Submitted:
15 May 2025
Posted:
16 May 2025
You are already at the latest version
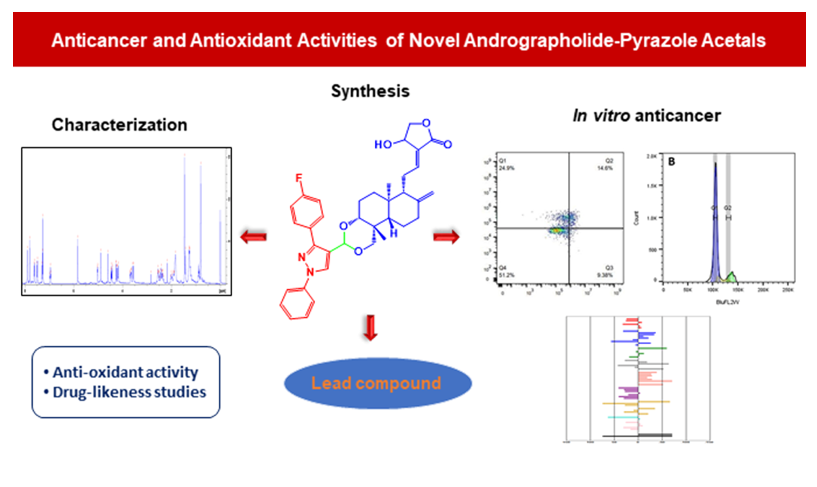
Abstract
Keywords:
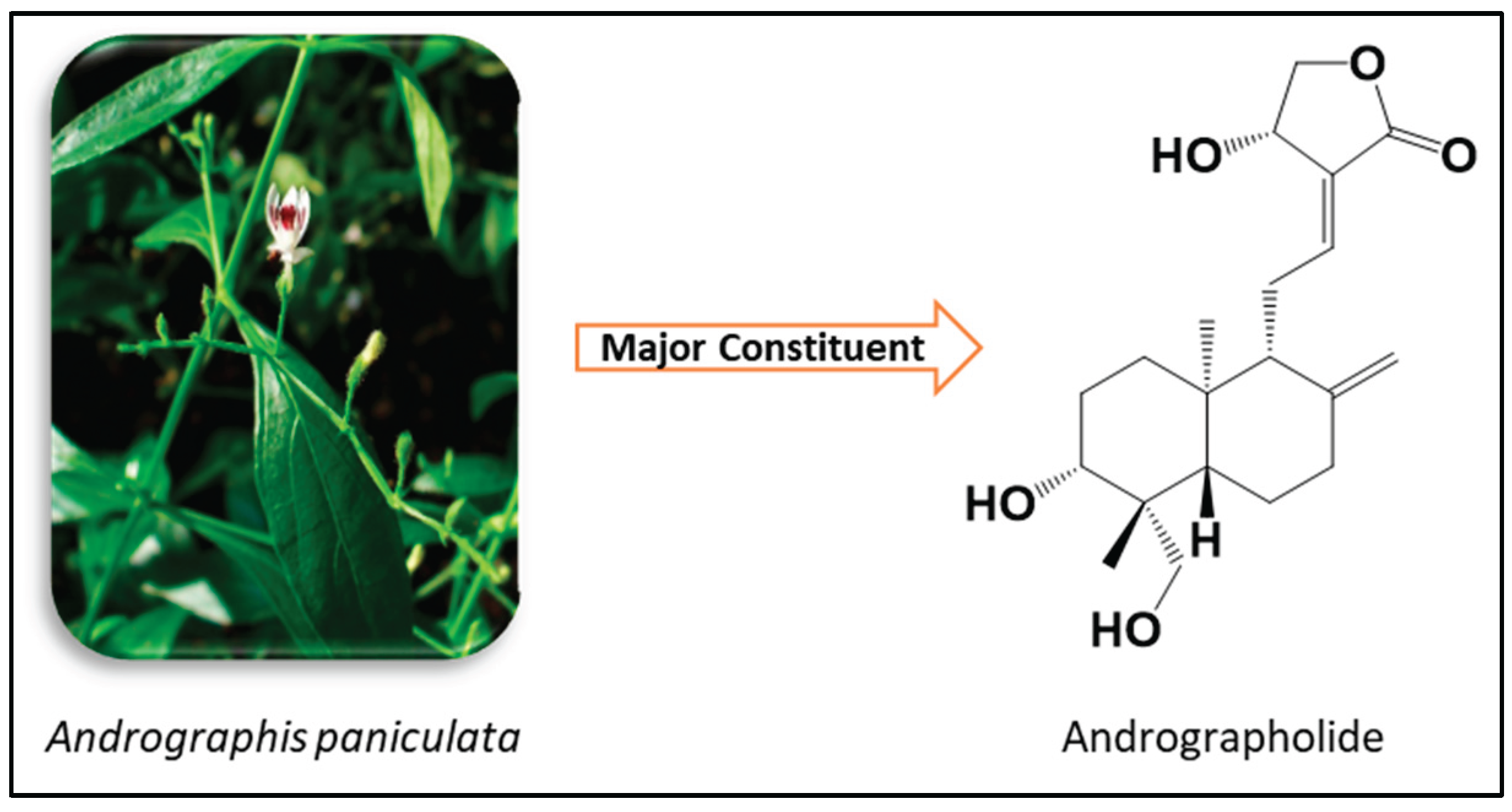
1. Introduction
2. Results and Discussion
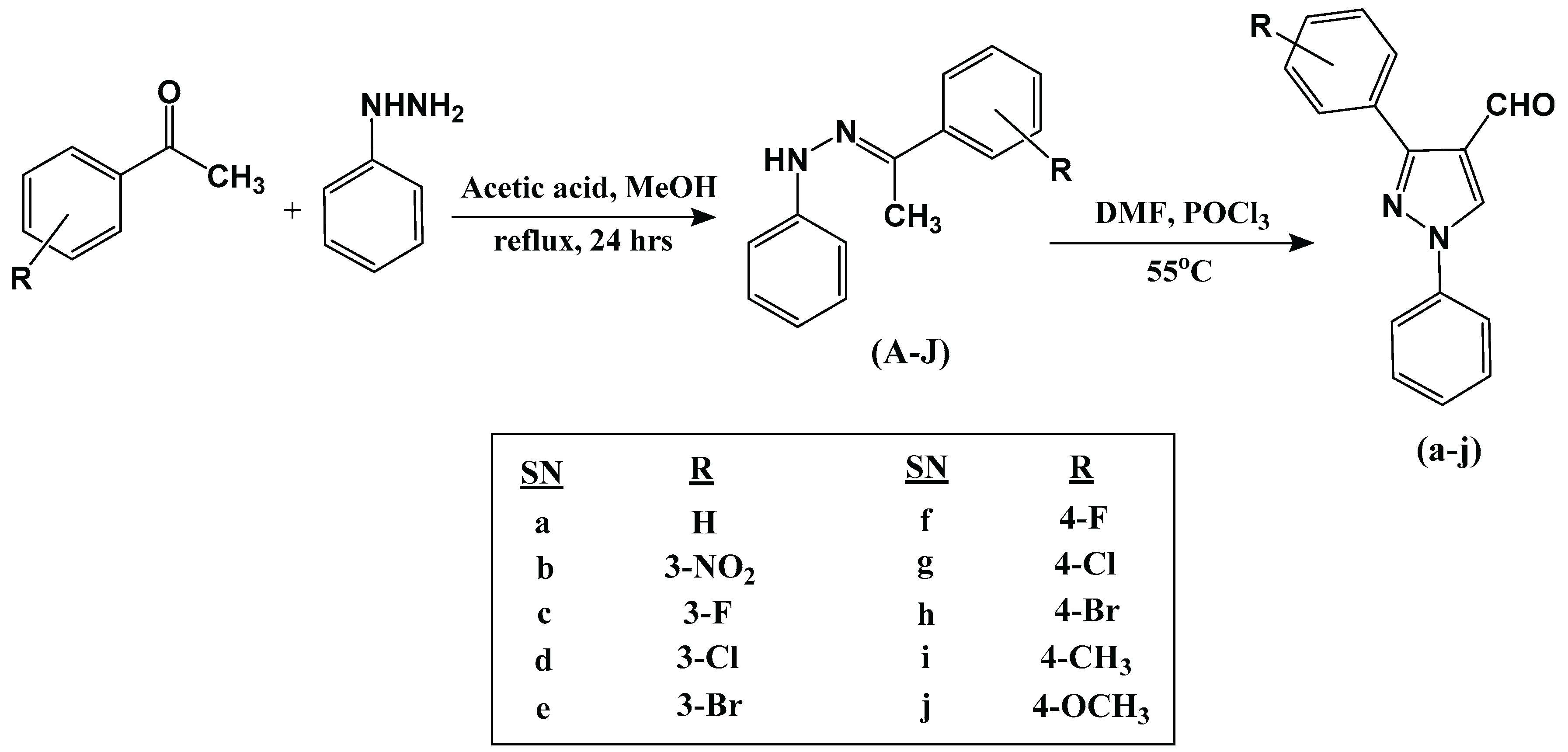
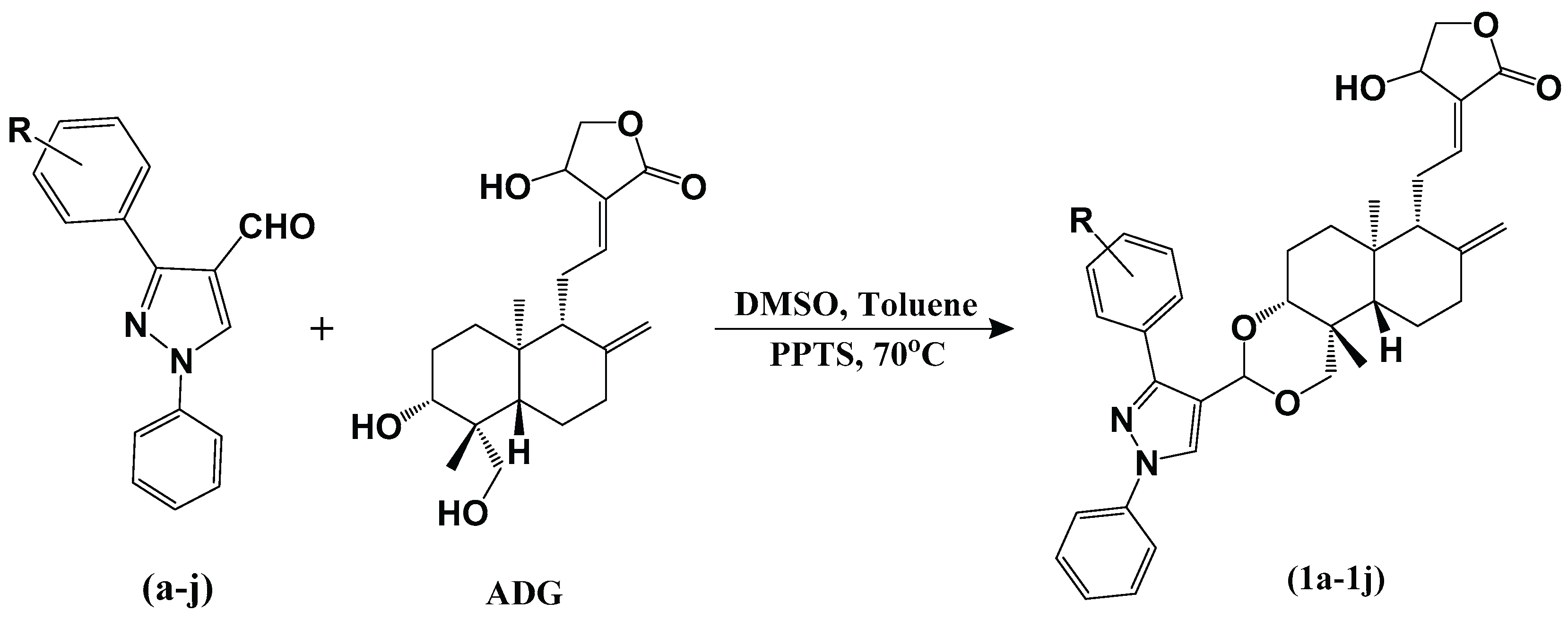
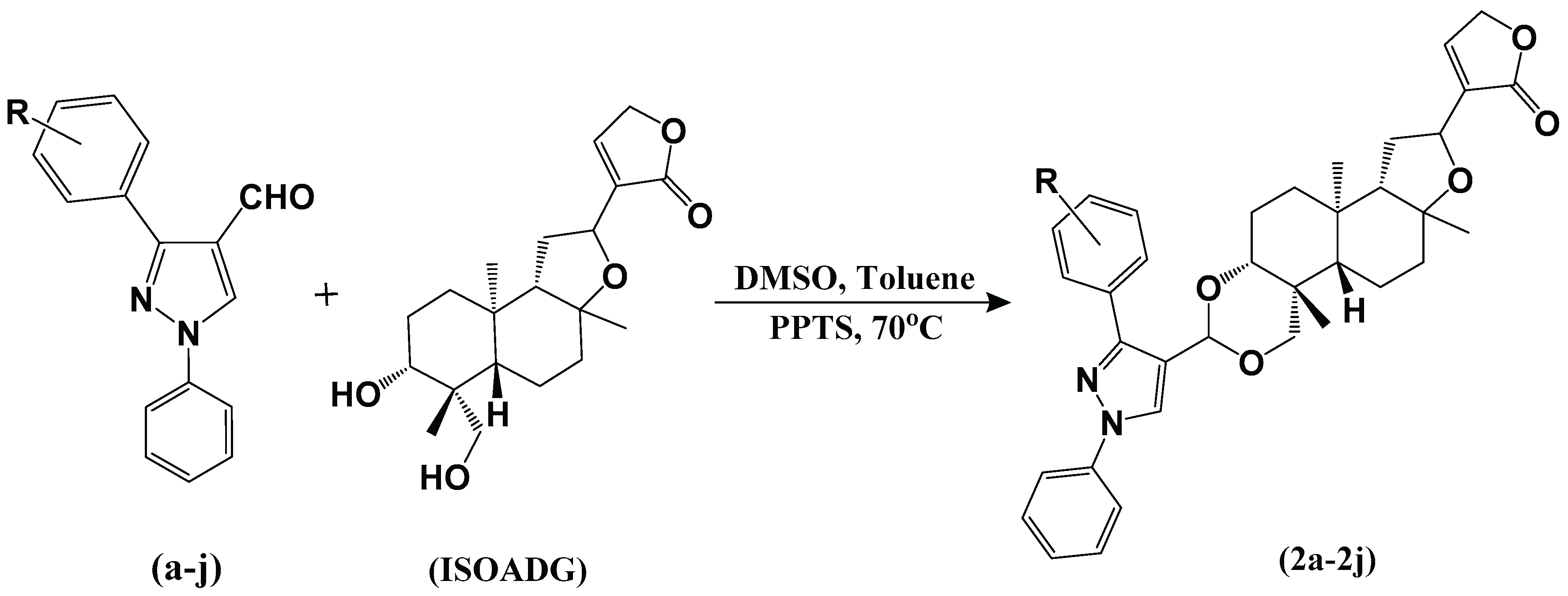
2.1. Chemistry
2.2. Characterization
2.3. In Vitro Anticancer Studies
2.3.1. NCI Screening
2.3.2. Single Dose Study
2.3.3. Five Dose Study
2.3.4. GI50 Values of the Compounds on Individual Cancer Types
2.3.5. NCI Compare Analysis
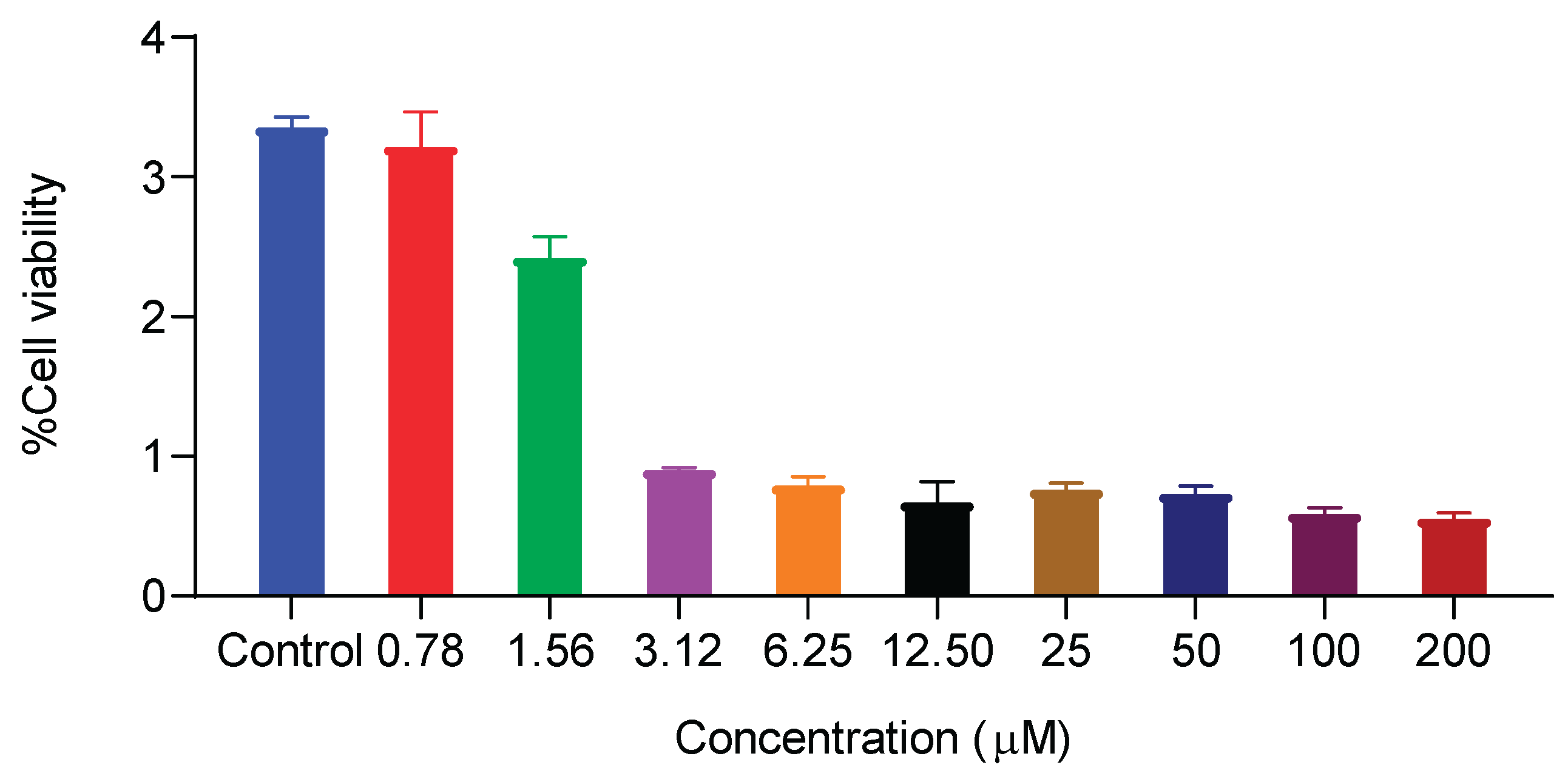
2.3.6. Effect of 1f on the Proliferation of MCF-10a and MDA-MB-231
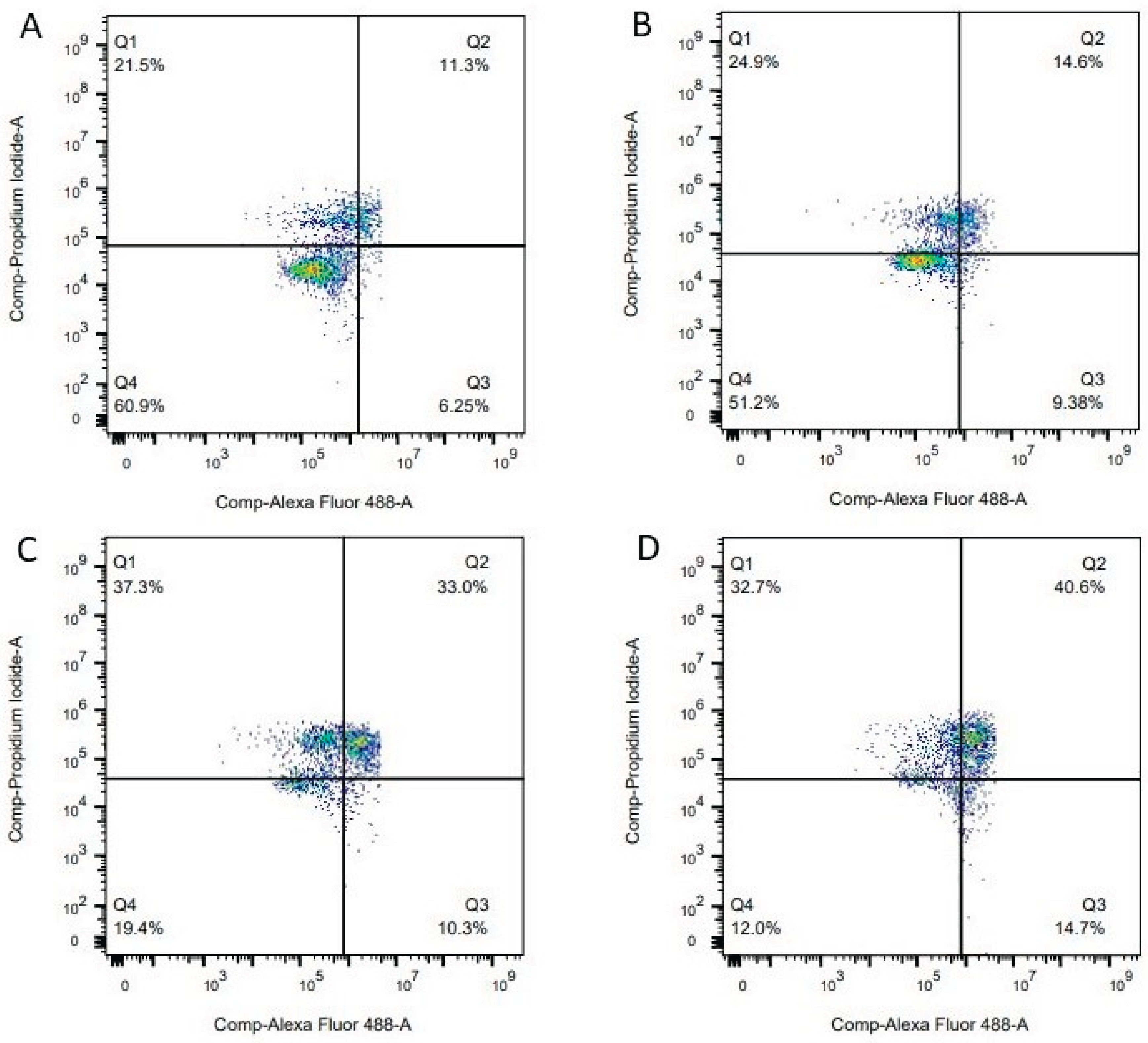
2.3.7. Effect of 1f on Cell Apoptosis Inducement
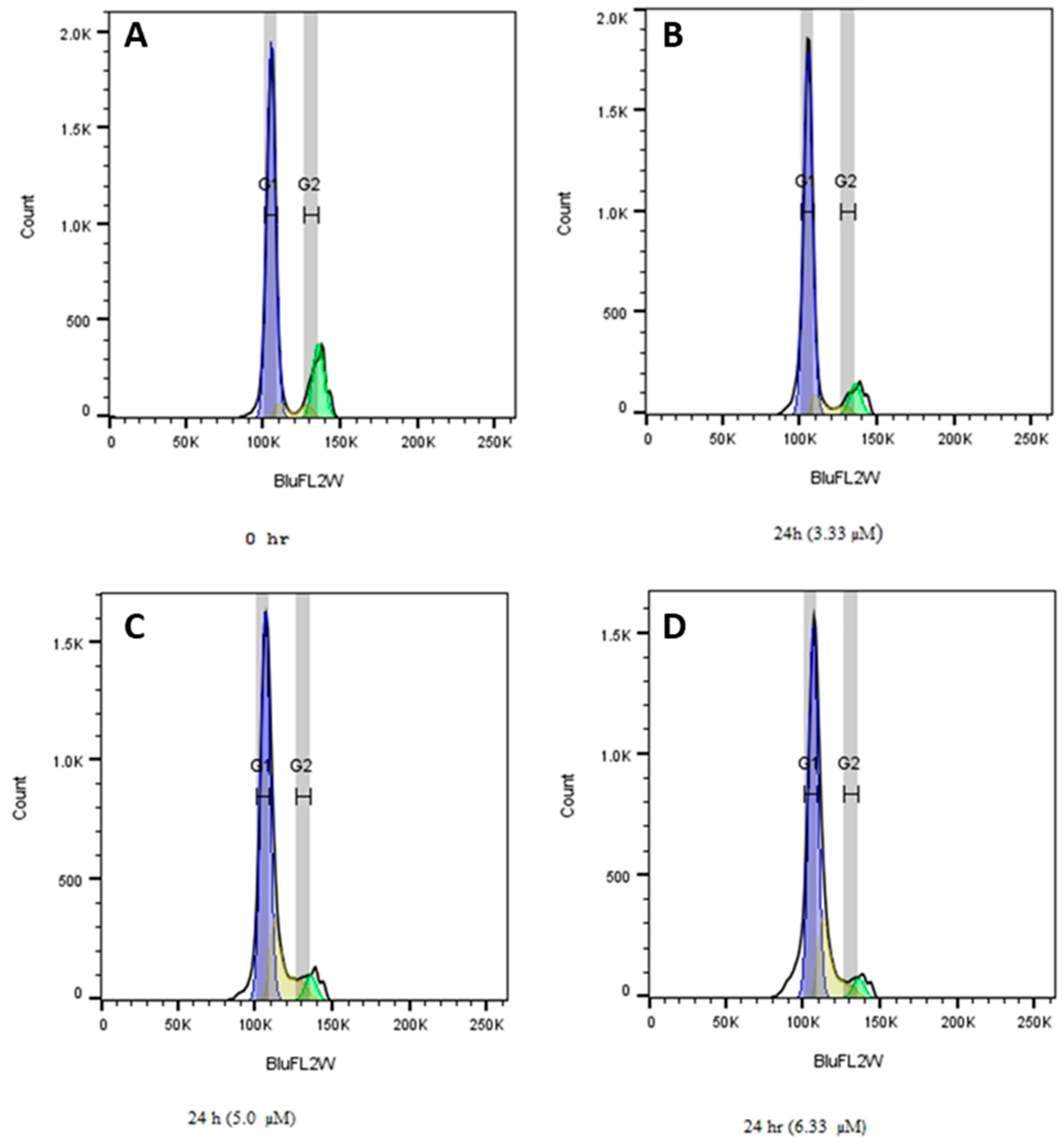
2.3.8. Effects of 1f on MDA-MB-231 Cell Cycle Distribution
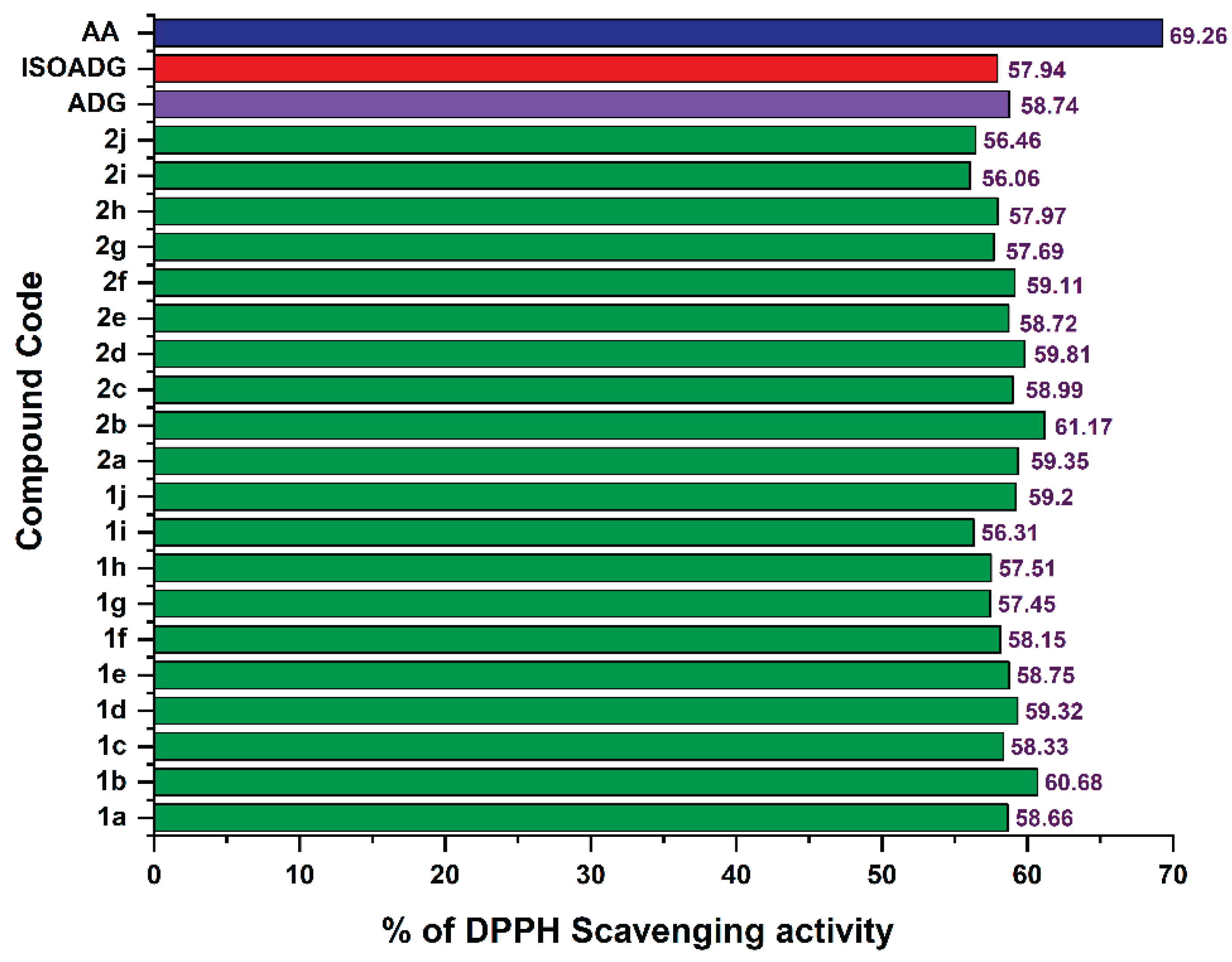
2.4. Antioxidant Activity
2.5. Drug-Likeness Studies for the Active Compounds
3. Experimental
3.1. Synthesis
3.1.1. Synthesis of Aryl Hydrazones (A-J)
3.1.2. Synthesis of 3-aryl-4-Pyrazole Carboxaldehydes (a-j)
3.1.3. Synthesis of 3,19-(N-phenyl-3-aryl-pyrazole) Acetals of Andrographolide (1a-1j)
3.1.4. Synthesis of Isoandrographolide and 3,19-(N-phenyl-3-aryl-pyrazole) Acetals of Isoandrographolide (2a-2j)
3.2. Instrumentation
3.3. In Vitro Anticancer Studies
3.3.1. Cell Proliferation Assay
3.3.2. Apoptotic Assay
3.3.3. Cell Cycle Assay
3.4. DPPH Assay
4. Conclusion
Supplementary Information
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability
Acknowledgments
Conflicts of Interest
References
- Majérus, M.-A. The Cause of Cancer: The Unifying Theory. Adv. Cancer Biol. - Metastasis 2022, 4, 100034. [Google Scholar] [CrossRef]
