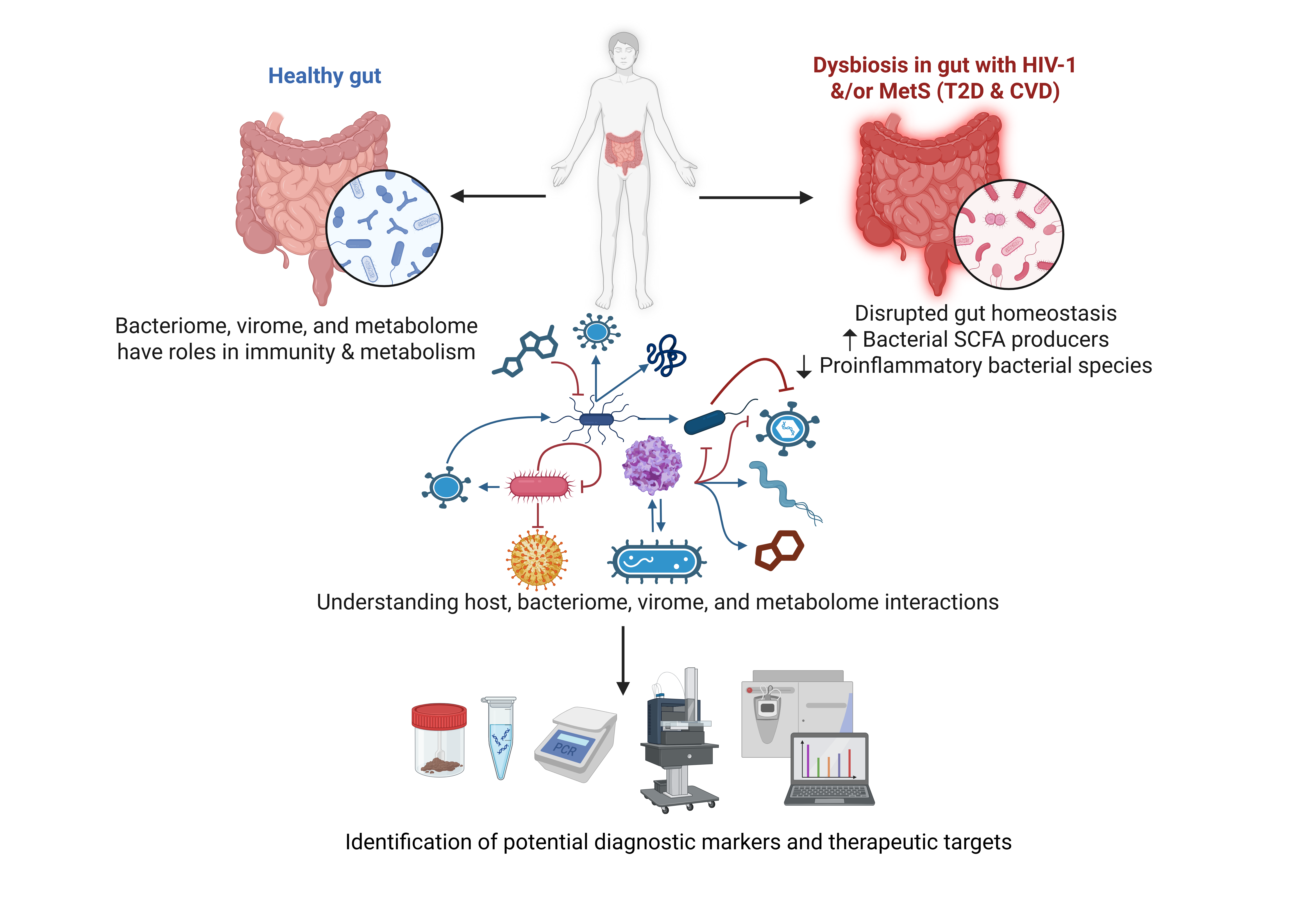
1. Introduction
The microbial communities residing in the human gut include bacteria, viruses, fungi and parasites that play various roles in keeping the human gut in balance, while its dysbiosis causes different harmful to injurious impacts [
1,
2]. The gut virome, a microbiome component, particularly bacterial phages, significantly influences human gut microbial ecology. The literature and the current evidence extensively discussed the role of various bacterial species in contributing to gut ecology and maintaining a balanced state of metabolism and immunity. However, there are other factors from the host side as well as from other microscopic entities that either regulate the gut environment directly or through these bacterial entities. A longitudinal study followed by a series of faecal samples for 6 months from a 2-year-old boy having atopic eczema (dermatitis) is an example of the same wherein 31 viral strains were identified, with a lytic crAssphage influencing its bacterial host by down-regulating aromatic amino acid metabolism, thus affecting the immune active aromatic amino acid (AAA) derivatives. These metabolic changes were also related to the remission of symptoms, depicting the interplay between virome dynamic, microbial metabolism and human host health outcomes [
3]. While earlier studies implicated genetic factors like mutations in the
filaggrin (
FLG) as major contributors to Atopic Dermatitis pathogenesis [
4,
5], the disease is now understood to be multifactorial and thus, the role of microbiota, both skin and gut, is expected to play a crucial role in its pathogenesis. Considering the role and interaction of these microbial entities with the host and modulating the entry and establishment of other pathogens in the gut environment is another area of exploration.
The growth and maturity of the host’s immune system and intestinal epithelium depend on the gut microbiota. This microbiota influences mucus layer characteristics, stimulates lymphoid structure formation, regulates the activation and differentiation of several lymphocyte populations, and maintains equilibrium in the synthesis of immunoglobulin A and antimicrobial peptides. By increasing nutrient sources, generating vital vitamins, and facilitating xenobiotic metabolism, the gut microbiota aids in host metabolism and adiposity. It also influences a variety of other host physiological factors, such as organ morphogenesis, intestinal vascularisation, tissue homeostasis, carcinogenesis, bone mass, and behaviour [
6].
The role of the gut microbiome is central to maintaining host health, and any sort of dysbiosis is supposedly linked to the development of disease conditions. Beyond atopic dermatitis, the gut microbiome plays a central role in immune homeostasis, and its dysregulation has been implicated in various diseases, including HIV-1 infection. The gut-associated lymphoid tissue (GALT) is key to human and simian immunodeficiency virus (HIV and SIV) infection, and HIV and SIV targets the gut at various levels, including immunological, structural, and microbiological [
7,
8].
HIV-1 viral replication occurring within CD4+ T lymphocytes, the primary target cells of virus, essentially contribute to the disease pathogenesis, however, HIV-1-induced chronic immune activation is shown to drive the overall disease progression [
9]. It is now believed that gut microbiome plays a central role in HIV-1 immunopathogenesis and associated chronic complications [
10,
11]. GALT is infected in the early stages of HIV-1 infection, which disrupts the gut integrity due to the massive depletion of CD4+ T lymphocytes, including T helper (Th) 17 and Th22 cells. Furthermore, HIV-1 induces enteropathy, mucosal inflammation, and intestinal epithelial cell damage. Indeed, such a damaged intestinal epithelial barrier is a major contributor of HIV-1-associated local and systemic inflammation [
12,
13]. At the cellular level, the structural damage of the gut barrier is a result of massive enterocyte apoptosis, decreased expression of tight junction proteins and increased intestinal permeability [
14]. Collectively, these abnormalities result in focal breaches of the gut barrier and subsequent microbial translocation at both local and systemic levels [
15,
16].
Microbial translocation regulates inflammation within the gut, which subsequently initiates the intestinal barrier dysfunction. As it is demonstrated that transcripts encoding inflammatory cytokines [such as tumor necrosis factor (TNF), interleukin 6 (IL-6), IL-10, and interferon-γ (IFN- γ)] are significantly upregulated in the colonic mucosa of HIV-1-infected individuals and persist in high levels despite antiretroviral therapy (ART) [
17,
18]. These inflammatory markers also serve as predictors of disease progression in untreated HIV-1-infected individuals [
19].
Of note, metagenomic analysis of people with gut inflammatory diseases has shown reduction in the microbial richness with low-gene counts (LGC) and altered intestinal microbiome (dysbiosis) [
20,
21,
22]. This review focuses on HIV-1 infection due to its higher hazard risk and significantly greater global prevalence compared to HIV-2. HIV-1 has been widely associated with an increased susceptibility to tuberculosis and a heightened risk of developing metabolic disorders [
23,
24,
25]. Importantly, HIV-1 infection is reported to cause intestinal microbiome dysbiosis [
26,
27,
28], which is implicated in not only worsening inflammatory response but also increasing susceptibility to HIV-1 infection [
28]. Accordingly, the relationship between intestinal microbiome and HIV-1-asociated gut mucosal pathogenesis is a two-way street, as HIV-1 associated changes of gut mucosa lead to dysbiosis and the dysbiosis subsequently disrupts intestinal homeostasis and further contribute to sustained HIV-1-associated immune activation and inflammation [
13,
29]. Indeed, an emphasis of the significance of intestinal microbiome has been concluded by studies focusing on rebalancing intestinal microbiome and mucosal barrier restoration in patients with AIDS to improve treatment outcomes [
30,
31]. Therefore, we aim to explore the interaction among HIV-1 infection, intestinal microbiome dysbiosis and mucosal barrier damage in HIV-1-infected patients, which is necessary for introduction of treatment strategy by recovering the balance of intestinal microecology and improving mucosal immunity.
The gut virome undergoes dynamic shifts throughout life, transitioning from an initially prophage-dominated state at birth to a transient lytic phase in infancy before stabilising with lysogeny in adulthood. These changes are influenced by bacterial abundance, diet, and host development. Early-life viral communities, including Microviridae and lytic p-crAssphage, play a crucial role in shaping bacterial populations, while prophages contribute to long-term microbiome stability. Such virome-bacteriome interactions may have significant implications for gut health and disease susceptibility [
32].
We hypothesize that shifts in gut virome composition contribute directly to the chronic inflammation observed in HIV-1 metabolic comorbidities through phage-host immune interactions
2. Diversity of the Components of Gut Microbiome
Before proceeding to a general microbiome profile of the human gut, it is essential to consider the variety and diversity in human geography, lifestyle, diet, age, sex, drug/antibiotic regimen, stress conditions [
33], which affect the gut microbiota. Thus, the microbiome also significantly varies from individual to individual [
1].
2.1. Communities Comprising Gut Microbiome
In general, healthy gut bacteriome comprises predominant Short- chain fatty acid (SCFA)-producing bacteria such as Ruminococcus obeum, Bifidobacterium longum, Roseburia intestinalis, Roseburia inulinivorans, Coprococcus comes, Akkermansia muciniphila, Eubacterium rectale, Bifidobacterium adolescentis, and Roseburia hominis. Faecalibacterium prausnitzii is a marker of gut health. SCFA has important functions such as maintaining gut integrity, and modulating immunity as discussed previously. Thus, an abundance of beneficial bacteria that contribute to metabolic health, low inflammation, and immune stability is evident in healthy gut microbiota. There is a balanced ratio of Firmicutes and Bacteroides (SCFA producers ) with a low abundance of pathogenic species and opportunistic microbes. Proteobacteria levels are regulated in a healthy gut, preventing dysbiosis [
34].
