Submitted:
13 May 2025
Posted:
15 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
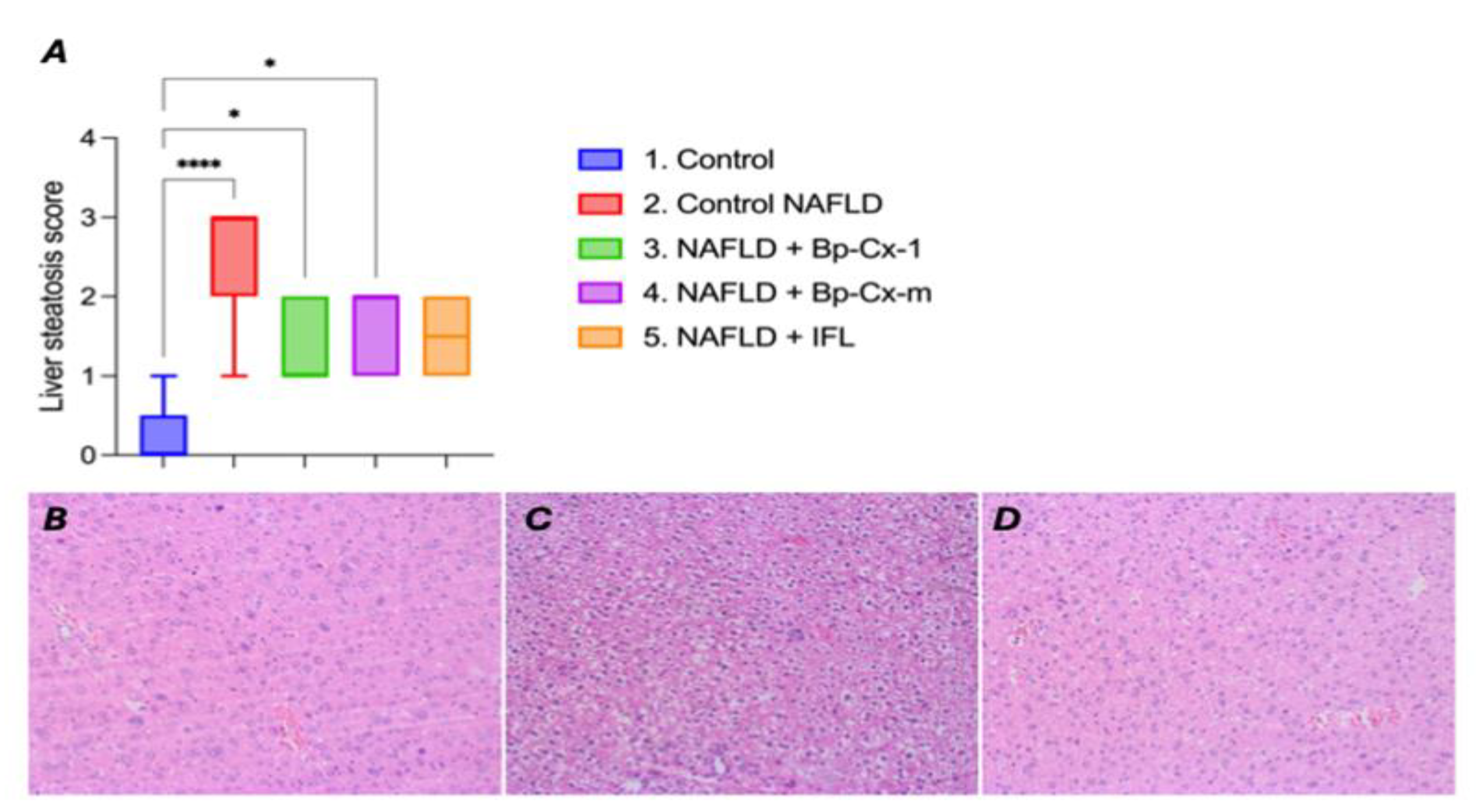
2.1. Hepatoprotective Effect In Vivo
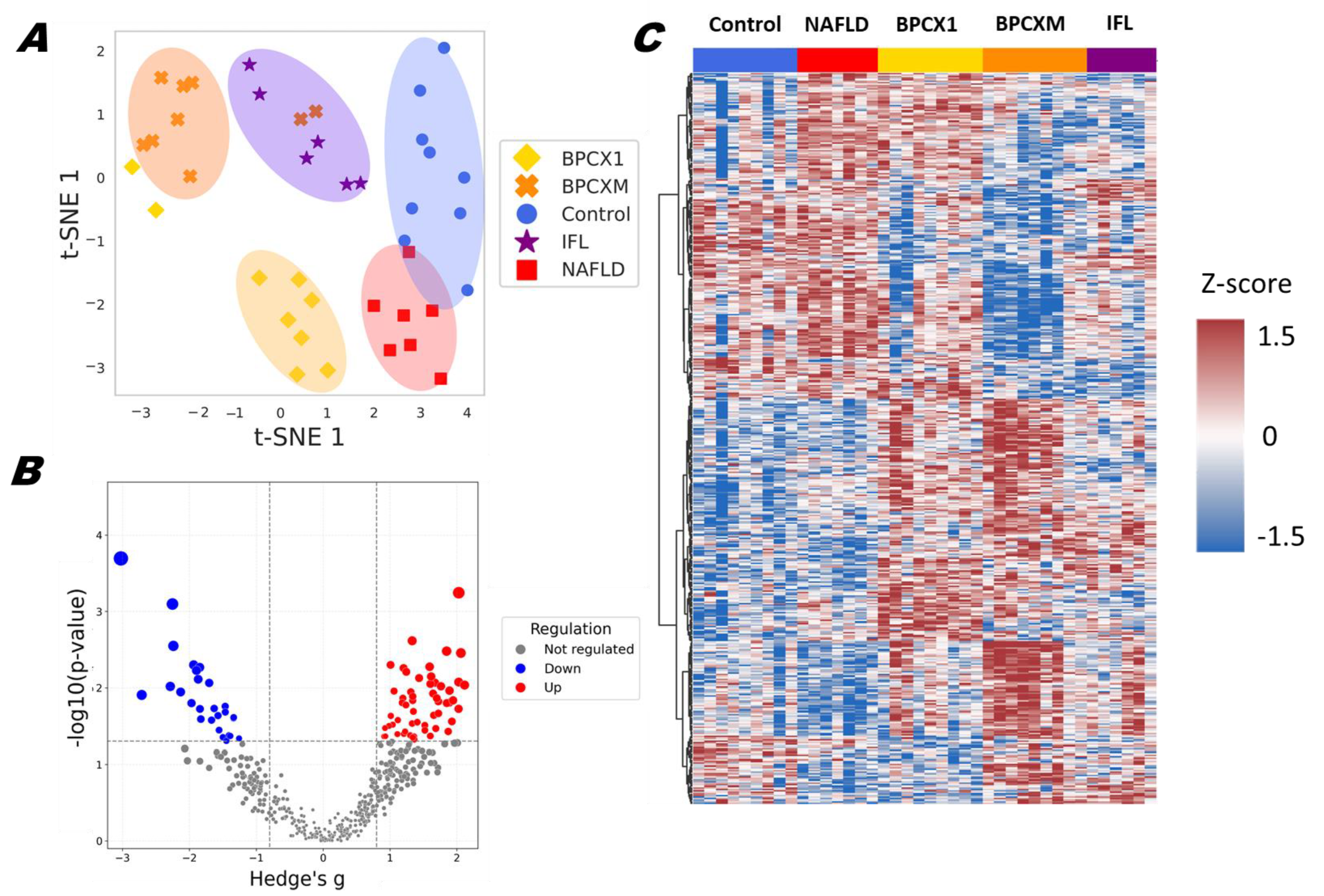
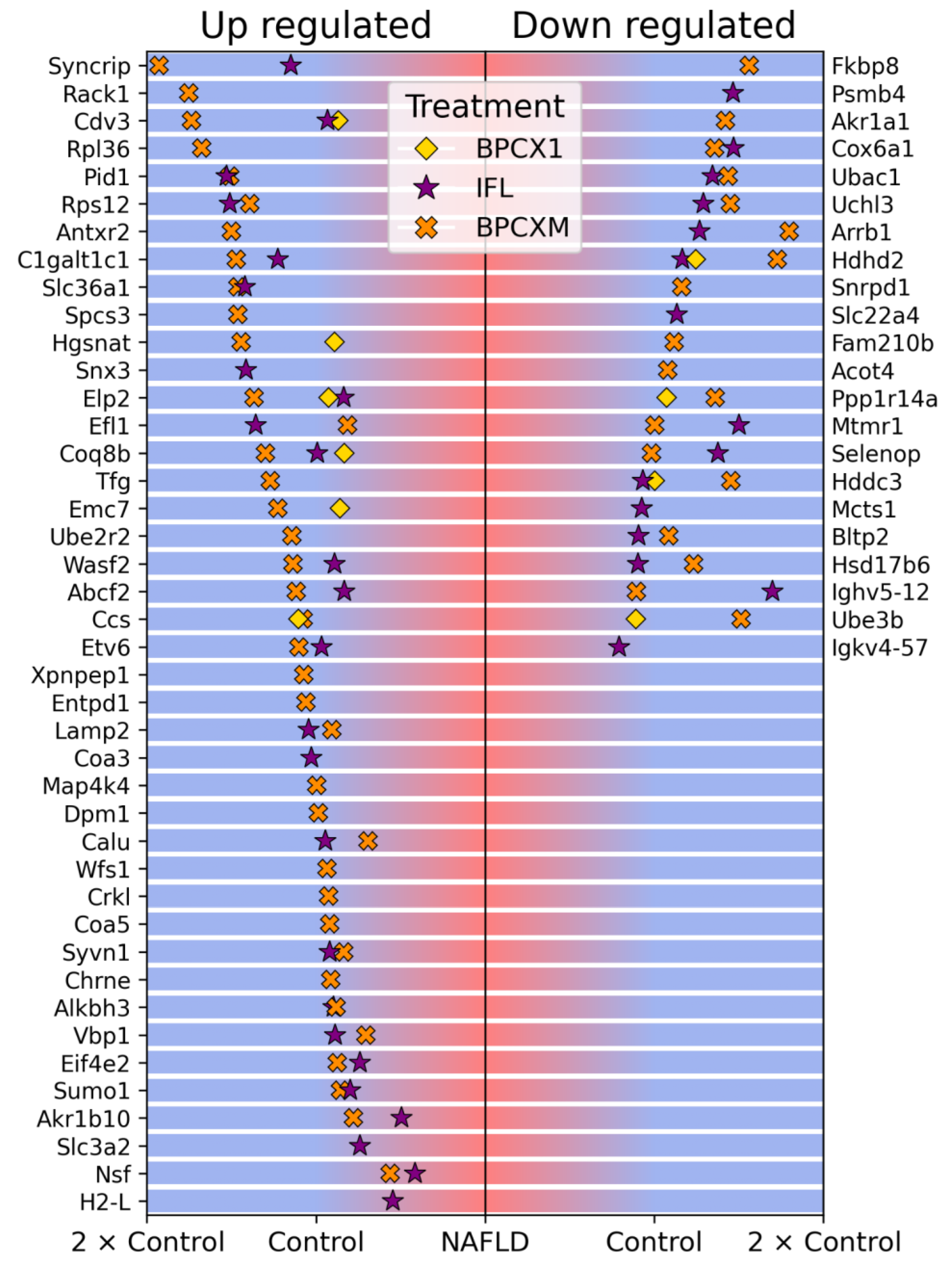
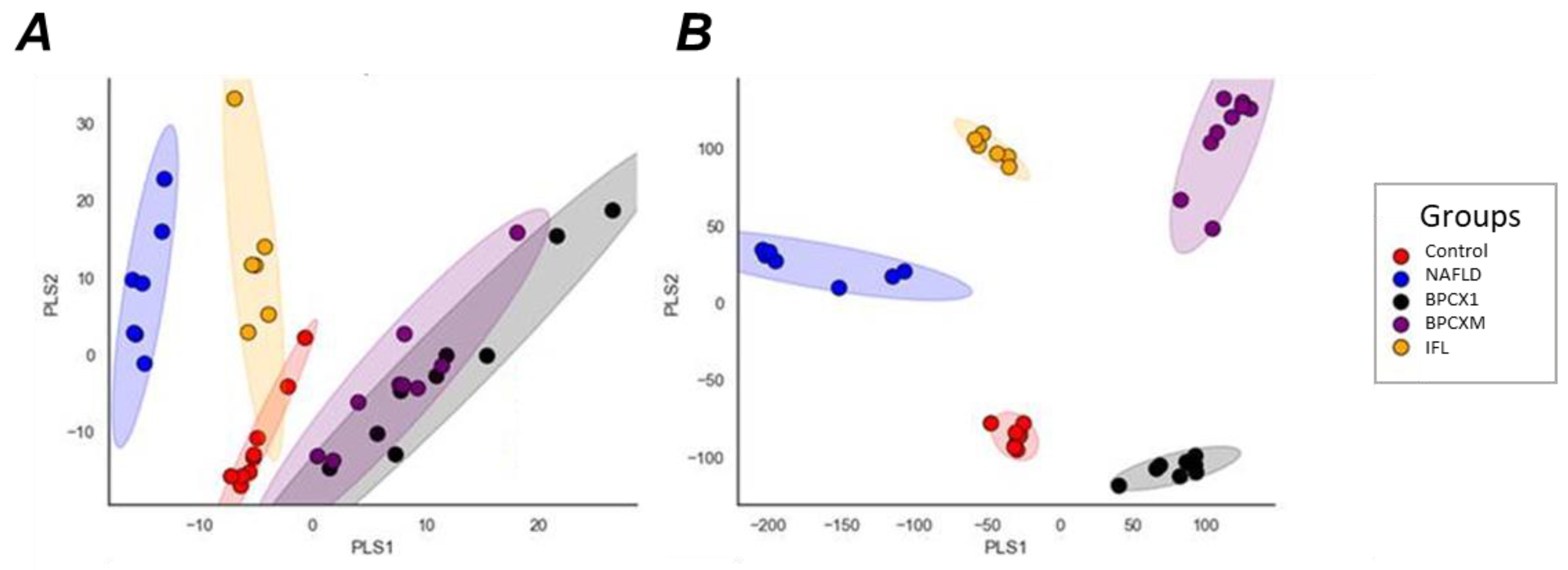
2.2. Proteomic Analysis
2.3. Results of Molecular Fingerprinting by FT-ICR MS
2.4. Evaluation of Potential Antigenotoxic Action of Nature-Derived Polyphenols In Vivo Study
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animal Studies
4.2.1. Experimental Design
4.2.2. DNA Comet Assay
4.3. Proteomic Analysis
4.3.1. Liver Samples Preparation
4.3.2. LC-MS/MS Analysis
4.3.3. Data Analysis
4.4. Molecular Fingerprinting by FTICR
4.4.1. Extraction of Metabolites
4.4.2. FTICR MS Analysis
4.4.3. Data Analysis
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- de Alwis, N.M.W.; Day, C.P. Non-Alcoholic Fatty Liver Disease: The Mist Gradually Clears. J. Hepatol. 2008, 48 Suppl 1, S104-12. [CrossRef]
- Yang, J.; Tian, C.; Liu, M.; Guo, H.; Lin, F.; Ding, Y.; Yao, W.; Zhang, J.; Fan, J.; Yu, C.; et al. Genetic Risk, BMI Status, BMI Change Patterns, and the Risk of Steatotic Liver Disease and Liver Enzyme Elevation in Chinese Adults. Nutrients 2024, 16. [CrossRef]
- Tilg, H.; Effenberger, M. From NAFLD to MAFLD: When Pathophysiology Succeeds. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 387–388. [CrossRef]
- Eslam, M.; Newsome, P.N.; Sarin, S.K.; Anstee, Q.M.; Targher, G.; Romero-Gomez, M.; Zelber-Sagi, S.; Wai-Sun Wong, V.; Dufour, J.-F.; Schattenberg, J.M.; et al. A New Definition for Metabolic Dysfunction-Associated Fatty Liver Disease: An International Expert Consensus Statement. J. Hepatol. 2020, 73, 202–209. [CrossRef]
