Submitted:
20 June 2025
Posted:
24 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
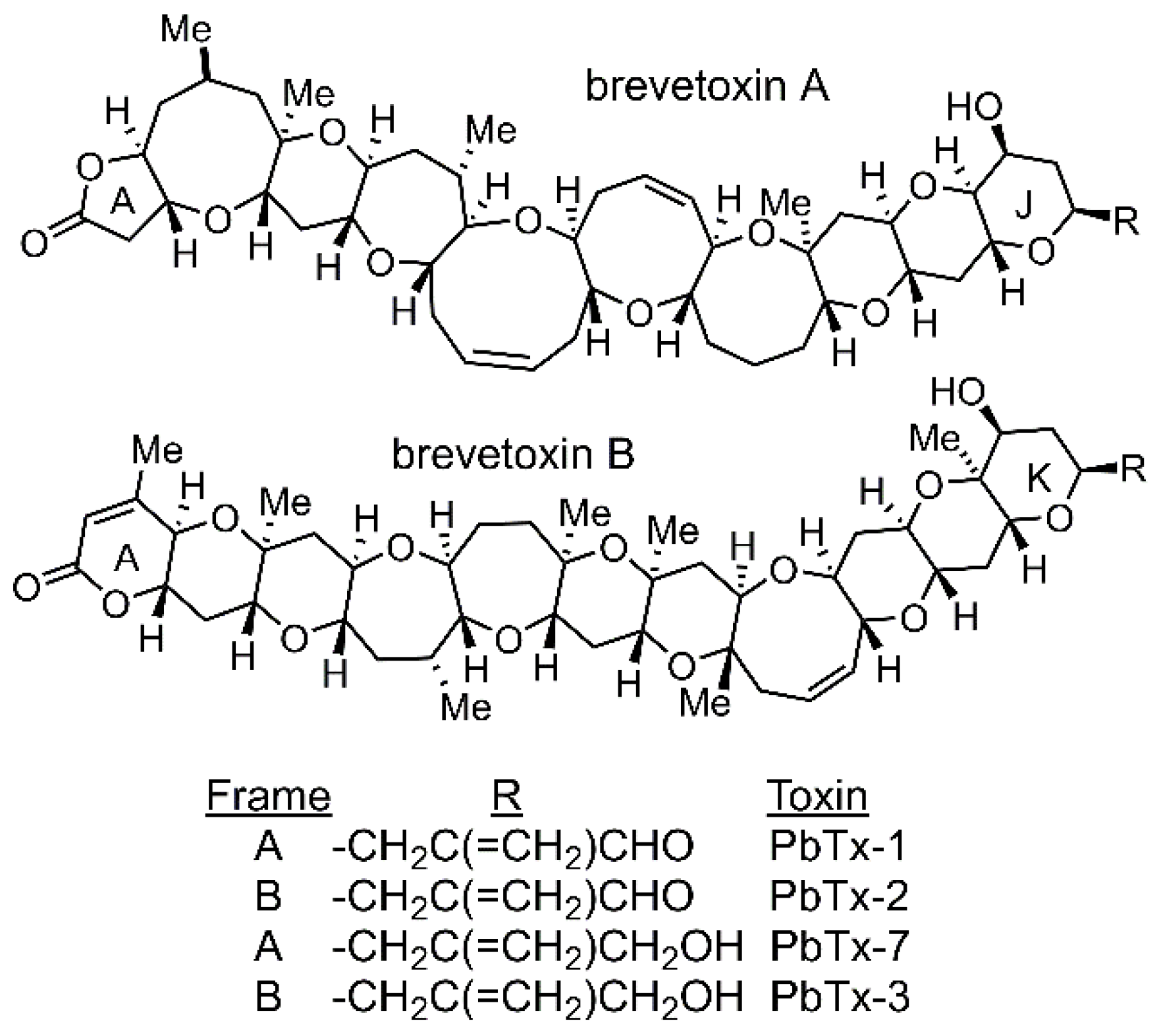
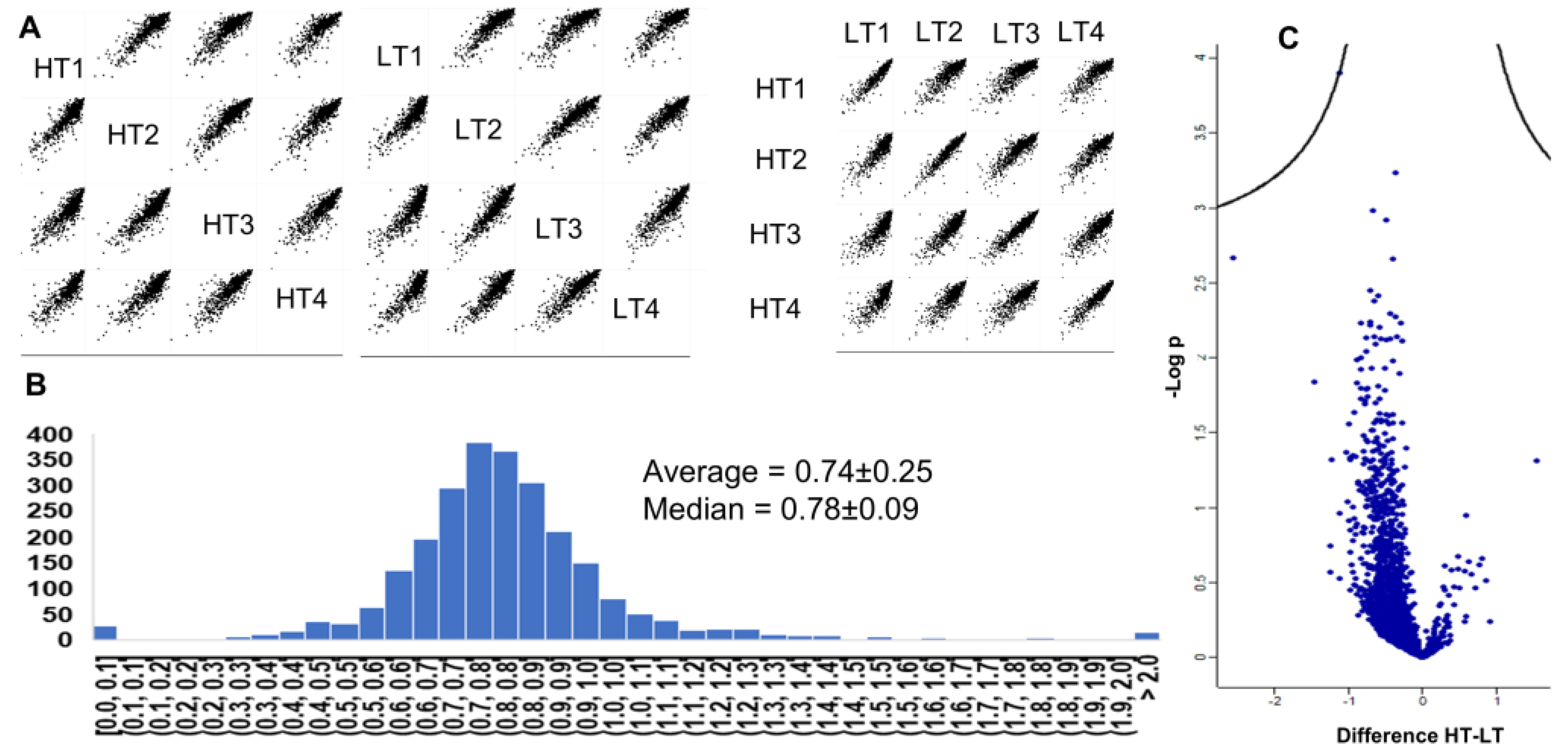
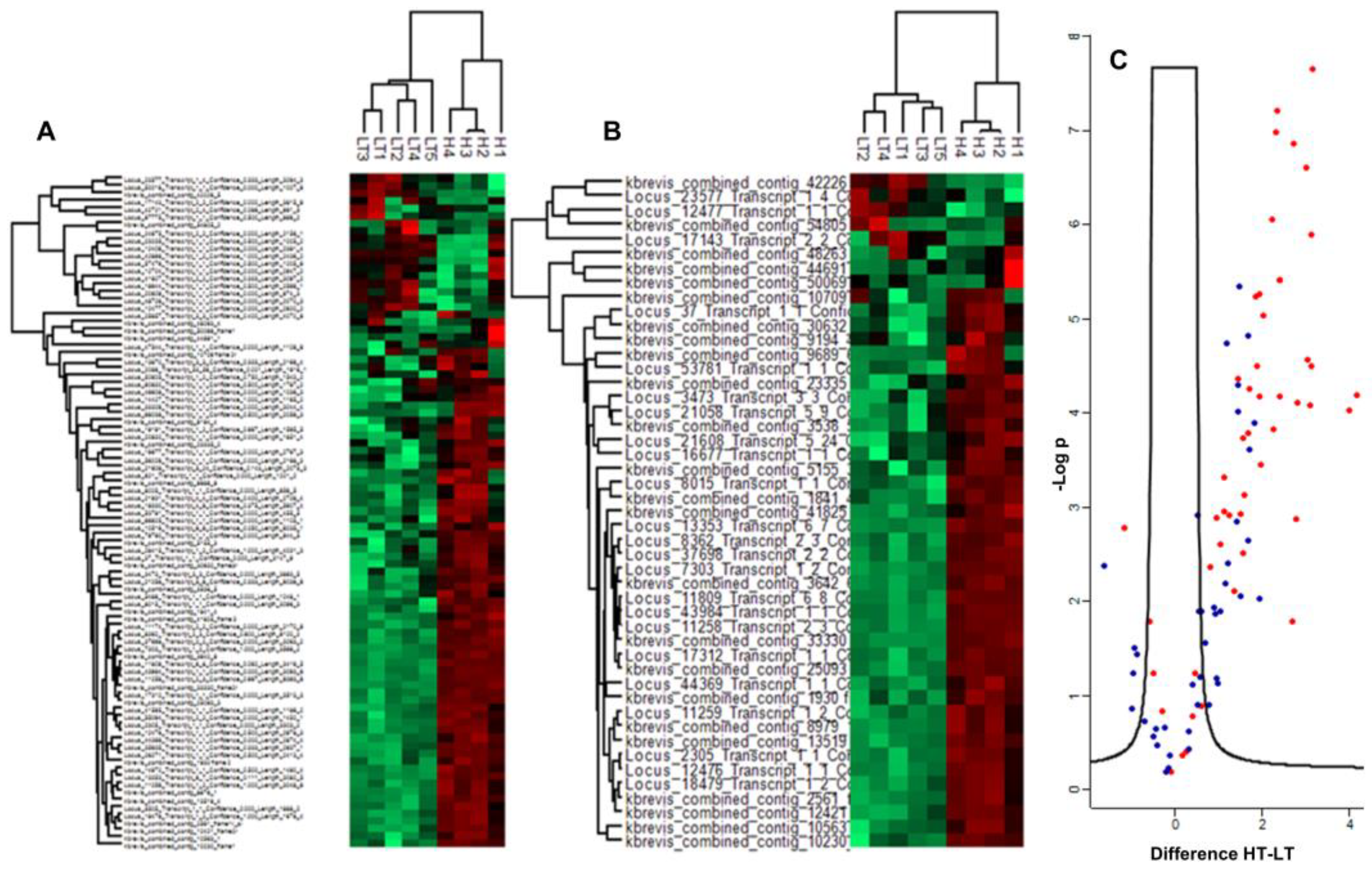
2.1. Differences in Redox Stoichiometry Between High and Low-Toxin K. brevis.
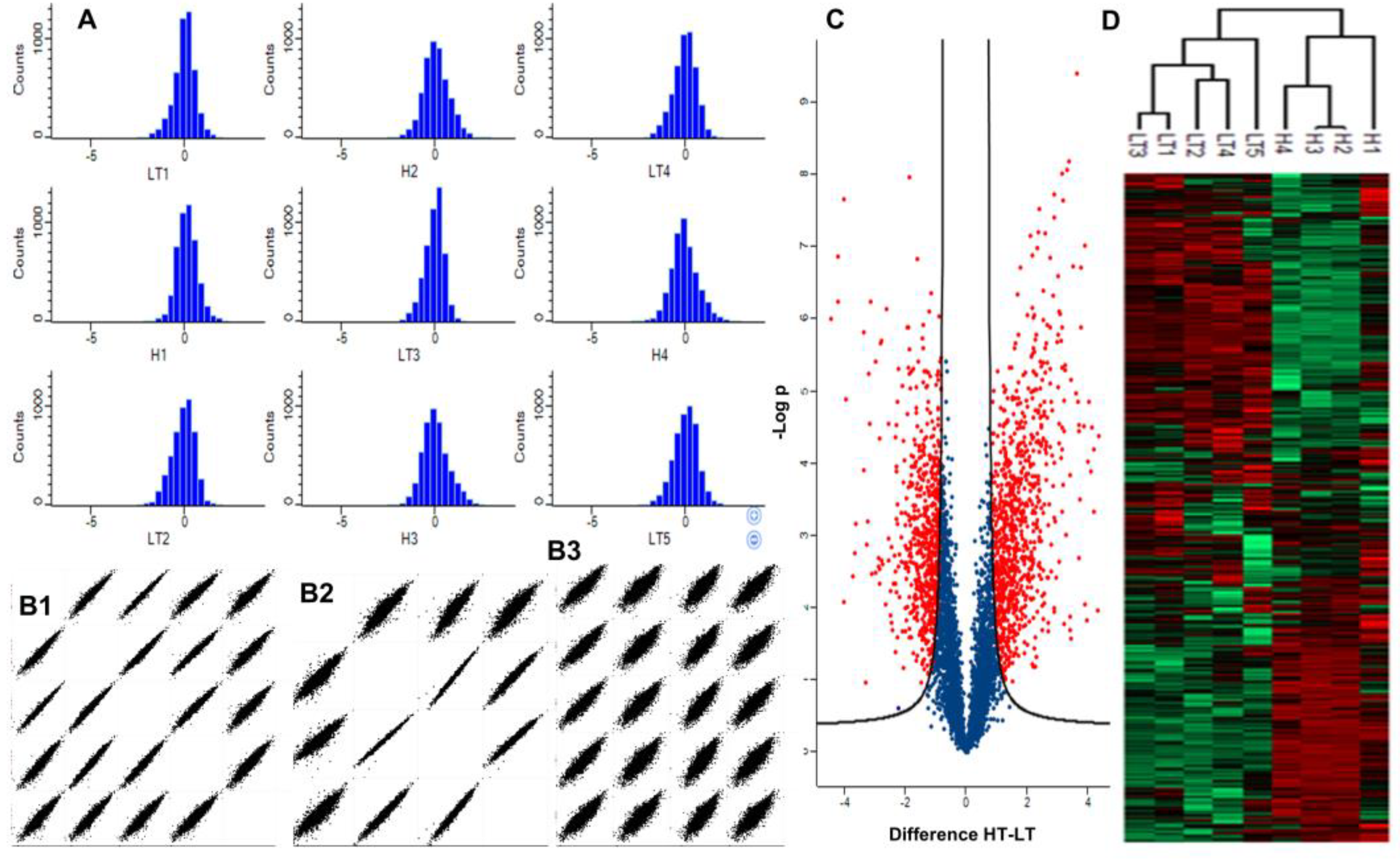
