Submitted:
19 August 2025
Posted:
21 August 2025
You are already at the latest version
Abstract
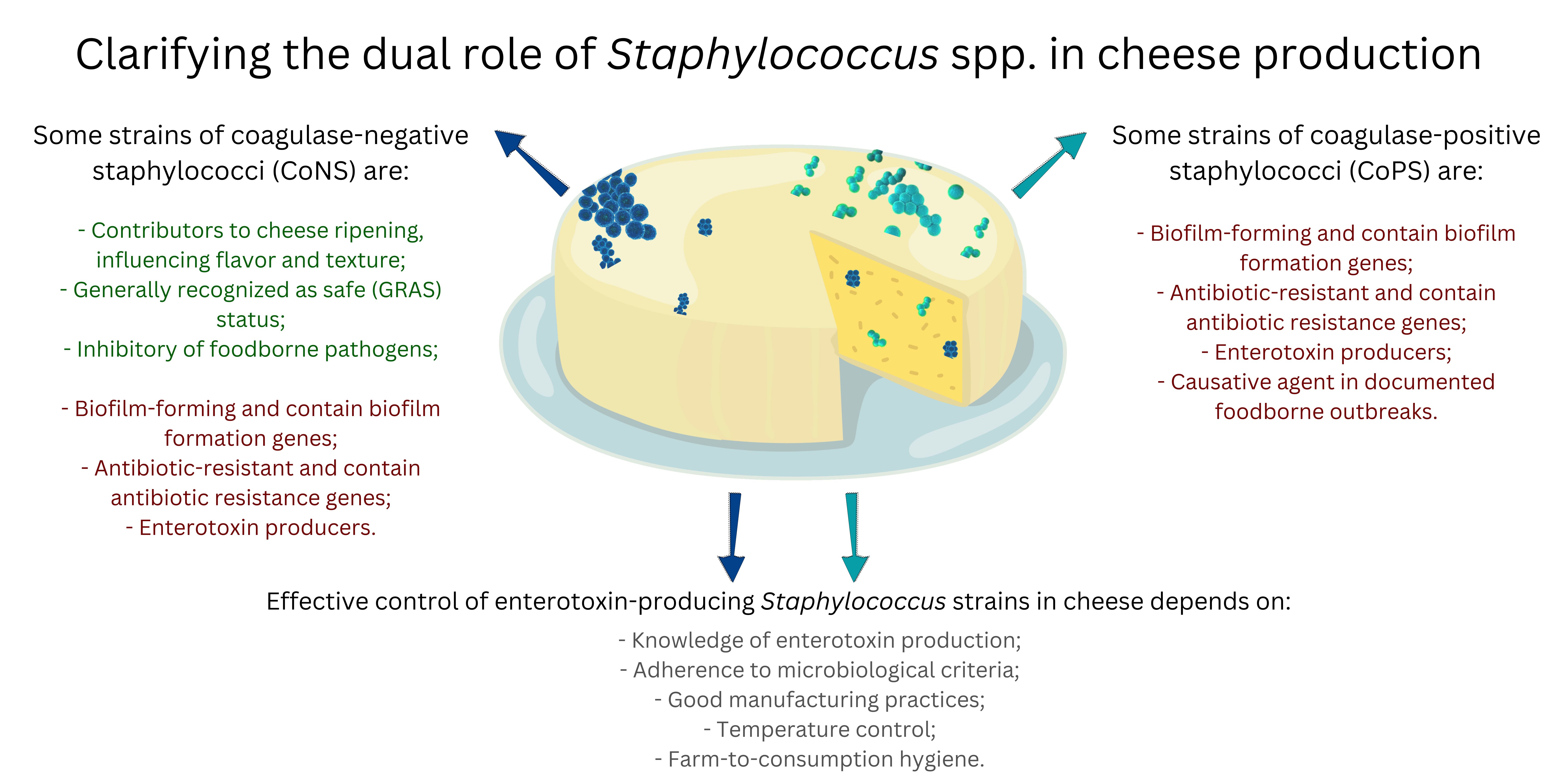
Staphylococcus spp. present a dual role during cheese production as some species may be pathogenic, while others bring beneficial characteristics to the final products. Coagulase-positive staphylococci (CoPS) species, particularly Staphylococcus aureus, are of concern due to their potential to produce enterotoxins commonly linked to foodborne outbreaks. These enterotoxins, encoded by a set of genes, can cause severe gastroenteritis, particularly vomiting. Many other members of the genus can harbor a wide range of genes encoding virulence factors and capability of forming biofilms on various surfaces. The alarming prevalence of antibiotic-resistant strains, including methicillin-resistant S. aureus (MRSA), further complicates their control. In contrast, some strains of coagulase-negative Staphylococcus species (CoNS) positively contribute to cheese ripening, influencing flavor and texture. Some strains are even considered safe for use in food production and have been studied as inhibitors of foodborne pathogens. Conversely, the expression of enterotoxin genes in some Staphylococcus species, particularly S. aureus, is regulated by different mechanisms including quorum sensing. Understanding enterotoxin gene expression in various environmental conditions, including during cheese production and ripening, can aid in developing targeted interventions. The risks posed by enterotoxin producing Staphylococcus in cheese are evident since numerous food poisoning outbreaks have been reported. Also concerning is the fact that several Staphylococcus species pose risks to animal health and livestock production. Effective control measures include adherence to microbiological criteria for CoPS and enterotoxin levels in cheeses with special attention to animal health, good manufacturing practices (GMP), temperature control, and strict hygiene protocols. This review highlights the need to balance the beneficial roles of CoNS in cheese production with the risks associated with virulent and enterotoxigenic strains of CoNS and CoPS.
Keywords:
1. Introduction
2. Coagulase-Positive Staphylococci (CoPS)
2.1. Staphylococcus aureus
2.2. Others Coagulase-Positive Staphylococci (CoPS)
3. Coagulase-Negative Staphylococci (CoNS)
Generally Recognized as Safe (GRAS) Status
4. Virulence Factors
4.1. Biofilm Formation
4.2. Antibiotic Resistance
4.3. Expression of Enterotoxins Genes
5. Staphylococcal Food Poisoning from Cheese Consumption
6. Control of Staphylococci in Cheeses
7. Microbiological Criteria for Staphylococcus and Enterotoxins in Cheeses
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CoNS | coagulase-negative Staphylococcus species |
| CoPS | Coagulase-positive staphylococci |
| CoPS | coagulase-positive |
| CoVS | coagulase-variable staphylococci |
| MRSA | methicillin-resistant S. aureus |
| MSSA | methicillin-susceptible S. aureus |
| SEs | staphylococcal enterotoxins |
| SFP | staphylococcal food poisoning |
| TSS-like | toxic shock syndrome |
| TSST-1 | like toxic shock syndrome toxin-1 |
References
- Silva, N.; Taniwaki, M.H.; Junqueira, V.C.A.; Silveira, N.; Okazaki, M.M.; Gomes, R.A.R. Microbiological Examination Methods of Food and Water, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Murray, E.; Draper, L.A.; Ross, R.P.; Hill, C. The advantages and challenges of using endolysins in a clinical setting. Viruses 2021, 13, 680. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, M.; Corbera, J.A.; Suárez-Bonnet, A.; Tejedor-Junco, M.T. Virulence factors in coagulase-positive staphylococci of veterinary interest other than Staphylococcus aureus. Vet. Q. 2020, 40, 118–131. [Google Scholar] [CrossRef]
- Becker, K.; David, M.Z.; Skov, R.L. Staphylococcus, Micrococcus, and other catalase-positive cocci. In Manual of Clinical Microbiology, 13th ed.; ASM Press: Washington, DC, USA, 2023. [Google Scholar] [CrossRef]
- Bhattacharya, C.; Harsha, P.; Gupta, S.; Roy, S. Isolation and characterization of bacterial isolates from agricultural soil at drug district. Indian J. Sci. Res. 2014, 4, 221–226. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Taxonomy browser: Staphylococcus.
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9. [Google Scholar] [CrossRef]
- Foster, T.J. Staphylococcus aureus—Molecular. In Encyclopedia of Dairy Sciences, 3rd ed.; Academic Press: London, UK, 2022. https://doi.org/10.1016/B978-0-08-100596-5.22969-7.aureus—Molecular. In Encyclopedia of Dairy Sciences, 3rd ed.; Academic Press: London, UK, 2022. [Google Scholar] [CrossRef]
- Casanova, C.; Iselin, L.; von Steiger, N.; Droz, S.; Sendi, P. Staphylococcus hyicus bacteremia in a farmer. J. Clin. Microbiol. 2011, 49. [Google Scholar] [CrossRef]
- da Silva, N.; Junqueira, V.C.A.; Silveira, N.F.A.; Taniwaki, M.H.; Gomes, R.A.R.; Okazaki, M.M. Manual de métodos de análise microbiológica de alimentos e água, 5th ed.; Blucher: São Paulo, Brazil, 2017. [Google Scholar]
- Santiago, M.C.L. Pesquisa de Staphylococcus coagulase positiva produtor da toxina 1 da síndrome do choque tóxico (TSST-1) em amostras de queijo Minas artesanal. Undergraduate Monograph, Federal University of Minas Gerais, Belo Horizonte, Brazil, 2019.
