Introduction
All cells in an organism originate from pre-existing cells, beginning with the fertilized egg and progressing to the blastocyst stage. Stem cells, possessing totipotency, have the capacity to differentiate into any cell type, including placental cells. Their key characteristics, proliferation and differentiation, enable them to multiply and specialize. The programming of stem cells relies heavily on epigenetic regulation to guide lineage-specific differentiation, which is especially critical for generating induced pluripotent stem cells (iPSCs) for applications such as cell replacement therapy [
1]. Epigenetic regulatory proteins function at distinct stages of differentiation, guiding stem cells toward lineage commitment. These finely tuned processes are crucial not only for development but also for advancing therapeutic strategies [
1].
Epigenetics refers to heritable changes in gene expression that occur without altering the DNA sequence. Key epigenetic mechanisms include DNA cytosine methylation, ATP-dependent nucleosome remodeling, histone variant substitution, numerous histone modifications, and regulation of gene expression by small non-coding RNAs [
2]. As stem cells begin to differentiate, epigenetic mechanisms undergo a significant shift—from initially suppressing genes associated with differentiation to actively promoting the expression of genes essential for specific cellular fates [
3]. Dysregulation of this process is frequently implicated in diseases such as cancer.
Histone modifications involve numerous post-translational changes, including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, formylation, butyrylation and others [
4]. Lysine residues within the core nucleosomal histones are the targets of many modifications, with acetylation playing a major role regulating nucleosome structure. Histone acetyltransferases (HATs, also called lysine acetyltransferases or KATs), including members of the p300/CBP, MYST (such as MOZ, Sas2, TIP60), and GNAT families, catalyze the acetylation of both histone and non-histone proteins [
5]. In stem cells, HATs play dual roles by maintaining pluripotency and promoting differentiation. By selectively activating or repressing gene expression, HAT complexes orchestrate the balance between self-renewal and differentiation [
6].
The MYST family of HATs, including MOF and MYST2/HBO1, exerts a significant influence on gene expression by acetylating histones to reshape chromatin structure and direct transcriptional patterns [
6]. Among these, the NuA4/TIP60 complex, a key member of the MYST family, is a major regulator of the epigenetic landscape controlling stem cell behavior.
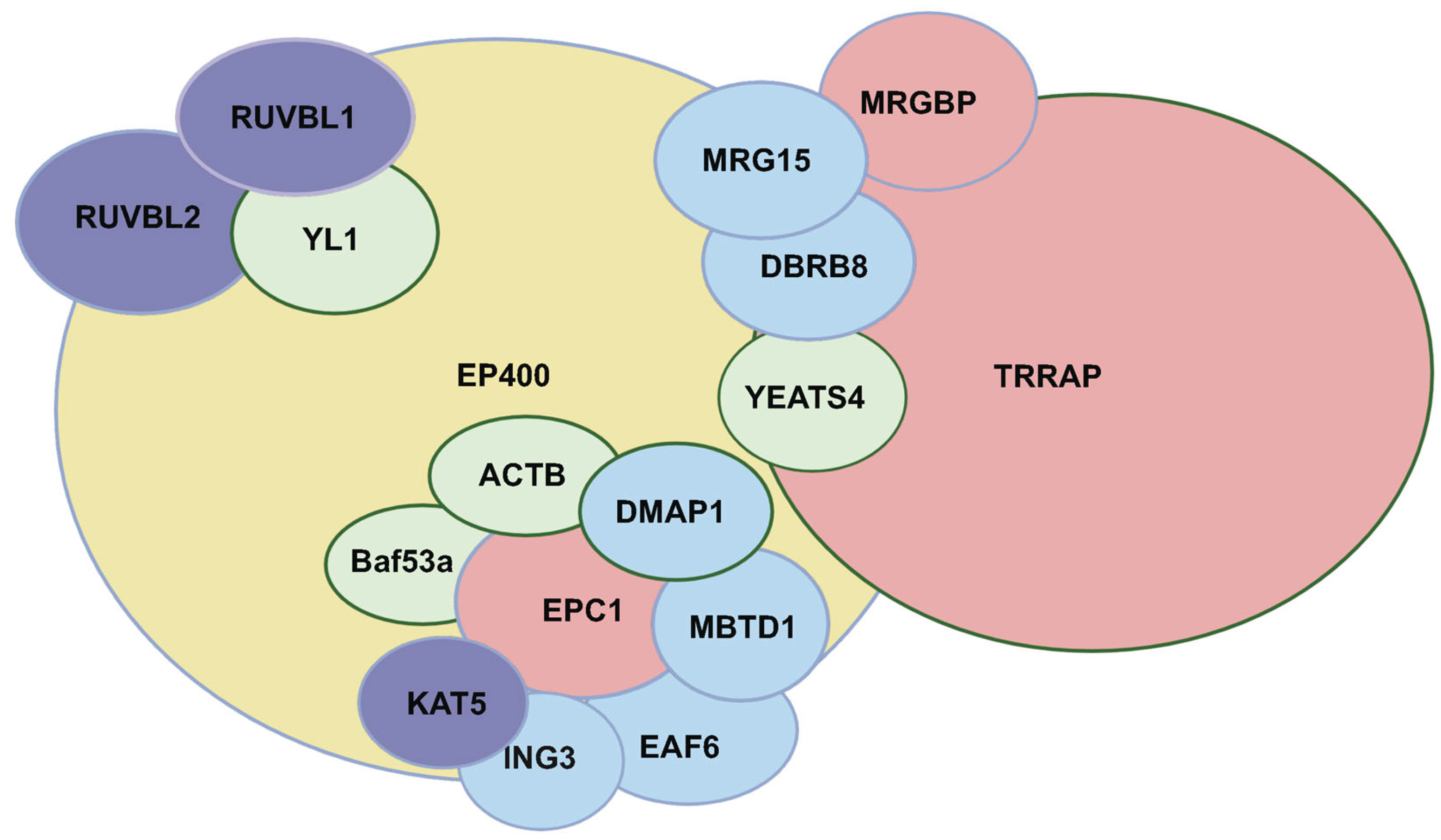
The human TIP60 complex is a large assembly (>1.7 megadaltons) composed of 17-20 subunits, including ACTB, ACTL6A, BRD8, DMAP1, EP400, EPC1, ING3, KAT5, MBTD1, MEAF6, MRG15, MRGBP, RUVBL1, RUVBL2, TRRAP, YEATS4 and YL1 (
Figure 1) [
7]. This complex regulates transcription, proliferation, and homologous recombination (HR)-mediated DNA repair. TIP60 acetylates histones H2A and H4 to regulate gene expression and facilitates the incorporation of variant histone H2A.Z through p400's ATP-dependent chromatin remodeling activity. Given these diverse roles, the TIP60 complex is vital for maintaining stem cell renewal and differentiation potential [
8].
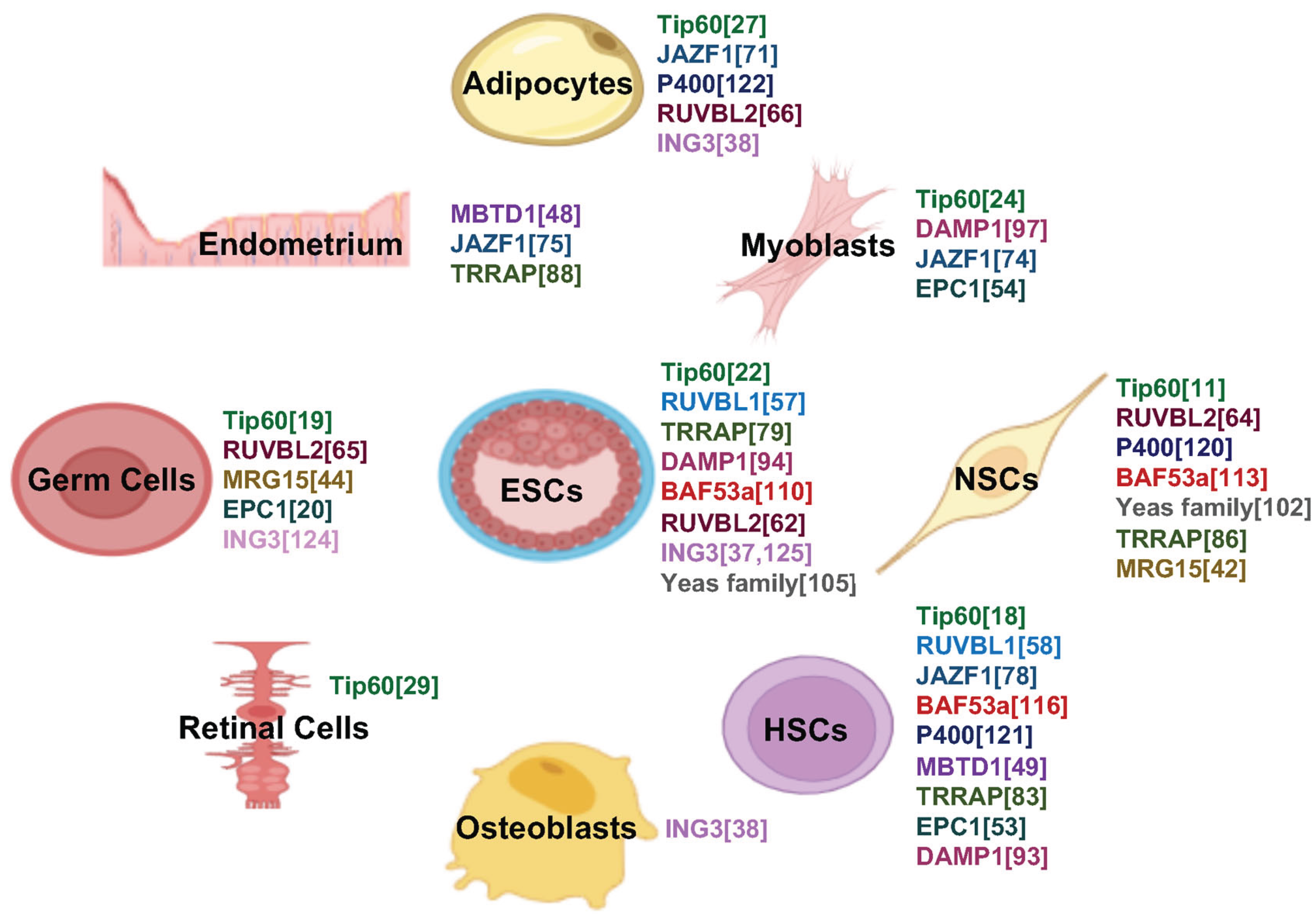
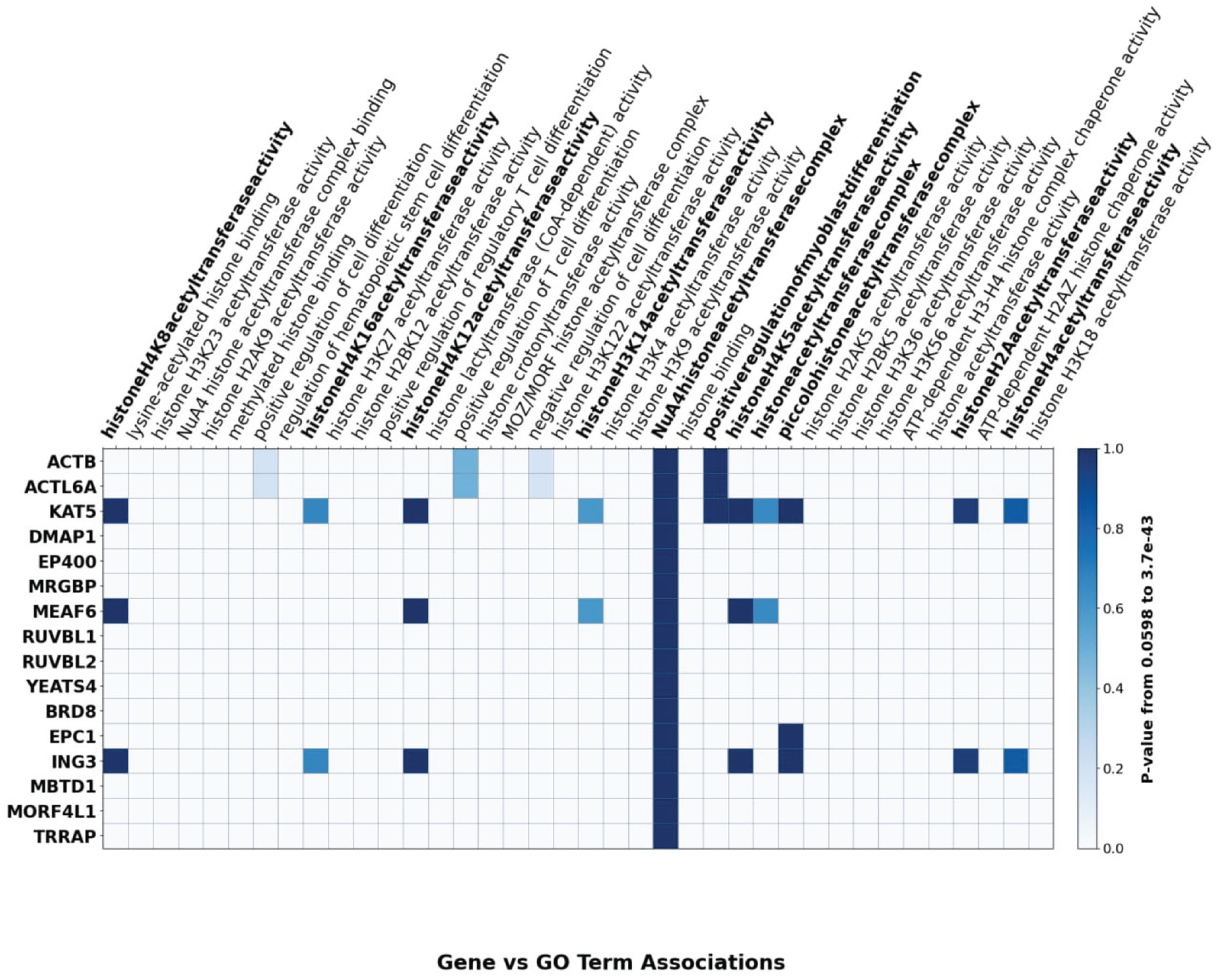
The differential roles of TIP60 complex subunits in lineage-specific differentiation are summarized in
Figure 2 that identifies the individual components that have been reported to contribute to the developmental pathways noted. A heatmap of TIP60 subunit–GO term associations further emphasizes biological processes involving TIP60 and its subunits, including histone acetyltransferase activity and chromatin regulation (
Figure 3).
