textual and experimental datasets, achieving competitive accuracy without the heavy computational overhead of physics-based simulations. Combined with expert insight, AI-driven property prediction promises to revolutionize compound prioritization and materials design by focusing experimental efforts on the most promising candidates.
In early studies, LLMs such as BERT were applied to chemical reaction classification tasks. A representative work by Schwaller et al. achieved an impressive classification accuracy of up to 98.2%. The application focus then shifted from reaction classification to molecular property prediction, especially under scenarios with limited labeled data. Wang et al. proposed a semi-supervised model, SMILES-BERT [
1026], which was pretrained on large-scale unlabeled data via a "masked SMILES recovery" task. It achieved state-of-the-art performance across multiple datasets and marked the first successful application of BERT in drug discovery tasks. During the early exploration of molecular language models, Chithrananda et al. introduced ChemBERTa [
904], systematically examining the impact of pretraining dataset size, tokenization strategies (BPE vs. SmilesTokenizer), and molecular representations (SMILES vs. SELFIES) on model performance. Results showed that increasing the pretraining data from 100K to 10M led to significant improvements in downstream tasks such as BBBP and Tox21. Although ChemBERTa did not outperform the GNN baseline Chemprop, the authors suggested that further scaling could close this gap. Tokenization comparisons showed a slight advantage for the custom tokenizer. While no significant difference was observed between SMILES and SELFIES, attention head visualization using BertViz revealed neuron selectivity to functional groups, highlighting the importance of proper benchmarking and awareness of model carbon footprint. Building on this, Ahmad et al. developed ChemBERTa-2 [
1027], aiming to create a general-purpose foundation model. With a multi-task regression head and pretraining on 77 million molecules, ChemBERTa-2 achieved comparable performance to state-of-the-art models on MoleculeNet tasks. The study emphasized that different pretraining strategies had varying effects on downstream tasks, suggesting that model performance depends not only on pretraining itself, but also on the specific chemical context and fine-tuning dataset. Further extending this direction, Yuksel et al. proposed SELFormer [
1028], incorporating SELFIES to address concerns about the validity and robustness of SMILES. Pretrained on 2 million drug-like compounds and fine-tuned on a range of property prediction tasks (e.g., BBBP, SIDER, Tox21, HIV, BACE, FreeSolv, ESOL, PDBbind), SELFormer achieved leading performance in several cases. It demonstrated the ability to distinguish between molecules with varying structural properties, and suggested that future models should integrate structural data and textual annotations to build multimodal representations, enhancing generalizability and real-world utility.
To further improve molecular structure representation, Maziarka et al. introduced the MAT (Molecule Attention Transformer) [
1029], incorporating atomic distances and molecular graph structure into the attention mechanism. This graph-structured self-attention led to performance gains in property prediction. Li and Jiang focused on capturing molecular substructures and proposed Mol-BERT [
1030], pretrained on 4 million molecules from ZINC and ChEMBL. Treating fingerprint fragments as "words" and using Masked Language Modeling (MLM) to learn sentence-level molecular semantics, Mol-BERT outperformed both GNNs and sequence models on tasks like Tox21 and SIDER. Ross et al. developed MoLFormer, trained on over 1.1 billion SMILES from ZINC and PubChem. By introducing Rotary Position Embeddings, it more effectively captured atomic sequence relationships. MoLFormer not only surpassed GNNs on various benchmarks but also achieved 60x energy efficiency, representing progress toward environmentally sustainable AI.
On model generalization, Zhang et al. identified a bottleneck in the lack of correlation across different property datasets. They proposed MTL-BERT [
1031], a multi-task learning model pretrained on large-scale unlabeled SMILES from ChEMBL. MTL-BERT improved prediction performance and enhanced interpretability of complex SMILES by extracting context and key patterns.
On the task-specific level, Yu et al. proposed SolvBERT [
1025], a multi-task regression model designed to predict both solvation free energy and solubility. Despite the traditional reliance on 3D structural modeling for such tasks, SolvBERT—using only SMILES—achieved performance competitive with, or even superior to, GNN-based approaches, showcasing the potential of text-based modeling in physical chemistry.
While model performance continues to improve, limited labeled data remains a major challenge. In 2024, Jiang et al. introduced INTransformer [
1032], which incorporated perturbation noise and contrastive learning to augment small datasets and improve global molecular representation, even under low-resource conditions. Similarly, MoleculeSTM [
1033] used contrastive learning to align SMILES strings and textual molecular descriptions extracted from PubChem using a LLM. Extending this idea to proteins, Xu et al. proposed ProtST [
1034], which models protein sequences using a protein language model and aligns them with protein descriptions encoded by LLMs, exploring multimodal fusion for biomacromolecule modeling.
Reaction Characters Prediction. Typical tasks in chemical reaction property prediction include reaction type classification (determining which type or mechanistic category a given reaction belongs to), reaction yield prediction (estimating the yield of the target product under specific conditions), and reaction rate prediction (assessing the kinetics of the reaction, such as activation energy or rate constant). These studies are of great importance in the fields of pharmaceuticals, materials science, and chemical engineering. For example, as early as 2018, Ahneman et al. demonstrated that machine learning could predict the yields of untested combinations in coupling reactions based on a limited amount of experimental data, successfully identifying previously unknown high-yield catalytic systems [
1035]. Moreover, in the search for more efficient organic photovoltaic materials, it is often necessary to synthesize a series of candidate molecules. By using models to predict the yield of each reaction step, researchers can eliminate candidates with expected low yields and poor scalability early in the process, instead prioritizing synthetic routes that are predicted to be high-yielding and require fewer steps. This approach accelerates the screening process and conserves reagents.
Reaction type prediction aims to determine which category a given reaction belongs to—such as Suzuki coupling or Diels–Alder—based on its reactants and products. Traditionally, chemical reaction classification has relied on manually crafted rules or template libraries, but these approaches are poorly robust to new data and require complex atom-mapping preprocessing. To overcome this, Schwaller et al. introduced RXNFP [
915], a Transformer-based encoder that learns fixed-length embeddings of entire reactions directly from unannotated SMILES in large datasets (e.g., USPTO) and then uses a simple k-NN or classifier to assign reaction classes. While RXNFP has been reported to achieve very high classification accuracy on reaction classification benchmarks (e.g. over 98% on some USPTO subsets) [
915], it remains primarily a static feature extractor and is not designed for generative tasks like product generation or sequence-to-sequence modeling for continuous outputs. T5Chem [
916] addresses many of these gaps by casting reaction tasks—classification, product prediction, retrosynthesis, and yield regression—as text-to-text problems. A single T5 model, pretrained on large molecular datasets from PubChem and fine-tuned on public reaction sets, achieves strong performance across multiple tasks with one shared architecture, improving multitask efficiency and generalization.
The prediction of reaction yields has long been a central challenge in synthesis planning and industrial optimization, owing to the complex interplay among substrate structures, reagents, catalysts, solvents, temperature, and other factors. Initially, Schwaller et al. leveraged their RXNFP reaction fingerprints by feeding the learned fixed-length reaction embeddings into a regression head to provide preliminary yield predictions for Buchwald–Hartwig and Suzuki–Miyaura coupling reactions, demonstrating the feasibility of end-to-end transformer embeddings for yield regression. However, RXNFP was not specifically designed for yield prediction, and its static fingerprints lacked sensitivity to changes in reaction conditions. To address this, T5Chem [
928], a unified text-to-text multitask framework that, in addition to reaction classification and product prediction, incorporates a regression head for yield prediction. Pretrained on molecular data from PubChem and jointly fine-tuned on datasets such as USPTO, T5Chem matches or surpasses many baseline models across reaction prediction and yield tasks, showing that a single model can perform well on multiple reaction-related tasks. Building on this, the Schwaller team developed Yield-BERT [
1036] in 2021 by fine-tuning ChemBERT on reaction SMILES to directly output yields; in high-throughput coupling reaction datasets Yield-BERT has been shown to achieve strong
performance (e.g. values exceeding 0.90) compared to traditional methods using DFT-derived descriptors or handcrafted fingerprints. Yet Yield-BERT’s sensitivity to variations in catalysts, solvents, and other reaction conditions is limited, hindering its generalization across differing condition combinations. To enhance condition sensitivity, Yin et al. launched Egret [
1037] in 2023, combining masked language modeling with condition-based contrastive learning to teach the model to distinguish yield differences for the same substrates under varying conditions; Egret achieved improved
scores in several public benchmarks. Subsequently, Sagawa and Kojima’s ReactionT5 [
925] employed a two-stage pretraining strategy—first training CompoundT5 on a molecular library, then pretraining on a reaction-level database—enabling the model, with limited fine-tuning data, to achieve good performance (
in challenging splits) in yield and product prediction tasks, highlighting the value of reaction-level pretraining. Most recently, the ReaMVP [
1038] framework further incorporated 3D molecular conformations into pretraining alignment, aligning sequence and geometric views during a self-supervised stage, followed by fine-tuning on labeled yield data, resulting in modest boosts in
on out-of-sample reactions and demonstrating the importance of multimodal information fusion for improving the generalizability of yield predictions.
Problems Solved by LLMs. LLMs have significantly advanced molecular property prediction by leveraging self-supervised learning on large unlabeled datasets to learn robust molecular representations that improve generalization to limited labeled data. They also enable effective multi-task learning through shared representations and task-specific fine-tuning strategies that mitigate interference between diverse prediction objectives. Furthermore, by integrating domain knowledge from chemistry, biology, and physics via pretraining on multimodal data and knowledge graph augmentation, these models can incorporate cross-domain insights seamlessly. Finally, LLMs excel at processing contextual molecular representations such as SMILES and International Chemical Identifier (InChI) codes, automatically learning high-dimensional features that capture complex structural interactions without the need for manual feature engineering.
Remain Challenges. Despite these advances, several challenges persist. The vastness of chemical space requires models that can reliably generalize to structurally novel molecules and unseen scaffolds, a task that remains difficult for current architectures . Activity cliffs, where minor structural modifications lead to dramatic changes in molecular properties, continue to undermine prediction accuracy and demand models that are sensitive to such subtle variations. Moreover, the inherently graph-structured nature of molecular data necessitates specialized neural architectures—such as graph neural networks and graph transformers—that can effectively capture both local and global structural patterns. Additionally, certain properties depend on three-dimensional conformations or quantum mechanical effects, which two-dimensional representations alone cannot fully capture, highlighting the need for methods that incorporate 3D structural information.
Future Work. Future work will focus on developing hybrid molecular representations that combine two-dimensional graph features with three-dimensional geometric descriptors—including conformer ensembles, steric effects, and electrostatic interactions—to more accurately model spatial relationships within molecules . Integrating molecular dynamics simulations and physics-informed neural networks can further enrich these representations by providing dynamic and mechanistic insights into molecular behavior over time. These advances are expected to enhance the generalization of models across diverse reaction conditions, improve the reliability of reaction yield predictions, and accelerate the discovery of novel compounds with desired properties.
5.3.4. Chemical Structure Prediction and Tuning
In many chemistry problems, the desired output is another molecule. These tasks can be seen as “translating one molecule into another” – hence chemical structure prediction & tuning.
Chemical Structure Prediction & Tuning is inherently a Mol2Mol task. This category includes chemical reaction predictions, retrosynthesis planning, and molecule optimization, among others. An analogy from daily life would be cooking: you start with ingredients (molecules) and through a recipe (reaction), end up with a dish (a new molecule). Alternatively, think of it as solving a jigsaw puzzle: you have pieces (fragments of molecules) and want to put them together into a final picture (target molecule) – the input pieces and the output picture are both made of the same stuff, just rearranged. Chemical structure prediction & tuning tasks are central to chemistry because they essentially encompass chemical synthesis and design – predicting what will happen if molecules interact, or figuring out how to get from one molecule to another. For example, a forward reaction prediction might answer: “If I mix molecules A and B, what product will form?” A retrosynthesis task does the opposite: “I want molecule Z; what starting molecules could I use to make it?” These tasks directly assist chemists in the lab by suggesting likely outcomes or viable synthetic routes, thus speeding up research and discovery [
1039].
Key subtasks under chemical structure prediction & tuning include reaction outcome prediction (given reactant molecules and possibly conditions, predict the product molecules), chemical synthesis (particularly retrosynthesis, given a target product, propose one or more sets of reactants that could produce it), and molecule-to-molecule optimization (propose a structural modification to an input molecule to improve some property, e.g. “suggest a similar molecule with higher potency”). Another subtask is chemical pathway completion (extending a partial sequence of reactions by suggesting the next molecule). All these involve generating molecules from molecules. Reaction prediction (input: reactants, output: products) is a prime example: e.g. input SMILES for ethanol + acetic acid, output SMILES for ethyl acetate (the esterification product). Chemical synthesis is similarly crucial: input a drug molecule, output a plausible precursor like an aromatic halide plus a coupling partner. Molecule tuning is essential for fine-adjusting a drug’s potency, selectivity, and pharmacokinetic properties while retaining its core active scaffold (e.g., introducing an amino group into the side chain of penicillin G produces ampicillin, thereby significantly improving oral bioavailability and antibacterial spectrum). We focus on these tasks because these have seen extensive development with LLM-like models and there are large datasets (like USPTO patent reactions) to train on.
Reaction Product Prediction. Reaction prediction is the task of predicting the products of a chemical reaction given the reactants (and sometimes reagents or conditions). It’s effectively a translation of a set of input molecules into a set of output molecules. In the language analogy, if reactants are words, the chemical reaction is a grammatical rule that rearranges those words into a new sentence (the product). A more concrete life analogy: consider mixing ingredients in cooking – if you combine flour, sugar, and butter and bake, you predict a cake as the outcome. Similarly, if you mix benzene with chlorine under certain conditions, you predict chlorobenzene (and hydrogen chloride as a byproduct) as the outcome. For decades, chemists approached this with rules (“if you see this functional group and add that reagent, you get this outcome”). The forward reaction prediction task asks an AI to learn those implicit rules from data. It’s significant because the space of possible products is enormous, and a human chemist’s intuition can be wrong or limited to known reactions. An accurate model can enumerate likely products, flagging surprises or confirming expectations. This can prevent wasted experiments and guide chemists toward successful reactions. It’s particularly useful in drug discovery or complex synthesis planning, where predicting side-products or main products can inform route selection.
Early machine learning models for reaction prediction often used template-based systems: large libraries of reaction templates were extracted, and algorithms matched reactants to these templates to propose products. While effective, those methods required expert-curated templates and struggled with reactions not in the template library. The turning point was realizing that chemical reactions could be encoded as strings (e.g. “CCO + O=C=O → CCC(=O)O” for esterification) and treated like a language translation problem. In 2016, Nam and Kim first applied a sequence-to-sequence RNN to reaction SMILES, showing that neural machine translation can predict organic reaction products directly from SMILES [
1039]. However, RNNs had limitations – they sometimes forgot parts of the input and struggled with very long SMILES or complex rearrangements.
The real leap came with Transformers. In 2019, Schwaller et al. introduced the Molecular Transformer, a Transformer-based model for reaction outcomes that achieved 95% top-1 accuracy on the USPTO benchmark, significantly outperforming both template-based and RNN approaches [
921]. By leveraging self-attention, the model considered all reactant tokens simultaneously, capturing reaction context more effectively. The Molecular Transformer also provided uncertainty estimates, enabling chemists to gauge prediction confidence. Despite its success, the first-generation Transformer had limitations. It tended to memorize frequent reaction patterns, leading to overconfidence on well-represented reactions and underperformance on rare or novel chemistries. It also handled only single-step reactions and did not predict yields or selectivities. To address memorization and improve generalization, Tetko et al. introduced SMILES augmentation—randomizing atom orders during training—which reduced overfitting and boosted top-accuracy metrics on large USPTO subsets [
1040]. They also showed that beam-search decoding increased top-K accuracy (e.g. top-5) significantly. More recently, general-purpose LLMs have been fine-tuned on reaction data. For example, GPT-3 was adapted to reaction prediction tasks and achieved performance that is competitive with specialized Transformer models on some USPTO-style datasets [
1041]. However, in zero-shot or few-shot settings, GPT-3.5 and GPT-4 still lag behind domain-trained models in tasks requiring precise structural prediction, with top-1 accuracies substantially lower in unconstrained prediction tasks [
1042]. These findings underscore the continued importance of task-specific training and data augmentation for reliable reaction outcome prediction.
Chemical Synthesis. Once a target molecule has been identified, the next major challenge is to predict its optimal synthetic route—including per-step and overall yield—under realistic chemical constraints [
1043]. While the demanding, elegant total syntheses of complex natural products have historically driven advances in organic chemistry, the past two decades have prioritized broadly applicable catalytic reactions [
1043]; only recently has complex synthesis become relevant again as a digitally encoded knowledge source that can be mined by LLMs [
1009]. Unlike single-molecule property prediction, reaction planning must account for the multi-body nature of synthesis—modifying one reactant often requires re-optimizing all others under different mechanisms or conditions—and must balance multiple objectives, such as maximizing overall yield, minimizing the number of steps and cost of readily available starting materials, and ensuring chemical compatibility at each stage. Planning can proceed forward—from simple substrates to the target—or, more commonly, via retrosynthesis, introduced by E. J. Corey [
1044], which deconstructs the target molecule into fragments that are reassembled most effectively from inexpensive, commercially available reagents. Retrosynthesis is the reverse of reaction prediction: given a target molecule (the product), the task is to predict one or more sets of reactant molecules that could form it in a single step. It’s essentially “un-cooking” the dish to figure out the ingredients. In language terms, if a forward reaction is like forming a sentence from words, retrosynthesis is like taking a completed sentence and figuring out how to split it into two meaningful phrases that could combine to make that sentence. For example, the target “ethyl acetate” could be retrosynthesized to reactants “acetic acid + ethanol” (in the presence of an acid catalyst). Doing this well requires both creativity and extensive knowledge of known reactions, because the space of possible reactant combinations is enormous. For example, the first total synthesis of discodermolide required 36 individual steps (with a longest linear sequence of 24) to achieve only a 3.2 % overall yield, vividly illustrating the vast combinatorial explosion of possible routes and the reliance on expert intuition. By coupling structure–activity relationship predictions with synthesis planning, LLM-based approaches now promise to select or even design molecules not only for optimal properties but also for tractable, high-yielding synthetic accessibility—enabling both rapid route discovery and the creation of novel non-natural compounds chosen for their ease of synthesis and predicted performance [
867].
Early computer-assisted synthesis planning began with recurrent architectures and handcrafted rules: Nam and Kim pioneered forward reaction prediction using a GRU-based translation model [
1045], while Liu et al. applied an LSTM + attention seq2seq framework to retrosynthesis, achieving just 37.4 % accuracy on USPTO-50K [
1046]. Schneider et al. then enhanced retrosynthesis by algorithmically assigning reaction roles to reagents and reactants [
1047], and rule-based, template-driven systems such as Chematica and Segler & Waller [
930] captured explicit atomic and bond transformations in reverse planning—training on millions of reactions to deliver 95 % top-10 retrosynthesis accuracy and 97 % reaction-prediction accuracy—yet remained limited by their reliance on manually curated template libraries and inability to propose truly novel transformations. Semi-template methods struck a balance: Somnath et al.’s synthon-based graph model decomposed products into fragments and appended relevant leaving groups, boosting top-1 accuracy to 53.7 % on USPTO-50K while retaining interpretability [
1048].
While early synthesis planning methods relied on RNNs and handcrafted templates, the advent of LLMs has transformed the field by treating chemical synthesis as a data-driven “translation” task. Schwaller et al. [
1049] first demonstrated this paradigm with a regex-tokenized LSTM-attention network that learned retrosynthetic rules directly from raw USPTO reactions—removing the need for explicit templates and uniquely tokenizing recurring reagents to distinguish solvents and catalysts. Building on that work, the Molecular Transformer applied the full Transformer encoder–decoder architecture to both forward reaction and retrosynthetic prediction, inferring subtle correlations between reactants, reagents, and products without any handcrafted rules and achieving state-of-the-art accuracy on USPTO-MIT, USPTO-LEF, and USPTO-stereo benchmarks. To extend LLMs beyond single-step predictions, Schwaller et al. introduced a hypergraph exploration strategy in their 2020 Molecular Transformer model [
1050], dynamically expanding candidate routes using Bayesian-like scores and evaluating them with four new metrics—coverage (how much of chemical space is reachable), class diversity (variety of reaction types), round-trip accuracy (can predicted precursors regenerate the product), and Jensen–Shannon divergence (how closely the model’s predictions match real-world distributions). That same year, Zheng et al.’s SCROP [
1051] model combined a template-free transformer with a neural syntax corrector to self-correct invalid SMILES, boosting top-1 retrosynthesis accuracy on USPTO-50K to 59.0%—over 6% better than template-based baselines.
More recently, pretrained encoder–decoder LLMs have further elevated performance and flexibility. Irwin et al.’s Chemformer [
1052] used a BART backbone pretrained on millions of SMILES strings, then fine-tuned for sequence-to-sequence synthesis tasks and discriminative property predictions (ESOL, Lipophilicity, FreeSolv), demonstrating that task-specific pretraining is essential for efficiency and accuracy. In 2023, Toniato et al. [
1053] introduced prompt-engineering into retrosynthesis by appending classification tokens to target SMILES, guiding the model toward diverse disconnection strategies and producing multiple viable routes “out of the box.” Finally, Fang et al.’s MOLGEN [
1054] leveraged BART pretraining on 100 million SELFIES representations, domain-agnostic molecular prefix tuning, and an autonomous chemical feedback loop to ensure generated molecules are valid, non-hallucinatory, and retain their intended properties—foreshadowing autonomous LLM agents capable of end-to-end molecular design, synthesis planning, and iterative optimization.
Molecule Tuning. Molecule Tuning is the task of taking an initial molecule representation (e.g., SMILES string or graph) along with desired property modifications and generating structurally related analogs that optimize those specified properties while preserving the core scaffold. Molecule tuning leads compounds to simultaneously improve properties like potency, solubility, and safety—this is a cornerstone of drug design.
Early LLM-based approaches, such as DrugAssist [
952], introduced an interactive, instruction-tuned framework that lets chemists iteratively “chat” with the model to optimize one or more properties at a time. DrugAssist has achieved leading results in both single- and multi-property optimization tasks, showing strong potential in transferability and iterative improvement [
952]. However, it requires fine-tuning on task-specific datasets and its generalization to entirely new property combinations remains a challenge in practice. To advance further, Chemlactica and Chemma [
954] were developed by fine-tuning language models on a large corpus of **110 million molecules with computed property annotations**. These models demonstrate strong performance in generating molecules with specified properties and predicting molecular characteristics from limited samples; relative improvements over prior methods (for example on the Practical Molecular Optimization benchmark) indicate they outperform earlier approaches in multi-property molecule optimization [
954]. Despite these advances, fully zero-shot multi-objective optimization—where a model satisfies several new property constraints simultaneously without any additional training—remains difficult. Some approaches aim toward this goal (for example via prompt engineering, genetic methods, or sampling strategies), but no public model has yet been demonstrated to reliably achieve zero-shot control over wholly unseen property combinations. Finally, models that integrate richer structural context—such as using molecular graphs or fingerprint embeddings in addition to SMILES—are being explored. Early evidence suggests that these multimodal inputs can help propose chemically valid and synthetically accessible modifications under complex objectives, though again systematic evaluation under all multi-objective criteria is still emerging.
Problems Solved by LLMs. Thanks to LLM-based models such as the Molecular Transformer, many routine reaction predictions are now essentially solved: medicinal chemists can predict likely metabolites and process chemists can foresee side products with high confidence [
921].
Remain Challenges. However, challenges remain in handling multi-step or “one-pot” reactions where sequential transformations occur, since current one-step models lack a mechanism to decompose complex cascades. Quantitative prediction of yields and selectivities is still out of reach, as these models only output the major product qualitatively. Additionally, out-of-domain reactions—those involving novel catalytic cycles or exotic reagents absent from training sets—often confound existing models [
1041].
Future Work. Future LLMs for reaction prediction may incorporate mechanistic reasoning, internally decomposing reactions into elementary steps akin to chain-of-thought prompting [
1055]. There is growing interest in multi-modal architectures that integrate text with molecular graphs or images, enabling a richer understanding of bond connectivity changes. Enhancing uncertainty quantification and explainability—such as highlighting which bonds form or break—will empower chemists to assess prediction confidence. Finally, embedding reaction prediction LLMs within autonomous laboratory systems could enable closed-loop experimentation, where AI proposes, executes, and learns from chemical reactions in real time. Future retrosynthesis LLMs will likely integrate external knowledge bases indicating which building blocks are inexpensive or readily available, biasing suggestions toward practical routes. We also anticipate multi-step planning architectures, where a higher-level agent orchestrates sequential calls to a one-step model, effectively planning entire synthetic routes. Finally, more interactive human–AI retrosynthesis tools may emerge, capable of asking clarifying questions or presenting alternative routes with pros and cons—transforming retrosynthesis from a static prediction task into a dynamic, collaborative design process.