- Bennett, R.W.; Hait, J.M.; Tallent, S.M. Staphylococcus aureus. In Guide to Foodborne Pathogens, 2nd ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Kinoshita, M.; Kobayashi, N.; Nagashima, S.; Ishino, M.; Otokozawa, S.; Mise, K.; Sumi, A.; Tsutsumi, H.; Uehara, N.; Watanabe, N.; Endo, M. Diversity of staphylocoagulase and identification of novel variants of staphylocoagulase gene in Staphylococcus aureus. Microbiol. Immunol. 2008, 52, 334–348. [Google Scholar] [CrossRef]
- Wan, M.T.; Lauderdale, T.L.; Chou, C.C. Characteristics and virulence factors of livestock-associated ST9 methicillin-resistant Staphylococcus aureus with a novel recombinant staphylocoagulase type. Vet. Microbiol. 2013, 162, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Shittu, A.O.; Lin, J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect. Dis. 2006, 6, 125. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Guadarrama, N.; Olivares-Cervantes, A.L.; Salinas, E.; Martínez, L.; Escorcia, M.; Oropeza, R.; Rosas, I. Presence of environmental coagulase-positive staphylococci, their clonal relationship, resistance factors and ability to form biofilm. Rev. Argent. Microbiol. 2017, 49, 15–23. [Google Scholar] [CrossRef]
- Foronda-García-Hidalgo, C. Staphylococcus and other catalase-positive cocci. In Rezaei, N. (Ed.), Encyclopedia of Infection and Immunity; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Wirth, J.S.; Saravanan, V.S. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 5926–5936. [Google Scholar] [CrossRef]
- Le Loir, Y.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003, 2, 63–76. [Google Scholar] [PubMed]
- Jay, J.M.; Loessner, M.J.; Golden, D.A. Staphylococcal gastroenteritis. In Modern Food Microbiology, 7th ed.; Springer Science and Business Media, Inc.: New York, NY, USA, 2005. [Google Scholar]
- McGuire, E.; Boyd, A.; Woods, K. Staphylococcus aureus bacteremia. Clin. Infect. Dis. 2020, 71, 2765–2766. [Google Scholar] [CrossRef] [PubMed]
- Grispoldi, L.; Karama, M.; Armani, A.; Hadjicharalambous, C.; Cenci-Goga, B.T. Staphylococcus aureus enterotoxin in food of animal origin and staphylococcal food poisoning risk assessment from farm to table. Ital. J. Anim. Sci. 2021, 20, 677–690. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins 2018, 10. [Google Scholar] [CrossRef]
- Barber, M. Methicillin-resistant staphylococci. J. Clin. Pathol. 1961, 14, 385–393. [Google Scholar] [CrossRef]
- Chalmers, S.J.; Wylam, M.E. Methicillin-resistant Staphylococcus aureus infection and treatment options. In Ji, Y. (Ed.), Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols, Methods in Molecular Biology, vol. 2069; Humana: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Morosini, M.I.; Cercenado, E.; Ardanuy, C.; Torres, C. Detección fenotípica de mecanismos de resistencia en microorganismos grampositivos. Enferm. Infecc. Microbiol. Clin. 2012, 30, 325–332. [Google Scholar] [CrossRef]
- Stefani, S.; Varaldo, P.E. Epidemiology of methicillin-resistant staphylococci in Europe. Clin. Microbiol. Infect. 2003, 9, 1179–1186. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; Ouakrim, D.A.; Oliveira, T.C.; Struelens, M.J.; Suetens, C.; Monnet, D.L.; Strauss, R.; Mertens, K.; Struyf, T.; Catry, B.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Surveillance of antimicrobial resistance in Europe. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017; ECDC: Stockholm, Sweden, 2018. [Google Scholar]
- Ferrasso, M.M.; Gonzalez, H.L.; Timm, C.D. Staphylococcus hyicus. Arq. Inst. Biol. 2015, 82, 1–6. [Google Scholar] [CrossRef]
- Food Standards Australia New Zealand. Staphylococcus aureus. Staphylococcus aureus. Food Standards Australia New Zealand (FSANZ) 2013.
- Loeffler, A.; Lloyd, D.H. Staphylococcus intermedius: A comparative review of its role in canine disease, zoonosis, and toxigenesis. Vet. Microbiol. 1994, 39, 225–245. [Google Scholar]
- Ruscher, C.; Lübke-Becker, A.; Wleklinski, C.G.; Şoba, A.; Wieler, L.H.; Walther, B. Prevalence of methicillin-resistant Staphylococcus pseudintermedius isolated from clinical samples of companion animals and equidaes. Vet. Microbiol. 2009, 136, 197–201. [Google Scholar] [CrossRef]
- Scherer, C.B.; Botoni, L.S.; Coura, F.M.; Silva, R.O.; dos Santos, R.D.; Heinemann, M.B.; Costa-Val, A.P. Frequency and antimicrobial susceptibility of Staphylococcus pseudintermedius in dogs with otitis externa. Cienc. Rural 2018, 48, e20170738. [Google Scholar] [CrossRef]
- Devriese, L.A.; Hermans, K.; Baele, M.; Haesebrouck, F. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet. Microbiol. 2009, 133, 206–207. [Google Scholar] [CrossRef]
- Sasaki, T.; Kikuchi, K.; Tanaka, Y.; Takahashi, N.; Kamata, S.; Hiramatsu, K. Reclassification of phenotypically identified Staphylococcus intermedius strains. J. Clin. Microbiol. 2007, 45, 2770–2778. [Google Scholar] [CrossRef]
- Morris, D.O.; Rook, K.A.; Shofer, F.S.; Rankin, S.C. Screening of Staphylococcus aureus, Staphylococcus intermedius, and Staphylococcus schleiferi isolates obtained from small companion animals for antimicrobial resistance: A retrospective review of 749 isolates (2003–2004). Vet. Dermatol. 2006, 17, 332–337. [Google Scholar] [CrossRef]
- Holt, D.C.; Holden, M.T.; Tong, S.Y.; Castillo-Ramirez, S.; Clarke, L.; Quail, M.A.; Currie, B.J.; Parkhill, J.; Bentley, S.D.; Feil, E.J.; Giffard, P.M. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol. Evol. 2011, 3, 881–895. [Google Scholar] [CrossRef]
- Schaumburg, F.; Alabi, A.S.; Köck, R.; Mellmann, A.; Kremsner, P.G.; Boesch, C.; Becker, K.; Leendertz, F.H.; Peters, G. Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ. Microbiol. Rep. 2012, 4, 141–146. [Google Scholar] [CrossRef]
- Gao, J.; Ferreri, M.; Yu, F.; Liu, X.; Chen, L.; Su, J.; Han, B. Molecular types and antibiotic resistance of Staphylococcus aureus isolates from bovine mastitis in a single herd in China. Vet. J. 2012, 192, 550–552. [Google Scholar] [CrossRef]
- Stavropoulou, D.A.; de Vuyst, L.; Leroy, F. Nonconventional starter cultures of coagulase-negative staphylococci to produce animal-derived fermented foods: A SWOT analysis. J. Appl. Microbiol. 2018, 125, 1570–1586. [Google Scholar] [CrossRef]
- Hammer, P.; Jordan, J.; Jacobs, C.; Klempt, M. Characterization of coagulase-negative staphylococci from brining baths in Germany. J. Dairy Sci. 2019, 102, 8734–8744. [Google Scholar] [CrossRef] [PubMed]
- Artini, M.; Papa, R.; Scoarughi, G.L.; Galano, E.; Barbato, G.; Pucci, P.; Selan, L. Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 2013, 114, 266–277. [Google Scholar] [CrossRef]
- Fernandes, P.B.; Reed, P.; Monteiro, J.M.; Pinho, M.G. Revisiting the role of VraTSR in Staphylococcus aureus response to cell wall-targeting antibiotics. J. Bacteriol. 2022, 204, e00162–22. [Google Scholar] [CrossRef]
- Gajewska, J.; Zakrzewski, A.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Meta-analysis of the global occurrence of S. aureus in raw cattle milk and artisanal cheeses. Food Control 2023, 147, 109603. [Google Scholar] [CrossRef]
- Morot-Bizot, S.C.; Leroy, S.; Talon, R. Staphylococcal community of a small unit manufacturing traditional dry fermented sausages. Int. J. Food Microbiol. 2006, 108, 210–217. [Google Scholar] [CrossRef]