- Cao, W.; Chen, H.-D.; Yu, Y.-W.; Li, N.; Chen, W.-Q. Changing Profiles of Cancer Burden Worldwide and in China: A Secondary Analysis of the Global Cancer Statistics 2020. Chin. Med. J. (Engl). 2021, 134, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Deo, S.V.S.; Sharma, J.; Kumar, S. GLOBOCAN 2020 Report on Global Cancer Burden: Challenges and Opportunities for Surgical Oncologists. Ann. Surg. Oncol. 2022, 29, 6497–6500. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA. Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Houghton, S.C.; Hankinson, S.E. Cancer Progress and Priorities: Breast Cancer. Cancer Epidemiol. biomarkers Prev. 2021, 30, 822–844. [Google Scholar] [CrossRef]
- Hong, R.; Xu, B. Breast Cancer: An Up-to-date Review and Future Perspectives. Cancer Commun. 2022, 42, 913–936. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New Drugs, New Toxicities: Severe Side Effects of Modern Targeted and Immunotherapy of Cancer and Their Management. Crit. Care 2017, 21, 1–11. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The Side Effects of Platinum-Based Chemotherapy Drugs: A Review for Chemists. Dalt. Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef]
- Iwamoto, T. Clinical Application of Drug Delivery Systems in Cancer Chemotherapy: Review of the Efficacy and Side Effects of Approved Drugs. Biol. Pharm. Bull. 2013, 36, 715–718. [Google Scholar] [CrossRef]
- Huang, M.; Lu, J.-J.; Ding, J. Natural Products in Cancer Therapy: Past, Present and Future. Nat. Products Bioprospect. 2021, 11, 5–13. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Qu, J.; Sun, Y.; Zhou, W. Role of Natural Products in Tumor Therapy from Basic Research and Clinical Perspectives. Acta Mater. Medica 2024, 3, 163–206. [Google Scholar] [CrossRef]
- Ouyang, L.; Luo, Y.; Tian, M.; Zhang, S.; Lu, R.; Wang, J.; Kasimu, R.; Li, X. Plant Natural Products: From Traditional Compounds to New Emerging Drugs in Cancer Therapy. Cell Prolif. 2014, 47, 506–515. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Basu, S.; Sarkar, N.; Ghosh, A.C. Advances in Cancer Therapy with Plant Based Natural Products. Curr. Med. Chem. 2001, 8, 1467–1486. [Google Scholar] [CrossRef]
