Submitted:
25 March 2025
Posted:
26 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methodology
2.1. Survey of Literature
2.2. Survey of Protein Data Bank
2.3. AlphaFold Predictions
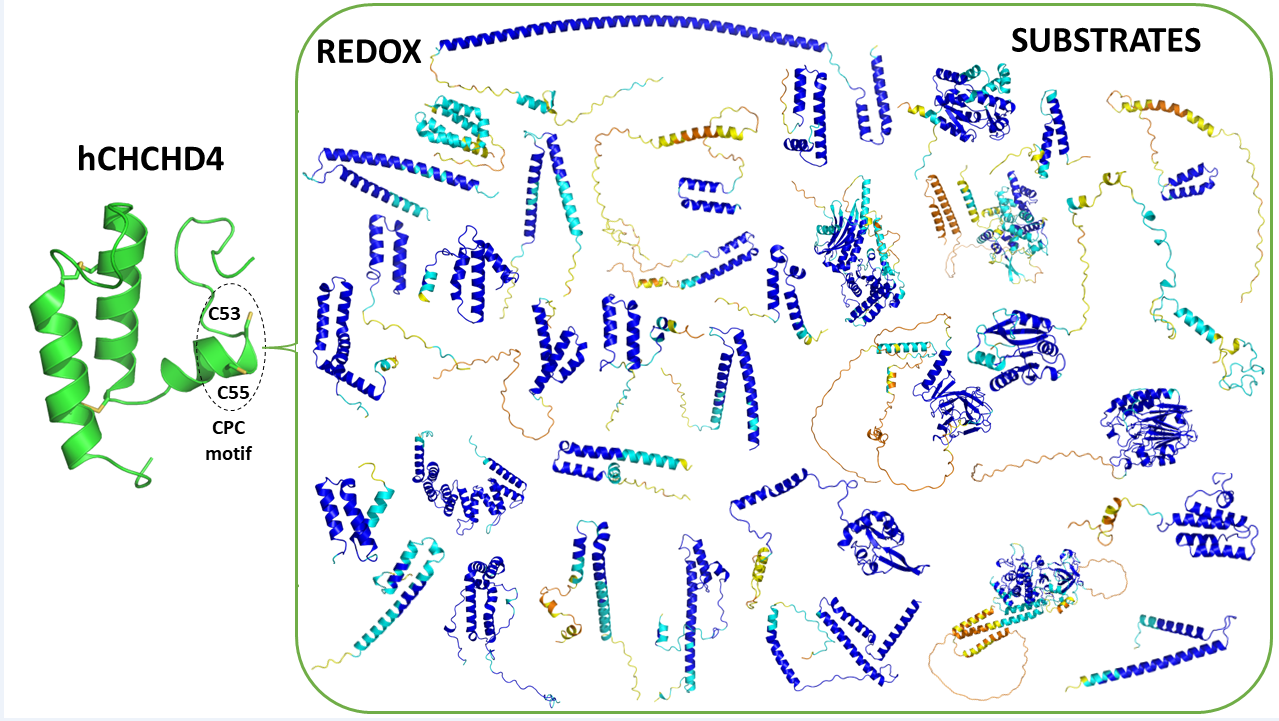
3. The hCHCHD4/yMia40 World
3.1. Evolution and Molecular Organization of hCHCHD4/yMia40
3.2. The Mitochondrial Functions of hCHCHD4/yMia40 and Their Mechanism of Action
3.3. Involvement of hCHCHD4 in Pathological States
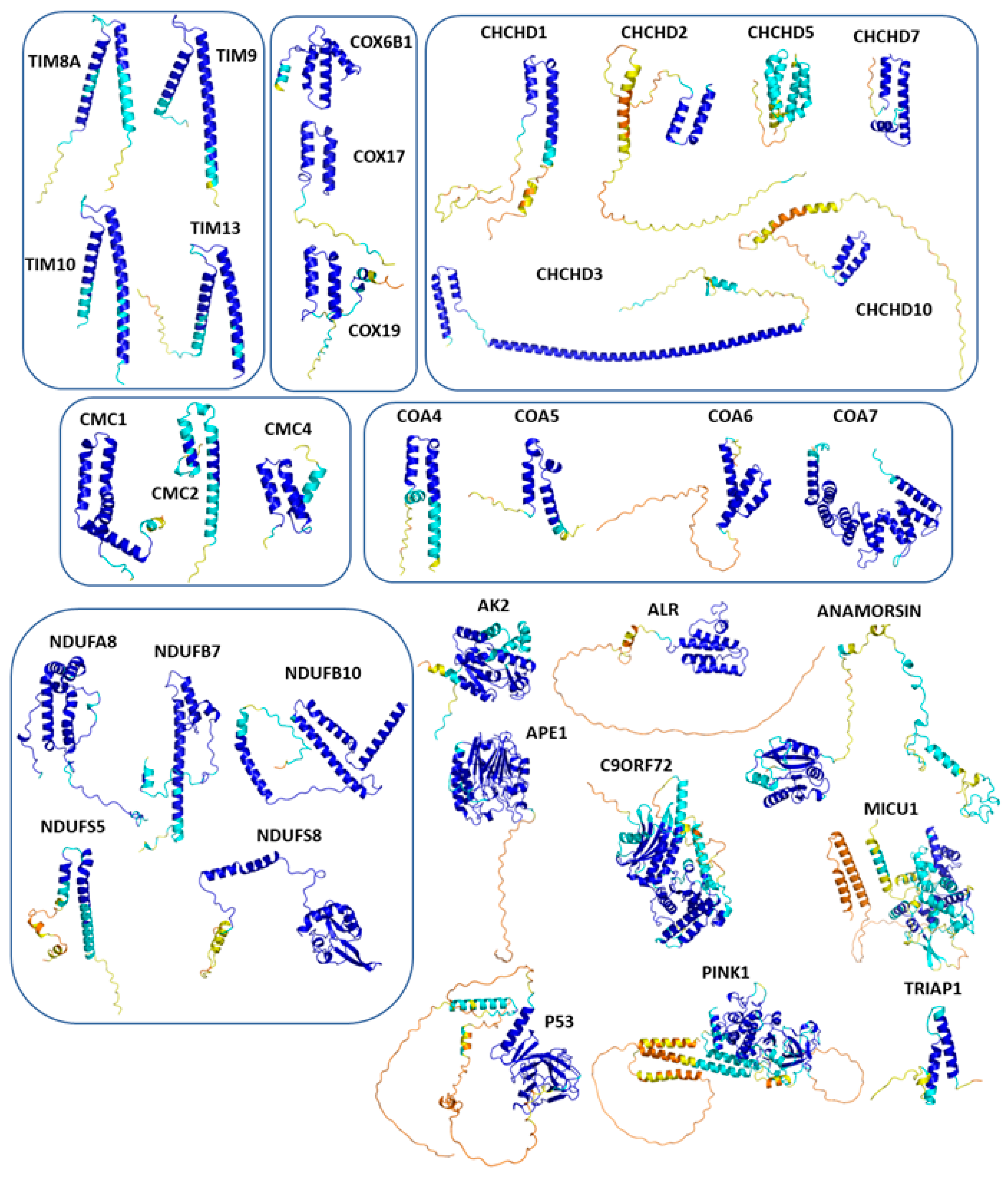
4. Redox Substrates of hCHCHD4/yMIA40
4.1. The TIMM Protein Family
4.2. The COX Protein Family
4.3. The CMC Protein Family
4.4. The CoA Protein Family
4.5. The CHCHD Protein Family
4.6. The NDUF_NADH Dehydrogenase Protein Family
4.7. The Human Adenylate Kinase 2
4.8. The Protein p53
4.9. The Protein TRIAP1
4.10. The Protein Anamorsin
4.11. The Protein C9orf72
4.12. The Protein APE1
4.13. The Protein PINK1
4.14. The Protein MICU1
4.15. The Thiol Oxidase ALR
|
Protein (Length, aa) |
Organism (UniProtKB) | PDB entry |
Method (Resolution, Å) |
Protein region (residues) | Notes | Reference |
| hCHCHD4 (142) |
H. sapiens (Q8N4Q1) |
2K3J | NMR | 41-105 | hCHCHD4 residues 1-40 and 106-142 are missing*. C53S mutation. |
[28] |
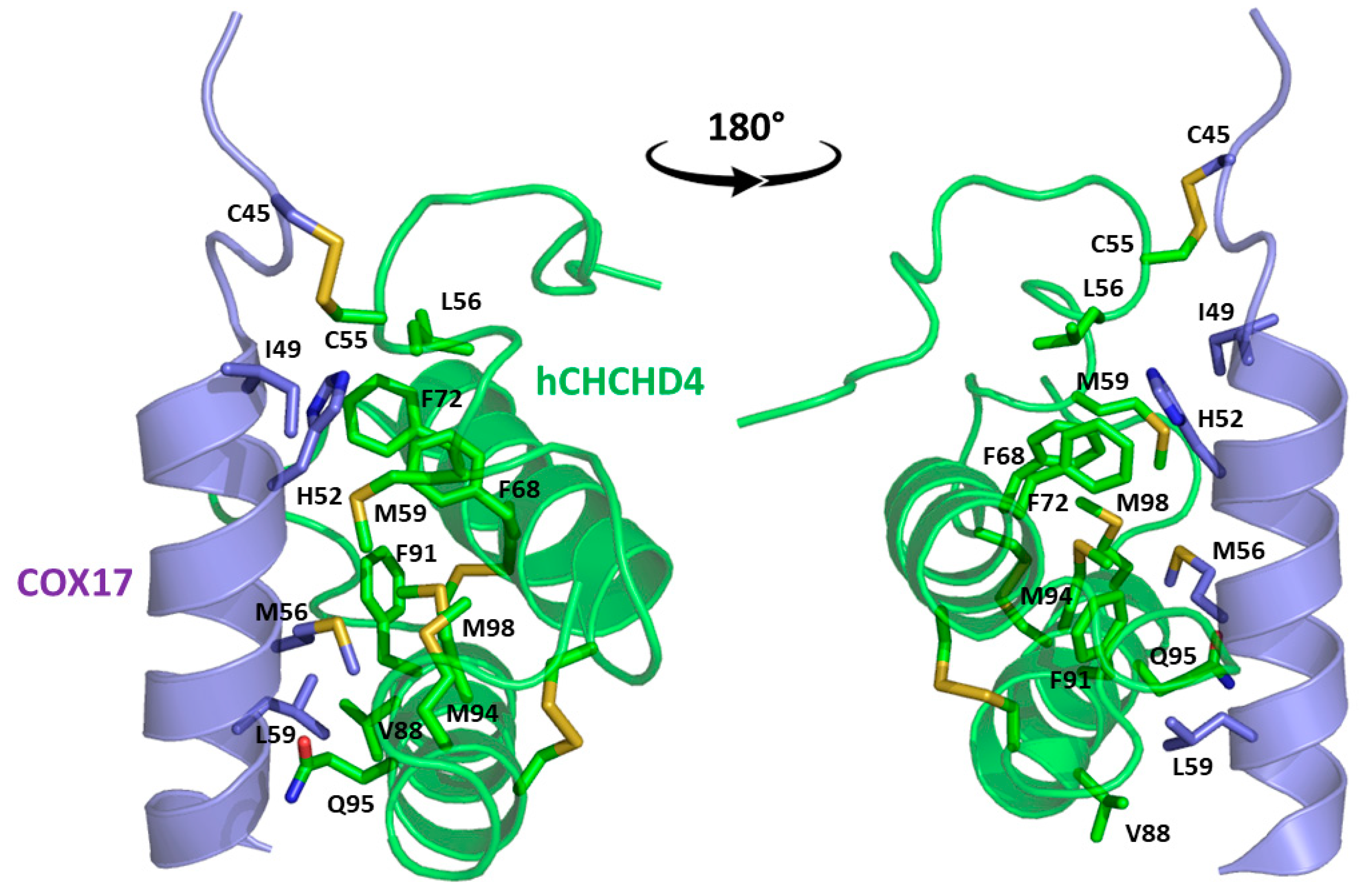
| 2L0Y | NMR | 46-105 | Complex of hCHCHD4 and human cytochrome c oxidase copper chaperone COX17 (UniProtKB Q14061, residues 1-63). hCHCHD4 residues 1-45 and 106-142 and COX17 residues 1-42 are missing*. C55S mutation in COX17. | [151] | ||
| yMia40 (403) |
S. cerevisiae (P36046) |
2ZXT | X-ray diffraction (3.00) | 284-353 | Fusion protein of maltose-binding periplasmic protein MMBP (UniProtKB P0AEX9, residues 27-392), Linker (NSSSVPGRGSIEGRPEF), yMia40 (residues 284-365). yMia40 residues 354-365 are missing*. | [63] |
| 3A3C | X-ray diffraction (2.50) | 284-353 | Fusion protein of MMBP (UniProtKB P0AEX9, residues 29-392), Linker (NSSSVPGRGSIEGRPEF), yMia40 (residues 284-365). yMia40 residues 354-365 are missing*. C296S, C298S mutations in yMia40. | |||
| mCHCHD4 (139) |
M. musculus (Q8VEA4) |
8QNS | X-ray diffraction (3.21) | 4-14 | Complex of apoptosis-inducing factor 1 AIF (UniProtKB Q9Z0X1, residues 101-612) and mCHCHD4 (residues 1-27). mCHCHD4 residues 1-3 and 15-27 are missing*. | [152] |
|
Substrate (UniprotKB) |
Disulfide bridges, docked Cys#, sequence | PDB structure* |
Superimposition (Exp vs AF) or AF3 model |
PAE of the AF3 model |
| Group I | ||||
| ALR (P55789) | C142-C145, C171-C188 #C71 or C74 123456789012345678901234567890 MAAPGERGRFHGGNLFFLPGGARSEMMDDLATDARGRGAGRRDAAASASTPAQAPTSDSPVAEDASRRRPCRACVDFKTWMRTQQKRDTK FREDCPPDREELGRHSWAVLHTLAAYYPDLPTPEQQQDMAQFIHLFSKFYPCEECAEDLRKRLCRNHPDTRTRACFTQWLCHLHNEVNRK LGKPDFDCSKVDERWRDGWKDGSCD |
Code: 3U5S Technique: X-ray crystallography (Resol. 1.50 Å) Chain: A Residues: 82-203 |
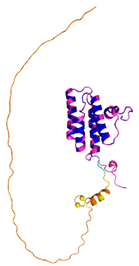 RMSD: 0.26 Å (114 Cα aligned) RMSD: 0.26 Å (114 Cα aligned) |
 |
| COX17 (Q14061) |
C26-C55, C36-C45 #C45 123456789012345678901234567890 MPGLVDSNPAPPESQEKKPLKPCCACPETK KARDACIIEKGEEHCGHLIEAHKECMRALG FKI |
Code: 2RNB Technique: NMR Chain: A Residues: 1-63 |
 RMSD: 0.64 Å (38 Cα aligned) RMSD: 0.64 Å (38 Cα aligned) |
 |
| TIMM9 (Q9Y5J7) | C28-C52, C32-C48 #C28 123456789012345678901234567890MAAQIPESDQIKQFKEFLGTYNKLTETCFLDCVKDFTTREVKPEETTCSEHCLQKYLKMTQRISMRFQEYHIQQNEALAAKAGLLGQPR |
Code: 2BSK Technique: X-ray crystallography (Resol. 3.30 Å) Chain: A Residues: 1-89 |
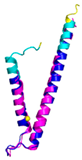 RMSD: 0.78 Å (67 Cα aligned) RMSD: 0.78 Å (67 Cα aligned) |
