Submitted:
11 June 2025
Posted:
12 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Overview of Satellite Image
1.2. Related Work
1.2.1. Overview of Clustering Algorithm
- Linear scalability with data volume;
- Support asynchronous parallel execution across independent data slices, enabling non-blocking processing of extremely large datasets (e.g. multiple 1 TB segments) by distributing computational task concurrently across multi-core or distributed systems;
- Flexibility to integrate algorithmic improvements such as advanced centroid initialization, iteration reduction, and outlier handling, as explored in this study.
1.2.2. Advancement in K-Means Clustering
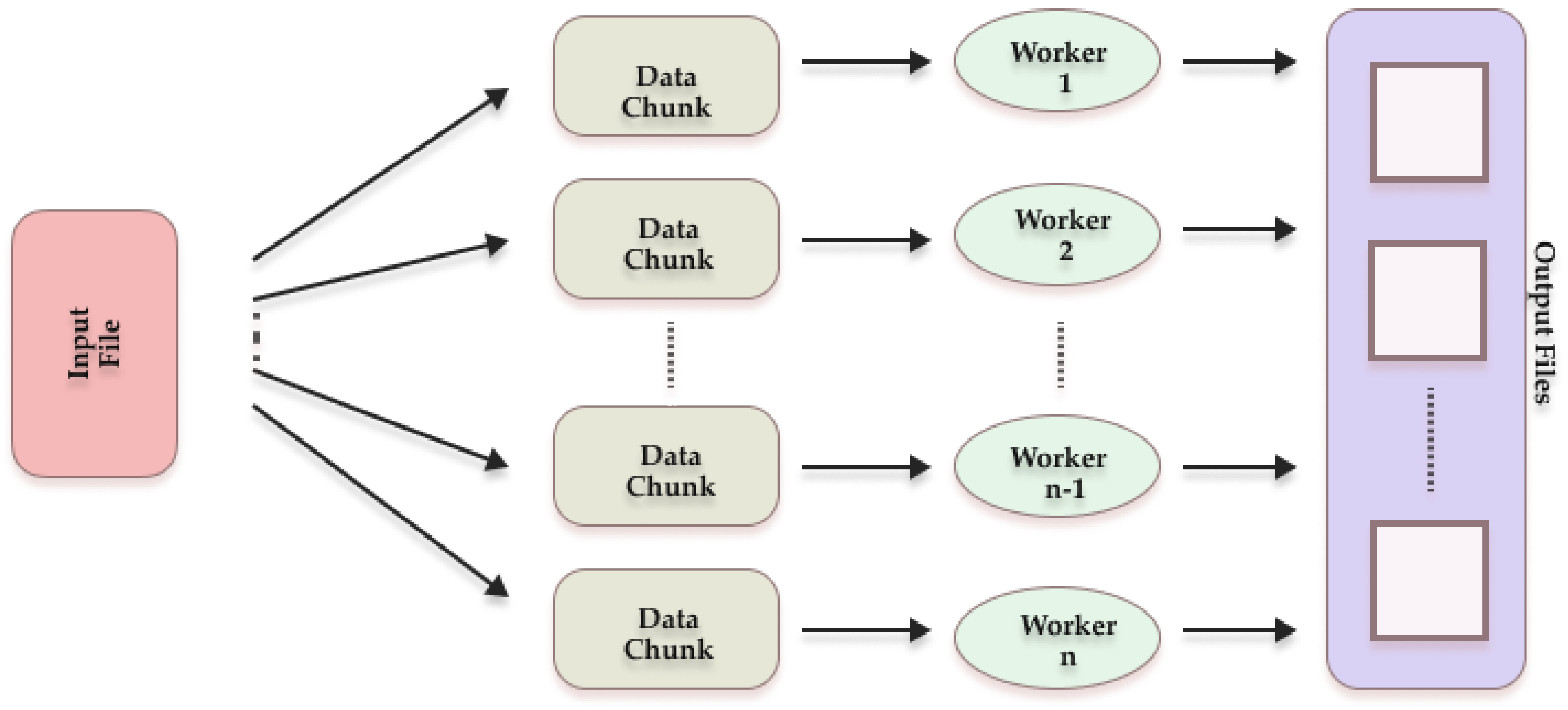
1.2.3. Parallelization Techniques in K-Means Cluster
1.2.4. Motivation and Contribution of This Study
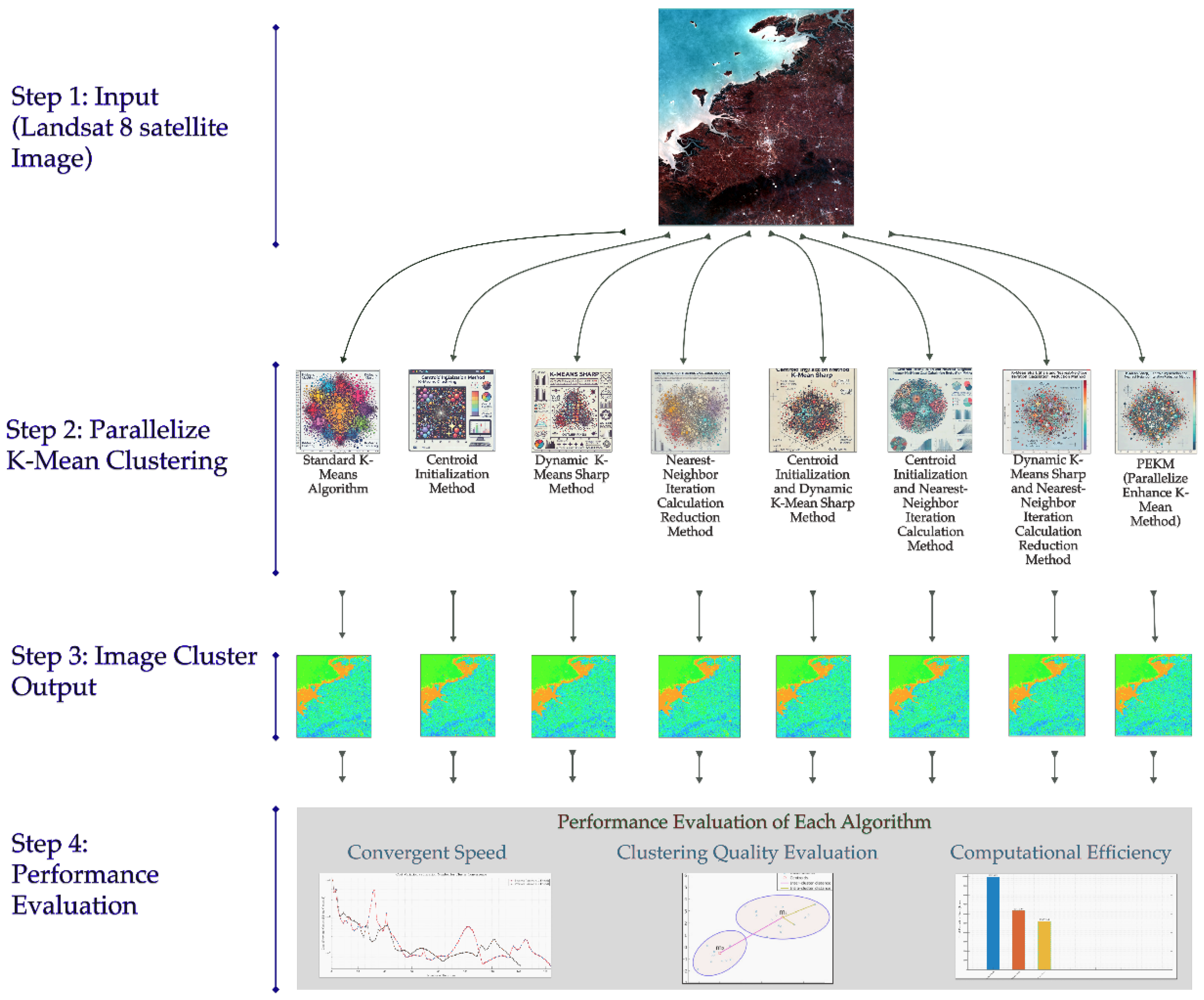
2. Materials
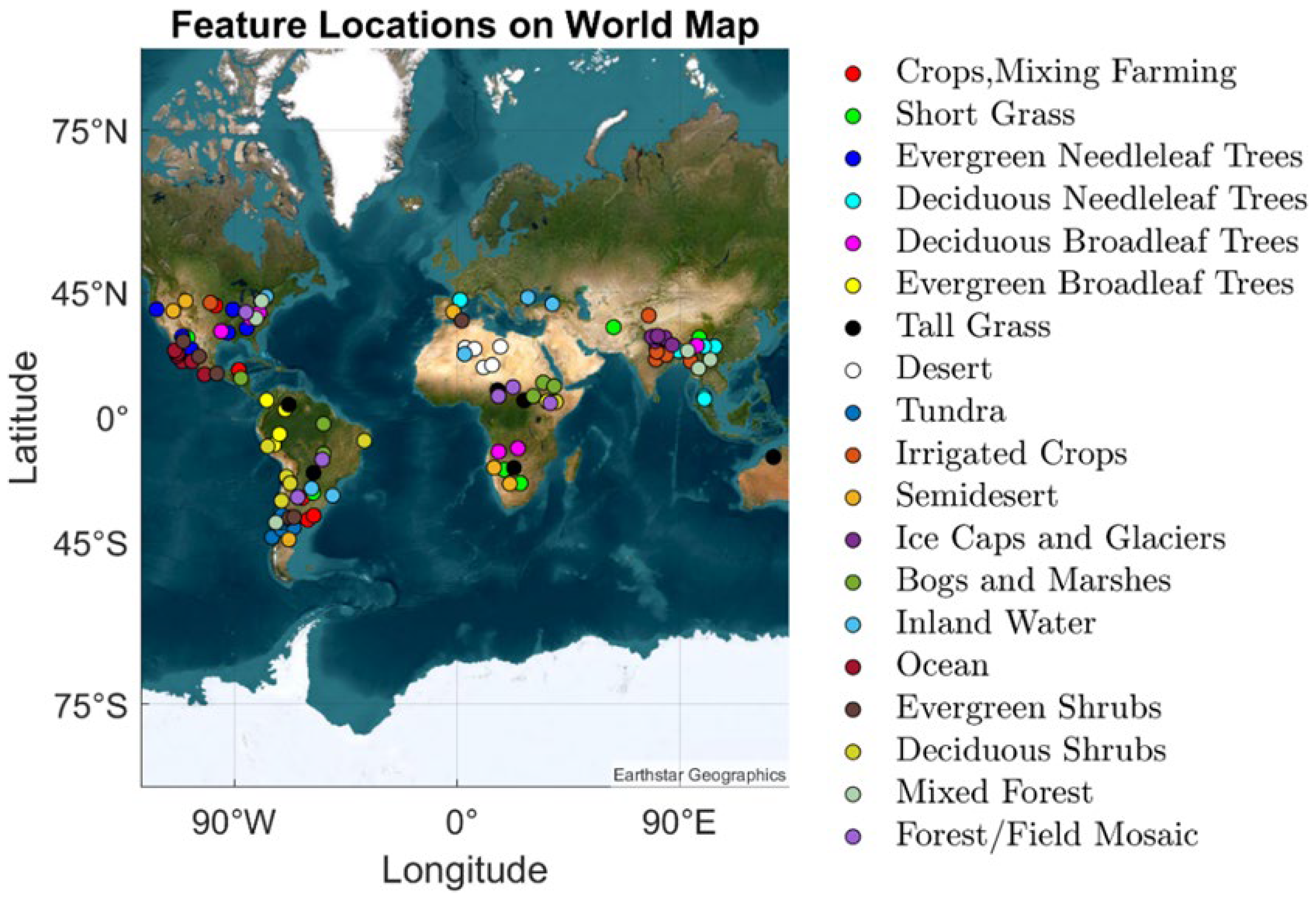
2.1. SDSU Dervied Data Product
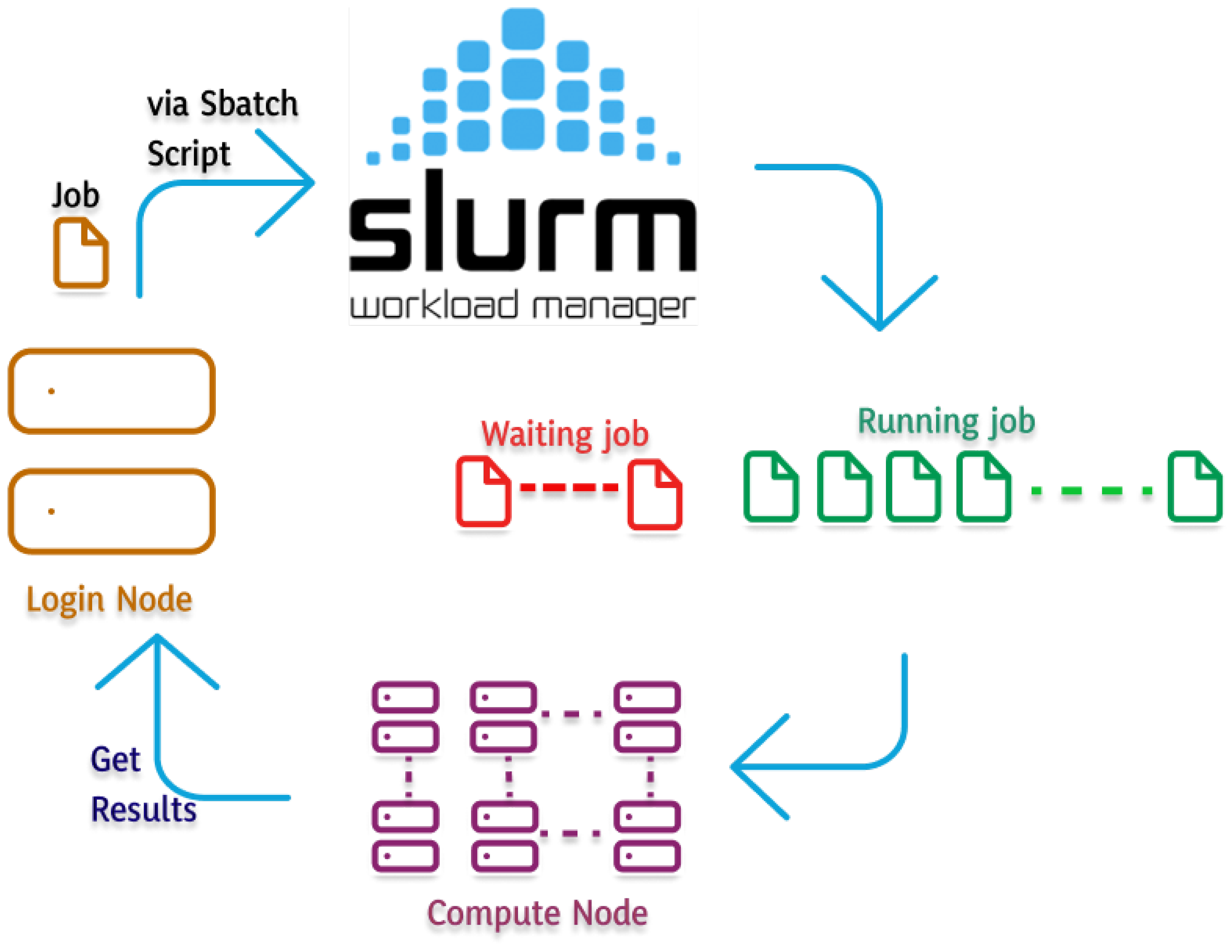
2.2. Hardware and Software Specifications
3. Methodology
3.1. Satellite Imagery Dataset
| Parameter | Values |
|---|---|
| Data cube Number | 114 |
| Data cube Size | 3712*3712*16 (~1 GB) |
| # Cluster (k) | 160 |
| Convergent Criteria | 0.0005 |
3.2. Algorithm Development
- Centroid Initialization Method
- Nearest-Neighbor Iteration Calculation Reduction Method
- Dynamic K-Means Sharp Method
3.2.1. Parallelize Standard K-Means Algorithms
| Algorithm 1: Pseudocode for K-Means Algorithm | |
| Input: | I= {I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of K distinct clusters |
| Step: | 1. Randomly select K initial cluster centers (centroids) from the 114 data cubes, where each centroid is represented by the matrix μmn as shown in Equation (1). 2. While max(|μmn,old - μmn,new |)<0.0005 do: 3. For (I=1: number_of_images)//for all images For (x=1: number_of_row) For (y=1: number_of_column)// for all pixel 4. Calculate the Euclidean distance between Pxyn and each centroid μmn using equation (2) 5. Assign Pxyn to the cluster with the nearest centroid (min(D(x,y,m))) 6. Update each centroid μmn: For each cluster m: 7. Calculate the new centroid using equation (3). 8. Check for convergence: 9. Calculate the maximum change in centroids: 10. DiffMean = max(|μmn,old - μmn_new|) 11. If DiffMean < 0.0005, then: Convergence achieved, exit loop 12. Else: Update centroids: μmn = μmn_new 13. Continue iteration End While |
| Algorithm 2: Pseudocode for Parallelization Standard K-Means algorithm | |
| Input: | I={I1, I2, I3,….,In} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of K distinct clusters |
