1. Introduction
The Adenoviridae family comprises a diverse group of viruses capable of infecting various vertebrates, including mammals, birds, fish, and reptiles [
1]. According to the 2023 International Committee on Taxonomy of Viruses (ICTV) classification, this family is divided into six genera:
Aviadenovirus,
Barthadenovirus (formerly
Atadenovirus),
Ichtadenovirus,
Mastadenovirus,
Siadenovirus, and
Testadenovirus. Among these,
Aviadenovirus,
Barthadenovirus, and
Siadenovirus are known to infect avian species, including chickens, turkeys, ducks, and geese.
Duck adenoviruses (DAdVs) are classified into two species: DAdV-A (also termed DAdV-1) and DAdV-B (comprising DAdV-2 to DAdV-6, with DAdV-6 being a proposed classification). While DAdV-A typically causes mild or asymptomatic infections in ducks, it can cross species barriers, infecting chickens and leading to Egg Drop Syndrome (EDS). In contrast, DAdV-B includes strains such as DAdV-2, DAdV-3, DAdV-4, and DAdV-5 (previously misclassified as fowl adenovirus 5), which share phylogenetic similarities with goose adenovirus 4 (GoAdV-4) (ICTV Global Report). Notably, DAdV-2 and DAdV-3, both isolated from Muscovy ducks, exhibit distinct genetic and pathogenic profiles. DAdV-2 possesses a single fiber gene and induces only mild symptoms, whereas DAdV-3 carries two fiber genes (fiber1 and fiber2) and demonstrates heightened virulence, causing severe hepatic and renal damage, including liver enlargement, hemorrhagic mottling, and kidney swelling [
2]. Although DAdV-4 has been identified in recent years, it has not been associated with significant disease outbreaks [
3].
Since its initial detection in 2014 [
4], DAdV-3 has emerged as a major threat to Muscovy duck farms across China, including provinces such as Guangdong, Fujian, Zhejiang, and Anhui [
5,
6,
7]. Outbreaks are characterized by high morbidity (40–55%) and mortality (35–43%) [
8], leading to substantial economic losses in the duck industry. The pathogenic mechanisms underlying DAdV-3’s heightened virulence remain unclear, necessitating further investigation.
Reverse genetics systems of Fowl adenoviruses have proven invaluable in studying viral pathogenesis, gene function, and host interactions. Successful applications in fowl adenoviruses—such as FAdV-1 (CELO strain) [
9], DAdV-3 [
10], FAdV-4 [
11,
12,
13], FAdV-8 [
14], FAdV-9 (A-2A strain) [
15]—have enabled genome manipulation either through direct in vitro modification or via infectious clones (e.g., FAdmids) generated by homologous recombination in
Escherichia coli.
Here, a DAdV-3 strain SD2019 was isolated and identified from the Muscovy duck farm, and we developed an reverse genetics system for DAdV-3 and the rescued virus was characterized in Muscovy ducks.
2. Materials and Methods
2.1. Cells, Plasmids and Reagents
Leghom male hepatocellular (LMH) cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco's Modified Eagle Medium: F12 (DMEM/F12) (Hyclone, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS) (Biowest, South America origin, Riverside, MO, USA), 100 U/ml of penicillin and 100 μg/ml of streptomycin (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China) at 37℃ in a 5% CO2 humidified incubator. DH5α competent cells were purchased from Shanghai Weidi Biotechnology Co., Ltd. The plasmid PET28a (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China) was engineered to include the kanamycin resistance (Kana) gene. The DAdV-3 monoclonal antibody 2F12 was generated and stored in our laboratory. The FITC-labeled sheep anti-mouse IgG antibodies (Jackson, Pennsylvania, USA), purchased from Shanghai Nonin Biological Technology Co., Ltd. The DAdV-3 SD2019 stran was isolated from livers of sick ducks in Shandong province and was passaged three times on the LMH cells by limited dilution method, aliquoted and stored at -80℃.
2.2. (. RT)-PCR Detection for Common Duck Viruses
The total RNAs and DNAs of livers and kidneys of sick Muscovy ducks were extracted using the viral RNA/DNA purification kit (Tiangen Biotec (Beijing) Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The RNAs were then reverse-transcribed into cDNAs following the manufacturer’s protocol (Tiangen, Beijing, China). To determine whether the samples were positive for common duck viruses, including duck parvovirus (DPV), goose parvovirus (GPV), duck enteritis virus (DEV), fowl adenovirus serotype 4 (FAdV-4), duck adenovirus serotype 3 (DAdV-3), duck circovirus (DuCV), avian influenza virus (AIV), duck Tembusu virus (DTMUV), duck hepatitis A virus type 1 (DHAV-1), duck hepatitis A virus type 3 (DHAV-3), Newcastle disease virus (NDV) and Novel duck reovirus (NDRV), the DNAs and cDNAs were used as templates for PCR test (Primers shown in
Table 1).