 |
| TIMM10 (P62072) | C29-C54, C33-C50 #C29 123456789012345678901234567890 MDPLRAQQLAAELEVEMMADMYNRMTSACHRKCVPPHYKEAELSKGESVCLDRCVSKYLDIHERMGKKLTELSMQDEELMKRVQQSSGPA |
Code: 2BSK Technique: X-ray crystallography (Resol. 3.30 Å) Chain: B Residues: 1-90 |
 RMSD: 0.90 Å (60 Cα aligned) RMSD: 0.90 Å (60 Cα aligned) |
 |
| Group IIa | ||||
| AK2 (P54819) |
C42-C92 #C42 123456789012345678901234567890 MAPSVPAAEPEYPKGIRAVLLGPPGAGKGTQAPRLAENFCVCHLATGDMLRAMVASGSELGKKLKATMDAGKLVSDEMVVELIEKNLETPLCKNGFLLDGFPRTVRQAEMLDDLMEKRKEKLDSVIEFSIPDSLLIRRITGRLIHPKSGRSYHEEFNPPKEPMKDDITGEPLIRRSDDNEKALKIRLQAYHTQTTPLIEYYRKRGIHSAIDASQTPDVVFASILAAFSKATCKDLVMFI |
Code: 2C9Y Technique: X-ray crystallography (Resol. 2.10 Å) Chain: A Residues: 1-239 |
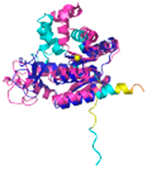 RMSD: 2.9 Å (180 Cα aligned) RMSD: 2.9 Å (180 Cα aligned) |
 |
| APE1 (P27695) |
C65-C93 #C93 123456789012345678901234567890 MPKRGKKGAVAEDGDELRTEPEAKKSKTAAKKNDKEAAGEGPALYEDPPDQKTSPSGKPATLKICSWNVDGLRAWIKKKGLDWVKEEAPD ILCLQETKCSENKLPAELQELPGLSHQYWSAPSDKEGYSGVGLLSRQCPLKVSYGIGDEEHDQEGRVIVAEFDSFVLVTAYVPNAGRGLV RLEYRQRWDEAFRKFLKGLASRKPLVLCGDLNVAHEEIDLRNPKGNKKNAGFTPQERQGFGELLQAVPLADSFRHLYPNTPYAYTFWTYM MNARSKNVGWRLDYFLLSHSLLPALCDSKIRSKALGSDHCPITLYLAL |
Code: 1E9N Technique: X-ray crystallography (Resol. 2.20 Å) Chain: A Residues: 2-318 |
 RMSD: 0.29 Å (256 Cα aligned) RMSD: 0.29 Å (256 Cα aligned) |
 |
| CHCHD2 (Q9Y6H1) |
C114-C144, C124-C134 #C144 123456789012345678901234567890 MPRGSRSRTSRMAPPASRAPQMRAAPRPAPVAQPPAAAPPSAVGSSAAAPRQPGLMAQMATTAAGVAVGSAVGHTLGHAITGGFSGGSNA EPARPDITYQEPQGTQPAQQQQPCLYEIKQFLECAQNQGDIKLCEGFNEVLKQCRLANGLA |
- | 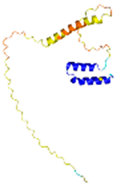 |
 |
| CHCHD3 (Q9NX63) |
C183-C214, C193-C204 #C193 123456789012345678901234567890 MGGTTSTRRVTFEADENENITVVKGIRLSENVIDRMKESSPSGSKSQRYSGAYGASVSDEELKRRVAEELALEQAKKESEDQKRLKQAKELDRERAAANEQLTRAILRERICSEEERAKAKHLARQLEEKDRVLKKQDAFYKEQLARLEERSSEFYRVTTEQYQKAAEEVEAKFKRYESHPVCADLQAKILQCYRENTHQTLKCSALATQYMHCVNHAKQSMLEKGG |
- |  |
 |
| CMC1 (Q7Z7K0) |
C31-C63, C41-C53 #C31 123456789012345678901234567890 MALDPADQHLRHVEKDVLIPKIMREKAKERCSEQVQDFTKCCKNSGVLMVVKCRKENSALKECLTAYYNDPAFYEECKMEYLKEREEFRK TGIPTKKRLQKLPTSM |
- |  |
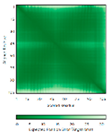 |
| COX19 (Q49B96) | C30-C61, C40-C51 #C51 123456789012345678901234567890 MSTAMNFGTKSFQPRPPDKGSFPLDHLGEC KSFKEKFMKCLHNNNFENALCRKESKEYLE CRMERKLMLQEPLEKLGFGDLTSGKSEAKK |
- |  |
 |
| MICU1 (Q9BPX6) | - #C463 123456789012345678901234567890 MFRLNSLSALAELAVGSRWYHGGSQPIQIRRRLMMVAFLGASAVTASTGLLWKRAHAESPPCVDNLKSDIGDKGKNKDEGDVCNHEKKTA DLAPHPEEKKKKRSGFRDRKVMEYENRIRAYSTPDKIFRYFATLKVISEPGEAEVFMTPEDFVRSITPNEKQPEHLGLDQYIIKRFDGKK ISQEREKFADEGSIFYTLGECGLISFSDYIFLTTVLSTPQRNFEIAFKMFDLNGDGEVDMEEFEQVQSIIRSQTSMGMRHRDRPTTGNTL KSGLCSALTTYFFGADLKGKLTIKNFLEFQRKLQHDVLKLEFERHDPVDGRITERQFGGMLLAYSGVQSKKLTAMQRQLKKHFKEGKGLT FQEVENFFTFLKNINDVDTALSFYHMAGASLDKVTMQQVARTVAKVELSDHVCDVVFALFDCDGNGELSNKEFVSIMKQRLMRGLEKPKD MGFTRLMQAMWKCAQETAWDFALPKQ |
Code: 6LB7 Technique: X-ray crystallography (Resol. 2.10 Å) Chain: C Residues: 97-444 |
 RMSD: 0.92 Å (270 Cα aligned) RMSD: 0.92 Å (270 Cα aligned) |
 |
| NDUFB10 (O96000) | C71-C78, C107-C119 #C107 123456789012345678901234567890 MPDSWDKDVYPEPPRRTPVQPNPIVYMMKAFDLIVDRPVTLVREFIERQHAKNRYYYYHRQYRRVPDITECKEEDIMCMYEAEMQWKRDYKVDQEIINIMQDRLKACQQREGQNYQQNCIKEVEQFTQVAKAYQDRYQDLGAYSSARKCLAKQRQRMLQERKAAKEAAAATS |
Code: 5XTD Technique: EM (Resol. 3.70 Å) Chain: d Residues: 1-171 |