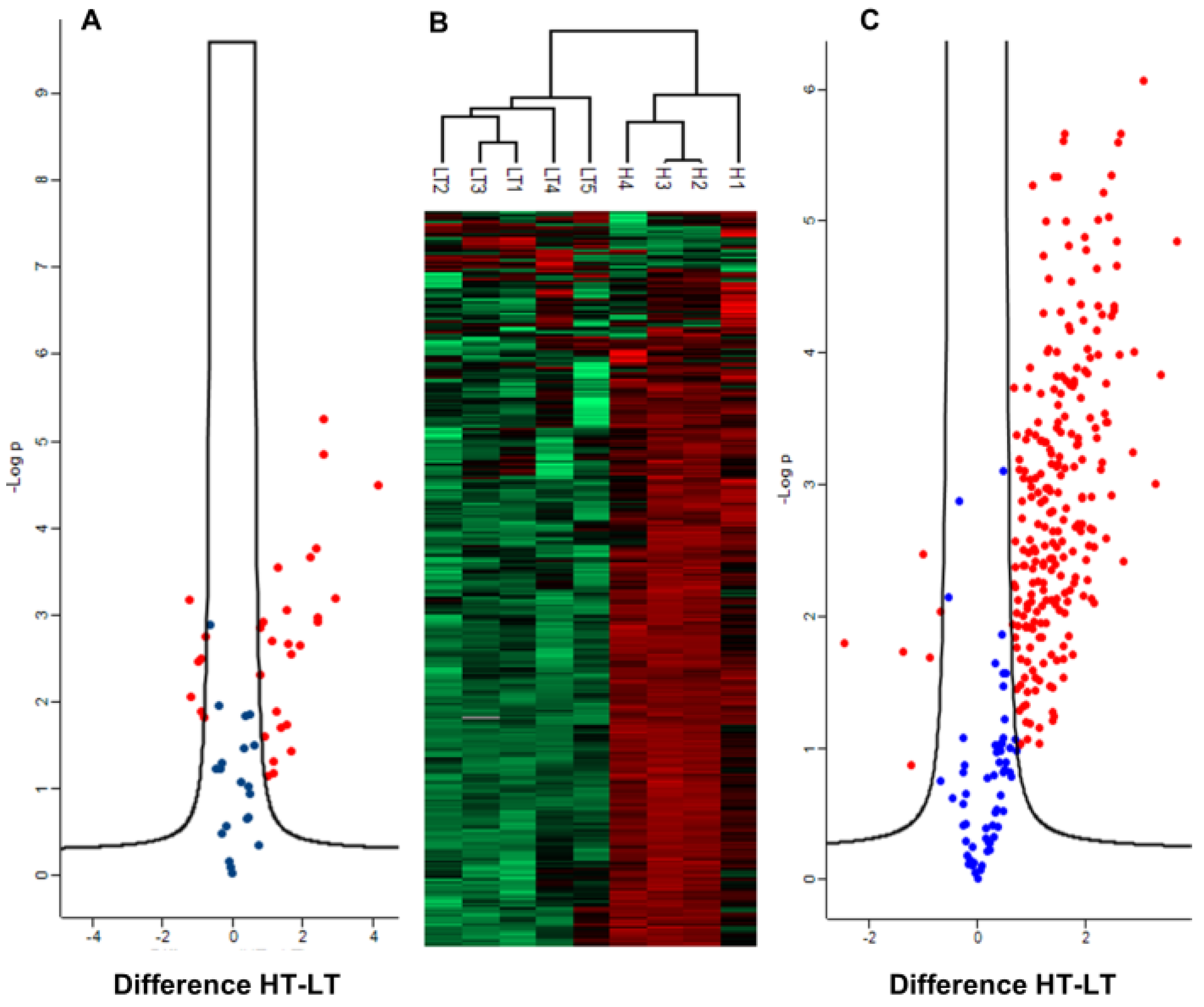
2.2. Differentially Abundant Proteins Between High and Low-Toxin K. brevis
3. Discussion
4. Materials and Methods
4.1. K. brevis Culture
4.2. Brevetoxin Extraction and Liquid Chromatography–Mass Spectrometry (LC–MS) Analysis
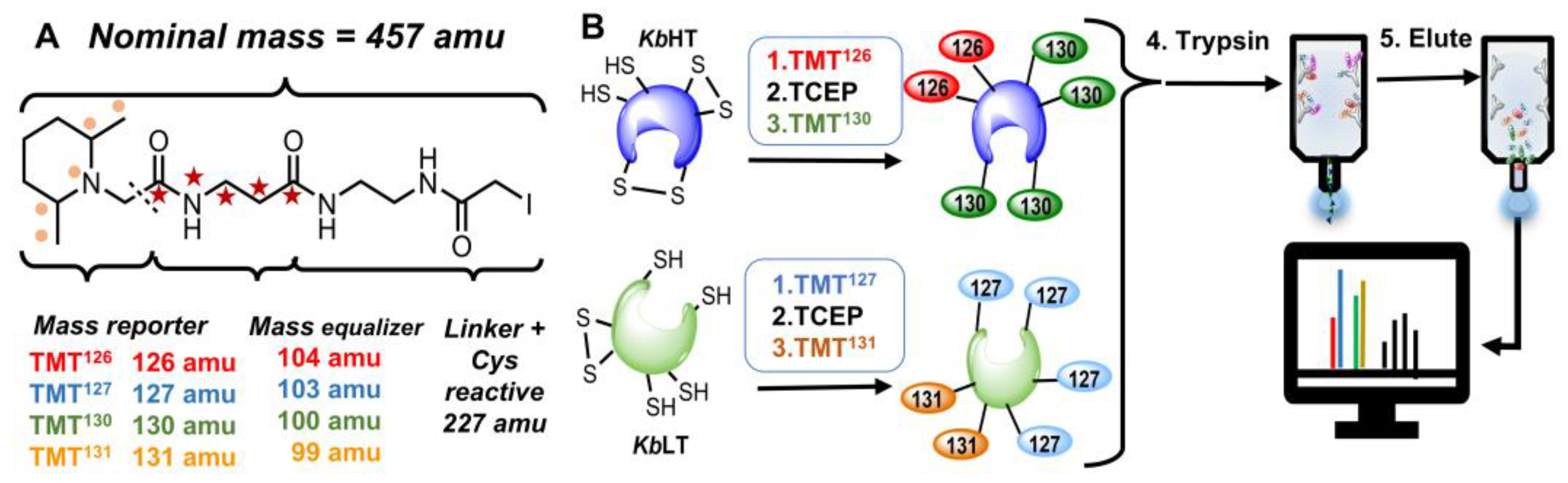
4.3. Peptide Isolation and Labeling
4.4. Liquid Chromatography–Mass Spectrometry (LC–MS) Analysis of Labeled Peptide Samples
4.5. Functional Annotation of the Identified Proteins
4.6. Analysis of Proteomic Data
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tominack, S.A.; Coffey, K.Z.; Yoskowitz, D.; Sutton, G.; Wetz, M.S. An Assessment of Trends in the Frequency and Duration of Karenia Brevis Red Tide Blooms on the South Texas Coast (Western Gulf of Mexico). PLOS ONE 2020, 15, e0239309. [CrossRef]
- Ecotoxicology of Marine Organisms; Duarte, B., Caçador, I., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, 2020; ISBN 978-1-138-03549-2.
