1. Introduction
Invasive fungal infections represent a persistent and growing threat to global public health, particularly among immunocompromised populations, where they are associated with significant morbidity and mortality. Among the most critical fungal pathogens are
Coccidioides immitis and
Coccidioides posadasii, the dimorphic fungi responsible for coccidioidomycosis, also known as "Valley fever". These fungi are endemic to arid regions of North and South America, where they thrive in soil and are transmitted to humans via inhalation of airborne arthroconidia [
1,
2,
3].
Coccidioidomycosis manifests as a spectrum of diseases, ranging from acute or chronic pulmonary infections to severe disseminated forms that can affect the central nervous system, blood, bones, and other organs [
4,
5,
6,
7]. The burden of coccidioidomycosis is substantial, with the Centers for Disease Control and Prevention (CDC) reporting 10,000 to 40,000 cases annually in the United States alone, predominantly in Arizona and California [
8]. Alarmingly, coccidioidomycosis has been associated with approximately 200 deaths per year from 1999 to 2021 [
9]. The global significance of
Coccidioides spp. as a public health threat was further emphasized in 2022 when the World Health Organization Antimicrobial Resistance Division included these pathogens in its First Fungal Priority Pathogens List (WHO FPPL) [
10]. Despite advances in antifungal therapies, treatment options for coccidioidomycosis remain limited, often requiring prolonged courses of antifungal agents such as azoles and Amphotericin B, which are associated with toxicity, drug interactions, and the potential for resistance development [
11,
12,
13]. This situation underscores the need for novel therapeutic agents with improved safety and potency profiles.
Large-scale screening techniques have revolutionized drug discovery by enabling the rapid and systematic evaluation of chemical libraries in search of bioactive compounds [
14,
15]. These platforms have proven valuable in identifying novel therapeutic agents, including both synthetic and natural products, against various pathogenic microorganisms [
16,
17,
18,
19,
20]. However, the use of antimicrobial screening for
Coccidioides spp. is inherently limited due to the requirement for Biosafety Level 3 (BSL-3) facilities, which introduce regulatory, logistical, operational, and cost barriers [
21,
22]. As a consequence, few studies have explored or implemented screening strategies for coccidioidomycosis, and existing methods are cumbersome and low-throughput. To address these challenges, we have taken advantage of the availability of an attenuated
C. posadasii mutant strain
∆cts2/∆ard1/∆cts3 (∆T), lacking expression of two chitinase genes and one D-arabinotol-2-dehydrogenase that can be handled under BSL-2 conditions [
23]. Here we report on the development and optimization of robust 96-well and 384-well plate screening methodologies facilitating safer, more efficient antifungal discovery for coccidioidomycosis and potentially other high-containment pathogens.
2. Materials and Methods
2.1. Fungal Cultures
The attenuated mutant strain
C. posadasii C735
∆cts2/∆ard1/∆cts3 (∆T) [
23], maintained under biosafety level 2 (BSL-2) conditions, was used for all experiments in this study. Arthroconidia were propagated on glucose yeast extract (GYE) agar plates (1% glucose, 0.5% yeast extract, 1.5% agar) and incubated at 30°C for 3 weeks. Arthroconidia were harvested by scraping the surface of the plates and homogenized via bead beating in phosphate-buffered saline (PBS). Hyphal fragments were removed by filtration through a 40-μm cell strainer, followed by washing and concentration of arthroconidia through centrifugation at 2,000 rpm for 5–10 minutes. The final pellet was resuspended in PBS, and cell concentration was quantified using a disposable hemocytometer. The viability of the arthroconidia and CFUs was validated by plating on GYE plates. To induce spherule differentiation, arthroconidia were inoculated into a chemically defined Converse medium at a density of 1–3 × 10
8 spores per 100 mL [
24]. Cultures were maintained in vented polypropylene flasks at 39°C, 10% CO
2, and incubated at 180 rpm for 24 hours for arthroconidia to grow into spherules (20-40 µM in diameter). Cultures were centrifuged at 2,000 rpm for 5–10 minutes to recover spherules and filtered through a 40-μm cell strainer to remove residual debris. Spherules were resuspended in RPMI-1640 medium without phenol red buffered with 2.0 g/L sodium bicarbonate (RPMI) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and adjusted to the desired concentration based on experimental conditions.