- Zell, C.; Resch, M.; Rosenstein, R.; Albrecht, T.; Hertel, C.; Götz, F. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int. J. Food Microbiol. 2008, 127, 246–251. [Google Scholar] [CrossRef]
- Heo, S.; Lee, J.H.; Jeong, D.W. Food-derived coagulase-negative Staphylococcus as starter cultures for fermented foods. Food Sci. Biotechnol. 2020, 29, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Martín, B.; Garriga, M.; Hugas, M.; Bover-Cid, S.; Veciana-Nogués, M.T.; Aymerich, T. Molecular, technological and safety characterization of Gram-positive catalase-positive cocci from slightly fermented sausages. Int. J. Food Microbiol. 2006, 107, 148–158. [Google Scholar] [CrossRef] [PubMed]
- McKay, A.M. Antimicrobial resistance and heat sensitivity of oxacillin-resistant, mecA-positive Staphylococcus spp. from unpasteurized milk. J. Food Prot. 2008, 71, 186–190. [Google Scholar] [CrossRef]
- Resch, M.; Nagel, V.; Hertel, C. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int. J. Food Microbiol. 2008, 127, 99–104. [Google Scholar] [CrossRef]
- Nanoukon, C.; Argemi, X.; Sogbo, F.; Orekan, J.; Keller, D.; Affolabi, D.; Schramm, F.; Riegel, P.; Baba-Moussa, L.; Prévost, G. Pathogenic features of clinically significant coagulase-negative staphylococci in hospital and community infections in Benin. Int. J. Med. Microbiol. 2017, 307, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Both, A.; Weibelberg, S.; Heilmann, C.; Rohde, H. Emergence of coagulase-negative staphylococci. Expert Rev. Anti-Infect. Ther. 2020, 18, 349–366. [Google Scholar] [CrossRef]
- Pereira, V.C.; Romero, L.C.; Pinheiro-Hubinger, L.; Oliveira, A.; Martins, K.B.; da Cunha, M.L.R.S. Coagulase-negative staphylococci: A 20-year study on the antimicrobial resistance profile of blood culture isolates from a teaching hospital. Braz. J. Infect. Dis. 2020, 24, 160–169. [Google Scholar] [CrossRef]
- Sender, G.; Pawlik, A.; Korwin-Kossakowska, A. Current concepts on the impact of coagulase-negative staphylococci causing bovine mastitis as a threat to human and animal health: A review. Anim. Sci. Pap. Rep. 2017, 35, 123–135. [Google Scholar]
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef]
- Madhusoodanan, J.; Seo, K.S.; Remortel, B.; Park, J.Y.; Hwang, S.Y.; Fox, L.K.; Park, Y.H.; Deobald, C.F.; Wang, D.; Liu, S.; Daugherty, S.C.; Gill, A.L.; Bohach, G.A.; Gill, S.R. An enterotoxin-bearing pathogenicity island in Staphylococcus epidermidis. J. Bacteriol. 2011, 193, 1854. [Google Scholar] [CrossRef]
- Seitter, M.; Nerz, C.; Rosenstein, R.; Götz, F.; Hertel, C. DNA microarray-based detection of genes involved in safety and technologically relevant properties of food-associated coagulase-negative staphylococci. Int. J. Food Microbiol. 2011, 145, 449–458. [Google Scholar] [CrossRef]
- Lina, G.; Bohach, G.A.; Nair, S.P.; Hiramatsu, K.; Jouvin-Marche, E.; Mariuzza, R. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 2004, 189, 2334–2336. [Google Scholar] [CrossRef]
- Cieza, M.Y.R.; Bonsaglia, E.C.R.; Rall, V.L.M.; Santos, M.V.; Silva, N.C.C. Staphylococcal enterotoxins: Description and importance in food. Pathogens 2024, 13, 676. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; Ercolini, D.; Pennacchia, C.; Fusco, V.; Casaburi, A.; Pepe, O.; Villani, F. PCR detection of staphylococcal enterotoxin genes in Staphylococcus spp. strains isolated from meat and dairy products: Evidence for new variants of seG and seI in S. aureus AB-8802. J. Appl. Microbiol. 2004, 97, 719–730. [Google Scholar] [CrossRef]
- Nunes, R.S.C.; de Souza, C.P.; Pereira, K.S.; Del Aguila, E.M.; Paschoalin, V.M.F. Identification and molecular phylogeny of coagulase-negative staphylococci isolates from Minas Frescal cheese in southeastern Brazil: Superantigenic toxin production and antibiotic resistance. J. Dairy Sci. 2016, 99, 2641–2653. [Google Scholar] [CrossRef]
- Andrade, A.P.C.; Borges, M.F.; de Figueiredo, E.A.T.; Arcuri, E.F. Diversity of Staphylococcus coagulase-positive and negative strains of coalho cheese and detection of enterotoxin encoding genes. Bol. Cent. Pesqui. Process. Aliment. 2018, 36, e57553. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Gajewska, J.; Wiśniewski, P.; Zadernowska, A. Enterotoxigenic potential of coagulase-negative staphylococci from ready-to-eat food. Pathogens 2020, 9, 734. [Google Scholar] [CrossRef]
- von Eiff, C.; Peters, G.; Heilmann, C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2002, 2, 677–685. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef]
- von Eiff, C.; Jansen, B.; Kohnen, W.; Becker, K. Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs 2005, 65, 179–214. [Google Scholar] [CrossRef]
- Kuroda, M.; Yamashita, A.; Hirakawa, H.; Kumano, M.; Morikawa, K.; Higashide, M.; Maruyama, A.; Inose, Y.; Matoba, K.; Toh, H.; Kuhara, S.; Hattori, M.; Ohta, T. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 2005, 102, 13272–13277. [Google Scholar] [CrossRef]
- Hammes, W.P.; Hertel, C. New developments in meat starter cultures. Meat Sci. 1998, 49 (Suppl. 1), S125–S138. [Google Scholar] [CrossRef]
- Barrière, C.; Centeno, D.; Lebert, A.; Leroy-Sétrin, S.; Berdagué, J.L.; Talon, R. Roles of superoxide dismutase and catalase of Staphylococcus xylosus in the inhibition of linoleic acid oxidation. FEMS Microbiol. Lett. 2001, 201, 181–185. [Google Scholar] [CrossRef]
- Jeong, D.W.; Lee, B.; Her, J.Y.; Lee, K.G.; Lee, J.H. Safety and technological characterization of coagulase-negative staphylococci isolates from traditional Korean fermented soybean foods for starter development. Int. J. Food Microbiol. 2016, 236, 9–16. [Google Scholar] [CrossRef]
- Götz, F. Staphylococcus carnosus: a new host organism for gene cloning and protein production. J. Appl. Bacteriol. 1990, 69 (Suppl. 19), 49S–53S. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, R.; Nerz, C.; Biswas, L.; Resch, A.; Raddatz, G.; Schuster, S.C.; Götz, F. Genome analysis of the meat starter culture bacterium Staphylococcus carnosus TM300. Appl. Environ. Microbiol. 2009, 75, 811–822. [Google Scholar] [CrossRef]
- Rosenstein, R.; Götz, F. Genomic differences between the food-grade Staphylococcus carnosus and pathogenic staphylococcal species. Int. J. Med. Microbiol. 2010, 300, 104–108. [Google Scholar] [CrossRef]
- Carnio, M.C.; Stachelhaus, T.; Francis, K.P.; Scherer, S. Pyridinyl polythiazole class peptide antibiotic micrococcin P1, secreted by foodborne Staphylococcus equorum WS2733, is biosynthesized nonribosomally. Eur. J. Biochem. 2001, 268, 6390–6401. [Google Scholar] [CrossRef]
- Deetae, P.; Bonnarme, P.; Spinnler, H.E.; Helinck, S. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 2007, 76, 1161–1171. [Google Scholar] [CrossRef]
- Ravyts, F.; Steen, L.; Goemaere, O.; Paelinck, H.; De Vuyst, L.; Leroy, F. The application of staphylococci with flavour-generating potential is affected by acidification in fermented dry sausages. Food Microbiol. 2010, 27, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.; Toldrá, F. Microbial enzymatic activities for improved fermented meats. Trends Food Sci. Technol. 2011, 22, 81–90. [Google Scholar] [CrossRef]
- Lee, J.H.; Heo, S.; Jeong, D.W. Genomic insights into Staphylococcus equorum KS1039 as a potential starter culture for the fermentation of high-salt foods. BMC Genomics 2018, 19, 4532. [Google Scholar] [CrossRef]