| Step: | Randomly select K initial cluster centers (centroids) from the 114 data cubes, where each centroid is represented by the matrix μmn as shown in Equation (1). While max(|μmn,old - μmn,new |) < 0.0005 do: Parfor each Data chunk I={I1, I2,I3,..,Id} For (x=1: number_of_rows) For (y=1: number_of_columns) Calculate the Euclidean distance between Pxyn and each centroid μmn using Equation 2. Assign Pxyn to the cluster with the nearest centroid (min(D(x,y,m))) Calculate the partial sums for each cluster for updating centroids End Parfor Combine partial sums from all chunks to calculate the new mean (centroid) for each cluster. Update each centroid μmn: For each cluster m: Calculate the new centroid using equation (3). Check for convergence: Calculate the maximum change in centroids: DiffMean = max(|μmn,old - μmn_new|) If DiffMean < 0.0005, then: Convergence achieved, exit while loop Else: Update centroids: μmn = μmn_new Continue iteration End While |
3.2.2. Parallelize Centroid Initialization Method
| Algorithm 3: Pseudocode for class initialization of parallel centroid initialization method | |
| Input: |
I={I1, I2, I3,….,In} // set of n data cube ( 114 data cube with size 3712*3712*16 ) K //Number of desired clusters |
| Output: | A set of K distinct clusters. |
| Step: | sampledpixelvalues ← 10*10 matrix of zeros (size: 100 x 16) counter← 1 For (i = 1 to 114): imagein ← read image from Imagefile Select only the first 16 layers of the image: imagein ← imagein(:,:,1:16) image ← imagein(:,:,5) // Select band 5 for operations Set grid size: gridsize ← 371 Calculate number of grid points: numpointX ← floor(size of image in dimension 1 / gridsize) numpointY ← floor(size of image in dimension 2 / gridsize) For each grid point, extract pixel values: For j = 1 to numpointX: For z = 1 to numpointY: Calculate x and y based on grid index: x ← (j - 1) * gridsize + 1 y ← (z - 1) * gridsize + 1 Assign sampled pixel values to sampledpix- elvalues [counter,:] ← imagein(x, y, :) Counter Increment Save the sampled pixel values: Sort the sample pixel values by the 5Th Column: columntosortby ← sampledpixelvalues(:, 5) sortedvalues, sortedindices ← sort(columntosortby) sortedMatrix ← sampledpixelvalues(sorted by sortedindices) Remove rows containing NaN values: rowswithnans ← any(isnan(sortedMatrix)) sortedMatrix ← sortedMatrix without rowswithnans Calculate the Initial centroid: row, column ← size(sortedMatrix) rows_per_group ← floor (row / 160) remainder ← mod (row, 160) InitialCluster ← 160 x column matrix of zeros Group rows and calculate mean for each group: start_index ← 1 For (i = 1 to 160): If i ≤ remainder set end_index ← start_index + rows_per_group Else Set end_index ← start_index + rows_per_group - 1 Currentgroup ← sortedMatrix[startindex:endindex, :] Calculate mean for the current group and store in InitialCluster[i, :] ← mean(currentgroup) Update startindex ← endindex + 1 Save the calculated initial centroids. |