2.2. Compounds
Niclosamide (NIC), Niclosamide ethanolamine (NEN), Hexachlorophene (HEX), and Amphotericin B (AmB) were procured from Sigma Aldrich (St. Louis, MO, USA). Antifungal solutions were prepared at 1 mM in dimethyl sulfoxide (DMSO), except for AmB, which was prepared as a stock solution at 250 µg/mL
2.3. Growth of C. posadasii in 96-Well Plates for Assay Optimization and Adaptation for Drug Screening
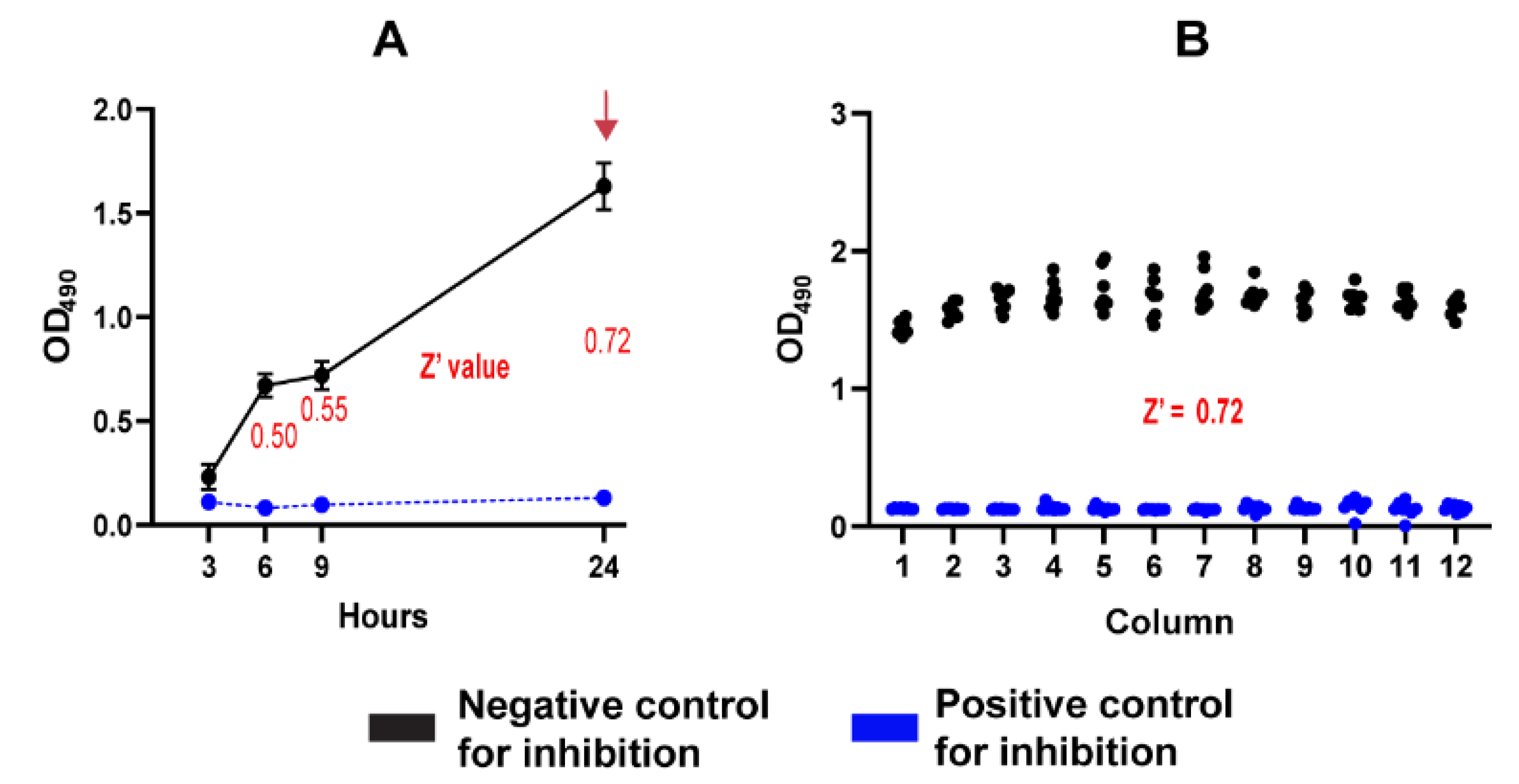
Spherule initials were prepared as previously described and adjusted to the desired working inoculum in RPMI medium. Two 96-well plate (Corning, New York, NY, USA) layouts were utilized: a positive control plate containing AmB at 10 µg/mL in all wells and a negative control plate containing DMSO at a final concentration of 1% (v/v). Each well of the control plates was seeded with the working inoculum. Plates were then incubated at 39°C with 10% CO2 for 24 hours. At the endpoint, 100 µL of 2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide (XTT) (Thermo Fisher Scientific) solution supplemented with menadione was added to each well, and the control plates were further incubated under the same conditions. XTT reduction was monitored at different timepoints by measuring absorbance at 490 nm (OD
490) using a microtiter plate reader (Synergy H1, Agilent BioTek, Santa Clara, CA, USA). Raw absorbance readings from the positive and negative control plates were used to calculate the Z' score statistics as previously described [
25,
26] as a measure of assay reliability and reproducibility.