- Ali Abdalla, Y.O.; Subramaniam, B.; Nyamathulla, S.; Shamsuddin, N.; Arshad, N.M.; Mun, K.S.; Awang, K.; Nagoor, N.H. Natural Products for Cancer Therapy: A Review of Their Mechanism of Actions and Toxicity in the Past Decade. J. Trop. Med. 2022, 2022, 5794350. [Google Scholar] [CrossRef]
- Wang, Y.; Zhong, J.; Bai, J.; Tong, R.; An, F.; Jiao, P.; He, L.; Zeng, D.; Long, E.; Yan, J. The Application of Natural Products in Cancer Therapy by Targeting Apoptosis Pathways. Curr. Drug Metab. 2018, 19, 739–749. [Google Scholar] [CrossRef]
- Huang, M.; Lu, J.-J.; Huang, M.-Q.; Bao, J.-L.; Chen, X.-P.; Wang, Y.-T. Terpenoids: Natural Products for Cancer Therapy. Expert Opin. Investig. Drugs 2012, 21, 1801–1818. [Google Scholar] [CrossRef]
- Demain, A.L.; Vaishnav, P. Natural Products for Cancer Chemotherapy. Microb. Biotechnol. 2011, 4, 687–699. [Google Scholar] [CrossRef]
- Hossain, M.S.; Urbi, Z.; Sule, A.; Rahman, K.M.H. Andrographis Paniculata (Burm. f.) Wall. Ex Nees: A Review of Ethnobotany, Phytochemistry, and Pharmacology. Sci. World J. 2014, 2014, 1–28. [Google Scholar] [CrossRef]
- Jiang, M.; Sheng, F.; Zhang, Z.; Ma, X.; Gao, T.; Fu, C.; Li, P. Andrographis Paniculata (Burm.f.) Nees and Its Major Constituent Andrographolide as Potential Antiviral Agents. J. Ethnopharmacol. 2021, 272, 113954. [Google Scholar] [CrossRef]
- Kandanur, S.G.S.; Tamang, N.; Golakoti, N.R.; Nanduri, S. Andrographolide: A Natural Product Template for the Generation of Structurally and Biologically Diverse Diterpenes. Eur. J. Med. Chem. 2019, 176, 513–533. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, T.; Hsieh, C.-Y.; Lee, J.-J.; Sheu, J.-R. Experimental and Clinical Pharmacology of Andrographis Paniculata and Its Major Bioactive Phytoconstituent Andrographolide. Evidence-Based Complement. Altern. Med. 2013, 2013, 846740. [Google Scholar] [CrossRef] [PubMed]
- Bharati, B.D.; Sharma, P.K.; Kumar, N.; Dudhe, R.; Bansal, V. Pharmacological Activity of Andrographis Paniculata: A Brief Review. Pharmacologyonline 2011, 2, 10. [Google Scholar]