| Algorithm 4: Pseudocode for main classification |
|
Parfor each Data chunk I={I1, I2,I3,..,Id} Reshape the image data into 2D array For (x=1:number_of_row) Calculate the Euclidean distance between Px,n and each centroid μmn using Equation 4 Assign Px,n to the cluster with the nearest centroid (min(D(x,m))) Calculate the partial sums for each cluster for updating centroids End Parfor |
3.2.3. Parallelized Nearest-Neighbor Iteration Calculation Reduction Method
| Algorithm 5: Pseudocode for Parallelize Nearest-Neighbor Iteration Calculation Reduction Method | |
| Input: | I={I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters. |
| Step: | Randomly select K initial cluster centers (centroids) from the 114 data cubes, where each centroid is represented by the matrix μmn as shown in Equation (1). While max(|μmn,old - μmn,new |) < 0.0005 do: Parfor each Data chunk I={I1, I2,I3,..,Id} Reshape the image data into 2D array For (x=1 : number_of_row) Calculate the Euclidean distance between Pxn and each centroid μmn using Equation 2. Reshape back to original dimension Assign Pxyn to the cluster with the nearest centroid (min(D(x,y,m))). Store the labels of 15 nearest cluster centers for pixel Pxyn in the arrays Cluster_Label[]. Separately store the distance of Pxyn to the nearest cluster in Closest_Dist[]. Set Cluster_Label[]=k(the indices of the 15 nearest clusters). Set Closest_Dist[x,y]=D(Pxyn, μmn ); (the minimum distance to the closest cluster). Calculate the partial sums for each cluster for updating centroids End Parfor Update each centroid μmn: For each cluster m: Calculate the new centroid using equation (3). Update centroids: μmn = μmn_new Repeat Parfor each Data chunk I={I1, I2,I3,..,Id} Reshape the image data into 2D array Calculate the distance between the pixel and the centroids of the 15 nearest clusters stored in Cluster_Label[]. Identify the rank of the currently assigned cluster within the 15 nearest clusters: If the currently assigned cluster is within the top 12 nearest clusters, no recomputation is required. If the currently assigned cluster is ranked between the 13th and 15th nearest clusters or the pixel has not been assigned, then go for the recomputation of pixel between all the cluster. End parfor; For each cluster m, (1<=m<=K), recalculate the centroid. Until the convergent criteria is met (previous centroid – new centroid) < 0.0005 |