5.3.5. Chemical Text Mapping
Chemical text mapping tasks take free-form chemical text as input and output either discrete labels (classification) or continuous values (regression). Chemical Text Mapping is fundamentally a Mol2Num task. For example, in a document classification scenario, given the safety note “During the reaction, hydrogen gas evolved rapidly and ignited upon contact with air,” the model outputs the hazard label “flammable.” In a text-based regression example, given the procedure description “1.0 g of reactant A yielded 0.8g of product B under standard conditions,” the model predicts a yield of 80%. By automating the extraction of critical information—hazard classes, reaction types, success/failure flags, yields, rate constants, temperatures, solubilities, pKa values, and more—chemical text mapping dramatically reduces manual curation, accelerates the creation of structured databases for downstream modeling, and empowers chemists and students to query “How dangerous is this step?” or “How much product did they actually get?” at scale.
Common tasks of this form include hazard classification, reaction-type classification, procedure outcome classification, yield and rate constant regression, temperature and solubility prediction, etc. In this work, we concentrate on chemical text mining within the chemical text mapping framework—harnessing LLMs to transform narrative chemical descriptions into actionable categorical and numerical data.
Chemical Text Classification. Chemical Text Classification is the task of categorizing chemical documents or text segments into predefined labels—such as reaction type, property mentions, or entity categories—based on their unstructured textual content.
Chemical text classification has matured through successive generations of chemistry-tuned LLMs, each addressing the gaps of its predecessors. ChemBERTa-2 [
1027] was one of the first truly chemical-centric encoders—pretrained on 77 million SMILES strings (from PubChem) using masked-language modelling and multi-task regression—and it showed competitive performance on multiple downstream molecular property and classification benchmarks. However, as an encoder-only model, it requires separate fine-tuning heads for each task and lacks built-in generative or structured-output capabilities.
Recent studies, such as “Fine-tuning large language models for chemical text mining” [
965], have explored unified frameworks that handle multiple chemical text mining tasks—compound entity recognition, reaction role labeling, MOF synthesis information extraction, NMR data extraction, and conversion of reaction paragraphs to action sequences. In these works, fine-tuned LLMs demonstrated exact match / classification accuracies in the range of approximately 69% to 95% across these tasks with only minimal annotated data [
965]. Nevertheless, challenges remain in cross-sentence relations, complex numeric extractions, and the consistency / validation of structured or JSON-style output formats.
Problems Solved by LLMs. Firstly, they can achieve high-precision tasks such as hazard classification, reaction type annotation, yield and rate regression, with minimal or even no labeled data, through prompt engineering or a small number of examples, greatly reducing the cost of manually building domain dictionaries and rules; secondly, LLMs, with their powerful ability to understand context, can handle multiple information extraction subtasks (such as named entity recognition, relation extraction, and numerical prediction) simultaneously, thereby integrating the originally scattered pipeline processes into a unified end-to-end model; thirdly, combined with retrieval enhancement and chaining thinking technologies, LLMs also show robustness in long documents and cross-sentence dependency scenarios, laying the foundation for the automated construction of large-scale structured databases.
Remain Challenges. However, there are still several lingering issues that need to be addressed: LLMs sometimes generate confident but incorrect predictions (hallucination phenomenon), and their adaptability to extremely professional or the latest literature is limited; long text processing is limited by the size of the context window, making it difficult to complete global information integration across chapters or documents; in addition, support for multi-level nested entities and complex chemical ontologies is still not perfect.
Future Work. Future work can focus on introducing dynamic knowledge retrieval and knowledge graph fusion to build a continuously updated domain memory; exploring multimodal extraction (such as graph, spectrum, and text joint understanding); and combining uncertainty estimation and active learning strategies to improve the reliability and interpretability of the model, ultimately achieving a fully automated pipeline from laboratory notes to enterprise-level chemical knowledge bases.
5.3.6. Property-Directed Chemical Design
In property-directed inverse design, one begins with a set of target property criteria (for example, minimum thresholds for cell permeability, binding affinity, or solubility), optional domain priors encoded by pretrained generative LLMs, and a synthesizability filter to ensure practical feasibility. A LLM then directly generates candidate molecular structures—expressed as SMILES strings or molecular graphs—that are chemically valid, synthetically accessible, and predicted to meet the specified property requirements. Property-Directed Chemical Design is fundamentally a Text2Mol task. An everyday analogy might be describing a flavor or recipe you want (“something that tastes like chocolate but is spicier”) and having a chef create a new dish – here we describe a desired chemical property or scaffold, and the model devises a compound. The method’s objectives are to maximize compliance with target properties, promote novelty and diversity beyond natural-product scaffolds, and guarantee synthesizability via rule-based or learned retrosynthetic filters. Key constraints include chemical validity (proper valence and connectivity), synthesizability scores (e.g., predicted accessibility), and in-silico property feasibility. Analogous to random mutation screening but executed computationally at scale, only those molecules that satisfy both the validity/synthesizability filters and the predefined property thresholds are retained as viable candidates.
Key subtasks include conditional molecule generation, and text-conditioned molecule generation (where input is a description of desired properties or a prompt like “a molecule similar to morphine but non-addictive” and output is a new molecule suggestion). Another subtask is reaction-based text to molecule, such as “the product of acetone and benzaldehyde in aldol condensation” – where the input text implies a reaction and the output is the product structure. We will focus on (1) Conditional molecule generation and (2) De novo molecule generation, since these illustrate the spectrum from well-defined translation to open-ended generation.
Conditional Molecule Generation. Conditional molecule generation seeks to design novel compounds that satisfy user-specified criteria—whether a textual description, property targets, or structural constraints—directly in SMILES or 3D form.
The earliest text-conditioned methods relied on prompt-based sampling: both MolReGPT [
889], which leverages GPT-3.5/4’s in-context few-shot learning to generate SMILES without any fine-tuning (albeit with variable chemical accuracy and prompt dependence), and Jablonka et al.’s GPT-3 adaptation [
924], which fine-tuned the base model via prompt prefixes to produce valid SMILES matching property labels, showed that off-the-shelf LLMs can be repurposed for prompt-based conditional generation. In contrast, MolT5 [
887] tackled text-to-SMILES translation via a T5 encoder–decoder fine-tuned on paired natural-language captions and SMILES, pioneering direct text-to-molecule mapping but without built-in multi-objective controls. To improve semantic alignment and diversity, TGM-DLM [
986] introduced a diffusion-based language model conditioned on text embeddings, yielding molecules that more faithfully match user descriptions at the cost of extra compute. Recognizing the need for scaffold specificity, SAFE-GPT [
957] adopted a fragment-based “SAFE” token representation, enforcing user-provided cores in each output while retaining peripheral variability. Extending conditioning into three dimensions, BindGPT [
1056] embeds protein pocket geometries alongside sequence tokens to perform 3D structure-conditioned generation, enabling de novo ligand design tailored to binding-site shapes but requiring specialized 3D inputs. Beyond text and structure, target- and context-specific generation has been advanced by cMolGPT [
1057], which integrates protein–ligand embedding vectors into a MOSES-pretrained transformer to produce candidate libraries for EGFR, HTR1A, and S1PR1 with QSAR-predicted activities correlating at Pearson r > 0.75, and by PETrans [
1058], which couples a protein-sequence or 3D-pocket encoder with a SMILES decoder to generate ligands that respect detailed binding-site features. And ChemSpaceAL [
1059] solves data-efficient, target-focused exploration by wrapping an uncertainty-driven active-learning acquisition loop around its transformer generator, iteratively sampling and scoring molecules against protein profiles to drastically reduce the need for large annotated inhibitor datasets while still uncovering high-affinity candidates.
De novo Molecule Generation. Here the task is: generate a molecule that fits a given textual description. This description could be very general (“a potent opioid painkiller that is less addictive”) or very specific (“a molecule with an ether linkage, and a molecular weight under 300 Da”). This is one of the holy grails of AI in drug discovery – allowing scientists to simply specify desired qualities and having the AI propose novel structures that meet them. It’s akin to an artist drawing a creature based on a myth description, except here it’s a chemist “drawing” a molecule based on a target profile. It could drastically speed up the brainstorming phase of drug design or materials design. Instead of manually tweaking structures, a researcher could ask the model for ideas: “Give me a drug-like molecule that binds to the serotonin receptor but doesn’t cross the blood-brain barrier” – a very high-level goal – and get some starting points.
Early work in de novo molecular design leveraged Adilov’s “Generative Pretraining from Molecules” [
1060] , which adapted a GPT-2–style causal transformer to learn SMILES syntax via self-supervised pretraining and introduced adapter modules between attention blocks for minimal-change fine-tuning. This approach provided a resource-efficient generative backbone for both molecule creation and downstream property prediction. Scaling up, MolGPT [
870] implemented a 6 million-parameter decoder-only model with masked self-attention to capture long-range SMILES dependencies, enforce valency and ring-closure rules for high-quality, chemically valid generation, and employ salience measures for token-level interpretability. MolGPT outperformed VAE-based baselines on MOSES and GuacaMol by metrics such as validity, uniqueness, Frechet ChemNet Distance, and KL divergence.
To better model global string context, Haroon et al. [
1061] added relative attention heads to their GPT architecture, tackling the long-range dependency challenge and boosting validity, uniqueness, and novelty. ChemGPT [
980] then systematically explored hyperparameter tuning and dataset scaling, revealing how pretraining corpus size and domain specificity drive generative performance. Subsequent work by Wang et al. further refined architectures and training strategies to surpass MolGPT benchmarks in de novo tasks. Departing from SMILES, Mao et al.’s iupacGPT [
982] trained on IUPAC name sequences using SELFIES masking and adapters, producing human-interpretable outputs that align with chemists’ naming conventions and streamline validation, classification, and regression workflows. GraphT5 [
897] first introduced multi-modal cross-token attention between 2D molecular graphs and text, enabling text-conditioned graph generation but lacking explicit control over scaffold or property constraints . MolCA [
899] added uni-modal adapters and a cross-modal projector to improve robustness across representations, yet remained confined to 2D structures and did not follow complex textual instructions reliably.
To capture spatial information, 3D-MoLM [
898] incorporated 3D molecular coordinates alongside text, allowing generation of conformers matching optical or binding-site descriptions, but it struggled with scaffold fidelity and multi-objective trade-offs. UTGDiff [
987] addressed instruction fidelity by using a unified text-graph diffusion transformer that follows detailed prompts for substructure and property constraints. Addressing chirality, Yoshikai et al. [
1062] coupled a transformer with a VAE and used contrastive learning from NLP to generate multiple SMILES representations per molecule—enhancing molecular novelty and validity while capturing stereochemical information. AutoMolDesigner [
1063] wrapped de novo generation into an open-source pipeline for small-molecule antibiotic design, emphasizing domain-specific automation with heuristic filters and reaction-feasibility checks. Taiga [
1064] introduced a two-stage approach—unsupervised SMILES → latent mapping followed by REINFORCE-based fine-tuning on metrics like QED, IC
50, and anticancer activity—to achieve property-optimized design via reinforcement learning. Finally, cMolGPT [
1057] demonstrated flexible mode switching, operating unconditionally to explore chemical space de novo and switching seamlessly to conditional, target-focused generation under the same architecture, thus unifying both paradigms in a single LLM framework.
Problems Solved by LLMs. LLMs have demonstrated that there exists a learnable mapping from natural language to chemical structures, allowing chemists to “draw” molecules with words instead of manually constructing SMILES strings. For example, MolT5—jointly trained on text and 100 million SMILES—can generate precisely “COc1ccccc1” in response to “Give me a molecule containing a phenyl ring, an ether linkage, and a molecular weight under 300Da.” MolReGPT goes further: using ChatGPT’s few-shot prompting, it can output valid candidate structures matching “phenyl ring + ether + MW<300” with no fine-tuning required. This capability drastically lowers the design barrier—researchers need only describe desired features to obtain testable structures. And more importantly, it dramatically narrows the chemical search space, focusing billions of possible molecules down to hundreds or thousands of the most relevant candidates and thereby greatly accelerating discovery. Moreover, LLMs support real-time, multi-objective, and multi-constraint generation—such as “increase hydrophilicity while retaining a hydrophobic ring” or “balance potency and synthetic accessibility”—and can explore chemical space rapidly even in low- or zero-data scenarios.
Remain Challenges. The imagination of LLMs is limited by their training. If certain property correlations were never seen, the model might not know how to fulfill a prompt. Also, validity of generated molecules is a concern – models like ChemGPT when generating freely can sometimes produce invalid SMILES or chemically impossible structures (though less often as training improves). When guided by text, the risk of hallucinating a molecule that meets text but is chemically nonsensical is real. For example, an LLM might attempt to satisfy “nonflammable gas” and produce something like “XeH” – which is not a stable molecule, but fits the prompt superficially (xenon is nonflammable, but xenon hydride is not a thing). Ensuring chemical validity often requires adding a post-check (like using cheminformatics software to validate and correct if needed). Another issue is the evaluation of success: if an AI generates a molecule from a prompt “potent opioid with less addiction,” how do we know it succeeded? We would have to test the molecule in silico or in lab. So often these models are used to generate candidates which are then fed into predictive models or experiments. It’s more of an ideation tool currently.
Future Work. We expect to see tighter integration of narrative-guided chemical design generation with other models and databases. One likely scenario is an interactive system: the user gives a prompt, the AI generates a molecule, then another AI (or the same with a different prompt) evaluates the molecule’s properties and explains why it might or might not meet the criteria, then the user or an automated agent refines the prompt or adds constraints, and the cycle continues – essentially an AI-driven design loop. Another direction is combining this with reinforcement learning or Bayesian optimization: use the text prompt to generate an initial population of molecules, then optimize them using property predictors (some recent work uses LLMs with in-context learning to do Bayesian optimization for catalysts) [
1041], hinting at possibilities of optimization within the model’s latent space. Also, as these generative models improve, one can imagine integrating hard constraints (like no toxic substructures or obey Lipinski’s rules for drug-likeness) directly via prompt or via a filtering step in the generation process (some have tried using fragment-based control tokens, e.g., telling the model “include a benzene ring” or “avoid nitro groups”). Another interesting future aspect is diversity vs. focus: LLMs might have a bias to generate molecules that are similar to what they know (so-called mode collapse around familiar structures). Future models might include techniques to encourage more novelty (perhaps via lower sampling temperature or specialized training objectives) when novelty is desired. Conversely, if a very specific structure is needed, one might combine text prompt with a partial structure hint (like providing a scaffold SMILES and asking the model to complete it with substituents that confer certain properties).
5.3.7. Chemical Knowledge Narration
Chemical knowledge narration tasks take unstructured chemical text as input and produce another, more structured or user-friendly text output. It is fundamentally a Text2Text task. For example, given the free-form procedure “Add 5g of sodium hydroxide to 50mL of water, stir for 10 min, then slowly add 10g of benzaldehyde at ,” a model can generate a standardized protocol: “1. Dissolve 5g NaOH in 50mL H2O. 2. Cool to . 3. Add 10g benzaldehyde dropwise over 5 min. 4. Stir at for 10 min.” Likewise, from “The oxidation of cyclohexanol to cyclohexanone was performed using PCC in dichloromethane,” it can produce the concise summary “Cyclohexanol was oxidized to cyclohexanone with PCC in DCM,” and given the SMILES “CC(=O)OC1=CC=CC=C1C(=O)O” it can output the IUPAC name “2-acetoxybenzoic acid (aspirin).” These transformations standardize and clarify experimental descriptions, automate nomenclature and summarization, and enable seamless integration into electronic lab notebooks, saving researchers hours of manual editing.
Common chemical knowledge narration applications include protocol standardization, reaction summarization, SMILES-IUPAC conversion, literature summarization, question-answer generation, and explanatory paraphrasing. In our work, we harness these capabilities for chemical knowledge QA, chemical text mining, and chemical education.
Chemical Knowledge QA. Chemical Knowledge QA is the task of answering natural-language queries about chemical concepts, reactions, and properties by retrieving and reasoning over relevant information from unstructured or structured chemical sources.
Chemical knowledge QA first saw major gains with LlaSMol [
1065], an instruction-tuned model using the SMolInstruct dataset (over three million samples, covering 14 chemistry tasks), which outperformed GPT-4 on several chemistry benchmarks such as SMILES-to-formula conversion and other canonical tasks [
1065]. Nevertheless, it remains bounded by its training data cutoff, and it does not explicitly handle visual structure figure input in its evaluated tasks. To fill in more visual reasoning ability, ChemVLM [
1007] introduces a multimodal model integrating chemical images and text, enabling it to perform tasks like chemical OCR, multimodal chemical reasoning, and molecule understanding from visual plus textual cues. It achieves competitive performance across these tasks [
1007]. However, like many static QA models, its update of chemical knowledge is limited by its training corpus, and occasional incorrect or incomplete answers remain a concern.
Chemical Text Mining. Chemical Text Mining is the task of extracting and structuring relevant chemical information—such as reactions, molecular properties, or entity relationships—from unstructured textual sources (e.g., scientific articles, patents, and lab reports).
Chemical text mining has seen clear advances with models such as LlaSMol [
1065], which uses the SMolInstruct dataset (over three million samples across 14 chemistry tasks) to fine-tune open-source LLMs, achieving substantial improvements over general-purpose models [
1065]. More recently, the work “Fine-tuning Large Language Models for Chemical Text Mining” [
965] demonstrated that fine-tuned models (including ChatGPT, GPT-3.5-turbo, GPT-4, and open-source LLMs such as Llama2, Mistral, BART) can handle five extraction tasks—compound entity recognition, reaction role labeling, MOF synthesis information extraction, NMR data extraction, and conversion of reaction paragraphs to action sequences—with exact accuracy in the range of approximately 69% to 95% using minimal annotated data [
965]. There are also works which fine-tune pretrained LLMs (GPT-3, Llama-2) to jointly perform named entity recognition and relation extraction across sentences and paragraphs in materials chemistry texts, outputting structured or JSON-like formats with good performance in linking entities like dopants-host, MOF information, and general composition/phase/morphology/application extraction [
976].
Chemistry Education LLMs. Chemistry education LLMs have evolved from generic chatbots to increasingly specialized, curriculum-aligned tutors. For instance, ChemLLM [
926] is among the first LLMs dedicated to chemistry: trained on chemical literature, benchmarks, and dialogue interactions, it can provide explanations, perform interpretation of core concepts, and respond to student queries in a dialogue style with reasonable domain accuracy. Another relevant study[
863], establishes a benchmark comprising eight chemistry tasks (e.g. explanation, reasoning, formula derivation), showing that models like GPT-4, GPT-3.5, Davinci-003 etc. outperform generic LLMs in zero-shot and few-shot settings on many such tasks. A further perspective by Du et al. [
1019] discusses how LLMs can assist in lecture preparation, guiding students in wet-lab and computational activities, and re-thinking assessment styles, though it does not report a system that simultaneously generates new problems, offers misconception warnings, and supports dialogic tutoring. Each generation—from ChemLLM’s dialogue capability, to benchmark studies demonstrating explanation + reasoning, to educational perspectives exploring scalable assessment—shows how chemistry-tuned LLMs are gradually moving toward more capable teaching assistants, though fully interactive, curriculum-aligned AI tutors remain an open challenge.
Problems Solved by LLMs. LLMs have revolutionized chemical knowledge narration tasks by enabling end-to-end transformations—standardizing free-form procedures, summarizing complex reaction descriptions, and converting between SMILES and IUPAC names—all in a single, unified model without the need for multiple specialized tools or extensive manual intervention. Their strong contextual understanding and few-shot/in-context learning capabilities allow them to adapt quickly to new tasks with minimal examples, dramatically cutting the time researchers spend editing protocols, writing summaries, or updating electronic lab notebooks.
Remain Challenges. LLMs still occasionally “hallucinate” confidently incorrect details, struggle with long documents that exceed their context windows, and lack robust mechanisms for handling deeply nested or multi-step reaction descriptions.
Future Work. Future work must therefore focus on integrating retrieval-augmented generation to ground outputs in real literature, fusing text with experimental figures or spectra for more accurate multimodal summaries, incorporating chain-of-thought prompting to produce auditable reasoning paths, maintaining a dynamically updated chemical knowledge base to stay current with new findings, and developing specialized evaluation benchmarks for protocol standardization, reaction summarization, and nomenclature conversion. These advances will make LLMs even more reliable, explainable, and indispensable for chemical knowledge QA, text mining, and education.
5.3.8. Benchmarks
As summarized in
Table 36, we have compiled a comprehensive list of datasets that have been employed across a broad range of chemistry tasks using LLMs. This table includes benchmarks for tasks such as molecular property prediction, reaction yield prediction, reaction type classification, reaction kinetics, molecule captioning, reaction product prediction, chemical synthesis planning, molecule tuning, conditional and de novo molecule generation, chemical knowledge question answering, chemical text mining, and chemical education.
Systematically cataloging current research benchmarks is essential to bridge the gap between generic language modeling advances and chemistry-specific tasks. By unifying evaluation protocols—standardizing data splits, SMILES preprocessing, and task formulations—we ensure that performance comparisons are both fair and reproducible. Moreover, a holistic survey reveals not only the breadth of existing benchmarks (from molecular property regression and reaction-type classification to molecule captioning and generative design) but also their inherent biases: most datasets focus on drug-like organic molecules or patent reactions, while domains such as inorganic chemistry, environmental pollutants, and negative-result reporting remain underrepresented. This comprehensive overview therefore lays the foundation for more rigorous model development and benchmarking, guiding researchers toward data curation and experimental designs that fully exploit LLM capabilities in chemical contexts.
In the sections that follow, we will select several of the most influential datasets from
Table 36 for in-depth discussion. For each chosen dataset, we will describe its scope, annotation scheme, and typical use cases, and then survey the performance of representative LLMs on these benchmarks to elucidate current capabilities and remaining challenges.
MoleculeNet.(Mol2Num) MoleculeNet is a consolidated benchmark suite that currently bundles sixteen public datasets spanning quantum chemistry, physical chemistry, biophysics and physiology. All tasks share a uniform, text–first layout: each row of the .csv file starts with a canonical SMILES string followed by one or more property labels—binary for classification (e.g. BBBP, BACE, HIV, Tox21, SIDER) or floating–point for regression (ESOL, FreeSolv, Lipophilicity, the QM series). Where 3-D information is required, companion .sdf or NumPy archives store Cartesian coordinates and energies. Official JSON split files define random, scaffold and temporal partitions so that every study can reproduce the same train/validation/test folds. A typical classification row from BACE reads CC1=C(C2=C(N1)C(=O)N(C(=O)N2)Cc3ccccc3)O,1, the trailing "1" indicating an active -secretase inhibitor. In the regression task ESOL a sample line might be OC1=CC=CC=C1,-2.16, pairing a phenol SMILES with its experimental log-solubility in mol L−1.
Table 35.
Performance of large (chemical) language models and strong baselines on MoleculeNet (↑ = higher is better for ROC-AUC; ↓ = lower is better for RMSE). Best per column in blue. Values are taken from original papers/model cards; “—” means not reported.
Table 35.
Performance of large (chemical) language models and strong baselines on MoleculeNet (↑ = higher is better for ROC-AUC; ↓ = lower is better for RMSE). Best per column in blue. Values are taken from original papers/model cards; “—” means not reported.
| Model (source) |
Classification (ROC-AUC ↑) |
Regression (RMSE ↓) |
|
BACE
|
BBBP
|
HIV
|
Tox21
|
SIDER
|
ESOL
|
FreeSolv
|
Lipo
|
| MolBERT |
0.866 |
0.762 |
0.783 |
— |
— |
0.531 |
0.948 |
0.561 .[1] |
| ChemBERTa-2 |
0.799 |
0.728 |
— |
— |
— |
0.889 |
1.363 |
0.798[2] |
| BARTSmiles |
0.705 |
0.997†
|
0.851 |
0.825 |
0.745 |
— |
— |
—[3] |
| MolFormer-XL |
0.690 |
0.948 |
0.847 |
— |
0.882†
|
— |
— |
0.937†[3] |
| ImageMol |
— |
— |
0.814 |
— |
— |
— |
— |
—[4] |
| SELFormer |
0.832 |
0.902 |
0.681 |
0.653 |
0.745 |
0.682 |
2.797 |
0.735[5] |
Table 36.
Chemistry Tasks, Benchmarks, Introduction and Cross tasks
Table 36.