- Bechard, A. The Economic Impacts of Harmful Algal Blooms on Tourism: An Examination of Southwest Florida Using a Spline Regression Approach. Nat. Hazards 2020, 104, 593–609. [CrossRef]
- Béchard, A. Economics Losses to Fishery and Seafood Related Businesses during Harmful Algal Blooms. Fish. Res. 2020, 230, 105678. [CrossRef]
- Patel, S.S.; Lovko, V.J.; Lockey, R.F. Red Tide: Overview and Clinical Manifestations. J. Allergy Clin. Immunol. Pract. 2020, 8, 1219–1223. [CrossRef]
- Diaz, R.E.; Friedman, M.A.; Jin, D.; Beet, A.; Kirkpatrick, B.; Reich, A.; Kirkpatrick, G.; Ullmann, S.G.; Fleming, L.E.; Hoagland, P. Neurological Illnesses Associated with Florida Red Tide (Karenia Brevis) Blooms. Harmful Algae 2019, 82, 73–81. [CrossRef]
- Hort, V.; Abadie, E.; Arnich, N.; Dechraoui Bottein, M.-Y.; Amzil, Z. Chemodiversity of Brevetoxins and Other Potentially Toxic Metabolites Produced by Karenia Spp. and Their Metabolic Products in Marine Organisms. Mar. Drugs 2021, 19, 656. [CrossRef]
- Lekan, D.K.; Tomas, C.R. The Brevetoxin and Brevenal Composition of Three Karenia Brevis Clones at Different Salinities and Nutrient Conditions. Harmful Algae 2010, 9, 39–47. [CrossRef]
- Jeglitsch, G.; Rein, K.; Baden, D.G.; Adams, D.J. Brevetoxin-3 (PbTx-3) and Its Derivatives Modulate Single Tetrodotoxin-Sensitive Sodium Channels in Rat Sensory Neurons. J. Pharmacol. Exp. Ther. 1998, 284, 516–525.
- Cohen, J.H.; Tester, P.A.; Forward, R.B. Sublethal Effects of the Toxic Dinoflagellate Karenia Brevis on Marine Copepod Behavior. J. Plankton Res. 2007, 29, 301–315. [CrossRef]
- Hong, J.; Talapatra, S.; Katz, J.; Tester, P.A.; Waggett, R.J.; Place, A.R. Algal Toxins Alter Copepod Feeding Behavior. PLoS ONE 2012, 7, e36845. [CrossRef]
- Waggett, R.; Hardison, D.; Tester, P. Toxicity and Nutritional Inadequacy of Karenia Brevis: Synergistic Mechanisms Disrupt Top-down Grazer Control. Mar. Ecol. Prog. Ser. 2012, 444, 15–30. [CrossRef]
- Kubanek, J.; Hicks, M.K.; Naar, J.; Villareal, T.A. Does the Red Tide Dinoflagellate Karenia Brevis Use Allelopathy to Outcompete Other Phytoplankton? Limnol. Oceanogr. 2005, 50, 883–895. [CrossRef]
- Poulson-Ellestad, K.; Mcmillan, E.; Montoya, J.P.; Kubanek, J. Are Offshore Phytoplankton Susceptible to Karenia Brevis Allelopathy? J. Plankton Res. 2014, 36, 1344–1356. [CrossRef]
- Poulson-Ellestad, K.L.; Jones, C.M.; Roy, J.; Viant, M.R.; Fernández, F.M.; Kubanek, J.; Nunn, B.L. Metabolomics and Proteomics Reveal Impacts of Chemically Mediated Competition on Marine Plankton. Proc. Natl. Acad. Sci. 2014, 111, 9009–9014. [CrossRef]
- Poulin, R.X.; Poulson-Ellestad, K.L.; Roy, J.S.; Kubanek, J. Variable Allelopathy among Phytoplankton Reflected in Red Tide Metabolome. Harmful Algae 2018, 71, 50–56. [CrossRef]
- Prince, E.K.; Myers, T.L.; Kubanek, J. Effects of Harmful Algal Blooms on Competitors: Allelopathic Mechanisms of the Red Tide Dinoflagellate Karenia Brevis. Limnol. Oceanogr. 2008, 53, 531–541. [CrossRef]
- Cassell, R.T.; Chen, W.; Thomas, S.; Liu, L.; Rein, K.S. Brevetoxin, the Dinoflagellate Neurotoxin, Localizes to Thylakoid Membranes and Interacts with the Light-Harvesting Complex II (LHCII) of Photosystem II. ChemBioChem 2015, 16, 1060–1067. [CrossRef]