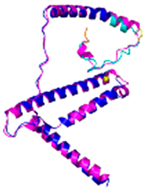 RMSD: 0.87 Å (156 Cα aligned) RMSD: 0.87 Å (156 Cα aligned) |
 |
| Group IIb | ||||
| Anamorsin (Q6FI81) | - 123456789012345678901234567890 MADFGISAGQFVAVVWDKSSPVEALKGLVDKLQALTGNEGRVSVENIKQLLQSAHKESSFDIILSGLVPGSTTLHSAEILAEIARILRPG GCLFLKEPVETAVDNNSKVKTASKLCSALTLSGLVEVKELQREPLTPEEVQSVREHLGHESDNLLFVQITGKKPNFEVGSSRQLKLSITK KSSPSVKPAVDPAAAKLWTLSANDMEDDSMDLIDSDELLDPEDLKKPDPASLRAASCGEGKKRKACKNCTCGLAEELEKEKSREQMSSQP KSACGNCYLGDAFRCASCPYLGMPAFKPGEKVLLSDSNLHDA |
Code: 4M7R Technique: X-ray crystallography (Resol. 1.80 Å) Chain: A Residues: 1-172 |
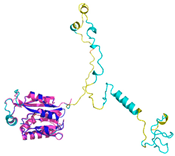 RMSD: 0.50 Å (150 Cα aligned) RMSD: 0.50 Å (150 Cα aligned) |
 |
| C9orf72 (Q96LT7) |
- 123456789012345678901234567890 MSTLCPPPSPAVAKTEIALSGKSPLLAATFAYWDNILGPRVRHIWAPKTEQVLLSDGEITFLANHTLNGEILRNAESGAIDVKFFVLSEKGVIIVSLIFDGNWNGDRSTYGLSIILPQTELSFYLPLHRVCVDRLTHIIRKGRIWMHKERQENVQKIILEGTERMEDQGQSIIPMLTGEVIPVMELLSSMKSHSVPEEIDIADTVLNDDDIGDSCHEGFLLNAISSHLQTCGCSVVVGSSAEKVNKIVRTLCLFLTPAERKCSRLCEAESSFKYESGLFVQGLLKDSTGSFVLPFRQVMY APYPTTHIDVDVNTVKQMPPCHEHIYNQRRYMRSELTAFWRATSEEDMAQDTIIYTDESFTPDLNIFQDVLHRDTLVKAFLDQVFQLKPGLSLRSTFLAQFLLVLHRKALTLIKYIEDDTQKGKKPFKSLRNLKIDLDLTAEGDLNIIMALAEKIKPGLHSFIFGRPFYTSVQERDVLMTF |
Code: 6LT0 Technique: EM (Resol. 3.20 Å) Chain: C Residues: 1-481 |
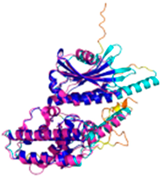 RMSD: 0.95 Å (364 Cα aligned) RMSD: 0.95 Å (364 Cα aligned) |
 |
| CHCHD1 (Q96BP2) | C45-C76, C55-C66 123456789012345678901234567890 MATPSLRGRLARFGNPRKPVLKPNKPLILANRVGERRREKGEATCITEMSVMMACWKQNEFRDDACRKEIQGFLDCAARAQEARKMRSIQETLGESGSLLPNKLNKLLQRFPNKPYLS |
Code: 6LT0 Technique: EM (Resol. 2.21 Å) Chain: A2 Residues: 1-118 |
 RMSD: 1.3 Å (110 Cα aligned) RMSD: 1.3 Å (110 Cα aligned) |
 |
| CHCHD5 (Q9BSY4) | C12-C44, C22-C34, C58-C89, C68-C79 123456789012345678901234567890 MQAALEVTARYCGRELEQYGQCVAAKPESWQRDCHYLKMSIAQCTSSHPIIRQIRQACAQPFEAFEECLRQNEAAVGNCAEHMRRFLQCAEQVQPPRSPATVEAQPLPAS |
Code: 2LQL Technique: NMR Chain: A Residues: 1-110 |
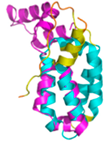 Residues 11-47 RMSD: 1.6 Å (35 Cα aligned)# Residues 11-47 RMSD: 1.6 Å (35 Cα aligned)#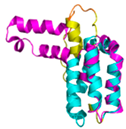 Residues 50-90 RMSD: 1.6 Å (36 Cα aligned) Residues 50-90 RMSD: 1.6 Å (36 Cα aligned) |
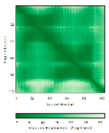 |
| CHCHD7 (Q9BUK0) |
C16-C47, C26-C37 123456789012345678901234567890 MPSVTQRLRDPDINPCLSESDASTRCLDENNYDRERCSTYFLRYKNCRRFWNSIVMQRRKNGVKPFMPTAAERDEILRAVGNMPY |
Code: 2LQT Technique: NMR Chain: A Residues: 1-85 |
 RMSD: 1.7 Å (69 Cα aligned) RMSD: 1.7 Å (69 Cα aligned) |
 |
| CHCHD10 (Q8WYQ3) | C102-C132, C112-C122 123456789012345678901234567890 MPRGSRSAASRPASRPAAPSAHPPAHPPPSAAAPAPAPSGQPGLMAQMATTAAGVAVGSAVGHVMGSALTGAFSGGSSEPSQPAVQQAPTPAAPQPLQMGPCAYEIRQFLDCSTTQSDLSLCEGFSEALKQCKYYHGLSSLP |
- | 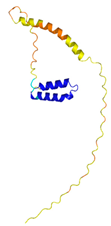 |
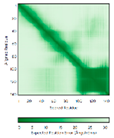 |
| CMC2 (Q9NRP2) |
C14-C47, C24-C37 123456789012345678901234567890 MHPDLSPHLHTEECNVLINLLKECHKNHNILKFFGYCNDVDRELRKCLKNEYVENRTKSREHGIAMRKKLFNPPEESEK |
- |  |
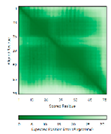 |
| CMC4 (P56277) |
C7-C38, C17-C28, C39-C50 123456789012345678901234567890 MPQKDPCQKQACEIQKCLQANSYMESKCQAVIQELRKCCAQYPKGRSVVCSGFEKEEEENLTRKSASK |
Code: 2HP8 Technique: NMR Chain: A Residues: 1-68 |
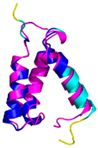 RMSD: 1.6 Å (60 Cα aligned) RMSD: 1.6 Å (60 Cα aligned) |
 |
| COA4 (Q9NYJ1) |
C34-C64, C44-C54 123456789012345678901234567890 MSTSVPQGHTWTQRVKKDDEEEDPLDQLISRSGCAASHFAVQECMAQHQDWRQCQPQVQAFKDCMSEQQARRQEELQRRQEQAGAHH |