2.4. Simulated Primary Screening Experiment Using the 96-Well Plate Assay
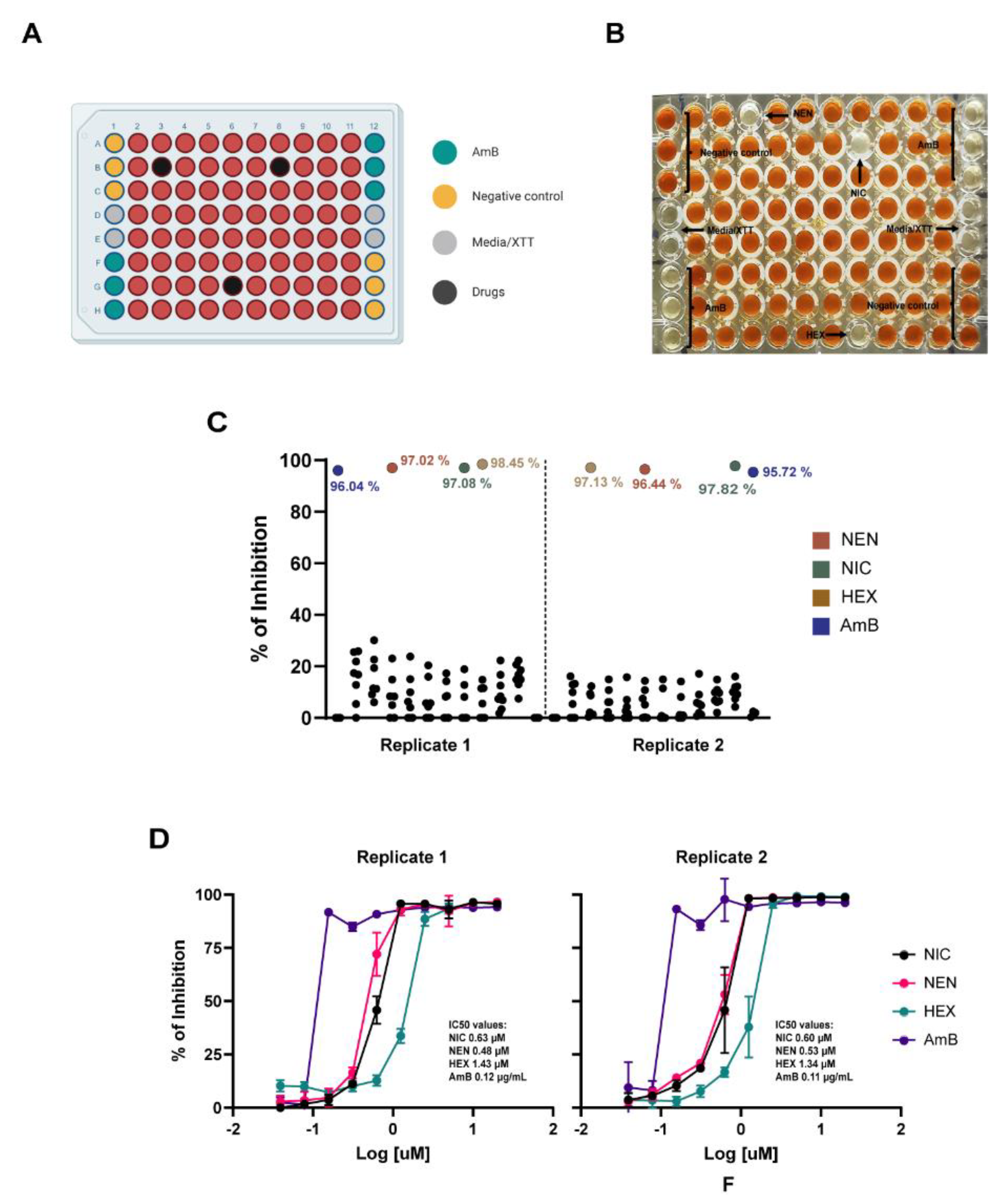
To validate the optimized antifungal screening protocol, a simulated primary screening experiment was conducted targeting C. posadasii spherules. A set of compounds with experimentally validated inhibitory activity against C. posadasii (NIC, NEN, and HEX) were selected. Duplicate "mock" screening library plates were prepared by spotting 1 µL of 1 mM stock drug solutions in DMSO into assigned wells of 96-well plates. Positive control wells contained amphotericin B (AmB) at a final concentration of 10 µg/mL, while negative and background controls included 1% DMSO and RPMI medium, respectively.
Briefly, a working inoculum of C. posadasii spherules (1 × 106 spherules/mL) in RPMI medium was prepared as described. Each well of the mock screening drug plates was seeded to a final volume of 100 µL and incubated at 39°C with 10% CO₂ for 24 hours. At the endpoint, 100 µL of XTT solution (0.5 mg/mL, supplemented with 40 µM menadione) was added to each well, followed by an additional 24-hour incubation under the same conditions. Absorbance at 490 nm was measured using a microplate reader. Raw absorbance values were normalized to calculate the percentage of inhibition, using positive and negative controls for reference. To standardize the results, percentage inhibition values exceeding 100% or falling below 0% were adjusted to 100% and 0%, respectively.
2.5. Dose-Response Experiments for Confirmation of Screening Results Using the 96-Well Plate Assay
To confirm the inhibitory activity of potential hit compounds identified during the simulated primary screening, a secondary screening experiment was conducted using a typical dose-response assay. Briefly, serial two-fold dilutions of NIC, NEN, HEX, and AmB were prepared in 96-well plates in 50 µL of RPMI medium, with