- Colon, R.; Wheater, M.; Joyce, E.J.; Ste.Marie, E.J.; Hondal, R.J.; Rein, K.S. The Marine Neurotoxin Brevetoxin (PbTx-2) Inhibits Karenia Brevis and Mammalian Thioredoxin Reductases by Targeting Different Residues. J. Nat. Prod. 2021, 84, 2961–2970. [CrossRef]
- Rein, K.S.; Borrone, J. Polyketides from Dinoflagellates: Origins, Pharmacology and Biosynthesis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999, 124, 117–131. [CrossRef]
- Hwang, S.; Lee, N.; Cho, S.; Palsson, B.; Cho, B.-K. Repurposing Modular Polyketide Synthases and Non-Ribosomal Peptide Synthetases for Novel Chemical Biosynthesis. Front. Mol. Biosci. 2020, 7, 87. [CrossRef]
- Kang, H.-S.; Kim, E.-S. Recent Advances in Heterologous Expression of Natural Product Biosynthetic Gene Clusters in Streptomyces Hosts. Curr. Opin. Biotechnol. 2021, 69, 118–127. [CrossRef]
- Li, S.; Li, Z.; Pang, S.; Xiang, W.; Wang, W. Coordinating Precursor Supply for Pharmaceutical Polyketide Production in Streptomyces. Curr. Opin. Biotechnol. 2021, 69, 26–34. [CrossRef]
- Yang, D.; Zhou, H.; Lee, S.Y. Production of Diversified Polyketides by Metabolic Engineering. Biochemistry 2021, 60, 3424–3426. [CrossRef]
- Riaz, S.; Sui, Z.; Niaz, Z.; Khan, S.; Liu, Y.; Liu, H. Distinctive Nuclear Features of Dinoflagellates with A Particular Focus on Histone and Histone-Replacement Proteins. Microorganisms 2018, 6, 128. [CrossRef]
- Wisecaver, J.H.; Hackett, J.D. Dinoflagellate Genome Evolution. Annu. Rev. Microbiol. 2011, 65, 369–387. [CrossRef]
- Lidie, K.B.; Van Dolah, F.M. Spliced Leader RNA-Mediated Trans -Splicing in a Dinoflagellate, Karenia Brevis. J. Eukaryot. Microbiol. 2007, 54, 427–435. [CrossRef]
- Zhang, H.; Campbell, D.A.; Sturm, N.R.; Lin, S. Dinoflagellate Spliced Leader RNA Genes Display a Variety of Sequences and Genomic Arrangements. Mol. Biol. Evol. 2009, 26, 1757–1771. [CrossRef]
- Roy, S.; Jagus, R.; Morse, D. Translation and Translational Control in Dinoflagellates. Microorganisms 2018, 6, 30. [CrossRef]
- Nimmo, I.C.; Barbrook, A.C.; Lassadi, I.; Chen, J.E.; Geisler, K.; Smith, A.G.; Aranda, M.; Purton, S.; Waller, R.F.; Nisbet, R.E.R.; et al. Genetic Transformation of the Dinoflagellate Chloroplast. eLife 2019, 8, e45292. [CrossRef]
- Te, M.R.; Lohuis; Miller, D.J. Genetic Transformation of Dinoflagellates ( Amphidinium and Symbiodinium ): Expression of GUS in Microalgae Using Heterologous Promoter Constructs. Plant J. 1998, 13, 427–435. [CrossRef]
- Gornik, S.G.; Maegele, I.; Hambleton, E.A.; Voss, P.A.; Waller, R.F.; Guse, A. Nuclear Transformation of a Dinoflagellate Symbiont of Corals. Front. Mar. Sci. 2022, 9, 1035413. [CrossRef]
- Sprecher, B.N.; Zhang, H.; Lin, S. Nuclear Gene Transformation in the Dinoflagellate Oxyrrhis Marina. Microorganisms 2020, 8, 126. [CrossRef]
- Van Dolah, F.M.; Kohli, G.S.; Morey, J.S.; Murray, S.A. Both Modular and Single-domain Type I Polyketide Synthases Are Expressed in the Brevetoxin-producing Dinoflagellate, Karenia Brevis (Dinophyceae). J. Phycol. 2017, 53, 1325–1339. [CrossRef]
- Kretzschmar, A.-L. Delving into the Genetic Code of Gambierdiscus: The Devil Is in the Detail. PhD Diss., 2019. Dissertation, University of Technology: Syndey, Australia, 2019.