- Irlinger, F.; Loux, V.; Bento, P.; Gibrat, J.F.; Straub, C.; Bonnarme, P.; Landaud, S.; Monnet, C. Genome sequence of Staphylococcus equorum subsp. equorum Mu2, isolated from a French smear-ripened cheese. J. Bacteriol. 2012, 194, 5141–5142. [Google Scholar] [CrossRef]
- Jeong, D.W.; Lee, J.H. Complete genome sequence of Staphylococcus succinus 14BME20 isolated from a traditional Korean fermented soybean food. Genome Announc. 2017, 5, e01731–16. [Google Scholar] [CrossRef]
- Figueiredo, A.M.S.; Mamizuka, E.M.; McCulloch, J.A. Staphylococcus aureus. In Microbiologia, 7th ed.; Alterthum, F., Ed.; Atheneu: São Paulo, Brazil, 2024. [Google Scholar]
- Donlan, R.M. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Araújo, P.; Lemos, M.; Mergulhão, F.; Melo, L.; Simões, M. Antimicrobial resistance to disinfectants in biofilms. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Formatex: Badajoz, Spain, 2011; pp. 826–834. [Google Scholar]
- Fontes, C.O.; Silva, V.L.; de Paiva, M.R.B.; Garcia, R.A.; Resende, J.A.; Ferreira-Machado, A.B.; Diniz, C.G. Prevalence, antimicrobial resistance, and virulence characteristics of mecA-encoding coagulase-negative staphylococci isolated from soft cheese in Brazil. J. Food Sci. 2013, 78, M1–M8. [Google Scholar] [CrossRef]
- Friedriczewski, A.B.; Gandra, E.Á.; da Conceição, R.D.C.D.S.; Cereser, N.D.; Moreira, L.M.; Timm, C.D. Biofilm formation by coagulase-positive Staphylococcus aureus isolated from mozzarella cheese elaborated with buffalo milk and its effect on sensitivity to sanitizers. Acta Sci. Vet. 2018, 46, 6. [Google Scholar] [CrossRef]
- Pineda, A.P.A.; Chacón, R.D.; Costa, T.G.; Campos, G.Z.; Munive Nuñez, K.V.; Ramos, R.C.Z.; Camargo, C.H.; Lacorte, G.A.; Silva, N.C.C.; Pinto, U.M. Molecular characterization and virulence potential of Staphylococcus aureus from raw milk artisanal cheeses. Int. Dairy J. 2025, 160, 106097. [Google Scholar] [CrossRef]
- Carvalho, L.G.; Alvim, M.M.A.; Fabri, R.L.; Apolônio, A.C.M. Staphylococcus aureus biofilm formation in Minas Frescal cheese packaging. Int. J. Dairy Technol. 2021, 74, 575–580. [Google Scholar] [CrossRef]
- , J. ; Chajęcka-Wierzchowska, W. Biofilm formation ability and presence of adhesion genes among coagulase-negative and coagulase-positive staphylococci isolates from raw cow's milk. Pathogens 2020, 9, 654. [Google Scholar] [CrossRef]
- Goetz, C.; Tremblay, Y.D.N.; Lamarche, D.; Blondeau, A.; Gaudreau, A.M.; Labrie, J.; Malouin, F.; Jacques, M. Coagulase-negative staphylococci species affect biofilm formation of other coagulase-negative and coagulase-positive staphylococci. J. Dairy Sci. 2017, 100, 6454–6464. [Google Scholar] [CrossRef]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics 2023, 12, 12. [Google Scholar] [CrossRef]
- Wu, X.; Wang, H.; Xiong, J.; Yang, G.X.; Hu, J.F.; Zhu, Q.; Chen, Z. Staphylococcus aureus biofilm: Formulation, regulatory, and emerging natural products-derived therapeutics. Biofilm 2024, 7, 100175. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026–19. [Google Scholar] [CrossRef]
- Cheng, C.S.; Jiang, T.; Zhang, D.W.; Wang, H.Y.; Fang, T.; Li, C.C. Attachment characteristics and kinetics of biofilm formation by Staphylococcus aureus on ready-to-eat cooked beef contact surfaces. J. Food Sci. 2023, 88, 2595–2610. [Google Scholar] [CrossRef] [PubMed]
- Wolz, C.; Goerke, C.; Landmann, R.; Zimmerli, W.; Fluckiger, U. Transcription of clumping factor A in attached and unattached Staphylococcus aureus in vitro and during device-related infection. Infect. Immun. 2002, 70, 2758–2762. [Google Scholar] [CrossRef]
- Sabat, A.; Melles, D.C.; Martirosian, G.; Grundmann, H.; Van Belkum, A.; Hryniewicz, W. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J. Clin. Microbiol. 2006, 44, 1135–1138. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- Maikranz, E.; Spengler, C.; Thewes, N.; Thewes, A.; Nolle, F.; Jung, P.; Bischoff, M.; Santen, L.; Jacobs, K. Different binding mechanisms of Staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale 2020, 12, 19267–19275. [Google Scholar] [CrossRef] [PubMed]
- Burel, C.; Dreyfus, R.; Purevdorj-Gage, L. Physical mechanisms driving the reversible aggregation of Staphylococcus aureus and response to antimicrobials. Sci. Rep. 2021, 11, 94457. [Google Scholar] [CrossRef] [PubMed]
- Sedarat, Z.; Taylor-Robinson, A.W. Biofilm formation by pathogenic bacteria: Applying a Staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens 2022, 11, 388. [Google Scholar] [CrossRef]
- Souza, M.A.M.; da Silva, G.C.; Lopes, U.; Rosa, J.N.; Bazzolli, D.M.S.; Pimentel-Filho, N.J. Characterisation of Staphylococcus aureus isolated from artisanal unripened cheeses produced in São Paulo State, Brazil. Int. Dairy J. 2024, 149, 105825. [Google Scholar] [CrossRef]
- Schroeder, K.; Jularic, M.; Horsburgh, S.M.; Hirschhausen, N.; Neumann, C.; Bertling, A.; Schulte, A.; Foster, S.; Kehrel, B.E.; Peters, G.; Heilmann, C. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS ONE 2009, 4. [Google Scholar] [CrossRef]
- Banner, M.A.; Cunniffe, J.G.; Macintosh, R.L.; Foster, T.J.; Rohde, H.; Mack, D.; Hoyes, E.; Derrick, J.; Upton, M.; Handley, P.S. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J. Bacteriol. 2007, 189, 2793–2804. [Google Scholar] [CrossRef] [PubMed]
- Corrigan, R.M.; Rigby, D.; Handley, P.; Foster, T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 2007, 153, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Rohde, H.; Burdelski, C.; Bartscht, K.; Hussain, M.; Buck, F.; Horstkotte, M.A.; Knobloch, J.K.M.; Heilmann, C.; Herrmann, M.; Mack, D. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 2005, 55, 1883–1895. [Google Scholar] [CrossRef]
- Reiter, K.C.; Villa, B.; Paim, T.G.S.; Sambrano, G.E.; Oliveira, C.F.; D’Azevedo, P.A. Enhancement of antistaphylococcal activities of six antimicrobials against sasG-negative methicillin-susceptible Staphylococcus aureus: An in vitro biofilm model. Diagn. Microbiol. Infect. Dis. 2012, 74, 101–105. [Google Scholar] [CrossRef]
- Savolainen, K.; Paulin, L.; Westerlund-Wikström, B.; Foster, T.J.; Korhonen, T.K.; Kuusela, P. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 2001, 69, 3013–3020. [Google Scholar] [CrossRef]
- Juuti, K.M.; Sinha, B.; Werbick, C.; Peters, G.; Kuusela, P.I. Reduced adherence and host cell invasion by methicillin-resistant Staphylococcus aureus expressing the surface protein Pls. J. Infect. Dis. 2004, 189, 1574–1584. [Google Scholar] [CrossRef]
- Hussain, M.; Schäfer, D.; Juuti, K.M.; Peters, G.; Haslinger-Löffler, B.; Kuusela, P.I.; Sinha, B. Expression of Pls (plasmin sensitive) in Staphylococcus aureus negative for pls reduces adherence and cellular invasion and acts by steric hindrance. J. Infect. Dis. 2009, 200, 107–117. [Google Scholar] [CrossRef]
- Artini, M.; Cellini, A.; Papa, R.; Tilotta, M.; Scoarughi, G.L.; Gazzola, S.; Fontana, C.; Tempera, G.; Cocconcelli, P.S.; Selan, L. Adhesive behaviour and virulence of coagulase negative staphylococci isolated from Italian cheeses. Int. J. Immunopathol. Pharmacol. 2015, 28, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, C.; Hartleib, J.; Hussain, M.S.; Peters, G. The multifunctional Staphylococcus aureus autolysin Aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 2005, 73, 4793–4802. [Google Scholar] [CrossRef]
