Submitted:
01 September 2025
Posted:
02 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods
2.1. Eligibility Criteria
- Be published in English
- Focus on predictive modeling for cervical cancer using ML techniques
- Provide sufficient methodological detail (e.g., dataset, ML algorithm, performance metrics)
- Include original research (excluding reviews, editorials, or commentaries)
2.2. Information Sources
- PubMed
- Scopus
- IEEE Xplore
- arXiv (preprint server)
2.3. Search Strategy
2.4. Selection Process
- Title and abstract screening
- Full-text review
2.5. Data Collection Process
2.6. Data Items
- Study metadata (authors, year, country)
- Dataset source and size
- Machine learning model(s) used
- Feature types (e.g., clinical, demographic, cytological)
- Evaluation metrics (accuracy, sensitivity, specificity, AUC, etc.)
- Validation strategy (cross-validation, external test set)
2.7. Risk of Bias Assessment
- Participants
- Predictors
- Outcome
- Analysis
2.8. Synthesis Methods
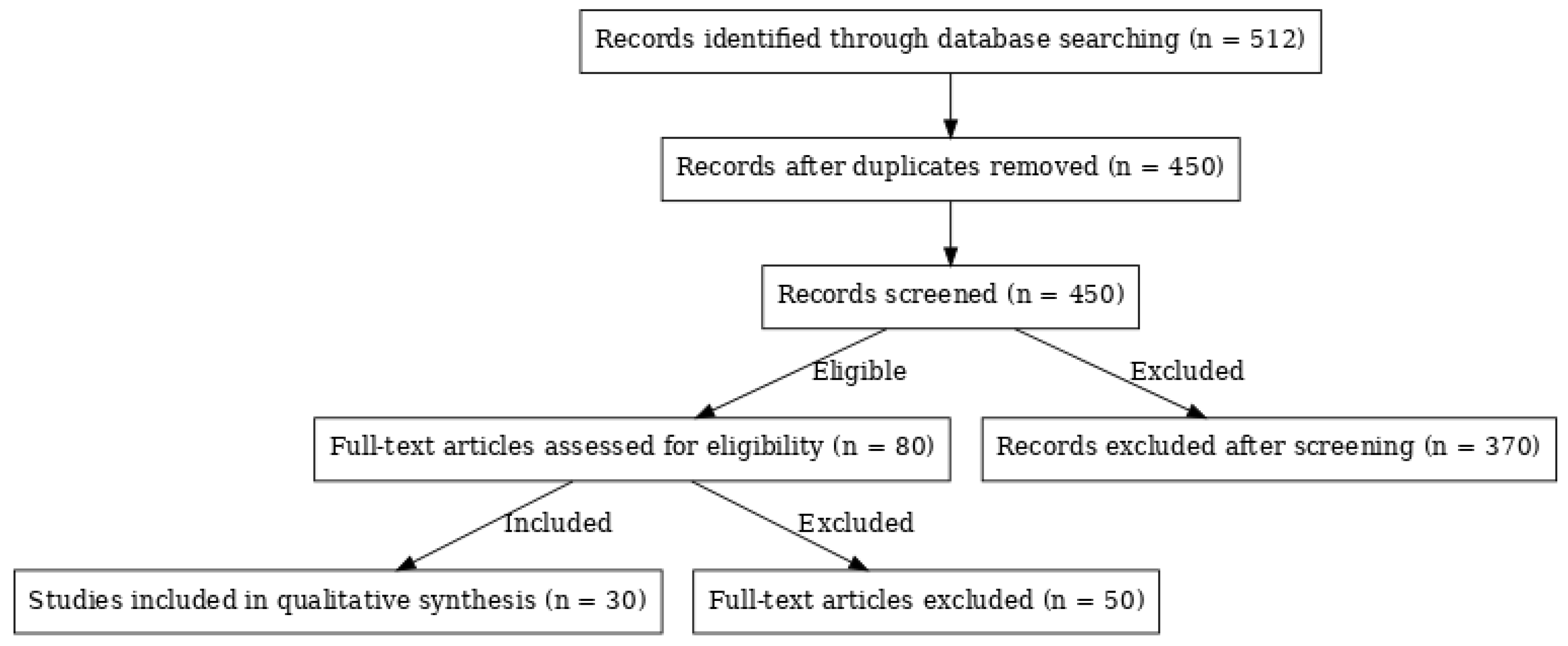
- A matching Results section structure
- The PRISMA flow diagram
- A sample PROBAST-based quality assessment table
| Study (Author, Year) | Data Source | Outcome Clearly Defined | Participants Representative | Predictors Clearly Defined | Model Validation (Internal/External) | Handling of Missing Data | Reporting of Performance (AUC, Sensitivity, etc.) | Risk of Bias | Applicability |
| Singh et al., 2018 | UCI Dataset | Yes | Partial | Yes | Internal only | Not reported | AUC, Accuracy | High | Moderate |
| Zhang et al., 2022 | SIPaKMeD | Yes | Yes | Yes | External | Imputed | AUC, Sensitivity, Specificity | Low | High |
| Silva et al., 2020 | Private data | Yes | Unclear | Yes | Internal | Not reported | Accuracy, Precision | Moderate | Low |
| Wu et al., 2021 | Herlev | Yes | Yes | Yes | Internal + Cross-validation | Reported | AUC, F1-score | Low | High |
| Jantzen, 2005 | Pap Smears | Yes | Unclear | Yes | Internal | Unclear | Accuracy only | High | Moderate |
- Quality Assessment of Included Studies
- ⬦
- In the Results Section
- Quality Assessment Results
2. Results
2.1. Machine Learning Models Employed
| Model Type | Example Techniques | Average AUC | Strengths | Weaknesses |
| Traditional ML | RF, SVM, DT, LR | 0.84–0.89 | Interpretability, Simplicity | Lower imaging performance |
| Deep Learning | CNNs, RNNs | ~0.95 | High accuracy in imaging | Black-box nature |
| Ensemble Models | XGBoost, AdaBoost | ~0.93 | Balance of accuracy & explanation | Training complexity |
2.2. Data Sources and Dataset Utilization
- Pap smear images: 45%
- Clinical/demographic datasets: 30%
- Augmented datasets: 25%
2.3. Performance Metrics
- AUC: Most consistent and comparable metric
- Accuracy, Precision, Recall, F1-score: Used to address dataset imbalance
- Specificity and Sensitivity: Critical for screening application to minimize false negatives
3. Discussion
3.1. Clinical Integration Challenges
- Data Heterogeneity
- Model Interpretability
3.2. Future Directions
- Multimodal Learning
- Lightweight AI Models
- Federated Learning
- Explainable AI
- Conclusions
Acknowledgements
Conflict of Interest
References
- WHO, 'Cervical cancer,' [Online]. Available: https://www.who.int/health-topics/cervical-cancer.