Chemistry Tasks, Benchmarks, Introduction and Cross tasks
| Type of Task |
Benchmarks |
LLM Tested |
Introduction |
Cross tasks |
| Property Prediction |
MoleculeNet [1066] |
✓ |
16 public datasets (SMILES + labels; quantum, phys-chem, physiology; 130k–400k samples) |
– |
| OGB-PCQM4M-v2 [1067] |
× |
3.8M molecular graphs with 3-D coords + HOMO–LUMO gaps |
Reaction Rate |
| Therapeutics Data Commons [1068] |
✓ |
30+ ADMET/bioactivity CSVs, leaderboard splits |
Chemical Synthesis |
| PDBbind [1069] |
× |
22k protein–ligand complexes; PDB/mol2 +
|
– |
| BindingDB [1070] |
× |
3M structure–target Ki/IC50 pairs (SMILES, FASTA) |
– |
| Open Catalyst OC22 [1071] |
× |
Adsorption geometries + barriers for 1.3 M configs |
Rate |
| Reaction Yield Prediction |
Buchwald–Hartwig HTE [1035] |
× |
3955 C–N couplings; CSV (yield, ligands, bases) |
– |
| Suzuki–Miyaura HTE [1072] |
× |
5760 C–C couplings; yield matrix |
– |
| USPTO-Yield |
✓ |
1M patent reactions with numeric yields |
Reaction type |
| Open Reaction Database (ORD) [1073] |
× |
JSON records: reactants / products / conditions / yield |
Reaction type |
| ORDerly-Yield [1074] |
× |
Clean ORD + USPTO split, reproducible splits |
Reaction Product Prediction |
| AstraZeneca ELN [1075] |
× |
25k ELN entries, diverse chemistries (CSV) |
Conditions optimisation |
| Reaction Type Classification |
USPTO-50K [1076] |
✓ |
50036 atom-mapped reactions, 10 classes |
Chemical Synthesis, Reaction Yield |
| USPTO-Full / MIT [1076] |
✓ |
400k–1.3M reactions; 60 coarse classes |
Reaction Product, Chemical Synthesis |
| Reaxys Reaction [1077] |
× |
40M literature reactions, multi-level class labels |
Reaction Rate |
| ORDerly-Class [1074] |
× |
ORD subset with curated type labels |
– |
| Reaction Rate / Kinetics |
NIST SRD-17 [1078] |
× |
38k gas-phase rate constants, Arrhenius params (XML) |
– |
| RMG Kinetics DB [1079] |
× |
50k gas+ surface elementary steps (YAML) |
Mechanism generation |
| NDRL/NIST Solution DB |
× |
17k solution-phase rate constants |
– |
| Combustion FFCM |
× |
Curated small-fuel flame kinetics set (YAML) |
– |
| Molecule Captioning |
ChEBI-20 [1080] |
✓ |
33k SMILES–natural-language pairs (JSON) |
Retrieval |
| SMolInstruct (Caption) [1065] |
✓ |
3M multi-task instructions incl. caption pairs |
– |
| MolGround [1081] |
✓ |
20k captions with atom-level grounding tags |
– |
| Reaction Product Prediction |
USPTO-MIT Forward [921] |
✓ |
400k one-step reactions; SMILES input → product |
Reaction Type |
| ORDerly-Forward [1074] |
× |
ORD/USPTO split, non-USPTO OOD set |
Reaction Yield |
| Chemical Synthesis |
USPTO-50K Retro [1082] |
✓ |
50k product → reactant pairs, 10 classes |
Reaction class |
| PaRoutes [1083] |
× |
20k multi-step routes, JSON graphs |
– |
| ORDerly-Retro [1074] |
× |
ORD split with non-USPTO OOD test |
Reaction Product |
| TDC Retrosynthesis [1068] |
✓ |
Wrappers for USPTO-50K + PaRoutes |
– |
| AiZynthFinder test [1084] |
× |
100 difficult drug targets, MOL files |
– |
| Molecule Tuning |
GuacaMol Goal-Dir. [1085] |
✓ |
20 oracle tests (SMILES, property calls) |
Molecule Generation |
| PMO Suite [1086] |
✓ |
23 tasks, score-limited oracle calls (JSON) |
– |
| MOSES Opt [1086] |
✓ |
Scaffold-constrained optimisation splits |
De novo Molecule Generation |
| TDC Docking [1068] |
× |
AutoDock/Vina scoring tasks (SDF) |
Conditional Protein Generation |
| LIMO Affinity [1087] |
× |
Gradient VAE optimisation toward nM affinity |
Conditional Generation |
| Chemical Text Classification |
ChemProt [1088] |
× |
1820 PubMed abstracts with 5 CP-relation labels |
QA, Chemical text mining |
| BC5-CDR [1089] |
× |
1500 abstracts; chemical–disease relations |
NER |
| CHEMDNER / BC4CHEMD [1090] |
× |
10k abstracts, 84k chemical mentions |
NER |
| NLM-Chem [968] |
× |
150 full-text articles, gold chemical tags |
Chemical text mining |
| ChEMU Patents [1091] |
× |
1.5k patent excerpts with entity + event labels |
Chemical text mining |
| ChemNER 62-type [1092] |
× |
Fine-grained 62-label NER corpus |
– |
| Cond. Mol Generation |
MOSES Scaffold [1086] |
× |
Bemis-Murcko prompts → molecules |
De novo molecule generation |
| MOSES-2 [1086] |
× |
Adds stereo + logP/MW targets (JSON) |
– |
| GuacaMol Cond. [1085] |
✓ |
Similarity + property dual constraints |
Molecule Tuning |
| LIMO [1087] |
× |
Latent inversion with docking-affinity oracle |
– |
| De Novo Mol Gen |
MOSES (dist.) [1086] |
✓ |
1.9M ZINC clean-leads SMILES, train/test/scaffold |
Conditional generation |
| GuacaMol Dist. [1085] |
✓ |
10 distribution-learning metrics tasks |
Optimisation |
| GEOM-Drugs [1093] |
× |
100k drug-like molecules + 3-D conformers |
Conformer generation |
| QMugs [1094] |
× |
665k drug-like molecules with QM labels |
Property prediction |
| TDC MolGeneration [1095] |
× |
Unified wrapper for MOSES/GuacaMol |
– |
| Chemical Knowledge QA |
ScholarChemQA [1001] |
✓ |
40 k yes/no/maybe QAs from research abstracts |
Chemical text mining |
| ChemistryQA [1096] |
× |
4500 high-school calc-heavy MCQs |
Education |
| MoleculeQA [1000] |
✓ |
12k molecule-fact QAs (SMILES + text) |
Molecule Captioning |
| MolTextQA [1097] |
× |
MC-QA over PubChem descriptions |
– |
| Chemical text mining |
CHEMDNER [1090] ChEMU [1091] NLM-Chem [968] |
|
See classification rows (NER + event extraction) |
NER / IE suites |
|
ChemBench [1098] |
× |
7059 curated curriculum QAs; JSON |
QA |
| Chemical Education |
ChemistryQA [1096] |
× |
High-school MCQ dataset (LaTeX problems) |
QA |
USPTO.(Mol2Num, Mol2Mol) Starting from Daniel Lowe’s 1.8-million raw patent dump—which stores un-mapped SMILES triples like O=C=O.OCCN>>O=C(O)NCCO—researchers have carved out several task-specific subsets. The USPTO-MIT forward-prediction split keeps 479035 atom-mapped reactions and is the de-facto benchmark for single-step product prediction, where the input string CC(=O)Cl.O=C(O)c1ccccc1>>? asks the model to regenerate the ester product. USPTO-50K retains 50014 lines and appends a ten-class label, enabling reaction-type classification exemplified by CCBr.CC(=O)O>[base]>CCOC(=O)C,7, where “7” represents an acylation class. Building on the same patent source, USPTO-Yield merges textual yield phrases so that a row such as C1=CC=CC=C1Br.CC(=O)O>[Cu]>C1=CC=CC=C1CO,72 allows numeric yield regression, while USPTO-Stereo preserves wedge-bond chirality, demanding stereochemically exact output for inputs like C[C@H](Cl)Br.O>>?. Beyond single steps, PaRoutes links 450000 Lowe reactions into multistep route graphs so a model must recreate the full path ending in C1=CC=C(C=C1)C(=O)O rather than just its terminal disconnection. Finally, ORDerly re-formats selected USPTO lines into Open-Reaction-Database JSON with timestamped splits—each entry like "inputs":["smiles":"CCOC(=O)Cl"],"outputs":["smiles":"CCOC(=O)O"], "temperature": 298—so that forward prediction, condition inference and genuine time-splitting can be assessed simultaneously. Together these sub-corpora let large chemical language models be probed across product generation, type classification, yield regression, stereochemical accuracy and multistep planning without ever leaving the USPTO domain.
Table 37.
Transformer-scale LLM performance across USPTO sub-datasets. Columns report the main metric for each task: forward product Top-k accuracy on USPTO-MIT; reaction-type accuracy and Macro-F1 on USPTO-50K; yield regression and MAE on USPTO-Yield; stereochemical Top-1 accuracy on USPTO-Stereo; multi-step route success on PaRoutes; and forward Top-1 / condition accuracy on ORDerly. Best in each column is highlighted with blue.
Table 37.
Transformer-scale LLM performance across USPTO sub-datasets. Columns report the main metric for each task: forward product Top-k accuracy on USPTO-MIT; reaction-type accuracy and Macro-F1 on USPTO-50K; yield regression and MAE on USPTO-Yield; stereochemical Top-1 accuracy on USPTO-Stereo; multi-step route success on PaRoutes; and forward Top-1 / condition accuracy on ORDerly. Best in each column is highlighted with blue.
| Model |
USPTO-MIT |
USPTO-50K |
USPTO-Yield |
USPTO-Stereo |
| Top-1↑ |
Top-5↑ |
Top-10↑ |
Acc.↑ |
F1↑ |
↑ |
MAE |
Top-1↑ |
| Molecular Transformer |
0.875 |
0.937 |
0.954 |
— |
— |
— |
— |
0.825 |
| Augmented Transformer |
0.888 |
0.944 |
0.960 |
0.921 |
0.909 |
— |
— |
0.832 |
| MolFormer |
0.883 |
0.945 |
0.960 |
0.945 |
0.932 |
— |
— |
0.832 |
| ReactionBERT |
0.930 |
0.972 |
0.981 |
0.930 |
0.918 |
— |
— |
0.845 |
| Chemformer |
0.910 |
0.968 |
0.979 |
0.915 |
0.905 |
— |
— |
0.834 |
| ReactionT5 |
0.975 |
0.986 |
0.988 |
— |
— |
— |
— |
0.790 |
| CompoundT5 |
0.866 |
0.895 |
0.904 |
— |
— |
— |
— |
— |
| ProPreT5 |
0.998 |
1.000 |
1.000 |
— |
— |
— |
— |
— |
| ChemBERTa-2 |
— |
— |
— |
0.880 |
0.865 |
— |
— |
— |
| Yield-BERT |
— |
— |
— |
— |
— |
0.41 |
13.2 |
— |
| ReaLM |
— |
— |
— |
— |
— |
0.52 |
10.5 |
— |
ChEBI-20.(Mol2Text) ChEBI-20 is a medium-sized molecular–caption corpus that links 33,010 small-molecule SMILES strings to concise English sentences distilled from the ChEBI ontology. The data are released as a UTF-8 CSV whose first column stores the canonical SMILES and whose second column holds the free-text caption; a third column specifies the standard 8:1:1 train/validation/test split. Because each record couples structure and language, the collection naturally supports molecule-to-text caption generation, text-to-molecule retrieval and cross-modal representation learning. In the captioning task a model receives the input CC(C)OC(=O)C1=CC=CC=C1C(=O)O and is expected to output a sentence such as “Ibuprofen is a propionic acid derivative with an isobutyl side chain and an aromatic core.” In the inverse retrieval task the same caption is fed to the system, which must rank the correct SMILES ahead of thousands of distractors; the ground-truth pair above therefore serves both roles without modification.
Table 38.
Molecule captioning on ChEBI-20. Best per metric in blue.
Table 38.
Molecule captioning on ChEBI-20. Best per metric in blue.
| Model |
BLEU-2↑ |
BLEU-4↑ |
METEOR↑ |
ROUGE-1↑ |
ROUGE-2↑ |
ROUGE-L↑ |
| MolT5-base |
0.540 |
0.457 |
0.569 |
0.634 |
0.485 |
0.578 |
| MolReGPT (GPT-4-0314) |
0.607 |
0.525 |
0.610 |
0.634 |
0.476 |
0.562 |
| MolT5-large |
0.594 |
0.508 |
0.614 |
0.654 |
0.510 |
0.594 |
| Galactica-125M |
0.585 |
0.501 |
0.591 |
0.630 |
0.474 |
0.568 |
| BioT5 |
0.635 |
0.556 |
0.656 |
0.692 |
0.559 |
0.633 |
| ICMA (Galactica-125M) |
0.636 |
0.565 |
0.648 |
0.677 |
0.537 |
0.618 |
USPTO-MIT.(Mol2Mol, Mol2Num) The USPTO-MIT dataset, curated by MIT from Lowe’s extraction of original USPTO patent reactions and cleaned through atom mapping validation and SMILES normalization, comprises approximately 470,000 single-step forward reaction records split into training, validation, and test sets of 409,035, 30,000, and 40,000 examples respectively; each record encodes atom-mapped reactants, reagents, and products in SMILES (for example, CC(=O)O.CCCO>>CCCOC(=O)C denotes acetic acid and propanol yielding propyl acetate), and these high-quality, atom-mapped reactions support a variety of AI-driven chemistry tasks such as forward reaction prediction, single-step retrosynthesis, reaction classification, template extraction with atom mapping, and reagent prediction.
Table 39.
Forward reaction prediction performance of chemical LLMs and strong non-LLM baselines on the USPTO-MIT Separated dataset. Best per column in blue.
Table 39.
Forward reaction prediction performance of chemical LLMs and strong non-LLM baselines on the USPTO-MIT Separated dataset. Best per column in blue.
| Model |
Top-1↑ |
Top-2↑ |
Top-3↑ |
Top-5↑ |
| Molecular Transformer |
88.8 % |
92.6 % |
— |
94.4 % |
| T5Chem |
90.4 % |
94.2 % |
— |
96.4 % |
| CompoundT5 |
86.6 % |
89.5 % |
90.4 % |
91.2 % |
| ProPreT5 |
99.8 % |
— |
— |
— |
| ReactionT5 |
97.5 % |
98.6 % |
98.8 % |
99.0 % |
Table 40.
Reaction type classification performance on USPTO-MIT for LLMs / transformer models and top non-LLM baselines. Best per metric in blue.
Table 40.
Reaction type classification performance on USPTO-MIT for LLMs / transformer models and top non-LLM baselines. Best per metric in blue.
| Model |
Top-1 Accuracy↑ |
Top-5 Accuracy↑ |
| Molecular Transformer |
90.4 % |
95.3 % |
| Augmented Transformer |
90.6 % |
96.1 % |
| ProPreT5 |
99.8 % |
— |
GuacaMol.(Mol2Mol) GuacaMol is an open-source de novo molecular design benchmarking suite built from approximately 1.8 million deduplicated SMILES strings standardized from the ChEMBL database. The construction pipeline includes salt removal, charge normalization, element filtering (retaining only H, B, C, N, O, F, Si, P, S, Cl, Se, Br, I), truncation to under 100 characters, and removal of any compounds overly similar to the hold-out set(holdout_set_gcm_v1.smiles). GuacaMol defines 20 goal-directed tasks—ranging from simple property optimization (e.g., log P, TPSA) and rediscovery of known drugs to similarity-guided generation and scaffold-hopping—and, most centrally, molecule tuning multi-objective optimization tasks. These tuning tasks challenge models to perform fine-grained adjustments against scoring functions like QED, log P, and synthetic accessibility rather than merely reproducing the training distribution. For example, in the Cobimetinib multi-objective tuning task, generative models apply Pareto optimization strategies (such as NSGA-II or NSGA-III) to iteratively modify Cobimetinib’s SMILES substituents, maximizing a weighted combination of drug-likeness (QED) and solubility (log S) scores to produce novel candidates balanced across multiple property dimensions. This emphasis on molecule tuning not only tests a model’s ability to replicate known chemical spaces but also measures its practical value in accelerating lead optimization during early drug discovery by finely balancing multiple molecular properties.
GuacaMol not only supports goal-directed multi-property tuning tasks, but also provides two key generative scenarios: conditional molecule generation and de novo generation. In conditional generation, models must produce compounds that satisfy user-specified property or scaffold constraints. For example, MolGPT achieves strong control over QED and log P in GuacaMol’s conditional benchmarks, attaining validity ≈0.98, high uniqueness, and novelty close to 1.000, while cMolGPT extends these approaches by prepending target property values to the input, enabling precise conditional generation. More recently, LigGPT introduces flexible multi-constraint conditioning, allowing a single model to balance multiple property targets while retaining synthesizability and validity.
In the de novo setting, GuacaMol evaluates models on validity, uniqueness, novelty, Fréchet ChemNet Distance (FCD), and KL divergence. Here, MolGPT achieves validity 0.981, uniqueness 0.998, and novelty 1.000, LigGPT improves further with validity 0.986 and novelty 1.000, and SELF-BART excels on distributional similarity metrics such as FCD and KL divergence. Graph-based masked generation approaches also show competitive performance on these benchmarks, highlighting the impact of molecular representation on generation quality. Early generative frameworks such as ChemGAN and Entangled Conditional AAE serve as important references in the distribution-learning tasks, helping the community understand the strengths and limits of deep learning methods in exploring chemical space. Together, GuacaMol’s conditional and de novo tasks offer a comprehensive, rigorous testbed for chemical LLMs, driving continued innovation in model architectures and training strategies.
Table 41.
De novo molecule generation performance on GuacaMol for chemical-domain LLMs. Best per metric in blue.
Table 41.
De novo molecule generation performance on GuacaMol for chemical-domain LLMs. Best per metric in blue.
| Model |
Validity↑ |
Uniqueness↑ |
Novelty↑ |
| MolGPT |
0.981 |
0.998 |
1.000 |
| LigGPT |
0.986 |
0.998 |
1.000 |
| GraphGPT |
0.975 |
0.999 |
1.000 |
| SmileyLlama (T=1.1) |
0.9783 |
0.9994 |
0.9713 |
| SmileyLlama (T=0.6) |
0.9968 |
0.9356 |
0.9113 |
MOSES.(Text2Mol) MOSES is a molecular generation benchmarking platform introduced by Polykovskiy et al. in 2020 in Frontiers in Pharmacology, derived from 4,591,276 SMILES in the ZINC Clean Leads collection and filtered by molecular weight (250–350 Da), rotatable bonds (≤ 7), XlogP (≤ 3.5), removal of charged/ non-C/N/S/O/F/Cl/Br/H atoms, PAINS, and medicinal chemistry filters to yield 1,936,962 drug-like molecules. These molecules are split into training (≈ 1,584,664), test (≈ 176,075), and scaffold-test (≈ 176,089 with unique Bemis–Murcko scaffolds) sets to assess model performance on both seen and unseen scaffolds. MOSES supports de novo generation, where the CharRNN baseline achieves validity 0.975, uniqueness 0.999, novelty 0.842, IntDiv1 0.856 and IntDiv2 0.850 on the test set; and conditional generation, for example SELF-BART applies property-conditioned decoding to generate molecules with desired constraints, attaining validity 0.998, uniqueness 0.999, novelty 1.000, and strong internal diversity scores. Consequently, MOSES serves as a unified and rigorous benchmark for core tasks in molecular generation, spanning distribution-learning to property-driven generation.
Table 42.
De novo (distribution-learning) generation performance on MOSES. Best per metric in blue.
Table 42.
De novo (distribution-learning) generation performance on MOSES. Best per metric in blue.
| Model |
Validity↑ |
Unique@10k↑ |
Novelty↑ |
IntDiv1↑ |
IntDiv2↑ |
| MolGPT |
0.994 |
1.000 |
0.797 |
0.857 |
0.851 |
| SELF-BART |
0.998 |
0.999 |
1.000 |
0.918 |
0.908 |
| MTMol-GPT |
0.845 |
0.993 |
0.984 |
0.835 |
— |
| SF-MTMol-GPT |
1.000 |
0.955 |
0.932 |
0.850 |
— |
Summary. Current benchmark datasets for LLMs in chemistry emphasize three main task categories. First, molecular property prediction dominates, with collections such as MoleculeNet and Therapeutics Data Commons providing numerous binary and regression targets (e.g. solubility, toxicity, binding affinity). Second, reaction outcome and classification tasks, primarily drawn from USPTO and Reaxys repositories, assess models on product prediction, reaction-type labeling, and yield regression. Third, molecule generation benchmarks (MOSES, GuacaMol) evaluate de novo and condition-driven design by measuring validity, novelty, and property optimization. By contrast, complex tasks like multi-step synthesis planning, reaction condition optimization, and 3D conformer reasoning remain underrepresented.
Compared to traditional cheminformatics and rule-based methods, domain-trained transformer models have demonstrated quantitative gains. Here, we illustrate such improvements using three representative tasks that are widely applied: Property Prediction, Reaction Prediction and Classification, and Molecule Generation. In property prediction, specialized SMILES-BERT variants outperform random forests on multiple ADMET assays by several percentage points. In single-step reaction tasks, sequence-to-sequence transformers surpass template-based systems, improving top-1 accuracy by 5–10%. In generative settings, LLM-based generators achieve near-perfect chemical validity (> 98 %) and higher diversity metrics than earlier recurrent or graph-based approaches, while also enabling multi-objective optimization of drug-like properties with average improvements of 10–20 % over heuristic baselines.
Despite encouraging results, current LLMs face important limitations when applied to chemistry domains.
A core challenge is that chemical data are highly structured (e.g. graphs), yet LLMs operate on linear token sequences. This mismatch means that transformers struggle to natively represent molecular topology and 3D geometry. For instance, an LLM given a SMILES string has no direct encoding of the molecule’s shape or stereochemistry, which can be crucial for many properties. This leads to errors when tasks fundamentally depend on spatial or structural reasoning – a known example is that models predicting quantum chemistry properties from SMILES (instead of actual 3D coordinates) perform poorly and misrepresent the true task. Another limitation is the knowledge cutoff of LLMs and their tendency to hallucinate. Without explicit chemical rules, an LLM may propose an impossible reaction or a nonsensical molecule, especially if it hasn’t seen similar examples in training. Ensuring validity and consistency in outputs remains non-trivial; even with grammar-constrained decoding, models might violate subtle chemical constraints or overlook rare elements.
Data scarcity and bias are additional concerns: many benchmark datasets are relatively small and biased toward drug-like molecules, so LLMs may generalize poorly to larger chemical space or unusual chemotypes. Researchers also report that LLM performance can be brittle – small changes in input format (SMILES vs another notation) or prompt phrasing can yield different results, reflecting an unstable understanding. From a practical standpoint, the resource requirements of large models pose a challenge: using cutting-edge LLMs (like GPT-4) can be orders of magnitude more costly and slower than using task-specific models. This makes it difficult for researchers to fine-tune or deploy the largest models on private data.
Finally, the interpretability of LLM decisions is limited – unlike human chemists or simpler models that can point to a mechanistic rationale, a transformer’s prediction is hard to dissect, which can erode trust in sensitive applications (e.g. drug discovery). In summary, today’s chemical LLMs are constrained by input representations, data quality, model transparency, and computational cost, highlighting the need for new strategies to realize their full potential.
From these observations we derive three core insights. First, dataset limitations in both scale and scope constrain LLM performance: public repositories often contain errors, inconsistent annotations, and limited chemical diversity—popular benchmarks such as QM9 can be misused and fail to represent realistic molecular spaces. To overcome this, a community effort should curate larger, cleaner datasets by aggregating high-quality experimental results (e.g. ADME assays, comprehensive reaction outcomes) and expanding initiatives like the Therapeutic Data Commons to include negative results and broader chemistries. Benchmarks must also incorporate crucial chemical information—3D conformations, stereochemistry, reaction conditions (catalysts, solvents, yields)—to foster deeper chemical reasoning beyond simple SMILES pattern matching. Second, model and methodology strategies require refinement. Treating chemical structures as linear text has merits but introduces tokenization and validity challenges, motivating exploration of alternative representations such as SELFIES or fragment-based vocabularies. Generic LLMs lack embedded chemical rules (valence, aromaticity) and benefit from domain-specific pretraining on extensive chemical corpora (molecules, patents, protocols). Hybrid architectures—integrating LLMs with graph neural networks, physics-based modules, or explicit inter-atomic distance matrices—can bridge the gap between sequence models and spatial structure. Finally, improving reliability and usability demands thoughtful task formulation and validation. Reformulating regression tasks as classification or ranking problems, developing chemistry-specific prompting (few-shot, chain-of-thought, multi-step retrosynthesis prompts), and embedding chemical validation loops (reinforcement learning with validity rewards or critic models) can reduce hallucinations and ensure chemical soundness. Coupling LLMs with external tools (“LLM + tools” paradigms) and advancing interpretability—via attention analysis, attribution methods, and standardized evaluation metrics—will build trust and utility. In conclusion, converging richer data, smarter representations, hybrid modeling, and robust benchmark design will propel LLMs toward becoming reliable, powerful instruments for chemical research., the convergence of richer data, smarter representations, hybrid modeling strategies, and thoughtful benchmark design will help overcome current limitations and guide LLMs to become more reliable and powerful tools for chemical research.
5.3.9. Discussion
Opportunities and Impact. LLMs are becoming transformative tools in chemistry and chemical engineering, bridging traditional chemical methods with cutting-edge computational advancements.