- Heilmann, C.; Thumm, G.; Chhatwal, G.S.; Hartleib, J.; Uekötter, A.; Peters, G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 2003, 149, 2769–2778. [Google Scholar] [CrossRef]
- Zoll, S.; Schlag, M.; Shkumatov, A.V.; Rautenberg, M.; Svergun, D.I.; Götz, F.; Stehle, T. Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J. Bacteriol. 2012, 194, 3789–3802. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Mangwani, N.; Kumari, S.; Das, S. Bacterial biofilms and quorum sensing: fidelity in bioremediation technology. Biotechnol. Genet. Eng. Rev. 2016, 32, 43–73. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [CrossRef]
- Herman-Bausier, P.; El-Kirat-Chatel, S.; Foster, T.J.; Geoghegan, J.A.; Dufrêne, Y.F. Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. mBio 2015, 6, e00413–15. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Nguyen, T.H.; Otto, M. The staphylococcal exopolysaccharide PIA – Biosynthesis and role in biofilm formation, colonization, and infection. Comput. Struct. Biotechnol. J. 2020, 18, 3324. [Google Scholar] [CrossRef] [PubMed]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Götz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67. [Google Scholar] [CrossRef] [PubMed]
- Cramton, S.E.; Ulrich, M.; Götz, F.; Döring, G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, Í.; Penadés, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Schaeffer, C.R.; Woods, K.M.; Longo, G.M.; Kiedrowski, M.R.; Paharik, A.E.; Büttner, H.; Christner, M.; Boissy, R.J.; Horswill, A.R.; Rohde, H.; Fey, P.D. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 2015, 83, 214–226. [Google Scholar] [CrossRef]
- Rohde, H.; Burandt, E.C.; Siemssen, N.; Frommelt, L.; Burdelski, C.; Wurster, S.; Scherpe, S.; Davies, A.P.; Harris, L.G.; Horstkotte, M.A.; Knobloch, J.K.M.; Ragunath, C.; Kaplan, J.B.; Mack, D. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 2007, 28, 1711–1720. [Google Scholar] [CrossRef]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; Dodson, R.J.; Daugherty, S.C.; Madupu, R.; Angiuoli, S.V.; Durkin, A.S.; Haft, D.H.; Vamathevan, J.; Khouri, H.; Utterback, T.; Fraser, C.M. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res. Int. 2015, 759348. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Gupta, P.; Sarkar, S.; Das, B.; Bhattacharjee, S.; Tribedi, P. Biofilm, pathogenesis and prevention – a journey to break the wall: a review. Arch. Microbiol. 2016, 198, 1–15. [Google Scholar] [CrossRef]
- Costerton, J.W. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Boles, B.R.; Horswill, A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011, 19, 449–455. [Google Scholar] [CrossRef]
- Vandana, D.S. Genetic regulation, biosynthesis and applications of extracellular polysaccharides of the biofilm matrix of bacteria. Carbohydr. Polym. 2022, 291, 119536. [Google Scholar] [CrossRef]
- Periasamy, S.; Joo, H.S.; Duong, A.C.; Bach, T.H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef]
- Cheung, G.Y.; Joo, H.S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins – critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Marinelli, P.; Pallares, I.; Navarro, S.; Ventura, S. Dissecting the contribution of Staphylococcus aureus α-phenol-soluble modulins to biofilm amyloid structure. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Pabst, B.; Pitts, B.; Lauchnor, E.; Stewart, P.S. Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob. Agents Chemother. 2016, 60, 6294–6301. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef]
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and small colony variants – an update on Staphylococcus aureus strategies toward drug resistance. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef]
- Conlon, B.P.; Rowe, S.E.; Gandt, A.B.; Nuxoll, A.S.; Donegan, N.P.; Zalis, E.A.; Clair, G.; Adkins, J.N.; Cheung, A.L.; Lewis, K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 2016, 1. [Google Scholar] [CrossRef]
- Hart, J.W.; Waigh, T.A.; Lu, J.R.; Roberts, I.S. Microrheology and spatial heterogeneity of Staphylococcus aureus biofilms modulated by hydrodynamic shear and biofilm-degrading enzymes. Langmuir 2019, 35, 3553–3561. [Google Scholar] [CrossRef]
- Rivera, E.S.; Weiss, A.; Migas, L.G.; Freiberg, J.A.; Djambazova, K.V.; Neumann, E.K.; Van de Plas, R.; Spraggins, J.M.; Skaar, E.P.; Caprioli, R.M. Imaging mass spectrometry reveals complex lipid distributions across Staphylococcus aureus biofilm layers. J. Mass Spectrom. Adv. Clin. Lab 2022, 26, 36–46. [Google Scholar] [CrossRef] [PubMed]
- França, A.; Gaio, V.; Lopes, N.; Melo, L.D.R. Virulence factors in coagulase-negative staphylococci. Pathogens 2021, 10, 1–46. [Google Scholar] [CrossRef]
- Teuber, M. Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 2001, 4, 493–499. [Google Scholar] [CrossRef]
- Wang, R.; Braughton, K.R.; Kretschmer, D.; Bach, T.H.; Queck, S.Y.; Li, M.; Kennedy, A.D.; Dorward, D.W.; Klebanoff, S.J.; Peschel, A.; DeLeo, F.R.; Otto, M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007, 13, 1510–1514. [Google Scholar] [CrossRef]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67. [Google Scholar] [CrossRef]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular mechanisms of drug resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanism of staphylococcal resistance to clinically relevant antibiotics. Drug Resist. Updat. 2024, 77, 101147. [Google Scholar] [CrossRef]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, R.A.C.; Ferreira, F.A.; Rubio Cieza, M.Y.; Silva, N.C.C.; Miotto, M.; Carvalho, M.M.; Bazzo, B.R.; Botelho, L.A.B.; Dias, R.S.; De Dea Lindner, J. Staphylococcus aureus isolated from traditional artisanal raw milk cheese from Southern Brazil: Diversity, virulence, and antimicrobial resistance profile. J. Food Prot. 2024, 87. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk, A.; Mlynarczyk, B.; Kmera-Muszynska, M.; Majewski, S.; Mlynarczyk, G. Mechanisms of the resistance and tolerance to beta-lactam and glycopeptide antibiotics in pathogenic gram-positive cocci. Mini Rev. Med. Chem. 2009, 9, 1527–1537. [Google Scholar] [CrossRef]
- Tulinski, P.; Fluit, A.C.; Wagenaar, J.A.; Mevius, D.; van de Vijver, L.; Duim, B. Methicillin-resistant coagulase-negative staphylococci on pig farms as a reservoir of heterogeneous staphylococcal cassette chromosome mec elements. Appl. Environ. Microbiol. 2012, 78, 299–304. [Google Scholar] [CrossRef]
- Miragaia, M. Factors contributing to the evolution of mecA-mediated β-lactam resistance in staphylococci: Update and new insights from whole genome sequencing (WGS). Front. Microbiol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Appelbaum, P.C.; Bozdogan, B. Vancomycin resistance in Staphylococcus aureus. Clin. Lab. Med. 2004, 24, 381–402. [Google Scholar] [CrossRef]
- Rehm, S.J.; Tice, A. Staphylococcus aureus: Methicillin-susceptible S. aureus to methicillin-resistant S. aureus and vancomycin-resistant S. aureus. Clin. Infect. Dis. 2010, 51 (Suppl. 2), S176–S182. [Google Scholar] [CrossRef]
- Saha, B.; Singh, A.K.; Ghosh, A.; Bal, M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 2008, 57 Pt 1, 72–79. [Google Scholar] [CrossRef]