- Marchesini, G.; Bugianesi, E.; Forlani, G.; Cerrelli, F.; Lenzi, M.; Manini, R.; Natale, S.; Vanni, E.; Villanova, N.; Melchionda, N.; et al. Nonalcoholic Fatty Liver, Steatohepatitis, and the Metabolic Syndrome. Hepatology 2003, 37, 917–923. [CrossRef]
- Ando, Y.; Jou, J.H. Nonalcoholic Fatty Liver Disease and Recent Guideline Updates. Clin. liver Dis. 2021, 17, 23–28. [CrossRef]
- Vernon, G.; Baranova, A.; Younossi, Z.M. Systematic Review: The Epidemiology and Natural History of Non-Alcoholic Fatty Liver Disease and Non-Alcoholic Steatohepatitis in Adults. Aliment. Pharmacol. Ther. 2011, 34, 274–285. [CrossRef]
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R. The Economic and Clinical Burden of Nonalcoholic Fatty Liver Disease in the United States and Europe. Hepatology 2016, 64, 1577–1586. [CrossRef]
- Powell, E.E.; Wong, V.W.-S.; Rinella, M. Non-Alcoholic Fatty Liver Disease. Lancet (London, England) 2021, 397, 2212–2224. [CrossRef]
- Hutchison, A.L.; Tavaglione, F.; Romeo, S.; Charlton, M. Endocrine Aspects of Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD): Beyond Insulin Resistance. J. Hepatol. 2023, 79, 1524–1541. [CrossRef]
- Younossi, Z.M.; Stepanova, M.; Afendy, M.; Fang, Y.; Younossi, Y.; Mir, H.; Srishord, M. Changes in the Prevalence of the Most Common Causes of Chronic Liver Diseases in the United States from 1988 to 2008. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2011, 9, 524-530.e1; quiz e60. [CrossRef]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Charlton, M.; Cusi, K.; Rinella, M.; Harrison, S.A.; Brunt, E.M.; Sanyal, A.J. The Diagnosis and Management of Nonalcoholic Fatty Liver Disease: Practice Guidance from the American Association for the Study of Liver Diseases. Hepatology 2018, 67, 328–357. [CrossRef]
- Martinou, E.; Pericleous, M.; Stefanova, I.; Kaur, V.; Angelidi, A.M. Diagnostic Modalities of Non-Alcoholic Fatty Liver Disease: From Biochemical Biomarkers to Multi-Omics Non-Invasive Approaches. Diagnostics (Basel, Switzerland) 2022, 12. [CrossRef]