Consistently, the gut virome is a heterogeneous composition of DNA and RNA viruses, including single-stranded (ss) and double-stranded (ds) forms. Phages occupy a significant proportion and dominate with about >90%, whereas eukaryotic viruses account for <10% [
35,
36]. Predominantly, DNA-genome phages like Caudovirecetes- dsDNA and Microviridae- ss DNA are found in the gut. CrAssphage from the Intestiviridae family is also a prevalent and stable coloniser in the gut, abundantly found in human faeces. A few viral families show a higher prevalence as, like Circoviridae, Myoviridae show a greater diversity of genus, and increased Podoviruses. In one study, the intestinal luminal content of both pigs and macaques showed increased Caudoviricetes), including crAss-like phages and ssDNA Microviridae, with a lower fraction of eukaryotic viruses (families Circoviridae, Astroviridae, Caliciviridae and Parvoviridae [
36].
Moreover, eukaryotic phages in the healthy gut are supposed to be less pathogenic, and even if such pathogenic groups are present, their activity is controlled by healthy immune conditions. They are latent without causing any general harm to the human host. Dormant DNA viral communities of herpesviruses, anelloviruses [
37], and adenoviruses are mostly found in the human gut, while RNA viruses are rare and often of plant origin. While commenting on the composition, one thing to note is the instability of RNA over DNA and possible bias towards analysis techniques to identify the RNA genetic material. Also, the gut bacteriome maintains homeostasis with the virome [
38].
The phages modulate bacterial population, and the phage-bacteria dynamics are central to the balance of the gut ecosystem, preventing dysbiosis. Stable colonisation of temperate phages is associated with a healthy gut with limited lytic activity [
36,
39].
2.2. Impact of Diet on Microbiome Communities
Since the introduction of different environmental cues for infants starts with diet as they age, the viewpoint of how diet impacts our microbiome, including bacteriome, virome, and phageome, is essential [
40].
Different regions have various dietary patterns: Western diets are rich in processed foods, and high-fat content, while the Indian diet includes more of spices, medicinal plants having anti-cancer spices. A vegan diet eliminates all the animal-derived protein sources, while a high non-vegetarian diet will ensure more protein-rich content, even the frequency and appetite is different with respect to different individuals. Likewise, the dietary differences in infants viz, formula-fed or breastfed, result in distinct gut phageome and bacteriome profiles. Specific food components, wheat and oats, promote beneficial bacteria like
Bifidobacterium species. Rice suppresses
Bifidobacterium,
Lactobacillus,
Ruminococcus, and
Bacteroides [
41]. The glucose homeostasis of T2DM patients was improved when a high-fibre diet was adopted. They also showed increased abundances of
Lactobacillus,
Bifidobacterium and
Akkermansia, while
Desulfovibrio,
Klebsiella and other opportunistic pathogens were decreased [
42].
A longitudinal study that evaluated viral sequence abundance revealed that gut virome is highly individual-specific, unique, and temporally stable unless exposed to different cues, as a response to which it is also altered [
43]. When mice models are fed with a high-fat diet (HFD), siphoviridae phage decreases while Microviridae shows an increased abundance, thus altering gut phage diversity. Specific dietary changes, including fructose and SCFA, can activate prophages from
Lactobacillus reuteri, thus triggering temperate phages to undergo lytic conversion, which justifies diet as one of the environmental cues, as mentioned previously. A high-fructose diet increases phage production, possibly through the ACK signalling pathway. One study reported spatial compartmentalisation between viruses present in mucosa and lumen of the gut, with both showing higher Caudovirales order members. HFD-induced mice show an increase in the virus families
Microviridae, Phycodnaviridae, and
Mimivirdae in the faecal virome. This study reported reduced lysogenic phages in the Siphoviridae family [
44,
45,
46].
3. Feed-Forward Cycle Between Gut Inflammation and Gut Dysbiosis in HIV-1
HIV-1 can replicate in GALT in early stage of infection and further mediate pronounced changes to intestinal barrier and its local immunity [
47,
48,
49,
50]. In GALT, HIV infection induces intestinal microbiota dysbiosis, which eventually makes way for worsening clinical symptoms and could promote the occurrence of comorbidities associated with metabolic disorders [
24,
25,
51]. The keys to these outcomes are damaged intestinal mucosal barrier and persistent inflammatory response [
52].
3.1. Multilevel Disruption of Gut Homeostasis in HIV-1 Infection on the Intestinal Barrier Integrity
Upon HIV-1 infection, the clinical condition begins when there is rapid and mass depletion of CD4+ T-cells as they serve as primary target cells of HIV-1 due to the availability of CCR5 receptors. The critical impact of HIV-1 on gut immunity is evident in GALT of humanised mice, too, as HIV-1 led to the depletion of immune cells by infecting nearby cells through cell-to-cell contact regions called virological synapses [
53].
The gut barrier is implicated in HIV-1 infection at the structural or anatomical level as well, as it leads to a phenomenon of ‘leaky gut’, which allows entry of microbial antigens and metabolites into the bloodstream. This induces the development of a sustained pro-inflammatory state, which further transforms into a chronic inflammation in treated HIV- patients as well. Immune reconstitution inflammatory syndrome (IRIS) in HIV- an infected individual was detected even after the anti-retroviral treatment (ART) introduction and achieving undetectable viral load, overall immune recovery remained incomplete, leaving the individual at higher risk of further complications [
54]. “Viro-immunological discordants” are a group of individuals who fail to recover their CD4+ T-cell count despite achieving viral suppression through ART [
55]. This might be related to other levels in which HIV-1 impacts the host, i.e., microbiological.
At the microbiological level, the gut bacteriome and virome profiles of a healthy individual differ distinctly from those of an HIV-infected individual. HIV-infected patients show a general increase in bacterial species associated with higher levels of proinflammatory cytokines. So, classes like
Negativicutes,
Bacilli, and
Coriobacteria are increased in abundance, whereas there is depletion in bacterial species associated with anti-inflammatory cytokines like
Clostridia [
51]. A general trend in the reduction of short-chain fatty acids (SCFAs) producing bacteria is seen as accompanied by an increase in species associated with an increase in inflammatory responses and inflammation [
56].
Similar evidence was provided from a study that investigated why some women living with HIV (WLHIV) continue to shed HIV in their genital tract despite effective viral suppression by ART. It was found that women having more shedding of HIV had greater fluctuations in their bacterial microbiome over time, suggesting an unstable microbial environment in their genital tract. Moreover, the virome profile also changes in different conditions A relation to virome was also seen as in women who did not shed HIV, the virome composition, particularly that of anelloviruses, showed changes over time, which could be due to a long-term ART regimen [
57].
Anelloviruses are small, single-stranded DNA viruses commonly found in the human gut. They are not known to be causing any disease yet, but there alteration is seen with diseased and healthy conditions. Advanced HIV infection is associated with elevated levels of human Anelloviridae sequences in the gut virome, especially in individuals with severe immunodeficiency (CD4 < 200). Longitudinal data show that short-term ART (24 months) significantly reduces Anelloviridae abundance. Notably, baseline detection of Anellovirus sequences independently predicts poor immune recovery during ART, suggesting potential use as a prognostic biomarker. However, whether this virome shift is causal or merely a correlate of immune dysfunction remains to be fully elucidated. [
58]. Bacteriophages comprise a vast proportion of gut virome, and their difference in the microbiome composition (beta diversity) shows altered composition in HIV-infected patients compared to healthy controls [
36]. An increase in phages belonging to the Caudoviricetes class (dsDNA) and a decrease in Malgrandaviricetes in the treatment-naive group were reported. Caudoviricetes are also increased in Inflammatory Bowel Disorder (IBD), indicating the possible link to increased inflammatory state and gut permeability [
59].
3.2. Multilevel Disruption of Gut Homeostasis in HIV-1 Infection on the Microbial Translocation
Indeed, the translocated microbial products from damaged/inflamed gut could directly stimulate the immune system, and there could be direct or indirect regulation by viruses. ART could induce partial restoration of the altered gut microbiome, but an exact reversion to a healthy state is not seen as evident by decreased alpha diversity in PLWH on long-term ART. Particularly, Enterobacteriaceae increases, which is associated with inflammation. The members of Enterobacteriaceae,
Aeromonas and
Prevotella are Gram-negative entities that have lipopolysaccharide (LPS) in their cell walls [
60].
LPS triggers inflammatory T-helper 1 lymphocyte (Th1) responses, inhibiting the differentiation of Th17 cells; thus, Interleukin-17 (IL-17) levels are reduced. IL-17 proves to be essential for the expression of tight junction proteins- claudin-1 and claudin-2. Its reduction worsens the gut integrity, promoting a leaky gut. It also induces a depletion of CD4+ T cells [
14].
Prevotella is seen to increase in HIV infection, although this observation is also seen in men who have sex with men (MSM) irrespective of their HIV status.
Negativicutes,
Bacilli, and
Coriobacteriia are also Gram-negative bacteria that have an outer membrane with LPS. They are positively correlated with interferon-γ (IFN-γ) and IL-1β plasma levels. Bacterial families like
Erysipelotrichaceae in
Bacilli and
Atopobiaceae in
Coriobacteriia are more abundant in the gut microbiota of HIV patients, and they are negatively correlated with anti-inflammatory cytokines -IL-19 and IL-35. Prevotella is also enriched in PWH [
61].
The enrichment of
Prevotella and the decrease in anti-inflammatory cytokines further emphasise the role of gut microbiota in influencing health outcomes for PWH [
51]. The enrichment of Prevotella and the decrease in anti-inflammatory cytokines further emphasise the role of gut microbiota in influencing health outcomes for PWH. The enriched bacterial species in HIV infection induce chemokines like- monocyte chemoattractant protein 1 (MCP-1/CCL2), MCP-4/CCL13, and macrophage inflammatory protein 1α (MIP-1α/CCL3), involved in the recruitment of monocytes to inflammation site, thus exacerbating the inflammation [
51]. A leaky gut makes microbial translocation easier and more convenient to the bloodstream and amplifies systemic inflammation, thus creating a vicious cycle of dysbiosis and immune activation in HIV-1 infection. At the same time, lower pathogenicity and slower disease progression of HIV-2 can be linked with preserved intestinal epithelial integrity [
51,
62]. Bacteroides are also Gram-negative bacteria, but they are essential SCFA producers along with Lachnospiraceae and Ruminococcaceae. SCFAs are beneficial for many reasons: butyrate is the primary energy source for gut epithelial cells. It promotes the expression of proteins forming tight junctions like occludin, claudins, and zonula occludens-1 (ZO-1). SCFA tends to suppress the production of pro-inflammatory cytokines and subsequently increase the production of anti-inflammatory cytokines by modulating signaling pathways. They are shown to stimulate mucus production by goblet cells and lower the luminal pH so that the acidic pH will inhibit the growth of pathogenic bacteria; finally, they also have a role in immune modulation as they can interact with G-protein-coupled receptors (GPCRs) [
63]. Since the discussion involves orders and families, different genera belonging to the same family or order may show different overall impacts. Hence, it is bound to show contrasting data availability from the same group of organisms, as observed in a few studies wherein the depletion of
Bacteroides members was associated with markers of immune disruption, T-cell activation, and chronic inflammation in HIV- infected individuals. The extent of bacterial dysbiosis generally correlates with the activity of the Kynurenine pathway of tryptophan catabolism and plasma concentration of IL-6 [
51].
Till now, the complex interplay of host metabolites, gut commensal bacterial communities, and inflammation in the host in the context of HIV has been examined. SIV has been widely recognised as a model for HIV research, with pathogenic SIV infection being associated with a change in the enteric virome, as 32 enteric viruses were identified during infection in one study. It reported adenovirus infection associated with enteritis and parvovirus viremia in animals with advanced AIDS, thus giving proof that viruses are also showing dysbiosis in SIV infection [
64].