- Okhuarobo, A.; Ehizogie Falodun, J.; Erharuyi, O.; Imieje, V.; Falodun, A.; Langer, P. Harnessing the Medicinal Properties of Andrographis Paniculata for Diseases and beyond: A Review of Its Phytochemistry and Pharmacology. Asian Pacific J. Trop. Dis. 2014, 4, 213–222. [Google Scholar] [CrossRef]
- Jadhav, A.K.; Karuppayil, S.M. Andrographis Paniculata (Burm. F) Wall Ex Nees: Antiviral Properties. Phyther. Res. 2021, 35, 5365–5373. [Google Scholar] [CrossRef]
- Lim, J.C.W.; Chan, T.K.; Ng, D.S.; Sagineedu, S.R.; Stanslas, J.; Wong, W.F. Andrographolide and Its Analogues: Versatile Bioactive Molecules for Combating Inflammation and Cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 300–310. [Google Scholar] [CrossRef]
- Wei, S.; Tang, Y.-B.; Hua, H.; Ohkoshi, E.; Goto, M.; Wang, L.-T.; Lee, K.-H.; Xiao, Z. Discovery of Novel Andrographolide Derivatives as Cytotoxic Agents. Bioorg. Med. Chem. Lett. 2013, 23, 4056–4060. [Google Scholar] [CrossRef]
- Suebsasana, S.; Pongnaratorn, P.; Sattayasai, J.; Arkaravichien, T.; Tiamkao, S.; Aromdee, C. Analgesic, Antipyretic, Anti-Inflammatory and Toxic Effects of Andrographolide Derivatives in Experimental Animals. Arch. Pharm. Res. 2009, 32, 1191–1200. [Google Scholar] [CrossRef]
- Liu, G.; Chu, H. Andrographolide Inhibits Proliferation and Induces Cell Cycle Arrest and Apoptosis in Human Melanoma Cells. Oncol. Lett. 2018, 15, 5301–5305. [Google Scholar] [CrossRef]
- Aromdee, C.; Sriubolmas, N.; Wiyakrutta, S.; Suebsasna, S.; Khunkitti, W. Effect of the Derivatives of Andrographolide on the Morphology of Bacillus Subtilis. Arch. Pharm. Res. 2011, 34, 71–77. [Google Scholar] [CrossRef]
- Rajagopal, S.; Kumar, R.A.; Deevi, D.S.; Satyanarayana, C.; Rajagopalan, R. Andrographolide, a Potential Cancer Therapeutic Agent Isolated from Andrographis Paniculata. J. Exp. Ther. Oncol. 2003, 3, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Berman, S.H.; Babish, J.G.; Ma, X.; Shinto, L.; Dorr, M.; Wells, K.; Wenner, C.A.; Standish, L.J. A Phase I Trial of Andrographolide in HIV Positive Patients and Normal Volunteers. Phyther. Res. 2000, 14, 333–338. [Google Scholar] [CrossRef]