- EASL-EASD-EASO Clinical Practice Guidelines for the Management of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2016, 64, 1388–1402. [CrossRef]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The Multiple-Hit Pathogenesis of Non-Alcoholic Fatty Liver Disease (NAFLD). Metabolism. 2016, 65, 1038–1048. [CrossRef]
- Cusi, K. Role of Insulin Resistance and Lipotoxicity in Non-Alcoholic Steatohepatitis. Clin. Liver Dis. 2009, 13, 545–563. [CrossRef]
- Manne, V.; Handa, P.; Kowdley, K. V Pathophysiology of Nonalcoholic Fatty Liver Disease/Nonalcoholic Steatohepatitis. Clin. Liver Dis. 2018, 22, 23–37. [CrossRef]
- Després, J.-P. Body Fat Distribution and Risk of Cardiovascular Disease: An Update. Circulation 2012, 126, 1301–1313. [CrossRef]
- Rabot, S.; Membrez, M.; Bruneau, A.; Gérard, P.; Harach, T.; Moser, M.; Raymond, F.; Mansourian, R.; Chou, C.J. Germ-Free C57BL/6J Mice Are Resistant to High-Fat-Diet-Induced Insulin Resistance and Have Altered Cholesterol Metabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 4948–4959. [CrossRef]
- Le Roy, T.; Llopis, M.; Lepage, P.; Bruneau, A.; Rabot, S.; Bevilacqua, C.; Martin, P.; Philippe, C.; Walker, F.; Bado, A.; et al. Intestinal Microbiota Determines Development of Non-Alcoholic Fatty Liver Disease in Mice. Gut 2013, 62, 1787–1794. [CrossRef]
- Velázquez, K.T.; Enos, R.T.; Bader, J.E.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; Carson, J.A.; Nagarkatti, P.S.; Nagarkatti, M.; Murphy, E.A. Prolonged High-Fat-Diet Feeding Promotes Non-Alcoholic Fatty Liver Disease and Alters Gut Microbiota in Mice. World J. Hepatol. 2019, 11, 619–637. [CrossRef]