2.3. Virus Isolation and Viral Amplification
The collected samples from the duck farm in Shandong province were weighed, and sterile PBS solution, supplemented with 1000 U/mL of penicillin and 1 mg/mL of streptomycin , was added at a ratio of 1 mL/g. The samples were ground using a multi-sample tissue grinder at 180 Hz for 3 minutes after steel balls were added. The samples were subsequently centrifuged at 5000 × g for 10 minutes and the supernatant was filtered through 0.22 µm filters. Exponentially growing LMH cells were seeded in a T25 cm² flask. One day later, when the cells reached 90% confluence, 1.5 mL of DMEM/F12 containing 2% FBS was inoculated with the sample supernatant. After adsorption for 90 minutes at 37°C, the supernatant was removed, and the cells were washed three times with 10 mM phosphate-buffered saline (PBS) and 5mL of fresh DMEM-F12, supplemented with 1% antibiotic-antimycotic and 2% FBS, was added. When 70-80% of the LMH cells exhibited complete cytopathic effects (CPEs), the cells and supernatant were frozen-thawed three times and were harvested. After the samples were centrifuged at 3300 × g for 15 minutes at 4°C, the supernatant was then collected and stored at -80°C. The viruses were purified three times in LMH cells using the limiting dilution method.
2.4. Phylogenetic Analysis
To investigate the evolutionary relationships of DAdV-3 SD2019 with other adenoviruses, the Hexon gene sequence of SD2019 was compared with those of 31 strains, retrieved from the NCBI database and the phylogenetic trees were constructed using the neighbor-joining (NJ) method in MEGA 11 software. The aligned Hexon gene sequences were analyzed to elucidate the phylogenetic relationships among these strains.
2.5. Construction of DAdV-3 Infectious Clone
The viral supernatant was ultra-centrifuged (100,000× g) in 30% sucrose in TNE buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA pH 8.0) at 4 °C for 2 h. The viral pellets were resuspended in TNE buffer followed by proteinase K treatment, DNA extraction with phenol, chloroform, and ethanol precipitation. A PCR product containing the ITR region,
Pac I cleavage site, kanamycin-resistant gene and origin (KAN-ORI) was amplified with the primer pairs DAdV-3 PacI-F/R(
Table 1). After gel purification, the KAN-ORI and DAdV-3 genomic DNA were took in a homologous recombination reaction, containing the 2μL of DAdV-3 genome, 1μL of KAN-ORI, 4 μL of 5× Exnase II buffer, 2 μL of Exnase II, 11 μL of ddH
2O, to construct DAdV-3 infectious clone pET-KanaR-DAdV-3 (pDAdV-3). After Incubating at 37°C for 30 min, the reaction solution was transformed into
E.coli Top10 competent cells. Positive clones were further identified and verified using the primer pairs pDAdV-3 Kana F/R (
Table 1).
2.6. Virus Rescue
Adenovirus plasmids including the whole DAdV-3 genome (pDAdV-3) were digested with the restriction enzyme PacI (NEB, USA ), and the linearized DNAs were recovered by ethanol precipitation and then the DNAs were elector transfected into LMH cells (130V, 950 uF,+∞, 2mm). The mixed solution was tranferred into a T25 flask with 5mL of 37℃-warmed DMEM/F12, supplemented with 10% FBS, and the supernatant was replaced with fresh DMEM-F12 containing with 2% FBS after 24 hours. The transfected cells were cultured in 37℃, with 5% CO2 for 6 days. The cells and the supernatant were frozen-thawed for three times, when 80% transfected cells showed apparent cytopathic effects (CPEs), and the cellular debris were removed by centrifugation at 5000 × g for 5 min. The supernatant was stored at −80 °C.
2.7. Indirect Immunofluorescence Assay (IFA)
LMH cells cultured on 6-well plates were infected with parental DAdV-3 SD2019 (wtSD2019) and rescued SD2019 (rSD2019) for IFA test. Briefly, LMH cells were inoculated with wtSD2019 or rSD2019 at a MOI of 0.01. At 72 hpi, the cell culture was removed and infected cells were fixed with 4% paraformaldehyde for 20 min at room temperature (RT) and washed with PBS. The plates were blocked with 10% BSA for 20min at RT and washed with PBS. Then the anti-DAdV-3 monoclonal antibody 2F12 (mAb, 1:200 dilution) was added into the wells and inoculated for 1h at 37 ℃. The wells were subsequently washed three times with PBS containing 1 % Tween-20 (PBST) and incubated with 100 μL FITC-conjugated goat anti-mouse IgG antibody (1:400 dilution, Invitrogen Carlsbab, CA, USA) for 1h at 37 ℃. After washing with PBS five times, the images of the cells were examined using a fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
2.8. Viral Growth Kinetics
To test the replication of parental and rescued DAdV-3 SD2019 strains in vitro, LMH cells were infected with wtSD2019 and rSD2019 at a MOI of 0.01), the cells were incubated with DMEM/F12 containing 2% FBS at 37℃ for 2 hours. After washing with PBS for 3 times, the cells were cultured at 37℃ for 6 days and the viral supernatant was harvested every 12 hours post infection and were subjected to virus titration on LMH cells to determine the virus titers.
2.9. Virus Titration
To determine the virus titers, duck tissue samples were weighed and homogenized in sterile PBS to yield 1:1 (ml/g) homogenates. Tissue homogenates or cell culture samples were clarified by centrifugation at 12,000 × g, 4℃ for 10 min, and undiluted and 10-fold serially diluted supernatants were titrated onto 70-80% confluent LMH cells in 96-well plates at 37℃ for 2 hours. After adsorption, cells were washed and cultured with DMEM/F12 with 2% FBS in a humidified chamber with 5% CO2 at 37℃. The lower limit of virus detection was 0.5 log10 TCID50 per 0.1 g tissue. The virus titer was calculated by the method of Reed and Muench.