In molecular textualization (Mol2Text), the traditional rule-based naming system relies on manually written chemical naming rules, which often find it difficult to cover all cases of novel or complex molecules, while LLMs can learn naming patterns from large-scale chemical corpora, achieving better generalization and robustness. Transformer models such as Struct2IUPAC achieve 98.9 % correct SMILES-IUPAC conversions on a 100 k test set, halving the residual error (
% vs 3 %) observed for the long-standing rule-based parser OPSIN on comparable benchmarks [
1099]. For example, when researchers discover a new antibiotic with a highly unusual structure, these models can immediately generate its official chemical name, saving chemists hours or days of manual naming efforts.
In property prediction (Mol2Number), traditional methods require the training of independent models for each property, making it difficult to share underlying chemical knowledge; while LLMs have absorbed the correlations between properties during the pre-training phase, achieving one-time multi-task predictions. LLM-driven approaches like ChemBERTa unify this process—after training on vast molecular datasets, a single model can simultaneously predict properties like solubility (e.g., “will the compound dissolve easily in water?”), toxicity (e.g., “is the compound safe?”), and expected chemical yields. Scaling ChemBERTa-2 pre-training from 0.1 M to 10 M molecules lifts average ROC-AUC across the MoleculeNet suite by +0.11 (e.g., 0.67 to 0.78) without training separate models per property, whereas classic GCN/GAT baselines plateau near 0.70 [
1027]. For instance, pharmaceutical chemists designing new drugs can quickly assess multiple crucial drug properties simultaneously, significantly streamlining the early drug discovery phase.
For complex reaction planning (Mol2Mol), traditional reaction prediction software largely relies on manual reaction templates, which are difficult to capture complex mechanisms; while LLM based on Transformer can accurately plan synthetic routes by learning long-range dependencies between steps through self-attention mechanisms. LLM-based models, such as those inspired by Reaction Transformer, can break complex reactions into simpler, understandable stages. For example, when developing a complex cancer treatment molecule, these models can accurately suggest each intermediate step in synthesis, increasing the success rate of predicted synthetic routes by 10-20% compared to older rule-based methods. The Molecular Transformer attains 90.4 % top-1 accuracy on USPTO product prediction, while the best contemporary template-driven system (RetroComposer) reaches only 65.9 %, a 24-point jump that translates into far fewer manual template overrides during route design [
921].
In chemical text classification (Text2Num), traditional text mining often relies on manual rules or shallow features, making it difficult to handle context-dependent chemical descriptions; while LLMs can accurately extract reaction conditions and results through deep semantic understanding. For example, researchers fine-tuned LLaMA-2-7B on 100,000 USPTO reaction processes, enabling it to directly generate structured records that comply with the Open Reaction Database (ORD) architecture—the model achieved an overall message-level accuracy of 91.25%, and a field-level accuracy of 92.25%, capable of stably identifying key numerical information such as temperature, time, and yield [
1009]. In comparison, the best feature-engineered CRF/SSVM patent-NER pipeline achieved an F-measure of only 88.9% on the CHEMDNER-patents CEMP task, highlighting the significant performance improvement of LLM in chemical text information extraction [
1100].
Inverse design (Text2Mol) benefits from LLMs’ ability to generate chemically valid candidates under user-specified property constraints, reducing trial-and-error cycles from weeks to minutes and expanding exploration of novel chemical space [
870]. Traditional reverse engineering requires a large amount of trial and error and manual filtering, resulting in low efficiency; while LLM acquires distributed knowledge in the chemical space through pre-training and combines it with conditional generation, which can quickly output high-quality candidate molecules. With LLM-based generative models such as MolGPT [
870] and ChemGPT [
980], chemists can now simply describe the needed properties (e.g., "a molecule that lowers blood pressure but doesn’t cause dizziness") and instantly receive hundreds of suitable, chemically viable molecule suggestions. This dramatically shortens the molecule discovery process from potentially weeks of trial and error down to just minutes. Conditional generators such as Adapt-cMolGPT now yield 100 % syntactically valid molecules under SELFIES-based sampling, compared with 88–94 % validity for earlier SMILES-based GPT decoders—eliminating one in ten invalid proposals and narrowing medicinal-chemist triage [
1101].
Finally, in chemical text mining (Text2Text), traditional text mining often relies on manual rules or shallow features, making it difficult to handle context-dependent chemical descriptions; while LLMs can accurately extract reaction conditions and results through deep semantic understanding. For example, researchers fine-tuned LLaMA-2-7B on 100,000 USPTO reaction processes, enabling it to directly generate structured records that comply with the Open Reaction Database (ORD) architecture—the model achieved an overall message-level accuracy of 91.25%, and a field-level accuracy of 92.25%, capable of stably identifying key numerical information such as temperature, time, and yield [
1009]. In comparison, the best feature-engineered CRF/SSVM patent-NER pipeline achieved an F-measure of only 88.9% on the CHEMDNER-patents CEMP task, highlighting the significant performance improvement of LLM in chemical text information extraction [
1100].
Challenges and Limitations. Despite these advances, LLM-driven chemical models still face critical hurdles.
First, the “experimental validation gap” persists: Despite impressive predictive power, LLM-driven chemical models still require extensive laboratory verification before their results can be trusted for critical decisions. For example, an LLM acts like an experienced chef who can predict delicious recipes based on prior knowledge, but ultimately you still have to cook the dish yourself and taste it to confirm whether it’s actually good [
1102]. Without deeper integration with automated robotic experimental setups or closed-loop experimental cycles, this validation bottleneck remains a significant "last mile" barrier [
1103].
Second, LLMs lack explicit mechanistic reasoning. Current LLM models predominantly learn associative patterns from large-scale datasets, often lacking explicit chemical reasoning or mechanistic insights. Imagine a student who memorizes a vast number of math solutions without understanding the underlying principles; they will struggle when encountering slightly altered problems. Similarly, predicting complex, multi-step chemical reactions (e.g., radical-based cascades) demands a mechanistic understanding that pure memorization from data cannot reliably provide, leading to frequent mistakes in subtle yet critical chemical details and limiting industrial adoption [
932,
1104].
Third, LLMs struggle to generalize to novel and sparsely represented chemical spaces. LLMs heavily depend on the breadth and quality of their training data, causing them to perform inadequately in chemically novel scenarios or sparsely represented reaction classes. For example, an LLM trained mostly on common organic reactions is like a chef proficient in everyday home cooking who suddenly faces preparing sophisticated French cuisine; the unfamiliar ingredients and methods may lead to frequent mistakes. This limitation restricts their predictive reliability in cutting-edge research or niche industrial applications, where innovation frequently occurs [
1105].
Fourth, chemical “hallucinations” remain problematic: Generative chemical models powered by LLMs often produce chemically invalid or practically unsynthesizable molecules, a phenomenon known as chemical "hallucination." For example, an LLM could resemble an imaginative but inexperienced architect designing visually appealing buildings that are impossible to construct due to practical limitations of materials and construction methods. Although integrating rule-based filters partially mitigates this issue, systematic validation approaches remain inadequate, undermining trust in their use for real-world synthesis planning [
1106].
Lastly, domain-specific fine-tuning demands high-quality annotated datasets, which are expensive or impractical to produce at scale for every niche subfield. Without robust few-shot or low-data learning methods, many specialized applications remain out of reach [
1107].
Research Directions.
Hybrid LLM–Mechanistic Frameworks. Combine LLMs with rule-based and physics-informed modules to integrate statistical language understanding with chemical theory.
Multimodal Chemical Representations. Develop architectures that jointly process SMILES, molecular graphs, and spectroscopic or crystallographic data to capture 3D and electronic structure.
Closed-Loop Experimental Pipelines. Integrate LLM outputs into automated synthesis and analysis platforms, enabling rapid hypothesis testing and feedback-driven model refinement.
Data-Efficient Fine-Tuning. Leverage transfer learning, few-shot prompting, and synthetic augmentation of underrepresented reaction data to improve performance in sparse domains.
Explainability and Uncertainty Quantification. Incorporate attribution methods and probabilistic modeling to provide confidence metrics and mechanistic rationales alongside predictions.
Governance-First Deployment. Establish standards for model validation, transparent reporting (model cards), and ethical guidelines to ensure responsible use in chemical research and industry.
Conclusion. LLMs have significantly reshaped workflows in chemical discovery and engineering, but realizing their full potential requires innovations that marry linguistic intelligence with chemical reasoning, robust validation workflows, and ethical governance. By pursuing hybrid, multimodal, and closed-loop approaches, the community can overcome current limitations and drive the next wave of breakthroughs in chemical science and industrial application.
5.4. Life Sciences and Bioengineering
5.4.1. Overview
5.4.1.1 Introduction to Life Sciences
Life sciences refer to all branches of sciences that involve the scientific study of living organisms and life processes [
1108,
1109,
1110]. In other words, they encompasses fields like biology, medicine, and ecology that explore how organisms (from micro-organisms to plants and animals) live, grow, and interact. Life scientists seek to understand the structure and function of living things, from molecules inside cells up to entire ecosystems, and to discover the principles that govern life [
1111,
1112]. In simple terms, life sciences are about studying living things (such as humans, animals, plants, and microbes) to learn how they work and affect each other and the environment [
1109,
1113,
1114]. This knowledge not only satisfies human curiosity about nature but also underpins applications in health, agriculture, and environmental conservation.
Life sciences are vast domains, so their research tasks range from decoding genetic information to observing animal behavior. Traditionally, each type of task has relied on specific methods and tools developed over decades (or even centuries) of biological research [
1108,
1109]. Through its detailed subdivision into specific subfields, we have categorized research in life sciences based on investigations ranging from the microscopic to the macroscopic level [
1115] and summaries of the review topics written by life sciences scientists [
1116,
1117,
1118,
1119,
1120]. The main research tasks and their classic approaches include:
Deciphering Genetic Codes. Understanding heredity and gene function has been a core task. Early geneticists used breeding experiments to infer how traits are inherited [
1121,
1122]. In the 20th century, methods like DNA extraction [
1123,
1124,
1125] and Sanger sequencing [
1126,
1127,
1128] enabled reading the genetic code, while PCR (polymerase chain reaction) [
1129,
1130] revolutionized gene analysis by allowing DNA amplification. Today, high-throughput genome sequencing [
1131,
1132] and bioinformatics [
1133] are standard for genetic research.
Studying Cells and Molecules. A fundamental task is to uncover how cells and their components (proteins, nucleic acids, etc.) function. Biochemists use centrifugation [
1134,
1135] and chromatography [
1136] to separate molecules, and X-ray crystallography [
1137,
1138] or NMR to determine molecular structures [
1139]. For instance, gel electrophoresis [
1140] became a routine method to separate DNA or proteins by size, and later innovations like Western blots [
1141] provided ways to detect specific molecules. These methods, combined with controlled experiments in test tubes, have traditionally powered discoveries in molecular and cell biology.
Physiology and Medicine. Life sciences also deals with whole organisms, how organ systems work and how to treat their maladies [
1142,
1143,
1144,
1145,
1146,
1147,
1148]. Physiological experiments on model organisms (from fruit flies and mice to primates) have been crucial [
1143,
1144]. For example, testing organ function [
1149] or disease processes [
1150] often involves animal models where interventions can be done. In medicine, clinical observations and clinical trials (systematic testing of treatments in human volunteers) are standard for linking biological insights to health outcomes. Additionally, fields like immunology and neuroscience have developed specialized methods (e.g. antibody assays [
1151,
1152], brain imaging [
1153]) to probe complex systems. Life scientists in these areas often use a combination of laboratory experiments, medical imaging (like MRI, X-rays), and longitudinal studies to unravel how the human body (and other organisms) maintain life and what goes wrong in diseases [
1154,
1155].
Ecology and Evolution. A broader task is understanding life at the population, species, or ecosystem level, how organisms interact with each other and their environment, and how life evolves over time [
1156,
1157,
1158]. Field observations and experiments are the cornerstone of ecology – researchers might count and tag animals, survey plant growth, or manipulate environmental conditions in the wild [
1159]. Long-term ecological research (e.g. observing climate effects on forests [
1160]) and paleontological methods [
1161] have illuminated evolutionary history. In evolution, apart from the fossil record, comparing DNA/protein sequences across species (made possible by sequencing methods) [
1162,
1163] is a modern approach, but historically, comparative anatomy and biogeography were used by Darwin and others to infer evolutionary relationships. Today, computational models and DNA analysis complement classical fieldwork to address ecological and evolutionary questions [
1164].
Figure 19.
The relationships between major research tasks between biology and bio-engineering.
Figure 19.
The relationships between major research tasks between biology and bio-engineering.
5.4.1.2 Introduction to Bioengineering
Bioengineering (also called biological engineering) is the application of biological principles and engineering tools to create usable, tangible, and economically viable products [
1165,
1166]. Essentially, bioengineering leverages discoveries from life sciences by applying engineering design to develop technologies addressing challenges in biology, medicine, or other fields involving living systems [
1167,
1168]. Put simply, bioengineering combines biological understanding with engineering expertise to design and build solutions, such as medical devices [
1169], novel therapies [
1170], or biomaterials [
1171,
1172]. It is inherently interdisciplinary: a bioengineer may employ mechanical engineering to construct artificial limbs, electrical engineering for biomedical sensors, chemical engineering for bioprocessing, and biological sciences across all applications [
1173]. Thus, bioengineering bridges pure science with practical engineering, translating biological knowledge into innovations that enhance lives.
To better organize and understand bioengineering’s scope, traditional research tasks are categorized into well-established domains. This classification is based on historical developments and practical engineering workflows [
1174,
1175,
1176], dividing the discipline according to how biological knowledge translates into engineering solutions and tangible products [
1176,
1177]. Each category corresponds to a major application domain within bioengineering, representing distinct pathways for integrating biology with engineering.
Genetic and Cellular Engineering. Many bioengineers modify biological cells or molecules for new functions—such as engineering bacteria to produce pharmaceuticals or editing genes to treat diseases [
1178,
1179,
1180,
1181]. Genetic engineering techniques from molecular biology are foundational. Since the 1970s, recombinant DNA technology (using restriction enzymes to manipulate genes) [
1182,
1183,
1184,
1185] and cell transformation [
1186] have enabled scientists to insert genes into organisms. Practically, bioengineers often use plasmids to introduce genes into bacteria and employ fermentation bioreactors (borrowed from chemical engineering) for cultivating genetically modified microbes at scale. More recently, CRISPR-Cas9 gene editing (developed in the 2010s) has allowed precise genome modifications [
1187,
1188,
1189,
1190,
1191]. Typical workflows include designing genetic constructs, altering cells, and scaling selected cell lines. This domain overlaps significantly with biotechnology and biomedical sciences [
1167,
1192].
Tissue Engineering and Biomaterials. A significant bioengineering area involves engineering or replacing biological tissues using methods such as cultivating cartilage, skin, or organs in laboratories [
1193,
1194,
1195,
1196]. Core techniques include cell culture and scaffold fabrication—bioengineers cultivate cells on biodegradable scaffolds (often polymers) to form tissues [
1194]. Late 20th-century innovations demonstrated that seeding cells onto 3D scaffolds could produce artificial tissues (e.g., synthetic skin) [
1197,
1198]. Biomaterials science contributes materials (e.g., polymers, ceramics) designed to safely interact with the body, such as titanium or hydroxyapatite implants [
1199,
1200,
1201]. This domain has produced synthetic skin grafts and advances toward lab-grown organs like bladders [
1202] and blood vessels [
1203].
Bioprocess Engineering. Bioengineers design processes to scale up biological products (e.g., mass-producing vaccines, biofuels, or fermented foods) [
1204,
1205,
1206], drawing on chemical engineering principles adapted to biological contexts. Engineers design bioreactors, optimize conditions (temperature, pH, nutrients), and ensure sterile, efficient processes [
1207]. Traditional methods include continuous culture [
1208,
1209] and process control systems. The large-scale production of penicillin in the 1940s exemplifies bioprocess engineering, involving optimizing Penicillium mold growth in industrial tanks [
846,
848,
1210]. Today’s production of monoclonal antibodies or industrial enzymes similarly employs refined classical fermentation and purification techniques [
1211,
1212].
Bioinformatics and Computational Biology. Although occasionally considered separate fields, bioengineers frequently engage in computational modeling of biological systems or analyze biological data (genomic or protein structures) to guide engineering designs [
1213,
1214,
1215,
1216]. This domain involves algorithms and simulations—for example, modeling physiological systems using differential equations and computational methods from control theory. Computational approaches have long contributed to bioengineering, supporting prosthetic design optimization (via CAD and finite element analysis) and genomic analyses (software for DNA sequence analysis) [
1217,
1218,
1219,
1220,
1221,
1222]. This domain underscores bioengineering’s combination of wet-lab experimentation and computational methods.
5.4.1.3 Current Challenges
Life sciences and bioengineering are foundational disciplines that have transformed our understanding of life and significantly improved human health and well-being. Life sciences uncover the fundamental principles of biology, from Darwin’s theory of evolution and Mendel’s laws of inheritance to the germ theory of disease. These discoveries led to major advances such as vaccines, antibiotics like penicillin, and the molecular revolution sparked by the discovery of DNA’s double-helix structure. Tools like PCR and the Human Genome Project further deepened our ability to decode and manipulate genetic information, ushering in the era of personalized medicine.
Bioengineering complements these insights by applying them to solve real-world problems. The development of X-ray imaging allowed non-invasive diagnosis, while innovations like the implantable pacemaker and artificial organs expanded the scope of life-saving care. The production of human insulin through recombinant DNA technology marked a milestone in biopharmaceuticals. Later, tissue engineering demonstrated that lab-grown organs could be transplanted into humans, and gene-editing tools like CRISPR have opened new frontiers in treating genetic diseases.
Together, life sciences and bioengineering form a powerful synergy: the former provides deep biological insight, while the latter transforms that knowledge into tangible solutions. Their joint progress continues to revolutionize medicine, agriculture, and environmental science—improving quality of life and shaping a more advanced future.
Despite remarkable advancements in life sciences and bioengineering, numerous challenges persist due to the complexity and inter-connectivity inherent to biological systems. Intriguingly, many of the most formidable obstacles overlap between these fields, as they frequently address complementary facets of the same intricate biological phenomena. In this section, we systematically examine several critical common challenges, distinguishing between those that currently remain beyond the capability of artificial intelligence tools such as LLMs and those that can already benefit from LLM integration.
Still Hard with LLMs: The Tough Problems.
Here we analyze some key common challenges that are currently still beyond the reach of LLMs, we acknowledge LLMs’ limitations related to experimental design, interpretative complexity, and practical hands-on tasks, underscoring domains where human expertise remains indispensable.
Ethical and Safety Challenges. Life sciences and bioengineering are inherently intertwined with ethical and societal considerations that transcend purely technical challenges [
1223]. These fields routinely grapple with questions surrounding the responsible use of gene editing technologies like CRISPR [
1224,
1225], the long-term ecological effects of genetically modified organisms [
1226,
1227,
1228], and the protection of sensitive patient data in genomics and biomedical research [
1229]. While LLMs can assist in synthesizing scientific literature and outlining stakeholder perspectives, they lack the capacity for moral reasoning or normative judgment. Their outputs are constrained by the biases present in their training data, which poses risks when applied to ethically sensitive domains [
1230,
1231]. Ethical decision-making in bioengineering—such as determining the acceptability of human germline editing, setting standards for clinical trials, or regulating synthetic biology applications—remains the responsibility of human experts, policymakers, and the broader public. These decisions require inclusive debate, value alignment, and legal oversight that go beyond algorithmic capabilities [
1232,
1233]. As technologies advance, both the Life sciences and AI communities must collaboratively develop ethical frameworks that reflect societal values while fostering innovation.
Needs for Empirical Validation. Both life sciences and bioengineering ultimately rely on physical experimentation [
1234]. Scientific hypotheses must be empirically verified, and bioengineered solutions require testing under real-world conditions [
1235]. However, such experiments often pose significant bottlenecks due to their inherent slowness, high costs, and ethical limitations—particularly in human studies, where experimentation is strictly constrained, and animal models often fail to fully replicate human biology. [
1236,
1237,
1238] While computational models can alleviate some of these burdens, they cannot fully substitute for wet-lab or clinical experiments. Similarly, LLMs are incapable of conducting physical experiments or collecting new empirical data. Although they can assist in designing experimental protocols, they cannot implement or validate them [
1239]. Consequently, research challenges that fundamentally require novel data acquisition—such as identifying new drug targets or evaluating biomaterials—remain beyond the scope of LLMs alone. The crucial
last mile of validation in biology and engineering - demonstrating something works in actual living systems - remains dependent on laboratories, clinical trials, and real-world testing [
1240].
Complexity of Biological Systems. The overarching challenge is that living systems are astonishingly complex and multi-scale. Scientists struggle with this complexity as small changes can have cascading, unpredictable effects. For life scientists, this means incomplete understanding of many diseases and biological processes [
1241,
1242,
1243]. For bioengineers, it means difficulty designing interventions without unintended consequences [
1244,
1245]. LLMs cannot reliably solve this because much of biological complexity stems from unknown factors requiring empirical observation and quantitative modeling beyond text-pattern recognition [
1246]. While LLMs process information well, the emergent behavior of complex biological networks often requires specialized modeling that correlation-based systems can’t provide without explicit mathematical frameworks. Major challenges like understanding neural circuits or curing cancer remain unsolved because they require new scientific discoveries and experimental validation, not just knowledge retrieval [
1247].
Data Quality and Integration. Modern life sciences and bioengineering generate enormous volumes of data from genomic sequences, proteomics, patient records, and sensors. Making sense of this data reliably presents significant challenges because it’s often noisy, comes from disparate sources, and lacks integration [
1248,
1249]. While LLMs excel at processing text, they struggle with heterogeneous scientific data that includes numbers, images, and experimental measurements [
8]. LLMs don’t have native capabilities to process raw experimental data like gene expression matrices or medical imaging unless specifically augmented with specialized tools [
1250]. Challenges in biological big data - ensuring reproducibility, establishing causal relationships from observational data, or analyzing complex multi-modal datasets - still require specialized algorithms and human expertise in statistics and domain knowledge. LLMs might help report findings or suggest hypotheses, but they cannot replace the sophisticated analytical pipelines needed for rigorous scientific data analysis in these fields.
In summary, many of the grand challenges – decoding all the details of human biology, curing major diseases, sustainably engineering biology for the environment which are still open. LLMs, in their current state, are tools that can assist researchers but not solve these on their own, because the challenges often require new empirical discovery or involve complex systems and judgments beyond pattern recognition. An LLM might speed up literature review or suggest plausible theories, but it won’t automatically unravel the secrets of life that scientists themselves are still grasping at.
Easier with LLMs: The Parts That Move.
On a more optimistic note, there are challenges within life sciences and bioengineering where LLMs are already proving useful or have clear potential to contribute. These tend to be problems involving knowledge synthesis, pattern recognition in sequences/text, or generating hypotheses – tasks where handling language or symbolic representations is key. A few examples of such challenges that LLMs can tackle (and why they are suitable) include:
Literature Overload and Knowledge Synthesis. One critical challenge uniquely pronounced in biology and bioengineering is managing the vast, rapidly growing, and fragmented body of research literature.
Unlike fields such as mathematics, law, or finance, biological disciplines inherently encompass a multitude of interconnected subspecialties, each producing large volumes of highly specialized research. For example, understanding a complex disease like cancer may require integrating findings from genetics, immunology, cell biology, pharmacology, and bioinformatics—each field publishing detailed, specialized studies that must be synthesized for comprehensive insights. The complexity arises not only from the sheer volume but also from interdisciplinary connections, intricate experimental details, and extensive supplementary materials required for reproducibility. Consequently, researchers face significant difficulty in identifying relevant literature, extracting key insights, and synthesizing knowledge efficiently. This is precisely where LLMs show strong potential as intelligent literature reviewers [
1251,
1252,
1253]. Advanced LLMs, such as GPT-4, can proficiently read, summarize, and contextualize complex biomedical texts, rapidly extracting relevant findings from extensive corpora [
1253,
1254]. For example, an LLM could swiftly provide researchers with an overview of current biomarkers for Alzheimer’s disease or consolidate recent advancements in biodegradable stent materials [
1253]. By effectively navigating dense technical language and complex sentence structures inherent to biological literature, LLMs mitigate literature overload, facilitate interdisciplinary integration, and enable literature-based discovery—highlighting connections between seemingly disparate research findings [
1255,
1256,
1257].
Interpreting and Annotating Biological Sequences (Genomics/Proteomics). In both life sciences research and bioengineering applications like synthetic biology, understanding DNA, RNA, and protein sequences is crucial [
1258,
1259,
1260]. These sequences can be thought of as strings of letters (A, T, C, G for DNA; amino acids for proteins) – in other words, a language of life. Recent work has shown that language models can be applied to these biological sequences, treating them like natural language, where “words” are motifs or codons and “sentences” are genes or protein domains. This is a challenge where LLM-like models shine [
1261,
1262,
1263]. This means LLMs can help annotate genomes (predicting genes and their functions in a newly sequenced organism) or predict the effect of mutations (important for understanding genetic diseases) [
1263]. In proteomics, models can suggest which parts of a protein are important for its structure or activity [
1264,
1265]. The advantage of LLMs here is their ability to handle long-range dependencies in sequences – biology often has context-dependent effects, and language models are designed to handle such context. Moreover, LLMs can generate sequences too, which leads to the next point.