For the logical extrapolation of this data to humans, the vast majority of phages and eukaryotic viruses must be considered as they outnumber the gut bacterial count [
38,
44]. So, the regulatory role of phages on bacteriome is vital to counteract and mitigate the gut dysbiosis that was discussed earlier. Viruses tend to colonise early after birth in the human gut. A study followed virome analysis of infant faecal samples from birth to age one and reported that most of the viral reads were of six families of bacteriophages, including five dsDNA virus families of the order Caudovirales with Siphoviridae and Podoviridae being the most abundant. Thirty-four eukaryotic virus families were found wherein animal viruses- Anelloviridae, Astroviridae, Caliciviridae, Genomoviridae, Parvoviridae, Picornaviridae, Reoviridae comprises 98% reads with one plant virus family- Virgaviridae [
65]. Another study reported that higher HIV replication leads to an abundance of Anellovirus, which correlates negatively with CD4+ T cell counts, although it’s important to note that short-term ART of 24 months was initially linked to a significant decrease in human Anelloviridae sequences. PLWH with severe immunodeficiency (CD4+ T cells <200 cells/µL) has an abundance of Anelloviridae, Adenoviridae, and Papillomaviridae [
58]. Pegivirus, on the other hand, is increased with CD4+ T cell counts and decreases as the HIV-1 viral load increases, thus indicating that gut eukaryotic viruses have different dynamics during infections [
66].
One thing to note is that Anelloviridae and Pegivirus are DNA and RNA viruses, respectively. Thus, ART could have acted on Pegivirus directly, leading to such effects and having some other mechanism of action to deal with Anellovirus. Hence, eukaryotic viruses could use direct and indirect approaches to interact with human hosts [
66]. This crosstalk is possible directly or indirectly mediated via metabolites. Another link of the Anelloviridae family, particularly the Torque teno virus (TTV), was found in colorectal cancer (CRC) as they were more abundant in CRC tissues than in healthy controls. Also, higher levels of TTV were found in intestinal lamina propria; thus, it can be inferred that Anelloviruses were consistently associated with diseased or inflammatory conditions [
67]. Bacteriophages also show dysbiosis, as seen by decreased Inoviridae sequences in stool samples of PLWH with CD4+ T cells <200 cells/µL [
68]. TTV miRNAs downregulate N-MYC interactor (NMI) proteins associated with the IFN pathway while ORF2 protein inhibits NF-κB translocation into the nucleus, reducing the inflammatory mediator’s [IL-6, IL-8, and Cyclo-oxygenase-2 (COX-2)] expression. TTV genogroup 4 with CpG motif [
69] can activate TLR9, stimulating pro-inflammatory cytokines- IFN-γ, IL-6, and IL-12 release. Thus, TTVs may exhibit both stimulatory and inhibitory CpG motifs, implicating their role in balancing inflammation. Dysregulated TTV activity may contribute to chronic immune activation, a hallmark of HIV disease progression [
70,
71,
72].
Additionally, variations in TTV miRNA profiles between HIV-positive individuals and healthy controls suggest their involvement in metabolic disorders and gut inflammation [
37,
69]. The increased prevalence of eukaryotic viruses in PLWH highlights their potential role in exacerbating gut dysbiosis and inflammation, promoting a feed-forward cycle of immune dysfunction and microbial imbalance. This aligns with evidence of viral expansion in stool samples from HIV-positive individuals, underscoring the need to explore the interplay between eukaryotic viruses and other gut microbiome components in shaping disease outcomes [
52].
3.3. Intestinal Dysbiosis Contribute to HIV-1- Associated Metabolic Changes
Notably, chronic inflammation, immune activation, and ART-associated metabolic changes are key factors predisposing people living with HIV (PLWH) to comorbidities [
73]. HIV-associated comorbidities related to metabolic disease primarily include cardiovascular disease (CVD) and diabetes, particularly type 2 diabetes (T2D) [
74]. A study on the gut dsDNA virome in school-aged children with normal weight (NW), obesity (O), and obesity with metabolic syndrome (OMS) revealed that phage richness and diversity increased in O and OMS. At the same time, the abundance of virus-like particles (VLP) remained unchanged across groups. The gut virome, dominated by Caudovirales, showed high inter-individual diversity [
75]. Further members of Proteobacteria order in HIV-infected individuals are increased, leading to a possibility of a decrease in Proteobacteria-infecting phages [
59]. Thus, there is evidence and possibility of the vital role of bacteriome-virome interactions in maintaining gut homeostasis. However, there are challenges characterising the gut virome: the unavailability of comprehensive data and viral sequences with their host specificity and interaction poses limitations that sometimes lead to biased and partial standards for the analysis of many gut viruses [
76]. Literature thus far, has investigated DNA viruses, which limits the applicability to varied forms of other viral genetic materials. Nevertheless, the attempt to understand the interplay of bacteriome, virome, and host factors will offer a unique opportunity to uncover disease-specific microbial patterns. Here, we discuss how bacteriome and virome, comprised of eukaryotic viruses and phages, are altered in HIV-associated co-morbidities related to metabolic disorders.
Moreover, metabolic syndrome includes many conditions like obesity, lipidemia, and insulin resistance. It was reported that the gut phage richness and diversity increased in obesity and obesity with metabolic syndrome compared to healthy individuals, suggesting disease-specific alterations [
75]. Obese patients with T2DM (type-2 diabetes mellitus) have decreased gut viral richness and diversity as compared to lean and healthy controls. Namely, Escherichia phage, Geobacillus phage, and Lactobacillus phage were predominant in obese subjects, accompanied by weakened viral-bacterial correlations as compared to lean controls [
77]. Compared to healthy controls, the abundance of Caudoviricetes phages over Microviridae is also reported in dysbiotic people. Alterations in the virome profile of individuals could be related to metabolic shifts, as seen when infection of murine Norovirus- eukaryotic virus was done in antibiotic-treated mice. The infection protected the mice against antibiotic-associated intestinal injuries and bacterial infection [
38]. One murine study provides evidence for a possible role of phages in activating gut mucosal immune responses, as in Caudovirales, which were shown to mediate TLR9-dependent activation of CD4 + T cells and increased intestinal inflammation by increasing the production of IFN-γ. While not very well-documented sources exist for gut-virome-mediated immune regulation, the interaction could be predicted to be direct and indirect [
78].
4. Common Microbiome Alterations in HIV-1 Infection & Type-2 Diabetes Mellitus (T2D)
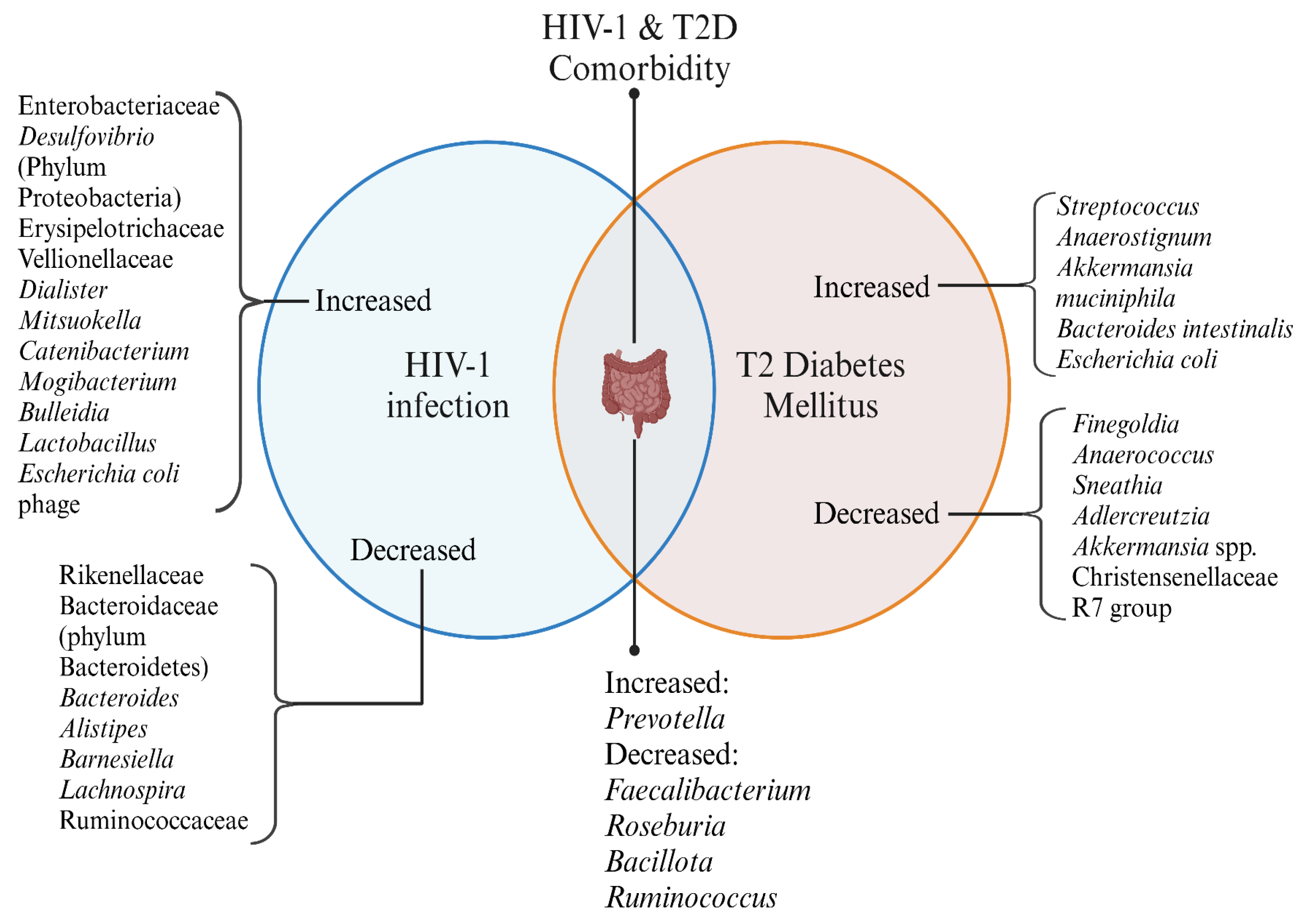
T2D, a chronic metabolic disease, is a public health burden often attributed to causes such as nutrition transitions, urbanisation, sedentary lifestyles and stress. It is characterised by polydipsia, polyuria, weight loss, and polyphagia as the primary clinical manifestations. The key pathological factors are insulin resistance and impaired pancreatic β cell function. The gut microbiota is highly affected by diabetes and co-morbidity of HIV & T2D. Butyrate producers such as
Finegoldia, Anaerococcus, Sneathia, and Adlercreutzia are decreased, implying SCFA importance. A presumed reduction in anti-inflammatory functions and impaired insulin sensitivity is evident in dysbiosis. Prediabetic individuals show more abundant
Streptococcus and
Anaerostignum [
79,
80].
In the gut of T2D patients, more
Akkermansia muciniphila, Bacteroides intestinalis, and
Escherichia coli are found. Also, the reduction in
Faecalibacterium and
Roseburia observed in T2D aligns with findings from HIV-1 infection. This implies the common microbial imbalances in both conditions that favour chronic inflammation and metabolic dysfunction [
81].
One study found significant differences in the abundance of 58 phage species in faecal samples between T2D patients and healthy controls. Phages specific to Enterobacteriaceae hosts, such as Brochothrix_phage_NF5, Enterococcus_phage_phiFL2A, Streptococcus_phage_PH10, and Streptococcus_phage_7201, showed significant variations between T2D patients and non-diabetic individuals, thus suggesting T2D may influence the composition of the gut phageome, potentially affecting gut health and microbial interactions [
82].
In T2D, members of the bacterial family Enterobacteriaceae increase along with the increase in bacteriophage- siphoviridae, podoviridae, myoviridae, and unclassified caudovirales families.