2.10. Duck Experiments
To test the virulence of parental and rescued SD2019 in Muscovy ducks, eighteen 21 day-old Muscovy ducks were randomly divided into 3 groups. Ducks in two of three groups were inoculated intramuscularly (i.m.) with 105.5 TCID50 of each virus ( wtSD2019 or rSD2019) at volume of 0.2 ml, respectively. Ducks in another group were inoculated with equal volume of sterile PBS. The ducks were monitored daily for clinical signs till 14 dpi. At 4 dpi, the liver and kidney samples of 3 ducks in each group were collected for qPCR test, respectively. Sera of another 3 ducks in each group were collected at 0, 7, and 14dpi for antibody detection.
2.11. The qPCR Assay
To quantify DAdV-3 DNA copies in duck tissue samples, a real-time qPCR assay was performed. The forward (5’-TGACATAAAGGGTGTGCTAG-3’) and reverse (5’-GGAGCTAGTGGATTGTAAG-3’) primers and probe (5’-FAM TCTTCTTTCAAACCATACAGTGG-BHQ1-3’) were designed based on the Hexon gene of DAdV-3 and were synthesized in Sangon Biotech (Shanghai) Co., Ltd. Tissue samples were homogenized in PBS at a ratio of 1:1 g/ml centrifugated at 12,000 × g for 10 min at 4 ˚C. The supernatant was used for DAdV-3 DNA quantification. Total DNAs were extracted using the Ezup Column Virus DNA purification Kit (Sangon Biotech, Shanghai, China). Add 12.5 μL of 2×Multiplex Probe qPCR Mix Plus, 0.5 μL of Forward primer (10μM), 0.5 μL of Reverse primer (10μM), 1μL of Probe (10μM), 2 μL of DNA template, 8.5 μL of Ultrapure water into a DNAase and RNAase-free tube and the real-time qPCR was performed on a Mastercycler ep realplex system (Eppendorf, Hamburg, Germany) using cycling conditions: 37℃,2min, 95℃ 30sec, (95℃ 10sec, 60℃, 30sec)×40cycles.
2.12. Antibody Detection
A blocking ELISA for detection of serum antibodies against DAdV-3 was developed and performed as previously described [
16]. Briefly, ELISA plates were coated with purified DAdV-3 antigen in 0.1 M carbonate–bicarbonate buffer (pH 9.6) and incubated overnight at 4 ˚C. Antigen-coated plates were washed with PBS (pH 7.4) containing 0.05 % Tween-20 (PBST), and blocked 5 % (w/v) skim milk dissolute in PBS for 1 h at 37 ˚C. 100 μl of serum sample (1:10 dilution) was added into the well and incubated at 37 ˚C for 1 h. After washing 3 times with PBST, mAb 2F12 (1:1000 dilution) was added and the plate was incubated at 37 ˚C for 1 h. After rinsing with PBST 3 times, HRP conjugated goat anti-mouse IgG (1:4000 dilution, Sigma, USA) was added,and the plate was incubated at 37 ˚C for 1 h. After rinsing with PBST 3 times, 100 μl of 3,3’, 5,5’-tetramethyl benzidine (TMB) was added, and the plate was incubated at RT for 10 min. The reaction was then stopped by adding 0.1 N sulfuric acid. The optical density (OD) was measured at 450 nm, and the percent inhibition (PI) value was determined using the formula: PI (%) = [1-(OD450 nm of test serum/OD450 nm of negative control serum) ] * 100 %. The serum was considered DAdV-3 positive when the PI value was ≥ 21.62 %. When the PI value was ≤ 16.79 %, the serum was considered negative.
2.13. Ethics Statement and Statistical Analysis
All animal experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. The protocol (SV-20240526-01) used in the study was approved by the Animal Care Committee of the Shanghai Veterinary Research Institute.
3. Results
3.1. Virus Isolation and Identification
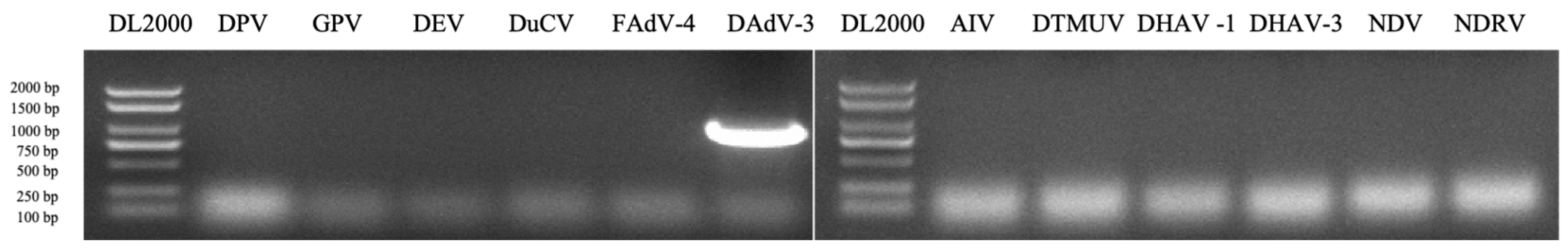
In 2019, an infectious disease outbreaks characterized by swelling and hemorrhagic livers and kidneys occurred in a Muscovy duck farm in Shandong. The liver and kidney samples collected from two diseased duck farms were homogenated and centrifuged, the supernatant was subjected to RT-PCR and PCR test. The results showed that the tissue samples were positive for DAdV-3(
Figure 1A), but were negative for other common duck viruses, including DPV, GPV, DEV, DuCV, FAdV-4, AIV, DTMUV, DHAV-1, DHAV-3, NDV and NDRV(
Figure 1). To isolate the virus, the samples were inoculated into LMH cells and the infected LMH cells showed CPEs at 4 days post infection, which suggested that the virus is virulent to LMH cells. To purify the virus, the infected cells and the supernatant were harvested and frozen-thawed 3 times, the mixtures were centrifuged at 12,000 × g at 4℃and the supernatant was purified 3 times in LMH cells by limiting dilution method. A DAdV-3 strain, isolated and named as SD2019, was further verified by sequencing.