Given its broad regulatory functions and epigenetic influence, increasing evidence supports TIP60’s involvement in directing cell fate decisions during differentiation. This review explores the role of the TIP60 complex in multiple differentiation pathways, focusing on its molecular mechanisms and interactions with lineage-specific transcriptional programs. We focus on TIP60 complex subunits with demonstrated roles in differentiation, without addressing those for which no such functions have yet been reported.
TIP60 and Differentiation
KAT5, the catalytic subunit of the TIP60 complex, is a 60 kD lysine acetyltransferase subunit that is evolutionarily conserved from yeast to humans. TIP60 interacts with numerous transcription factors and co-regulators, playing critical roles in apoptosis, DNA damage response, cell cycle control, and transcriptional regulation [
9]. It functions as a co-activator for transcription factors such as HIV Tat and MyoD, facilitating histone acetylation at target promoters. It also interacts with CREB, STAT3, ZEB, and p73, further influencing gene expression [
9,
10].
TIP60 is essential in multiple differentiation processes, including neurodevelopment, hematopoiesis, germ cell maturation, adipogenesis, and myogenesis (
Figure 2). In the sections that follow, we examine TIP60's roles in the context of lineage-specific differentiation.
TIP60 and neuronal differentiation. Studies using neural stem cell (NSC)-specific TIP60 knockout mice have shown that TIP60 is critical for neurodevelopment. Its absence leads to disrupted NSC proliferation, microcephaly, impaired neuronal migration, and increased GFAP-positive glial cells. TIP60 is required for neurogenesis, NSC maintenance, and the neurogenesis-to-gliogenesis transition [
9].
TIP60 is recruited to chromatin through its interaction with neuronal lineage–determining transcription factors, which enables histone H2A.Z acetylation and promotes neuronal fate [
11]. Bivalent chromatin domains play a key role in regulating development in pluripotent stem cells, marking important gene promoters and enhancers ready for activation. Recent studies suggest that TIP60-mediated H2A.Z acetylation might influence the transition from bivalency to monovalent domains during differentiation, particularly in the activation of bivalent genes and the acquisition of H3K4me3 marks [
11]. Increasing evidence suggests that H3K4me3 primarily prevents active repression rather than directly activating genes. Its absence leads to increased H3K27me3 and DNA methylation in these regions, disrupting differentiation as genes involved in neuronal differentiation cannot be activated [
11,
12]. Mutations in the KAT5 gene, encoding TIP60, are associated with a neurodevelopmental syndrome characterized by progressive cerebellar atrophy, developmental delays, and intellectual disabilities [
13,
14].
Eichele et al. reported parallels between TIP60 deficiency and the CK-p25 neurodegeneration model, noting hypoacetylation of H4K12 and dysregulation of transcription in TIP60-deficient mice. Genes linked to immunity and cell cycle were upregulated, while those associated with learning and plasticity were downregulated. This imbalance leads to caspase-3 activation, β-actin fragmentation in CA1 neurites, and neuronal loss [
15]. Thus, TIP60 emerges as a key guardian of neuronal health by mitigating early imbalances in TIP60 HAT/ histone deacetylase (HDAC) activity observed in various neurodegenerative diseases. When TIP60/HDAC homeostasis is disrupted, synaptic anomalies and cognitive decline emerge in the preclinical stages of these diseases. Elevating cellular levels of specific neural HATs, notably TIP60, serves as a protective measure, restoring acetylation equilibrium and preserving the essential gene expression profiles vital for neuroplasticity and neural well-being [
10,
16].
TIP60 and hematopoietic stem cells (HSCs). In hematopoiesis, TIP60 plays a regulatory role in both normal development and malignancy. Chromosomal translocations in some acute myeloid leukemia (AML) cases produce a fusion gene known as ZMYND11-MBTD1 (ZM), which recruits TIP60 to stemness-related gene regions, including MYC, HoxA, Meis1, and Sox4. This recruitment promotes continuous HSC self-renewal and contributes to leukemogenesis [
17]. Using conditional knockout models, Daniel et al. demonstrated that TIP60 is essential for regulating MYC target gene expression during both embryonic and adult stages of HSC maintenance. TIP60 colocalizes with c-Myc, and its deletion suppresses genes responsible for cell cycle control and DNA repair; two fundamental processes for sustaining HSC function [
18].
TIP60 and germ cell differentiation. In female
drosophila germ cells, TIP60 functions in conjunction with Nipped-A to regulate the differentiation of germline stem cells (GSCs). GSCs maintain the ability to self-renew or differentiate, and genes such as bag of marbles and benign gonial cell neoplasm (bgcn) are key to this process. Nipped-A, through its association with the TIP60 complex, promotes cell cycle progression and the expression of bgcn, facilitating the transition of GSC daughter cells toward differentiation. Loss of Nipped-A results in an accumulation of undifferentiated GSC daughters, a defect that can be rescued by bgcn overexpression. This highlights TIP60's role in coordinating cell cycle regulation with differentiation cues in germ cells [
19].
TIP60 and spermiogenesis. During spermiogenesis, the final phase of sperm development, TIP60 plays an essential role. This process involves replacing histones with protamines to achieve chromatin condensation, and histone hyperacetylation is a critical preparatory step. Specifically, TIP60 and its associated complex member Epc1 are necessary for this histone hyperacetylation. Disruption of either gene results in abnormal spermatid morphology and impaired chromatin remodeling, indicating important functional roles in genome compaction during male germ cell differentiation [
20].
TIP60 and embryonic stem cell (ESC) differentiation. ESCs are widely used to study pluripotency and differentiation. In the absence of TIP60-p400 complex components, ESCs fail to differentiate properly, forming small, irregular embryoid bodies and lacking activation of lineage-specific markers [
21]. In TIP60-knockout mice, embryos exhibit abnormalities at the blastocyst stage, with impaired hatching and failure to progress post-implantation [
22].
Interestingly, TIP60 has both HAT-dependent and HAT-independent functions in ESCs. While its KAT activity is necessary for differentiation into mesoderm and endoderm, TIP60 also represses differentiation genes by reducing chromatin accessibility at their promoters—independent of its enzymatic function. It maintains compact promoter regions through its binding, not through acetylation. ESCs with TIP60 KAT mutations can still self-renew but display differentiation defects. This dual role highlights TIP60's complex regulatory influence on ESC fate [
23].
TIP60 and myoblasts. Myoblasts are precursor cells that commit to the skeletal muscle lineage and eventually fuse to form multinucleated myotubes. TIP60 plays an essential role in this process by activating MyoD's transcriptional activity, a key driver of myogenesis. In C2C12 myoblasts, knockdown of TIP60 impaired the transition from proliferating myoblasts to differentiated myotubes, while ectopic TIP60 expression enhanced MyoD-mediated gene activation. TIP60 physically interacts with MyoD via its chromo- and Zn-finger domains, which are critical for effective transcriptional activation. This interaction facilitates MyoD binding to the myogenin promoter, an early marker of muscle differentiation, and promotes chromatin remodeling by increasing histone H3 and H4 acetylation at the promoter site [
24].
In addition to its role in MyoD activation, TIP60's influence extends to the regulation of other transcription factors such as SOX4, which also play critical roles in myoblast differentiation. TIP60 specifically acetylates SOX4 at lysine 95, enabling its binding to target gene promoters and the activation of muscle-specific genes. Furthermore, SOX4 initially associates with the co-repressor HDAC1, and during C2C12 differentiation, HDAC1 dissociates, allowing SOX4 to be acetylated by TIP60. This acetylation is essential for the recruitment of SOX4 to the promoter of caldesmon 1, a gene involved in actin cytoskeleton organization and muscle contraction, ensuring its full transcriptional activation. This study identified a molecular switch between the co-activator TIP60 and co-repressor HDAC1 that governs SOX4’s transcriptional activity for precisely regulating gene expression and muscle cell differentiation [
25].
TIP60 and adipogenesis. Peroxisome proliferator-activated receptor γ (PPARγ) is an important regulator in adipocytes, affecting adipogenesis, lipid storage, thermogenesis, and insulin sensitivity. In response to metabolic cues, PPARγ recruits different cofactors, forming dynamic transcriptional complexes for specific signal transduction [
26]. In adipogenesis, TIP60 plays a central role by influencing the activity of PPARγ. This collaboration enhances PPARγ's functions, including gene regulation and the process of turning preadipocytes into mature fat cells [
27]. Moreover, the stability of TIP60, carefully maintained by ubiquitin-specific protease 7 (USP7), becomes especially important during the early stages of adipocyte development, particularly when cells undergo mitotic clonal expansion. TIP60's dual role thus spans both early and later phases of fat cell maturation [
28].
TIP60 and retinal differentiation. TIP60 functions as a co-activator in the transcriptional regulation of rod photoreceptor differentiation in the retina. TIP60 interacts with Nrl, a bZIP transcription factor, to enhance expression of rod-specific genes like rhodopsin and Ppp2r5c. This interaction is facilitated by the phosphorylation of Nrl at Ser50 by JNK1, which enables TIP60 to acetylate histones at the promoters of these genes, thereby promoting their transcription [
29].
ING3 and Differentiation
The INhibitor of Growth family of proteins (ING1-5) all function as stoichometric targeting components of HAT and HDAC complexes by serving as epigenetic readers of the histone H3K4me3 mark through their plant homeodomain [
30]. The ING1&2 proteins also act as receptors and transducers of stress-activated phosphoinositides, linking stress to transcriptional repression [
31] These repressive functions, through targeting the Sin3a HDAC complex are likely to underlie their function as type II tumor suppressor genes due to their frequent inactivation in malignancies [
32]. ING3 was initially discovered through an in silico computational approach [
33]. Like other ING proteins [
34], ING3 affects TP53-mediated transcription [
35], exerting control over the cell cycle and apoptosis. ING3 exhibits elevated expression in cells undergoing rapid proliferation, suggesting a positive role in cellular growth and self-renewal [
36]. ING3 is also required for development where its loss causes severe growth restriction and embryonic death. The most pronounced effects are on ectoderm-derived tissues, resulting in a developmental defect in neural tube closure and the inability to form intact primary brain vesicles in mid-gestation homozygous knockout embryos [
37].
ING3 and mesenchymal stem cell differentiation. A study conducted by Chesi et al. [
38] indicated a role for ING3 in differentiation of mesenchymal cells. ING3 knockdown cells displayed reduced osteoblast differentiation, indicated by decreased ALP expression and loss of calcium phosphate mineral deposition. These cells also showed enhanced adipogenic differentiation, with increased Oil Red O staining and higher expression of the adipogenic transcription factor C/EBP alpha. ING3 may regulate osteoblast differentiation by controlling osteogenic gene expression through targeting the NuA4 complex and by supporting cell cycle exit via activating p53 target gene expression. ING3 loss disrupts these processes and instead promotes adipogenic differentiation without affecting proliferation [
38].
ING3 and oocyte maturation. Beyond its role in mesenchymal stem cell and skeletal differentiation, ING3 is essential for oocyte maturation, a process defined by asymmetric cell division. It may exert this function through the mammalian target of rapamycin (mTOR) pathway, as ING3 knockdown decreases the expression of mTOR and its downstream targets, including cell division control protein 42 (Cdc42), Ras-related C3 botulinum toxin substrate 1 (Rac1), and Ras homolog family member A (RhoA). The absence of ING3 results in symmetric division, a feature commonly seen in aging mouse oocytes, highlighting its role in regulating genes needed for proper oocyte maturation [
39].
MRG15 and Differentiation
MRG15 (MORF4-Related Gene on chromosome 15) is a conserved chromatin-associated protein that plays a key role in epigenetic regulation by associating with histone-modifying complexes such as TIP60. It contains an N-terminal chromodomain that binds to H3K36me2/3 (histone H3 lysine 36 di/trimethylation), linking it to transcriptionally active chromatin. Its C-terminal MRG domain mediates interactions with partners like hMOF, Rb, and MRGBP, facilitating gene regulation, DNA repair, and cell cycle control. MRG15 is essential for embryonic development and neural stem cell maintenance [
40]. Depleting MRG15 in Drosophila causes embryonic lethality, hampers DNA repair, and disrupts cellular differentiation. In mice, Mrg15 null cells exhibit reduced proliferation, leading to early senescence [
41].