- Chen, J.X.; Xue, H.J.; Ye, W.C.; Fang, B.H.; Liu, Y.H.; Yuan, S.H.; Yu, P.; Wang, Y.Q. Activity of Andrographolide and Its Derivatives against Influenza Virus in Vivo and in Vitro. Biol. Pharm. Bull. 2009, 32, 1385–1391. [Google Scholar] [CrossRef]
- Yu, B.C.; Chen, W.C.; Cheng, J.T. Antihyperglycemic Effect of Andrographolide in Streptozotocin-Induced Diabetic Rats. Planta Med. 2003, 69, 1075–1079. [Google Scholar]
- Wang, Z.; Yu, P.; Zhang, G.; Xu, L.; Wang, D.; Wang, L.; Zeng, X.; Wang, Y. Design, Synthesis and Antibacterial Activity of Novel Andrographolide Derivatives. Bioorg. Med. Chem. 2010, 18, 4269–4274. [Google Scholar] [CrossRef]
- Hu, J.; Li, Y.; Xie, X.; Song, Y.; Yan, W.; Luo, Y.; Jiang, Y. The Therapeutic Potential of Andrographolide in Cancer Treatment. Biomed. Pharmacother. 2024, 180, 117438. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Attar, R.; Sabitaliyevich, U.Y.; Alaaeddine, N.; de Sousa, D.P.; Xu, B.; C. Cho, W. The Prowess of Andrographolide as a Natural Weapon in the War against Cancer. Cancers (Basel). 2020, 12. [Google Scholar] [CrossRef]
- Lim, J.C.W.; Chan, T.K.; Ng, D.S.W.; Sagineedu, S.R.; Stanslas, J.; Wong, W.S.F. Andrographolide and Its Analogues: Versatile Bioactive Molecules for Combating Inflammation and Cancer. Clin. Exp. Pharmacol. Physiol. 2012, 39, 300–310. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tripathi, S.; Shukla, A.; Oh, S.H.; Kim, H.M. Andrographolide and Analogues in Cancer Prevention. Front Biosci (Elite Ed) 2015, 7, 255–266. [Google Scholar] [CrossRef]
- Malik, Z.; Parveen, R.; Parveen, B.; Zahiruddin, S.; Khan, M.A.; Khan, A.; Massey, S.; Ahmad, S.; Husain, S.A. Anticancer Potential of Andrographolide from Andrographis Paniculata (Burm. f.) Nees and Its Mechanisms of Action. J. Ethnopharmacol. 2021, 272, 113936. [Google Scholar] [CrossRef]