- Zhu, W.; Clark, N.C.; McDougal, L.K.; Hageman, J.; McDonald, L.C.; Patel, J.B. Vancomycin-resistant Staphylococcus aureus isolates associated with Inc18-like vanA plasmids in Michigan. Antimicrob. Agents Chemother. 2008, 52, 452–457. [Google Scholar] [CrossRef]
- Limbago, B.M.; Kallen, A.J.; Zhu, W.; Eggers, P.; McDougal, L.K.; Albrecht, V.S. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J. Clin. Microbiol. 2014, 52, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar] [PubMed]
- Cameron, D.R.; Howden, B.P.; Peleg, A.Y. The interface between antibiotic resistance and virulence in Staphylococcus aureus and its impact upon clinical outcomes. Clin. Infect. Dis. 2011, 53, 576–582. [Google Scholar] [CrossRef]
- Alkuraythi, D.M.; Alkhulaifi, M.M.; Binjomah, A.Z.; Alarwi, M.; Mujallad, M.I.; Alharbi, S.A.; Alshomrani, M.; Gojobori, T.; Alajel, S.M. Comparative genomic analysis of antibiotic resistance and virulence genes in Staphylococcus aureus isolates from patients and retail meat. Front. Cell. Infect. Microbiol. 2023, 13, 1339339. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Dadashi, M.; Chegini, Z.; van Belkum, A.; Mirzaii, M.; Khoramrooz, S.S.; Darban-Sarokhalil, D. The global prevalence of daptomycin, tigecycline, quinupristin/dalfopristin, and linezolid-resistant Staphylococcus aureus and coagulase-negative staphylococci strains: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2020, 9, 56. [Google Scholar] [CrossRef]
- Podkowik, M.; Bystroń, J.; Bania, J. Genotypes, antibiotic resistance, and virulence factors of staphylococci from ready-to-eat food. Foodborne Pathog. Dis. 2012, 9, 91–93. [Google Scholar] [CrossRef]
- Klempt, M.; Franz, C.M.A.P.; Hammer, P. Characterization of coagulase-negative staphylococci and macrococci isolated from cheese in Germany. J. Dairy Sci. 2022, 105, 7951–7958. [Google Scholar] [CrossRef]
- Gajewska, J.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Occurrence and Characteristics of Staphylococcus aureus strains along the production chain of raw milk cheeses in Poland. Molecules 2022, 27. [Google Scholar] [CrossRef]
- Allaion, J.R.; Barrionuevo, K.G.; Grande Burgos, M.J.; Gálvez, A.; Franco, B.D.G.M. Staphylococcus aureus from Minas artisanal cheeses: Biocide tolerance, antibiotic resistance and enterotoxin genes. Appl. Sci. 2022, 12. [Google Scholar] [CrossRef]
- Fox, E.M.; Jiang, Y.; Tinoco, M.B. Staphylococcus aureus – Dairy. In Encyclopedia of Dairy. In Encyclopedia of Dairy Sciences, 3rd ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 123–135. [Google Scholar]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Enterotoxin-producing Staphylococcus aureus. In Molecular Medical Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2015; pp. 1234–1245. [Google Scholar]
- Alibayov, B.; Baba-Moussa, L.; Sina, H.; Zdeňková, K.; Demnerová, K. Staphylococcus aureus mobile genetic elements. Mol. Biol. Rep. 2014, 41, 5005–5018. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sánchez, S.; Ramos, I.M.; Rodríguez-Pérez, M.; Poveda, J.M.; Seseña, S.; Palop, M.L. Lactic acid bacteria as biocontrol agents to reduce Staphylococcus aureus growth, enterotoxin production and virulence gene expression. LWT 2022, 170, 114025. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front. Microbiol. 2018, 9, 436. [Google Scholar] [CrossRef]
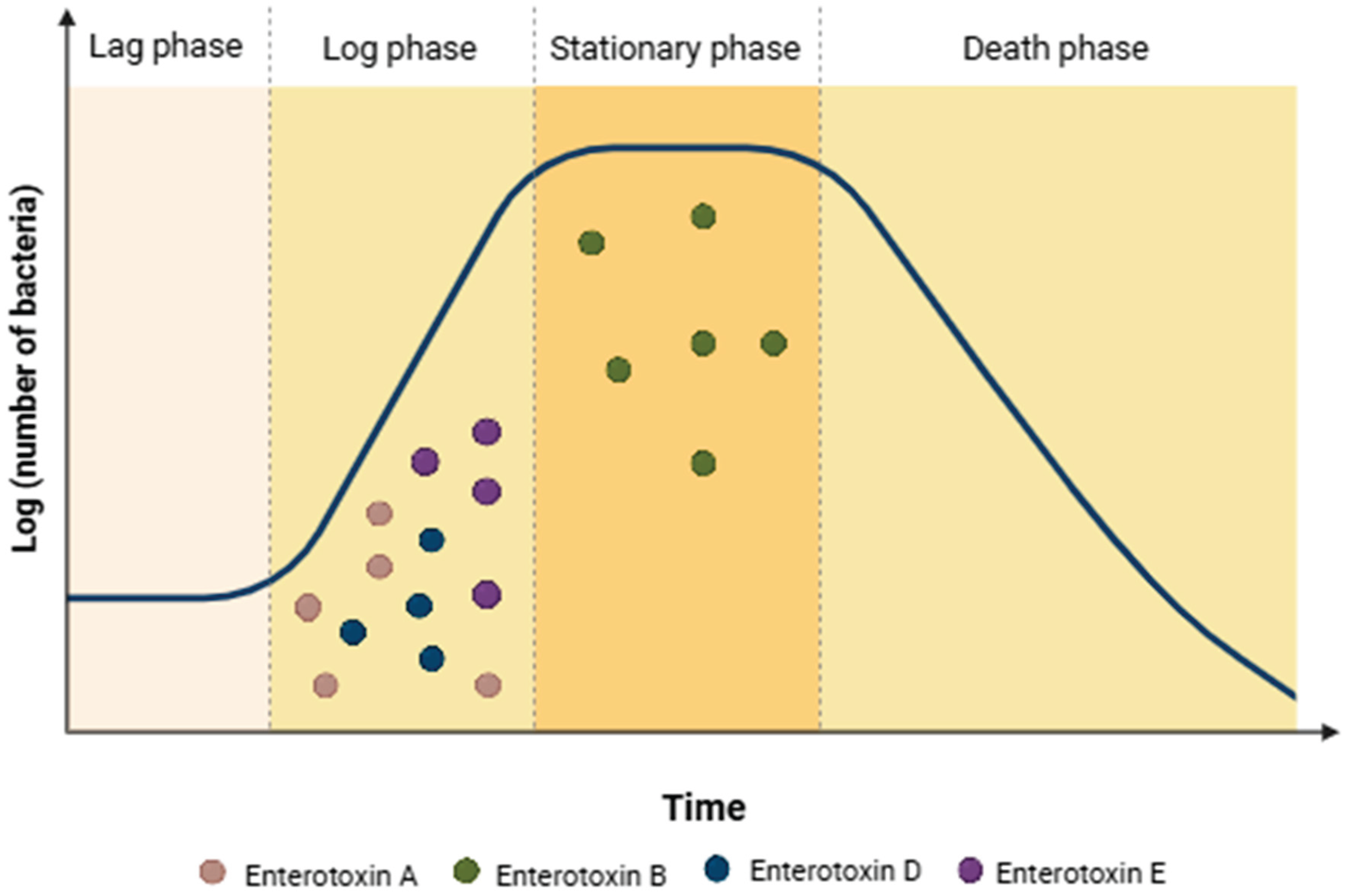
- Derzelle, S.; Dilasser, F.; Duquenne, M.; Deperrois, V. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 2009, 26, 896–904. [Google Scholar] [CrossRef]
- Zeaki, N.; Johler, S.; Skandamis, P.N.; Schelin, J. The role of regulatory mechanisms and environmental parameters in staphylococcal food poisoning and resulting challenges to risk assessment. Front. Microbiol. 2019, 10, 1307. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef]
- Thomas, M.S.; Wigneshweraraj, S. Regulation of virulence gene expression. Virulence 2014, 5, 832. [Google Scholar] [CrossRef]
- Olson, M.E.; Nygaard, T.K.; Ackermann, L.; Watkins, R.L.; Zurek, O.W.; Pallister, K.B.; Griffith, S.; Kiedrowski, M.R.; Flack, C.E.; Kavanaugh, J.S.; Kreiswirth, B.N.; Horswill, A.R.; Voyich, J.M. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 2013, 81, 1316–1324. [Google Scholar] [CrossRef]
- Ulrich, M.; Bastian, M.; Cramton, S.E.; Ziegler, K.; Pragman, A.A.; Bragonzi, A.; Memmi, G.; Wolz, C.; Schlievert, P.M.; Cheung, A.; Döring, G. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 2007, 65, 1276–1287. [Google Scholar] [CrossRef]
- Walker, J.N.; Crosby, H.A.; Spaulding, A.R.; Salgado-Pabón, W.; Malone, C.L.; Rosenthal, C.B.; Schlievert, P.M.; Boyd, J.M.; Horswill, A.R. The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis. PLoS Pathog. 2013, 9, e1003819. [Google Scholar] [CrossRef]
- Hauserman, M.R.; Sullivan, L.E.; James, K.L.; Ferraro, M.J.; Rice, K.C. Response of Staphylococcus aureus physiology and Agr quorum sensing to low-shear modeled microgravity. J. Bacteriol. 2024, 206, e00272–24. [Google Scholar] [CrossRef]
- Benson, M.A.; Lilo, S.; Nygaard, T.; Voyich, J.M.; Torres, V.J. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J. Bacteriol. 2012, 194, 4355–4365. [Google Scholar] [CrossRef]
- Choueiry, F.; Xu, R.; Zhu, J. Adaptive metabolism of Staphylococcus aureus revealed by untargeted metabolomics. J. Proteome Res. 2022, 21, 470–481. [Google Scholar] [CrossRef]
- Howden, B.P.; Giulieri, S.G.; Lung, T.W.F.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 2023, 21, 380. [Google Scholar] [CrossRef]
- Bastos, C.P.; Bassani, M.T.; Mata, M.M.; Lopes, G.V.; da Silva, W.P. Prevalence and expression of staphylococcal enterotoxin genes in Staphylococcus aureus isolated from food poisoning outbreaks. Can. J. Microbiol. 2017, 63, 834–840. [Google Scholar] [CrossRef]
- Johler, S.; Giannini, P.; Jermini, M.; Hummerjohann, J.; Baumgartner, A.; Stephan, R. Further evidence for staphylococcal food poisoning outbreaks caused by egc-encoded enterotoxins. Toxins 2015, 7, 997–1004. [Google Scholar] [CrossRef]
- Fricker, C.R.; Tompkin, R.B. Staphylococcus aureus. In Compendium of methods for the microbiological examination of foods, 4th ed.; Vanderzant, C., Splittstoesser, D.F., Eds.; APHA Press: [local], [país], 2005. [Google Scholar]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Jiang, D. A dose response model for Staphylococcus aureus. Sci. Rep. 2021, 11, 12279. [Google Scholar] [CrossRef]
- Sabioni, J.G.; Hirooka, E.Y.; de Souza, M.L. Intoxicação alimentar por queijo Minas contaminado com Staphylococcus aureus. Rev. Saúde Pública 1988, 22, 458–461. [Google Scholar] [CrossRef]
- Pereira, M.L.; do Carmo, L.S.; dos Santos, E.J.; Pereira, J.L.; Bergdoll, M.S. Enterotoxin H in staphylococcal food poisoning. J. Food Prot. 1996, 59, 559–561. [Google Scholar] [CrossRef]
- do Carmo, L.S.; Dias, R.S.; Linardi, V.R.; Sena, M.J.; Santos, D.A.; Faria, M.E.; Pena, E.C.; Jett, M.; Heneine, L.G. Food poisoning due to enterotoxigenic strains of Staphylococcus present in Minas cheese and raw milk in Brazil. Food Microbiol. 2002, 19, 9–14. [Google Scholar] [CrossRef]
- Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde e Ambiente. Surtos de doenças de transmissão hídrica e alimentar – informe, [: da Saúde, 2024.