MRG15 and neuronal differentiation. Chen et al. reported that MRG15 functions in NPC proliferation and differentiation during neurogenesis. Loss of MRG15 in embryonic brains leads to thinner neuroepithelia and increased apoptosis in vivo, while NPCs from Mrg15-deficient embryos show reduced proliferation and impaired neuronal differentiation in vitro, without increased apoptosis. These defects are likely due to MRG15’s role in chromatin remodeling through its association with TIP60 HAT and HDAC complexes, which regulate gene expression necessary for NPC fate decisions. Interestingly, although MRG15 knockdown in ESCs shows minimal effects—possibly due to compensation by its homolog MRGX—its absence significantly affects tissue-specific precursor cells like NPCs. MRG15-deficient NPCs fail to properly attach and differentiate, suggesting cell-autonomous defects that may extend to other tissues [
42].
MRG15 also helps maintain genomic stability by regulating the DNA damage response. Loss of MRG15 leads to activation of p53, upregulation of p21, increased 53BP1 DNA damage foci, and impaired DNA repair, even without external insults [
43]. These effects may be mediated through disruption of the TIP60 complex or the BRCA2-PALB2 pathway since both are involved in DNA repair. MRG15 may also suppress p21 expression through transcriptional repression mechanisms.
MRG15 and germ cell differentiation. Some researchers have shown that that MRG-1, a protein containing a chromodomain and a homologue of mammalian MRG15 helps balance stem cell proliferation and differentiation in
C. elegans. Proteasome-mediated protein degradation regulates MRG-1 levels, and its interaction with the E3 ubiquitin ligase RFP-1 is involved in this regulation. Reduced proteasome activity or loss of RFP-1 expression leads to elevated MRG-1 levels and stem cell over-proliferation while MRG-1 absence results in germ cell loss. This suggests that MRG-1 likely influences the expression of proteins necessary for proliferation and represses those necessary for differentiation, thereby affecting the balance between the two processes. Notably, MRG15 exhibits similar functions in murine neurospheres suggesting conservation of its role in regulating stem cell proliferation and differentiation across species [
44].
MRG15 also functions during spermatogenesis since MRG15 loss in postnatal male germ cells results in spermatogenesis being blocked at the round spermatid stage. This arrest does not disrupt meiotic division or histone acetylation, but it exerts a significant influence on the epigenetic control of pre-mRNA splicing. Genes like transition nuclear protein 2 (Tnp2) are particularly affected by the absence of MRG15, leading to errors in splicing. These splicing errors cause an accumulation of multinucleated round spermatids and inflammation in the epididymides [
45].
MBTD1 and Differentiation
Mbt Domain Containing 1 (MBTD1), also known as Hemp (hematopoietic expressed mammalian polycomb) is a nuclear protein with a Cys2-Cys2 zinc finger domain and four malignant brain tumor (MBT) repeats. Originally discovered in mouse fetal liver, MBTD1 is involved in regulating gene expression by binding methylated lysine residues, especially H4K20 [
46]. In the context of DNA double-strand break (DSB) repair, MBTD1 competes with 53BP1, a DNA repair factor, for binding to the H4K20me mark at DSB sites and, in association with TIP60/KAT5, acetylates histone H4 and H2AK15. This destabilizes 53BP1 interactions and prevents its recruitment, ultimately directing the cellular response toward error-free HR repair of DSBs [
47].
MBTD1 and endometrial stromal cell differentiation. MBTD1 affects differentiation during endometrial stromal cell decidualization. This differentiation process is needed for successful pregnancy since deficiencies in it can lead to early miscarriages. MBTD1 is upregulated during decidualization and functions in progesterone-driven human and mouse endometrial stromal cell differentiation by promoting DNA double-strand break repair, which is critical during decidualization when DNA damage levels can increase. This suggests that MBTD1 contributes to genomic stability during the differentiation process, potentially protecting against DNA damage [
48].
MBTD1 and HSCs. MBTD1 also contributes to the development and maintenance of hematopoietic stem cells. During fetal development, it is highly expressed in early fetal liver HSCs, where it supports their proliferation, survival, and repopulating capacity. Loss of MBTD1 during this stage results in a significant reduction in HSC numbers, impaired colony formation, and decreased engraftment potential. These defects are associated with reduced cell proliferation, increased apoptosis, and dysregulation of key hematopoietic genes, including Sox4, Stat3, and KLF6 [
49].
In adult HSCs, MBTD1 helps preserve quiescence and long-term function. Conditional knockout studies in mice show that MBTD1 promotes the expression of FOXO3a, a transcription factor that activates cell cycle inhibitors such as p57 and p21. Without MBTD1, HSCs lose their quiescent state, enter the cell cycle more frequently, and become less capable of coping with physiological stress. MBTD1 exerts its effects as part of the TIP60 histone acetyltransferase complex, likely regulating gene expression through chromatin remodeling. This epigenetic regulation is vital for maintaining HSC stability and preventing premature HSC exhaustion [
46].
EPC1 and Differentiation
EPC1 (enhancer of polycomb homolog 1) is a conserved chromatin regulator and a member of the polycomb group (PcG) protein family involved in epigenetic gene silencing and maintenance of cellular identity. As a non-catalytic but essential component of the NuA4/TIP60 histone acetyltransferase complex, EPC1 promotes transcription, DNA repair, and cell cycle regulation by coordinating histone acetylation. Its yeast homolog, Epl1, plays a similar role in chromatin remodeling. EPC1 also modulates gene expression as a transcriptional co-regulator of E2F1 and interacts with complexes like PRC2. While EPC1 is implicated in developmental processes such as hematopoiesis and muscle differentiation, it is also associated with cancer progression, suppressing apoptosis and contributing to drug resistance through interactions with TIP60 and EZH2 [
50,
51].
Studies in Drosophila development have illuminated the involvement of EPC in processes related to differentiation and the determination of stem cell fate. For example, activation of the JNK (Jun amino-terminal kinase) signaling pathway in imaginal disc cells during regeneration leads to the downregulation of EPC, facilitating wound healing and contributing to the formation of major adult fly structures. Furthermore, in multipotent hematopoietic progenitors, EPC operates downstream of JNK to initiate cellular differentiation [
52].
EPC1 and germ cell differentiation. EPC1 also promotes histone hyperacetylation during spermatid differentiation. This process is fundamental because as mentioned, it allows for the replacement of histones with transition proteins and protamines, which ultimately leads to genome compaction during spermiogenesis. Genetic ablation of either Epc1 or TIP60 disrupts this hyperacetylation process, impairing histone replacement and causing aberrant spermatid development. This acetylation creates a nuclear gradient, with faster acetylation near the apical pole and slower in the caudal regions that are needed for spermatid nucleus maturation [
20].
EPC1 and HSCs. EPC1 regulates hematopoietic stem and progenitor cell (HSPC) development by linking epigenetic modulation to metabolic control. Loss of EPC1 or its paralog EPC2 impairs HSPC proliferation and reduces histone H3 acetylation, impacting hematopoiesis. Among downstream targets, DLST (dihydrolipoamide S-succinyltransferase), an essential enzyme in the tricarboxylic acid (TCA) cycle, shows expression patterns closely tied to histone H3 acetylation. This regulation appears to be mediated through EPC1's collaboration with chromatin modifiers such as KATs and BRPF1. Since the TCA cycle is known to support both the emergence and differentiation of HSPCs, EPC1 likely serves as an integrator of metabolic activity and chromatin state. Furthermore, EPC1 may enhance DLST transcription by recruiting transcription factors like SRF or FOXR2 to its promoter, suggesting a context-specific mechanism that fine-tunes gene expression to sustain blood stem cell formation and expansion [
53].
EPC1 and myoblast differentiation. Epc1 helps initiate myoblast differentiation by promoting skeletal muscle lineage commitment. Identified as a novel binding partner of the transcriptional co-regulator Hop, Epc1 is transiently upregulated during the early stages of H9c2 myoblast differentiation, preceding the induction of myogenic transcription factors such as myogenin, MyoD, Myf5, and Myf6. Its expression correlates with morphological changes like cell elongation and myotube formation, which are significantly impaired when Epc1 is silenced and enhanced when it is overexpressed. The functional interaction between Epc1 and Hop appears essential for optimal activation of muscle-specific genes, likely through modulation of chromatin structure or transcription factor accessibility [
54]. Moreover, EPC1 interacts with SRF, a regulator of muscle differentiation and cell survival, to activate SRF/SRE-dependent genes by recruiting p300, which in turn induces skeletal muscle differentiation [
55].
RUVBL1 and Differentiation
RUVBL1, also known as Pontin is an evolutionarily conserved chromatin remodeling protein with ATPase and helicase functions that is involved in several complexes including Ino80, Swr1, and TIP60. It forms hexamers and affects transcription, DNA repair and embryonic development, partly through its interactions with transcription factors like c-Myc and p53 [
56].
RUVBL1 and ESCs. Pontin is involved in maintaining ESC identity by acting as a transcriptional coactivator for Oct4, a core pluripotency factor. Loss of Pontin severely disrupts ESC self-renewal and compromises the expression of genes needed for maintaining pluripotency. Genome-wide analyses reveal that Pontin and Oct4 share many targets, including protein-coding genes and a distinct group of long intergenic non-coding RNAs (lincRNAs) that help repress differentiation. Pontin binds to these genomic regions in an Oct4-dependent manner and is needed for recruiting the histone acetyltransferase p300 to promote transcription. Although Oct4 binding remains intact without Pontin, the lack of p300 recruitment impairs transcriptional activation and destabilizes the ESC state. Through coordinated control of both coding and non-coding genes, Pontin regulates chromatin-based networks that preserve ESC pluripotency [
57].
Other studies in mice have shown that loss of Pontin results in failure of embryos to progress beyond the blastocyst stage, due to impaired proliferation of the pluripotent inner cell mass. This suggests that Pontin helps sustain ESC viability and pluripotency, likely through its role in regulating transcriptional programs and chromatin structure necessary for stem cell function [
58].
Pontin acts in retinoic acid (RA)-induced differentiation of embryonic stem cells by acting as a transcriptional co-activator. Upon RA stimulation, Pontin is recruited to RA response elements (RAREs) in coordination with RARα, particularly at the HOX gene clusters, which are key regulators of embryonic patterning and cell fate. This recruitment is associated with increased transcriptional activity, as evidenced by enhanced H3K36me3 marks at HOX loci. Depletion of Pontin significantly reduces RA-driven HOX gene expression, indicating that Pontin is needed for the full activation of RA-responsive developmental genes [
56].
RUVBL1 and HSCs. Pontin plays a role in the survival and function of HSCs. Studies using conditional deletion of the gene encoding Pontin (Ruvbl1) in hematopoietic tissues revealed that its absence leads to bone marrow failure due to loss of HSCs by apoptosis. This role of Pontin in HSCs parallels its function in early embryonic development, where it supports the proliferation of pluripotent cells. Moreover, Pontin’s interaction with factors like c-Myc may contribute to its function in sustaining the hematopoietic system, highlighting its importance in stem cell maintenance and potential implications for diseases such as cancer [
58].
RUVBL1 and epidermal differentiation. Pontin also supports the survival and differentiation of basal epidermal cells during embryogenesis. Conditional deletion of Ruvbl1 in the mouse epidermis leads to profound defects in skin architecture, including the near-complete loss of the keratin-14-positive basal layer and failure to form skin appendages such as hair follicles. These abnormalities are evident as early as embryonic day 12.5 and culminate in severe skin barrier dysfunction and perinatal lethality. While the precise molecular mechanisms remain unclear, the phenotype suggests disrupted cell proliferation and survival, potentially involving impaired regulation of the p53 signaling pathway and transcriptional dysregulation of epidermal differentiation programs [
59].
RUVBL2 and Differentiation
Reptin, a member of the AAA+ ATPase family (ATPases associated with diverse cellular activities), contains conserved Walker A and B motifs that bind ATP, arginine fingers, sensor domains, and shares limited homology with the bacterial RuvB ATP-dependent DNA helicase. Frequently found in complex with Pontin (RUVBL1), Reptin (RUVBL2) can also function independently or even exert opposing effects on gene regulation compared to Pontin. Reptin interacts with transcription factors, co-regulators, and SUMO-modifying enzymes, thereby contributing to transcriptional regulation, DNA damage repair, and ribonucleoprotein complex biogenesis [
60]. Moreover, Reptin regulates oncogenic signaling by interacting with key oncogenic proteins such as β-catenin and c-Myc, influencing cancer cell proliferation and survival [
61].
RUVBL2 and ESCs. RUVBL2 helps maintain pluripotency and self-renewal in hESCs. Its expression is significantly higher in hESCs than in mesenchymal stem cells, indicating a strong link to the unique properties of embryonic cells. As part of chromatin remodeling complexes, RUVBL2 interacts with β-catenin, c-Myc, and Pontin (RUVBL1), regulators of gene expression programs that preserve the undifferentiated state. Its elevated levels support active Wnt signaling and genomic stability, suggesting that RUVBL2 not only marks the embryonic state but also acts as a regulator of stem cell plasticity and lineage decisions, providing insights into potential strategies for controlling differentiation and improving regenerative therapies [
62].