- Kandanur, S.G.S.; Kundu, S.; Cadena, C.; Juan, H.S.; Bajaj, A.; Guzman, J.D.; Nanduri, S.; Golakoti, N.R. Design, Synthesis, and Biological Evaluation of New 12-Substituted-14-Deoxy-Andrographolide Derivatives as Apoptosis Inducers. Chem. Pap. 2019, 73, 1669–1675. [Google Scholar] [CrossRef]
- Elsheikh, M.A.; Rizk, S.A.; Elnaggar, Y.S.R.; Abdallah, O.Y. Nanoemulsomes for Enhanced Oral Bioavailability of the Anticancer Phytochemical Andrographolide: Characterization and Pharmacokinetics. AAPS PharmSciTech 2021, 22, 1–12. [Google Scholar] [CrossRef]
- Ebenezer, O.; Shapi, M.; Tuszynski, J.A. A Review of the Recent Development in the Synthesis and Biological Evaluations of Pyrazole Derivatives. Biomedicines 2022, 10. [Google Scholar] [CrossRef]
- Ansari, A.; Ali, A.; Asif, M. Biologically Active Pyrazole Derivatives. New J. Chem. 2017, 41, 16–41. [Google Scholar] [CrossRef]
- Bennani, F.E.; Doudach, L.; Cherrah, Y.; Ramli, Y.; Karrouchi, K.; Ansar, M.; Faouzi, M.E.A. Overview of Recent Developments of Pyrazole Derivatives as Anticancer Agents in Different Cell Lines. Bioorg. Chem. 2020, 97, 103470. [Google Scholar] [CrossRef]
- Kumar, H.; Saini, D.; Jain, S.; Jain, N. Pyrazole Scaffold: A Remarkable Tool in the Development of Anticancer Agents. Eur. J. Med. Chem. 2013, 70, 248–258. [Google Scholar] [CrossRef]
- Prabhu, V.V.; Guruvayoorappan, C. Anti-Inflammatory and Anti-Tumor Activity of the Marine Mangrove Rhizophora Apiculata. J. Immunotoxicol. 2012, 9, 341–352. [Google Scholar] [CrossRef]
- Rokkam, S.K.; Bhujel, M.; Jain, D.; Sripada, L.; Nanduri, S.; Bajaj, A.; Golakoti, N.R. Synthesis of Novel Pyrazole Acetals of Andrographolide and Isoandrographolide as Potent Anticancer Agents. RSC Adv. 2024, 14, 26625–26636. [Google Scholar] [CrossRef]
- Robert, M. Silverstein, Francis X. Webster, David J. Kiemle, D.L.B. Spectrometric Identification of Organic Compounds, 8th Edition, 2014. [Google Scholar]
- Pavia, D.L.; Lampman, G.M.; Kriz, G.S. Introduction to Spectroscopy-a Guide for Students of Organic Chemistry; Thomson Learning, Inc., 2001.