In diseased conditions, the abundance of Caudovirales phages, including Streptococcus phage PH10, Brochothrix phage NF5, Streptococcus phage 7201, Enterococcus phage phiFL2A, Escherichia phage, Geobacillus phage, and Lactobacillus phage is significantly increased. Also, there is an altered association and interaction among humans, bacteriome and virome in such diseased conditions [
82].
Lactobacillus and
Prevotella are elevated in both HIV-1 infection and T2D, suggesting that these microbial groups play a role in the dysbiosis observed in these conditions. The abundance of Prevotella species, in particular, is possibly linked to insulin resistance; thus, similar phenomena are seen in co-morbid conditions of HIV and T2D.
Bacteroides species show decreased abundance in HIV-1 infection and an increase in T2D, suggesting that the imbalance of
Bacteroides may contribute significantly to dysbiosis in both conditions. Changes in gut virome in HIV-T2D comorbid individuals are seen as Podoviridae phages targeting Escherichia, Clostridium, and Siphoviridae phages infecting
Lactobacillus, Pseudomonas, and
Staphylococcus show an elevated abundance. These virome changes may contribute to altered microbial composition, reduced SCFA production, and chronic low-grade inflammation, all of which are linked to the progression of metabolic disorders like T2D in HIV-positive individuals. The metabolism is also influenced by gut microbiota, as evidenced by an elevation of the kynurenine/tryptophan ratio (KTR) in the tryptophan metabolism pathway. KTR indicates indoleamine-2,3-dioxygenase (IDO) activation, which facilitates the transformation of tryptophan into kynurenine, reducing tryptophan availability and altering gut and systemic immunity. Kynurenine shows bactericidal properties, but its accumulation also promotes regulatory T-cell proliferation and apoptosis of T-effector cells, thus compromising host immunity in HIV-1 infection. In conditions such as T2D, chronic inflammation driven by microbial dysbiosis leads to a similar elevation in KTR, further linking microbial changes to metabolic disturbances [
83,
84].
Figure 1.
Gut Microbial Alterations in HIV-1 infection and T2D. This Venn diagram represents the microbial shifts observed in HIV-1 infection and Type 2 Diabetes (T2D), along with their comorbid states. Created in
https://BioRender.com.
Figure 1.
Gut Microbial Alterations in HIV-1 infection and T2D. This Venn diagram represents the microbial shifts observed in HIV-1 infection and Type 2 Diabetes (T2D), along with their comorbid states. Created in
https://BioRender.com.
HIV-1 infection and T2D exhibit distinct microbial changes, with some overlaps in their dysbiosis patterns. The comorbid state of HIV-1 and T2D presents unique alterations, highlighting potential microbial contributions to disease progression and metabolic health.
5. Common Microbiome in HIV-1 Infection & Cardiovascular Diseases (CVD)
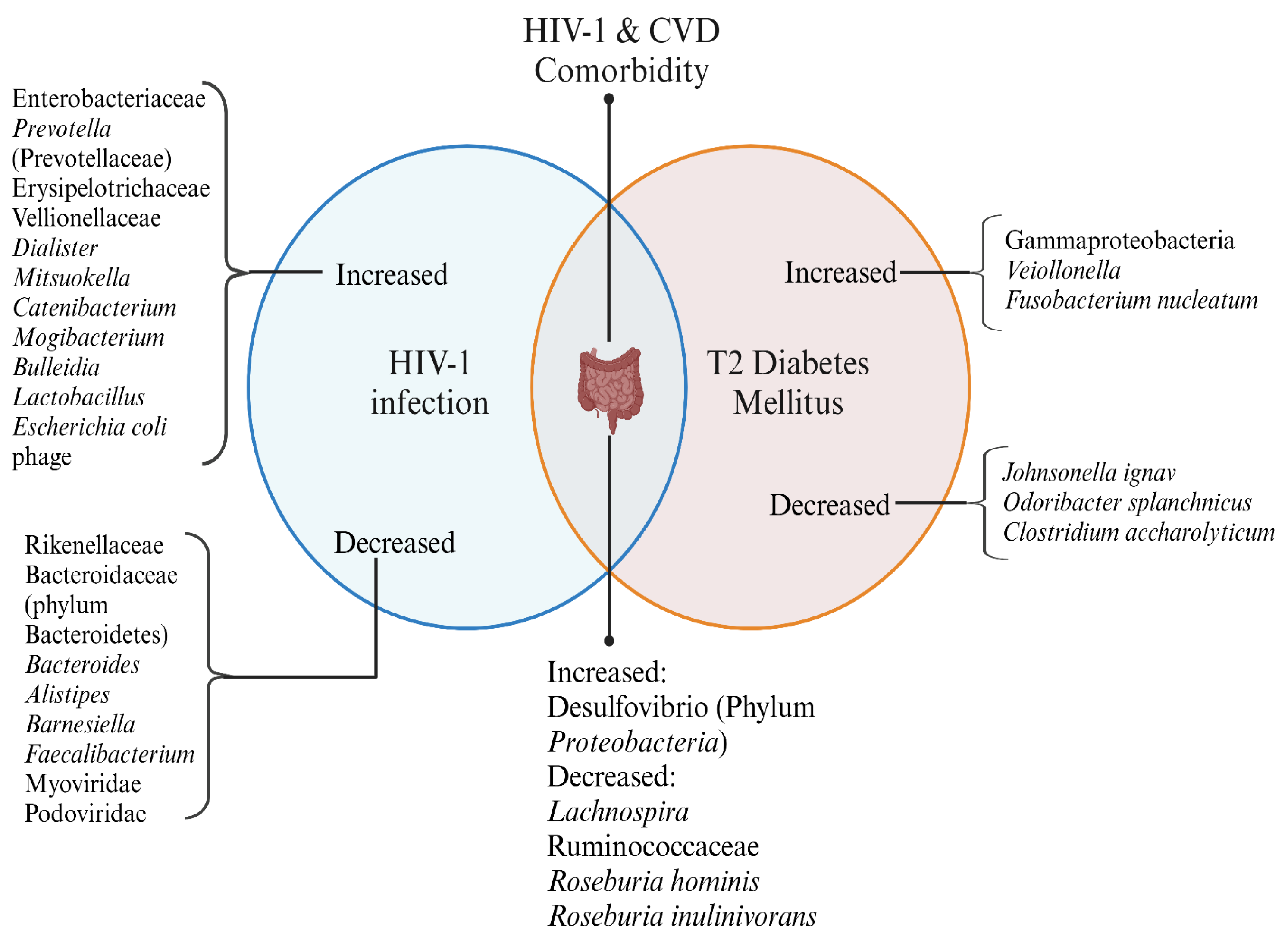
Obesity and high lipid content increase the chances of an individual to develop cardiovascular diseases. Similar to diabetes, cardiovascular diseases are also a subset of conditions accompanied by gut microbiota changes. A marked depletion of beneficial bacteria from families like Lachnospiraceae and Ruminococcaceae produces SCFA-like butyrate. In contrast, the pro-inflammatory taxa like
Gammaproteobacteria, Desulfovibrionaceae, and Ruminococcus gnavus are seen. One study showed distinct gut dysbiosis in patients with obstructive coronary artery disease (CAD), including
Ruminococcus gnavus and
Veillonella. These are known producers of imidazole propionate (ImP), which shows higher levels in plasma in patients with obstructive CAD. The gut microbial metabolites, thus, are related to diseased conditions [
85].
The reduced SCFA production and increased lipopolysaccharide are seen in the general population with CAD, which is common in the pathology of HIV. Studies of microbial metabolites such as ImP and their association with gut integrity, inflammation, and cardiovascular outcomes in PLWH could illuminate novel pathways linking gut dysbiosis to HIV-associated comorbidities, including heart failure, diabetes, and systemic metabolic disturbances. One study reported an increase of gut bacterial species, Fusobacterium nucleatum, to be associated with plaque formation, implying its role as a pathogenic factor in atherosclerosis development. Other gut bacterial species such as
Roseburia hominis, Roseburia inulinivorans, Johnsonella ignava, Odoribacter splanchnicus, and
Clostridium saccharolyticum are decreased with plaque formation in PLWH, highlighting their possible protective role [
86]. A study on gut microbiota composition of abdominal obesity individuals showed a decrease in
Akkermansia; thus, the factors that could lead to CVD are also associated with altered gut bacteriome. However, there is not much evidence available about the role of viruses or bacteriophages in CVD. [
87].
Figure 2.
Gut Microbial Alterations in HIV-1 infection and CVD. This Venn diagram represents the microbial shifts observed in HIV-1 infection and Cardiovascular Diseases (CVD), along with their comorbid states. Created in
https://BioRender.com.
Figure 2.
Gut Microbial Alterations in HIV-1 infection and CVD. This Venn diagram represents the microbial shifts observed in HIV-1 infection and Cardiovascular Diseases (CVD), along with their comorbid states. Created in
https://BioRender.com.
HIV-1 infection and CVD exhibit distinct microbial changes, with some overlaps in their dysbiosis patterns. The comorbid state of HIV-1 and CVD presents unique alterations, highlighting potential microbial contributions to disease progression and metabolic health.
6. Putative Crosstalk Between Bacteriome and Virome in HIV-Associated Metabolic Disorders
Considering crosstalk among various components of the microbiome, viz, bacteriome, virome, and Considering crosstalk among various components of the microbiome, viz, the bacteriome, virome, and phageome, with the human host, is essential when assuming the whole consortia in the human gut, as they basically work as an ecosystem with all the components functioning in accordance. In HIV- 1 infection, such a crosstalk is affected due to the dysbiotic community of organisms evident in our discussion. A study followed Mendelian randomisation and mediation analyses to conclude the cause-effect relationship between HIV and bacterial dysbiosis concluded that increased
Proteobacteria and
Ruminococcaceae, UCG013 is related to an increased risk of HIV infection, while
Clostridium sensu stricto and
Erypsipelotrichaceae were generally associated with less disease severity [
88]. As a matter of fact, phages could regulate their bacterial host by following two well-known cycles- lytic or lysogenic. Additionally, some phages could exist as pseudo-lysogens, in which the phage genome exists in bacterial cells as a plasmid-like episomal construct without any integration or replication. For instance, Plasmaviridae could lead to a replication mode that can bud out without lysis and death of the bacterial host. Phages outnumber their bacterial host by a huge margin, as discussed previously. So, the remarkable adaptability of phages to change gut environments is understood in different diseased conditions. In PLWH, a depletion of beneficial bacteria (e.g., SCFA producers) suggests a possible increase in the abundance of phages infecting them. On the other hand, since pro-inflammatory species are predominant, the phages infecting them should decrease in the same setup. Haemophilus and Sellimonas supposedly mediate the regulation of specific metabolites, potentially influenced by phage-host interactions [
88].
Another definitive example is
Bacteroides phage B40-8 of the family
Siphoviridae infecting
Bacteroides fragilis- an important SCFA producer and immune modulator. This phage-bacterium interplay is likely affected in PLWH, presumably showing increased phage activity, further contributing to the depletion of the beneficial
Bacteroides population.
Prevotella, which is often increased in PLWH, produces LPS that may fuel systemic inflammation, worsening HIV-1 infection. Reduced populations of phages infecting
Prevotella could signify an imbalance favouring pathogenic bacterial expansion [
89].