- Hui, S.T.; Parks, B.W.; Org, E.; Norheim, F.; Che, N.; Pan, C.; Castellani, L.W.; Charugundla, S.; Dirks, D.L.; Psychogios, N.; et al. The Genetic Architecture of NAFLD among Inbred Strains of Mice. Elife 2015, 4, e05607. [CrossRef]
- Chella Krishnan, K.; Kurt, Z.; Barrere-Cain, R.; Sabir, S.; Das, A.; Floyd, R.; Vergnes, L.; Zhao, Y.; Che, N.; Charugundla, S.; et al. Integration of Multi-Omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-Alcoholic Fatty Liver Disease. Cell Syst. 2018, 6, 103-115.e7. [CrossRef]
- Ritze, Y.; Bárdos, G.; Claus, A.; Ehrmann, V.; Bergheim, I.; Schwiertz, A.; Bischoff, S.C. Lactobacillus Rhamnosus GG Protects against Non-Alcoholic Fatty Liver Disease in Mice. PLoS One 2014, 9, e80169. [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L. V; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15718–15723. [CrossRef]
- Chen, D.; Wang, Y.; Yang, J.; Ou, W.; Lin, G.; Zeng, Z.; Lu, X.; Chen, Z.; Zou, L.; Tian, Y.; et al. Shenling Baizhu San Ameliorates Non-Alcoholic Fatty Liver Disease in Mice by Modulating Gut Microbiota and Metabolites. Front. Pharmacol. 2024, 15, 1343755. [CrossRef]
- Kim, H.-J.; Jeon, H.-J.; Kim, D.-G.; Kim, J.-Y.; Shim, J.-J.; Lee, J.-H. Lacticaseibacillus Paracsei HY7207 Alleviates Hepatic Steatosis, Inflammation, and Liver Fibrosis in Mice with Non-Alcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2024, 25. [CrossRef]
- Wei, Y.; Pan, T.; Zhao, Y.; Chen, Z.; Wu, L.; Fang, S.; Wang, X.; Wang, X.; Chen, D.; Chen, Y. Nicotine Aggravates High-Fat Diet-Induced Non-Alcoholic Fatty Liver Disease in Mice via Inhibition of CISD3. Int. Immunopharmacol. 2024, 142, 113067. [CrossRef]
- Kim, D.-G.; Krenz, A.; Toussaint, L.E.; Maurer, K.J.; Robinson, S.-A.; Yan, A.; Torres, L.; Bynoe, M.S. Non-Alcoholic Fatty Liver Disease Induces Signs of Alzheimer’s Disease (AD) in Wild-Type Mice and Accelerates Pathological Signs of AD in an AD Model. J. Neuroinflammation 2016, 13, 1. [CrossRef]