- |  |
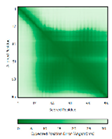 |
| COA5 (Q86WW8) |
- 123456789012345678901234567890 MPKYYEDKPQGGACAGLKEDLGACLLQSDCVVQEGKSPRQCLKEGYCNSLKYAFFECKRSVLDNRARFRGRKGY |
- | 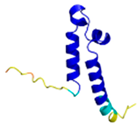 |
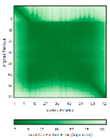 |
| COA6 (Q5JTJ3) |
C58-C90, C68-C79 123456789012345678901234567890 MGPGGPLLSPSRGFLLCKTGWHSNRLLGDCGPHTPVSTALSFIAVGMAAPSMKERQVCWGARDEYWKCLDENLEDASQCKKLRSSFESSC PQQWIKYFDKRRDYLKFKEKFEAGQFEPSETTAKS |
Code: 6PCE Technique: X-ray crystallography (Resol. 1.65 Å) Chain: A Residues: 50-119 |
 RMSD: 0.43 Å (50 Cα aligned) RMSD: 0.43 Å (50 Cα aligned) |
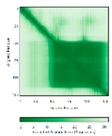 |
| COA7 (Q96BR5) | C28-C37, C62-C71, C100-C111, C142-C150, C179-C187 123456789012345678901234567890 MAGMVDFQDEEQVKSFLENMEVECNYHCYHEKDPDGCYRLVDYLEGIRKNFDEAAKVLKFNCEENQHSDSCYKLGAYYVTGKGGLTQDLK AAARCFLMACEKPGKKSIAACHNVGLLAHDGQVNEDGQPDLGKARDYYTRACDGGYTSSCFNLSAMFLQGAPGFPKDMDLACKYSMKACD LGHIWACANASRMYKLGDGVDKDEAKAEVLKNRAQQLHKEQQKGVQPLTFG |
Code: 7MQZ Technique: X-ray crystallography (Resol. 2.39 Å)Chain: A Residues: 1-231 |
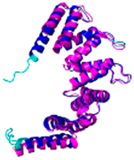 RMSD: 1.4 Å (202 Cα aligned) RMSD: 1.4 Å (202 Cα aligned) |
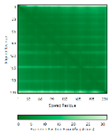 |
| COX6B1 (P14854) | C30-C65, C40-C54 123456789012345678901234567890 MAEDMETKIKNYKTAPFDSRFPNQNQTRNCWQNYLDFHRCQKAMTAKGGDISVCEWYQRVYQSLCPTSWVTDWDEQRAEGTFPGKI |
Code: 5Z62Technique: EM (Resol. 3.60 Å) Chain: H Residues: 5-86 |
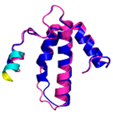 RMSD: 0.84 Å (73 Cα aligned) RMSD: 0.84 Å (73 Cα aligned) |
 |
| NDUFA8 (P51970) | C36-C66, C46-C56, C78-C110, C88-C100 123456789012345678901234567890 MPGIVELPTLEELKVDEVKISSAVLKAAAHHYGAQCDKPNKEFMLCRWEEKDPRRCLEEGKLVNKCALDFFRQIKRHCAEPFTEYWTCIDYTGQQLFRHCRKQQAKFDECVLDKLGWVRPDLGELSKVTKVKTDRPLPENPYHSRPRPDPSPEIEGDLQPATHGSRFYFWTK |
Code: 5XTD Technique: EM (Resol. 3.70 Å) Chain: u Residues: 4-172 |
 RMSD: 0.71 Å (149 Cα aligned) RMSD: 0.71 Å (149 Cα aligned) |
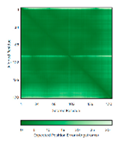 |
| NDUFB7 (P17568) | C59-C90, C69-C80 123456789012345678901234567890 MGAHLVRRYLGDASVEPDPLQMPTFPPDYGFPERKEREMVATQQEMMDAQLRLQLRDYCAHHLIRLLKCKRDSFPNFLACKQERHDWDYCEHRDYVMRMKEFERERRLLQRKKRREKKAAELAKGQGPGEVDPKVAL |
Code: 5XTD Technique: EM (Resol. 3.70 Å) Chain: v Residues: 1-137 |
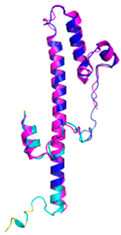 RMSD: 0.84 Å (96 Cα aligned) RMSD: 0.84 Å (96 Cα aligned) |
 |
| NDUFS5 (O43920) |
C33-C66, C43-C56 123456789012345678901234567890 MPFLDIQKRFGLNIDRWLTIQSGEQPYKMAGRCHAFEKEWIECAHGIGYTRAEKECKIEYDDFVECLLRQKTMRRAGTIRKQRDKLIKEGKYTPPPHHIGKGEPRP |
Code: 5XTD Technique: EM (Resol. 3.70 Å) Chain: h Residues: 2-105 |
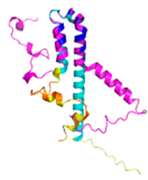 RMSD: 0.61 Å (34 Cα aligned) RMSD: 0.61 Å (34 Cα aligned) |
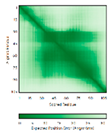 |
| NDUFS8 (O00217) |
- 123456789012345678901234567890 MRCLTTPMLLRALAQAARAGPPGGRSLHSSAVAATYKYVNMQDPEMDMKSVTDRAARTLLWTELFRGLGMTLSYLFREPATINYPFEKGP LSPRFRGEHALRRYPSGEERCIACKLCEAICPAQAITIEAEPRADGSRRTTRYDIDMTKCIYCGFCQEACPVDAIVEGPNFEFSTETHEE LLYNKEKLLNNGDKWEAEIAANIQADYLYR |
Code: 5XTD Technique: EM (Resol. 3.70 Å) Chain: B Residues: 35-210 |
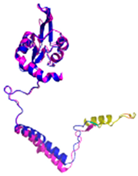 RMSD: 1.3 Å (155 Cα aligned) RMSD: 1.3 Å (155 Cα aligned) |
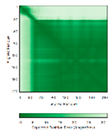 |
| p53 (P04637) |
- 123456789012345678901234567890 MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGPDEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD |
Code: 7XZZ Technique: EM (Resol. 4.07 Å) Chain: M Residues: 1-393 |