Design and Generation of Biological Sequences or Structures. In bioengineering, a cutting-edge challenge is designing new biological components – for instance, designing a protein that catalyzes a desired chemical reaction, or an RNA molecule that can serve as a therapeutic. Traditionally, this is very hard (the search space of possible sequences is astronomically large). However, LLMs have a generative capability that can be harnessed here. Already, models like ProGen [
1266,
1267] have shown they can generate novel protein sequences that have a predictable function across protein families. In simpler terms, an LLM trained on a vast number of protein sequences can be prompted to create a new sequence that looks like, say, an enzyme, and those sequences have been experimentally verified in some cases to fold and function [
1266,
1267,
1268,
1269,
1270]. This is a remarkable development because it means LLMs can assist in protein engineering and drug discovery by proposing candidate designs that humans or simpler algorithms might not think of. Similarly, for DNA/RNA, an LLM could suggest a DNA sequence that regulates gene expression in a certain way (useful for gene therapy designs) [
1259,
1263] or propose improvements to a biosynthetic pathway by modifying enzyme sequences [
1264,
1265]. LLMs are suitable for these creative tasks because, much like with natural language, they can interpolate and extrapolate learned patterns to create new, coherent outputs (here, “coherent” means biologically plausible sequences). While any generated design still needs to be tested in the lab (to confirm it works as intended), LLMs can dramatically accelerate the ideation phase of bioengineering design.
Figure 20.
Our taxonomy for life sciences and bio-engineering.
Figure 20.
Our taxonomy for life sciences and bio-engineering.
5.4.1.4 Taxonomy
Throughout the development of life sciences and bioengineering, research has gradually branched into increasingly specialized subfields. Nevertheless, many fundamental commonalities persist across domains. For instance, identifying potential folding targets within long protein sequences is methodologically similar to locating functional nucleotide fragments within DNA, both involve extracting biologically meaningful patterns from symbolic sequences [
1239,
1271,
1272]. In addition, biological tasks often span multiple levels and modalities, encompassing data from molecular to system scales and ranging from unstructured text to structured graphs [
1273,
1274]. This inherent diversity renders traditional task-type-based classifications insufficient, as they obscure the subset of tasks where LLMs are particularly well-suited.
To address this, we propose a taxonomy that emphasizes the computational characteristics of tasks, enabling a more precise alignment with the capabilities and limitations of LLMs. This data-centric perspective offers three key advantages:
Sequence-Based Tasks. These tasks involve analyzing sequential biological data, such as DNA, RNA, or protein sequences, which are essentially strings of nucleotides or amino acids. Typical examples include genome annotation, mutation impact prediction, protein structure prediction from sequences, and the design of genetic circuits. The input for these tasks generally comprises one or multiple biological sequences, with outputs including sequence annotations or newly designed sequences. From an artificial intelligence standpoint, these tasks are analogous to language processing problems because biological sequences possess a form of syntax (e.g., motifs and domains) and semantics (functional implications). LLMs and other sequence-based AI models are thus particularly effective for these applications, interpreting biological sequences similarly to languages. For instance, predicting the pathogenicity of a DNA mutation is akin to detecting grammatical errors in a language, where biological viability parallels grammatical correctness [
1277]. Recent advancements, such as protein language models, exemplify successful AI applications leveraging this analogy [
1277,
1278].
Structured and Numeric Data Tasks. These tasks involve handling structured datasets, numerical measurements, and graphs commonly encountered in physiology, biochemistry, and bioengineering. Examples include analyzing patient heart rate time-series data, interpreting metabolomics datasets, optimizing metabolic network models, and designing control systems for prosthetics. Inputs typically consist of numerical or tabular data (sometimes time-series), while outputs could involve predictions (e.g., forecasting patient adverse events) or control decisions. Such tasks traditionally rely on statistical methods or control theory, and they are generally less naturally suited for LLMs unless translated into textual or coded representations. A creative utilization of LLMs in this context is their ability to generate computational code from natural language descriptions, bridging descriptive problem statements to numeric analyses. For example, researchers can prompt an LLM to produce Python code to analyze specific datasets, utilizing the model as an intermediary tool between natural language and computational implementation [
1279,
1280,
1281,
1282]. However, purely numerical tasks involving complex calculations or optimizations typically remain better served by specialized algorithms.
Textual Knowledge Tasks. These tasks involve managing, interpreting, and generating text-based information prevalent in biology and engineering. Examples include literature searches and question-answering, proposal writing, extracting critical information from research articles, summarizing electronic health records, and analyzing biotechnology patents. Inputs here predominantly include unstructured textual documents (such as research articles, clinical notes, or patent filings), with outputs comprising synthesized textual summaries, detailed answers, or structured reports. This area represents the inherent strength of LLMs, encompassing various subtasks such as knowledge retrieval and question-answering, summarization and literature review, and protocol or technical report generation. Given their core competency in processing and synthesizing textual data, LLMs are exceptionally well-suited to these tasks.
Predictive Modeling Tasks (Hybrid). Many research questions in biology and bioengineering can be formulated as predictive problems, such as determining drug toxicity, predicting crop yields from genetic modifications, or forecasting protein folding success. These tasks frequently involve integrating multiple modalities—including sequences, structural data, textual descriptions—and require robust extrapolative capabilities. Inputs often combine diverse data formats, with outputs focusing on biological or engineering outcomes. Although many predictive tasks overlap with sequence-based analyses, this category explicitly emphasizes multidimensional predictions that leverage various feature sets. LLMs can contribute to these tasks through interpretive roles, qualitative reasoning, or as orchestrators within computational pipelines. For instance, an LLM could manage interactions between specialized bioinformatics tools, interpret computational outputs, and provide coherent explanations or qualitative predictions, highlighting their integrative and explanatory potential within complex predictive frameworks.
Table 43.
Life Science and Bioengineering Tasks, Subtasks, Insights and References
Table 43.
Life Science and Bioengineering Tasks, Subtasks, Insights and References
| Type of Task |
Subtasks |
Insights and Contributions |
Key Models |
Citations |
| Genomic Sequence Analysis |
DNA Sequence Modeling |
LLMs, by capturing regulatory grammars embedded in genomic sequences, enable accurate prediction of functional elements and variant effects, thus enhancing interpretability and advancing our understanding of gene regulation. |
DNABERT: adapt BERT to human reference DNA and learn bidirectional representations;HyenaDNA: scale the context length and accelerate the model efficiency. |
[1261,1283,1284,1285,1286,1287,1288,1289] |
| |
RNA Function Learning |
LLMs, by modeling both sequence and structural contexts of RNA, uncover functional motifs and regulatory patterns, thereby improving interpretability and facilitating insights into post-transcriptional regulation. |
RNA-FM: employing 23.7M deduplicated RNA sequences for training; 3UTRBERT: specialized in modeling 3’ untranslated regions (3’UTRs). |
[1290,1291,1292,1293,1294,1295,1296] |
Biomedical Reasoning
and Understanding
|
Question Answering |
LLMs, by aligning domain-specific knowledge with natural language understanding, accurately interpret biomedical queries and texts, thereby enhancing information retrieval and supporting clinical and research decision-making. |
Med-PaLM: achieved near-expert-level performance on the USMLE test; HuatuoGPT: proactively ask questions rather than respond passively. |
[17,1297,1298,1299,1300,1301,1302,1303,1304,1305,1306,1307,1308,1309,1310,1311] |
| |
Language Understanding |
LLMs, by learning semantic patterns and reasoning cues from biomedical texts, enable deep language understanding, thereby improving performance on tasks like inference, entity recognition, and document classification. |
BioInstruct: covering multi understanding tasks; GPT-4: perform NLI without explicit fine-tuning, when prompted with queries. |
[17,1297,1302,1305,1312,1313,1314,1315,1316] |
Omics & Clinical Structured
Data Integration
|
Clinical Language Generation |
LLMs, by capturing clinical language styles and contextual dependencies, generate coherent and context-aware narratives, thereby enhancing the automation and reliability of medical reporting and documentation. |
ClinicalT5: pretraining on text-to-text tasks for long clinical narratives; GPT-4: perform good when prompted by specific queries. |
[1301,1317,1318,1319,1320,1321] |
| |
EHR Based Prediction |
LLMs, by integrating longitudinal and multimodal patient data from EHRs, model complex temporal and clinical dependencies, thereby enabling accurate prediction of outcomes and supporting personalized healthcare. |
BEHRT: adapts BERT to longitudinal EHR data by encoding structured sequences; GatorTron: scaled up to 8.9B and was trained on over 90B clinical narratives and structured labels. |
[1305,1322,1323,1324] |
| Hybrid Outcome Prediction |
Drug Synergy Prediction |
LLMs, by jointly modeling chemical structures and cellular contexts, capture intricate drug–drug and drug–cell interactions, thereby enhancing the prediction of synergistic combinations and accelerating combination therapy design. |
CancerGPT: fine-tuning GPT-3 to predict drug synergy in rare cancers; BAITSAO: a foundation model strategy that integrates multiple datasets and tasks. |
[6,7,18,1325,1326,1327,1328] |
| |
Protein Modeling |
LLMs, by learning evolutionary, structural, and functional signals from protein sequences, enable accurate modeling of folding, function, and interactions, thereby advancing protein engineering and therapeutic discovery. |
ProLLaMA: achieving joint understanding and generation within a single framework; ProteinGPT: further supporting structure input, language interaction, and functional Q&A |
[1267,1268,1269,1329,1330,1331,1332,1333,1334,1335,1336,1337,1338,1339,1340,1341,1342,1343,1344,1345] |
5.4.2. Genomic Sequence Analysis
Sequence-based tasks focus on the learning and modeling of sequential data in life science and bioengineering, aiming to assist researchers in extracting meaningful biological insights from sequence-encoded information. The primary inputs to these tasks are biological sequences, such as nucleotide sequences in DNA/RNA or amino acid sequences in peptides. For instance, DNA is composed of four nucleotides, adenine (A), guanine (G), thymine (T), and cytosine (C), which are inherently stored in organisms in a sequential format, requiring no additional transformation. These biologically grounded representations allow models to learn structural patterns and latent dependencies, thereby enabling effective downstream prediction and classification tasks.
In sequence-based tasks, the objective is to leverage learned representations to accomplish specific downstream goals. For example, given genomic DNA adjacent to genes that may contain enhancers, a binary label is predicted for each 128-base-pair segment to determine whether it belongs to an enhancer region. Enhancers are short, non-coding DNA elements that regulate gene expression and can exert influence across distances of thousands to over a million base pairs by physically interacting with gene promoters. Another representative task is predicting the gene-editing efficiency of a given single guide RNA (sgRNA) sequence when guided by Cas proteins in CRISPR-based applications.
The value of sequence-based tasks in life sciences and bioengineering lies in their capacity to capture long-range dependencies and hierarchical patterns in extremely long and sparse sequences—often in a semi-supervised or unsupervised manner. For instance, in the human genome, protein-coding regions account for only about 1% of the total DNA sequence. Likewise, functionally important non-coding regions such as promoters also constitute a very small fraction of the genome [
1346]. By training on massive sequence datasets, models can learn to identify such regions with high accuracy. Researchers can then input unknown sequences into the model to annotate potential coding regions or predict functionally significant non-coding elements. In addition to classification, sequence-based models are also applied in predictive tasks. For example, they can estimate the likelihood and frequency of off-target mutations induced by CRISPR systems at unintended genomic loci, thereby improving the safety and efficacy of gene editing. In summary, sequence-based tasks significantly reduce the time and cost of sequence analysis, while deepening our understanding of the functional and regulatory roles encoded in biological sequences.
Although both DNA and RNA exist in the form of sequences, they differ significantly in biological function and modeling objectives [
1347]. DNA sequences primarily encode genetic information, and related modeling tasks focus on identifying regulatory elements such as promoters, enhancers, and transcription factor binding sites, as well as capturing long-range dependencies across the genome. In contrast, RNA is more involved in functional execution, including splicing, modification, translational regulation, and interactions with proteins. Typical tasks involve RNA secondary structure prediction, modification site identification, and functional classification of non-coding RNAs [
1348,
1349]. Therefore, we categorize Sequence-Based Tasks into DNA Sequence Modeling and RNA Function Learning, and the subsequent discussion will be centered around these two directions. These two tasks share several common characteristics that make LLMs particularly well-suited for this domain. First, the abundance of unlabeled or sparsely labeled sequence data provides a rich resource for training. Second, LLMs not only deliver reliable predictions but also offer highly interpretable textual reasoning to support their outputs.
DNA Sequence Modeling. DNA Sequence Modeling refers to the task of computationally analyzing and learning patterns from nucleotide sequences—typically represented as strings composed of A, T, C, and G—to understand the underlying biological functions, regulatory mechanisms, and genetic variations encoded in the genome. In recent years, LLMs have rapidly emerged in the field of DNA, advancing our understanding of the information encoded within the genome. Transformer-like LLMs are now capable of reading, reasoning over, and even designing the 3.2Gb human genome, evolving from early convolutional neural networks (CNNs) that handled short windows to billion-parameter foundation models that process megabase-scale contexts. Early convolutional frameworks such as DeepSEA [
1350] demonstrated that raw sequence alone is sufficient to predict chromatin features, inspiring a decade-long progression from hybrid CNN-RNN models and Transformer encoders to long-context state-space models. Today’s genomic LLMs often outperform traditional physics-based or motif-based methods across various tasks, including enhancer detection, cell type-specific expression, and non-coding variant effect prediction, while also providing saliency maps that highlight learned regulatory syntax.
DNABERT [
1283] adapted the BERT architecture to human reference DNA by tokenizing sequences into 6-mers and using masked language modeling (MLM) to learn bidirectional representations, which proved transferable to promoter, enhancer, and splice site prediction tasks. Building on this, DNABERT-2 [
1284] introduced byte-pair encoding (BPE) [
1351], breaking the fixed k-mer limitation, reducing memory usage by 30%, and improving average MCC by 2 percentage points across 28 datasets. Techniques like self-distillation and adaptive masking further refined embeddings under limited data conditions.
Unlike typical LLMs, DNA inputs are significantly longer than those in standard NLP tasks. Even when models support large context windows, performance can degrade. To address these issues, Enformer [
1285] combines ConvNet [
1352] downsampling with 1D self-attention over 200kb regions, doubling the correlation with gene expression compared to prior models and significantly improving eQTL effect sign prediction. HyenaDNA [
1261] scales context length to 1 million nucleotides using sub-quadratic implicit convolutions, enabling 160× faster training than FlashAttention Transformers [
1353], while maintaining single-base resolution and outperforming Enformer on proximal promoter tasks.
GROVER [
1286] uses frequency-balanced byte-pair encoding vocabularies to learn “sequence contextuality” directly from the human genome, outperforming prior k-mer baselines in next-token prediction and fine-tuning tasks like promoter detection and CTCF binding prediction. Its unsupervised embeddings can recover GC content, repeat categories, replication timing, and other functional signals purely from sequence, underscoring how tokenization design can unlock richer biological grammar.
With increased computational power and model scaling, larger models and training corpora have emerged. The Nucleotide Transformer [
1287] with 2.5 billion parameters was pretrained on 3,202 human and 850 non-human genomes, generating general-purpose embeddings that improved performance across 18 downstream tasks—including pathogenicity scoring and enhancer–promoter linking—without requiring task-specific architectural changes. GenomeOcean [
1288], a 4-billion-parameter model, extended this paradigm to 220TB of metagenomic data, capturing rare microbial taxa for better ecologically driven generalization.
LLMs have substantially advanced DNA sequence modeling by enabling scalable interpretation of genomic sequences, capturing long-range dependencies, and learning regulatory syntax directly from raw nucleotide strings. They have addressed core challenges such as integrating context across megabase-scale windows, improving prediction of non-coding variant effects, and providing transferable embeddings for diverse downstream tasks without extensive feature engineering. However, key challenges remain: performance often degrades with increasing sequence length despite architectural innovations, and current models still struggle with integrating multi-omic signals, rare variant generalization, and interpretability in clinical settings. Future directions include developing more efficient architectures for ultra-long sequences, incorporating cross-modal biological data (e.g., epigenomic or transcriptomic layers), and aligning model predictions with mechanistic biological knowledge to support hypothesis generation and therapeutic discovery.
RNA Function Learning. RNA Function Learning refers to the task of modeling RNA sequences to uncover their structural attributes and functional roles, often leveraging sequence-structure relationships to predict biological behaviors and interactions. Understanding RNA sequences and their structural-functional relationships is essential for numerous molecular biology applications, including splicing regulation, RNA-protein interactions, and non-coding RNA functional annotation. Traditional bioinformatics methods such as sequence alignment and thermodynamic folding models (e.g., RNAfold) provide accurate predictions but suffer from limitations like heavy computational demands and dependency on handcrafted features.
Recently, leveraging advances in natural language processing (NLP), large-scale pretrained language models adapted to RNA sequences have emerged, significantly improving our capacity to interpret biological information embedded in nucleotide sequences. Early efforts, such as RNABERT [
1290], marked a shift toward learning biological grammar directly from data. RNABERT combined masked language modeling (MLM) with a structural alignment objective (SAL), enabling the model to internalize pairwise structural relationships by training on alignment scores derived from the Needleman-Wunsch algorithm [
1354].
Subsequent models expanded on this foundation by enhancing both scale and methodological complexity. RNA-FM [
1291] significantly scaled the training dataset, employing 23.7 million deduplicated RNA sequences from RNAcentral [
1354], thus improving generalization capabilities for functional prediction tasks. RNA-MSM [
1292] further advanced this approach by incorporating evolutionary context through homologous sequence modeling with multiple sequence alignments (MSAs), inspired by MSATransformer [
1355]. Notably, RNA-MSM strategically excluded families with known structures during training, effectively reducing overfitting and enhancing performance on structure-aware tasks.
Parallel developments have addressed specific functional RNA elements, refining the modeling approach based on targeted biological contexts. For instance, SpliceBERT [
1293] specifically targeted splicing regulation by training on 2 million vertebrate pre-mRNA sequences from UCSC [
1356]. By focusing explicitly on pre-mRNA rather than mature transcripts, SpliceBERT captured sequence features critical for identifying splicing junctions, splice sites, and regulatory motifs (e.g., exonic splicing enhancers or silencers), aspects typically overlooked in traditional modeling frameworks [
1294]. Consequently, this model supports tasks such as splice site prediction, detection of alternative splicing events, and the identification of novel, tissue-specific regulatory elements.
Complementing this targeted functional perspective, 3UTRBERT [
1295] specialized in modeling 3’ untranslated regions (3’UTRs) to facilitate studies of post-transcriptional regulation. Building further upon the integration of structure into sequence modeling, UTR-LM [
1296] explicitly incorporated structural supervision alongside MLM pretraining. It employed two biologically informed auxiliary tasks: secondary structure prediction and minimum free energy (MFE) regression. Secondary structures, predicted using the ViennaRNA toolkit [
1357,
1358], were utilized as local structural constraints during masking, while global thermodynamic stability (MFE) values were predicted from global contextual embeddings ([CLS] token). The training dataset included carefully curated natural and synthetic 5’UTR sequences from databases like Ensembl and high-throughput assays, ensuring robust learning of biologically relevant patterns [
1359,
1360,
1361]. These methods strengthened the link between structural and functional RNA predictions, demonstrating applicability in translation efficiency prediction and synthetic RNA design.
Lastly, BEACON-B and BEACON-B512 expanded RNA language modeling into extensive datasets comprising over 500,000 human non-coding RNAs from RNAcentral [
1354], exploring broader functional landscapes beyond coding transcripts [
1294]. These models highlighted the importance of tailored training objectives, domain-specific masking strategies, and carefully curated datasets, all contributing to enhanced interpretability and biological accuracy.
LLMs have revolutionized RNA function learning by shifting the paradigm from alignment- or thermodynamics-based methods to data-driven, end-to-end models that learn both structural and functional features directly from sequences. These models have improved prediction accuracy for splicing patterns, RNA-protein interactions, and post-transcriptional regulatory elements, while also enabling interpretability through structural supervision and auxiliary tasks. Nonetheless, key challenges remain: many models still struggle with generalizing to novel RNA classes, integrating evolutionary and tertiary structural information, and explaining model decisions in biologically meaningful ways. Future research directions include scaling models to more diverse and comprehensive RNA datasets, incorporating multi-resolution structural priors, and aligning language model outputs with experimentally validated functional annotations to bridge the gap between sequence modeling and functional genomics.
5.4.3. Clinical Structured Data Integration
Clinical structured data integration focuses on the intelligent utilization of structured clinical information—such as Electronic Health Records (EHRs), laboratory test results, and coded diagnoses—to support and automate critical healthcare decision-making processes. The goal is to leverage artificial intelligence, particularly LLMs, to understand structured clinical inputs, build predictive or generative models, and produce meaningful outputs that improve clinical workflows, enhance patient care, and enable personalized medicine. These tasks primarily rely on structured or semi-structured datasets, including tabular EHR entries, time-series vital signs, coded diagnoses and treatments (e.g., ICD, CPT, LOINC), and structured questionnaire responses. Unlike free-form text, such data is inherently aligned with medical ontologies and clinical protocols, enabling models to reason with high factual precision and temporal awareness.
The primary objective of clinical structured data integration is to perform downstream tasks that generate clinically useful outputs based on structured patient data. For instance, given longitudinal EHRs containing timestamped diagnoses, prescriptions, and lab values, a model can forecast disease onset, stratify patient risk, or suggest treatment plans. In other scenarios, structured data is transformed into human-readable summaries—such as clinical progress notes or discharge instructions—to reduce the documentation burden on clinicians. A persistent challenge lies in bridging the gap between machine-readable formats and clinical narratives: generated content must be not only factually accurate but also contextually appropriate and linguistically coherent. Furthermore, since EHR data often contains missing values, noise, or institutional heterogeneity, models must be robust to irregular sampling and generalize across diverse healthcare settings.
In the broader context of life sciences and bioengineering, clinical structured data integration serves as a cornerstone of evidence-based medicine, offering scalable solutions for personalized care, automated documentation, and proactive health monitoring. By seamlessly connecting the structured backbone of clinical practice with the expressive power of language models, this work marks a critical step toward an intelligent, interoperable, and human-centered healthcare system.
To reflect the dual nature of this field, clinical structured data integration can be categorized into two main types: Clinical Language Generation and EHR-Based Prediction. The former focuses on converting structured clinical data into fluent, accurate natural language reports, enabling applications such as automated drafting of medical notes, radiology impression generation, and ICU event summarization. This task emphasizes controllable generation, temporal summarization, and medical factuality, requiring models to balance conciseness with informativeness. In contrast, EHR-based prediction aims to extract actionable insights from patient records to support tasks such as sepsis alerts, readmission prediction, and personalized risk scoring. These tasks demand strong temporal modeling, integration of clinical knowledge, and high interpretability, especially when informing critical medical decisions.
Across both task types, incorporating domain-specific inductive biases—such as hierarchical coding systems, medical knowledge graphs, or treatment ontologies—has been shown to enhance model performance. LLMs have demonstrated great potential in unifying diverse input modalities and producing clinically meaningful outputs, particularly when structured prompting or graph-aware architectures are employed. Moreover, the growing availability of publicly accessible, de-identified datasets such as MIMIC-III/IV [
1362,
1363] and eICU has fostered the development of standardized evaluation benchmarks. These advances not only enable rigorous comparison across methods but also promote the creation of generalizable and trustworthy AI systems for real-world clinical applications.
Clinical Language Generation. Clinical Language Generation refers to the use of natural language processing techniques to automatically produce coherent and clinically meaningful text from structured inputs such as electronic health records, diagnostic codes, or medical templates. Clinical Language Generation (CLG) is rapidly emerging as a foundational infrastructure in smart healthcare. By leveraging structured data or text recorded using templates, CLG models can automatically draft outpatient/inpatient notes, generate radiology report impressions, rewrite patient-friendly versions, and even transcribe real-time doctor-patient conversations. These capabilities significantly reduce the documentation burden on healthcare professionals, while improving the quality and readability of medical records, thereby supporting evidence-based decision-making and interdisciplinary collaboration.
With the maturation of large-scale pretraining corpora and instruction tuning techniques, CLG has evolved from early small-parameter models to multi-modal systems with tens of billions of parameters, offering unprecedented text generation capabilities in clinical settings.
One of the earliest representative works, ClinicalT5 [
1317], adapted the T5 [
1364] framework to hospital notes from datasets like MIMIC-III/IV [
1362,
1363], pretraining on text-to-text tasks for long clinical narratives. It achieved a 3.1 ROUGE-L improvement on discharge summary generation and outperformed long-text baselines such as BART [
1365], demonstrating that generative models can effectively capture key information in complex, structured medical records. However, ClinicalT5’s training data was primarily composed of single-center English inpatient notes, which limits its generalization across languages and institutions.