RUVBL2 and NSCs. RUVBL2 also directs embryonic stem cells toward a neuroectodermal fate. Overexpression of RUVBL2 in mouse embryonic stem cells promotes neuroectodermal differentiation by upregulating specific markers such as Pax6 and Nestin, while not inducing mesodermal or endodermal markers. This effect is observed during both spontaneous and directed differentiation, indicating a strong tendency toward neural lineage specification. In vivo, RUVBL2 is predominantly expressed in the ectodermal regions of early mouse embryos, further supporting its involvement in neuroectoderm development [
63]. Similarly, during neural differentiation of human embryonic stem cells, RUVBL2 expression increases significantly and is accompanied by enhanced interactions with nuclear partners such as Pontin, β-catenin, and ATF-2. These interactions likely contribute to transcriptional regulation and the chromatin remodeling required for neural commitment, suggesting a conserved role for RUVBL2 across species in promoting early neural and neuroectodermal differentiation [
64].
RUVBL2 and germ cell differentiation. RUVBL2 contributes to the post-transcriptional regulation of gene expression in round spermatids. As part of a diverse RNA-binding proteome highly expressed before spermatid elongation, RUVBL2 is involved in controlling mRNA stability and preventing premature translation of key transcripts. This regulation ensures that specific mRNAs remain repressed or properly processed until the later stages of sperm development, supporting the accurate timing of protein production required for spermatid elongation and maturation. Through these functions, RUVBL2 helps orchestrate the molecular remodeling necessary for producing functional and fertile spermatozoa [
65].
RUVBL2 and adipogenesis. RUVBL2 functions in both the nucleus and cytoplasm to support adipocyte development and metabolic function. As a direct downstream target of PPARγ, RUVBL2 expression is upregulated during 3T3-L1 pre-adipocyte differentiation, which in turn promotes adiponectin polymerization and secretion; key processes for improving insulin sensitivity and lipid metabolism [
66]. Beyond its nuclear functions, RUVBL2 accumulates in the cytosol after differentiation, where it interacts with AS160 to enhance its phosphorylation. This interaction facilitates insulin-stimulated GLUT4 translocation to the cell surface, boosting glucose uptake into adipocytes and supporting energy storage. Altogether, by regulating adiponectin dynamics and insulin-responsive glucose transport, RUVBL2 reinforces both adipocyte maturation and metabolic health, underscoring its importance in adipose tissue development and insulin sensitivity [
67].
JAZF1 and Differentiation
Juxtaposed with another zinc finger gene 1 (JAZF1) is a member of the C2H2-type zinc finger protein family that is recognized for its unique zinc finger domains facilitating specific DNA binding and transcriptional regulation. Initially identified in endometrial stromal tumors, JAZF1 functions in various cellular functions such as cell proliferation, differentiation, apoptosis, and metabolic regulation [
68,
69]. Recent genome-wide ChIP-seq analyses reveal a strong association between JAZF1 and H2A.Z acetylation at more than 1,000 regulatory sites, primarily within intronic regions. Beyond its established link to tumorigenesis, JAZF1 also regulates fundamental processes such as ribosome biogenesis. This regulatory activity appears largely cell-type independent, suggesting that JAZF1 acts as a broad, context-independent modulator of cellular function [
70].
JAZF1 and adipogenesis. JAZF1 regulates adipose differentiation through its interactions with nuclear receptors and adipokines. During the differentiation of 3T3-L1 preadipocytes, JAZF1 expression rises markedly, paralleling adipocytokine visfatin levels, suggesting a functional link. JAZF1 enhances visfatin transcription indirectly by upregulating PPARα and PPARβ/δ, which in turn promote visfatin expression, as shown by experiments using receptor antagonists and siRNAs. While visfatin facilitates adipocyte differentiation and triglyceride accumulation, JAZF1 paradoxically reduces lipid accumulation in mature adipocytes, likely by suppressing TAK1 and PPARγ expression and by promoting pathways that favor fatty acid oxidation through PPARα and PPARβ/δ activation [
71,
72].
Studies using mouse embryonic fibroblasts and 3T3-L1 cells have shown that loss of JAZF1 severely disrupts the formation of mature fat cells, indicating its role in proper adipose development. In mice with partial JAZF1 deficiency, reduced fat mass and altered glucose regulation further confirm its importance [
73].
JAZF1 and myoblast differentiation. JAZF1 plays a dual role in skeletal muscle development by encouraging myoblast proliferation and hindering their differentiation into mature muscle fibers. Mechanistically, JAZF1 represses expression of transcription factors such as myocyte enhancer factor 2C (MEF2C) and myogenic regulatory factor (MRF4), which are crucial for activating myogenic gene programs and maintaining muscle fiber integrity. Additionally, JAZF1 indirectly reduces AMPD1 expression through these factors, affecting downstream AMPK signaling and potentially linking muscle function to metabolic regulation. Overexpression of JAZF1 leads to sustained cell cycle progression and delays differentiation, suggesting a possible role in muscle-related tumorigenesis when aberrantly regulated. Conversely, reducing JAZF1 levels accelerates myogenic gene expression, affecting muscle cell fate [
74].
JAZF1 and multiciliated cell differentiation. The airway mucociliary escalator that is essential for respiratory health, depends on a balance between secretory and multiciliated cells to clear inhaled debris. Disruptions in this system can lead to chronic infections and serious lung diseases. JAZF1 plays a significant role in promoting multiciliated cell differentiation, as demonstrated through overexpression and knock-down experiments in mouse airway models. Together with Fibronectin type 3 and ankyrin repeat domains 1 (Fank1), JAZF1 acts to regulate this process. JAZF1 and Fank1 likely function downstream of IL6 signaling and upstream of the multiciliated cell transcription factor Foxj1, emphasizing roles in coordinating multiciliogenesis [
75].
JAZF1 and β-cell differentiation. Disruption of JAZF1 in iPSCs derived from knockout mice leads to reduced size and impaired differentiation of insulin-producing β-cells, resulting in decreased insulin production and insulin resistance [
72]. Although JAZF1 deficiency does not markedly affect early iPSC maintenance, it alters differentiation and proliferation capacity, as evidenced by smaller teratomas and lower expression of proliferation markers. Overexpression studies in various cell types, including cancer and cardiac microvascular cells, suggest that JAZF1 can promote proliferation through the Akt signaling pathway. In iPSCs, JAZF1 deficiency also leads to lower insulin and C-peptide levels in differentiated β-cells, accompanied by changes in key endoderm markers such as GATA6 and α-fetoprotein, which are critical for proper β-cell development. These findings indicate that JAZF1 regulates endodermal gene expression during β-cell differentiation, thereby influencing pancreatic insulin secretion, glucose homeostasis, and overall β-cell function in mice [
76].
JAZF1 and decidualization. Human endometrial stromal cells (hEnSCs) undergo decidualization, a differentiation process needed for tissue remodeling and embryo implantation. The Polycomb repressive complex 2 (PRC2) subunit SUZ12 plays a role in this process by trimethylating histone H3 lysine 27 (H3K27me3). The JAZF1-SUZ12 fusion protein, found in low-grade endometrial stromal sarcoma (LG-ESS), disrupts PRC2 composition and its chromatin occupancy, leading to reduced H3K27me3 and increased H4 acetylation at polycomb target genes. In hEnSCs, JAZF1-SUZ12 upregulates genes involved in cell adhesion, characteristic of decidual cells, while repressing genes associated with immune clearance and senescence. These changes suggest that JAZF1-SUZ12 drives an abnormal decidualization program that promotes cell retention within the endometrium and may contribute to tumorigenesis in low-grade endometrial stromal sarcoma [
77].
JAZF1 and Treg differentiation. JAZF1 and PPAR-γ were examined for their roles in Treg differentiation, inflammation, and insulin resistance using a transgenic mouse model [
78]. Notably, JAZF1 transgenic mice exhibited markers indicative of improved insulin sensitivity, compared to wild-type mice. Furthermore, JAZF1 overexpression correlated with elevated levels of CD4+, CD25+, and FOXP3+ markers associated with Treg differentiation, while PPAR-γ inhibition led to reduced expression of these markers. Inflammatory markers (TNF-α, IL-1β, IL-6) were decreased in JAZF1 transgenic groups, with concomitant increases in anti-inflammatory cytokines (IL-10, TGF-β). Conversely, PPAR-γ inhibition reversed these trends. The collaborative action of JAZF1 and PPAR-γ was implicated in promoting Treg differentiation and regulating insulin resistance by modulating pro- and anti-inflammatory cytokine expression. These findings suggest a synergistic role for JAZF1 and PPAR-γ in mitigating insulin resistance, suggesting potential as therapeutic targets for metabolic diseases.
TRRAP and Differentiation
The Transformation/Transcription Domain-Associated Protein (TRRAP) is a 3,859 amino acid protein with a molecular weight of 434 kD in the Ataxia-Telangiectasia Mutated (ATM) superfamily [
79]. TRRAP is a phosphoinositide 3 kinase-related kinase (PIKK) and a pseudokinase as it lacks the typical catalytic residues found in related kinases [
80]. TRRAP is typically found as a monomer but is also a component of larger macromolecular assemblies, including the Spt-Ada-Gcn5 acetyltransferase complex (known as SAGA in yeast and mammals) and the nucleosome acetyltransferase of H4 complex (NuA4 in yeast and TIP60 in mammals). In mammals, TRRAP interacts with the transcription factors E2F1, c-Myc, p53, LXR, FoxO3, and β-catenin, resulting in the activation of their target genes [
81,
82]. Mechanistically, in collaboration with additional complex components, TRRAP recruits HATs such KAT5 and transcription factors (TFs) to chromatin, leading to increased histone acetylation and the initiation of target gene transcription [
79].
TRRAP and HSCs. Conditional TRRAP deletion in bone marrow stem/progenitor cells revealed that TRRAP removal led to a depletion of hematopoietic stem and progenitor cells, paradoxically without impacting the stem cell niche [
83]. TRRAP-deficient hematopoietic stem/progenitor cells exhibited increased apoptosis independent of p53, indicating that the loss of these cells caused bone marrow failure, resulting in the severe phenotype and high mortality in TRRAP-conditional knockout (CKO) mice. This suggests that TRRAP may regulate genes within the hematopoietic stem cell niche that are essential for the survival of stem/progenitor cells. TRRAP overexpression also increased the stability of the NANOG protein by inhibiting its ubiquitination that is typically regulated by FBXW8, an E3 ubiquitin ligase. Since NANOG is essential for maintaining the cancer stem cell (CSC)-like traits of colon cancer spheroids [
84], TRRAP may promote cancer spheroid development.
The Wnt pathway is vital for embryonic development and stem cell self-renewal, but excessive activation is associated with various human cancers. Central to this pathway is β-catenin. TRRAP interacts with Skp1/SCF, promoting the ubiquitination of β-catenin. Deletion of TRRAP reduces β-catenin ubiquitination, slows degradation, and causes β-catenin accumulation, ultimately leading to hyperactive Wnt signaling [
81,
82].
TRRAP and neuronal differentiation. The TRRAP-HAT-Sp1 axis also influences neural arborisation and protects neural cells by regulating the Sp1 pathway, which in turn governs microtubule dynamics [86]. Deleting TRRAP has detrimental effects on the self-renewal and differentiation capacity of adult neural stem cells (aNSCs) in both mice and cell cultures. Deacetylating the K639 residue of Sp1 allowed Sp1 to bind to chromatin at target gene promoters, a process normally hindered by TRRAP. In the absence of TRRAP, K639 on Sp1 becomes acetylated by HATs independent of TRRAP, preventing Sp1 from binding to chromatin and repressing gene expression related to adult neurogenesis. Additionally, TRRAP deletion destabilizes Sp1, reducing its transcription-activating capability. This destabilization may be attributed to the deacetylation of other residues influenced by TRRAP-HAT that have yet to be identified, or because TRRAP itself is necessary for maintaining Sp1's stability as a scaffold [87].
TRRAP and ESCs. Depleting TRRAP in ESCs led to unforeseen differentiation, likely attributed to TRRAP's critical role in HAT-mediated chromatin remodeling and its regulation of essential stemness marker genes such as Nanog, Oct4, and Sox2. Maintaining high TRRAP levels impedes ESC differentiation, underscoring the importance of reducing TRRAP expression for transcriptional reprogramming and ESC differentiation [
79].
TRRAP and multiciliated cell differentiation. TRRAP operates between the Notch2-mediated determination of basal progenitor cell fate and the subsequent regulation of Multicilin to control the differentiation of multiciliated cells (MCCs). It binds to promoters and controls the expression of a set of genes involved in MCC differentiation and function, including several genes associated with human ciliopathies [88]. Through a kinome-wide RNA interference approach, TRRAP was also identified as a regulator of brain tumor-initiating cell (BTIC) differentiation. Knocking down the expression of the adaptor protein TRRAP significantly promoted BTIC differentiation in culture, sensitized the cells to apoptotic signals, and negatively affected cell cycle progression [89]. Down regulation of TRRAP-dependent hTERT expression by Myc/Max was also found to contribute to the differentiation of HL60 cells after exposure to DMSO [90].
DMAP1 and Differentiation
Dnmt1-associated protein 1 (DMAP1), one of the components of both the NuA4 and Swr1/SRCAP complexes, was originally identified as a molecule interacting with DNMT1 that co-localized with PCNA and DNMT1 at DNA replication foci during S phase in human cells. The DMAP1–DNMT1 complex was also reported to interact with the p33ING1–Sin3–HDAC complex and localize in pericentric heterochromatin to maintain heterochromatin structure and histone modifications in late S phase, affecting cell differentiation capabilities [91,92].