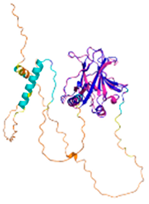 RMSD: 0.28 Å (180 Cα aligned) RMSD: 0.28 Å (180 Cα aligned) |
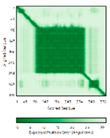 |
| PINK1 (Q9BXM7) | - 123456789012345678901234567890 MAVRQALGRGLQLGRALLLRFTGKPGRAYGLGRPGPAAGCVRGERPGWAAGPGAEPRRVGLGLPNRLRFFRQSVAGLAARLQRQFVVRAW GCAGPCGRAVFLAFGLGLGLIEEKQAESRRAVSACQEIQAIFTQKSKPGPDPLDTRRLQGFRLEEYLIGQSIGKGCSAAVYEATMPTLPQ NLEVTKSTGLLPGRGPGTSAPGEGQERAPGAPAFPLAIKMMWNISAGSSSEAILNTMSQELVPASRVALAGEYGAVTYRKSKRGPKQLAP HPNIIRVLRAFTSSVPLLPGALVDYPDVLPSRLHPEGLGHGRTLFLVMKNYPCTLRQYLCVNTPSPRLAAMMLLQLLEGVDHLVQQGIAH RDLKSDNILVELDPDGCPWLVIADFGCCLADESIGLQLPFSSWYVDRGGNGCLMAPEVSTARPGPRAVIDYSKADAWAVGAIAYEIFGLV NPFYGQGKAHLESRSYQEAQLPALPESVPPDVRQLVRALLQREASKRPSARVAANVLHLSLWGEHILALKNLKLDKMVGWLLQQSAATLL ANRLTEKCCVETKMKMLFLANLECETLCQAALLLCSWRAAL |
Code: 8UYF^ Technique: EM (Resol. 2.75 Å) Chain: A Residues: 3-463 |
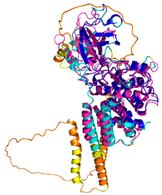 RMSD: 2.4 Å^ (314 Cα aligned) |
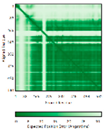 |
| TIMM8A (O60220) | C43-C66, C47-C62 123456789012345678901234567890 MDSSSSSSAAGLGAVDPQLQHFIEVETQKQRFQQLVHQMTELCWEKCMDKPGPKLDSRAEACFVNCVERFIDTSQFILNRLEQTQKSKPV FSESLSD |
- |  |
 |
| TIMM13 (Q9Y5L4) | C46-C69, C50-C65 123456789012345678901234567890 MEGGFGSDFGGSGSGKLDPGLIMEQVKVQIAVANAQELLQRMTDKCFRKCIGKPGGSLDNSEQKCIAMCMDRYMDAWNTVSRAYNSRLQR ERANM |
- |  |
 |
| TRIAP1 (O43715) |
C8-C47, C18-C37 123456789012345678901234567890 MNSVGEACTDMKREYDQCFNRWFAEKFLKGDSSGDPCTDLFKRYQQCVQKAIKEKEIPIEGLEFMGHGKEKPENSS |
Code: 6I3V Technique: X-ray crystallography (Resol. 1.98 Å) Chain: A Residues: 1-67 |
 RMSD: 1.7 Å (49 Cα aligned) |
 |
5. The Structural Basis of the hCHCHD4 Redox-Regulated Substrate Recognition
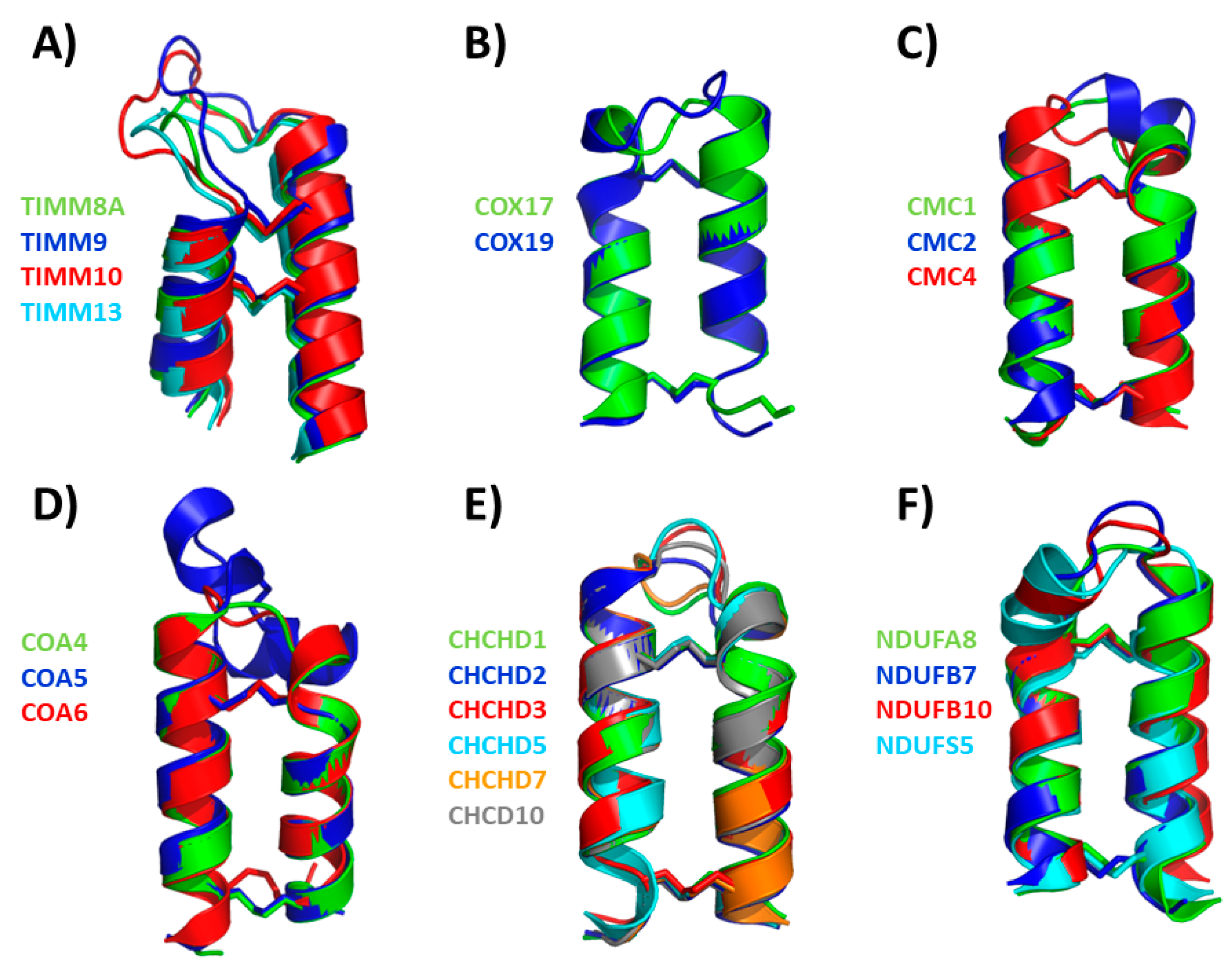
6. Extensive Analysis of the Structures of hCHCHD4 Substrates: The Contribution of AlphaFold
6.1. The TIMM Protein Family
6.2. The COX Protein Family
6.3. The CMC Protein Family
6.4. The CoA Protein Family
6.5. The CHCHD Protein Family
6.6. The NDUF_NADH Dehydrogenase Protein Family
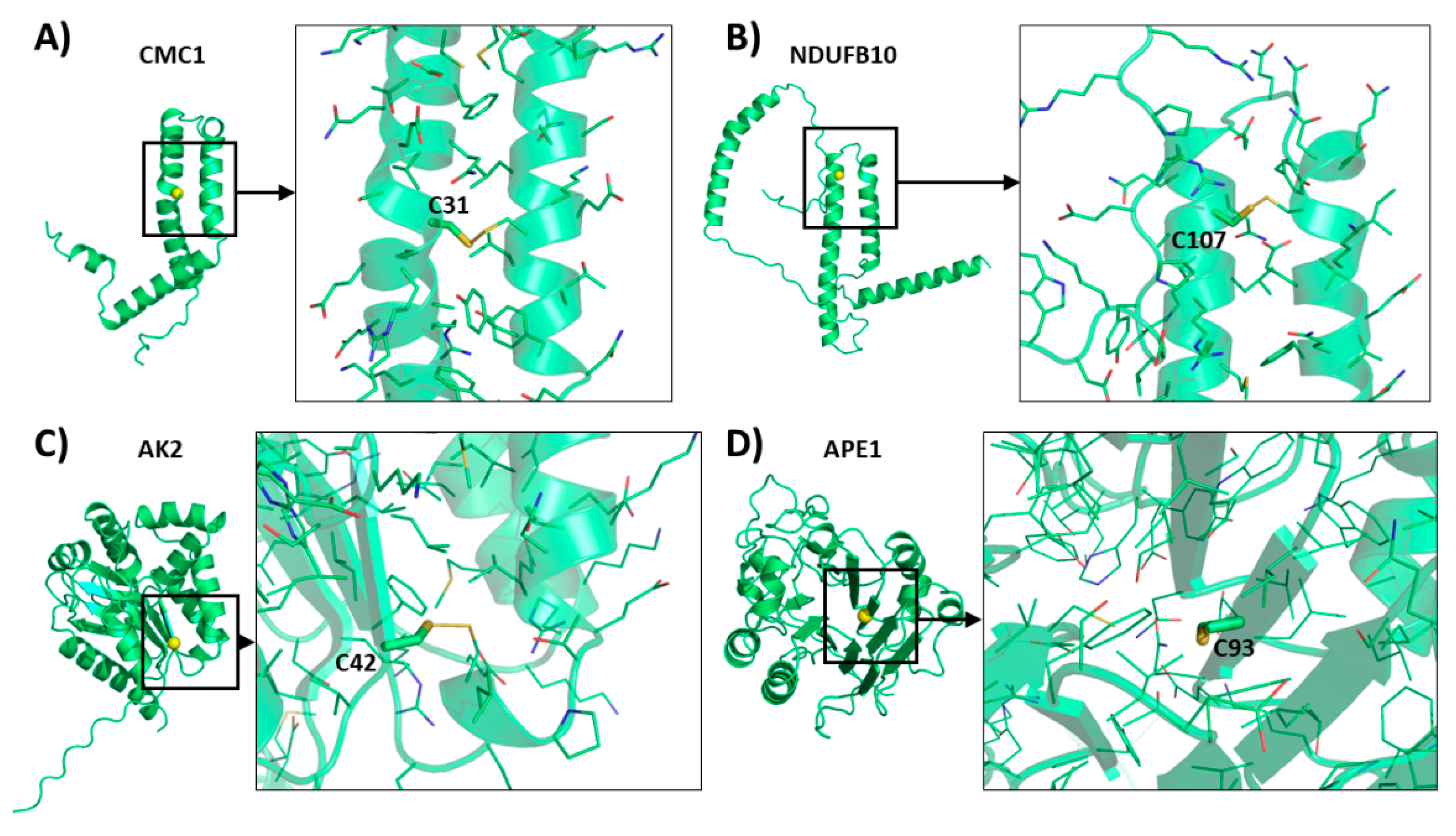
6.7. Other Substrates
7. Conclusions and Perspectives
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levy, E.D.; Erba, E.B.; Robinson, C.V.; Teichmann, S.A. Assembly Reflects Evolution of Protein Complexes. Nature 2008, 453, 1262–1265. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Zhang, W.-B. Chemical Topology and Complexity of Protein Architectures. Trends Biochem. Sci. 2018, 43, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabási, A.-L. Interactome Networks and Human Disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Arkin, M.R.; Tang, Y.; Wells, J.A. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality. Chem. Biol. 2014, 21, 1102–1114. [Google Scholar] [CrossRef]
- Nooren, I.M.A.; Thornton, J.M. Structural Characterisation and Functional Significance of Transient Protein–Protein Interactions. J. Mol. Biol. 2003, 325, 991–1018. [Google Scholar] [CrossRef]
- Mintseris, J.; Weng, Z. Structure, Function, and Evolution of Transient and Obligate Protein–Protein Interactions. Proc. Natl. Acad. Sci. 2005, 102, 10930–10935. [Google Scholar] [CrossRef]
- Ivanov, A.A.; Khuri, F.R.; Fu, H. Targeting Protein–Protein Interactions as an Anticancer Strategy. Trends Pharmacol. Sci. 2013, 34, 393–400. [Google Scholar] [CrossRef]
- Monti, A.; Vitagliano, L.; Caporale, A.; Ruvo, M.; Doti, N. Targeting Protein–Protein Interfaces with Peptides: The Contribution of Chemical Combinatorial Peptide Library Approaches. Int. J. Mol. Sci. 2023, 24, 7842. [Google Scholar] [CrossRef]