- Naasani, I. COMPARE Analysis, a Bioinformatic Approach to Accelerate Drug Repurposing against Covid-19 and Other Emerging Epidemics. SLAS Discov. Adv. Sci. Drug Discov. 2020, 26, 345–351. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Igney, F.H.; Krammer, P.H. Death and Anti-Death: Tumour Resistance to Apoptosis. Nat. Rev. Cancer 2002, 2, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.A. Apoptosis Induced by Anticancer Drugs. Cancer Metastasis Rev. 1992, 11, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The Biochemistry of Apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Klaunig, J.E. Oxidative Stress and Cancer. Curr. Pharm. Des. 2018, 24, 4771–4778. [Google Scholar] [CrossRef]
- Sosa, V.; Moliné, T.; Somoza, R.; Paciucci, R.; Kondoh, H.; LLeonart, M.E. Oxidative Stress and Cancer: An Overview. Ageing Res. Rev. 2013, 12, 376–390. [Google Scholar] [CrossRef]
- Liou, G.Y.; Storz, P. Reactive Oxygen Species in Cancer. Free Radic. Res. 2010, 44, 479–496. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Ganesan, A. The Impact of Natural Products upon Modern Drug Discovery. Curr. Opin. Chem. Biol. 2008, 12, 306–317. [Google Scholar] [CrossRef]
- Kumar Rokkam, S.; Mas-Rosario, J.A.; Joshi, B.P.; Joshi, M.; Choudhury, A.R.; Kar, S.; Golakoti, N.R.; Farkas, M.E. Diarylidene-N-Methyl-4-Piperidones and Spirobibenzopyrans as Antioxidant and Anti-Inflammatory Agents. Chem. Biodivers. 2023, 20. [Google Scholar] [CrossRef]
| Cancer | Sub Panel | GI50 (µM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 1d | 1e | 1f | 1g | 1h | 1i | 1j | ||
| Leukemia | CCRF-CEM | 0.59 | 1.14 | 2.06 | 1.32 | 0.70 | 0.40 | 0.92 | 0.48 | 8.40 | 0.64 |
| HL-60(TB) | 2.32 | 1.73 | 0.76 | 1.38 | 1.17 | 0.64 | 2.67 | 0.67 | 2.17 | 1.08 | |
| K-562 | 2.97 | 2.16 | 2.14 | 2.41 | 2.89 | 2.34 | 3.67 | 2.07 | 3.82 | 1.90 | |
| MOLT-4 | 1.97 | 2.87 | 0.69 | 1.14 | 0.91 | 0.60 | 0.88 | 0.61 | 2.11 | 1.71 | |
| RPMI-8226 | 2.02 | 2.45 | 2.63 | 1.38 | 2.75 | 0.49 | 2.46 | 1.28 | 8.60 | 0.92 | |
| SR | 3.79 | 3.46 | 2.20 | 1.29 | 1.77 | 1.70 | 2.53 | 1.69 | 2.74 | 1.74 | |
| Non-Small Cell Lung | A549/ATCC | 2.49 | 3.62 | 3.88 | 2.82 | 3.07 | 2.14 | 3.57 | 3.10 | 3.09 | 2.39 |
| EKVX | 2.07 | 2.75 | 2.21 | 2.21 | 2.10 | 2.06 | 2.24 | 2.21 | 2.13 | 1.96 | |
| HOP-62 | 1.87 | 3.15 | 1.74 | 1.78 | 2.08 | 1.62 | 2.36 | 1.80 | 2.12 | 2.65 | |
| HOP-92 | 1.53 | 1.86 | 1.53 | 1.56 | 1.75 | 1.29 | 1.50 | 1.38 | 1.43 | 1.47 | |
| NCI-H226 | 1.95 | 2.38 | 1.67 | 1.76 | 1.75 | 1.51 | 1.55 | 1.36 | 1.47 | 1.72 | |
| NCI-H23 | 1.69 | 1.83 | 1.79 | 1.75 | 1.90 | 1.70 | 1.73 | 1.71 | 1.76 | 1.71 | |
| NCI-H322M | 3.35 | 1.76 | 4.22 | 3.26 | 6.06 | 3.34 | 1.02 | 4.20 | 4.92 | 2.94 | |
| NCI-H460 | 1.66 | 2.27 | 1.81 | 1.68 | 1.78 | 1.69 | 1.79 | 1.59 | 1.83 | 1.74 | |
| NCI-H522 | 1.90 | 1.75 | 1.52 | 1.54 | 1.50 | 1.95 | 1.58 | 1.69 | 1.75 | 1.73 | |
| Colon | COLO 205 | 2.01 | 2.41 | 2.06 | 1.98 | 2.09 | 1.48 | 2.30 | 1.70 | 1.97 | 1.95 |
| HCC-2998 | 1.68 | 1.92 | 1.73 | 1.81 | 1.72 | 1.76 | 1.88 | 1.83 | 1.63 | 1.70 | |
| HCT-116 | 1.61 | 1.99 | 3.48 | 1.04 | 1.24 | 0.39 | 1.20 | 0.94 | 3.91 | 1.40 | |
| HCT-15 | 1.72 | 1.74 | 1.57 | 1.58 | 1.60 | 1.61 | 1.59 | 1.60 | 1.59 | 1.58 | |
| HT29 | 2.37 | 1.82 | 1.70 | 1.49 | 1.66 | 2.38 | 1.69 | 2.16 | 1.90 | 1.70 | |
| KM12 | 2.06 | 1.99 | 2.26 | 1.67 | 1.83 | 1.84 | 2.29 | 1.68 | 3.01 | 1.66 | |
| SW-620 | 1.73 | 2.21 | 5.71 | 1.53 | 1.72 | 4.49 | NT | NT | 5.42 | 1.69 | |
| CNS | SF-268 | 3.20 | 7.65 | 2.27 | 1.60 | 2.02 | 2.36 | 2.39 | 2.11 | 2.61 | 1.67 |
| SF-295 | 1.64 | 1.68 | 1.52 | 1.42 | 1.36 | 1.55 | 1.50 | 1.54 | 1.58 | 1.43 | |
| SF-539 | 1.84 | 1.74 | 1.77 | 1.63 | 1.78 | 1.77 | 1.72 | 1.76 | 1.79 | 1.69 | |
| SNB-19 | 1.85 | 3.64 | NT | 2.24 | 2.17 | 1.44 | 1.40 | 1.47 | 1.36 | 2.46 | |
| SNB-75 | 1.36 | 7.34 | NT | 1.12 | 1.57 | 1.66 | 1.69 | 1.51 | 1.71 | 1.15 | |