DMAP1 and HSCs. The Mat1-mediated transcriptional repressor (MMTR)/DMAP1 is a transcription corepressor involved in chromatin remodeling, cell cycle regulation, DNA double-strand break repair, and tumor suppression. DMAP1 functions in the repair of damaged DNA in mammals that is linked to stem cell aging, causing HSC depletion over time. DMAP1-knockdown induced by DMAP1-specific shRNA severely compromised the proliferative capacity of HSCs in vitro and long-term repopulating capacity of HSCs in recipient mice [93].
DMAP1 and ESCs. MMTR/DMAP1, along with other TIP60-p400 complex proteins, binds the promoters of differentiation commitment genes in mouse ESCs and controls gene expression during differentiation [94]. A Dmap1-null allele was generated to study the role of DMAP1 in development. The results suggest that Dmap1−/− mice died during preimplantation and the DMAP1 somatic form of DNA methyltransferase 1 (DNMT1s) and the DMAP1 oocyte form (DNMT1o) were needed for normal development [95]. An RNA interference (RNAi) screen performed in mouse ES cells showed that reduction of the expression of individual components of the TIP60-p400 complex, including DMAP1, caused a loss of characteristic ES cell morphology and activation of genes associated with cell differentiation [96].
DMAP1 and myoblast differentiation. Utilizing CRISPR-KO screening, Yu et al. uncovered a role for DMAP1 in mediating both progenitor induction and differentiation of chemically induced smooth muscle progenitor cells via regulation of the methylation of the promoters of Nkx2-5 and Cdh1, respectively [97].
YEATS Family and Differentiation
The most comprehensively described post-translational modifications of histones are acetylation, methylation, phosphorylation, ubiquitination, and sumoylation. These modifications are often identified in conserved protein domains such as bromodomains, chromodomains, PHD fingers, and YEATs domains, which are present in various chromatin-modifying complexes. The "YEATS domain" originates from the initial identification of five proteins (Yaf9, ENL, AF9, Taf14 and Sas5) possessing it. It is found in a family of readers of histone acylation and is present in over 100 proteins across 50 organisms. Among humans, AF9, ENL, and GAS41 stand out as the three most extensively studied YEATS domain proteins [98]. The YEATS domain of AF9 (also called MLLT3) exhibits robust binding to acetylated histone H3K9 with a comparatively weaker affinity for acetylated H3K27 and H3K18 [99]. The AF9 protein interacts with DOT1L, MLL, and MPc3, and affects cell differentiation. Different studies suggest that mutations in the human AF9 gene are linked to neurodevelopmental disorders, such as intellectual disability, epilepsy, and ataxia [100,101].
YEATS family and neuronal differentiation. AF9 affects the neural differentiation of hESCs and the activation of genes involved in neurodevelopment [102]. It physically interacts with TET2, and together they bind to gene loci associated with neural cell targets. This binding facilitates the conversion of 5mC to 5hmC and activates genes related to neurodevelopment. AF9 also appears to direct TET2 to occupy neural gene loci by recognizing motifs containing AAC [102]. The Af9 null mutation was observed to impact the development of the axial skeleton, indicating that Af9 contributes to embryo-patterning processes, likely by regulating Hox genes. Heterozygous Af9 mutant mice displayed normal fertility and showed no segment anomalies. However, complete disruption of the gene in homozygous mutants resulted in an anterior transformation of the cervical and thoracic regions, leading to early postnatal lethality [103].
Af9 also functions in the maturation of the forebrain, revealing that the expression of Af9 safeguards against the premature exhaustion of progenitor cells during corticogenesis [100]. This study indicated that AF9 hinders the expression of Tbr1 in neurons and modifies H3K79 dimethylation at the Tbr1 transcriptional start site, concurrent with a reduction in RNA polymerase II levels. In Af9 mutants, elevated TBR1 expression is linked to an increase in the TBR1-mediated regulation of the NMDAR subunit Nr1 [104].
YEATS family and ESCs. YEATS4 (also called Gas41) is the human counterpart of the yeast Yaf9 protein. Both proteins share a common structural arrangement, featuring an N-terminal YEATS domain followed by A and B boxes at the C-terminus. Silencing Gas41 in mESCs resulted in increased expression of many genes, with a limited number downregulated (1082 versus 92). The upregulated genes were enriched in various developmental processes. Comparative analysis with control mESCs through gene set enrichment analysis (GSEA) indicated that the upregulated genes following Gas41 knockdown exhibited significant enrichment in the embryoid body differentiation signature but displayed a negative correlation for the ESC signature. Additionally, Gas41 knockdown induced differentiation phenotypes in mESCs, including flattened morphology and reduced alkaline phosphatase activity. These findings suggest that YEATS4/Gas41 plays a role in preserving mESC identity by acting as a suppressor of developmental genes [105,106].
BAF53a and Differentiation
The Brg1/Brm-associated factors (Baf) complex, alternatively recognized as the mammalian SWI/SNF ATP-dependent chromatin-remodeling complex, plays a role in influencing the differentiation of both embryonic and adult stem cells. Actl6a (also known as Baf53a or Arp4) is a shared subunit present in multiple complexes, such as esBAF, npBAF, INO80, and TIP60-p400. Its expression is observed in various stem/progenitor cells, including neural progenitor cells, hematopoietic stem cells, epidermal progenitor cells, and ES cells [107,108].
BAF53a and ESCs. Baf53a can sustain a progenitor state in epidermal cells by inhibiting KLF4, an activator of differentiation. Conditional KO of the Baf53a gene in epidermal cells resulted in cell cycle exit, terminal differentiation, and hypoplasia, whereas ectopic expression of Baf53a repressed Klf4 expression and enhanced the epidermal progenitor state [109]. Actl6a has the capability to protect mESCs from differentiating into primitive endoderm (PrE). Specifically, Actl6a can interact with Nanog and Sox2, facilitating Nanog's binding to pluripotency-related genes like Oct4 and Sox2. Moreover, Actl6a is able to bind to the promoters of PrE regulators like Sall4 and Fgf4, suppressing their expression and impeding the differentiation of mESCs into primitive endoderm [110]. In hypoxic conditions, HIF-1α, a subunit of hypoxia-inducible factor, preserves the pluripotency of pluripotent stem cells (PSCs). Cui and colleagues [111] discovered that disrupting HIF-1α expression led to a decline in self-renewal and pluripotency in human induced pluripotent stem cells (hiPSCs). The knockdown of HIF-1α hindered the expression of Actl6a and the acetylation of histone H3K9 (H3K9ac). Knocking down Actl6a resulted in reduced levels of H3K9ac and compromised pluripotency in hiPSCs, impacting their differentiation into endoderm. Actl6a exhibited increased expression in fully pluripotent stem cell lines. ATP stimulation enhanced both Actl6a expression and histone acetylation. Knocking down Actl6a resulted in decreased pluripotency in ESCs, and the addition of ATP did not restore this reduction. Additionally, suppressing ATP production through rotenone treatment diminished the pluripotency of ESCs. Together, these findings indicate that the level of mitochondrially generated ATP influences stem cell pluripotency through Actl6a-mediated histone acetylation [112].
BAF53a and neuronal differentiation. Intellectual disability is linked to heterozygous variations in ACTL6A [
48]. Baf45a and Baf53a function as subunits within the neural stem/progenitor BAF (npBAF) complex in neural stem/progenitor cells. As neural progenitors exit the cell cycle, these subunits are further replaced by the analogous BAF45b, BAF45c, and BAF53b proteins. The presence of BAF45a/53a subunits is essential for both the initiation and continuation of neural progenitor proliferation. Disrupting the switch between these subunits hampers the process of neuronal differentiation [113].
Yoo and colleagues demonstrated that the transition is facilitated through the suppression of BAF53a by miR-9* and miR-124. They identified that the repression of BAF53a is governed by specific sequences in the untranslated region, corresponding to the recognition sites for miR-9* and miR-124, which are selectively active in post-mitotic neurons. The mutation of these sites resulted in the sustained expression of BAF53a and impaired activity-dependent dendritic outgrowth in neurons. Moreover, the overexpression of miR-9* and miR-124 in neural progenitors led to reduced proliferation [114].
Park and coworkers recognized the significance of ACTL6a in linking the recognition of axonal caliber with the transcriptional program of myelination. Exposure to large caliber axons or nanofibers enhances the nuclear levels of ACTL6A in Schwann cells (SC), leading to the removal of repressive histone marks and thereby promoting myelination. In the absence of Actl6a, Schwann cells are unable to synchronize caliber recognition and the production of myelin [115].
BAF53a and HSCs. The conditional knockout (cKO) of Baf53a in HSCs led to mice experiencing bone marrow failure, aplastic anemia, and death. Analysis of cell counts revealed a reduction in mature hematopoietic cells, including macrophages, granulocytes, erythrocytes, B cells, and T cells, as well as in HSCs/progenitor cell fractions such as common myeloid progenitors, megakaryo-erythrocyte progenitors, granulocyte-monocyte progenitors and so on in the bone marrow of Baf53a cKO mice. These findings indicate the involvement of Baf53a in the proliferation and survival of hematopoietic cells [116].
The long non-coding RNA uc.291, containing a highly conserved element, was demonstrated to engage in a physical interaction with ACTL6A, influencing chromatin remodeling for the facilitation of cellular differentiation. When uc.291 was depleted, ACTL6A exhibited binding to promoters of differentiation genes, impeding the BAF complex from targeting them and thereby hindering the activation of terminal differentiation genes. Conversely, in the presence of uc.291, the inhibitory effect of ACTL6A was alleviated, enabling chromatin modifications that foster the expression of differentiation genes [117].
p400 and Differentiation
p400/mammalian Domino, which was identified as an interaction partner for adenovirus E1A and a myeloid-specific transcription factor, MZF-2A, is an SWR1-class chromatin-remodeling ATPase that is homologous to the yeast Swr1p and Drosophila Domino proteins. The p400 protein, like its homologs in other eukaryotes, catalyzes ATP-dependent incorporation of histone H2A variant H2A.Z into chromatin via exchange of H2A-H2B dimers within nucleosomes for free H2A.Z-H2B dimers. Interestingly, p400 was recently shown to incorporate histone H3 variant H3.3 into chromatin. H2A.Z and H3.3 are often enriched near gene regulatory regions, consistent with a role for p400 (like TIP60) as a co-activator of transcription. However, p400 also appears to repress transcription in some contexts as well as promote DNA repair in concert with TIP60 [23,118].
p400 and neuronal differentiation. Elsesser and coworkers targeted the deletion of Ep400 during distinct phases of mouse oligodendrocyte development. Despite the normal development of Ep400-deficient oligodendrocyte precursors, significant impairment was observed in terminal differentiation and myelination processes. This impairment is attributed to the mechanistic involvement of Ep400, which interacts with the transcription factor Sox10, binds to regulatory regions of the Myrf gene, and is essential for inducing this central transcriptional regulator within the myelination program [119]. Moreover, the deletion of Ep400 specifically in Schwann cells (SCs) in mice led to defects in the late stages of SC development and peripheral myelination, stemming from the aberrant expression of developmental regulators [120].
p400 and HSCs. The targeted mutation of p400/mDomino in mice resulted in early embryonic lethality accompanied by significant deficiencies in yolk sac hematopoiesis. In embryos with the mDomino mutation, there was a notable decrease in the expressions of embryonic globin genes and a globin chaperone gene. Conversely, there was a substantial increase in the expression of a specific set of Hox genes. These findings indicate a role for p400/mDomino in the epigenetic regulation of genes under developmental control [121]. Fujii and colleagues explored the involvement of the p400/mDomino gene in adult bone marrow hematopoiesis and embryonic fibroblast cell-cycle progression by generating a conditional knock-out mouse model. This led to a sudden decline in nucleated cells within the bone marrow, affecting committed myeloid and erythroid cells, as well as hematopoietic progenitor and stem cells. Additionally, the absence of p400/mDomino in MEFs resulted in pronounced growth inhibition. Analysis of DNA microarray uncovered that the deletion of p400/mDomino from MEFs led to impaired expression of numerous cell-cycle-regulatory genes, including G2/M-specific genes regulated by the transcription factors FoxM1 and c-Myc [118].
p400 and adipogenesis. Following hormonal stimulation, the expression of Brd8, p400, TIP60, H2A.Z, and PPARγ increases in 3T3-L1 preadipocytes. Subsequently, the PPARγ/RXRα heterodimer forms a complex with Brd8 and p400, binding to PPAR-response elements (PPREs). In this process, p400 incorporates H2A.Z into chromatin, leading to transcriptional activation by recruiting RNAPII. Additionally, TIP60 is recruited to the complex by Brd8, promoting histone acetylation and thereby further enhancing transcription [122].
Summary
The NuA4/TIP60 complex is a cornerstone of epigenetic regulation, orchestrating cellular differentiation by integrating histone acetylation and chromatin remodeling with lineage-specific transcriptional programs. Its subunits exhibit diverse and sometimes overlapping roles, collectively contributing to proper development and tissue homeostasis. From maintaining neural stem cell pools to influencing germ cell maturation and hematopoietic differentiation, the multiple biological roles of the complex is evident across numerous biological contexts.
Disruptions in its functionality are implicated in many disorders, emphasizing its role in maintaining cellular and organismal integrity. Understanding the mechanistic details of how individual subunits contribute to the complex’s overall functionality will provide new insights into the regulation of differentiation and offer potential therapeutic targets for diseases stemming from differentiation defects. Future studies should focus on the interplay between the NuA4/TIP60 complex and other epigenetic regulators to uncover broader networks governing cellular identity and plasticity. Additionally, exploring its role in disease models may pave the way for precision medicine approaches aimed at modulating differentiation pathways for therapeutic benefit.