3.2.4. Parallelize Dynamic K-Means Sharp Method
Outlier Detection
| Algorithm 6: Pseudocode for Parallelize Dynamic K-Means Sharp Method | |
| Input: | I={I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters. |
| Step: | Randomly select K initial cluster centers (centroids) from the 114 data cubes, where each centroid is represented by the matrix μmn as shown in Equation (1). While max (|μmn,old - μmn,new |)<0.0005 do: Parfor each Data chunk I={I1, I2,I3,..,Id} Reshape the image data into 2D array For (x=1 : number_of_row) Calculate the Euclidean distance between Pxn and each centroid μmn using Equation (3). Reshape back to original dimension Assign pixel Pxyn to the cluster with the nearest centroid (min(D(x,y,m))). Calculate the partial sums for each cluster for updating centroids END Parfor Update each centroid μmn: For each cluster m: Calculate the new centroid using equation (3). Update centroids: μmn = μmn_new Calculate Spatial Standard deviation For each data chunk, calculate the sum of squared difference for each band of clusters squared_diff=(Meantile-NewCluster)2 Accumulate the sum across all data cubes. Compute the standard deviation for each cluster and band: std_values=sqrt(sum_squared_diff/count) Repeat Parfor loop to update pixel assignments: Compute inliers and outliers based on the spatial standard deviation: For each cluster and band: get pixels in the current cluster and compute the absolute difference pixels and the cluster mean for each band. inliers are those within 3*std_value. inliers=abs(cluster_pixel(:,feature)- mean_value)<=3*std_value Update inlier count and calculate partial sums for the next centroid update: Inlier_count(cluster,band)+=sum(inliers) MeanCluster(cluster,band)+=sum(cluster_pixel(inliers,band)) Outlier count: Outlier_count(cluster,band)=total_pixel_in_cluster-inlier_count(cluster,band) End Parfor Update the cluster centroids based on inliers using equation 5. Until the convergent criteria is met (previous centroid – new centroid) < 0.0005 |
3.2.5. Integrated Variant Comparisons
| Combination Name | Centroid Initialization Algorithm | Nearest-Neighbor Iteration Calculation Reduction Algorithm | Dynamic K-Means Sharp Algorithm |
|---|---|---|---|
| Parallel Combination 1 | 1 | 0 | 1 |
| Parallel Combination 2 | 0 | 1 | 1 |
| Parallel Combination 3 | 1 | 1 | 0 |
| Parallel Enhanced K-Means (PEKM) | 1 | 1 | 1 |
| Algorithm 7: Centroid Initialization and Dynamic K-Means Sharp Method | |