- Pan, X.; Wang, P.; Luo, J.; Wang, Z.; Song, Y.; Ye, J.; Hou, X. Adipogenic Changes of Hepatocytes in a High-Fat Diet-Induced Fatty Liver Mice Model and Non-Alcoholic Fatty Liver Disease Patients. Endocrine 2015, 48, 834–847. [CrossRef]
- Wang, W.; Zhao, J.; Gui, W.; Sun, D.; Dai, H.; Xiao, L.; Chu, H.; Du, F.; Zhu, Q.; Schnabl, B.; et al. Tauroursodeoxycholic Acid Inhibits Intestinal Inflammation and Barrier Disruption in Mice with Non-Alcoholic Fatty Liver Disease. Br. J. Pharmacol. 2018, 175, 469–484. [CrossRef]
- Zheng, S.; Hoos, L.; Cook, J.; Tetzloff, G.; Davis, H.J.; van Heek, M.; Hwa, J.J. Ezetimibe Improves High Fat and Cholesterol Diet-Induced Non-Alcoholic Fatty Liver Disease in Mice. Eur. J. Pharmacol. 2008, 584, 118–124. [CrossRef]
- Lou, D.; Fang, Q.; He, Y.; Ma, R.; Wang, X.; Li, H.; Qi, M. Oxymatrine Alleviates High-Fat Diet/Streptozotocin-Induced Non-Alcoholic Fatty Liver Disease in C57BL/6 J Mice by Modulating Oxidative Stress, Inflammation and Fibrosis. Biomed. Pharmacother. 2024, 174, 116491. [CrossRef]
- Tang, N.; Ji, L.; Shi, X.; Xiong, Y.; Xiong, X.; Zhao, H.; Song, H.; Wang, J.; Zhang, L.; You, S.; et al. Effects of Ganjianglingzhu Decoction on Lean Non-Alcoholic Fatty Liver Disease in Mice Based on Untargeted Metabolomics. Pharmaceuticals (Basel). 2024, 17. [CrossRef]
- Wu, Y.; Yin, W.; Hao, P.; Chen, Y.; Yu, L.; Yu, X.; Wu, Y.; Li, X.; Wang, W.; Zhou, H.; et al. Polysaccharide from Panax Japonicus C.A. Mey Prevents Non-Alcoholic Fatty Liver Disease Development Based on Regulating Liver Metabolism and Gut Microbiota in Mice. Int. J. Biol. Macromol. 2024, 260, 129430. [CrossRef]
- Ghani, I.; An, Y.; Qiao, Q.; He, S.; Li, Z. Polyphenols from Foxtail Millet Improve Non-Alcoholic Fatty Liver Disease by Regulating Intestinal Microbiome in Mice. Foods (Basel, Switzerland) 2024, 13. [CrossRef]