- Todd, E.C.D. Foodborne and waterborne disease in Canada — 1980 annual summary. J. Food Prot. 1987, 50, 420–428. [Google Scholar] [CrossRef]
- Wieneke, A.A.; Roberts, D.; Gilbert, R.J. Staphylococcal food poisoning in the United Kingdom, 1969–90. Epidemiol. Infect. 1993, 110, 519–531. [Google Scholar] [CrossRef]
- Bone, F.J.; Bogie, D.; Morgan-Jones, S.C. Staphylococcal food poisoning from sheep milk cheese. Epidemiol. Infect. 1989, 103, 449–458. [Google Scholar] [CrossRef]
- Kérouanton, A.; Hennekinne, J.A.; Letertre, C.; Petit, L.; Chesneau, O.; Brisabois, A.; De Buyser, M.L. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int. J. Food Microbiol. 2007, 115, 369–375. [Google Scholar] [CrossRef]
- Ostyn, A.; de Buyser, M.L.; Guillier, F.; Groult, J.; Félix, B.; Salah, S.; Delmas, G.; Hennekinne, J.A. First evidence of a food poisoning outbreak due to staphylococcal enterotoxin type E, France, 2009. Euro Surveill. 2010, 15, 10–13. [Google Scholar] [CrossRef]
- Filipello, V.; Bonometti, E.; Campagnani, M.; Bertoletti, I.; Romano, A.; Zuccon, F.; Campanella, C.; Losio, M.N.; Finazzi, G. Investigation and follow-up of a staphylococcal food poisoning outbreak linked to the consumption of traditional hand-crafted alm cheese. Pathogens 2020, 9, 1064. [Google Scholar] [CrossRef]
- Bianchi, D.M.; Romano, A.; Tramuta, C.; Distasio, P.; Decastelli, L. A foodborne outbreak caused by staphylococcal enterotoxins in cheese sandwiches in northern Italy. Ital. J. Food Sci. 2024, 36, 136–141. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442.
- Kumagai, Y.; Pires, S.M.; Kubota, K.; Asakura, H. Attributing human foodborne diseases to food sources and water in Japan using analysis of outbreak surveillance data. J. Food Prot. 2020, 83, 2087–2094. [Google Scholar] [CrossRef]
- Schelin, J.; Wallin-Carlquist, N.; Cohn, M.T.; Lindqvist, R.; Barker, G.C.; Rådström, P. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2011, 2, 580. [Google Scholar] [CrossRef]
- Emiliano, J.V.S.; Fusieger, A.; Camargo, A.C.; Rodrigues, F.F.C.; Nero, L.A.; Perrone, Í.T.; Carvalho, A.F. Staphylococcus aureus in dairy industry: Enterotoxin production, biofilm formation, and use of lactic acid bacteria for its biocontrol. Foodborne Pathog. Dis. 2024, 21, [ID]. [Google Scholar] [CrossRef]
- Pineda, A.P.A.; Campos, G.Z.; Pimentel-Filho, N.J.; Franco, B.D.G.M.; Pinto, U.M. Brazilian artisanal cheeses: Diversity, microbiological safety, and challenges for the sector. Front. Microbiol. 2021, 12, 666922. [Google Scholar] [CrossRef]
- Lacorte, G.A.; Cruvinel, L.A.; Ávila, M.P.; Dias, M.F.; Pereira, A.A.; Nascimento, A.M.A.; Franco, B.D.G.M. Investigating the influence of food safety management systems (FSMS) on microbial diversity of Canastra cheeses and their processing environments. Food Microbiol. 2022, 105, 104023. [Google Scholar] [CrossRef]
- Finger, J.A.F.F.; Lima, E.M.F.; Coelho, K.S.; Behrens, J.H.; Landgraf, M.; Franco, B.D.G.M.; Pinto, U.M. Adherence to food hygiene and personal protection recommendations for prevention of COVID-19. Trends Food Sci. Technol. 2021, 112, 847–852. [Google Scholar] [CrossRef]
- Madera, C.; Montero, I.; Pérez, M.D.; Álvarez-Ordoñez, A.; Prieto, M. Evaluation of bacteriophage cocktails for biocontrol of Staphylococcus aureus in milk. Microorganisms 2024, 12, 347. [Google Scholar]
- El-Deeb, W.M.; Fayez, M.; Ibrahim, M.A. Use of probiotics to reduce intramammary infection by Staphylococcus aureus in dairy cows. Vet. World 2023, 16, 48–56. [Google Scholar]
- Doležalová, M.; Sedlaříková, M.; Krátký, M.; Neužil, J. Magnetic microbots for selective removal of Staphylococcus aureus from milk. Food Control 2023, 150, 109734. [Google Scholar]
- Kérouanton, A.; Derzelle, S.; Tenenhaus-Aziza, F. Advances in detection of staphylococcal enterotoxins in dairy products: Molecular and rapid methods. Front. Microbiol. 2024, 15, 1223450. [Google Scholar]
- Food Standards Australia New Zealand. Compendium of microbiological criteria for food. First published October 2016; last updated March 2022. Food Standards Australia New Zealand (FSANZ), 2022.
- Brazil, Ministry of Agriculture, Livestock, and Supply. Normative Instruction No. 161, of July 1, 2022. Official Gazette of the Union: Section 1, 2022.
- Health Canada. Draft – Table of microbiological criteria for food. Government of Canada, 2023.
- World Bank. Comparison of microbiological criteria in food products in the EU and China, The World Bank Group, 2017.
- European Commission. Guidance on cheese as raw material in the manufacture of food products; EDA/EUCOLAIT: [cidade], [país], 2018. [Google Scholar]
- U. S. Food and Drug Administration. Compliance Policy Guide Sec. 527.300 Dairy Products - Microbial Contaminants and Alkaline Phosphatase Activity; U.S. Food and Drug Administration: [cidade], EUA, 2010. [Google Scholar]
| Coagulase-positive staphylococci (CoPS) species | |
| Staphylococcus argenteus | Staphylococcus intermedius |
| Staphylococcus aureus | Staphylococcus lutrae |
| Staphylococcus coagulans | Staphylococcus pseudintermedius |
| Staphylococcus cornubiensis | Staphylococcus schweitzeri |
| Staphylococcus delphini | - |
| Coagulase-negative staphylococci (CoNS) species | |
| Staphylococcus americanisciuri | Staphylococcus leei |
| Staphylococcus argensis | Staphylococcus lloydii |
| Staphylococcus arlettae | Staphylococcus lugdunensis |
| Staphylococcus auricularis | Staphylococcus lyticans |
| Staphylococcus borealis | Staphylococcus marylandisciuri |
| Staphylococcus brunensis | Staphylococcus massiliensis |
| Staphylococcus caeli | Staphylococcus microti |
| Staphylococcus caledonicus | Staphylococcus muscae |
| Staphylococcus canis | Staphylococcus nepalensis |
| Staphylococcus capitis | Staphylococcus pasteuri |
| Staphylococcus caprae | Staphylococcus petrasii |
| Staphylococcus carnosus | Staphylococcus pettenkoferi |
| Staphylococcus casei | Staphylococcus piscifermentans |
| Staphylococcus chromogenes | Staphylococcus pragensis |
| Staphylococcus cohnii | Staphylococcus pseudolugdunensis |
| Staphylococcus condimenti | Staphylococcus pseudoxylosus |
| Staphylococcus croceilyticus | Staphylococcus ratti |
| Staphylococcus debuckii | Staphylococcus rostri |
| Staphylococcus devriesei | Staphylococcus saccharolyticus |
| Staphylococcus durrellii | Staphylococcus saprophyticus |
| Staphylococcus edaphicus | Staphylococcus schleiferi |
| Staphylococcus epidermidis | Staphylococcus shinii |
| Staphylococcus equorum | Staphylococcus simiae |
| Staphylococcus felis | Staphylococcus simulans |
| Staphylococcus gallinarum | Staphylococcus succinus |
| Staphylococcus haemolyticus | Staphylococcus taiwanensis |
| Staphylococcus hominis | Staphylococcus ureilyticus |
| Staphylococcus hsinchuensis | Staphylococcus warneri |
| Staphylococcus kloosii | Staphylococcus xylosus |
| Coagulase-variable staphylococci (CoVS) species | |
| Staphylococcus agnetis | Staphylococcus roterodami |
| Staphylococcus hyicus | Staphylococcus singaporensis |
| SEs/SEls/ TSST-1 |
Gene | Genetic element | Superantigenic activity | Emetic activity | Source |
|---|---|---|---|---|---|
| SEA | sea | Prophage | Yes | Yes | Food poisoning, dairy products, human, bovine, caprine, ovine |