Author Contributions
Conceptualization, FH and KR; writing-original draft preparation, FH and AN; writing, review and editing, FH, AN, and KR; figure preparation, FH, AN and MSL; supervision and fund acquisition, K.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by CIHR grant PJT-178099 awarded to K. Riabowol.
Acknowledgments
The authors would like to thank colleagues Drs. Ana Nikolic and Jack (Li-Fang) Chu for valuable suggestions and discussions that informed aspects of this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Srinageshwar, B.; Maiti, P.; Dunbar, G.L.; Rossignol, J. Role of Epigenetics in Stem Cell Proliferation and Differentiation: Implications for Treating Neurodegenerative Diseases. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Wu, H.; Sun, Y.E. Epigenetic Regulation of Stem Cell Differentiation. Pediatr. Res. 2006, 59, 21R–5R. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem Cells: Past, Present, and Future. Stem Cell Res. Ther. 2019, 10, 68. [Google Scholar] [CrossRef]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-Translational Modifications of Histones: Mechanisms, Biological Functions, and Therapeutic Targets. MedComm 2023, 4, e292. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hematopoiesis. Front. Oncol. 2015, 5, 108. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Dantas, A.; Riabowol, K. Histone Acetyltransferases and Stem Cell Identity. Cancers (Basel). 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, L.; Yu, Z.; Yu, J.; Ren, Y.; Wang, Q.; Xu, Y. Structure of the Human TIP60 Complex. Nat. Commun. 2024, 15, 7092. [Google Scholar] [CrossRef] [PubMed]
- Barry, R.M.; Sacco, O.; Mameri, A.; Stojaspal, M.; Kartsonis, W.; Shah, P.; De Ioannes, P.; Hofr, C.; Côté, J.; Sfeir, A. Rap1 Regulates TIP60 Function during Fate Transition between Two-Cell-like and Pluripotent States. Genes Dev. 2022, 36, 313–330. [Google Scholar] [CrossRef]
- Tominaga, K.; Sakashita, E.; Kasashima, K.; Kuroiwa, K.; Nagao, Y.; Iwamori, N.; Endo, H. Tip60/KAT5 Histone Acetyltransferase Is Required for Maintenance and Neurogenesis of Embryonic Neural Stem Cells. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Kang, E.-J.; Choi, K.-H. New Molecular Bridge between RelA/P65 and NF-ΚB Target Genes via Histone Acetyltransferase TIP60 Cofactor. J. Biol. Chem. 2012, 287, 7780–7791. [Google Scholar] [CrossRef]
- Janas, J.A.; Zhang, L.; Luu, J.H.; Demeter, J.; Meng, L.; Marro, S.G.; Mall, M.; Mooney, N.A.; Schaukowitch, K.; Ng, Y.H.; et al. Tip60-Mediated H2A.Z Acetylation Promotes Neuronal Fate Specification and Bivalent Gene Activation. Mol. Cell 2022, 82, 4627–4646.e14. [Google Scholar] [CrossRef]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.-P.; Allis, C.D.; et al. DNMT3L Connects Unmethylated Lysine 4 of Histone H3 to de Novo Methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Humbert, J.; Salian, S.; Makrythanasis, P.; Lemire, G.; Rousseau, J.; Ehresmann, S.; Garcia, T.; Alasiri, R.; Bottani, A.; Hanquinet, S.; et al. De Novo KAT5 Variants Cause a Syndrome with Recognizable Facial Dysmorphisms, Cerebellar Atrophy, Sleep Disturbance, and Epilepsy. Am. J. Hum. Genet. 2020, 107, 564–574. [Google Scholar] [CrossRef]
- Shen, E.; Shulha, H.; Weng, Z.; Akbarian, S. Regulation of Histone H3K4 Methylation in Brain Development and Disease. Philos. Trans. R. Soc. London. Ser. B, Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Urban, I.; Kerimoglu, C.; Sakib, M.S.; Wang, H.; Benito, E.; Thaller, C.; Zhou, X.; Yan, J.; Fischer, A.; Eichele, G. TIP60/KAT5 Is Required for Neuronal Viability in Hippocampal CA1. Sci. Rep. 2019, 9, 16173. [Google Scholar] [CrossRef]
- Beaver, M.; Bhatnagar, A.; Panikker, P.; Zhang, H.; Snook, R.; Parmar, V.; Vijayakumar, G.; Betini, N.; Akhter, S.; Elefant, F. Disruption of Tip60 HAT Mediated Neural Histone Acetylation Homeostasis Is an Early Common Event in Neurodegenerative Diseases. Sci. Rep. 2020, 10, 18265. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Galbo, P.M.J.; Gong, W.; Storey, A.J.; Tsai, Y.-H.; Yu, X.; Ahn, J.H.; Guo, Y.; Mackintosh, S.G.; Edmondson, R.D.; et al. ZMYND11-MBTD1 Induces Leukemogenesis through Hijacking NuA4/TIP60 Acetyltransferase Complex and a PWWP-Mediated Chromatin Association Mechanism. Nat. Commun. 2021, 12, 1045. [Google Scholar] [CrossRef]
- Numata, A.; Kwok, H.S.; Zhou, Q.-L.; Li, J.; Tirado-Magallanes, R.; Angarica, V.E.; Hannah, R.; Park, J.; Wang, C.Q.; Krishnan, V.; et al. Lysine Acetyltransferase Tip60 Is Required for Hematopoietic Stem Cell Maintenance. Blood 2020, 136, 1735–1747. [Google Scholar] [CrossRef]
- McCarthy, A.; Deiulio, A.; Martin, E.T.; Upadhyay, M.; Rangan, P. Tip60 Complex Promotes Expression of a Differentiation Factor to Regulate Germline Differentiation in Female Drosophila. Mol. Biol. Cell 2018, 29, 2933–2945. [Google Scholar] [CrossRef]
- Dong, Y.; Isono, K.-I.; Ohbo, K.; Endo, T.A.; Ohara, O.; Maekawa, M.; Toyama, Y.; Ito, C.; Toshimori, K.; Helin, K.; et al. EPC1/TIP60-Mediated Histone Acetylation Facilitates Spermiogenesis in Mice. Mol. Cell. Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.B.; Hung, J.-H.; Hickman, T.L.; Coles, A.H.; Carey, J.F.; Weng, Z.; Chu, F.; Fazzio, T.G. Hdac6 Regulates Tip60-P400 Function in Stem Cells. Elife 2013, 2, e01557. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fisher, J.B.; Koprowski, S.; McAllister, D.; Kim, M.-S.; Lough, J. Homozygous Disruption of the Tip60 Gene Causes Early Embryonic Lethality. Dev. Dyn. an Off. Publ. Am. Assoc. Anat. 2009, 238, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Acharya, D.; Hainer, S.J.; Yoon, Y.; Wang, F.; Bach, I.; Rivera-Pérez, J.A.; Fazzio, T.G. KAT-Independent Gene Regulation by Tip60 Promotes ESC Self-Renewal but Not Pluripotency. Cell Rep. 2017, 19, 671–679. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Kang, E.-J.; Choi, K.-H. Tip60 Regulates Myoblast Differentiation by Enhancing the Transcriptional Activity of MyoD via Their Physical Interactions. FEBS J. 2011, 278, 4394–4404. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.-M.; Kim, J.-W.; Kim, C.-H.; An, J.-H.; Johnson, A.; Song, P.I.; Rhee, S.; Choi, K.-H. KAT5-Mediated SOX4 Acetylation Orchestrates Chromatin Remodeling during Myoblast Differentiation. Cell Death Dis. 2015, 6, e1857. [Google Scholar] [CrossRef]
- Ma, X.; Wang, D.; Zhao, W.; Xu, L. Deciphering the Roles of PPARγ in Adipocytes via Dynamic Change of Transcription Complex. Front. Endocrinol. (Lausanne). 2018, 9, 473. [Google Scholar] [CrossRef]
- van Beekum, O.; Brenkman, A.B.; Grøntved, L.; Hamers, N.; van den Broek, N.J.F.; Berger, R.; Mandrup, S.; Kalkhoven, E. The Adipogenic Acetyltransferase Tip60 Targets Activation Function 1 of Peroxisome Proliferator-Activated Receptor Gamma. Endocrinology 2008, 149, 1840–1849. [Google Scholar] [CrossRef]
- Gao, Y.; Koppen, A.; Rakhshandehroo, M.; Tasdelen, I.; van de Graaf, S.F.; van Loosdregt, J.; van Beekum, O.; Hamers, N.; van Leenen, D.; Berkers, C.R.; et al. Early Adipogenesis Is Regulated through USP7-Mediated Deubiquitination of the Histone Acetyltransferase TIP60. Nat. Commun. 2013, 4, 2656. [Google Scholar] [CrossRef]
- Kim, J.-W.; Jang, S.-M.; Kim, C.-H.; An, J.-H.; Choi, K.-H. Transcriptional Activity of Neural Retina Leucine Zipper (Nrl) Is Regulated by c-Jun N-Terminal Kinase and Tip60 during Retina Development. Mol. Cell. Biol. 2012, 32, 1720–1732. [Google Scholar] [CrossRef]
- Dantas, A.; Al Shueili, B.; Yang, Y.; Nabbi, A.; Fink, D.; Riabowol, K. Biological Functions of the ING Proteins. Cancers (Basel). 2019, 11. [Google Scholar] [CrossRef]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear Phosphatidylinositol-5-Phosphate Regulates ING2 Stability at Discrete Chromatin Targets in Response to DNA Damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef]
- Gou, W.-F.; Yang, X.-F.; Shen, D.-F.; Zhao, S.; Sun, H.-Z.; Luo, J.-S.; Zheng, H.-C. Immunohistochemical Profile of ING3 Protein in Normal and Cancerous Tissues. Oncol. Lett. 2017, 13, 1631–1636. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Hara, Y.; Riabowol, K. Different HATS of the ING1 Gene Family. Trends Cell Biol. 2002, 12, 532–538. [Google Scholar] [CrossRef]
- Thalappilly, S.; Feng, X.; Pastyryeva, S.; Suzuki, K.; Muruve, D.; Larocque, D.; Richard, S.; Truss, M.; von Deimling, A.; Riabowol, K.; et al. The P53 Tumor Suppressor Is Stabilized by Inhibitor of Growth 1 (ING1) by Blocking Polyubiquitination. PLoS One 2011, 6, e21065. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A Novel PHD-Finger Motif Protein, P47ING3, Modulates P53-Mediated Transcription, Cell Cycle Control, and Apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef]
- Nabbi, A.; Almami, A.; Thakur, S.; Suzuki, K.; Boland, D.; Bismar, T.A.; Riabowol, K. ING3 Protein Expression Profiling in Normal Human Tissues Suggest Its Role in Cellular Growth and Self-Renewal. Eur. J. Cell Biol. 2015, 94, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Fink, D.; Yau, T.; Nabbi, A.; Wagner, B.; Wagner, C.; Hu, S.M.; Lang, V.; Handschuh, S.; Riabowol, K.; Rülicke, T. Loss of Ing3 Expression Results in Growth Retardation and Embryonic Death. Cancers (Basel). 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Chesi, A.; Wagley, Y.; Johnson, M.E.; Manduchi, E.; Su, C.; Lu, S.; Leonard, M.E.; Hodge, K.M.; Pippin, J.A.; Hankenson, K.D.; et al. Genome-Scale Capture C Promoter Interactions Implicate Effector Genes at GWAS Loci for Bone Mineral Density. Nat. Commun. 2019, 10, 1260. [Google Scholar] [CrossRef]
- Suzuki, S.; Nozawa, Y.; Tsukamoto, S.; Kaneko, T.; Imai, H.; Minami, N. ING3 Is Essential for Asymmetric Cell Division during Mouse Oocyte Maturation. PLoS One 2013, 8, e74749. [Google Scholar] [CrossRef]
- Zhang, P.; Du, J.; Sun, B.; Dong, X.; Xu, G.; Zhou, J.; Huang, Q.; Liu, Q.; Hao, Q.; Ding, J. Structure of Human MRG15 Chromo Domain and Its Binding to Lys36-Methylated Histone H3. Nucleic Acids Res. 2006, 34, 6621–6628. [Google Scholar] [CrossRef]
- Tominaga, K.; Kirtane, B.; Jackson, J.G.; Ikeno, Y.; Ikeda, T.; Hawks, C.; Smith, J.R.; Matzuk, M.M.; Pereira-Smith, O.M. MRG15 Regulates Embryonic Development and Cell Proliferation. Mol. Cell. Biol. 2005, 25, 2924–2937. [Google Scholar] [CrossRef]
- Chen, M.; Takano-Maruyama, M.; Pereira-Smith, O.M.; Gaufo, G.O.; Tominaga, K. MRG15, a Component of HAT and HDAC Complexes, Is Essential for Proliferation and Differentiation of Neural Precursor Cells. J. Neurosci. Res. 2009, 87, 1522–1531. [Google Scholar] [CrossRef]