concentrations ranging from 40 µM to 0.0005 µM. Each well was then seeded with 50 µL of a working spherule inoculum (2 × 106 spherules/mL), halving the compound concentrations. Plate incubation and XTT assays were conducted using the same conditions described in the screening assay above. OD490 values were measured using a microplate reader. As described previously, the percentage of growth inhibition was calculated and normalized relative to the positive (AmB) and negative (DMSO) controls for inhibition. Afterward, the inhibitory concentration required to reduce metabolic activity by 50% (IC50) was determined by fitting the normalized data to the variable slope Hill equation using GraphPad Prism (version v10.4.1). Dose-response assays were performed in duplicate, with three technical replicates per treatment at each concentration.
2.6. Growth of C. posadasii in 384-Well Plates for Assay Miniaturization, Optimization and Adaptation for Drug Screening
To miniaturize the multi-well plate protocol for use in a 384-well plate format, optimal conditions for the different parameters were extrapolated and adapted from our initial experiments using the lower-density 96-well plates. Briefly, a spherule working inoculum was prepared in RPMI medium, and dispensed into each well of the 384-well plates (Corning). Wells containing AmB (10 µg/mL) served as positive controls, while wells with non-drug media were used as negative controls. Following seeding, plates were incubated at 39°C with 10% CO₂ for 24 hours. At the endpoint, an XTT solution supplemented with menadione was added to each well, and plates were further incubated under the same conditions. XTT-reduction was monitored at the indicated time points by measuring absorbance at 490 nm using a microplate reader. Absorbance readings were subsequently used to calculate the Z' score statistics, as previously described [
27].