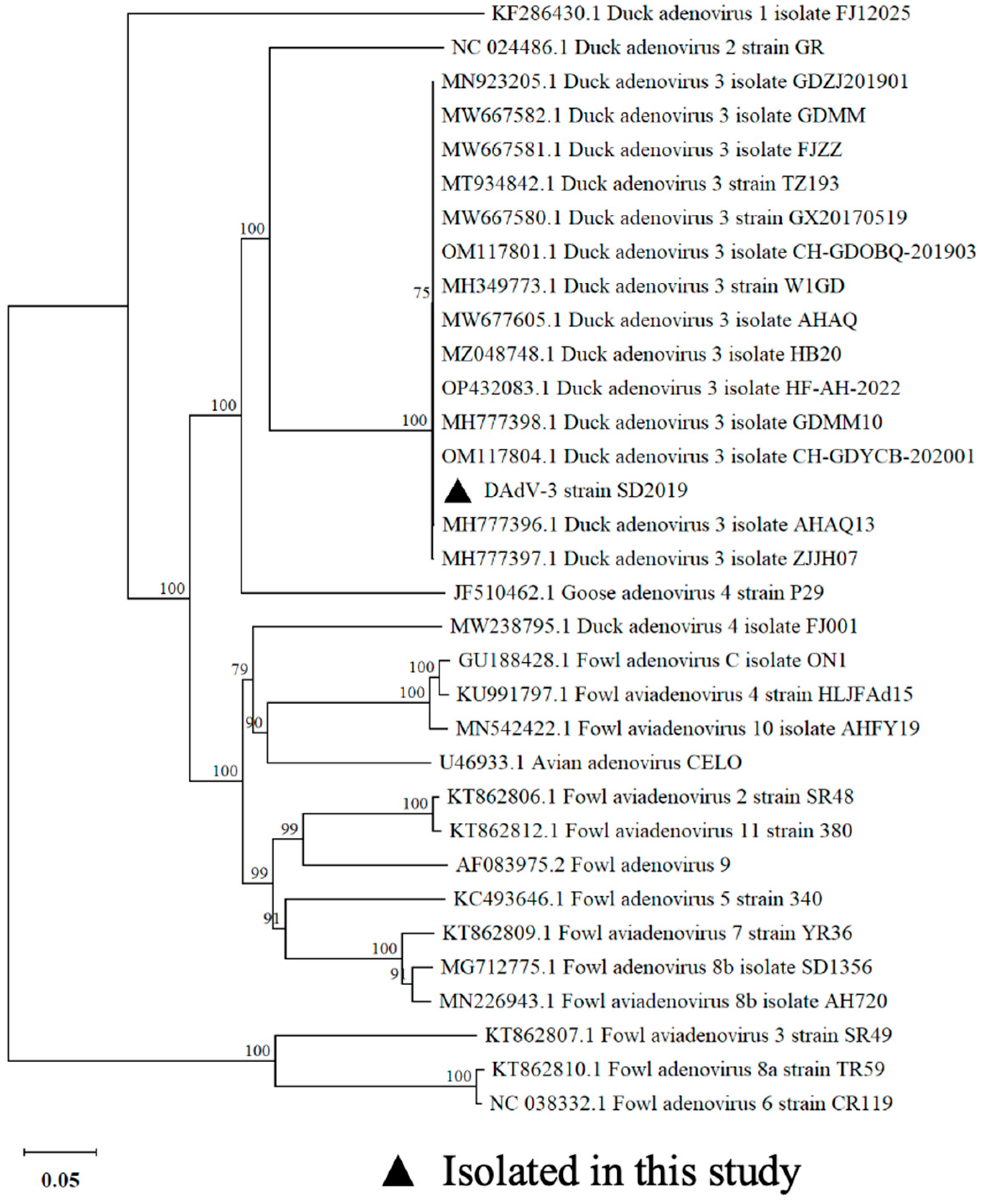
3.2. Phylogenetic Analysis
To verify the virus, the Hexon gene sequence of the DAdV-3 SD2019 was amplified for sequencing. To determine the phylogenetic relationship of the DAdV-3 SD2019 and other reported fowl adenoviruses, the Hexon genes, including DAdV-3 2019 and other 31 representative fowl adenoviruses, were used for further analysis. A phylogenetic tree was generated by using the neighbor-joining (NJ) method in MEGA 11.0 version software. The phylogenetic analysis showed that DAdV-3 SD2019 was grouped into DAdV-3, and shared a high level of sequence identity (98.5%-100%) with other previous reported DAdV-3 strains (
Figure 2).