- Bechtel, T.J.; Weerapana, E. From Structure to Redox: The Diverse Functional Roles of Disulfides and Implications in Disease. PROTEOMICS 2017, 17, 1600391. [Google Scholar] [CrossRef]
- Al-Habib, H.; Ashcroft, M. CHCHD4 (MIA40) and the Mitochondrial Disulfide Relay System. Biochem. Soc. Trans. 2021, 49, 17–27. [Google Scholar] [CrossRef]
- Chatzi, A.; Manganas, P.; Tokatlidis, K. Oxidative Folding in the Mitochondrial Intermembrane Space: A Regulated Process Important for Cell Physiology and Disease. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2016, 1863, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Staples, O.; Thomas, L.W.; Briston, T.; Robson, M.; Poon, E.; Simões, M.L.; El-Emir, E.; Buffa, F.M.; Ahmed, A.; et al. Human CHCHD4 Mitochondrial Proteins Regulate Cellular Oxygen Consumption Rate and Metabolism and Provide a Critical Role in Hypoxia Signaling and Tumor Progression. J. Clin. Invest. 2012, 122, 600–611. [Google Scholar] [CrossRef]
- Zhuang, J.; Wang, P.; Huang, X.; Chen, X.; Kang, J.-G.; Hwang, P.M. Mitochondrial Disulfide Relay Mediates Translocation of P53 and Partitions Its Subcellular Activity. Proc. Natl. Acad. Sci. 2013, 110, 17356–17361. [Google Scholar] [CrossRef]
- Thomas, L.W.; Staples, O.; Turmaine, M.; Ashcroft, M. CHCHD4 Regulates Intracellular Oxygenation and Perinuclear Distribution of Mitochondria. Front. Oncol. 2017, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, T.; Xie, C.; Xu, Y.; Zhou, K.; Rodriguez, J.; Han, W.; Wang, X.; Kroemer, G.; Modjtahedi, N.; et al. Haploinsufficiency in the Mitochondrial Protein CHCHD4 Reduces Brain Injury in a Mouse Model of Neonatal Hypoxia-Ischemia. Cell Death Dis. 2017, 8, e2781–e2781. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, C.; Arena, G.; Nedara, K.; Edwards, R.; Brenner, C.; Tokatlidis, K.; Modjtahedi, N. AIF Meets the CHCHD4/Mia40-Dependent Mitochondrial Import Pathway. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 2020, 1866, 165746. [Google Scholar] [CrossRef]
- Thomas, L.W.; Stephen, J.M.; Ashcroft, M. CHCHD4 Regulates the Expression of Mitochondrial Genes That Are Essential for Tumour Cell Growth. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 2024, 1870, 167282. [Google Scholar] [CrossRef]
- Hangen, E.; Féraud, O.; Lachkar, S.; Mou, H.; Doti, N.; Fimia, G.M.; Lam, N.; Zhu, C.; Godin, I.; Muller, K.; et al. Interaction between AIF and CHCHD4 Regulates Respiratory Chain Biogenesis. Mol. Cell 2015, 58, 1001–1014. [Google Scholar] [CrossRef]
- Doti, N.; Reuther, C.; Scognamiglio, P.L.; Dolga, A.M.; Plesnila, N.; Ruvo, M.; Culmsee, C. Inhibition of the AIF/CypA Complex Protects against Intrinsic Death Pathways Induced by Oxidative Stress. Cell Death Dis. 2014, 5, e993–e993. [Google Scholar] [CrossRef]
- Modjtahedi, N.; Hangen, E.; Gonin, P.; Kroemer, G. Metabolic Epistasis among Apoptosis-Inducing Factor and the Mitochondrial Import Factor CHCHD4. Cell Cycle 2015, 14, 2743–2747. [Google Scholar] [CrossRef]