- Brzhozovskiy, A.G.; Semenov, S.D.; Zherebker, A.Y.; Bugrova, A.E.; Yurova, M.N.; Zhernov, Y. V; Kovaleva, O.A.; Semenov, A.L.; Abroskin, D.P.; Kruglov, S.S.; et al. Hepatoprotective Activity of Nature-Derived Polyphenols Studied by Mass Spectrometry Based Multi-OMICS Approach. Int. J. Mol. Sci. 2025, 26. [CrossRef]
- Sarycheva, A.; Perminova, I. V; Nikolaev, E.N.; Zherebker, A. Formulae Differences Commence a Database for Interlaboratory Studies of Natural Organic Matter. Environ. Sci. Technol. 2023, 57, 6238–6247. [CrossRef]
- Zherebker, A.; Babcock, O.; Pereira, D.L.; D’Aronco, S.; Filippi, D.; Soldà, L.; Michoud, V.; Gratien, A.; Cirtog, M.; Cantrell, C.; et al. Decreasing the Uncertainty in the Comparison of Molecular Fingerprints of Organic Aerosols with H/D Exchange Mass Spectrometry. Environ. Sci. Technol. 2024, 58, 20468–20479. [CrossRef]
- Zhou, B.; Luo, Y.; Bi, H.; Zhang, N.; Ma, M.; Dong, Z.; Ji, N.; Zhang, S.; Wang, X.; Liu, Y.; et al. Amelioration of Nonalcoholic Fatty Liver Disease by Inhibiting the Deubiquitylating Enzyme RPN11. Cell Metab. 2024, 36, 2228-2244.e7. [CrossRef]
- Baek, J.-H.; Kim, M.S.; Jung, H.R.; Hwang, M.-S.; Lee, C.-H.; Han, D.H.; Lee, Y.-H.; Yi, E.C.; Im, S.-S.; Hwang, I.; et al. Ablation of the Deubiquitinase USP15 Ameliorates Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Exp. Mol. Med. 2023, 55, 1520–1530. [CrossRef]
- Kanayama, H.O.; Tamura, T.; Ugai, S.; Kagawa, S.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Demonstration That a Human 26S Proteolytic Complex Consists of a Proteasome and Multiple Associated Protein Components and Hydrolyzes ATP and Ubiquitin-Ligated Proteins by Closely Linked Mechanisms. Eur. J. Biochem. 1992, 206, 567–578. [CrossRef]