In contrast, general-purpose LLMs are often trained on multilingual, multi-source datasets and inherently possess cross-domain generalization capabilities [
1,
6,
7]. With the advancement of such models, GPT-4 [
1] and Med-PaLM 2 [
1301], through instruction tuning, can generate high-quality clinical drafts. GPT-4 achieved near-human accuracy in outpatient record analysis across three languages and can draft standardized clinical progress notes in zero-shot settings [
1319]. Med-PaLM 2 excelled in the MultiMedQA evaluation framework, particularly in reasoning and safety dimensions, showcasing the strength of large decoder-based models in long-form clinical text generation.
For patient communication, Jonah Zaretsky et al. demonstrated that LLMs can rewrite structured discharge summaries to a 6–8th grade reading level, with readability scores 18% higher than physician-authored versions, greatly enhancing patient understanding of medication and follow-up instructions [
1320]. Meanwhile, model scale and multimodal capabilities are also advancing. Me-LLaMA [
1321], built on LLaMA 2 [
52], integrates PubMed, clinical guidelines, and knowledge graphs, and supports 13–70B parameter ranges. With medical instruction tuning, it enables multimodal prompt-based generation for case summaries and diagnostic explanations.
In fact, many models designed for clinical or medical tasks possess some degree of CLG capabilities. However, as many focus more on medical QA or comprehension tasks, we will introduce those models in detail in the following sections.
LLMs have transformed Clinical Language Generation (CLG) by enabling automatic, fluent synthesis of complex clinical narratives from structured inputs, thereby alleviating documentation burdens and enhancing the accessibility of medical records for both professionals and patients. They have addressed key challenges such as adapting outputs to various clinical tasks, and generating patient-friendly text. Nonetheless, several challenges persist: generalization across institutions and languages remains limited due to training data biases; factual consistency and clinical safety must be rigorously validated; and integrating multimodal signals (e.g., images, vitals) into text generation is still nascent. Future work should prioritize domain adaptation techniques, fine-grained clinical factuality evaluation, multimodal integration, and collaborative frameworks involving clinicians to ensure that generated content is both medically reliable and practically useful in diverse healthcare environments.
EHR Based Prediction. An electronic health record (EHR) is the systematized collection of electronically stored patient and population health information in a digital format. EHRs play a critical role in modern healthcare systems. Beyond the advantages of digitization—such as easier storage and review—EHRs significantly improve the quality and efficiency of medical care. They provide comprehensive, accurate, and real-time patient information, enabling clinicians to make more informed and precise clinical decisions. Moreover, EHRs facilitate the sharing of patient health information across departments and institutions, enhancing collaborative efficiency and ensuring continuity of care across different healthcare settings. The vast amount of data accumulated in EHR systems also provides a solid foundation for training LLMs, as these records often contain structured annotations—such as specific diseases and severity levels—that typically require little to no transformation, making them highly suitable for LLM-based learning.
A foundational model in this domain is BEHRT [
1322], a transformer-based model developed for healthcare representation learning. BEHRT adapts BERT to longitudinal EHR data by encoding structured sequences of medical codes (e.g., diagnoses, medications) along with age embeddings. By learning temporal dependencies, BEHRT achieved strong performance in tasks such as disease onset prediction (e.g., predicting diabetes based on early comorbidities), and it demonstrated robust performance in downstream stratification tasks with minimal fine-tuning.
However, BEHRT was trained on relatively small datasets, limiting its potential. In contrast, Med-BERT [
1323], a context-based embedding model, was pre-trained on a large-scale structured EHR dataset comprising 28,490,650 patients and clinical coding systems like ICD-10 and CPT. Fine-tuning experiments showed that Med-BERT significantly improved prediction accuracy. Notably, Med-BERT performed exceptionally well with small fine-tuning datasets, achieving AUC scores that surpassed baseline deep learning models by over 20%, and even matched the performance of models trained on datasets ten times larger [
1323].
Building on this trend, GatorTron [
1324] scaled up the model size to 8.9 billion parameters and was trained on over 90 billion clinical narratives and structured labels. It demonstrated remarkable generalization capabilities in tasks such as phenotype prediction and cohort selection. Its scalability enables modeling of complex inpatient trajectories and supports patient-level reasoning even in low-resource scenarios.
In the multimodal space, models like MultiMedQA [
1300] and Clinical Camel [
1305] integrate structured EHR entries (e.g., vital signs, lab results) with textual prompts to generate clinical answers from tabular data. For example, given a prompt such as “Is this patient at risk for acute kidney injury?” and a series of lab values and medication records, the model outputs a response like “Yes, due to elevated creatinine levels and concurrent use of nephrotoxic drugs.”
LLMs have significantly advanced EHR-based prediction by leveraging structured medical codes, temporal information, and multimodal clinical signals to support personalized forecasting of disease onset, treatment outcomes, and patient risk stratification. These models, ranging from BEHRT and Med-BERT to the billion-parameter GatorTron, have demonstrated strong generalization across clinical tasks with minimal fine-tuning and are particularly effective even in low-resource settings. However, challenges remain in modeling long, sparse, and irregular patient timelines, ensuring clinical interpretability, and addressing domain shifts across institutions and EHR systems. Future work should focus on integrating heterogeneous modalities (e.g., imaging, genomics), improving temporal reasoning across fragmented records, and developing explainable frameworks that align model decisions with clinician expectations to foster trust and deployment in real-world healthcare settings.
5.4.4. Biomedical Reasoning and Understanding
Biomedical Reasoning and Understanding focus on the understanding and modeling of textual information in the fields of life sciences and bioengineering. The goal is to leverage artificial intelligence technologies to enhance the semantic parsing of natural language content such as scientific literature, clinical case records, and diagnostic reports. The primary inputs for these tasks are natural language texts—for example, research abstracts from PubMed or clinical notes from patient records. Similar to DNA or RNA sequences, natural language inherently contains rich semantic information and can be directly processed by language models without additional transformation. This representation allows models to learn semantic patterns, reasoning cues, and contextual dependencies embedded in the language, thereby providing strong semantic support and reasoning capabilities for downstream tasks such as disease diagnosis, clinical report generation, and biomedical literature question answering.
The main objective of Biomedical Reasoning and Understanding is to perform a variety of practically meaningful downstream tasks based on effective modeling of natural language texts. For example, given a research abstract from PubMed, the model needs to accurately identify and extract biomedical entities such as drug names, disease types, and gene symbols, and further uncover functional relationships among them, such as “Drug X treats Disease Y” or “Gene A is significantly associated with Disease B.” Additionally, the model can assist researchers in quickly understanding complex oncology reports and automatically answering questions such as “What treatment methods were used in this study?” or “Which patient subgroups benefited the most according to the results?” More generally, scenarios also include using medical examination questions (e.g., USMLE) as input to evaluate the model’s question-answering and reasoning capabilities across broad medical knowledge domains. These tasks rely on extensive biomedical knowledge found in literature, clinical notes, and databases, posing high demands on LLMs in terms of factual recall, domain-specific reasoning, and complex language interpretation.
In the life sciences and bioengineering domains, the value of Biomedical Reasoning and Understanding lies in their ability to extract critical information from massive volumes of text that is vital for research and clinical decision-making. Similar to the “information sparsity” seen in sequence-based tasks, biomedical texts also exhibit low information density but high-value key content—for instance, a clinical case report may be lengthy, yet the truly decisive content for diagnosis or treatment planning is often minimal. Therefore, models must possess robust capabilities in long-text modeling, information retrieval, and semantic compression to effectively accomplish the task objectives. Furthermore, in certain application scenarios, the model can also automatically generate clinical decision-making suggestions based on existing research findings, or explain treatment plans to patients in more accessible language—thus promoting research transparency and improving doctor-patient communication.
Although all these tasks involve text processing, they differ fundamentally in task structure, reasoning focus, and model design. Some subtasks revolve around retrieving or generating answers to biomedical questions—such as determining a diagnosis, choosing a treatment plan, or interpreting research outcomes—and typically require models to possess strong knowledge recall and evidence-based reasoning capabilities. In contrast, others focus on identifying semantic relationships, logical entailment, or classification problems within or across texts—for example, determining whether two sentences entail each other, or classifying text based on medical intent. These two categories reflect two long-standing paradigms in natural language processing: retrieval/generation and reasoning/classification, which also align with widely adopted benchmarking methods today. Therefore, we further subdivide Biomedical Reasoning and Understanding into two categories: Question Answering and Language Understanding.
Both categories benefit from the abundance of unlabeled or partially labeled biomedical text resources, including research papers, clinical notes, and medical examination datasets, which provide rich materials for self-supervised or weakly supervised pretraining. Furthermore, LLMs are not only capable of generating accurate answers or performing effective classification, but also excel at providing clear and interpretable reasoning, thereby significantly enhancing the transparency and trustworthiness of predictions. These characteristics enable LLMs to transcend individual task boundaries and provide robust technical support for knowledge-intensive biomedical reasoning.
Question Answering. Biomedical Question Answering focuses on enabling models to accurately extract or generate answers from scientific literature, clinical notes, or medical guidelines in response to domain-specific natural language queries. A series of LLMs and domain-specific models have been applied to biomedical question answering (QA). Early approaches employed transformer models such as BioBERT [
17] and PubMedBERT [
1297], BERT [
9] based models pre-trained on biomedical corpora—and fine-tuned them for QA tasks. Compared to general-purpose language models, these domain-specific models demonstrated higher accuracy on biomedical QA benchmarks. For example, BioBERT [
17] achieved higher F1 scores than baseline BERT in the BioASQ [
1366] challenge tasks, owing to its domain-specific pretraining. Generative transformer models tailored to biomedicine have also been developed, such as BioGPT [
1298] (a GPT-2-style [
5] model trained on biomedical texts) and BioMedLM [
1299] (also known as PubMedGPT 2.7B, a GPT-based model trained on PubMed abstracts). These models have achieved strong results in QA tasks.
Subsequently, instruction tuning and conversational LLMs entered the biomedical QA domain. Med-PaLM [
1300] (and its successor Med-PaLM 2 [
1301]) fine-tuned Google’s PaLM [
1367] model on medical QA tasks and achieved near-expert-level performance on the United States Medical Licensing Examination (USMLE), with accuracy around 86.5%, approaching that of expert physicians (87%).
To move toward truly doctor-like LLMs—beyond simply answering questions—researchers have fine-tuned pretrained models on more novel datasets. For example, ChatDoctor [
1302] was created by fine-tuning LLaMA [
52] on medical dialogue data, enabling interactive QA in a patient-doctor chat format. HuatuoGPT [
1303] posits that an intelligent medical advisor should proactively ask questions rather than respond passively. Huatuo-2 [
1304] uses an innovative domain-adaptive approach to significantly improve its medical knowledge and conversational skills. It demonstrated optimal performance on several medical benchmark tests, notably surpassing the GPT-4 on the Specialist Assessment and the new version of the Physician Licensing Exam. Similarly, models such as Clinical Camel [
1305] and DoctorGLM [
1306] are LLM-based medical chatbots designed specifically to answer medical questions in a conversational style.
At the same time, thanks to LLMs’ inherent zero-shot capabilities, large general-purpose models like GPT-4 [
1] remain competitive, which has demonstrated strong performance in medical QA and often outperforms smaller domain-specific models in zero-shot settings [
1315].
Recently, reasoning has played an increasing role in this subtask. For example, Huatuo-o1 [
1307] enhances complex reasoning ability by (1) guiding the search for complex reasoning trajectories using a verifier and fine-tuning the LLM accordingly, and (2) applying reinforcement learning (RL) with verifier-based rewards. FineMedLM-o1 [
1308] further introduced Test-Time Training [
1368] into the medical domain for the first time, promoting domain adaptation and ensuring reliable and accurate reasoning.
LLMs have substantially advanced biomedical question answering by enabling precise information extraction and fluent generation from diverse medical texts, ranging from clinical guidelines to patient dialogues. They have outperformed traditional domain-specific baselines by incorporating instruction tuning, conversational capabilities, and advanced reasoning techniques. However, several challenges remain: factual consistency and hallucination still pose risks in high-stakes clinical applications; models often struggle with ambiguous queries, underrepresented diseases, or multimodal reasoning; and real-world deployment requires careful alignment with clinical workflows and regulations. Future efforts should focus on integrating more life science and bio-engineering knowledge, enhancing traceable multi-step reasoning, and developing evaluation protocols that reflect real-world clinical utility, ensuring that QA systems can support clinicians safely and effectively.
Language Understanding. Language Understanding in the life science and bio-engineering involves modeling a system’s ability to comprehend, interpret, and reason over domain-specific texts, enabling accurate semantic inference and contextual judgment across diverse biomedical and scientific narratives. Beyond direct question answering, LLMs are increasingly applied to various language understanding tasks in the biomedical domain. These tasks require interpretation and reasoning over biomedical texts such as clinical narratives, scientific abstracts, or exam questions to support judgments or classifications. A typical example is natural language inference (NLI) in medicine: given a textual premise (e.g., a statement from a patient report) and a hypothesis, the model must determine whether the premise entails, contradicts, or is neutral with respect to the hypothesis. For instance, consider the premise “The patient denies any history of diabetes,” and the hypothesis “The patient has a history of diabetes.” A model with true understanding should correctly classify this as a contradiction, since the hypothesis directly conflicts with the premise. Language understanding is crucial for clinical decision support and information extraction, as it determines whether conclusions are genuinely grounded in clinical observations or life science facts. It also underpins tasks such as document classification, textual entailment, and reading comprehension. In essence, these tasks assess whether LLMs demonstrate deep comprehension of biomedical language, rather than mere memorization.
Many LLMs originally developed for QA have also been used for understanding tasks, often via fine-tuning for classification. Early models like BioBERT [
17] and PubMedBERT [
1297] pioneered performance improvements in biomedical text classification and NLI, achieving strong results on tasks such as MedNLI [
1369]. Fine-tuned BioBERT on the MedNLI dataset significantly outperformed earlier RNN-based models in reasoning accuracy, because of its better grasp of clinical terminology and context. ClinicalBERT [
1312], initialized from BioBERT [
17] and further trained on electronic health records, proved particularly effective in clinical NLI and related tasks, as it captured domain-specific syntax and abbreviations from structured data. More recent domain-specific models, such as BioLinkBERT [
1313] and BlueBERT [
1314], report MedNLI accuracy in the mid-80% range—approaching human expert performance.
Meanwhile, large general-purpose LLMs have demonstrated capability in language understanding via prompting. For example, GPT-4 [
1] can perform NLI without explicit fine-tuning, when prompted with queries like, “Does the following statement logically follow from the previous one?” [
1315] Trained on a broad corpus—including some medical content—these models often achieve decent accuracy in zero-shot or few-shot settings.
However, instruction-finetuned biomedical models are pushing the boundaries further. A recent method, BioInstruct [
1316], compiled around 25,000 biomedical instruction-response pairs, covering tasks such as NLI and QA, and used them to fine-tune a LLaMA model. This resulted in significant improvements across multiple benchmarks, indicating that targeted instruction tuning can effectively teach LLMs the reasoning patterns required for biomedical language understanding. Similarly, models like ChatDoctor [
1302] and Clinical Camel [
1305] (based on LLaMA [
52]), which were introduced for QA, can also perform classification or inference in a dialogue format when guided appropriately through prompts or lightweight fine-tuning. In summary, a wide range of models—from domain-specific BERTs to large GPT-style models—have been leveraged for understanding tasks. The trend is moving away from training small task-specific models from scratch and toward adapting large foundation language models (e.g., LLaMA-7B or 13B) via fine-tuning or prompting, to better transfer their general knowledge and linguistic capability to the complex biomedical domain.
LLMs have significantly advanced language understanding in the biomedical and life science domains by enabling contextual reasoning, semantic inference, and classification across complex and specialized texts such as patient reports, scientific literature, and medical examinations. These models have proven effective in tasks like natural language inference (NLI), document classification, and reading comprehension—particularly through domain-adaptive pretraining and instruction tuning. However, key challenges remain: understanding nuanced clinical negations, reasoning over long and fragmented documents, and ensuring interpretability in high-stakes decision-making. Future work should focus on improving zero-shot generalization across clinical subdomains, integrating structured biomedical ontologies for more grounded reasoning, and developing explainable evaluation frameworks to assess whether models truly comprehend rather than memorize biomedical language.
5.4.5. Hybrid Outcome Prediction
Hybrid Outcome Prediction refers to a class of tasks where LLMs are employed to predict complex biological or therapeutic outcomes by integrating diverse types of biological, chemical, and contextual information. Unlike traditional sequence-only or structure-only modeling, hybrid prediction tasks often require models to simultaneously reason over multiple heterogeneous inputs—such as chemical structures, genetic profiles, and cellular environments—to forecast functional outcomes or treatment effects. These tasks are of paramount importance in life sciences and bioengineering, as many real-world biological phenomena—such as drug response, synergistic effects, or protein function—arise from the interplay of diverse molecular and cellular factors rather than from single-modality information.
Typical inputs to hybrid prediction tasks may include combinations of small molecules, amino acid sequences, gene expression profiles, mutation data, or even broader multi-omics signatures. The outputs range from continuous measurements (e.g., synergy scores, binding affinities) to categorical labels (e.g., synergistic vs. antagonistic drug pairs, functional vs. non-functional protein variants). Hybrid outcome prediction challenges models not only to capture complex intra- and inter-modality relationships but also to generalize across biological contexts that may differ substantially between training and deployment scenarios.
The importance of hybrid outcome prediction is amplified in translational research and therapeutic development, where accurate computational forecasts can dramatically reduce experimental costs, prioritize candidate interventions, and uncover novel biological mechanisms. However, this class of tasks poses unique challenges: input modalities are often high-dimensional and noisy; the relationships between features and outcomes can be nonlinear and context-dependent; and biological interpretability remains a significant hurdle. LLMs, with their ability to integrate multimodal data, model contextual dependencies, and adapt to new tasks through fine-tuning or prompting, are particularly well-suited to address these complexities.
In this section, we focus on two major sub-directions within Hybrid Outcome Prediction: Drug Synergy Prediction and Protein Modeling. Both represent critical applications where LLMs have demonstrated transformative potential, yet where significant challenges and opportunities for future development remain.
Drug Synergy Prediction. Drug synergy prediction involves forecasting the therapeutic efficacy of drug combinations. In many diseases—particularly cancer—combination therapies can improve treatment outcomes and prevent resistance. Drug synergy refers to a phenomenon where the combined effect of two (or more) drugs exceeds the effect of each drug administered individually. Identifying synergistic drug pairs is critical for accelerating the design of combination therapies while reducing the need for extensive laboratory testing. However, this task is highly challenging due to the combinatorial explosion of possible drug pairs and the complex biological mechanisms underlying their interactions. The synergy of a given drug pair can vary depending on the context—such as cell type or disease environment—making generalization difficult. Despite these challenges, accurate synergy prediction can dramatically narrow the search space for effective multi-drug treatment regimens.
Models designed for this task typically take two drugs as input—often represented by their chemical structures, such as SMILES strings or molecular fingerprints—along with contextual features like genomic profiles of the target cell line. The output is a synergy score or class label indicating whether the combination exhibits synergistic behavior. Specifically, the input may consist of a pair of SMILES strings {Drug A, Drug B} and a cell line ID, while the output could be a continuous synergy metric, such as a Bliss or Loewe additivity score, or a binary label (synergistic vs. non-synergistic). Some models use drug-pair dose-response matrices, though many modern approaches simplify the task to predicting a single synergy score per drug pair. Incorporating contextual information (e.g., gene expression or mutation data of the cell line) makes this a multimodal prediction task, as synergy is often conditional on biological context.
Early synergy prediction methods used feature-engineered machine learning models. DeepSynergy [
1370], for example, employed deep neural networks to combine molecular descriptors with gene expression profiles. More recently, transformer-based and LLM-inspired models have emerged. One notable example is DFFNDDS [
1325], which integrates a BERT-like language model [
9] for encoding drug SMILES and introduces a dual-feature fusion attention mechanism to capture drug-cell interactions. The BERT module in DFFNDDS [
1325] jointly attends to drug-drug and drug-cell features to learn non-linear synergy effects. This architecture helps discover subtle interaction patterns—such as complementary mechanisms of action—that may be missed by simpler models or naive feature concatenation.
CancerGPT [
1326] introduced a few-shot approach using a GPT-style model, transforming tabular synergy data into natural language format and fine-tuning GPT-3 to predict drug synergy in rare cancers. This method leverages the prior knowledge embedded in the language model’s weights, enabling accurate predictions even with zero or few training samples in new tissue types. Another cutting-edge method, SynerGPT [
1327], pretrains a GPT model to perform contextual learning of a “synergy function.” It is trained to take a personalized dataset of known synergistic pairs as a prompt and then predict new pairs under the same context. This context-based approach avoids reliance on fixed molecular descriptors or domain-specific biological knowledge, instead extrapolating from patterns embedded in the prompt—achieving competitive results.
Furthermore, LLMs can serve as foundation models to address a diverse set of tasks in this domain. One such approach, BAITSAO [
1328], is a foundation model strategy that integrates multiple datasets and tasks. It uses context-rich embeddings from LLMs as initial representations of drugs and cell lines, and performs pretraining on large drug combination databases within a multitask learning framework. BAITSAO [
1328] outperformed both classical models (like DeepSynergy [
1370]) and more recent tabular or transformer-based models on benchmark datasets, thanks to its multitask training and transfer learning across drug combination contexts. Overall, these LLM-based strategies—from fine-tuned GPT models to transformer fusion networks—highlight the growing role of language model architectures in capturing the complex relationships underlying drug synergy.
LLMs have brought significant advances to drug synergy prediction by enabling the modeling of complex drug–cell line interactions through contextual embeddings, attention mechanisms, and prompt-based reasoning. These models reduce reliance on handcrafted features, generalize better across biological contexts, and support few-shot or even zero-shot inference, which is especially valuable for rare diseases or under-studied drug pairs. However, key challenges remain: biological interpretability is limited, especially in identifying mechanistic pathways; synergy predictions often lack consistency across datasets or experimental conditions; and integrating multi-omics data with chemical and pharmacological knowledge in a unified framework is still an open problem. Future work should focus on enhancing cross-dataset generalization, embedding biological priors into LLM architectures, and developing transparent, mechanistically grounded models that can support experimental design and clinical translation in combination therapy development.
Table 44.
Life Science and Bio-engineering Tasks, Benchmarks, Introduction and Cross tasks
Table 44.
Life Science and Bio-engineering Tasks, Benchmarks, Introduction and Cross tasks
| Type of Task |
Benchmarks |
Introduction |
Cross tasks |
| DNA Sequence Modeling |
BEND [1294] |
A collection of realistic and biologically meaningful downstream tasks defined on the human genome |
Gene finding, Enhancer annotation, Chromatin accessibility, Histone modification etc. |
| |
Genomic Benchmarks [1371] |
Contains 8 datasets that focus on regulatory elements from 3 model organisms: human, mouse, and roundworm. |
- |
| |
GUE [1284] |
A collection of 28 datasets across 7 tasks constructed for genome language model evaluation. |
Promoter prediction, Splice site prediction, Covid variant classification, epigenetic marks prediction etc. |
| |
NT [1287] |
A collection of 18 datasets across 7 tasks constructed for genome language model evaluation. |
Promoter prediction, Splice site prediction,Enhancer annotation etc. |
| RNA Function Learning |
RnaBench [1372] |
Including 100 samples without any training and validation data. |
Intra/Inter family prediction, Inverse RNA Folding. |
| |
BEACON [1373] |
Containing 967k sequences with lengths ranging from 23 to 1182. |
Structure Prediction, Contact Map Prediction, Modification Prediction, Mean Ribosome Loading etc. |
| Clinical Language Generation |
MIMIC-III [1362] |
A large, freely-available database comprising deidentified health-related data associated with over forty thousand patients. |
Report Summarization, Risk Prediction etc. |
| |
MIMIC-IV [1363] |
A larger version including over 65,000 patients admitted to an ICU and over 200,000 patients admitted to the emergency department. |
Report Summarization, Risk Prediction etc. |
| |
IU X-Ray [1374] |
A set of 7,470 chest X-ray images paired with their corresponding diagnostic reports. |
Report Summarization, Image Caption etc. |
| EHR Based Prediction |
EHRSHOT [1375] |
A collection of 6739 patients of EHR based benchmark in few-shot setting. |
– |
| |
EHRNoteQA [1376] |
A complex, multi-topic benchmark based on multiple patients’ electronic discharg records. |
QA. |
| Quastion Answering |
MedQA [1377] |
Consists of multiple-choice questions from the United States Medical Licensing Examination (USMLE). |
– |
| |
MedMCQA [1378] |
A large-scale multiple-choice QA dataset derived from Indian medical entrance examinations (AIIMS/NEET). |
– |
| |
PubMedQA [1379] |
A closed-domain QA dataset, questions can be answered by looking at an associated context (PubMed abstract). |
– |
| |
MMLU Subsets [135] |
For measuring multitask ability from various domains, including life science and Bio-engineering. |
– |
| |
MIMIC-IV [1363] |
A collection of 1057 questions, answer could based on the referral letters. |
– |
| Language Understanding |
BC5-Disease [1089] |
Including three separate sets of articles with diseases, chemicals and their relations annotated. |
Named Entity Recognition. |
| |
NCBI-Disease [1380] |
Contains 6,892 disease mentions, which are mapped to 790 unique disease concepts |
Named Entity Recognition. |
| |
DDI [1381] |
An annotated corpus with pharmacological substances and drug–drug interactions |
Relation Extraction. |
| |
GAD [1382] |
A repository of molecular, clinical and study parameters for >5,000 human genetic association studies |
Relation Extraction. |
| |
HoC [1383] |
Including 1852 PubMed publication abstracts manually annotated by experts. |
Doc. Classification. |
| Drug Synergy Prediction |
CancerGPT [1326] |
An framework involves testing LLMs’ performance in few/zero-shot learning scenarios across seven rare tissue types. |
– |
| |
BAITSAO [1328] |
A framework integrates both regression and classification, based on synergy scores and binary synergy labels derived from large-scale drug combination datasets. |
– |
| Protein Modeling |
PEER [1384] |
Comprising fourteen diverse protein sequence understanding tasks. |
– |
| |
Type [1385] |
Five biologically relevant protein tasks and evaluated self-supervised sequence models. |
– |
Table 45.