| Input: |
I= {I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters |
| Step: | Phase 1: Identify the initial cluster centroids by using Algorithm 3 Phase 2: Update centroid by using Algorithm 6 |
| Algorithm 8: Dynamic K-Means Sharp and Nearest-Neighbor Iteration Calculation Reduction Method | |
| Input: |
I={I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters |
| Step: | Phase 1: Assign each data point to the appropriate cluster by Algorithm 5. Phase 2: Update centroid by using Algorithm 6 |
| Algorithm 9: Centroid Initialization and Nearest-Neighbor Iteration Calculation Reduction Method | |
| Input: |
I={I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters |
| Step: | Phase 1: Identify the initial cluster centroids by using Algorithm 3 Phase 2: Assign each data point to the appropriate cluster by Algorithm 5 |
Parallel Enhanced K-Means Method (PEKM)
| Algorithm 10: Centroid Initialization Method, Dynamic K-Means Sharp, and Nearest-Neighbor Iteration Calculation Reduction Method | |
| Input: |
I={I1, I2, I3,….,Id} // set of d data cube ( 114 data cube with size 3712*3712*16 ) K // Number of desired clusters |
| Output: | A set of k clusters |
| Step: | Phase 1: Determine the initial centroids of the clusters by using Algorithm 3 Phase 2: Assign each data point to the appropriate cluster by Algorithm 5. Phase 3: Update centroid by using Algorithm 6 |
3.3. Convergent Criteria
- Reaching a predetermined maximum number of iterations.
- Having fewer pixel reassignments per iteration than a set threshold.
- Centroid shifts fall below a specified distance threshold during an update cycle.
3.4. Performance Metrics
3.4.1. Convergence Speed
3.4.2. Clustering Quality
- Vij : The jth valid pixel in the ith cluster.
- Cik : The centroid value for the kth band of the ith cluster.
- Ni : The number of valid pixels in the ith cluster.
- C : Total number of clusters.
- B : Total number of bands.
3.4.3. Computational Efficiency
3.5. Comparative Analysis
4. Evaluation and Experimental Results
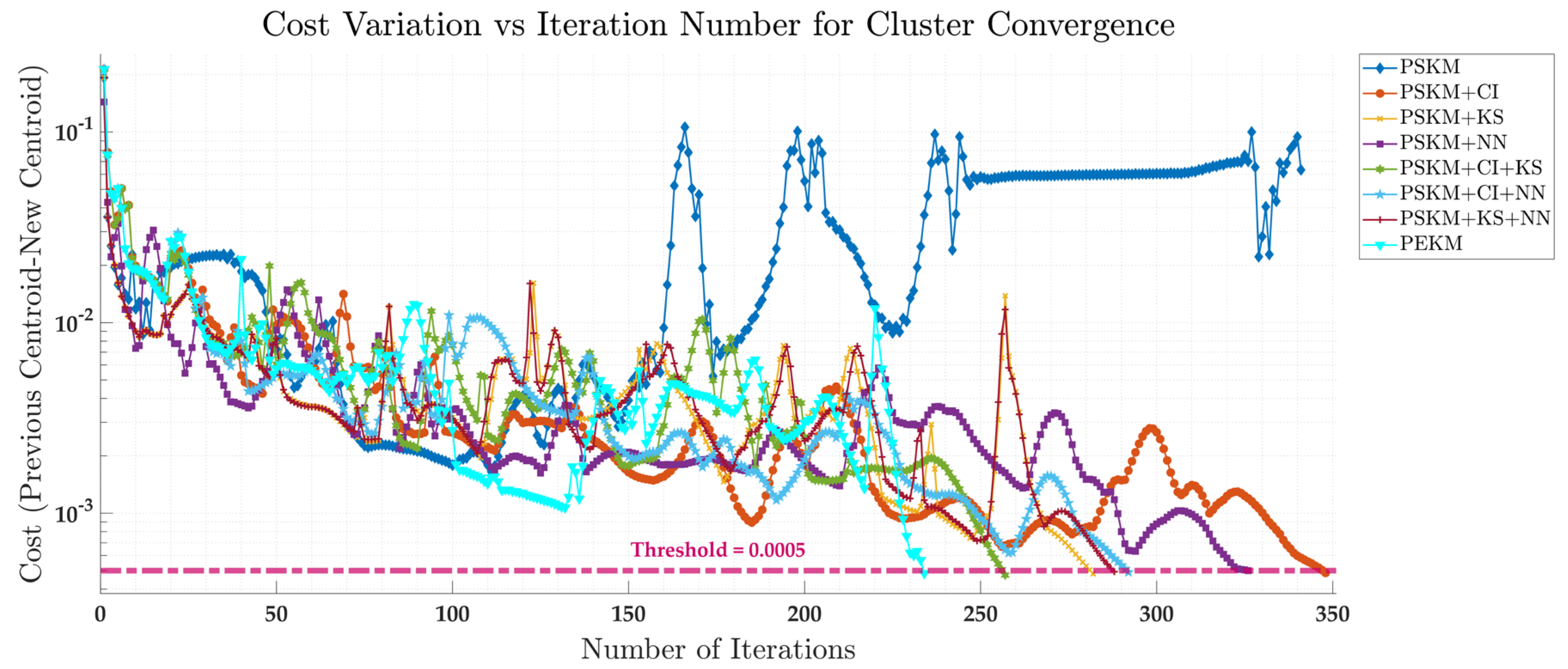
4.1. Convergent Speed Comparison
| Method Abbreviation | Full Method Description |
|---|---|
| PSKM | Parallelized Standard K-Means Algorithm |
| PSKM+CI | Parallelized Centroid Initialization Method |
| PSKM+KS | Parallelized Dynamic K-Means Sharp Method |
| PSKM + NN | Parallelized Nearest-Neighbor Iteration Calculation Reduction Method |
| PSKM + CI + KS | Parallelized Centroid Initialization and Dynamic K-Means Sharp Method |
| PSKM + CI +NN | Parallelized Centroid Initialization and Nearest-Neighbor Iteration Calculation Reduction Method |