- Sano, Y.; Furuta, A.; Setsuie, R.; Kikuchi, H.; Wang, Y.-L.; Sakurai, M.; Kwon, J.; Noda, M.; Wada, K. Photoreceptor Cell Apoptosis in the Retinal Degeneration of Uchl3-Deficient Mice. Am. J. Pathol. 2006, 169, 132–141. [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Superoxide Anion Chemistry-Its Role at the Core of the Innate Immunity. Int. J. Mol. Sci. 2023, 24. [CrossRef]
- Esposito, L.A.; Kokoszka, J.E.; Waymire, K.G.; Cottrell, B.; MacGregor, G.R.; Wallace, D.C. Mitochondrial Oxidative Stress in Mice Lacking the Glutathione Peroxidase-1 Gene. Free Radic. Biol. Med. 2000, 28, 754–766. [CrossRef]
- Esworthy, R.S.; Chu, F.F.; Paxton, R.J.; Akman, S.; Doroshow, J.H. Characterization and Partial Amino Acid Sequence of Human Plasma Glutathione Peroxidase. Arch. Biochem. Biophys. 1991, 286, 330–336. [CrossRef]
- Sun, C.; Guo, Y.; Cong, P.; Tian, Y.; Gao, X. Liver Lipidomics Analysis Revealed the Novel Ameliorative Mechanisms of L-Carnitine on High-Fat Diet-Induced NAFLD Mice. Nutrients 2023, 15. [CrossRef]
- Semenov, A.L.; Gubareva, E.A.; Ermakova, E.D.; Dorofeeva, A.A.; Tumanyan, I.A.; Radetskaya, E.A.; Yurova, M.N.; Aboushanab, S.A.; Kanwugu, O.N.; Fedoros, E.I.; et al. Astaxantin and Isoflavones Inhibit Benign Prostatic Hyperplasia in Rats by Reducing Oxidative Stress and Normalizing Ca/Mg Balance. Plants (Basel, Switzerland) 2021, 10. [CrossRef]
- Collins, A.; Møller, P.; Gajski, G.; Vodenková, S.; Abdulwahed, A.; Anderson, D.; Bankoglu, E.E.; Bonassi, S.; Boutet-Robinet, E.; Brunborg, G.; et al. Measuring DNA Modifications with the Comet Assay: A Compendium of Protocols. Nat. Protoc. 2023, 18, 929–989. [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural Networks and Interference Correction Enable Deep Proteome Coverage in High Throughput. Nat. Methods 2020, 17, 41–44. [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [CrossRef]