As discussed earlier, the gut virome shifts a lot as an individual age. Initially, the neonatal gut virome is colonised by prophages at birth, but the low bacterial abundance limits the lysogenic maintenance, so transiently, the lytic phages take over the abundance status. The virus-to-bacteria ratio rises as bacterial and viral densities increase [
90]. The maturation, ageing, and diet changes lead to the colonization of a diverse community of prophage-harbouring obligate anaerobes. So, the gut virome now comprises temperate phages that maintain overall lysogenic stability in the system, as seen in adult gut microbiomes [
90]. By the age of 2, the Microviridae family of virulent phages are most prevalent, shifting from Caudovirales predominance at birth. p-crAssphage (lytic phages) infecting Bacteroidetes become widespread among 2 to 5-year-old children. Lytic bacteria may infect dominant bacteria, causing cyclical shifts in bacterial abundance as seen in interactions between bifidoprophages and Bifidobacterium in preterm infants [
91,
92].
The lytic phage phenotype often becomes more prominent during environmental stress and gut inflammation due to associated changes in the host’s microbiome and immune system. Although the whole mechanism is still under investigation, there is evidence to confirm that gut virome shifts from a lysogenic to a lytic state in diseased conditions [
93]. Whether the transformation is a cause or effect of disease and how phages change during disease is still a topic of further research. Also noted here is the age-dependent lytic-to-lysogenic shift. In adulthood, the gut virome shift dominated by prophages offers advantages like metabolic benefits, antibiotic resistance, and increased bacterial competitiveness, as evident in
Vibrio cholerae and its prophage named CTXΦ [
91].
Furthermore, there could be three applicable driving forces to understand the presumable shift from a lysogenic to a lytic state in diseased conditions.
Diversity-generating retroelements (DGRs): As evident in Lak phages with genomes exceeding 540 kilobase (kb), hence also called megaphages, the adaptability could be due to hypervariability caused by DGRs. DGRs could modify tail fibre proteins to alter host tropism, thus enabling one temperate phage-
Bacteroides dorei Hankyphage to infect a broad range of 13
Bacteroides species.
Bordetella phage BPP-1 is an other example of the same. Such DGR-containing phages are commonly integrated as prophages in bacterial hosts from groups- Bacteroidetes, Firmicutes, and Proteobacteria. The lysogenic state predominating in the human gut is consistent with the “piggyback-the-winner” hypothesis, wherein phages benefit from maintaining prophage status during bacterial host replication. These prophages remain functional, producing viral particles and facilitating interactions within the microbial community, as evident by viral metagenomics [
94,
95,
96].
Dynamic genomic inversions (DGI): Recent research has shown DGI in bacteria such as Bacteroides fragilis illustrating phage-bacteria co-evolution. DGI could alter gene expression and affect host immunity, as seen in IBD. One study identified multiple invertible regions where a particular orientation was correlated with IBD as the promoter of polysaccharide A (PSA) of
B. fragilis was mostly oriented ‘OFF’ in IBD patients, correlating with the increased B. fragilis-associated bacteriophages. To further prove this, in mice colonised with a healthy human microbiota and
B. fragilis, induction of colitis caused a decline of PSA in the “OFF” orientation. Thus, the lysogenic to lytic conversion in HIV and associated co-morbidities related to metabolic diseases could be predicted due to DGI. Dynamic bacterial phase variations due to bacteriophages and host inflammation signify functional plasticity during diseased conditions [
97].
Holin-anti-holin secretion system: Balancing lysogeny and lysis is highly dependent on stressors and environmental cues. Gram-positive gut bacteria
Lactobacillus, Bifidobacterium, and
Enterococcus often harbour prophages. One mechanism for releasing these temperate phages to initiate the lytic cycle could be the holin-anti-holin system. Holins create pores in bacterial plasma membrane, enabling lysins to enter the peptidoglycan layer and lyse the host bacterial cell. But the timing is also of utmost importance as the shift will be due to stresses like nutrient depletion, oxidative stress, or other dysbiosis caused by diseased conditions like HIV. To ensure the release of holins, anti-holin and anti-holin-like proteins are in place as gatekeepers releasing holins under specific conditions only. For Gram-negative bacteria such as Prevotella and Bacteroides, the lysis is a bit tricky due to an outer membrane (OM). Phages could use Spanins to disrupt OM and lipases to degrade lipids, thus weakening the cell envelope. Once the OM is compromised, holins and endolysins complete the lytic cycle transition. The transition from temperate to lytic form in
Lactococcus lactis MG1363 involves coordinated resonance between 2 prophages- TP712 (holin protein) and CAP integrated into the bacterial genome. Mitomycin C treatment causes environmental stress during the transition. The CAP prophage induces lytic proteins- CAP endolysin, while TP712 creates pores, thus exemplifying the synergistic lysis and explaining the lytic phenotype of lysogens [
98].
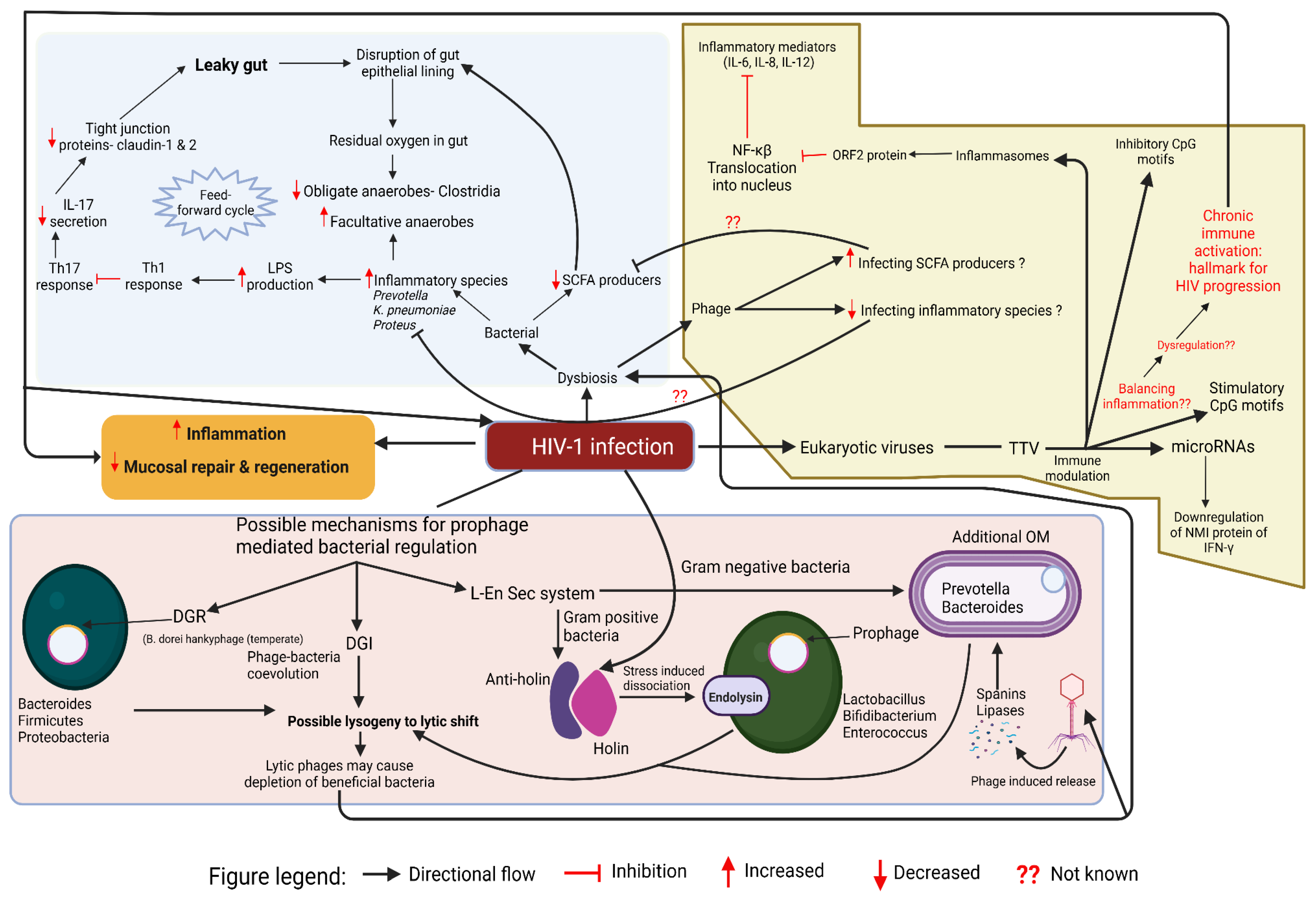
Figure 3.
Putative Crosstalk Between HIV-1 Infection, Gut Microbiome, and Virome in HIV Disease Progression. Created in
https://BioRender.com.
Figure 3.
Putative Crosstalk Between HIV-1 Infection, Gut Microbiome, and Virome in HIV Disease Progression. Created in
https://BioRender.com.
This schematic diagram shows the putative interplay between HIV-1 infection, gut bacteria, phages, and eukaryotic viruses, driving gut dysbiosis, chronic inflammation, and immune modulation. Leaky gut arises from epithelial barrier disruption, reduced tight junction proteins, and altered immune responses, leading to increased LPS production and inflammatory species expansion. A decline in SCFA producers worsens dysbiosis, while eukaryotic viruses like TTV modulate inflammation via CpG motifs, microRNAs, and NF-κB signaling. Phages influence microbial composition by infecting SCFA producers and inflammatory species. The lower panel highlights prophage-mediated bacterial regulation, where lysogeny and lytic shifts deplete beneficial bacteria, while stress-induced prophage activation leads to bacterial lysis through holins, endolysins, and lipases.
7. Search Strategy and Selection Criteria
This review was conducted based on a comprehensive literature search focusing on gut microbiome-virome interactions in HIV-1 infection and associated metabolic disorders, including Type 2 Diabetes Mellitus (T2DM) and Cardiovascular Disease (CVD). The search was performed using scientific databases such as PubMed, Scopus, and Google Scholar. A combination of Medical Subject Headings (MeSH) terms and keywords were used, including: “Gut microbiome in HIV”, “HIV and gut virome”, “phage therapy in metabolic disorders”, “microbial translocation in HIV”, “gut dysbiosis in cardiovascular disease”, “gut dysbiosis in type 2 diabetes mellitus”, “intestinal microbiota alterations in HIV”, “eukaryotic viruses and metabolic disorders”, and “bacteriophage-host interactions in gut health.” Manuscripts that were from the last 5 years and assessed incident clinical events, or included meta-analyses comprehensively summarising existing data were preferred. Relevant articles were screened by title and abstract, and full texts of selected studies were reviewed for final inclusion. Reference lists of key papers were also examined to identify additional relevant literature. Only papers published in English were reviewed.
8. Conclusions
The gut microbiome is critical in maintaining metabolic, cardiovascular, and immune homeostasis. In people living with HIV, gut microbial dysbiosis is characterised by reduced diversity, depletion of beneficial SCFA-producing bacteria (e.g., Lachnospiraceae, Ruminococcaceae), and enrichment of pro-inflammatory taxa (e.g., Ruminococcus gnavus, Gammaproteobacteria). The gut virome analysis precludes the complexity and challenges, particularly in analysing the strain-specific interactions and estimating RNA viruses. Advancement in metagenomic sequencing and bioinformatics is required for detecting unknown viruses. We highlighted the intricate interplay between the human host, gut bacteriome, and virome comprising bacteriophages and eukaryotic viruses in healthy and diseased conditions of metabolic disorders associated with HIV-1 infection. The feed-forward cycle is critically discussed to observe the vicious, never-ending “feed” for prolonged inflammation, which is supposedly the commonality that joins these comorbidities. The regulatory mechanisms for adapting lytic or lysogenic cycles also depend on the host’s environment. The observed alterations in phage-host dynamics, bacterial composition, and virome diversity in HIV and comorbidities imply the essentiality of considering the virome’s potential as a modulator of disease. Future research should focus on unraveling the precise mechanisms through which gut viruses contribute to disease pathogenesis and exploring phage-driven precision therapies for restoring gut homeostasis in HIV-1 and associated metabolic disorders.