| PSKM + KS + NN | Parallelized Dynamic K-Means Sharp and Nearest-Neighbor Iteration Calculation Reduction Method |
| PEKM | Parallelized Enhance K-Means Method |
| Method | Iterations till convergence |
|---|---|
| PSKM+CI | 348 iterations |
| PSKM+KS | 282 iterations |
| PSKM + NN | 326 iterations |
| PSKM + CI + KS | 257 iterations |
| PSKM + CI +NN | 292 iterations |
| PSKM + KS + NN | 288 iterations |
| PEKM | 234 iterations |
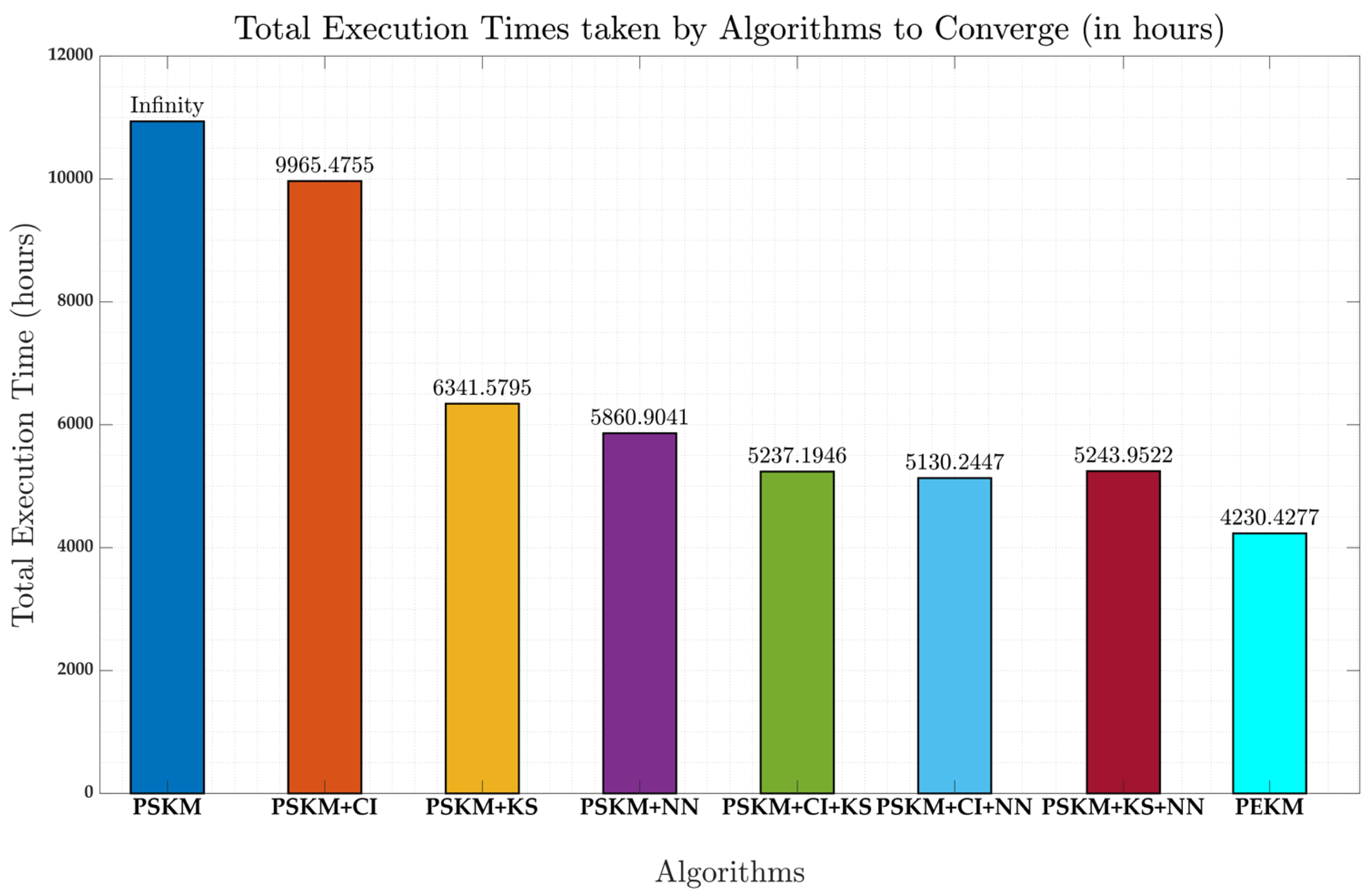
4.2. Computation Efficiency Comparison
| Method | Hours till convergence |
|---|---|
| PSKM+CI | 9965.47 hours |
| PSKM+KS | 6341.58 hours |
| PSKM + NN | 5860.90 hours |
| PSKM + CI + KS | 5237.19 hours |
| PSKM + CI +NN | 5130.24 hours |
| PSKM + KS + NN | 5243.95 hours |
| PEKM | 4230.43 hours |
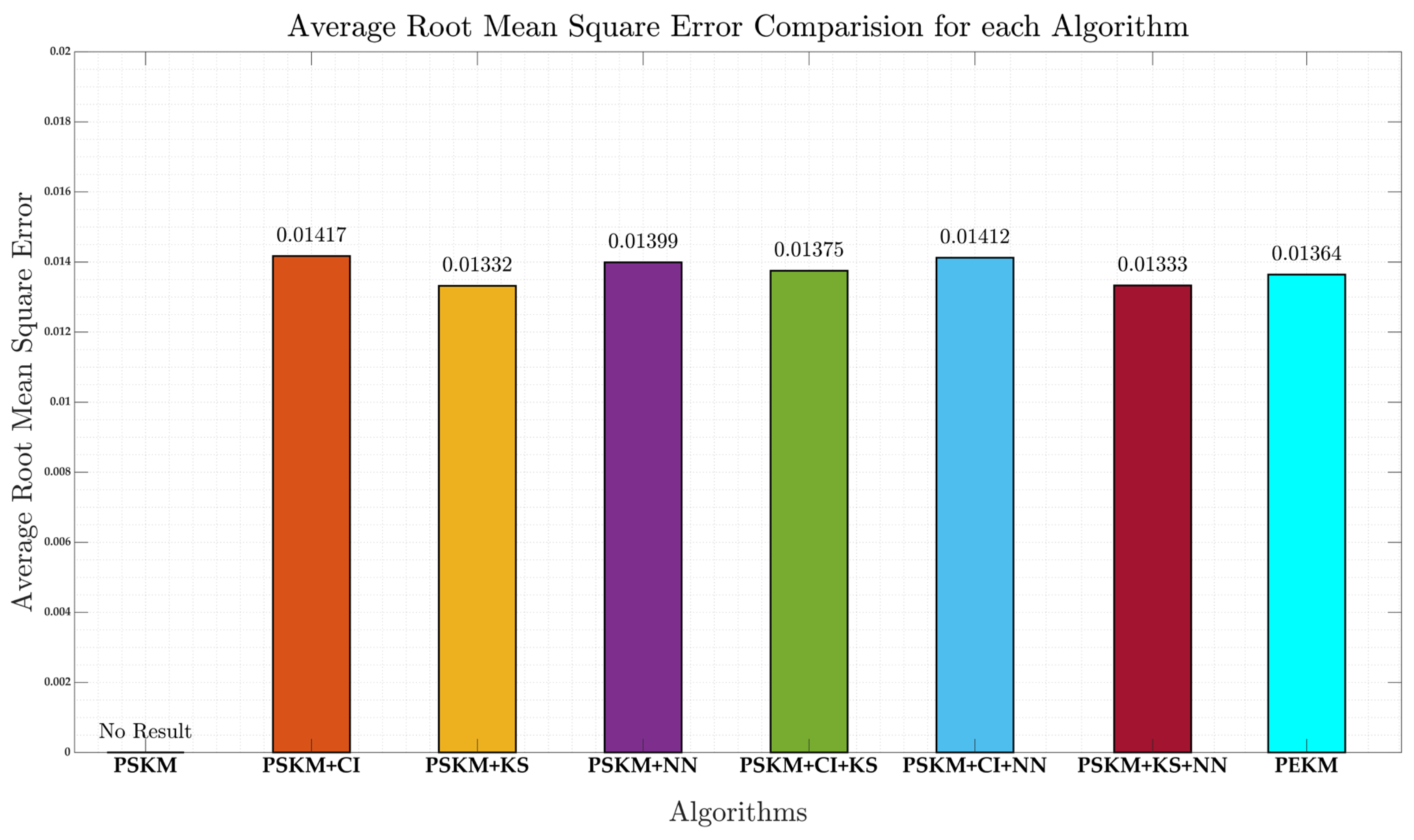
4.3. Clustering Quality Evaluation
| Method | RMSE |
|---|---|
| PSKM+CI | 0.01417 |
| PSKM+KS | 0.01332 |
| PSKM + NN | 0.01399 |
| PSKM + CI + KS | 0.01375 |
| PSKM + CI +NN | 0.01412 |
| PSKM + KS + NN | 0.01333 |
| PEKM | 0.01364 |
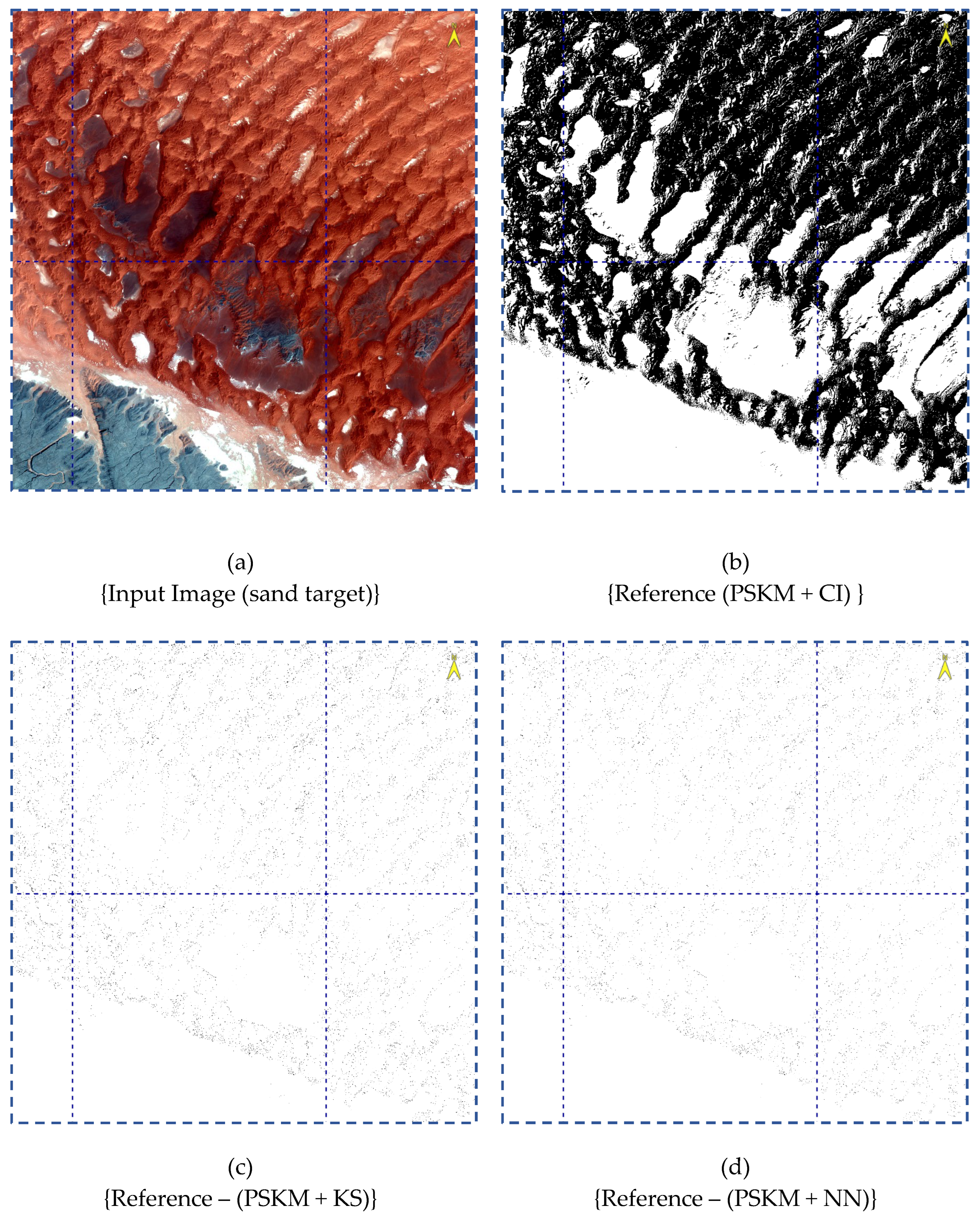
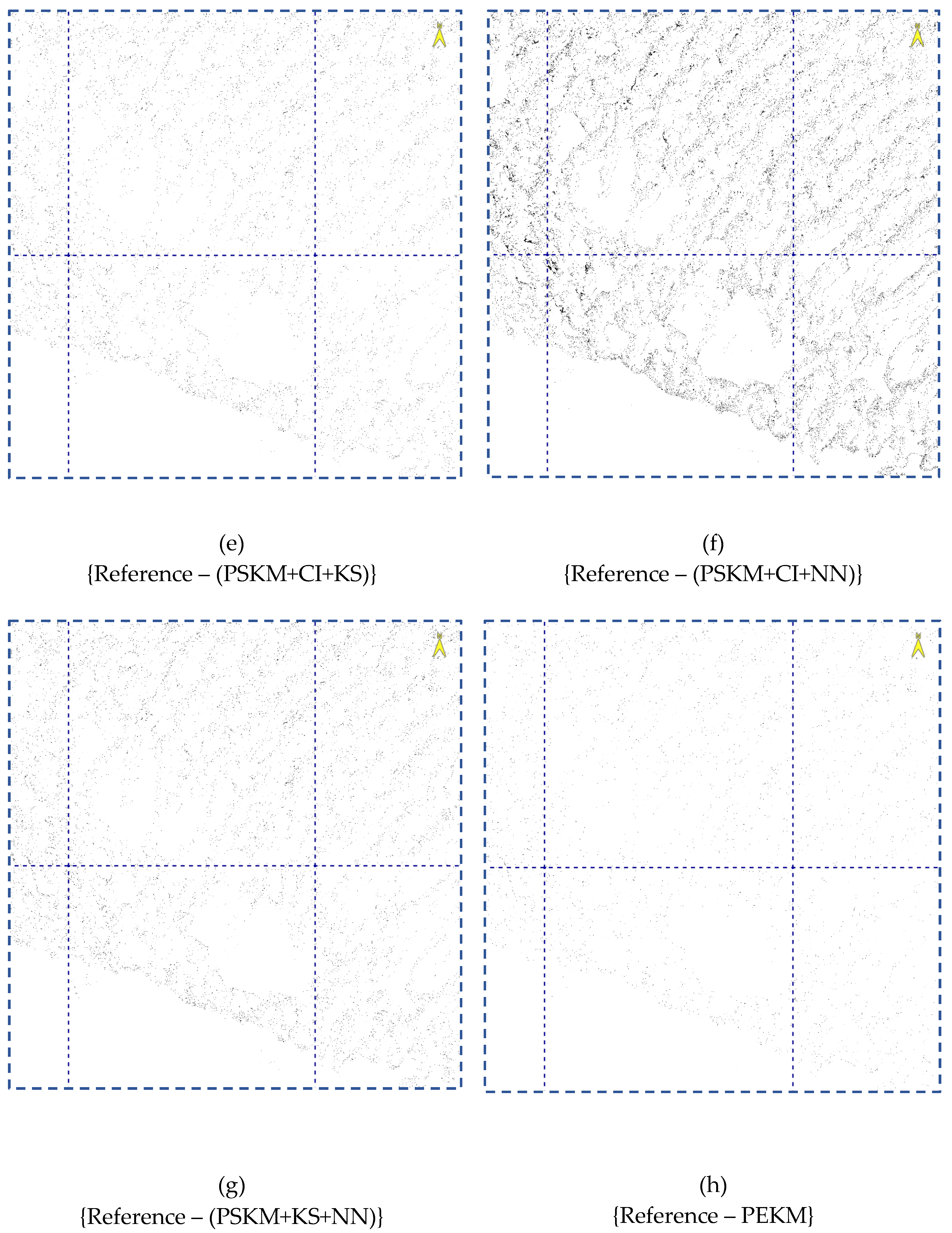
4.4. Image Cluster Output
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ao, S.I. World Congress on Engineering : WCE 2012 : 4-6 July, 2012, Imperial College London, London, U.K; Newswood Ltd.: International Association of Engineers, 2012; ISBN 9789881925138. [Google Scholar]
- Wulder, M.A.; Roy, D.P.; Radeloff, V.C.; Loveland, T.R.; Anderson, M.C.; Johnson, D.M.; Healey, S.; Zhu, Z.; Scambos, T.A.; Pahlevan, N.; et al. Fifty Years of Landsat Science and Impacts. Remote Sens Environ 2022, 280, 113195. [Google Scholar] [CrossRef]
- Dutta, S.; Das, M. Remote Sensing Scene Classification under Scarcity of Labelled Samples—A Survey of the State-of-the-Arts. Comput Geosci 2023, 171. [Google Scholar] [CrossRef]
- Shrestha, M.; Leigh, L.; Helder, D. Classification of North Africa for Use as an Extended Pseudo Invariant Calibration Sites (EPICS) for Radiometric Calibration and Stability Monitoring of Optical Satellite Sensors. Remote Sens (Basel) 2019, 11. [Google Scholar] [CrossRef]
- Fajardo Rueda, J.; Leigh, L.; Teixeira Pinto, C.; Kaewmanee, M.; Helder, D. Classification and Evaluation of Extended Pics (Epics) on a Global Scale for Calibration and Stability Monitoring of Optical Satellite Sensors. Remote Sens (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Babawuro Usman. (Satellite Imagery Land Cover Classification Using K-Means Clustering Algorithm: Computer Vision for Environmental Information Extraction), 2013.