- McKinney, W. Data Structures for Statistical Computing in Python. Proc. 9th Python Sci. Conf. 2010, 1, 56–61. [CrossRef]
- Friedjungová, M.; Vašata, D.; Balatsko, M.; Jiřina, M. Missing Features Reconstruction Using a Wasserstein Generative Adversarial Imputation Network. Comput. Sci. – ICCS 2020 20th Int. Conf. Amsterdam, Netherlands, June 3–5, 2020, Proceedings, Part IV 2020, 12140, 225–239.
- Tonoyan, N.M.; Chagovets, V. V; Starodubtseva, N.L.; Tokareva, A.O.; Chingin, K.; Kozachenko, I.F.; Adamyan, L. V; Frankevich, V.E. Alterations in Lipid Profile upon Uterine Fibroids and Its Recurrence. Sci. Rep. 2021, 11, 11447. [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open Source Software for Rapid Proteomics Tools Development. Bioinformatics 2008, 24, 2534–2536. [CrossRef]
- Goloborodko, A.A.; Levitsky, L.I.; Ivanov, M. V; Gorshkov, M. V Pyteomics--a Python Framework for Exploratory Data Analysis and Rapid Software Prototyping in Proteomics. J. Am. Soc. Mass Spectrom. 2013, 24, 301–304. [CrossRef]
- Volikov, A.; Rukhovich, G.; Perminova, I. V NOMspectra: An Open-Source Python Package for Processing High Resolution Mass Spectrometry Data on Natural Organic Matter. J. Am. Soc. Mass Spectrom. 2023, 34, 1524–1527. [CrossRef]
- Zielinski, A.T.; Kourtchev, I.; Bortolini, C.; Fuller, S.J.; Giorio, C.; Popoola, O.A.M.; Bogialli, S.; Tapparo, A.; Jones, R.L.; Kalberer, M. A New Processing Scheme for Ultra-High Resolution Direct Infusion Mass Spectrometry Data. Atmos. Environ. 2018, 178, 129–139. [CrossRef]
- Kozhinov, A.N.; Zhurov, K.O.; Tsybin, Y.O. Iterative Method for Mass Spectra Recalibration via Empirical Estimation of the Mass Calibration Function for Fourier Transform Mass Spectrometry-Based Petroleomics. Anal. Chem. 2013, 85, 6437–6445. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