- Chen, M.; Pereira-Smith, O.M.; Tominaga, K. Loss of the Chromatin Regulator MRG15 Limits Neural Stem/Progenitor Cell Proliferation via Increased Expression of the P21 Cdk Inhibitor. Stem Cell Res. 2011, 7, 75–88. [Google Scholar] [CrossRef]
- Gupta, P.; Leahul, L.; Wang, X.; Wang, C.; Bakos, B.; Jasper, K.; Hansen, D. Proteasome Regulation of the Chromodomain Protein MRG-1 Controls the Balance between Proliferative Fate and Differentiation in the C. Elegans Germ Line. Development 2015, 142, 291–302. [Google Scholar] [CrossRef]
- Iwamori, N.; Tominaga, K.; Sato, T.; Riehle, K.; Iwamori, T.; Ohkawa, Y.; Coarfa, C.; Ono, E.; Matzuk, M.M. MRG15 Is Required for Pre-MRNA Splicing and Spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E5408–15. [Google Scholar] [CrossRef] [PubMed]
- Takubo, K.; Htun, P.W.; Ueda, T.; Sera, Y.; Iwasaki, M.; Koizumi, M.; Shiroshita, K.; Kobayashi, H.; Haraguchi, M.; Watanuki, S.; et al. MBTD1 Preserves Adult Hematopoietic Stem Cell Pool Size and Function. Proc. Natl. Acad. Sci. U. S. A. 2023, 120, e2206860120. [Google Scholar] [CrossRef]
- Zhang, H.; Devoucoux, M.; Song, X.; Li, L.; Ayaz, G.; Cheng, H.; Tempel, W.; Dong, C.; Loppnau, P.; Côté, J.; et al. Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex. Cell Rep. 2020, 30, 3996–4002.e4. [Google Scholar] [CrossRef] [PubMed]
- Chadchan, S.B.; Maurya, V.K.; Krekeler, G.L.; Jungheim, E.S.; Kommagani, R. A Role for Malignant Brain Tumor Domain-Containing Protein 1 in Human Endometrial Stromal Cell Decidualization. Front. cell Dev. Biol. 2020, 8, 745. [Google Scholar] [CrossRef] [PubMed]
- Honda, H.; Takubo, K.; Oda, H.; Kosaki, K.; Tazaki, T.; Yamasaki, N.; Miyazaki, K.; Moore, K.A.; Honda, Z.; Suda, T.; et al. Hemp, an Mbt Domain-Containing Protein, Plays Essential Roles in Hematopoietic Stem Cell Function and Skeletal Formation. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 2468–2473. [Google Scholar] [CrossRef]
- Searle, N.E.; Torres-Machorro, A.L.; Pillus, L. Chromatin Regulation by the NuA4 Acetyltransferase Complex Is Mediated by Essential Interactions Between Enhancer of Polycomb (Epl1) and Esa1. Genetics 2017, 205, 1125–1137. [Google Scholar] [CrossRef]
- Wang, Y.; Alla, V.; Goody, D.; Gupta, S.K.; Spitschak, A.; Wolkenhauer, O.; Pützer, B.M.; Engelmann, D. Epigenetic Factor EPC1 Is a Master Regulator of DNA Damage Response by Interacting with E2F1 to Silence Death and Activate Metastasis-Related Gene Signatures. Nucleic Acids Res. 2015, 44, 117–133. [Google Scholar] [CrossRef]
- Searle, N.E.; Pillus, L. Critical Genomic Regulation Mediated by Enhancer of Polycomb. Curr. Genet. 2018, 64, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, X.; Li, L.; Tai, Z.; Li, G.; Liu, J.-X. EPC1/2 Regulate Hematopoietic Stem and Progenitor Cell Proliferation by Modulating H3 Acetylation and DLST. iScience 2024, 27, 109263. [Google Scholar] [CrossRef]
- Kee, H.J.; Kim, J.-R.; Nam, K.-I.; Park, H.Y.; Shin, S.; Kim, J.C.; Shimono, Y.; Takahashi, M.; Jeong, M.H.; Kim, N.; et al. Enhancer of Polycomb1, a Novel Homeodomain Only Protein-Binding Partner, Induces Skeletal Muscle Differentiation. J. Biol. Chem. 2007, 282, 7700–7709. [Google Scholar] [CrossRef]
- Kim, J.-R.; Kee, H.J.; Kim, J.-Y.; Joung, H.; Nam, K.-I.; Eom, G.H.; Choe, N.; Kim, H.-S.; Kim, J.C.; Kook, H.; et al. Enhancer of Polycomb1 Acts on Serum Response Factor to Regulate Skeletal Muscle Differentiation. J. Biol. Chem. 2009, 284, 16308–16316. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Zhang, Z.; Zboril, E.; Wetzel, M.D.; Xu, X.; Zhang, W.; Chen, L.; Liu, Z. Pontin Functions as A Transcriptional Co-Activator for Retinoic Acid-Induced HOX Gene Expression. J. Mol. Biol. 2021, 433, 166928. [Google Scholar] [CrossRef]
- Boo, K.; Bhin, J.; Jeon, Y.; Kim, J.; Shin, H.-J.R.; Park, J.-E.; Kim, K.; Kim, C.R.; Jang, H.; Kim, I.-H.; et al. Pontin Functions as an Essential Coactivator for Oct4-Dependent LincRNA Expression in Mouse Embryonic Stem Cells. Nat. Commun. 2015, 6, 6810. [Google Scholar] [CrossRef]
- Bereshchenko, O.; Mancini, E.; Luciani, L.; Gambardella, A.; Riccardi, C.; Nerlov, C. Pontin Is Essential for Murine Hematopoietic Stem Cell Survival. Haematologica 2012, 97, 1291–1294. [Google Scholar] [CrossRef]
- Dafinger, C.; Benzing, T.; Dötsch, J.; Schermer, B.; Liebau, M.C. Targeted Deletion of Ruvbl1 Results in Severe Defects of Epidermal Development and Perinatal Mortality. Mol. Cell. Pediatr. 2021, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, W.; Liu, H.; Yan, L.; Jiao, W.; Li, D.; Tang, Y.; Gu, G.; Xu, Z. Overexpression of Reptin in Renal Cell Carcinoma Contributes to Tumor Malignancies and Its Inhibition Triggers Senescence of Cancer Cells. Urol. Oncol. 2013, 31, 1358–1366. [Google Scholar] [CrossRef]
- Bauer, A.; Chauvet, S.; Huber, O.; Usseglio, F.; Rothbächer, U.; Aragnol, D.; Kemler, R.; Pradel, J. Pontin52 and Reptin52 Function as Antagonistic Regulators of Beta-Catenin Signalling Activity. EMBO J. 2000, 19, 6121–6130. [Google Scholar] [CrossRef]
- Jaishankar, A.; Barthelery, M.; Freeman, W.M.; Salli, U.; Ritty, T.M.; Vrana, K.E. Human Embryonic and Mesenchymal Stem Cells Express Different Nuclear Proteomes. Stem Cells Dev. 2009, 18, 793–802. [Google Scholar] [CrossRef]
- Hong, S.; Jo, J.; Kim, H.J.; Lee, J.E.; Shin, D.H.; Lee, S.-G.; Baek, A.; Shim, S.H.; Lee, D.R. RuvB-Like Protein 2 (Ruvbl2) Has a Role in Directing the Neuroectodermal Differentiation of Mouse Embryonic Stem Cells. Stem Cells Dev. 2016, 25, 1376–1385. [Google Scholar] [CrossRef]
- Barthéléry, M.; Jaishankar, A.; Salli, U.; Vrana, K.E. Reptin52 Expression during in Vitro Neural Differentiation of Human Embryonic Stem Cells. Neurosci. Lett. 2009, 452, 47–51. [Google Scholar] [CrossRef]
- Chapman, K.M.; Powell, H.M.; Chaudhary, J.; Shelton, J.M.; Richardson, J.A.; Richardson, T.E.; Hamra, F.K. Linking Spermatid Ribonucleic Acid (RNA) Binding Protein and Retrogene Diversity to Reproductive Success. Mol. Cell. Proteomics 2013, 12, 3221–3236. [Google Scholar] [CrossRef]
- Zhu, D.; Xu, L.; Wei, X.; Xia, B.; Gong, Y.; Li, Q.; Chen, X. PPARγ Enhanced Adiponectin Polymerization and Trafficking by Promoting RUVBL2 Expression during Adipogenic Differentiation. Gene 2021, 764, 145100. [Google Scholar] [CrossRef]
- Xie, X.; Chen, Y.; Xue, P.; Fan, Y.; Deng, Y.; Peng, G.; Yang, F.; Xu, T. RUVBL2, a Novel AS160-Binding Protein, Regulates Insulin-Stimulated GLUT4 Translocation. Cell Res. 2009, 19, 1090–1097. [Google Scholar] [CrossRef]
- Fu, H.; Zhou, F.; Yuan, Q.; Zhang, W.; Qiu, Q.; Yu, X.; He, Z. MiRNA-31-5p Mediates the Proliferation and Apoptosis of Human Spermatogonial Stem Cells via Targeting JAZF1 and Cyclin A2. Mol. Ther. Nucleic Acids 2019, 14, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Mameri, A.; Côté, J. JAZF1: A Metabolic Actor Subunit of the NuA4/TIP60 Chromatin Modifying Complex. Front. cell Dev. Biol. 2023, 11, 1134268. [Google Scholar] [CrossRef] [PubMed]
- Procida, T.; Friedrich, T.; Jack, A.P.M.; Peritore, M.; Bönisch, C.; Eberl, H.C.; Daus, N.; Kletenkov, K.; Nist, A.; Stiewe, T.; et al. JAZF1, A Novel P400/TIP60/NuA4 Complex Member, Regulates H2A.Z Acetylation at Regulatory Regions. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.; Xiao, D.; Gong, W.; Liu, H.; Liu, J.; Zhou, H.; Liu, Z. JAZF1 Can Regulate the Expression of Lipid Metabolic Genes and Inhibit Lipid Accumulation in Adipocytes. Biochem. Biophys. Res. Commun. 2014, 445, 673–680. [Google Scholar] [CrossRef]
- Ming, G.; Li, X.; Yin, J.; Ai, Y.; Xu, D.; Ma, X.; Liu, Z.; Liu, H.; Zhou, H.; Liu, Z. JAZF1 Regulates Visfatin Expression in Adipocytes via PPARα and PPARβ/δ Signaling. Metabolism. 2014, 63, 1012–1021. [Google Scholar] [CrossRef]
- Jeong, J.; Jang, S.; Park, S.; Kwon, W.; Kim, S.-Y.; Jang, S.; Ko, J.; Park, S.J.; Lim, S.-G.; Yoon, D.; et al. JAZF1 Heterozygous Knockout Mice Show Altered Adipose Development and Metabolism. Cell Biosci. 2021, 11, 161. [Google Scholar] [CrossRef]
- Yuasa, K.; Aoki, N.; Hijikata, T. JAZF1 Promotes Proliferation of C2C12 Cells, but Retards Their Myogenic Differentiation through Transcriptional Repression of MEF2C and MRF4-Implications for the Role of Jazf1 Variants in Oncogenesis and Type 2 Diabetes. Exp. Cell Res. 2015, 336, 287–297. [Google Scholar] [CrossRef]
- Johnson, J.-A.; Watson, J.K.; Nikolić, M.Z.; Rawlins, E.L. Fank1 and Jazf1 Promote Multiciliated Cell Differentiation in the Mouse Airway Epithelium. Biol. Open 2018, 7. [Google Scholar] [CrossRef]
- Park, S.J.; Kwon, W.; Park, S.; Jeong, J.; Kim, D.; Jang, S.; Kim, S.-Y.; Sung, Y.; Kim, M.O.; Choi, S.-K.; et al. Jazf1 Acts as a Regulator of Insulin-Producing β-Cell Differentiation in Induced Pluripotent Stem Cells and Glucose Homeostasis in Mice. FEBS J. 2021, 288, 4412–4427. [Google Scholar] [CrossRef]
- Tavares, M.; Khandelwal, G.; Muter, J.; Viiri, K.; Beltran, M.; Brosens, J.J.; Jenner, R.G. JAZF1-SUZ12 Dysregulates PRC2 Function and Gene Expression during Cell Differentiation. Cell Rep. 2022, 39, 110889. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Hao, P.; Du, H. Regulatory T Cells Differentiation in Visceral Adipose Tissues Contributes to Insulin Resistance by Regulating JAZF-1/PPAR-γ Pathway. J. Cell. Mol. Med. 2023, 27, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.-K.; Wang, Z.-Q. Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Detilleux, D.; Raynaud, P.; Pradet-Balade, B.; Helmlinger, D. The TRRAP Transcription Cofactor Represses Interferon-Stimulated Genes in Colorectal Cancer Cells. Elife 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Elías-Villalobos, A.; Fort, P.; Helmlinger, D. New Insights into the Evolutionary Conservation of the Sole PIKK Pseudokinase Tra1/TRRAP. Biochem. Soc. Trans. 2019, 47, 1597–1608. [Google Scholar] [CrossRef]
- Fusi, L.; Paudel, R.; Meder, K.; Schlosser, A.; Schrama, D.; Goebeler, M.; Schmidt, M. Interaction of Transcription Factor FoxO3 with Histone Acetyltransferase Complex Subunit TRRAP Modulates Gene Expression and Apoptosis. J. Biol. Chem. 2022, 298, 101714. [Google Scholar] [CrossRef]
- Loizou, J.I.; Oser, G.; Shukla, V.; Sawan, C.; Murr, R.; Wang, Z.-Q.; Trumpp, A.; Herceg, Z. Histone Acetyltransferase Cofactor Trrap Is Essential for Maintaining the Hematopoietic Stem/Progenitor Cell Pool. J. Immunol. 2009, 183, 6422–6431. [Google Scholar] [CrossRef]