- H.E. Yasin, E.; Kornel, C. Evaluating Satellite Image Classification: Exploring Methods and Techniques. In Geographic Information Systems - Data Science Approach; IntechOpen, 2024. [Google Scholar]
- Kaewmanee, M.; Leigh, L.; Shah, R.; Gross, G. Inter-Comparison of Landsat-8 and Landsat-9 during On-Orbit Initialization and Verification (OIV) Using Extended Pseudo Invariant Calibration Sites (EPICS): Advanced Methods. Remote Sens (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Shah, R.; Leigh, L.; Kaewmanee, M.; Pinto, C.T. Validation of Expanded Trend-to-Trend Cross-Calibration Technique and Its Application to Global Scale. Remote Sens (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Yin, L.; Lv, L.; Wang, D.; Qu, Y.; Chen, H.; Deng, W. Spectral Clustering Approach with K-Nearest Neighbor and Weighted Mahalanobis Distance for Data Mining. Electronics (Switzerland) 2023, 12. [Google Scholar] [CrossRef]
- Ni, L.; Manman, P.; Qiang, W. A Spectral Clustering Algorithm for Non-Linear Graph Embedding in Information Networks. Applied Sciences (Switzerland) 2024, 14. [Google Scholar] [CrossRef]
- Ran, X.; Xi, Y.; Lu, Y.; Wang, X.; Lu, Z. Comprehensive Survey on Hierarchical Clustering Algorithms and the Recent Developments; Springer Netherlands, 2023; Vol. 56, ISBN 0123456789. [Google Scholar]
- Zhang, X.; Shen, X.; Ouyang, T. Extension of DBSCAN in Online Clustering: An Approach Based on Three-Layer Granular Models. Applied Sciences (Switzerland) 2022, 12. [Google Scholar] [CrossRef]
- Dinh, T.; Hauchi, W.; Lisik, D.; Koren, M.; Tran, D.; Yu, P.S.; Torres-Sospedra, J. Data Clustering: An Essential Technique in Data Science. 2024. [Google Scholar]
- Chaudhry, M.; Shafi, I.; Mahnoor, M.; Vargas, D.L.R.; Thompson, E.B.; Ashraf, I. A Systematic Literature Review on Identifying Patterns Using Unsupervised Clustering Algorithms: A Data Mining Perspective. Symmetry (Basel) 2023, 15, 1–44. [Google Scholar] [CrossRef]
- Miao, S.; Zheng, L.; Liu, J.; Jin, H. K-Means Clustering Based Feature Consistency Alignment for Label-Free Model Evaluation. IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops 2023, 2023, 3299–3307. [Google Scholar] [CrossRef]
- Al-Sabbagh, A.; Hamze, K.; Khan, S.; Elkhodr, M. An Enhanced K-Means Clustering Algorithm for Phishing Attack Detections. Electronics (Switzerland) 2024, 13, 1–18. [Google Scholar] [CrossRef]
- Ahmed, M.; Seraj, R.; Islam, S.M.S. The K-Means Algorithm: A Comprehensive Survey and Performance Evaluation. Electronics (Switzerland) 2020, 9, 1–12. [Google Scholar] [CrossRef]
- Ikotun, A.M.; Ezugwu, A.E.; Abualigah, L.; Abuhaija, B.; Heming, J. K-Means Clustering Algorithms: A Comprehensive Review, Variants Analysis, and Advances in the Era of Big Data. Inf Sci (N Y) 2023, 622, 178–210. [Google Scholar] [CrossRef]
- Rana, M.; Rahman, A.; Smith, D. Hierarchical Semi-Supervised Approach for Classifying Activities of Workers Utilising Indoor Trajectory Data. Internet of Things (The Netherlands) 2024, 28, 101386. [Google Scholar] [CrossRef]
- Dash, M.; Liu, H.; Scheuermann, P.; Tan, K.L. Fast Hierarchical Clustering and Its Validation. Data Knowl Eng 2003, 44, 109–138. [Google Scholar] [CrossRef]
- Shi, K.; Yan, J.; Yang, J. A Semantic Partition Algorithm Based on Improved K-Means Clustering for Large-Scale Indoor Areas. ISPRS Int J Geoinf 2024, 13. [Google Scholar] [CrossRef]
- Degirmenci, A.; Karal, O. Efficient Density and Cluster Based Incremental Outlier Detection in Data Streams. Inf Sci (N Y) 2022, 607, 901–920. [Google Scholar] [CrossRef]
- Spectral Clustering for Large Scale Datasets (Part 1) | by Guava | Medium. Available online: https://medium.com/%40guava1427/spectral-clustering-for-large-scale-datasets-part-1-874571887610 (accessed on 15 April 2025).
- Sreedhar, C.; Kasiviswanath, N.; Chenna Reddy, P. Clustering Large Datasets Using K-Means Modified Inter and Intra Clustering (KM-I2C) in Hadoop. J Big Data 2017, 4. [Google Scholar] [CrossRef]
- Capó, M.; Pérez, A.; Lozano, J.A. An Efficient K -Means Clustering Algorithm for Massive Data. 2018. [Google Scholar]
- Jin, S.; Cui, Y.; Yu, C. A New Parallelization Method for K-Means.
- Honggang, W.; Jide, Z.; Hongguang, L.; Jianguo, W. Parallel Clustering Algorithms for Image Processing on Multi-Core CPUs. In Proceedings of the Proceedings - International Conference on Computer Science and Software Engineering, CSSE 2008; 2008; Vol. 3, pp. 450–453. [Google Scholar]
- Zhang, Y.; Xiong, Z.; Mao, J.; Ou, L. The Study of Parallel K-Means Algorithm.