2.7. Simulated Primary Screening Using the 384-Well Plate Assay
Once optimal conditions for the 384-well plate assay were determined, a simulated primary screening was performed by preparing “mock” screening library plates containing NIC, NEN, and HEX. Mock screening library plates were prepared in duplicate by spotting 1 µL of 0.5 mM stock drug solutions in DMSO into assigned wells of 384-well plates. Positive control wells contained AmB at a final concentration of 10 µg/mL, while negative and background controls included DMSO and RPMI medium, respectively. Plates were then seeded with a working inoculum of C. posadasii spherules (1 × 106 spherules/mL) in RPMI medium, reaching a final volume of 50 uL per well. Plates were then incubated at 39°C with 10% CO₂ for 24 hours. After the incubation, 30 µL of XTT solution (0.5 mg/mL, supplemented with 40 µM menadione) was added to each well, followed by an additional 24-hour incubation. OD490 was monitored, and values were normalized to calculate the percentage of inhibition using positive and negative controls for reference.
2.8. Dose-Response Experiments for Confirmation of Screening Results Using the 384-Well Plate Assay
After identifying “potential hit” compounds from the mock screening library, dose-response assays were conducted to characterize their antifungal activity further. Serial two-fold dilutions of NIC, NEN, HEX, and AmB were prepared in 25 µL of RPMI. Each well was then seeded with 25 µL of a spherule working inoculum, resulting in final drug concentrations ranging from 20 µM to 0.0005 µM. Plates were incubated at 39°C with 10% CO₂ for 24 hours. Afterward, 30 µL of XTT solution (0.5 mg/mL, supplemented with 40 µM menadione) was added to each well, and plates were further incubated under the same conditions. OD490 values were obtained, and IC50 values were determined by fitting the normalized OD490 data into the variable slope Hill equation using GraphPad Prism. Experiments were performed in duplicate, with three technical replicates per treatment.
4. Discussion
The primary antifungal agents for coccidioidomycosis, such as azoles and polyenes, are limited in their effectiveness and often have significant limitations that hinder long-term use. These limitations include mainly drug toxicity, adverse side effects, and the potential for developing resistance [
33,
34]. These challenges underscore the need to discover and develop novel antifungal agents with reduced toxicity and improved therapeutic profiles [
35,
36]. One of the main strategies to overcome such challenges is using large-scale screening, including high-throughput screening technologies [
37]. For instance, by systematically applying HTS technology, it is possible to evaluate hundreds to millions of compounds for specific antifungal activity [
38]. Moreover, we can experimentally narrow the chemical space for those exhibiting specific desired properties by sourcing these compounds from specialized libraries [
39]. For example, it is possible to screen molecules from different libraries, such as FDA-approved drugs, de novo compounds with drug-like properties, diffusible blood-brain barrier molecules, etc.
Despite their widespread application in antimicrobial drug discovery, including antifungals, their use for targeting
Coccidioides spp. remains limited. A significant barrier to their implementation for
Coccidioides spp. is the requirement for Biosafety Level 3 (BSL-3) facilities, which are necessary to handle these pathogens safely [
21]. This requirement introduces considerable logistical and operational complexities for screening under high containment levels, including stringent regulatory oversight, specialized infrastructure, and increased costs, hindering their broader application in coccidioidomycosis research and limiting the discovery of new antifungal agents for this neglected disease [
23]. To our knowledge, there have been limited antifungal large-scale screening efforts against
Coccidioides spp. Recently, Mead
et al. reported screening the 1,280-compound LOPAC library, identifying active inhibitory compounds on
Coccidioides arthroconidia [
40]. The screening protocol relied on measuring OD
600 over 120 h, with fungal growth being the primary readout. Most recently, our group reported on the screening of compound libraries in search of potential antifungals that are effective against
C. posadasii spherule initials [
29]. The screening used an XTT-colorimetric technique to assess the metabolic activity of fungal elements in 96-well microtiter plates, identifying 254 potential drugs that inhibited more than 70% of the metabolism of the cells. However, the associated methodology was relatively cumbersome, performed under BSL-3 conditions, and consisted of many steps that significantly limited the utility and overall throughput of these screening techniques. At the same time, we also reported on the possibility of using an attenuated mutant strain of
C. posadasii (C735
∆cts2/∆ard1/∆cts3, or
∆T) as a surrogate for the wild-type BSL-3 strain in this type of screening assays [
23]. Thus, we posited that this attenuated strain, which can be handled under BSL-2 conditions, constitutes an ideal tool allowing for the manipulation of different experimental parameters for the optimization of the screening methodologies, thereby potentially circumventing the limitations associated with working in high containment (BSL-3) environments.