3.3. Generation of DAdV-3 Infectious Clone
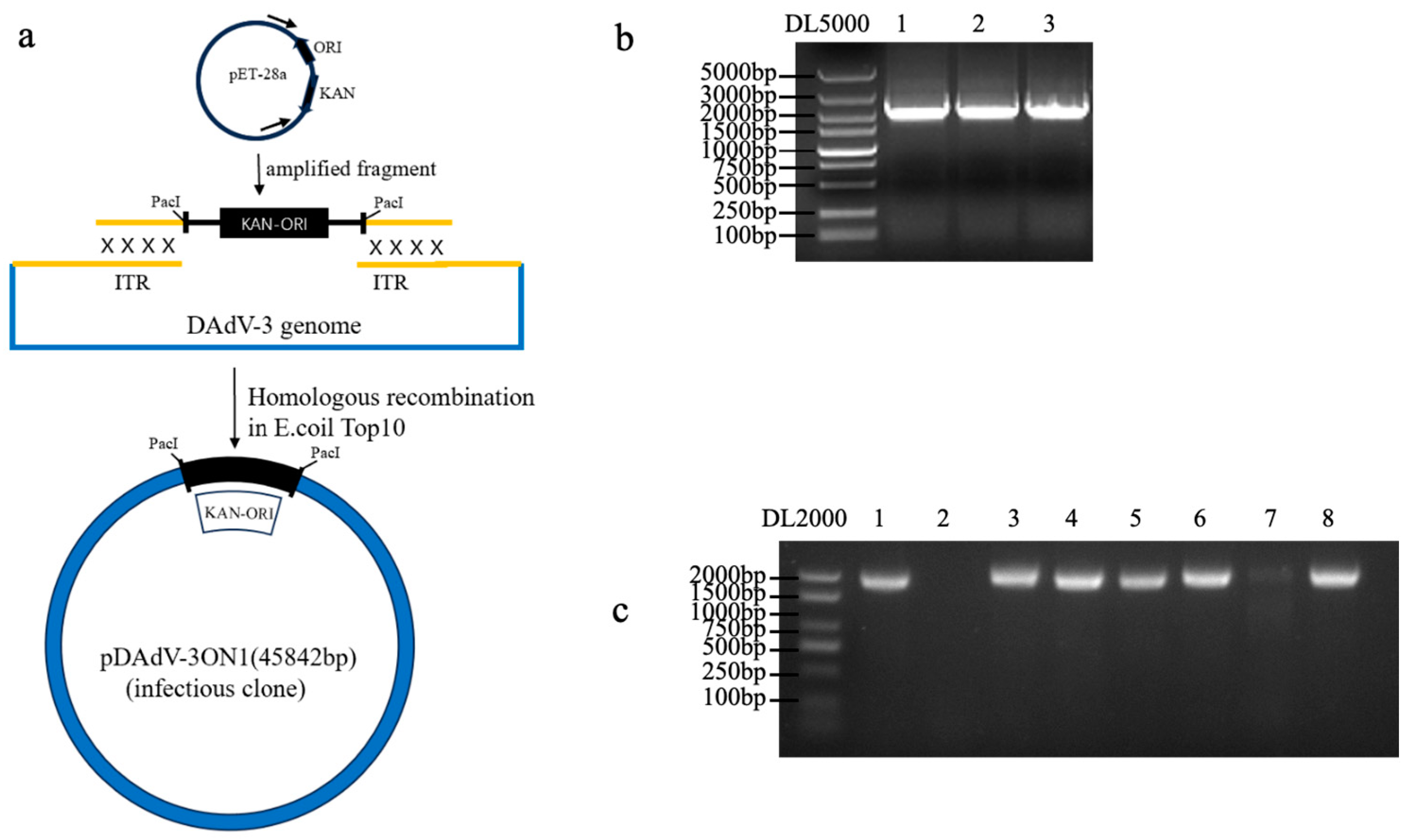
To generate the infectious clone of DAdV-3 (
Figure 3a), the
Pac I cleavage site, kanamycin-resistant gene and origin (KAN-ORI) sequences were amplified (
Figure 3b) and were recombinated with viral DNAs. Then the plasmid was circularized through homologous recombination and transformed into
E. coli Top10 competent cells. After 24 hours of incubation, clones grown on kanamycin-resistant LB plates were screened via PCR for positive recombinants (
Figure 3c). A plasmid containing the full-length DAdV-3 genome was successfully constructed through homologous recombination and designated pDAdV-3.