| SEA | sea | Prophage | Yes | Yes | Food poisoning, dairy products, human, bovine, caprine, ovine |
| SEB | seb | SaPI3, chromosome, plasmid | Yes | Yes | Food poisoning, dairy products, human, bovine, caprine, ovine |
| SEC | sec | SaPI* | Yes | Yes | Food poisoning, dairy products, human, bovine, caprine, ovine |
| SEC-1 | sec | SaPI | Yes | Yes | Human |
| SEC-2 | sec | SaPI | Yes | Not evaluated | Human |
| SEC-3 | sec | SaPI | Yes | Yes | Human |
| SED | sed | Plasmid | Yes | Yes | Food poisoning, bovine |
| SEE | see | Prophage (hypothetical location) | Yes | Yes | Food poisoning, unpasteurized milk soft cheese |
| SEG | seg | egc1, egc2, egc3, egc4 | Yes | Yes | Bovine |
| SEH | seh | Transposon | Yes | Yes | Empyema human |
| SEI | sei | egc1, egc2, egc3 | Yes | Yes | Mastitis cows, humans |
| SElJ | selj | Plasmid | Yes | Not evaluated | Epidemiologically implicated in food poisoning |
| SElK | selk | SaPI1, SaPI3, SaPI5, SaPIbov1, prophages | Yes | Not evaluated | Human |
| SElL | sell | SaPIn1, SaPIm1, SaPImw2, SaPIbov1 | Yes | Yes | Human |
| SElM | selm | egc1, egc2 | Yes | Yes | Bovine |
| SElN | seln | egc1, egc2, egc3, egc4 | Yes | Yes | Human |
| SElO | selo | egc1, egc2, egc3, egc4, transposon | Yes | Yes | Human |
| SElP | selp | Prophage | Yes | Yes | Human, ulcer |
| SElQ | selq | SaPI1, SaPI3, SaPI5, prophage | Yes | Yes | Human |
| SElR | selr | Plasmid | Yes | Yes | Human |
| SElS | sels | Plasmid | Yes | Yes | Not found |
| SElT | selt | Plasmid | Yes | Yes | Not found |
| SElU | selu | egc2, egc3 | Yes | Not evaluated | Human |
| SElV | selv | egc4 | Yes | Not evaluated | Not found |
| SElW | selw | egc4 | Yes | Not evaluated | Human |
| SElX | selx | Chromosome | Yes | Not evaluated | Milk, raw meat, human |
| SElY | sely | Chromosome | Yes | Not evaluated | Human |
| SElZ | selz | Chromosome | Not evaluated | Not evaluated | Bovine |
| TSST-1 | tst/TssT | Chromosome | Yes | No | Human |
| Antibiotic class | Resistance gene | Encoded protein |
|---|---|---|
| Aminoglycoside | aacA-aphD | 6'-aminoglycoside N-acetyltransferase/2''-aminoglycoside phosphotransferase |
| aadA2 | Spectinomycin 9-adenylyltransferase | |
| aadA5 | Aminoglycoside-3'-adenylyltransferase | |
| ant(4’)-Ia | Aminoglycoside adenyltransferase | |
| aph(2’’)-Ih | Aminoglycoside 2’’-phosphotransferase | |
| aph(3’)-IIIa | Aminoglycoside 3’-phosphotransferase | |
| Beta-lactam | mecA | Penicillin-binding protein 2a (PBP2a) |
| mecA1 | ||
| blaZ | Beta-lactamase | |
| blaTEM | ||
| Diaminopyrimidine | dfrG | Dihydrofolate reductase |
| Diaminopyrimidine | dfrG | Dihydrofolate reductase |
| Fusidane | fusB | 2-domain zinc-binding protein |
| fusC | ||
| Glycopeptide | bleO | Bleomycin resistant proteins |
| vanA | Vancomycin/teicoplanin A-type resistance protein | |
| Lincosamide | lnuA | Lincosamide nucleotidyltransferase |
| Lincosamide/ macrolide/ streptogramin | ermC | rRNA adenine N-6-methyltransferase |
| Lincosamide/ pleuromutilin/ streptogramin/ | salA | Iron-sulfur cluster carrier protein |
| vgaA-lc | ABC transporter | |
| Macrolide | mphC | Macrolide 2’- phosphotransferase |
| Macrolide/streptogramin | msrA | Peptide methionine sulfoxide reductase |
| Nucleoside | sat-4 | Streptothricin N-acetyltransferase and streptothricin |
| Phenicol | fexA | Chloramphenicol/florfenicol exporter |
| cmlA1 | Bcr/CflA family efflux transporter | |
| Phosphonic acid | fosB-saur | Metallothiol transferase |
| Quinolone | gyrA | DNA gyrase subunit A |
| Tetracycline | tetK | Tetracycline resistance protein |
| tetL | ||
| tet38 | Tetracycline efflux MFS transporter | |
| Trimethoprim | dfrA12 | Dihydrofolate reductase |
| dfr17 |
| Year | Location | Product | Enterotoxin type | Symptoms | Number of patients (deaths) | Reference |
| 1980 | Canada | Curd Cheese | SEA, SEC | Nausea, vomiting, abdominal cramps and diarrhea | 62 (0) | [196] |
| 1981 | United Kingdom | Halloumi cheese | SEA | Nausea, vomiting, abdominal cramps and diarrhea | 4 (0) | [198] |
| 1981 | France | Raw milk semi-hard cheese | SEA | Unknown | 4 (0) | [199] |
| 1983 | France | Raw milk semi-hard cheese | SEA, SED | Vomiting and abdominal cramps | 20 (0) | [199] |
| 1983 | France | Raw milk soft cheese | Absent | Vomiting and diarrhea | 4 (0) | [199] |
| 1985 | France | Soft cheese | SEB | Vomiting and diarrhea | 2 (0) | [199] |
| 1985 | France | Soft cheese | SEB | Vomiting and diarrhea | 3 (0) | [199] |
| 1985 | United Kingdom | Raw ewe's milk cheese | SEA | Nausea, vomiting, abdominal cramps and diarrhea | 27 (0) | [197] |
| 1986 | France | Sheep’s milk cheese | SEB | Unknown | Unknown | [199] |
| 1988 | Brazil | Fresh Minas cheese | SEA, SEB, SED, SEE | Nausea, vomiting, abdominal cramps and diarrhea | 4 (0) | [192] |
| 1995 | Brazil | Minas cheese | SEH | Vomiting and diarrhea | 7 (0) | [193] |
| 1997 | France | Raw milk cheese | Present but not specified | Unknown | 43 (0) | [199] |
| 1998 | France | Raw milk cheese | Present but not specified | Vomiting, abdominal cramps and diarrhea | 47 (0) | [199] |
| 1998 | France | Raw milk semi-hard cheese | Absent | Vomiting and abdominal cramps | 10 (0) | [199] |
| 1999 | Brazil | Minas cheese | SEA, SEB, SEC | Vomiting, dizziness, chills, headaches and Diarrhea, | 378 (0) | [194] |
| 2000 | France | Raw sheep’s milk cheese | SEA | Unknown | Unknown | [199] |
| 2001 | France | Sliced soft cheese | SEA | Nausea, vomiting, abdominal cramps and diarrhea | 2 (0) | [199] |
| 2001 | France | Raw milk semi-hard cheese | SED | Vomiting | 17 (0) | [199] |
| 2002 | France | Raw sheep’s milk cheese | SEA | Nausea, vomiting, abdominal cramps and diarrhea | 43 (0) | [199] |
| 2007 | Switzerland | Robiola cheese | SEG, SEI, SEM, SEN, SEO | Nausea, vomiting, abdominal cramps and diarrhea (in some cases) | 5 (0) | [188] |
| 2009 | France | Soft cheese | SEE | Nausea, vomiting, abdominal cramps and diarrhoea and fever (in some cases) | 23 (0) | [200] |
| 2014 | Switzerland | Tomme cheese | SEA, SED | Vomiting, Abdominal cramps, severe diarrhea and fever | 14 (0) | [188] |
| 2018 | Italy | Alm cheese | SED | Vomiting, abdominal cramps and diarrhea | 3 (0) | [201] |
| 2022 | Italy | Raw milk cheese | SEA, SEB, SEC, SED | Vomiting and diarrhea, headaches | 8 (0) | [202] |
| Country or region | Microbiological criteria | ||||||
|---|---|---|---|---|---|---|---|
| CoPS or enterotoxins | n | c | m | M | Notes | Reference | |
| Australia | CoPS | 5 | 2 | 100 | 1000 | All types of cheese | [214] |
| Brazil | CoPS | 5 | 2 | 100 | 1000 | All types of cheese | [215] |
| Staphylococcus Enterotoxin (SE) | 5 | 0 | absence | - | All types of cheese | ||
| Canada | S. aureus | 5 | 2 | 1000 | 10,000 | Cheese made from an unpasteurized source | [216] |
| China | S. aureus | 5 | 2 | 100 | 1000 | All types of cheese | [217] |
| European Union | CoPS | 5 | 2 | 10,000 | 100,000 | Cheese made from raw milk | [218] |
| European Union | CoPS | 5 | 2 | 100 | 1000 | Cheese made from mild heat treated milk | [218] |
| European Union | CoPS | 5 | 2 | 10 | 100 | Unriped soft cheese made with pasteurized milk | [218] |
| United States | S. aureus | - | - | - | 10,000 | All dairy products | [219] |
| SE | - | - | not detected | - | All dairy products | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