| U251 | 1.84 | 1.74 | 1.81 | 1.59 | 1.69 | 2.36 | 2.39 | 2.11 | 2.61 | 1.66 | |
| Melanoma | LOX IMVI | 1.54 | 1.72 | 1.44 | 1.61 | 1.64 | 1.32 | 1.58 | 1.55 | 1.43 | 1.57 |
| MALME-3M | 1.73 | 1.78 | 1.87 | 1.47 | 1.87 | 1.58 | 1.82 | 1.60 | 2.46 | 1.54 | |
| M14 | 1.81 | 2.08 | 1.75 | 1.69 | 1.86 | 1.50 | 1.84 | 1.71 | 1.76 | 1.72 | |
| MDA-MB-435 | 1.86 | 3.03 | 1.50 | 1.56 | 1.72 | 1.68 | 1.65 | 1.67 | 1.61 | 1.54 | |
| SK-MEL-2 | 1.74 | 1.78 | 1.79 | 1.57 | 1.78 | 1.69 | 1.77 | 1.76 | 1.79 | 1.57 | |
| SK-MEL-28 | 1.77 | 2.04 | 1.77 | 1.80 | 1.73 | 1.66 | 1.78 | 1.73 | 1.74 | 1.94 | |
| SK-MEL-5 | 1.40 | 1.63 | 1.65 | 1.63 | 1.54 | 1.62 | 1.70 | 1.60 | 1.59 | 1.54 | |
| UACC-257 | 1.90 | 1.80 | 1.93 | 1.77 | 1.77 | 1.65 | 1.80 | 1.70 | 1.74 | 1.64 | |
| UACC-62 | 1.65 | 1.69 | 1.65 | 1.71 | 1.68 | 1.58 | 1.68 | 1.58 | 1.64 | 1.91 | |
| Ovarian | IGROV1 | 1.77 | 2.08 | 2.07 | 1.39 | 2.20 | 1.84 | 4.29 | 1.67 | 3.95 | 1.37 |
| OVCAR-3 | 1.82 | 1.87 | 1.95 | 1.60 | 1.69 | 2.02 | 1.78 | 1.52 | 1.95 | 1.50 | |
| OVCAR-4 | 2.31 | 4.05 | 1.74 | 2.09 | 2.49 | 1.75 | 2.03 | 1.89 | 1.92 | 1.72 | |
| OVCAR-5 | 1.86 | 2.06 | 1.92 | 1.78 | 1.85 | 1.74 | 1.98 | 1.93 | 1.93 | 1.86 | |
| OVCAR-8 | 1.81 | 1.90 | 1.77 | 1.53 | 1.76 | 1.48 | 1.95 | 1.55 | 1.67 | 1.35 | |
| NCI/ADR-RES | 4.75 | 2.98 | 1.77 | 1.88 | 2.15 | 1.54 | 1.83 | 1.83 | 1.65 | 1.94 | |
| SK-OV-3 | 1.77 | 2.08 | 2.70 | 5.86 | 4.16 | 1.88 | 3.04 | 2.03 | 3.47 | 3.44 | |
| Renal | 786-0 | 2.03 | 3.04 | 1.78 | 2.31 | 2.59 | 1.60 | 1.99 | 2.07 | 1.77 | 2.30 |
| A498 | 2.34 | 1.90 | 4.13 | 2.90 | 4.33 | 3.31 | 5.20 | 4.35 | 4.03 | 2.63 | |
| ACHN | 1.64 | 1.89 | 1.80 | 1.64 | 1.61 | 1.66 | 1.89 | 2.07 | 1.83 | 1.52 | |
| CAKI-1 | 1.86 | 2.81 | 1.56 | 1.32 | 1.42 | 1.58 | 1.63 | 1.59 | 1.71 | 1.26 | |
| RXF 393 | 1.40 | 1.62 | 1.64 | 1.05 | 1.18 | 1.60 | 1.79 | 1.71 | 1.49 | 0.89 | |
| SN12C | 1.87 | 2.93 | 1.59 | 1.65 | 1.57 | 1.55 | 1.67 | 1.51 | 1.66 | 1.98 | |
| TK-10 | 1.78 | 2.22 | 1.41 | 2.14 | 2.12 | 1.27 | 1.33 | 1.19 | 1.33 | 1.95 | |
| UO-31 | 1.35 | 1.82 | 1.78 | 1.30 | 1.45 | 1.60 | 1.99 | 2.07 | 1.77 | 1.22 | |
| Prostate | PC-3 | 1.95 | 2.75 | 1.97 | 2.07 | 2.29 | 1.55 | 2.07 | 2.03 | 1.92 | 2.18 |
| DU-145 | 2.05 | 4.11 | 1.97 | 1.88 | 2.12 | 1.82 | 2.05 | 1.77 | 2.05 | 2.04 | |
| Breast | MCF7 | 2.00 | 2.64 | 1.63 | 1.48 | 1.77 | 1.43 | 1.51 | 1.49 | 1.57 | 1.46 |
| MDA-MB-231 | 1.72 | 1.84 | 1.72 | 1.49 | 1.68 | 1.68 | 1.68 | 1.59 | 1.70 | 1.47 | |
| HS 578T | 2.31 | 2.69 | 1.66 | 1.91 | 1.94 | 1.51 | 1.75 | 1.52 | 1.85 | 1.86 | |
| BT-549 | 1.64 | 1.94 | 1.30 | 1.57 | 1.46 | 1.31 | 1.49 | 1.47 | 1.20 | 1.65 | |
| T-47D | 1.42 | 1.76 | 1.39 | 1.63 | 2.29 | 1.19 | 1.31 | 1.18 | 1.14 | 1.80 | |
| MDA-MB-468 | 1.69 | 1.76 | 1.63 | 1.74 | 1.82 | 1.53 | 2.00 | 1.93 | 1.85 | 1.71 | |
| Compound | Pearson Correlation Coefficient (pcc) | Marketed drug | Mechanism of action | |
|---|---|---|---|---|
| 1f | GI50 | 0.51 | Nelarabine | Inhibits DNA elongation, apoptosis, and cellular destruction. |
| 1h | GI50 | 0.60 | Melphalan | Inhibits DNA synthesis or transcription. |
| 1j |
GI50 |
0.50 |
Trisenox 3 |
DNA fragmentation characteristic of apoptosis in NB4 human promyelocytic leukemia cells. |
| Time | G1 (%) | S (%) | G2 (%) |
|---|---|---|---|
| 0 hr | 74.96 | 2.62 | 22.41 |
| 24 hr (3.33 µM) | 83.53 | 3.26 | 13.19 |
| 24 hr (5.00 µM) | 80.50 | 8.29 | 11.2 |
| 24 hr (6.66 µM) | 82.43 | 8.64 | 8.91 |
| Compound | *MW | LogP | HBD | HBA | TPSA |
|---|---|---|---|---|---|
| Ia | 580.29 | 5.69 | 1 | 7 | 82.81 |
| Ib | 625.28 | 5.55 | 1 | 10 | 125.95 |
| Ic | 598.28 | 5.87 | 1 | 7 | 82.81 |
| Id | 614.25 | 6.28 | 1 | 7 | 82.81 |
| Ie | 658.20 | 6.38 | 1 | 7 | 82.81 |
| If | 598.28 | 6.35 | 1 | 7 | 78.88 |
| Ig | 614.25 | 6.27 | 1 | 7 | 82.81 |
| Ih | 658.20 | 6.38 | 1 | 7 | 82.81 |
| Ii | 594.31 | 6.09 | 1 | 7 | 82.81 |
| Ij | 610.30 | 5.71 | 1 | 8 | 92.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