3.4. Virus Rescue
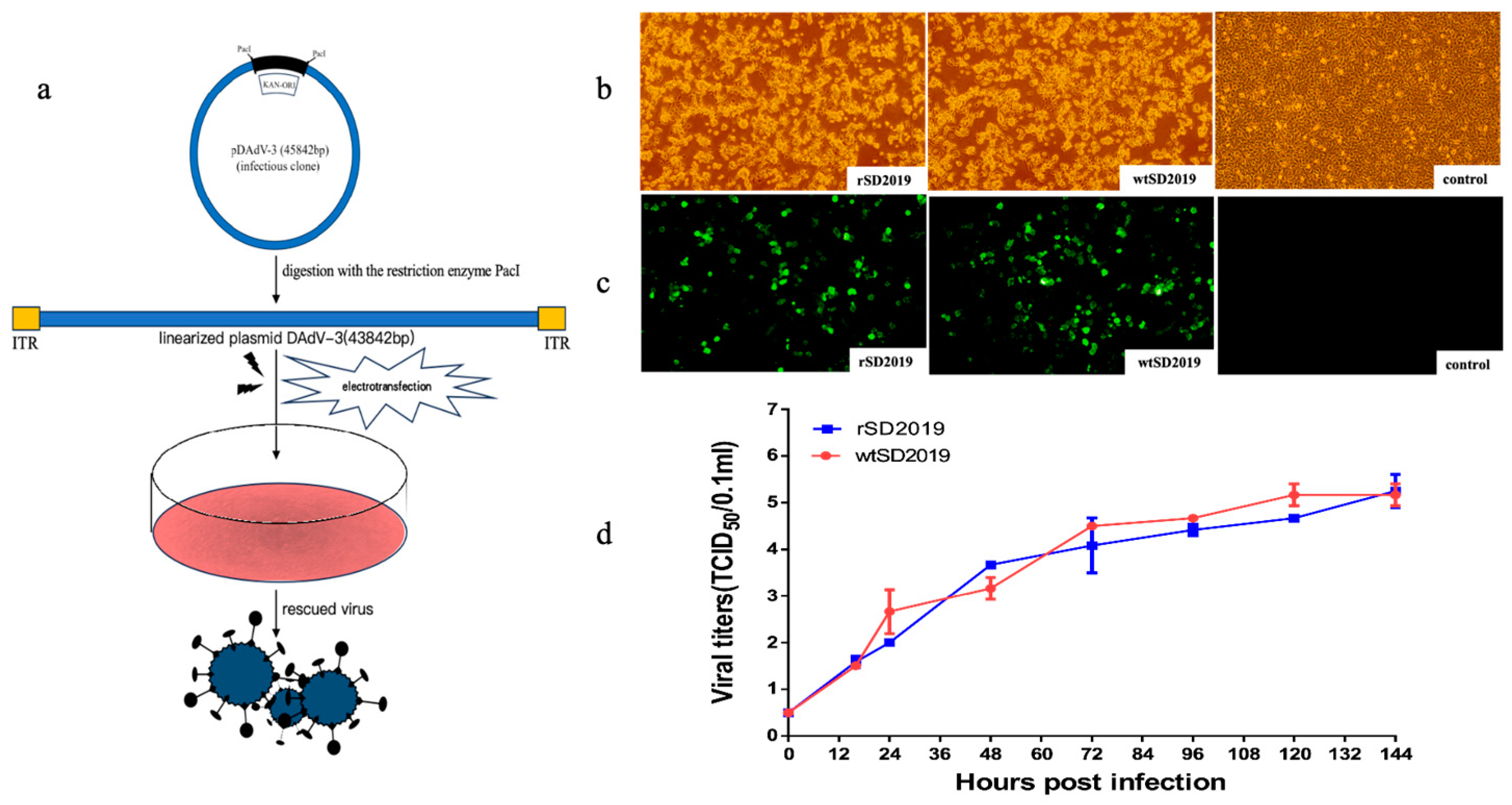
To rescue DAdV-3 SD2019, the plasmids containing whole genome of DAdV-3 SD2019 (pDAdV-3) were digested by the
PacI, and the linearized DNAs were then electro transfected into LMH cells (
Figure 4a). Detectable CPEs were appeared at 96 hours post-transfection, while no CPEs were found in untransfected cells (
Figure 4b). The transfected cells and supernatant were harvested and used for IFA test. Fresh LMH cells infected with the rescued virus showed obvious specific green fluorescence at 72 hours post-infection, while no fluorescence was detected in uninfected cells (
Figure 4c). The rescued viruses passage 2 (P2) were used for PCR test and sequencing. The results showed rDAdV-3 SD2019 P2 was positive for DAdV-3 and the sequence analysis confirmed the rescued virus have no unexpected mutations (data not shown). The results indicated that the virus was successfully rescued.
3.5. Growth Curves of rSD2019 and wtSD2019 in LMH Cells
To determine the replication kinetics of rSD2019 and wtSD2019, LMH cells, cultured in T25 flasks, were inoculated with different viruses at multiplicities of infection of 0.01, respectively. The supernatant was collected at every 12 hours post-infection and titrated. Compared with that of wtSD2019, the rSD2019 grew similarly within 144 hours post-infection (
Figure 4d).