9. Future Perspective
Existing studies on alterations in the human gut microbiome across various diseases suggest a strong link between gut microbiome dysbiosis, including virome imbalance, and disease pathophysiology. However, the field is still emerging and multiple key questions remain unanswered. Few studies explore the causal relationship between gut microbial communities and disease, limiting our understanding of how these interactions may promote or inhibit disease onset and progression. Specifically, the correlation versus causation dynamic between gut bacteriome and phageome dysbiosis in different diseases remains poorly defined. Future research must shift from correlative observations to mechanistic investigations to unravel the causal relationships between alterations in gut microbial communities, particularly the bacteriome and phageome, and disease onset, progression, or resolution. Virome research faces significant challenges due to limited computational tools and incomplete reference databases, which are biased toward known viruses, predominantly DNA viruses. Consequently, the gut virome remains underexplored, with more than 50% of viral sequences identified by metagenomic sequencing remaining unclassified. This gap hinders the accuracy and reliability of generalisations drawn from current studies. The RNA virome has received less attention than the DNA virome. The integration of longitudinal multi-omics approaches, improved viral annotation techniques, and robust in vitro and in vivo models will be pivotal in advancing virome research. These developments hold promise not only for enhancing diagnostic precision but also for guiding the design of next-generation microbiome-based interventions. In particular, faecal virome transplants (FVT) and targeted phage therapies may emerge as refined therapeutic strategies to restore microbial balance and treat disease with higher specificity. Moreover, the potential of the faecal virome and phageome as non-invasive biomarkers for disease diagnostics and prognostics deserves deeper exploration. As our understanding of the virome matures, it may serve as a critical component in the personalized medicine landscape, offering new avenues for early detection and therapeutic modulation of disease.
Author Contributions
KS: Review of Literature, manuscript draft writing, HN: Manuscript Editing, data validation, final approval, TI: Supervision and Manuscript Editing, VN: Conceptualization, Supervision and Manuscript Editing. All authors participated in data acquisition, analysis, and interpretation. All authors reviewed the manuscript and gave final approval for submission.
Conflicts of Interest
All authors declare no competing interests.
References
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Li, P.; Roos, S.; Luo, H.; Ji, B.; Nielsen, J. Metabolic Engineering of Human Gut Microbiome: Recent Developments and Future Perspectives. Metab. Eng. 2023, 79, 1–13. [Google Scholar] [CrossRef]
- Chu, Y.; Meng, Q.; Yu, J.; Zhang, J.; Chen, J.; Kang, Y. Strain-Level Dynamics Reveal Regulatory Roles in Atopic Eczema by Gut Bacterial Phages. Microbiol. Spectr. 2023, 11, e04551-22. [Google Scholar] [CrossRef] [PubMed]
- Schuler, C.F.; Tsoi, L.C.; Billi, A.C.; Harms, P.W.; Weidinger, S.; Gudjonsson, J.E. Genetic and Immunological Pathogenesis of Atopic Dermatitis. J. Invest. Dermatol. 2024, 144, 954–968. [Google Scholar] [CrossRef] [PubMed]
- Arehart, C.H.; Daya, M.; Campbell, M.; Boorgula, M.P.; Rafaels, N.; Chavan, S.; David, G.; Hanifin, J.; Slifka, M.K.; Gallo, R.L.; et al. Polygenic Prediction of Atopic Dermatitis Improves with Atopic Training and Filaggrin Factors. J. Allergy Clin. Immunol. 2022, 149, 145–155. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The Gut Microbiota — Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Bays, D.J.; Savage, H.P.; Bäumler, A.J. The Human Gut Microbiome in Health and Disease: Time for a New Chapter? Infect. Immun. 2024, 92, e00302-24. [Google Scholar] [CrossRef]
- Fakharian, F.; Thirugnanam, S.; Welsh, D.A.; Kim, W.-K.; Rappaport, J.; Bittinger, K.; Rout, N. The Role of Gut Dysbiosis in the Loss of Intestinal Immune Cell Functions and Viral Pathogenesis. Microorganisms 2023, 11, 1849. [Google Scholar] [CrossRef]
- Mazzuti, L.; Turriziani, O.; Mezzaroma, I. The Many Faces of Immune Activation in HIV-1 Infection: A Multifactorial Interconnection. Biomedicines 2023, 11, 159. [Google Scholar] [CrossRef]
- Sandler, N.G.; Douek, D.C. Microbial Translocation in HIV Infection: Causes, Consequences and Treatment Opportunities. Nat. Rev. Microbiol. 2012, 10, 655–666. [Google Scholar] [CrossRef]
- Klatt, N.R.; Chomont, N.; Douek, D.C.; Deeks, S.G. Immune Activation and HIV Persistence: Implications for Curative Approaches to HIV Infection. Immunol. Rev. 2013, 254, 326–342. [Google Scholar] [CrossRef]
- Brenchley, J.M.; Douek, D.C. Microbial Translocation across the GI Tract. Annu. Rev. Immunol. 2012, 30, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.M.; Frank, D.N.; Wilson, C.C. The Gut Microbiome and HIV-1 Pathogenesis: A Two-Way Street. AIDS 2016, 30, 2737–2751. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Tato, C.M.; Joyce-Shaikh, B.; Gulen, M.F.; Cayatte, C.; Chen, Y.; Blumenschein, W.M.; Judo, M.; Ayanoglu, G.; McClanahan, T.K.; et al. Interleukin-23-Independent IL-17 Production Regulates Intestinal Epithelial Permeability. Immunity 2015, 43, 727–738. [Google Scholar] [CrossRef]
- Nazli, A.; Chan, O.; Dobson-Belaire, W.N.; Ouellet, M.; Tremblay, M.J.; Gray-Owen, S.D.; Arsenault, A.L.; Kaushic, C. Exposure to HIV-1 Directly Impairs Mucosal Epithelial Barrier Integrity Allowing Microbial Translocation. PLoS Pathog. 2010, 6, e1000852. [Google Scholar] [CrossRef]
- Mohan, M.; Aye, P.P.; Borda, J.T.; Alvarez, X.; Lackner, A.A. Gastrointestinal Disease in Simian Immunodeficiency Virus-Infected Rhesus Macaques Is Characterized by Proinflammatory Dysregulation of the Interleukin-6-Janus Kinase/Signal Transducer and Activator of Transcription3 Pathway. Am. J. Pathol. 2007, 171, 1952–1965. [Google Scholar] [CrossRef]
- Schulbin, H.; Bode, H.; Stocker, H.; Schmidt, W.; Zippel, T.; Loddenkemper, C.; Engelmann, E.; Epple, H.-J.; Arastéh, K.; Zeitz, M. Cytokine Expression in the Colonic Mucosa of Human Immunodeficiency Virus-Infected Individuals before and during 9 Months of Antiretroviral Therapy. Antimicrob. Agents Chemother. 2008, 52, 3377–3384. [Google Scholar] [CrossRef] [PubMed]
- Tulkens, J.; Vergauwen, G.; Van Deun, J.; Geeurickx, E.; Dhondt, B.; Lippens, L.; De Scheerder, M.-A.; Miinalainen, I.; Rappu, P.; De Geest, B.G.; et al. Increased Levels of Systemic LPS-Positive Bacterial Extracellular Vesicles in Patients with Intestinal Barrier Dysfunction. Gut 2020, 69, 191–193. [Google Scholar] [CrossRef]
- Marchetti, G.; Cozzi-Lepri, A.; Merlini, E.; Bellistrì, G.M.; Castagna, A.; Galli, M.; Verucchi, G.; Antinori, A.; Costantini, A.; Giacometti, A.; et al. Microbial Translocation Predicts Disease Progression of HIV-Infected Antiretroviral-Naive Patients with High CD4+ Cell Count. AIDS Lond. Engl. 2011, 25, 1385–1394. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.-M.; Kennedy, S.; et al. Richness of Human Gut Microbiome Correlates with Metabolic Markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary Intervention Impact on Gut Microbial Gene Richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Shoaie, S.; Ghaffari, P.; Kovatcheva-Datchary, P.; Mardinoglu, A.; Sen, P.; Pujos-Guillot, E.; de Wouters, T.; Juste, C.; Rizkalla, S.; Chilloux, J.; et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015, 22, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Wejse, C.; Patsche, C.B.; Kühle, A.; Bamba, F.J.V.; Mendes, M.S.; Lemvik, G.; Gomes, V.F.; Rudolf, F. Impact of HIV-1, HIV-2, and HIV-1+2 Dual Infection on the Outcome of Tuberculosis. Int. J. Infect. Dis. 2015, 32, 128–134. [Google Scholar] [CrossRef]
- Ang, L.W.; Ng, O.T.; Boudville, I.C.; Leo, Y.S.; Wong, C.S. An Observational Study of the Prevalence of Metabolic Syndrome in Treatment-Experienced People Living with HIV in Singapore. PLOS ONE 2021, 16, e0252320. [Google Scholar] [CrossRef] [PubMed]
- Jantarapakde, J.; Phanuphak, N.; Chaturawit, C.; Pengnonyang, S.; Mathajittiphan, P.; Takamtha, P.; Dungjun, N.; Pinyakorn, S.; Pima, W.; Prasithsirikul, W.; et al. Prevalence of Metabolic Syndrome Among Antiretroviral-Naive and Antiretroviral-Experienced HIV-1 Infected Thai Adults. AIDS Patient Care STDs 2014, 28, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Rhodes, M.E.; Neff, C.P.; Fontenot, A.P.; Campbell, T.B.; Palmer, B.E. HIV-Induced Alteration in Gut Microbiota: Driving Factors, Consequences, and Effects of Antiretroviral Therapy. Gut Microbes 2014, 5, 562–570. [Google Scholar] [CrossRef]