- Verma, A.; Kohli, G.S.; Harwood, D.T.; Ralph, P.J.; Murray, S.A. Transcriptomic Investigation into Polyketide Toxin Synthesis in Ostreopsis (Dinophyceae) Species. Environ. Microbiol. 2019, 21, 4196–4211. [CrossRef]
- Hardison, D.R.; Sunda, W.G.; Shea, D.; Litaker, R.W. Increased Toxicity of Karenia Brevis during Phosphate Limited Growth: Ecological and Evolutionary Implications. PLoS ONE 2013, 8, e58545. [CrossRef]
- Heil, C.A.; Dixon, L.K.; Hall, E.; Garrett, M.; Lenes, J.M.; O’Neil, J.M.; Walsh, B.M.; Bronk, D.A.; Killberg-Thoreson, L.; Hitchcock, G.L.; et al. Blooms of Karenia Brevis (Davis) G. Hansen & Ø. Moestrup on the West Florida Shelf: Nutrient Sources and Potential Management Strategies Based on a Multi-Year Regional Study. Harmful Algae 2014, 38, 127–140. [CrossRef]
- Errera, R.M.; Bourdelais, A.; Drennan, M.A.; Dodd, E.B.; Henrichs, D.W.; Campbell, L. Variation in Brevetoxin and Brevenal Content among Clonal Cultures of Karenia Brevis May Influence Bloom Toxicity. Toxicon 2010, 55, 195–203. [CrossRef]
- Van Dolah, F.M.; Morey, J.S.; Milne, S.; Ung, A.; Anderson, P.E.; Chinain, M. Transcriptomic Analysis of Polyketide Synthases in a Highly Ciguatoxic Dinoflagellate, Gambierdiscus Polynesiensis and Low Toxicity Gambierdiscus Pacificus, from French Polynesia. PLOS ONE 2020, 15, e0231400. [CrossRef]
- Monroe, E.A.; Johnson, J.G.; Wang, Z.; Pierce, R.K.; Van Dolah, F.M. CHARACTERIZATION AND EXPRESSION OF NUCLEAR-ENCODED POLYKETIDE SYNTHASES IN THE BREVETOXIN-PRODUCING DINOFLAGELLATE KARENIA BREVIS 1. J. Phycol. 2010, 46, 541–552. [CrossRef]
- Butterfield, D.A.; Perluigi, M. Redox Proteomics: A Key Tool for New Insights into Protein Modification with Relevance to Disease. Antioxid. Redox Signal. 2017, 26, 277–279. [CrossRef]
- Day, N.J.; Gaffrey, M.J.; Qian, W.-J. Stoichiometric Thiol Redox Proteomics for Quantifying Cellular Responses to Perturbations. Antioxidants 2021, 10, 499. [CrossRef]
- Pham, T.K.; Buczek, W.A.; Mead, R.J.; Shaw, P.J.; Collins, M.O. Proteomic Approaches to Study Cysteine Oxidation: Applications in Neurodegenerative Diseases. Front. Mol. Neurosci. 2021, 14, 678837. [CrossRef]
- Knoke, L.R.; Leichert, L.I. Global Approaches for Protein Thiol Redox State Detection. Curr. Opin. Chem. Biol. 2023, 77, 102390. [CrossRef]
- Colon, R.; Rein, K.S. Essential Components of the Xanthophyll Cycle Differ in High and Low Toxin Karenia Brevis. Harmful Algae 2021, 103, 102006. [CrossRef]
- Galperin, M.Y.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Microbial Genome Analysis: The COG Approach. Brief. Bioinform. 2019, 20, 1063–1070. [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [CrossRef]
- Ødum, M.T.; Teufel, F.; Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Winther, O.; Nielsen, H. DeepLoc 2.1: Multi-Label Membrane Protein Type Prediction Using Protein Language Models. Nucleic Acids Res. 2024, 52, W215–W220. [CrossRef]
- Cox, J.; Mann, M. 1D and 2D Annotation Enrichment: A Statistical Method Integrating Quantitative Proteomics with Complementary High-Throughput Data. BMC Bioinformatics 2012, 13, S12. [CrossRef]
- Khater, S.; Gupta, M.; Agrawal, P.; Sain, N.; Prava, J.; Gupta, P.; Grover, M.; Kumar, N.; Mohanty, D. SBSPKSv2: Structure-Based Sequence Analysis of Polyketide Synthases and Non-Ribosomal Peptide Synthetases. Nucleic Acids Res. 2017, 45, W72–W79. [CrossRef]
- Bachmann, B.O.; Ravel, J. Chapter 8 Methods for In Silico Prediction of Microbial Polyketide and Nonribosomal Peptide Biosynthetic Pathways from DNA Sequence Data. In Methods in Enzymology; Elsevier, 2009; Vol. 458, pp. 181–217 ISBN 978-0-12-374588-0.