- Macqueen, J. SOME METHODS FOR CLASSIFICATION AND ANALYSIS OF MULTIVARIATE OBSERVATIONS.
- Parveen, S.; Yang, M. Lasso-Based k-Means ++ Clustering. 2025. [Google Scholar]
- Khan, A.A.; Bashir, M.S.; Batool, A.; Raza, M.S.; Bashir, M.A. K-Means Centroids Initialization Based on Differentiation Between Instances Attributes. 2024; 2024. [Google Scholar] [CrossRef]
- Chan, J.Y.K.; Leung, A.P.; Xie, Y. Efficient High-Dimensional Kernel k-Means++ with Random Projection. Applied Sciences (Switzerland) 2021, 11. [Google Scholar] [CrossRef]
- Li, H.; Sugasawa, S.; Katayama, S. Adaptively Robust and Sparse K-Means Clustering. Transactions on Machine Learning Research 2024, 2024, 1–29. [Google Scholar]
- Olukanmi, P.O.; Twala, B. K-Means-Sharp: Modified Centroid Update for Outlier-Robust k-Means Clustering. In Proceedings of the 2017 Pattern Recognition Association of South Africa and Robotics and Mechatronics International Conference, PRASA-RobMech 2017; Institute of Electrical and Electronics Engineers Inc., 2017; pp. 14–19. [Google Scholar]
- Zhao, J.; Bao, Y.; Li, D.; Guan, X. An Improved K-Means Algorithm Based on Contour Similarity. Mathematics 2024, 12. [Google Scholar] [CrossRef]
- Yao, X.; Chen, Z.; Gao, C.; Zhai, G.; Zhang, C. ResAD: A Simple Framework for Class Generalizable Anomaly Detection. 2024; 1–25. [Google Scholar]
- Wu, S.; Zhai, Y.; Liu, J.; Huang, J.; Jian, Z.; Dai, H.; Di, S.; Chen, Z.; Cappello, F. TurboFFT: A High-Performance Fast Fourier Transform with Fault Tolerance on GPU. 2024; 1–13. [Google Scholar]
- Shi, N.; Liu, X.; Guan, Y. Research on K-Means Clustering Algorithm: An Improved k-Means Clustering Algorithm. In Proceedings of the 3rd International Symposium on Intelligent Information Technology and Security Informatics, IITSI 2010; 2010; pp. 63–67. [Google Scholar]
- Wang, J.; Wang, J.; Ke, Q.; Zeng, G.; Li, S.; Valley, S. 人脸识别方面很好的国际会议 Fast Approximate k -Means via Cluster. 2012; 3037–3044. [Google Scholar]
- Moodi, F.; Saadatfar, H. An Improved K-Means Algorithm for Big Data. IET Software 2022, 16, 48–59. [Google Scholar] [CrossRef]
- Mussabayev, R.; Mussabayev, R. Superior Parallel Big Data Clustering Through Competitive Stochastic Sample Size Optimization in Big-Means. 2024. [Google Scholar] [CrossRef]
- Rashmi, C.; Chaluvaiah, S.; Kumar, G.H. An Efficient Parallel Block Processing Approach for K -Means Algorithm for High Resolution Orthoimagery Satellite Images. In Proceedings of the Procedia Computer Science; Elsevier B.V., 2016; Vol. 89, pp. 623–631. [Google Scholar]
- Jin, R.; Yang, G.; Agrawal, G. Shared Memory Parallelization of Data Mining Algorithms: Techniques, Programming Interface, and Performance.
- Cuomo, S.; De Angelis, V.; Farina, G.; Marcellino, L.; Toraldo, G. A GPU-Accelerated Parallel K-Means Algorithm. Computers and Electrical Engineering 2019, 75, 262–274. [Google Scholar] [CrossRef]
- Bellavita, J.; Pasquali, T.; Del Rio Martin, L.; Vella, F.; Guidi, G. Popcorn: Accelerating Kernel K-Means on GPUs through Sparse Linear Algebra. Association for Computing Machinery, 2025; Vol. 1, ISBN 9798400714436. [Google Scholar]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-Scale Geospatial Analysis for Everyone. Remote Sens Environ 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Dickinson, R.E.; Henderson-Sellers, A.; Kennedy, P.J.; Wilson, M.F. NCAR/TN-257+STR Biosphere-Atmosphere Transfer Scheme (BATS) for the NCAR Community Climate Model. 1986. [Google Scholar]
- Shahapure, K.R.; Nicholas, C. Cluster Quality Analysis Using Silhouette Score. Proceedings - 2020 IEEE 7th International Conference on Data Science and Advanced Analytics, DSAA 2020, 2020; 747–748. [Google Scholar] [CrossRef]
- Syahputri, Z.; Sutarman, S.; Siregar, M.A.P. Determining The Optimal Number of K-Means Clusters Using The Calinski Harabasz Index and Krzanowski and Lai Index Methods for Groupsing Flood Prone Areas In North Sumatra. Sinkron 2024, 9, 571–580. [Google Scholar] [CrossRef]
- K C, M.; Leigh, L.; Pinto, C.T.; Kaewmanee, M. Method of Validating Satellite Surface Reflectance Product Using Empirical Line Method. Remote Sens (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Fajardo Rueda, J.; Leigh, L.; Kaewmanee, M.; Byregowda, H.; Teixeira Pinto, C. Derivation of Hyperspectral Profiles for Global Extended Pseudo Invariant Calibration Sites (EPICS) and Their Application in Satellite Sensor Cross-Calibration. US Geological Survey Open-File Report 2025, 17, 1–34. [Google Scholar] [CrossRef]
- Fajardo Rueda, J.; Leigh, L.; Teixeira Pinto, C. Identification of Global Extended Pseudo Invariant Calibration Sites (EPICS) and Their Validation Using Radiometric Calibration Network (RadCalNet). Remote Sens (Basel) 2024, 16. [Google Scholar] [CrossRef]
- Alshari, E.A.; Gawali, B.W. Development of Classification System for LULC Using Remote Sensing and GIS. Global Transitions Proceedings 2021, 2, 8–17. [Google Scholar] [CrossRef]
- Yang, C.; Li, Y.; Cheng, F. Accelerating K-Means on GPU with CUDA Programming. IOP Conf Ser Mater Sci Eng 2020, 790. [Google Scholar] [CrossRef]
- Han, S.; Lee, J. Parallelized Inter-Image k-Means Clustering Algorithm for Unsupervised Classification of Series of Satellite Images. Remote Sens (Basel) 2024, 16. [Google Scholar] [CrossRef]
- Andoni, A.; Indyk, P.; Razenshteyn, I. Approximate Nearest Neighbor Search in High Dimensions. Proceedings of the International Congress of Mathematicians, ICM 2018 2018, 4, 3305–3336. [Google Scholar] [CrossRef]
- Shindler, M.; Wong, A.; Meyerson, A. Fast and Accurate κ-Means for Large Datasets. Advances in Neural Information Processing Systems 24: 25th Annual Conference on Neural Information Processing Systems 2011, NIPS 2011, 2011. [Google Scholar]
- Spalding-Jamieson, J.; Robson, E.W.; Zheng, D.W. Scalable K-Means Clustering for Large k via Seeded Approximate Nearest-Neighbor Search. 2025. [Google Scholar]
- Peng, K.; Leung, V.C.M.; Huang, Q. Clustering Approach Based on Mini Batch Kmeans for Intrusion Detection System over Big Data. IEEE Access 2018, 6, 11897–11906. [Google Scholar] [CrossRef]
- Jourdan, B.; Schwartzman, G. Mini-Batch Kernel $k$-Means. 2024; 1–20. [Google Scholar]
- Newling, J. Nested Mini-Batch K-Means. 2015; 1–9. [Google Scholar]
- Aggarwal, C.C.; Hinneburg, A.; Keim, D.A. On the Surprising Behavior of Distance Metrics in High Dimensional Space. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 2001, 1973, 420–434. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