- Modjtahedi, N.; Tokatlidis, K.; Dessen, P.; Kroemer, G. Mitochondrial Proteins Containing Coiled-Coil-Helix-Coiled-Coil-Helix (CHCH) Domains in Health and Disease. Trends Biochem. Sci. 2016, 41, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Mordas, A.; Tokatlidis, K. The MIA Pathway: A Key Regulator of Mitochondrial Oxidative Protein Folding and Biogenesis. Acc. Chem. Res. 2015, 48, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, A.J.; Riemer, J. Mitochondrial Disulfide Relay and Its Substrates: Mechanisms in Health and Disease. Cell Tissue Res. 2017, 367, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Costanzo, L.D.; Christie, C.; Duarte, J.M.; Dutta, S.; et al.; wwPDB consortium Protein Data Bank: The Single Global Archive for 3D Macromolecular Structure Data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing Structure Coverage for over 214 Million Protein Sequences. Nucleic Acids Res. 2024, 52, D368–D375. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cefaro, C.; Ciofi-Baffoni, S.; Gallo, A.; Martinelli, M.; Sideris, D.P.; Katrakili, N.; Tokatlidis, K. MIA40 Is an Oxidoreductase That Catalyzes Oxidative Protein Folding in Mitochondria. Nat. Struct. Mol. Biol. 2009, 16, 198–206. [Google Scholar] [CrossRef]
- Sideris, D.P.; Petrakis, N.; Katrakili, N.; Mikropoulou, D.; Gallo, A.; Ciofi-Baffoni, S.; Banci, L.; Bertini, I.; Tokatlidis, K. A Novel Intermembrane Space–Targeting Signal Docks Cysteines onto Mia40 during Mitochondrial Oxidative Folding. J. Cell Biol. 2009, 187, 1007–1022. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cefaro, C.; Cenacchi, L.; Ciofi-Baffoni, S.; Felli, I.C.; Gallo, A.; Gonnelli, L.; Luchinat, E.; Sideris, D.; et al. Molecular Chaperone Function of Mia40 Triggers Consecutive Induced Folding Steps of the Substrate in Mitochondrial Protein Import. Proc. Natl. Acad. Sci. 2010, 107, 20190–20195. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Calderone, V.; Cefaro, C.; Ciofi-Baffoni, S.; Gallo, A.; Kallergi, E.; Lionaki, E.; Pozidis, C.; Tokatlidis, K. Molecular Recognition and Substrate Mimicry Drive the Electron-Transfer Process between MIA40 and ALR. Proc. Natl. Acad. Sci. 2011, 108, 4811–4816. [Google Scholar] [CrossRef] [PubMed]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A Local Superposition-Free Score for Comparing Protein Structures and Models Using Distance Difference Tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Dickson-Murray, E.; Nedara, K.; Modjtahedi, N.; Tokatlidis, K. The Mia40/CHCHD4 Oxidative Folding System: Redox Regulation and Signaling in the Mitochondrial Intermembrane Space. Antioxidants 2021, 10, 592. [Google Scholar] [CrossRef]
- Hofmann, S.; Rothbauer, U.; Mühlenbein, N.; Baiker, K.; Hell, K.; Bauer, M.F. Functional and Mutational Characterization of Human MIA40 Acting During Import into the Mitochondrial Intermembrane Space. J. Mol. Biol. 2005, 353, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Chacinska, A.; Guiard, B.; Müller, J.M.; Schulze-Specking, A.; Gabriel, K.; Kutik, S.; Pfanner, N. Mitochondrial Biogenesis, Switching the Sorting Pathway of the Intermembrane Space Receptor Mia40. J. Biol. Chem. 2008, 283, 29723–29729. [Google Scholar] [CrossRef]
- Chacinska, A.; Pfannschmidt, S.; Wiedemann, N.; Kozjak, V.; Sanjuán Szklarz, L.K.; Schulze-Specking, A.; Truscott, K.N.; Guiard, B.; Meisinger, C.; Pfanner, N. Essential Role of Mia40 in Import and Assembly of Mitochondrial Intermembrane Space Proteins. EMBO J. 2004, 23, 3735–3746. [Google Scholar] [CrossRef]
- Terziyska, N.; Lutz, T.; Kozany, C.; Mokranjac, D.; Mesecke, N.; Neupert, W.; Herrmann, J.M.; Hell, K. Mia40, a Novel Factor for Protein Import into the Intermembrane Space of Mitochondria Is Able to Bind Metal Ions. FEBS Lett. 2005, 579, 179–184. [Google Scholar] [CrossRef]