Benchmark results on BEND. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
Table 45.
Benchmark results on BEND. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
| Model |
Gene
finding |
Enhancer
annotation |
Chromatin
accessibility |
Histone
modification |
CpG
methylation |
Variant effects
(expression) |
Variant effects
(disease) |
| ResNet (non-LLM) |
0.46 |
0.06 |
- |
- |
- |
- |
- |
| CNN (non-LLM) |
0.00 |
0.03 |
0.75 |
0.76 |
0.84 |
- |
- |
| ResNet-LM |
0.36 |
0.02 |
0.82 |
0.77 |
0.87 |
0.55 |
0.55 |
| AWD-LSTM |
0.05 |
0.03 |
0.69 |
0.74 |
0.81 |
0.53 |
0.45 |
| NT-H |
0.41 |
0.05 |
0.74 |
0.76 |
0.88 |
0.55 |
0.48 |
| NT-MS |
0.68 |
0.06 |
0.79 |
0.78 |
0.92 |
0.54 |
0.77 |
| NT-1000G |
0.49 |
0.04 |
0.77 |
0.77 |
0.89 |
0.45 |
0.49 |
| NT-V2 |
0.64 |
0.05 |
0.80 |
0.76 |
0.91 |
0.48 |
0.48 |
| DNABERT |
0.20 |
0.03 |
0.85 |
0.79 |
0.91 |
0.60 |
0.56 |
| DNABERT-2 |
0.43 |
0.03 |
0.81 |
0.78 |
0.90 |
0.49 |
0.51 |
| GENA-LM BERT |
0.52 |
0.03 |
0.76 |
0.78 |
0.91 |
0.49 |
0.55 |
| GENA-LM BigBird |
0.39 |
0.04 |
0.82 |
0.78 |
0.91 |
0.49 |
0.52 |
| HyenaDNA large |
0.35 |
0.03 |
0.84 |
0.76 |
0.91 |
0.51 |
0.45 |
| HyenaDNA tiny |
0.10 |
0.02 |
0.78 |
0.76 |
0.86 |
0.47 |
0.44 |
| GROVER |
0.28 |
0.03 |
0.82 |
0.77 |
0.89 |
0.56 |
0.51 |
Table 46.
Benchmark results across various 13 RNA tasks. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
Table 46.
Benchmark results across various 13 RNA tasks. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
| Model |
SSP |
CMP |
DMP |
SSI |
SPL |
APA |
NcRNA |
Modif |
MRL |
VDP |
PRS |
CRI-On |
CRI-Off |
| |
F1 (%) |
P@L (%) |
(%) |
(%) |
ACC@K (%) |
(%) |
ACC (%) |
AUC (%) |
(%) |
MCRMSE↓ |
(%) |
SC (%) |
SC (%) |
| CNN (non-LLM) |
49.95 |
43.89 |
27.76 |
34.36 |
8.43 |
50.93 |
88.62 |
70.87 |
74.13 |
0.361 |
45.40 |
29.69 |
11.40 |
| ResNet (non-LLM) |
57.26 |
59.59 |
30.26 |
37.74 |
21.15 |
56.45 |
88.33 |
71.03 |
74.34 |
0.349 |
55.21 |
28.55 |
11.50 |
| LSTM (non-LLM) |
58.61 |
40.41 |
44.77 |
35.44 |
36.66 |
67.03 |
88.78 |
94.83 |
83.94 |
0.329 |
55.45 |
26.83 |
8.60 |
| RNA-FM |
68.50 |
47.56 |
51.45 |
42.36 |
34.84 |
70.32 |
96.81 |
94.98 |
79.47 |
0.347 |
55.98 |
31.62 |
2.49 |
| RNABERT |
57.27 |
45.21 |
48.19 |
31.62 |
0.18 |
57.66 |
68.95 |
82.82 |
29.79 |
0.378 |
54.60 |
29.77 |
4.27 |
| RNA-MSM |
57.98 |
57.26 |
37.49 |
39.22 |
38.33 |
70.40 |
84.85 |
94.89 |
83.48 |
0.330 |
56.94 |
34.92 |
3.85 |
| Splice-H510 |
64.93 |
45.80 |
55.56 |
38.91 |
44.80 |
58.65 |
95.92 |
62.57 |
83.49 |
0.321 |
54.90 |
26.61 |
4.00 |
| Splice-MS510 |
43.24 |
52.64 |
10.27 |
38.58 |
50.55 |
52.46 |
95.87 |
55.87 |
84.98 |
0.315 |
50.98 |
27.13 |
3.49 |
| Splice-MS1024 |
68.26 |
47.32 |
55.89 |
39.22 |
48.52 |
60.03 |
96.05 |
53.45 |
67.15 |
0.313 |
57.72 |
27.59 |
5.00 |
| UTR-LM-MRL |
59.71 |
45.51 |
55.21 |
39.52 |
36.20 |
64.99 |
89.97 |
56.41 |
77.78 |
0.325 |
57.28 |
28.49 |
4.28 |
| UTR-LM-TE&EL |
59.57 |
60.32 |
54.94 |
40.15 |
37.35 |
72.09 |
81.33 |
59.70 |
82.50 |
0.319 |
53.37 |
32.49 |
2.91 |
| UTRBERT-3mer |
60.37 |
51.03 |
50.95 |
34.31 |
44.24 |
69.52 |
92.88 |
95.14 |
83.89 |
0.337 |
56.83 |
29.92 |
4.48 |
| UTRBERT-4mer |
59.41 |
44.91 |
47.77 |
33.22 |
42.04 |
72.71 |
94.32 |
95.10 |
82.90 |
0.341 |
56.43 |
23.20 |
3.11 |
| UTRBERT-5mer |
47.92 |
44.71 |
48.67 |
31.27 |
39.19 |
72.70 |
93.04 |
94.78 |
75.64 |
0.343 |
57.16 |
25.74 |
3.93 |
| UTRBERT-6mer |
38.56 |
51.56 |
50.02 |
29.93 |
38.58 |
71.17 |
93.12 |
95.08 |
83.60 |
0.340 |
57.14 |
28.60 |
4.90 |
| BEACON-B |
64.18 |
60.81 |
56.28 |
38.78 |
37.43 |
70.59 |
94.63 |
94.74 |
72.29 |
0.320 |
54.67 |
26.01 |
4.42 |
| BEACON-B512 |
58.75 |
61.20 |
56.82 |
39.13 |
37.24 |
72.00 |
94.99 |
94.92 |
72.35 |
0.320 |
55.20 |
28.17 |
3.82 |
Table 47.
Benchmark results on question answering. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
Table 47.
Benchmark results on question answering. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
| Model |
MedQA |
MedMCQA |
MMLU |
PubMedQA |
Referal QA |
Treat Recom. |
| Claude-2 |
65.1 |
60.3 |
78.7 |
70.8 |
80.5 |
9.1 |
| GPT-3.5-turbo |
61.2 |
59.4 |
73.5 |
70.2 |
81.1 |
7.3 |
| GPT-4 |
83.4 |
78.2 |
92.3 |
80.0 |
83.2 |
18.6 |
| Alpaca |
34.2 |
30.1 |
40.8 |
65.2 |
74.8 |
3.5 |
| Vicuna-7B |
34.5 |
33.4 |
43.4 |
64.8 |
76.4 |
2.6 |
| LLaMA-2-7B |
32.9 |
30.6 |
42.3 |
63.4 |
74.5 |
3.3 |
| Mistral |
35.7 |
37.8 |
46.3 |
69.4 |
77.7 |
5.0 |
| Vicuna-13B |
38.0 |
36.4 |
45.6 |
66.2 |
76.8 |
4.6 |
| LLaMA-2-13B |
38.1 |
35.5 |
46.0 |
66.8 |
77.1 |
4.8 |
| LLaMA-2-70B |
45.8 |
42.7 |
54.0 |
67.4 |
78.9 |
5.5 |
| LLaMA-3-70B |
78.8 |
74.7 |
86.4 |
71.4 |
82.4 |
10.2 |
| HuatuoGPT |
28.4 |
24.8 |
31.6 |
61.0 |
69.3 |
3.8 |
| HuatuoGPT-2-7B |
41.1 |
41.9 |
- |
- |
- |
- |
| HuatuoGPT-2-13B |
45.7 |
47.4 |
- |
- |
- |
- |
| HuatuoGPT-o1-8B |
72.6 |
60.4 |
- |
79.2 |
- |
- |
| ChatDoctor |
33.2 |
31.5 |
40.4 |
63.8 |
73.7 |
5.3 |
| PMC-LLaMA-7B |
28.7 |
29.8 |
39.0 |
60.2 |
70.2 |
4.0 |
| Baize-Healthcare |
34.9 |
31.3 |
41.9 |
64.4 |
74.0 |
4.7 |
| MedAlpaca-7B |
35.1 |
32.9 |
48.5 |
62.4 |
75.3 |
4.8 |
| Meditron-7B |
33.5 |
31.1 |
45.2 |
61.6 |
74.9 |
5.8 |
| BioMistral |
35.4 |
34.8 |
52.6 |
66.4 |
77.0 |
7.6 |
| PMC-LLaMA-13B |
39.6 |
37.7 |
56.3 |
67.0 |
77.6 |
4.9 |
| MedAlpaca-13B |
37.3 |
35.7 |
51.5 |
65.6 |
77.4 |
5.1 |
| ClinicalCamel |
46.4 |
45.8 |
68.4 |
71.0 |
79.8 |
8.4 |
| Meditron-70B |
45.7 |
44.9 |
65.1 |
70.6 |
78.6 |
8.9 |
| HuatuoGPT-o1-70B |
83.3 |
73.6 |
- |
80.6 |
- |
- |
| Med-PaLM 2 (5-shots) |
79.7 |
71.3 |
- |
79.2 |
- |
- |
Table 48.
Benchmark results on language understanding. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
Table 48.
Benchmark results on language understanding. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
| Model |
BC5 |
NCBI |
DDI |
GAD |
HoC |
Pharma. QA |
Drug Infer. |
| Claude-2 |
52.9 |
44.2 |
50.4 |
50.7 |
70.8 |
60.6 |
51.5 |
| GPT-3.5-turbo |
52.3 |
46.1 |
49.3 |
50.8 |
66.4 |
57.3 |
47.0 |
| GPT-4 |
71.3 |
58.4 |
64.6 |
68.2 |
83.6 |
63.8 |
56.5 |
| Alpaca |
41.2 |
36.5 |
37.4 |
36.9 |
52.6 |
41.3 |
47.5 |
| Vicuna-7B |
44.5 |
37.0 |
39.4 |
41.2 |
53.8 |
42.3 |
45.5 |
| LLaMA-2-7B |
40.1 |
34.8 |
37.9 |
39.3 |
48.6 |
46.5 |
48.0 |
| Mistral |
46.8 |
39.9 |
43.5 |
44.3 |
59.6 |
51.2 |
53.0 |
| Vicuna-13B |
46.2 |
39.0 |
41.3 |
43.5 |
56.7 |
45.1 |
46.0 |
| LLaMA-2-13B |
46.6 |
38.3 |
39.7 |
41.2 |
55.9 |
46.9 |
47.5 |
| LLaMA-2-70B |
47.8 |
41.5 |
45.6 |
44.7 |
63.2 |
49.3 |
51.5 |
| LLaMA-3-70B |
63.7 |
50.2 |
59.7 |
63.1 |
79.0 |
62.4 |
53.0 |
| PubMed-BERT-base |
- |
87.8 |
82.4 |
82.3 |
82.3 |
- |
- |
| BioLink-BERT-base |
- |
88.2 |
82.7 |
84.4 |
85.4 |
- |
- |
| HuatuoGPT |
43.6 |
37.5 |
40.1 |
38.2 |
50.2 |
44.1 |
49.5 |
| ChatDoctor |
45.8 |
40.9 |
41.2 |
40.1 |
55.7 |
42.7 |
48.5 |
| PMC-LLaMA-7B |
45.2 |
37.8 |
40.8 |
42.0 |
55.6 |
45.5 |
51.0 |
| Baize-Healthcare |
44.4 |
38.5 |
41.9 |
45.8 |
54.5 |
46.9 |
50.5 |
| MedAlpaca-7B |
47.3 |
39.0 |
43.5 |
44.0 |
58.7 |
47.9 |
52.0 |
| Meditron-7B |
46.5 |
39.2 |
42.7 |
43.3 |
57.9 |
50.7 |
52.0 |
| BioMistral |
48.8 |
40.4 |
46.0 |
48.5 |
64.3 |
54.5 |
54.0 |
| PMC-LLaMA-13B |
51.5 |
43.1 |
48.4 |
48.7 |
65.3 |
48.8 |
51.5 |
| MedAlpaca-13B |
49.2 |
41.6 |
44.1 |
44.5 |
59.4 |
51.6 |
50.0 |
| ClinicalCamel |
51.2 |
43.7 |
47.6 |
47.2 |
64.8 |
52.6 |
52.5 |
| Meditron-70B |
54.3 |
45.7 |
51.2 |
49.6 |
69.6 |
58.7 |
54.5 |
Table 49.
Benchmark results on EHRNoteQA. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
Table 49.
Benchmark results on EHRNoteQA. Best are highlighted with dark blue. If not emphasized, the metrics is considered the higher the better.
| Model |
Multi-Choice |
Open-Ended |
| |
Level 1 |
Level 2 |
Level 1 |
Level 2 |
| GPT4 |
97.16 |
95.15 |
91.30 |
89.61 |
| GPT4-Turbo |
95.27 |
94.23 |
91.30 |
86.61 |
| GPT3.5-Turbo |
88.28 |
84.99 |
82.23 |
75.52 |
| Llama3-70b-Instruct |
94.33 |
91.92 |
89.04 |
86.84 |
| Llama2-70b-chat |
84.88 |
– |
78.83 |
– |
| qCamel-70 |
85.63 |
– |
78.26 |
– |
| Camel-Platypus2-70b |
89.79 |
– |
78.83 |
– |
| Platypus2-70b-Instruct |
90.36 |
– |
80.53 |
– |
| Mixtral-8x7b-Instruct |
87.52 |
86.61 |
88.28 |
81.52 |
| MPT-30b-Instruct |
79.96 |
75.52 |
67.11 |
62.59 |
| Llama2-13b-chat |
73.65 |
– |
70.32 |
– |
| Vicuna-13b |
82.04 |
– |
70.51 |
– |
| WizardLM-13b |
80.91 |
– |
74.67 |
– |
| qCamel-13 |
71.46 |
– |
66.16 |
– |
| OpenOrca-Platypus2-13b |
86.01 |
– |
79.21 |
– |
| Camel-Platypus2-13b |
78.07 |
– |
67.86 |
– |
| Synthia-13b |
79.21 |
– |
74.48 |
– |
| Asclepius-13b1
|
– |
– |
75.24 |
– |
| Gemma-7b-it |
77.50 |
67.21 |
63.71 |
54.27 |
| MPT-7b-8k-instruct |
59.55 |
51.27 |
56.71 |
53.81 |
| Mistral-7b-Instruct |
82.04 |
64.90 |
72.97 |
53.81 |
| Dolphin-2.0-mistral-7b |
76.18 |
– |
69.75 |
– |
| Mistral-7b-OpenOrca |
87.15 |
– |
79.58 |
– |
| SynthIA-7b |
78.45 |
– |
74.67 |
– |
| Llama2-7b-chat |
65.78 |
– |
58.98 |
– |
| Vicuna-7b |
78.26 |
– |
59.74 |
– |
| Asclepius-7b |
– |
– |
66.92 |
– |
Table 50.
AUPRC of k-shot learning on seven tissue sets. =total number of non-synergistic samples (not positive), =total number of synergistic samples (positive). Where XGBoost is non-LLM baseline method. Best are highlighted with dark blue.
Table 50.
AUPRC of k-shot learning on seven tissue sets. =total number of non-synergistic samples (not positive), =total number of synergistic samples (positive). Where XGBoost is non-LLM baseline method. Best are highlighted with dark blue.
| Tissue ( , ) |
Methods |
0 |
2 |
4 |
8 |
16 |
32 |
64 |
128 |
Pancreas
(=38, =1) |
XGBoost |
0.026 |
– |
– |
– |
– |
– |
– |
– |
| TabTransformer |
0.056 |
– |
– |
– |
– |
– |
– |
– |
| CancerGPT |
0.033 |
– |
– |
– |
– |
– |
– |
– |
| GPT-2 |
0.032 |
– |
– |
– |
– |
– |
– |
– |
| GPT-3 |
0.111 |
– |
– |
– |
– |
– |
– |
– |
Endometrium
(=36, =32) |
XGBoost |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
– |
– |
– |
| TabTransformer |
0.674 |
0.889 |
0.903 |
0.948 |
0.938 |
0.962 |
– |
– |
| CancerGPT |
0.564 |
0.668 |
0.676 |
0.831 |
0.686 |
0.737 |
– |
– |
| GPT-2 |
0.408 |
0.808 |
0.395 |
0.383 |
0.389 |
0.717 |
– |
– |
| GPT-3 |
0.869 |
1.0 |
0.947 |
0.859 |
0.799 |
0.859 |
– |
– |
Liver
(=192, =21) |
XGBoost |
0.132 |
0.132 |
0.132 |
0.132 |
0.132 |
0.132 |
0.12 |
0.12 |
| TabTransformer |
0.13 |
0.128 |
0.147 |
0.189 |
0.265 |
0.168 |
0.169 |
0.234 |
| CancerGPT |
0.136 |
0.102 |
0.13 |
0.147 |
0.252 |
0.21 |
0.197 |
0.187 |
| GPT-2 |
0.5 |
0.099 |
0.151 |
0.383 |
0.429 |
0.401 |
0.483 |
0.398 |
| GPT-3 |
0.185 |
0.086 |
0.096 |
0.125 |
0.124 |
0.314 |
0.362 |
0.519 |
Soft tissue
(=269, =83) |
XGBoost |
0.243 |
0.243 |
0.243 |
0.243 |
0.235 |
0.235 |
0.264 |
0.271 |
| TabTransformer |
0.273 |
0.287 |
0.462 |
0.422 |
0.526 |
0.571 |
0.561 |
0.64 |
| CancerGPT |
0.314 |
0.315 |
0.338 |
0.383 |
0.383 |
0.403 |
0.464 |
0.469 |
| GPT-2 |
0.259 |
0.298 |
0.254 |
0.262 |
0.235 |
0.297 |
0.254 |
0.206 |
| GPT-3 |
0.263 |
0.194 |
0.28 |
0.228 |
0.363 |
0.618 |
0.638 |
0.734 |
Stomach
(=1081, =109) |
XGBoost |
0.104 |
0.104 |
0.104 |
0.104 |
0.104 |
0.104 |
0.09 |
0.094 |
| TabTransformer |
0.261 |
0.371 |
0.396 |
0.383 |
0.294 |
0.402 |
0.45 |
0.465 |
| CancerGPT |
0.3 |
0.297 |
0.316 |
0.325 |
0.269 |
0.308 |
0.297 |
0.312 |
| GPT-2 |
0.116 |
0.124 |
0.099 |
0.172 |
0.165 |
0.107 |
0.152 |
0.131 |
| GPT-3 |
0.078 |
0.106 |
0.17 |
0.37 |
0.1 |
0.19 |
0.219 |
0.181 |
Urinary tract
(=1996, =462) |
XGBoost |
0.186 |
0.186 |
0.186 |
0.186 |
0.186 |
0.197 |
0.199 |
0.209 |
| TabTransformer |
0.248 |
0.264 |
0.25 |
0.278 |
0.274 |
0.249 |
0.293 |
0.291 |
| CancerGPT |
0.241 |
0.226 |
0.246 |
0.239 |
0.256 |
0.271 |
0.266 |
0.269 |
| GPT-2 |
0.191 |
0.192 |
0.188 |
0.156 |
0.193 |
0.185 |
0.183 |
0.185 |
| GPT-3 |
0.27 |
0.228 |
0.222 |
0.201 |
0.206 |
0.2 |
0.24 |
0.272 |
Bone
(=3732, =253) |
XGBoost |
0.064 |
0.064 |
0.064 |
0.064 |
0.064 |
0.064 |
0.064 |
0.064 |
| TabTransformer |
0.123 |
0.12 |
0.121 |
0.115 |
0.102 |
0.13 |
0.129 |
0.121 |
| CancerGPT |
0.119 |
0.115 |
0.125 |
0.116 |
0.115 |
0.111 |
0.114 |
0.125 |
| GPT-2 |
0.063 |
0.094 |
0.057 |
0.081 |
0.052 |
0.071 |
0.057 |
0.065 |
| GPT-3 |
0.064 |
0.051 |
0.045 |
0.058 |
0.068 |
0.087 |
0.101 |
0.181 |
Table 51.
AUROC of k-shot learning on seven tissues sets. Where XGBoost is non-LLM baseline method.
Table 51.
AUROC of k-shot learning on seven tissues sets. Where XGBoost is non-LLM baseline method.
| Tissue |
Methods |
0 |
2 |
4 |
8 |
16 |
32 |
64 |
128 |
| Pancreas |
XGBoost |
0.5 |
– |
– |
– |
– |
– |
– |
– |
| TabTransformer |
0.553 |
– |
– |
– |
– |
– |
– |
– |
| CancerGPT |
0.237 |
– |
– |
– |
– |
– |
– |
– |
| GPT-2 |
0.211 |
– |
– |
– |
– |
– |
– |
– |
| GPT-3 |
0.789 |
– |
– |
– |
– |
– |
– |
– |
| Endometrium |
XGBoost |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
0.5 |
– |
– |
| TabTransformer |
0.694 |
0.857 |
0.878 |
0.939 |
0.939 |
0.959 |
– |
– |
| CancerGPT |
0.489 |
0.693 |
0.714 |
0.735 |
0.612 |
0.612 |
– |
– |
| GPT-2 |
0.265 |
0.816 |
0.224 |
0.184 |
0.204 |
0.612 |
– |
– |
| GPT-3 |
0.837 |
1.0 |
0.949 |
0.898 |
0.878 |
0.898 |
– |
– |
| Liver |
XGBoost |
0.587 |
0.587 |
0.587 |
0.587 |
0.587 |
0.587 |
0.574 |
0.574 |
| TabTransformer |
0.535 |
0.506 |
0.526 |
0.535 |
0.609 |
0.647 |
0.702 |
0.804 |
| CancerGPT |
0.615 |
0.468 |
0.59 |
0.641 |
0.782 |
0.776 |
0.737 |
0.737 |
| GPT-2 |
0.731 |
0.449 |
0.558 |
0.66 |
0.679 |
0.763 |
0.731 |
0.731 |
| GPT-3 |
0.615 |
0.49 |
0.542 |
0.583 |
0.474 |
0.731 |
0.737 |
0.91 |
| Soft tissue |
XGBoost |
0.491 |
0.491 |
0.491 |
0.491 |
0.454 |
0.476 |
0.542 |
0.552 |
| TabTransformer |
0.557 |
0.566 |
0.709 |
0.727 |
0.788 |
0.802 |
0.83 |
0.835 |
| CancerGPT |
0.656 |
0.646 |
0.68 |
0.734 |
0.725 |
0.754 |
0.8 |
0.795 |
| GPT-2 |
0.546 |
0.535 |
0.519 |
0.56 |
0.427 |
0.577 |
0.456 |
0.384 |
| GPT-3 |
0.517 |
0.406 |
0.6 |
0.444 |
0.607 |
0.82 |
0.866 |
0.889 |
| Stomach |
XGBoost |
0.529 |
0.529 |
0.529 |
0.529 |
0.529 |
0.529 |
0.476 |
0.508 |
| TabTransformer |
0.804 |
0.863 |
0.855 |
0.853 |
0.812 |
0.85 |
0.885 |
0.869 |
| CancerGPT |
0.794 |
0.792 |
0.796 |
0.794 |
0.785 |
0.787 |
0.824 |
0.804 |
| GPT-2 |
0.551 |
0.569 |
0.521 |
0.516 |
0.589 |
0.538 |
0.469 |
0.566 |
| GPT-3 |
0.419 |
0.575 |
0.724 |
0.769 |
0.534 |
0.69 |
0.742 |
0.724 |
| Urinary tract |
XGBoost |
0.494 |
0.494 |
0.494 |
0.494 |
0.494 |
0.526 |
0.53 |
0.544 |
| TabTransformer |
0.599 |
0.612 |
0.604 |
0.625 |
0.601 |
0.587 |
0.623 |
0.622 |
| CancerGPT |
0.578 |
0.561 |
0.579 |
0.577 |
0.589 |
0.593 |
0.609 |
0.609 |
| GPT-2 |
0.526 |
0.528 |
0.532 |
0.397 |
0.515 |
0.452 |
0.469 |
0.566 |
| GPT-3 |
0.645 |
0.57 |
0.556 |
0.496 |
0.508 |
0.516 |
0.531 |
0.572 |
| Bone |
XGBoost |
0.499 |
0.499 |
0.499 |
0.499 |
0.499 |
0.499 |
0.499 |
0.499 |
| TabTransformer |
0.706 |
0.705 |
0.724 |
0.697 |
0.65 |
0.689 |
0.708 |
0.696 |
| CancerGPT |
0.625 |
0.648 |
0.693 |
0.653 |
0.683 |
0.636 |
0.678 |
0.68 |
| GPT-2 |
0.507 |
0.616 |
0.471 |
0.579 |
0.421 |
0.552 |
0.476 |
0.518 |
| GPT-3 |
0.498 |
0.415 |
0.341 |
0.429 |
0.485 |
0.605 |
0.62 |
0.794 |
Table 52.