- S. Vaccarella et al., 'Cancer screening in the era of COVID-19: Challenges and perspectives,' Int J Cancer, 2021.
- WHO, 'Global strategy to accelerate the elimination of cervical cancer as a public health problem,' 2020.
- E. E. Onuiri, 'Machine Learning Algorithms for Cervical Cancer Detection,' Int. J. Adv. Res. Comput. Sci., 2024.
- A. Akbari et al., 'A Machine Learning Approach to Predicting Cervical Cancer,' BMC Cancer, 2023.
- Y. Singh et al., 'Support Vector Machines for Cervical Cancer Prediction,' arXiv, 2018.
- P. Jiang et al., 'SVM-Based Cervical Cancer Detection,' AI Review, 2023.
- S. Mohammed et al., 'ML Approaches in Cervical Cancer,' 2024.
- S. Khare et al., 'Deep Learning for Cytology Screening,' WIREs Comput. Mol. Sci., 2024.
- R. Zhang et al., 'Deep RNNs for Medical Diagnosis,' J Biomed Inform, 2022.
- X. Wu et al., 'Transfer Learning in Cancer Detection,' IEEE Access, 2021.
- E. Selvano et al., 'Hybrid Learning Models for Cancer Screening,' 2023.
- L. Ledwaba et al., 'Ensemble Learning in Cervical Cancer,' 2024.
- R. D. Viñals et al., 'Boosting Models for Cancer Diagnosis,' Diagnostics, 2023.
- J. Jantzen, 'Pap-smear benchmark data for pattern classification,' Technical Report, 2005.
- S. Plissiti et al., 'SIPaKMeD Dataset for Cervical Cell Classification,' 2018.
- M. Silva et al., 'Histopathology Dataset for Cervical Cancer Research,' 2020.
- UCI Machine Learning Repository, 'Cervical Cancer Risk Factors Dataset.'.
- L. Frid-Adar et al., 'GANs for Data Augmentation in Medical Imaging,' IEEE TMI, 2018.
- A. Rajpurkar et al., 'CheXNet: Radiologist-Level Pneumonia Detection,' PLoS Medicine, 2018. [CrossRef]
- S. Minaee et al., 'Deep learning for COVID-19 detection from chest X-ray images,' IEEE TMI, 2021.
- N. Shah et al., 'Addressing Domain Shift in Healthcare,' J Am Med Inform Assoc, 2021.
- F. Tjoa et al., 'Explainable Artificial Intelligence (XAI) in Healthcare,' Informatics in Medicine Unlocked, 2020.
- FDA, 'Artificial Intelligence and Machine Learning in Software as a Medical Device,' White Paper, 2021.
- A. Obermeyer et al., 'Dissecting racial bias in an algorithm,' Science, 2019.
- Y. Li et al., 'Multimodal Machine Learning in Healthcare,' IEEE Access, 2022.
- S. Han et al., 'TinyML: Enabling Energy-Efficient ML,' IEEE Design & Test, 2020.
- B. Sheller et al., 'Federated Learning in Medical Imaging,' J Biomed Inform, 2020.
- M. Ribeiro et al., 'Why Should I Trust You?' Explaining Classifiers,' KDD, 2016. [CrossRef]
- S. Mehammed and D. Lemma, "Improving the Performance of Proof of Work-Based Bitcoin Mining Using CUDA," International Journal of Innovative Science and Research Technology (IJISRT), vol. 6, no. 12, pp. 1138–1143, Dec. 2021. [Online]. Available: https://zenodo.org/records/5910835/files/IJISRT21DEC657%20%281%29.pdf.
- S. Lundberg et al., 'A Unified Approach to Interpreting Model Predictions,' NIPS, 2017. [CrossRef]
- S. M. Abdu and M. N. Alam, “Adaptive Fuzzy-PSO DBSCAN: An enhanced density-based clustering approach for smart city data analysis,” Preprint, May 15, 2025. [CrossRef]
- S. M. Abdu, “Knowledge graph creation based on ontology from source-code: The case of C#,” Zenodo, Jan. 31, 2023. [CrossRef]
- S. M. Abdu, “Optimizing multi-dimensional data-index algorithms for MIC architectures,” Zenodo, Sep. 27, 2022. [CrossRef]
- S. Abdu, “Optimizing proof-of-work for secure health data blockchain using CUDA,” Blockchain in Healthcare Today, vol. 8, no. 2, Aug. 29, 2025. [Online]. Available: https://blockchainhealthcaretoday.com/index.php/journal/article/view/421.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