- Lu, W.; Feng, Y.; Jing, F.; Han, Y.; Lyu, N.; Liu, F.; Li, J.; Song, X.; Xie, J.; Qiu, Z.; et al. Association Between Gut Microbiota and CD4 Recovery in HIV-1 Infected Patients. Front. Microbiol. 2018, 9, 1451. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, H.; Cole, M.; Morris, A.; Martinson, J.; Mckay, H.; Mimiaga, M.; Margolick, J.; Fitch, A.; Methe, B.; et al. Signature Changes in Gut Microbiome Are Associated with Increased Susceptibility to HIV-1 Infection in MSM. Microbiome 2021, 9, 237. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Somsouk, M. HIV and the Gut Microbiota: Composition, Consequences, and Avenues for Amelioration. Curr. HIV/AIDS Rep. 2019, 16, 204–213. [Google Scholar] [CrossRef]
- Crakes, K.R.; Santos Rocha, C.; Grishina, I.; Hirao, L.A.; Napoli, E.; Gaulke, C.A.; Fenton, A.; Datta, S.; Arredondo, J.; Marco, M.L.; et al. PPARα-Targeted Mitochondrial Bioenergetics Mediate Repair of Intestinal Barriers at the Host–Microbe Intersection during SIV Infection. Proc. Natl. Acad. Sci. 2019, 116, 24819–24829. [Google Scholar] [CrossRef]
- Irvine, S.L.; Hummelen, R.; Hekmat, S.; W. N. Looman, C.; Habbema, J.D.F.; Reid, G. Probiotic Yogurt Consumption Is Associated With an Increase of CD4 Count Among People Living With HIV/AIDS. J. Clin. Gastroenterol. 2010, 44, e201–e205. [Google Scholar] [CrossRef]
- Dikareva, E.; Matharu, D.; Lahtinen, E.; Kolho, K.-L.; De Vos, W.M.; Salonen, A.; Ponsero, A.J. An Extended Catalog of Integrated Prophages in the Infant and Adult Faecal Microbiome Shows High Prevalence of Lysogeny. Front. Microbiol. 2023, 14, 1254535. [Google Scholar] [CrossRef] [PubMed]
- Ritz, N.L.; Draper, L.A.; Bastiaanssen, T.F.S.; Turkington, C.J.R.; Peterson, V.L.; Van De Wouw, M.; Vlckova, K.; Fülling, C.; Guzzetta, K.E.; Burokas, A.; et al. The Gut Virome Is Associated with Stress-Induced Changes in Behaviour and Immune Responses in Mice. Nat. Microbiol. 2024, 9, 359–376. [Google Scholar] [CrossRef]
- Hassan, N.E.; El Shebini, S.M.; El-Masry, S.A.; Ahmed, N.H.; Kamal, A.N.; Ismail, A.S.; Alian, K.M.; Mostafa, M.I.; Selim, M.; Afify, M.A.S. Brief Overview of Dietary Intake, Some Types of Gut Microbiota, Metabolic Markers and Research Opportunities in Sample of Egyptian Women. Sci. Rep. 2022, 12, 17291. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Luo, M.; Fan, J.; Xiong, L. Multiomics Analysis Reveals Gut Virome–Bacteria–Metabolite Interactions and Their Associations with Symptoms in Patients with IBS-D. Viruses 2024, 16, 1054. [Google Scholar] [CrossRef]
- Cao, Z.; Sugimura, N.; Burgermeister, E.; Ebert, M.P.; Zuo, T.; Lan, P. The Gut Virome: A New Microbiome Component in Health and Disease. eBioMedicine 2022, 81, 104113. [Google Scholar] [CrossRef]
- Kaczorowska, J.; van der Hoek, L. Human Anelloviruses: Diverse, Omnipresent and Commensal Members of the Virome. FEMS Microbiol. Rev. 2020, 44, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Pargin, E.; Roach, M.J.; Skye, A.; Papudeshi, B.; Inglis, L.K.; Mallawaarachchi, V.; Grigson, S.R.; Harker, C.; Edwards, R.A.; Giles, S.K. The Human Gut Virome: Composition, Colonization, Interactions, and Impacts on Human Health. Front. Microbiol. 2023, 14, 963173. [Google Scholar] [CrossRef]
- Li, N.; Li, Y.; Huang, Z.; Cao, Z.; Cao, C.; Gao, X.; Zuo, T. Faecal Phageome Transplantation Alleviates Intermittent Intestinal Inflammation in IBD and the Timing of Transplantation Matters: A Preclinical Proof-of-Concept Study in Mice. Gut 2024, gutjnl-2024-333598. [Google Scholar] [CrossRef]
- Tobin, C.A.; Hill, C.; Shkoporov, A.N. Factors Affecting Variation of the Human Gut Phageome. Annu. Rev. Microbiol. 2023, 77, 363–379. [Google Scholar] [CrossRef]
- Ren, C.; Hong, B.; Zhang, S.; Yuan, D.; Feng, J.; Shan, S.; Zhang, J.; Guan, L.; Zhu, L.; Lu, S. Autoclaving-Treated Germinated Brown Rice Relieves Hyperlipidemia by Modulating Gut Microbiota in Humans. Front. Nutr. 2024, 11, 1403200. [Google Scholar] [CrossRef]
- Chen, L.; Liu, B.; Ren, L.; Du, H.; Fei, C.; Qian, C.; Li, B.; Zhang, R.; Liu, H.; Li, Z.; et al. High-Fiber Diet Ameliorates Gut Microbiota, Serum Metabolism and Emotional Mood in Type 2 Diabetes Patients. Front. Cell. Infect. Microbiol. 2023, 13, 1069954. [Google Scholar] [CrossRef]
- Laursen, M.F. Gut Microbiota Development: Influence of Diet from Infancy to Toddlerhood. Ann. Nutr. Metab. 2021, 77, 21–34. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, R. The Human Gut Phageome: Composition, Development, and Alterations in Disease. Front. Microbiol. 2023, 14, 1213625. [Google Scholar] [CrossRef]
- Mihindukulasuriya, K.A.; Mars, R.A.T.; Johnson, A.J.; Ward, T.; Priya, S.; Lekatz, H.R.; Kalari, K.R.; Droit, L.; Zheng, T.; Blekhman, R.; et al. Multi-Omics Analyses Show Disease, Diet, and Transcriptome Interactions With the Virome. Gastroenterology 2021, 161, 1194–1207.e8. [Google Scholar] [CrossRef]
- Littlejohn, P.T.; Metcalfe-Roach, A.; Cardenas Poire, E.; Holani, R.; Bar-Yoseph, H.; Fan, Y.M.; Woodward, S.E.; Finlay, B.B. Multiple Micronutrient Deficiencies in Early Life Cause Multi-Kingdom Alterations in the Gut Microbiome and Intrinsic Antibiotic Resistance Genes in Mice. Nat. Microbiol. 2023, 8, 2392–2405. [Google Scholar] [CrossRef] [PubMed]
- Mehandru, S.; Poles, M.A.; Tenner-Racz, K.; Manuelli, V.; Jean-Pierre, P.; Lopez, P.; Shet, A.; Low, A.; Mohri, H.; Boden, D.; et al. Mechanisms of Gastrointestinal CD4+ T-Cell Depletion during Acute and Early Human Immunodeficiency Virus Type 1 Infection. J. Virol. 2007, 81, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Epple, H.-J.; Schneider, T.; Troeger, H.; Kunkel, D.; Allers, K.; Moos, V.; Amasheh, M.; Loddenkemper, C.; Fromm, M.; Zeitz, M.; et al. Impairment of the Intestinal Barrier Is Evident in Untreated but Absent in Suppressively Treated HIV-Infected Patients. Gut 2009, 58, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.J.; Nazli, A.; Rojas, O.L.; Chege, D.; Alidina, Z.; Huibner, S.; Mujib, S.; Benko, E.; Kovacs, C.; Shin, L.Y.Y.; et al. A Role for Mucosal IL-22 Production and Th22 Cells in HIV-Associated Mucosal Immunopathogenesis. Mucosal Immunol. 2012, 5, 670–680. [Google Scholar] [CrossRef]
- Kök, A.; Hocqueloux, L.; Hocini, H.; Carrière, M.; Lefrou, L.; Guguin, A.; Tisserand, P.; Bonnabau, H.; Avettand-Fenoel, V.; Prazuck, T.; et al. Early Initiation of Combined Antiretroviral Therapy Preserves Immune Function in the Gut of HIV-Infected Patients. Mucosal Immunol. 2015, 8, 127–140. [Google Scholar] [CrossRef]
- Ishizaka, A.; Koga, M.; Mizutani, T.; Parbie, P.K.; Prawisuda, D.; Yusa, N.; Sedohara, A.; Kikuchi, T.; Ikeuchi, K.; Adachi, E.; et al. Unique Gut Microbiome in HIV Patients on Antiretroviral Therapy (ART) Suggests Association with Chronic Inflammation. Microbiol. Spectr. 2021, 9, e00708-21. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Wu, N.; Jin, C. Intestinal Microbiota Dysbiosis Promotes Mucosal Barrier Damage and Immune Injury in HIV-Infected Patients. Can. J. Infect. Dis. Med. Microbiol. 2023, 2023, 1–15. [Google Scholar] [CrossRef]
- Ladinsky, M.S.; Kieffer, C.; Olson, G.; Deruaz, M.; Vrbanac, V.; Tager, A.M.; Kwon, D.S.; Bjorkman, P.J. Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue. PLoS Pathog. 2014, 10, e1003899. [Google Scholar] [CrossRef]
- Drain, P.K.; Mosam, A.; Gounder, L.; Gosnell, B.; Manzini, T.; Moosa, M.-Y.S. Recurrent Giant Molluscum Contagiosum Immune Reconstitution Inflammatory Syndrome (IRIS) after Initiation of Antiretroviral Therapy in an HIV-Infected Man. Int. J. STD AIDS 2014, 25, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Obeagu, E.I.; Obeagu, G.U. Utilization of Immunological Ratios in HIV: Implications for Monitoring and Therapeutic Strategies. Medicine (Baltimore) 2024, 103, e37354. [Google Scholar] [CrossRef]
- Serrano-Villar, S.; Sainz, T.; Ma, Z.-M.; Utay, N.S.; Wook-Chun, T.; Mann, S.; Kashuba, A.D.; Siewe, B.; Albanese, A.; Troia-Cancio, P.; et al. Effects of Combined CCR5/Integrase Inhibitors-Based Regimen on Mucosal Immunity in HIV-Infected Patients Naïve to Antiretroviral Therapy: A Pilot Randomized Trial. PLOS Pathog. 2016, 12, e1005381. [Google Scholar] [CrossRef]
- Kaelin, E.A.; Mitchell, C.; Soria, J.; Rosa, A.L.; Ticona, E.; Coombs, R.W.; Frenkel, L.M.; Bull, M.E.; Lim, E.S. Longitudinal Cervicovaginal Microbiome and Virome Alterations during ART and Discordant Shedding in Women Living with HIV 2024.