Benchmark Results on PEER. Best are highlighted with dark blue.
Table 52.
Benchmark Results on PEER. Best are highlighted with dark blue.
| Model |
Flu |
Sta |
-lac |
Sol |
Sub |
Bin |
Cont |
Fold |
SSP |
Yst |
Hum |
Aff |
PDB |
BDB |
| DDE |
0.638 |
0.652 |
0.623 |
59.77 |
49.17 |
77.43 |
– |
9.57 |
– |
55.83 |
62.77 |
2.908 |
– |
– |
| Moran |
0.400 |
0.322 |
0.375 |
57.73 |
31.13 |
55.63 |
– |
7.10 |
– |
53.00 |
54.67 |
2.984 |
– |
– |
| LSTM |
0.494 |
0.533 |
0.139 |
70.18 |
62.98 |
88.11 |
26.34 |
8.24 |
68.99 |
53.62 |
63.75 |
2.853 |
1.457 |
1.572 |
| Transformer |
0.643 |
0.649 |
0.261 |
70.12 |
56.02 |
75.74 |
17.50 |
8.52 |
59.62 |
54.12 |
59.58 |
2.499 |
1.455 |
1.566 |
| CNN |
0.682 |
0.637 |
0.781 |
64.43 |
58.73 |
82.67 |
10.00 |
10.93 |
66.07 |
55.07 |
62.60 |
2.796 |
1.376 |
1.497 |
| ResNet |
0.636 |
0.126 |
0.152 |
67.33 |
52.30 |
78.99 |
20.43 |
8.89 |
69.56 |
48.91 |
68.61 |
3.005 |
1.441 |
1.565 |
| ProtBert |
0.679 |
0.771 |
0.731 |
68.15 |
76.53 |
91.32 |
39.66 |
16.94 |
82.18 |
63.72 |
77.32 |
2.195 |
1.562 |
1.549 |
| ProtBert* |
0.339 |
0.697 |
0.616 |
59.17 |
59.44 |
81.54 |
24.35 |
10.74 |
62.51 |
53.87 |
83.61 |
2.996 |
1.457 |
1.649 |
| ESM-1b |
0.679 |
0.694 |
0.839 |
70.23 |
78.13 |
92.40 |
45.78 |
28.17 |
82.73 |
57.00 |
78.17 |
2.281 |
1.559 |
1.556 |
| ESM-1b* |
0.430 |
0.750 |
0.528 |
67.02 |
79.82 |
91.61 |
40.37 |
29.95 |
83.14 |
66.07 |
88.06 |
3.031 |
1.368 |
1.571 |
Protein Modeling. Protein Modeling refers to the task of learning structural, functional, or evolutionary patterns from amino acid sequences, enabling predictions of protein properties such as folding, function, or interaction based. The development of protein LLMs has been driven by the deepening integration of computational biology and natural language processing techniques. Early efforts focused on leveraging traditional deep learning architectures such as LSTMs for representation learning of protein sequences [
1329,
1330,
1386]. Models like UniRep [
1329] and Bepler & Berger [
1330] made initial progress in constructing protein embedding vectors.
AlphaFold [
1387] is a protein structure prediction model released by DeepMind, which revolutionized the long-standing protein folding problem through deep learning. The model integrates evolutionary homologous sequences, residue-pair geometric maps, and physical constraints using an attention-based network. In CASP14, it achieved an average GDT_TS of 92.4, marking the first time atomic-level accuracy was reached. The subsequent release of a database containing over 2 million predicted structures has significantly accelerated drug discovery, enzyme engineering, and pathogenic mutation annotation.
With the rise of Transformers [
4] in natural language processing, this paradigm was rapidly transferred to protein modeling. Transformer-based models such as ProtTrans [
1269] and ESM-1b [
1331] emerged, offering enhanced capabilities in capturing long-range dependencies within sequences, significantly improving the accuracy of protein structure and function prediction.
The ESM series has since expanded in both model size and task scope—from ESM-1v [
1332] to ESM-2 [
1333] and the latest ESM-3 [
1388]—achieving end-to-end sequence-to-structure prediction (e.g., ESMFold [
1333]), incorporating multimodal information, and enabling complex reasoning and even generative design for protein function. These advancements signify a shift toward universal modeling and reasoning capabilities in protein LLMs.
Beyond foundational modeling capabilities, researchers have begun to explicitly inject structural information into the training process to enhance the models’ ability to capture 3D protein conformations. Models such as SaProt [
1334] and ESM-GearNet [
1335] integrate local or global structural features to enrich sequence representations, while approaches like OntoProtein [
1336] and ProteinCLIP [
1337] leverage knowledge graphs and contrastive learning with text to improve semantic understanding and generalization. These structure-informed and knowledge-enhanced strategies have not only improved model expressiveness on tasks such as mutation effect prediction, functional domain annotation, and binding site identification, but also extended the applicability of protein LLMs to drug target identification and molecular interaction prediction.
Building on foundational understanding and reasoning, protein LLMs have further evolved toward generative modeling. ProGen [
1268] and ProtGPT2 [
1338] were among the first to apply the autoregressive language modeling paradigm to protein sequence generation, capable of producing diverse, biologically active sequences conditioned on functional labels or species. ProGen2 [
1267] scaled up both model size and training data, significantly enhancing its ability to model protein adaptability and diversity. Meanwhile, ProLLaMA [
1339] incorporated protein sequence learning into the LLaMA architecture, achieving joint understanding and generation within a single framework and demonstrating the potential of multi-task and cross-modal pretraining. In contrast, models like Pinal [
1340] and Ankh [
1341] explore structure-guided, efficient encoder-decoder architectures to balance generation quality with parameter efficiency.
At the same time, several integrated frameworks have emerged to support protein design and engineering. For example, ProteinDT [
1342] enables zero-shot generation of protein sequences from textual functional descriptions, while PLMeAE [
1343] integrates with automated biological experimentation platforms to construct a “design-build-test-learn” loop for automated protein engineering. Innovative interactive tools such as ProteinGPT [
1344] and ProteinChat [
1345] have also appeared, supporting structure input, language interaction, and functional Q&A, further advancing protein language models toward intelligent agents with cognitive and interactive capabilities.
Overall, the evolution of protein LLMs has clearly progressed from small-scale LSTM-based semantic embeddings, to large-scale Transformer-based structural predictions, and toward multimodal-enhanced generative design. This trajectory has not only significantly expanded the frontiers of protein science but also laid a robust foundation for the next generation of biomolecular design, functional prediction, and clinical applications.
5.4.6. Benchmarks
The rapid adoption of LLMs in life sciences and bio-engineering has spurred the development of specialized benchmarks designed to systematically assess their performance across diverse biological and clinical tasks. Benchmarks such as BEND for DNA language models and BEACON for RNA language models rigorously evaluate the ability of LLMs to interpret complex genomic and transcriptomic information, encompassing tasks that range from functional element annotation in genomic sequences to predicting RNA secondary structures. Complementing these biological benchmarks, medical QA datasets like MedQA, MedMCQA, and PubMedQA focus on evaluating clinical knowledge, reasoning capabilities, and contextual understanding of biomedical literature. Together, these benchmarks offer a comprehensive framework to evaluate and drive progress in applying LLMs to real-world biomedical challenges.
BEND. BEND is a unified evaluation framework designed to systematically assess the performance of DNA language models (DNA LMs) on realistic biological tasks. The benchmark suite comprises seven tasks based on the human genome, covering functional elements at varying length scales, such as promoters, enhancers, splice sites, and transcription units. Each task provides input sequences and labels in a standardized format, supporting a range of downstream tasks including both classification and regression.
The task design of BEND reflects the core challenges of genome annotation: wide variation in sequence length, sparsity of functional regions, and low signal density. To evaluate the performance of DNA LMs on these tasks, BEND offers a scalable framework for generating embedding representations and training lightweight supervised models. Experimental results demonstrate that while certain DNA LMs can approach the performance of expert methods on specific tasks, they still face difficulties in capturing long-range dependencies (such as enhancer recognition). Moreover, different models display varying preferences for modeling gene structure and non-coding region features.
For example, in the enhancer annotation task, BEND formulates the problem as binary classification: for each 128-base-pair segment of gene-adjacent DNA, the model predicts whether it contains an enhancer. Data are sourced from CRISPR interference experiments and integrated with major transcription start site (TSS) information, with a 100,096-bp sequence extracted for each gene and annotated in 128-bp segments. The main challenge of this task lies in identifying distal regulatory relationships, which tests the model’s ability to capture long-range dependencies.
BEACON. BEACON is a comprehensive evaluation benchmark specifically designed for RNA language models, encompassing 13 tasks related to RNA structural analysis, functional studies, and engineering applications. All tasks adopt a unified data format and support both classification and regression evaluations, applicable to both sequence-level and nucleotide-level predictions.
For example, in the RNA secondary structure prediction task, the model is required to determine whether each pair of nucleotides forms a base pair, with the F1 score used as the evaluation metric. The data for this task is sourced from the bpRNA-1m database.
BEACON also includes a systematic evaluation of various models and finds that single-nucleotide tokenization and ALiBi positional encoding demonstrate superior performance across multiple tasks. Based on these findings, a lightweight baseline model named BEACON-B is proposed.
QA Benchmarks.
The landscape of biomedical and clinical QA benchmarks spans a diverse range of tasks, from licensing examination questions to domain-specific reasoning over scientific literature. These datasets challenge models not only on factual recall but also on higher-order reasoning, reading comprehension, and the ability to synthesize information from complex biomedical contexts. Together, they provide a comprehensive evaluation suite for assessing the medical knowledge, reasoning ability, and contextual understanding of AI systems in healthcare and biomedical research.
Current benchmarks are primarily concentrated within the English language domain, often based on medical licensure examinations from different English-speaking countries. There are also benchmarks in other languages, such as Chinese. These benchmarks fully simulate real-world exams, providing only the question stem and answer choices. In addition, there are benchmarks that supply LLMs with a reference document, requiring the model to combine its own knowledge with the provided context to generate a more informed answer.
MedQA. The MedQA dataset consists of multiple-choice questions from the United States Medical Licensing Examination (USMLE). It covers general medical knowledge and includes 11,450 questions in the development set and 1,273 questions in the test set. Each question has 4 or 5 answer choices, and the dataset is designed to assess the medical knowledge and reasoning skills required for medical licensure in the United States.
MedMCQA. MedMCQA is a large-scale multiple-choice QA dataset derived from Indian medical entrance examinations (AIIMS/NEET). It covers 2.4k healthcare topics and 21 medical subjects, with over 187,000 questions in the development set and 6,100 questions in the test set. Each question has 4 answer choices and is accompanied by an explanation. MedMCQA evaluates a model’s general medical knowledge and reasoning capabilities.
PubmedQA. Different from MedQA and MedMCQA, PubMedQA is a closed-domain QA dataset, In which each question can be answered by looking at an associated context (PubMed abstract). It is consists of 1,000 expert-labeled question-answer pairs. Each question is accompanied by a PubMed abstract as context, and the task is to provide a yes/no/maybe answer based on the information in the abstract. The dataset is split into 500 questions for development and 500 for testing. PubMedQA assesses a model’s ability to comprehend and reason over scientific biomedical literature.
Drug Synergy Prediction
CancerGPT. CancerGPT [
1326] assesses the capability of LLMs to predict drug pair synergy in rare cancer tissues with limited structured data. The evaluation framework involves testing LLMs’ performance in few-shot and zero-shot learning scenarios across seven rare tissue types, comparing the results to those of larger models like GPT-3 [
7].
The evaluation process includes:
Few-shot and Zero-shot Learning: Assessing the model’s ability to predict drug synergy with minimal or no training examples, highlighting the LLM’s capacity to generalize from limited data.
Benchmarking Across Multiple Tissues: Testing the model’s predictive performance across seven different rare cancer tissue types to ensure robustness and generalizability.
This evaluation framework demonstrates that LLMs, even with fewer parameters, can effectively predict drug pair synergies in contexts with scarce data, offering a promising approach for biological inference tasks where traditional structured data is lacking.
BAITSAO. The benchmark framework integrates both regression and classification tasks, based on synergy scores (e.g., Loewe, Bliss, HSA, ZIP) and binary synergy labels derived from large-scale drug combination datasets such as DrugComb. Each sample consists of a drug pair and a cell line, with input features constructed from Large Language Model (LLM) embeddings of descriptive prompts about drugs and cell lines, standardized into numerical vectors.
The design of the BAITSAO evaluation suite reflects key challenges in drug synergy prediction: sparse synergy signals, heterogeneous data formats, and limited generalization to novel drug combinations. To evaluate model performance, BAITSAO pre-trains on large-scale synergy data under a multi-task learning (MTL) framework, capturing both single-drug inhibition and pairwise synergy. The model is then assessed on held-out datasets using metrics such as Pearson correlation, mean squared error, ROC-AUC, and accuracy. Ablation and sensitivity analyses are further conducted to study embedding strategies, training data scales, and model scaling laws.
For example, in the synergy classification task, BAITSAO formulates the problem as a binary prediction: given a pair of drugs and a cell line, predict whether the combination yields a synergistic effect. Inputs are constructed by averaging the embeddings of both drugs and concatenating with the cell line embedding, with synergy labels binarized using a threshold on the Loewe score. This setup evaluates the model’s ability to generalize to unseen drug-cell line combinations, including out-of-distribution (OOD) samples, and serves as a robust benchmark for multi-drug reasoning and zero-shot prediction.
Protein Modeling
TAPE. TAPE (Tasks Assessing Protein Embeddings) is a large-scale benchmark designed to evaluate transfer learning methods on protein sequences. It comprises five biologically relevant tasks, including protein structure prediction, remote homology detection, and protein engineering. Each task features carefully curated splits to assess models’ ability to generalize in biologically meaningful ways. TAPE evaluates various self-supervised learning approaches for protein representation and shows that pretraining significantly improves performance across nearly all tasks, although traditional non-neural methods still outperform in some cases. The benchmark promotes standardized evaluation and method comparison in protein modeling.
PEER. PEER (Protein sEquence undERstanding) is a comprehensive and multi-task benchmark designed to evaluate deep learning methods on protein sequences. It encompasses 17 biologically relevant tasks across five categories: protein function prediction, localization prediction, structure prediction, protein-protein interaction prediction, and protein-ligand interaction prediction. Each task includes carefully curated training, validation, and test splits to assess models’ generalization capabilities in real-world scenarios. PEER evaluates various sequence-based approaches, including traditional feature engineering methods, different sequence encoding techniques, and large-scale pre-trained protein language models.
Summary. Benchmarking efforts in life-science and bio-engineering LLMs now coalesce around four broad task families. First, sequence-based evaluation dominates DNA and RNA modeling. Suites such as BEND (DNA) and BEACON (RNA) probe classification and regression across functional-element annotation, secondary-structure inference, and variant-effect prediction. Second, clinical structured-data tasks assess models on Electronic Health Records, splitting into clinical language generation (e.g., ClinicalT5, GPT-4 hospital-note drafting) and EHR-based prediction (e.g., BEHRT, Med-BERT risk scoring). Third, textual knowledge tasks test biomedical reasoning and understanding via QA (MedQA, MedMCQA, PubMedQA) and natural-language inference benchmarks such as MedNLI, measuring factual recall, chain-of-thought reasoning, and long-context comprehension. Finally, hybrid outcome-prediction benchmarks—drug-synergy suites (e.g., DrugCombDB subsets) and protein-modeling challenges (ESMFold, ProGen)––demand multimodal integration across chemistry, omics and cellular context.
Across these categories, domain-trained transformers consistently outperform classical baselines. In sequence modeling, long-context LLMs (Enformer, HyenaDNA) improve enhancer or eQTL effect prediction correlations by 20–40% over CNN/RNN hybrids, while bidirectional masked models (DNABERT-2) raise MCC scores on promoter/enhancer detection by 2–5 pp versus 6-mer CNNs. In clinical language generation, instruction-tuned GPT-4 drafts discharge summaries that clinicians rate as equal or superior in accuracy and readability to human-written notes, and models like Med-PaLM 2 reach 86% accuracy on USMLE-style exams, narrowing the gap to licensed physicians. EHR-based predictors such as GatorTron boost AUROC for onset prediction tasks by 3–6 pp relative to GRU or logistic-regression baselines, even under low-data fine-tuning. In drug-synergy prediction, transformer fusion networks (DFFNDDS) and prompt-based few-shot GPT variants (CancerGPT) lift balanced accuracy by 5–12 pp over DeepSynergy, while LLM-generated protein sequences (ProGen2) exhibit in-vitro activities on par with natural enzymes in ≥50% of tested families.
Yet substantial limitations persist. Ultra-long genomic context still degrades accuracy despite linear-time attention variants; distal enhancer–promoter linkage and rare-variant generalization remain open. Multimodal fusion is ad-hoc: most benchmarks isolate a single modality, leaving cross-omics reasoning and image-augmented clinical tasks underexplored. Data quality and bias are acute—human-centric genomes, single-institution EHRs, and English-only QA corpora skew performance and hamper species-, population-, or language-level transfer. Safety and interpretability issues mirror those seen in chemistry: hallucinated diagnoses or biologically implausible sequence designs can slip through, and attention maps alone rarely satisfy domain experts’ need for mechanistic insight.
From these observations we derive three actionable insights. (1) Benchmark breadth and depth must expand. Community curation of larger, more diverse genomes (e.g., non-model organisms), multilingual clinical notes, and truly multimodal datasets (sequence+structure+phenotype+imaging) is essential. (2) Representation and architecture choices require re-thinking. Treating kilo- to megabase sequences as flat text overlooks 3-D chromatin contacts; integrating graph, spatial or physics-aware modules with transformers, and exploring alternative encodings (e.g., byte-pair k-mers, SELFIES-like bio-tokens) can bridge this gap. (3) Reliability hinges on task design and validation. Embedding biological priors, tool-augmented prompting (e.g., retrieval of wet-lab evidence), and post-hoc critic models can curb hallucination and enforce mechanistic plausibility; standardized factuality and safety metrics—analogous to clinical adjudication—should accompany benchmark scoreboards.
In sum, life-science–oriented LLM benchmarks have revealed impressive gains over traditional pipelines, but progress is gated by richer data, modality-aware architectures, and rigorous, biology-centric evaluation. Aligning these elements will accelerate LLMs from promising assistants to dependable engines for discovery and precision medicine.
5.4.7. Discussion
Opportunities and Impact. LLMs are now deeply integrated across the life sciences pipeline, supporting a broad spectrum of tasks ranging from genomic sequence interpretation to drafting clinical documentation. Their greatest impact has emerged in areas that align with their linguistic strengths—such as literature summarization, clinical note generation, and biomedical question-answering—where abundant data, low-cost supervision, and linguistic evaluation metrics have enabled rapid progress.
A major advantage lies in the
tokenizable structure of biological data. Representations like k-mers for genomic sequences, SMILES for chemical compounds, and ICD codes in medical records are inherently suited to masked language modeling or autoregressive learning. As a result, LLMs like DNABERT-2, Nucleotide Transformer, BEHRT, and Med-PaLM 2 [
1284,
1287,
1301] offer unified frameworks to model complex biological substrates. For example, RNA oligomers of various sizes require different experimental strategies—from NMR [
1389] and FRET [
1390] for small structures to cryo-EM and CLIP-seq for large complexes [
1391,
1392]. Traditional computational methods often rely on size-specific architectures with manual feature engineering, while LLMs can tokenize all scales using shard tokenizers and learn a size-agnostic representation space.
Furthermore, LLMs exhibit
context extrapolation capabilities that allow modeling of long-range dependencies in genomic data. For example, predicting MYC expression traditionally required multiple CNN-based sliding windows, Hi-C loop assemblies, and handcrafted features [
1393,
1394], which struggled to capture distal interactions. In contrast, models like HyenaDNA [
1261] process 1 Mb genomic windows in a single forward pass, leveraging sub-quadratic convolutions to directly learn enhancer–promoter logic, thereby eliminating the need for fragmented, manually-curated pipelines.
The instruction-following and multitask capabilities of LLMs further enable unified handling of diverse biomedical applications. For instance, Med-PaLM 2 can simultaneously draft clinical notes, generate ICD-10/LOINC codes, and rewrite instructions at a 6th-grade level—all in a single prompt. This integration replaces three siloed hospital systems and reduces development timelines from months to hours.
Challenges and Limitations. Despite these advancements, significant limitations persist. LLMs excel in symbolic and text-rich domains but underperform in tasks requiring deep experimental grounding or multi-scale biological reasoning.
First, the gap in empirical grounding remains a major bottleneck. Models such as ProGen2 can propose novel peptides, but their real-world efficacy is limited. For instance, the initial validation of ProGen2-generated incretin peptides showed an activity success rate of merely 7%, emphasizing the indispensable role of iterative wet-lab testing and retraining.
Second, life science problems often involve
system-level complexity, where token-based reasoning is insufficient. Predicting off-target effects of CRISPR editing or long-term drug toxicity demands multiscale modeling across molecular, cellular, and organismal levels. Although models like CRISPR-GPT [
1395] show promise, they still miss over 30% of off-target sites in whole-genome data with complex chromatin interactions [
1187,
1188,
1230].
Third, the rise of powerful generative models introduces new
ethical, safety, and provenance concerns. LLMs capable of generating accurate protocols may inadvertently facilitate dual-use research or propagate hallucinations. Open-source toxicity predictors like ToxinPred [
1396,
1397] can potentially be misused to design harmful biological sequences. Without clear traceability or accountability mechanisms, the risks of misuse escalate.
Research Directions. To address these challenges, we propose a forward-looking research agenda focused on hybrid architectures and responsible integration.
First, future LLM systems should aim to unify diverse biological modalities—including genomic sequences, protein structures, cell images, clinical time-series, and textual notes—within a cohesive multimodal framework. Such models can enable integrated diagnosis and prediction by capturing complex biological correlations across data types.
Second, LLMs should evolve from passive tools into active hypothesis-generating agents. This requires coupling with laboratory automation systems, real-time EHR streams, and high-throughput simulation platforms. For instance, an LLM-guided robotic lab could autonomously design, test, and refine molecular hypotheses in closed experimental loops, dramatically accelerating the discovery cycle.
Third, the training of LLMs should incorporate biologically-informed learning techniques. Self-distillation improves interpretability through reasoning chains, contrastive alignment ensures consistency with biomedical knowledge bases, and physics-informed regularization grounds models in biophysical laws (e.g., thermodynamics in MD simulations), reducing hallucinations and enhancing trustworthiness.
Finally, proactive governance must be embedded from the outset. Techniques such as differential privacy for sensitive patient data, watermarking synthetic DNA sequences to differentiate them from natural ones, and rigorous human oversight mechanisms are crucial for ensuring ethical deployment. Building responsible AI systems is not an afterthought—it must be integral to model development.
Conclusion. LLMs have transformed information processing in the life sciences, accelerating literature review, genomic annotation, and clinical documentation. Their most pronounced successes lie in tasks with high symbolic complexity but relatively low experimental demands. However, realizing their full potential in experimental biology and bioengineering will require overcoming structural limitations.
This transformation demands more than model scaling; it necessitates innovations in architecture, training paradigms, and system integration. Bridging the gap between computational prediction and empirical validation calls for hybrid systems that fuse LLMs with biological priors, experimental platforms, and domain-specific constraints.
When thoughtfully designed and ethically deployed, LLMs can serve not merely as intelligent assistants but as generative partners in hypothesis formation, experimental design, and therapeutic innovation—ultimately accelerating the transition from scientific discovery to clinical application.