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [CrossRef]
- Zhang, M.; Zhang, H.; Li, Q.; Gao, Y.; Guo, L.; He, L.; Zang, S.; Guo, X.; Huang, J.; Li, L. Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella Piezotolerans. J. Agric. Food Chem. 2021, 69, 2316–2324. [CrossRef]
- Skellam, E. Biosynthesis of Fungal Polyketides by Collaborating and Trans -Acting Enzymes. Nat. Prod. Rep. 2022, 39, 754–783. [CrossRef]
- Li, J.; Yang, S.; Wu, Y.; Wang, R.; Liu, Y.; Liu, J.; Ye, Z.; Tang, R.; Whiteway, M.; Lv, Q.; et al. Alternative Oxidase: From Molecule and Function to Future Inhibitors. ACS Omega 2024, acsomega.3c09339. [CrossRef]
- Saha, B.; Borovskii, G.; Panda, S.K. Alternative Oxidase and Plant Stress Tolerance. Plant Signal. Behav. 2016, 11, e1256530. [CrossRef]
- Vishwakarma, A.; Tetali, S.D.; Selinski, J.; Scheibe, R.; Padmasree, K. Importance of the Alternative Oxidase (AOX) Pathway in Regulating Cellular Redox and ROS Homeostasis to Optimize Photosynthesis during Restriction of the Cytochrome Oxidase Pathway in Arabidopsis Thaliana. Ann. Bot. 2015, 116, 555–569. [CrossRef]
- Jacobs, H.T.; Ballard, J.W.O. What Physiological Role(s) Does the Alternative Oxidase Perform in Animals? Biochim. Biophys. Acta BBA - Bioenerg. 2022, 1863, 148556. [CrossRef]
- Edrich, E.S.M.; Duvenage, L.; Gourlay, C.W. Alternative Oxidase – Aid or Obstacle to Combat the Rise of Fungal Pathogens? Biochim. Biophys. Acta BBA - Bioenerg. 2024, 1865, 149031. [CrossRef]
- Schnaufer, A.; Clark-Walker, G.D.; Steinberg, A.G.; Stuart, K. The F 1 -ATP Synthase Complex in Bloodstream Stage Trypanosomes Has an Unusual and Essential Function: F 1 -ATPase in Bloodstream Stage Trypanosomes. EMBO J. 2005, 24, 4029–4040. [CrossRef]
- Gelhaye, E.; Rouhier, N.; Navrot, N.; Jacquot, J.P. The Plant Thioredoxin System. Cell. Mol. Life Sci. 2005, 62, 24–35. [CrossRef]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The Unprecedented Versatility of the Plant Thioredoxin System. Trends Plant Sci. 2017, 22, 249–262. [CrossRef]
- Montrichard, F.; Alkhalfioui, F.; Yano, H.; Vensel, W.H.; Hurkman, W.J.; Buchanan, B.B. Thioredoxin Targets in Plants: The First 30 Years. J. Proteomics 2009, 72, 452–474. [CrossRef]
- Chen, H.; Wang, J.; Zhuang, Y.; Yu, W.; Liu, G. Reduced Fitness and Elevated Oxidative Stress in the Marine Copepod Tigriopus Japonicus Exposed to the Toxic Dinoflagellate Karenia Mikimotoi. Antioxidants 2022, 11, 2299. [CrossRef]
- Deng, Y.; Wang, K.; Hu, Z.; Hu, Q.; Tang, Y.Z. Toxic and Non-Toxic Dinoflagellates Host Distinct Bacterial Communities in Their Phycospheres. Commun. Earth Environ. 2023, 4, 263. [CrossRef]
- Chen, W.; Colon, R.; Louda, J.W.; Del Rey, F.R.; Durham, M.; Rein, K.S. Brevetoxin (PbTx-2) Influences the Redox Status and NPQ of Karenia Brevis by Way of Thioredoxin Reductase. Harmful Algae 2018, 71, 29–39. [CrossRef]
- Lillig, C.H.; Berndt, C. Glutaredoxins in Thiol/Disulfide Exchange. Antioxid. Redox Signal. 2013, 18, 1654–1665. [CrossRef]
- Hall, E.R.; Heil, C.A.; Frankle, J.D.; Klass, S.; Devillier, V.; Lovko, V.; Toyoda, J.H.; Pierce, R. Mitigation of Karenia Brevis Cells and Brevetoxins Using Curcumin, a Natural Supplement. Water 2024, 16, 1458. [CrossRef]
- AziziHariri, P.; Hossain, I.; Burni, F.; Raghavan, S.R.; Lovko, V.J.; McLean, T.I.; John, V.T. A Simple Method to Clear Harmful Algal Blooms: Sprayable Foams with Algaecides and Flocculants. ACS EST Water 2025, acsestwater.5c00111. [CrossRef]
- Fallon, T.R.; Shende, V.V.; Wierzbicki, I.H.; Auber, R.P.; Gonzalez, D.J.; Wisecaver, J.H.; Moore, B.S. Giant Polyketide Synthase Enzymes Biosynthesize a Giant Marine Polyether Biotoxin 2024.
- Atkinson, H.J.; Babbitt, P.C. An Atlas of the Thioredoxin Fold Class Reveals the Complexity of Function-Enabling Adaptations. PLoS Comput. Biol. 2009, 5, e1000541. [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [CrossRef]
- Yang, X.; Liu, Z.; Zhang, Y.; Shi, X.; Wu, Z. Dinoflagellate–Bacteria Interactions: Physiology, Ecology, and Evolution. Biology 2024, 13, 579. [CrossRef]
- Hallegraeff, G.M.; Anderson, D.M.; Cembella, A.D. Manual on Harmful Marine Microalgae; Monographs on oceanographic methodology; UNESCO publ: Paris, 2003; ISBN 978-92-3-103871-6.
- Gates, J.A.; Wilson, W.B. THE TOXICITY OF GONYAULAX MONILATA HOWELL TO MUGIL CEPHALUS. Limnol. Oceanogr. 1960, 5, 171–174. [CrossRef]
- Ryan, D.E.; Pepper, A.E.; Campbell, L. De Novo Assembly and Characterization of the Transcriptome of the Toxic Dinoflagellate Karenia Brevis. BMC Genomics 2014, 15, 888. [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [CrossRef]
- Aguilan, J.T.; Kulej, K.; Sidoli, S. Guide for Protein Fold Change and p -Value Calculation for Non-Experts in Proteomics. Mol. Omics 2020, 16, 573–582. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