- Boukadida, C.; Peralta-Prado, A.; Chávez-Torres, M.; Romero-Mora, K.; Rincon-Rubio, A.; Ávila-Ríos, S.; Garrido-Rodríguez, D.; Reyes-Terán, G.; Pinto-Cardoso, S. Alterations of the Gut Microbiome in HIV Infection Highlight Human Anelloviruses as Potential Predictors of Immune Recovery. Microbiome 2024, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Villoslada-Blanco, P.; Pérez-Matute, P.; Íñiguez, M.; Recio-Fernández, E.; Jansen, D.; De Coninck, L.; Close, L.; Blanco-Navarrete, P.; Metola, L.; Ibarra, V.; et al. Impact of HIV Infection and Integrase Strand Transfer Inhibitors-Based Treatment on the Gut Virome. Sci. Rep. 2022, 12, 21658. [Google Scholar] [CrossRef]
- Zhang, Y.; Andreu-Sánchez, S.; Vadaq, N.; Wang, D.; Matzaraki, V.; Van Der Heijden, W.A.; Gacesa, R.; Weersma, R.K.; Zhernakova, A.; Vandekerckhove, L.; et al. Gut Dysbiosis Associates with Cytokine Production Capacity in Viral-Suppressed People Living with HIV. Front. Cell. Infect. Microbiol. 2023, 13, 1202035. [Google Scholar] [CrossRef]
- Belda, E.; Capeau, J.; Zucker, J.-D.; Chatelier, E.L.; Pons, N.; Oñate, F.P.; Quinquis, B.; Alili, R.; Fellahi, S.; Katlama, C.; et al. Major Depletion of Insulin Sensitivity-Associated Taxa in the Gut Microbiome of Persons Living with HIV Controlled by Antiretroviral Drugs. BMC Med. Genomics 2024, 17, 209. [Google Scholar] [CrossRef]
- Marsch, P.; Rajagopal, N.; Nangia, S. Biophysics of Claudin Proteins in Tight Junction Architecture: Three Decades of Progress. Biophys. J. 2024, 123, 2363–2378. [Google Scholar] [CrossRef] [PubMed]
- Blaak, E.E.; Canfora, E.E.; Theis, S.; Frost, G.; Groen, A.K.; Mithieux, G.; Nauta, A.; Scott, K.; Stahl, B.; Van Harsselaar, J.; et al. Short Chain Fatty Acids in Human Gut and Metabolic Health. Benef. Microbes 2020, 11, 411–455. [Google Scholar] [CrossRef]
- Handley, S.A.; Thackray, L.B.; Zhao, G.; Presti, R.; Miller, A.D.; Droit, L.; Abbink, P.; Maxfield, L.F.; Kambal, A.; Duan, E.; et al. Pathogenic Simian Immunodeficiency Virus Infection Is Associated with Expansion of the Enteric Virome. Cell 2012, 151, 253–266. [Google Scholar] [CrossRef]
- Taboada, B.; Morán, P.; Serrano-Vázquez, A.; Iša, P.; Rojas-Velázquez, L.; Pérez-Juárez, H.; López, S.; Torres, J.; Ximenez, C.; Arias, C.F. The Gut Virome of Healthy Children during the First Year of Life Is Diverse and Dynamic. PLOS ONE 2021, 16, e0240958. [Google Scholar] [CrossRef]
- Liu, K.; Li, Y.; Xu, R.; Zhang, Y.; Zheng, C.; Wan, Z.; Li, H.; Yang, Z.; Jin, X.; Hao, P.; et al. HIV-1 Infection Alters the Viral Composition of Plasma in Men Who Have Sex with Men. mSphere 2021, 6, e00081-21. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jin, Y.; Chen, B.; Lin, A.; Wang, E.; Xu, F.; Hu, G.; Xiao, C.; Liu, H.; Hou, X.; et al. Exploring the Relationship between the Gut Mucosal Virome and Colorectal Cancer: Characteristics and Correlations. Cancers 2023, 15, 3555. [Google Scholar] [CrossRef] [PubMed]
- Zuppi, M.; Hendrickson, H.L.; O’Sullivan, J.M.; Vatanen, T. Phages in the Gut Ecosystem. Front. Cell. Infect. Microbiol. 2022, 11, 822562. [Google Scholar] [CrossRef]
- Lapa, D.; Del Porto, P.; Minosse, C.; D’Offizi, G.; Antinori, A.; Capobianchi, M.R.; Visco-Comandini, U.; McPhee, F.; Garbuglia, A.R.; Zaccarelli, M. Clinical Relevance of Torque Teno Virus (TTV) in HIV/HCV Coinfected and HCV Monoinfected Patients Treated with Direct-Acting Antiviral Therapy. J. Clin. Med. 2021, 10, 2092. [Google Scholar] [CrossRef]
- Del Rosal, T.; García-García, M.L.; Casas, I.; Iglesias-Caballero, M.; Pozo, F.; Alcolea, S.; Bravo, B.; Rodrigo-Muñoz, J.M.; Del Pozo, V.; Calvo, C. Torque Teno Virus in Nasopharyngeal Aspirate of Children With Viral Respiratory Infections. Pediatr. Infect. Dis. J. 2023, 42, 184–188. [Google Scholar] [CrossRef]
- Mancuso, R.; Saresella, M.; Hernis, A.; Agostini, S.; Piancone, F.; Caputo, D.; Maggi, F.; Clerici, M. Torque Teno Virus (TTV) in Multiple Sclerosis Patients with Different Patterns of Disease. J. Med. Virol. 2013, 85, 2176–2183. [Google Scholar] [CrossRef]
- Rocchi, J.; Ricci, V.; Albani, M.; Lanini, L.; Andreoli, E.; Macera, L.; Pistello, M.; Ceccherini-Nelli, L.; Bendinelli, M.; Maggi, F. Torquetenovirus DNA Drives Proinflammatory Cytokines Production and Secretion by Immune Cells via Toll-like Receptor 9. Virology 2009, 394, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Webel, A.R.; Schexnayder, J.; Cioe, P.A.; Zuñiga, J.A. A Review of Chronic Comorbidities in Adults Living With HIV: State of the Science. J. Assoc. Nurses AIDS Care 2021, 32, 322–346. [Google Scholar] [CrossRef] [PubMed]
- Nix, L.M.; Tien, P.C. Metabolic Syndrome, Diabetes, and Cardiovascular Risk in HIV. Curr. HIV/AIDS Rep. 2014, 11, 271–278. [Google Scholar] [CrossRef]
- Bikel, S.; López-Leal, G.; Cornejo-Granados, F.; Gallardo-Becerra, L.; García-López, R.; Sánchez, F.; Equihua-Medina, E.; Ochoa-Romo, J.P.; López-Contreras, B.E.; Canizales-Quinteros, S.; et al. Gut dsDNA Virome Shows Diversity and Richness Alterations Associated with Childhood Obesity and Metabolic Syndrome. iScience 2021, 24, 102900. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Peng, Y. Ten Computational Challenges in Human Virome Studies. Virol. Sin. 2024, 39, 845–850. [Google Scholar] [CrossRef]
- Yang, K.; Niu, J.; Zuo, T.; Sun, Y.; Xu, Z.; Tang, W.; Liu, Q.; Zhang, J.; Ng, E.K.W.; Wong, S.K.H.; et al. Alterations in the Gut Virome in Obesity and Type 2 Diabetes Mellitus. Gastroenterology 2021, 161, 1257–1269.e13. [Google Scholar] [CrossRef]
- Geng, S.-T.; Zhang, Z.-Y.; Wang, Y.-X.; Lu, D.; Yu, J.; Zhang, J.-B.; Kuang, Y.-Q.; Wang, K.-H. Regulation of Gut Microbiota on Immune Reconstitution in Patients With Acquired Immunodeficiency Syndrome. Front. Microbiol. 2020, 11, 594820. [Google Scholar] [CrossRef]
- Rathi, K.K.; Kumari, N.; Javaid, M.D.; Saleem, U.; Mortensen, E.; Zhou, Y.; Maheshwari, N. Gut Microbiome and Prediabetes - a Review. Front. Bacteriol. 2023, 2, 1242297. [Google Scholar] [CrossRef]
- Chang, W.-L.; Chen, Y.-E.; Tseng, H.-T.; Cheng, C.-F.; Wu, J.-H.; Hou, Y.-C. Gut Microbiota in Patients with Prediabetes. Nutrients 2024, 16, 1105. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Chen, Q.; Ma, X.; Li, C.; Shen, Y.; Zhu, W.; Zhang, Y.; Guo, X.; Zhou, J.; Liu, C. Enteric Phageome Alterations in Patients With Type 2 Diabetes. Front. Cell. Infect. Microbiol. 2021, 10, 575084. [Google Scholar] [CrossRef] [PubMed]
- Hoel, H.; Hove-Skovsgaard, M.; Hov, J.R.; Gaardbo, J.C.; Holm, K.; Kummen, M.; Rudi, K.; Nwosu, F.; Valeur, J.; Gelpi, M.; et al. Impact of HIV and Type 2 Diabetes on Gut Microbiota Diversity, Tryptophan Catabolism and Endothelial Dysfunction. Sci. Rep. 2018, 8, 6725. [Google Scholar] [CrossRef] [PubMed]
- Rim, S.; Vedøy, O.B.; Brønstad, I.; McCann, A.; Meyer, K.; Steinsland, H.; Hanevik, K. Inflammation, the Kynurenines, and Mucosal Injury during Human Experimental Enterotoxigenic Escherichia Coli Infection. Med. Microbiol. Immunol. (Berl.) 2024, 213, 2. [Google Scholar] [CrossRef]
- Trøseid, M.; Molinaro, A.; Gelpi, M.; Vestad, B.; Kofoed, K.F.; Fuchs, A.; Køber, L.; Holm, K.; Benfield, T.; Ueland, P.M.; et al. Gut Microbiota Alterations and Circulating Imidazole Propionate Levels Are Associated With Obstructive Coronary Artery Disease in People With HIV. J. Infect. Dis. 2024, 229, 898–907. [Google Scholar] [CrossRef]
- Wang, Z.; Peters, B.A.; Bryant, M.; Hanna, D.B.; Schwartz, T.; Wang, T.; Sollecito, C.C.; Usyk, M.; Grassi, E.; Wiek, F.; et al. Gut Microbiota, Circulating Inflammatory Markers and Metabolites, and Carotid Artery Atherosclerosis in HIV Infection. Microbiome 2023, 11, 119. [Google Scholar] [CrossRef]
- Peters, B.A.; Burk, R.D.; Kaplan, R.C.; Qi, Q. The Gut Microbiome, Microbial Metabolites, and Cardiovascular Disease in People Living with HIV. Curr. HIV/AIDS Rep. 2023, 20, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, J.; Han, D. Causal Relationships between Gut Microbiota, Plasma Metabolites, and HIV Infection: Insights from Mendelian Randomization and Mediation Analysis. Virol. J. 2024, 21, 204. [Google Scholar] [CrossRef]
- Campbell, D.E.; Ly, L.K.; Ridlon, J.M.; Hsiao, A.; Whitaker, R.J.; Degnan, P.H. Infection with Bacteroides Phage BV01 Alters the Host Transcriptome and Bile Acid Metabolism in a Common Human Gut Microbe. Cell Rep. 2020, 32, 108142. [Google Scholar] [CrossRef]
- Avellaneda-Franco, L.; Dahlman, S.; Barr, J.J. The Gut Virome and the Relevance of Temperate Phages in Human Health. Front. Cell. Infect. Microbiol. 2023, 13, 1241058. [Google Scholar] [CrossRef]
- Shamash, M.; Maurice, C.F. Phages in the Infant Gut: A Framework for Virome Development during Early Life. ISME J. 2022, 16, 323–330. [Google Scholar] [CrossRef]
- Pavia, G.; Marascio, N.; Matera, G.; Quirino, A. Does the Human Gut Virome Contribute to Host Health or Disease? Viruses 2023, 15, 2271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sharma, S.; Tom, L.; Liao, Y.-T.; Wu, V.C.H. Gut Phageome—An Insight into the Role and Impact of Gut Microbiome and Their Correlation with Mammal Health and Diseases. Microorganisms 2023, 11, 2454. [Google Scholar] [CrossRef] [PubMed]
- Benler, S.; Cobián-Güemes, A.G.; McNair, K.; Hung, S.-H.; Levi, K.; Edwards, R.; Rohwer, F. A Diversity-Generating Retroelement Encoded by a Globally Ubiquitous Bacteroides Phage. Microbiome 2018, 6, 191. [Google Scholar] [CrossRef]
- Guo, H.; Arambula, D.; Ghosh, P.; Miller, J.F. Diversity-Generating Retroelements in Phage and Bacterial Genomes. Microbiol. Spectr. 2014, 2, 2.6.25. [Google Scholar] [CrossRef]
- Medhekar, B.; Miller, J.F. Diversity-Generating Retroelements. Curr. Opin. Microbiol. 2007, 10, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Carasso, S.; Zaatry, R.; Hajjo, H.; Kadosh-Kariti, D.; Ben-Assa, N.; Naddaf, R.; Mandelbaum, N.; Pressman, S.; Chowers, Y.; Gefen, T.; et al. Inflammation and Bacteriophages Affect DNA Inversion States and Functionality of the Gut Microbiota. Cell Host Microbe 2024, 32, 322–334.e9. [Google Scholar] [CrossRef]
- Escobedo, S.; Wegmann, U.; Pérez De Pipaon, M.; Campelo, A.B.; Stentz, R.; Rodríguez, A.; Martínez, B. Resident TP712 Prophage of Lactococcus Lactis Strain MG1363 Provides Extra Holin Functions to the P335 Phage CAP for Effective Host Lysis. Appl. Environ. Microbiol. 2021, 87, e01092-21. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).