As our main target for drug discovery is spherule initials, we decided to use the existing XTT protocol described recently by our group as a starting point. Therefore, we modified key parameters to suit our experimental setup, aiming to streamline the experimental workflow and increase the throughput of potential screening efforts. To establish a robust and reproducible high-throughput screening assay, we evaluated different experimental parameters, including initial inoculum size, menadione concentration, and XTT-reduction time. By optimizing key parameters, our protocol increases the efficiency of spherule production and the throughput for screening thousands of compounds per experimental run. Of note, the experimental conditions initially established in this study address a potential limitation associated with producing sufficient spherule numbers in vitro. Spherules derived from arthroconidia only enlarge in cell size without cell division over 5-7 days. Furthermore, a mature spherule at 5-7 days post inoculation can produce 300-800 endospores, which form clusters, making it challenging to normalize cell numbers for this assay [
41]. It is reasonable to use spherule initials and mature spherules before endosporulation for these screening assays; however, preparing a large quantity of spherules is a critical limiting step for these assays. Thus, compared to our original report [
29] our optimized protocol uses 1/10th of the concentration of cells and ½ the volume in the initial inoculum per well. Thus, by itself, this has the potential to increase the throughput already by a factor of 20, even when using the lower-density 96-well microtiter plate format. Additionally, fulfilling one of our initial objectives, the optimized protocol also minimizes the number of experimental steps, particularly those that are more labor-intensive and time-consuming (i.e., washings, centrifugation, filtration, etc.) and thereby more prone to introducing operator-related error, which could potentially compromise the reproducibility of the technique [
42]. Overall, we found that the best conditions were using a working inoculum of 1x106 spherules/mL, an initial menadione concentration of 40 µM, and an XTT-reduction time of 24 h. Using the optimal experimental conditions, we consistently obtained Z’ score values exceeding 0.6 across multiple plates and experimental days for both well-plate formats. It is particularly crucial for primary screening at single compound concentrations, as this step depends on high Z’ values to identify active compounds that warrant further evaluation through dose-response analysis [
30]. It is also important to note that this protocol can be readily adapted to other BSL-2 and BSL-3
Coccidioides strains, although minor experimental adjustments may be required to optimize conditions for each specific strain.
To further validate the developed protocols, we performed a primary mock screening to assess whether potential hits could be accurately identified using our assay. The compounds chosen for this screening were a subset previously known to show activity against
Coccidioides spherules [
29]. Using both 96-well and 384-well plate formats, we successfully identified three active “hit” compounds. These hits displayed consistent inhibition percentages across replicates in both formats, demonstrating the robustness of the protocols. Moreover, the inhibition values were similar between the two plate formats, highlighting their interchangeability and adaptability for laboratories with varying equipment. Finally, we conducted follow-up experiments to simulate a typical secondary confirmation screening. Through dose-response assays, we calculated the IC
50 values for NIC, NEN, HEX, and AmB, which proved to be highly reproducible across different days. Furthermore, the IC
50 values were comparable between the 96-well and 384-well plate formats, further validating the reliability and repeatability of the protocols.
Overall, in this study, we established and optimized robust protocols for large-scale screening, including “true” high throughput screening (HTS), enabling the safe and accessible evaluation of compound libraries against Coccidioides spherule initials. To enhance accessibility and scalability, we adapted the protocol into both 96-well and 384-well plate formats, ensuring compatibility with a wide range of laboratory setups. Furthermore, by using the attenuated C. posadasii C735 ∆cts2/∆ard1/∆cts3 (∆T) strain, our protocol enables drug screening campaigns to be performed under BSL-2 facilities. This advancement not only accelerates the identification of novel antifungal agents for coccidioidomycosis but also provides a scalable framework for future research on this and other high-containment pathogens.
Author Contributions
AV-R, conceptualization, formal analysis, investigation, methodology, writing – original draft, writing – review and editing; J-JY, conceptualization, formal analysis, validation, writing – review and editing; CYH, conceptualization, formal analysis, writing – review and editing, funding acquisition; JL-R, conceptualization, formal analysis, writing – review and editing, funding acquisition. All authors have read and agreed to the published version of the manuscript.