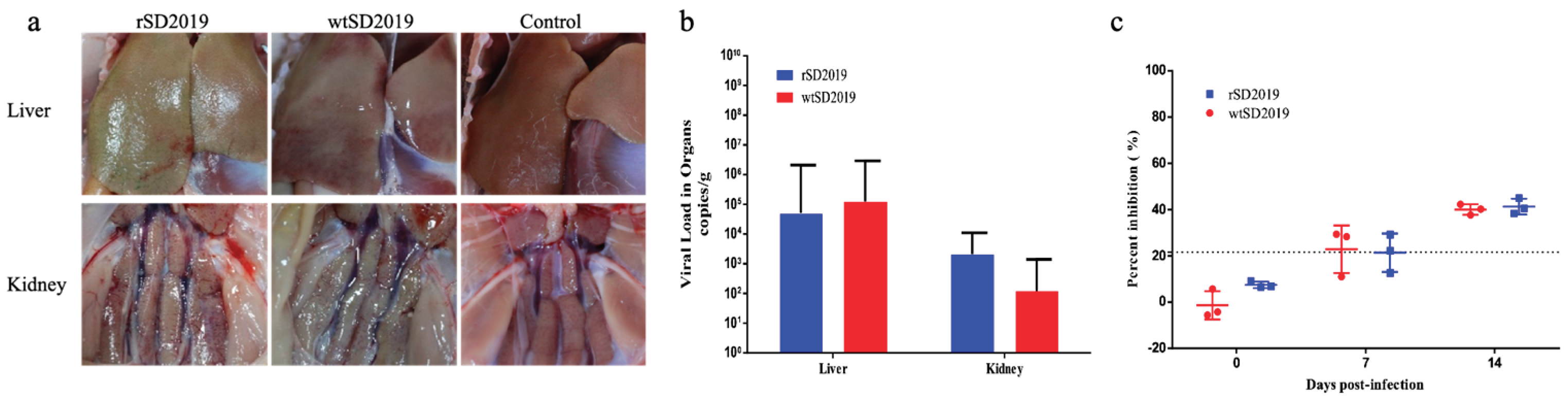
3.6. Pathogenicity of rSD2019 and wtSD2019 in Muscovy Ducks
To evaluate and compare the pathogenesis of the rescued and parental SD2019 in ducks, each six 21-day-old Muscovy ducks were inoculated intramuscularly with 105.5 TCID50 of rSD2019 or wtSD2019, respectively. No ducks died during the entire experiment.
At 4 dpi, the livers and kidneys of infected ducks showed hemorrhage in both rSD2019 and wtSD2019 groups (
Figure 5A). Viral DNA copies in different tissue samples were quantified by real-time qPCR at 4 dpi and all the livers and kidneys tested were positive for DAdV-3 DNA. In liver and kidney samples, the virus titers did not significantly differ between rSD2019 and wtSD2019 groups, respectively (
Figure 5B). Two of three ducks inoculated with either the parental wtSD20199 or its rescued strain rSD2019 showed seroconversion at 7 dpi. and the sera of the last one was positive in each group (
Figure 5C). All the results indicated that the rSD2019 replicate similarly compared with that of wtSD2019 in Muscovy ducks.
4. Discussion
The first isolation of a novel duck adenovirus type 3 (DAdV-3) strain from infected Muscovy ducks in China was reported in 2014 [
17]. Since then, DAdV-3 infections have been increasingly affected duck farms, emerging as a significant threat to the poultry industry with considerable economic impacts [
18,
19,
20].
The pathogenicity of different DAdV-3 strains differs in Muscovy ducks. The DAdV-3 TZ193 strain caused characteristic lesions of swelling as well as hemorrhagic liver and kidney in the infected ducklings and the mortality rate of TZ193 in 5-day-old domestic ducks was 100% [
20]. FJGT01 was highly virulent for ten-day-old Muscovy ducklings, with a mortality rate of 60%, which started on day 5 after inoculation. Meanwhile, gross lesions showed signs of swelling and hemorrhage in the liver and the kidneys of infected ducklings [
19]. The morbidity of CH-GD-12-2014 to one-day-old SPF Shaoxing ducks and Muscovy ducks is both 100%, but the mortality to these two kinds of ducks differs, CH-GD-12-2014 caused 7 out of 25 one-day-old SPF Shaoxing ducks‘ deaths, while no infected Muscovy ducks died [
17]. In another study, DAdV-3 CH-GD-12-2014 caused death of 10-week-old ducks [
21]. Interestingly, the HF-AH-2020 has very low pathogenicity to Muscovy ducks, but can induce a high level of cellular immunity [
22]. In this study, we isolated a wtDAdV-3 SD2019 strain on LMH cells, and the Hexon gene sequence shared 98.7%–100% similarities with that of other DAdV-3 reference strains. Considering to establish a challenge model at an appreciate age for immunized ducks in the future, the 21-day-old Muscovy ducks were infected with wtSD2019 strain. However, we found that wtSD2019 was not fetal to 21-day-old Muscovy ducks. Besides, the lesions of the livers and kidneys seemed to be relatively mild at 4 dpi, which indicated that the wtSD2019 also has low pathogenicity to Muscovy ducks compared to some other reported strains.
To further study the mechanism of different virulence of DAdV-3 strains in waterfowl, a reverse genetics system for DAdV-3 should be established. Here, we have generated the DAdV-3 infectious clone and rescue the virus (rSD2019) successfully. The rescued virus exhibited identical CPE formation, immunological reactivity and replication capacity to wild-type virus, demonstrating both the reliability of our reverse genetics system and the preserved infectivity of the cloned genome.