- Kang, K.-T.; Shin, M.-J.; Moon, H.-J.; Choi, K.-U.; Suh, D.-S.; Kim, J.-H. TRRAP Enhances Cancer Stem Cell Characteristics by Regulating NANOG Protein Stability in Colon Cancer Cells. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, M.G.; Sawan, C.; Ouzounova, M.; Murr, R.; Herceg, Z. HAT Cofactor TRRAP Mediates Beta-Catenin Ubiquitination on the Chromatin and the Regulation of the Canonical Wnt Pathway. Cell Cycle 2008, 7, 3908–3914. [Google Scholar] [CrossRef]
- Tapias, A.; Lázaro, D.; Yin, B.-K.; Rasa, S.M.M.; Krepelova, A.; Kelmer Sacramento, E.; Grigaravicius, P.; Koch, P.; Kirkpatrick, J.; Ori, A.; et al. HAT Cofactor TRRAP Modulates Microtubule Dynamics via SP1 Signaling to Prevent Neurodegeneration. Elife 2021, 10. [Google Scholar] [CrossRef]
- Yin, B.-K.; Lázaro, D.; Wang, Z.-Q. TRRAP-Mediated Acetylation on Sp1 Regulates Adult Neurogenesis. Comput. Struct. Biotechnol. J. 2023, 21, 472–484. [Google Scholar] [CrossRef]
- Wang, Z.; Plasschaert, L.W.; Aryal, S.; Renaud, N.A.; Yang, Z.; Choo-Wing, R.; Pessotti, A.D.; Kirkpatrick, N.D.; Cochran, N.R.; Carbone, W.; et al. TRRAP Is a Central Regulator of Human Multiciliated Cell Formation. J. Cell Biol. 2018, 217, 1941–1955. [Google Scholar] [CrossRef]
- Charles, N.A.; Holland, E.C. TRRAP and the Maintenance of Stemness in Gliomas. Cell Stem Cell 2010, 6, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Bi, K.; Tang, T.; Wang, J.; Zhang, Y.; Zhang, W.; Ren, H.; Bai, H.; Wang, Y. Down-Regulation of TRRAP-Dependent HTERT and TRRAP-Independent CAD Activation by Myc/Max Contributes to the Differentiation of HL60 Cells after Exposure to DMSO. Int. Immunopharmacol. 2006, 6, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Rountree, M.R.; Bachman, K.E.; Baylin, S.B. DNMT1 Binds HDAC2 and a New Co-Repressor, DMAP1, to Form a Complex at Replication Foci. Nat. Genet. 2000, 25, 269–277. [Google Scholar] [CrossRef]
- Negishi, M.; Chiba, T.; Saraya, A.; Miyagi, S.; Iwama, A. Dmap1 Plays an Essential Role in the Maintenance of Genome Integrity through the DNA Repair Process. Genes Cells 2009, 14, 1347–1357. [Google Scholar] [CrossRef]
- Koizumi, T.; Negishi, M.; Nakamura, S.; Oguro, H.; Satoh, K.; Ichinose, M.; Iwama, A. Depletion of Dnmt1-Associated Protein 1 Triggers DNA Damage and Compromises the Proliferative Capacity of Hematopoietic Stem Cells. Int. J. Hematol. 2010, 91, 611–619. [Google Scholar] [CrossRef]
- Lee, Y.J.; Son, S.H.; Lim, C.S.; Kim, M.Y.; Lee, S.W.; Lee, S.; Jeon, J.; Ha, D.H.; Jung, N.R.; Han, S.Y.; et al. MMTR/Dmap1 Sets the Stage for Early Lineage Commitment of Embryonic Stem Cells by Crosstalk with PcG Proteins. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Mohan, K.N.; Ding, F.; Chaillet, J.R. Distinct Roles of DMAP1 in Mouse Development. Mol. Cell. Biol. 2011, 31, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- Fazzio, T.G.; Huff, J.T.; Panning, B. An RNAi Screen of Chromatin Proteins Identifies Tip60-P400 as a Regulator of Embryonic Stem Cell Identity. Cell 2008, 134, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.S.L.; Palano, G.; Lim, C.; Moggio, A.; Drowley, L.; Plowright, A.T.; Bohlooly-Y, M.; Rosen, B.S.; Hansson, E.M.; Wang, Q.-D.; et al. CRISPR-Knockout Screen Identifies Dmap1 as a Regulator of Chemically Induced Reprogramming and Differentiation of Cardiac Progenitors. Stem Cells 2019, 37, 958–972. [Google Scholar] [CrossRef]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS Domain Proteins: A Diverse Family with Many Links to Chromatin Modification and Transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.R.; Li, W.; et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef]
- Pramparo, T.; Grosso, S.; Messa, J.; Zatterale, A.; Bonaglia, M.C.; Chessa, L.; Balestri, P.; Rocchi, M.; Zuffardi, O.; Giorda, R. Loss-of-Function Mutation of the AF9/MLLT3 Gene in a Girl with Neuromotor Development Delay, Cerebellar Ataxia, and Epilepsy. Hum. Genet. 2005, 118, 76–81. [Google Scholar] [CrossRef]
- Striano, P.; Elia, M.; Castiglia, L.; Galesi, O.; Pelligra, S.; Striano, S. A t(4;9)(Q34;P22) Translocation Associated with Partial Epilepsy, Mental Retardation, and Dysmorphism. Epilepsia 2005, 46, 1322–1324. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, X.; Wang, R.; Li, Y.; Yu, F.; Yang, X.; Song, L.; Xu, G.; Chin, Y.E.; Jing, N. AF9 Promotes HESC Neural Differentiation through Recruiting TET2 to Neurodevelopmental Gene Loci for Methylcytosine Hydroxylation. Cell Discov. 2015, 1, 15017. [Google Scholar] [CrossRef]
- Collins, E.C.; Appert, A.; Ariza-McNaughton, L.; Pannell, R.; Yamada, Y.; Rabbitts, T.H. Mouse Af9 Is a Controller of Embryo Patterning, like Mll, Whose Human Homologue Fuses with Af9 after Chromosomal Translocation in Leukemia. Mol. Cell. Biol. 2002, 22, 7313–7324. [Google Scholar] [CrossRef]
- Büttner, N.; Johnsen, S.A.; Kügler, S.; Vogel, T. Af9/Mllt3 Interferes with Tbr1 Expression through Epigenetic Modification of Histone H3K79 during Development of the Cerebral Cortex. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 7042–7047. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-C.; Zhao, D.; Shi, J.; Peng, D.; Guan, H.; Li, Y.; Huang, Y.; Wen, H.; Li, W.; Li, H.; et al. Gas41 Links Histone Acetylation to H2A.Z Deposition and Maintenance of Embryonic Stem Cell Identity. Cell Discov. 2018, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, S.; Li, X.; Li, X.D. YEATS Domains as Novel Epigenetic Readers: Structures, Functions, and Inhibitor Development. ACS Chem. Biol. 2023, 18, 994–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, B.; Li, W.; Ma, L.; Zheng, D.; Li, L.; Yang, W.; Chu, M.; Chen, W.; Mailman, R.B.; et al. Transcriptional Repression by the BRG1-SWI/SNF Complex Affects the Pluripotency of Human Embryonic Stem Cells. Stem cell reports 2014, 3, 460–474. [Google Scholar] [CrossRef]
- Zhu, B.; Ueda, A.; Song, X.; Horike, S.-I.; Yokota, T.; Akagi, T. Baf53a Is Involved in Survival of Mouse ES Cells, Which Can Be Compensated by Baf53b. Sci. Rep. 2017, 7, 14059. [Google Scholar] [CrossRef]
- Bao, X.; Tang, J.; Lopez-Pajares, V.; Tao, S.; Qu, K.; Crabtree, G.R.; Khavari, P.A. ACTL6a Enforces the Epidermal Progenitor State by Suppressing SWI/SNF-Dependent Induction of KLF4. Cell Stem Cell 2013, 12, 193–203. [Google Scholar] [CrossRef]
- Lu, W.; Fang, L.; Ouyang, B.; Zhang, X.; Zhan, S.; Feng, X.; Bai, Y.; Han, X.; Kim, H.; He, Q.; et al. Actl6a Protects Embryonic Stem Cells from Differentiating into Primitive Endoderm. Stem Cells 2015, 33, 1782–1793. [Google Scholar] [CrossRef]
- Cui, P.; Zhang, P.; Zhang, Y.; Sun, L.; Cui, G.; Guo, X.; Wang, H.; Zhang, X.; Shi, Y.; Yu, Z. HIF-1α/Actl6a/H3K9ac Axis Is Critical for Pluripotency and Lineage Differentiation of Human Induced Pluripotent Stem Cells. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 5740–5753. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, P.; Li, Y.; Feng, G.; Tong, M.; Guo, L.; Li, T.; Liu, L.; Li, W.; Zhou, Q. Mitochondrially Produced ATP Affects Stem Cell Pluripotency via Actl6a-Mediated Histone Acetylation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 1891–1902. [Google Scholar] [CrossRef]
- Lessard, J.; Wu, J.I.; Ranish, J.A.; Wan, M.; Winslow, M.M.; Staahl, B.T.; Wu, H.; Aebersold, R.; Graef, I.A.; Crabtree, G.R. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron 2007, 55, 201–215. [Google Scholar] [CrossRef]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-Mediated Switching of Chromatin-Remodelling Complexes in Neural Development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef]
- Park, H.-J.; Tsai, E.; Huang, D.; Weaver, M.; Frick, L.; Alcantara, A.; Moran, J.J.; Patzig, J.; Melendez-Vasquez, C. V; Crabtree, G.R.; et al. ACTL6a Coordinates Axonal Caliber Recognition and Myelination in the Peripheral Nerve. iScience 2022, 25, 104132. [Google Scholar] [CrossRef]
- Krasteva, V.; Buscarlet, M.; Diaz-Tellez, A.; Bernard, M.-A.; Crabtree, G.R.; Lessard, J.A. The BAF53a Subunit of SWI/SNF-like BAF Complexes Is Essential for Hemopoietic Stem Cell Function. Blood 2012, 120, 4720–4732. [Google Scholar] [CrossRef] [PubMed]
- Panatta, E.; Lena, A.M.; Mancini, M.; Smirnov, A.; Marini, A.; Delli Ponti, R.; Botta-Orfila, T.; Tartaglia, G.G.; Mauriello, A.; Zhang, X.; et al. Long Non-Coding RNA Uc.291 Controls Epithelial Differentiation by Interfering with the ACTL6A/BAF Complex. EMBO Rep. 2020, 21, e46734. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Ueda, T.; Nagata, S.; Fukunaga, R. Essential Role of P400/MDomino Chromatin-Remodeling ATPase in Bone Marrow Hematopoiesis and Cell-Cycle Progression. J. Biol. Chem. 2010, 285, 30214–30223. [Google Scholar] [CrossRef] [PubMed]
- Elsesser, O.; Fröb, F.; Küspert, M.; Tamm, E.R.; Fujii, T.; Fukunaga, R.; Wegner, M. Chromatin Remodeler Ep400 Ensures Oligodendrocyte Survival and Is Required for Myelination in the Vertebrate Central Nervous System. Nucleic Acids Res. 2019, 47, 6208–6224. [Google Scholar] [CrossRef]
- Fröb, F.; Sock, E.; Tamm, E.R.; Saur, A.-L.; Hillgärtner, S.; Williams, T.J.; Fujii, T.; Fukunaga, R.; Wegner, M. Ep400 Deficiency in Schwann Cells Causes Persistent Expression of Early Developmental Regulators and Peripheral Neuropathy. Nat. Commun. 2019, 10, 2361. [Google Scholar] [CrossRef]
- Ueda, T.; Watanabe-Fukunaga, R.; Ogawa, H.; Fukuyama, H.; Higashi, Y.; Nagata, S.; Fukunaga, R. Critical Role of the P400/MDomino Chromatin-Remodeling ATPase in Embryonic Hematopoiesis. Genes Cells 2007, 12, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Couture, J.-P.; Nolet, G.; Beaulieu, E.; Blouin, R.; Gévry, N. The P400/Brd8 Chromatin Remodeling Complex Promotes Adipogenesis by Incorporating Histone Variant H2A.Z at PPARγ Target Genes. Endocrinology 2012, 153, 5796–5808. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jin, J.; Tomomori-Sato, C.; Sato, S.; Sorokina, I.; Parmely, T.J.; Conaway, R.C.; Conaway, J.W. Identification of New Subunits of the Multiprotein Mammalian TRRAP/TIP60-Containing Histone Acetyltransferase Complex. J. Biol. Chem. 2003, 278, 42733–42736. [Google Scholar] [CrossRef] [PubMed]
- Awe, J.P.; Byrne, J.A. Identifying Candidate Oocyte Reprogramming Factors Using Cross-Species Global Transcriptional Analysis. Cell. Reprogram. 2013, 15, 126–133. [Google Scholar] [CrossRef]
- Song, Y.; Hou, G.; Diep, J.; Ooi, Y.S.; Akopyants, N.S.; Beverley, S.M.; Carette, J.E.; Greenberg, H.B.; Ding, S. Inhibitor of Growth Protein 3 Epigenetically Silences Endogenous Retroviral Elements and Prevents Innate Immune Activation. Nucleic Acids Res. 2021, 49, 12706–12715. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).