In summary, a duck adenovirus type 3 strain SD2019 was isolated and an infectious clone, based on full length of the SD2019, was successfully established. The establishment of the reliable reverse genetics system for DAdV-3 provides a foundation for future studies of pathogenicity basis of DAdV-3.
Author Contributions
Conceptualization, Haipeng Lu, Mei Tang and Zejun Li; Data curation, Zhifei Zhang, Mi Wu and Chunxiu Yuan; Formal analysis, Xue Pan; Funding acquisition, Zejun Li; Investigation, Qinfang Liu, Qiaoyang Teng and Bangfeng Xu; Methodology, Minghao Yan; Software, Haipeng Lu; Supervision, Fenglong Wang and Zejun Li; Validation, Dawei Yan; Writing – original draft, Mei Tang and Dawei Yan; Writing – review & editing, Qiaoyang Teng, Qinfang Liu, Fenglong Wang and Zejun Li.
Funding
This research was funded by the National Key Research and Development Program of China (2023YFD1800600).
Institutional Review Board Statement
All animal experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of China. The protocol (SHVRI-SZ-20240526-01) used in the study was approved by the Animal Care Committee of the Shanghai Veterinary Research Institute.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Greber, U.F., Adenoviruses - Infection, pathogenesis and therapy. FEBS Lett, 2020. 594(12): p. 1818-1827.
- Tan, Y., et al., Isolation, characterization, evaluation of pathogenicity, and immunomodulation through interferon production of duck adenovirus type-3 (DAdV-3). Poult Sci, 2024. 103(3): p. 103411. [CrossRef]
- Huang, Y., et al., Isolation and partial genetic characterization of a new duck adenovirus in China. Vet Microbiol, 2020. 247: p. 108775. [CrossRef]
- Zhang, X., et al., Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology, 2016. 493: p. 12-21. [CrossRef]
- Shi, S., et al., Isolation and characterization of duck adenovirus 3 circulating in China. Arch Virol, 2019. 164(3): p. 847-851. [CrossRef]
- Shi, X., et al., Isolation and pathogenic characterization of duck adenovirus 3 mutant circulating in China. Poult Sci, 2022. 101(1): p. 101564. [CrossRef]
- Yin, L., et al., Epidemiological investigation of duck adenovirus 3 in southern China, during 2018-2020. Avian Pathol, 2022. 51(2): p. 171-180. [CrossRef]
- Wan, C., et al., Development of a TaqMan-based real-time PCR for detecting duck adenovirus 3. J Virol Methods, 2018. 261: p. 86-90. [CrossRef]
- François, A., et al., Construction of avian adenovirus CELO recombinants in cosmids. J Virol, 2001. 75(11): p. 5288-301. [CrossRef]
- Wen, Y., et al., Construction and immune evaluation of the recombinant duck adenovirus type 3 delivering capsid protein VP1 of the type 1 duck hepatitis virus. Poult Sci, 2023. 102(12): p. 103117. [CrossRef]
- Pei, Y., et al., Fowl Adenovirus 4 (FAdV-4)-Based Infectious Clone for Vaccine Vector Development and Viral Gene Function Studies. Viruses, 2018. 10(2). [CrossRef]
- Pan, Q., et al., Development of a Novel Avian Vaccine Vector Derived From the Emerging Fowl Adenovirus 4. Front Microbiol, 2021. 12: p. 780978. [CrossRef]
- Tang, Z., et al., Generation of an artificially attenuated fowl adenovirus 4 viral vector using the reverse genetics system based on full-length infectious clone. Vet Res, 2025. 56(1): p. 62. [CrossRef]
- Johnson, M.A., et al., A recombinant fowl adenovirus expressing the S1 gene of infectious bronchitis virus protects against challenge with infectious bronchitis virus. Vaccine, 2003. 21(21-22): p. 2730-6. [CrossRef]
- Pei, Y., et al., Rapid generation of fowl adenovirus 9 vectors. J Virol Methods, 2015. 223: p. 75-81. [CrossRef]
- Li, X., et al., Development of a blocking ELISA for detection of serum neutralizing antibodies against newly emerged duck Tembusu virus. PLoS One, 2012. 7(12): p. e53026. [CrossRef]
- Zhang, X., et al., Molecular characterization, phylogeny analysis and pathogenicity of a Muscovy duck adenovirus strain isolated in China in 2014. Virology, 2016. 493: p. 12-21. [CrossRef]
- Yin, L., et al., Epidemiological investigation of duck adenovirus 3 in southern China, during 2018-2020. Avian Pathol, 2022. 51(2): p. 171-180. [CrossRef]
- Shi, S., et al., Isolation and characterization of duck adenovirus 3 circulating in China. Arch Virol, 2019. 164(3): p. 847-851. [CrossRef]
- Shi, X., et al., Isolation and pathogenic characterization of duck adenovirus 3 mutant circulating in China. Poult Sci, 2022. 101(1): p. 101564. [CrossRef]
- Wen, Y., et al., Construction and immune evaluation of the recombinant duck adenovirus type 3 delivering capsid protein VP1 of the type 1 duck hepatitis virus. Poult Sci, 2023. 102(12): p. 103117. [CrossRef]
- Tan, Y., et al., Isolation, characterization, evaluation of pathogenicity, and immunomodulation through interferon production of duck adenovirus type-3 (DAdV-3). Poult Sci, 2024. 103(3): p. 103411. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).