- Terziyska, N.; Grumbt, B.; Kozany, C.; Hell, K. Structural and Functional Roles of the Conserved Cysteine Residues of the Redox-Regulated Import Receptor Mia40 in the Intermembrane Space of Mitochondria. J. Biol. Chem. 2009, 284, 1353–1363. [Google Scholar] [CrossRef]
- Naoé, M.; Ohwa, Y.; Ishikawa, D.; Ohshima, C.; Nishikawa, S.; Yamamoto, H.; Endo, T. Identification of Tim40 That Mediates Protein Sorting to the Mitochondrial Intermembrane Space. J. Biol. Chem. 2004, 279, 47815–47821. [Google Scholar] [CrossRef]
- Susin, S.A.; Lorenzo, H.K.; Zamzami, N.; Marzo, I.; Snow, B.E.; Brothers, G.M.; Mangion, J.; Jacotot, E.; Costantini, P.; Loeffler, M.; et al. Molecular Characterization of Mitochondrial Apoptosis-Inducing Factor. Nature 1999, 397, 441–446. [Google Scholar] [CrossRef]
- Hangen, E.; Blomgren, K.; Bénit, P.; Kroemer, G.; Modjtahedi, N. Life with or without AIF. Trends Biochem. Sci. 2010, 35, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.; Mascanzoni, F.; Farina, B.; Dolga, A.M.; Monti, A.; Caporale, A.; Culmsee, C.; Fattorusso, R.; Ruvo, M.; Doti, N. Design, Optimization, and Structural Characterization of an Apoptosis-Inducing Factor Peptide Targeting Human Cyclophilin A to Inhibit Apoptosis Inducing Factor-Mediated Cell Death. J. Med. Chem. 2021, 64, 11445–11459. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Xie, C.; Li, T.; Sun, Y.; Wang, Y.; Xu, Y.; Li, K.; Zhang, S.; Zhou, K.; Wang, Y.; et al. Inhibiting the Interaction between Apoptosis-Inducing Factor and Cyclophilin A Prevents Brain Injury in Neonatal Mice after Hypoxia-Ischemia. Neuropharmacology 2020, 171, 108088. [Google Scholar] [CrossRef]
- Candé, C.; Vahsen, N.; Kouranti, I.; Schmitt, E.; Daugas, E.; Spahr, C.; Luban, J.; Kroemer, R.T.; Giordanetto, F.; Garrido, C.; et al. AIF and Cyclophilin A Cooperate in Apoptosis-Associated Chromatinolysis. Oncogene 2004, 23, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Buettner, S.; Ghezzi, D.; Zeviani, M.; Bano, D.; Nicotera, P. Loss of Apoptosis-Inducing Factor Critically Affects MIA40 Function. Cell Death Dis. 2015, 6, e1814–e1814. [Google Scholar] [CrossRef]
- Modjtahedi, N.; Kroemer, G. CHCHD4 Links AIF to the Biogenesis of Respiratory Chain Complex I. Mol. Cell. Oncol. 2016, 3, e1074332. [Google Scholar] [CrossRef]
- Brosey, C.A.; Shen, R.; Tainer, J.A. NADH-Bound AIF Activates the Mitochondrial CHCHD4/MIA40 Chaperone by a Substrate-Mimicry Mechanism. EMBO J. 2025, 44, 1220–1248. [Google Scholar] [CrossRef]
- Salscheider, S.L.; Gerlich, S.; Cabrera-Orefice, A.; Peker, E.; Rothemann, R.A.; Murschall, L.M.; Finger, Y.; Szczepanowska, K.; Ahmadi, Z.A.; Guerrero-Castillo, S.; et al. AIFM1 Is a Component of the Mitochondrial Disulfide Relay That Drives Complex I Assembly through Efficient Import of NDUFS5. EMBO J. 2022, 41, e110784. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Riemer, J. Apoptosis Inducing Factor and Mitochondrial NADH Dehydrogenases: Redox-Controlled Gear Boxes to Switch between Mitochondrial Biogenesis and Cell Death. Biol. Chem. 2021, 402, 289–297. [Google Scholar] [CrossRef]
- Murschall, L.M.; Gerhards, A.; MacVicar, T.; Peker, E.; Hasberg, L.; Wawra, S.; Langer, T.; Riemer, J. The C-Terminal Region of the Oxidoreductase MIA40 Stabilizes Its Cytosolic Precursor during Mitochondrial Import. BMC Biol. 2020, 18, 96. [Google Scholar] [CrossRef]
- Wischhof, L.; Scifo, E.; Ehninger, D.; Bano, D. AIFM1 beyond Cell Death: An Overview of This OXPHOS-Inducing Factor in Mitochondrial Diseases. eBioMedicine 2022, 83, 104231. [Google Scholar] [CrossRef] [PubMed]
- Sztolsztener, M.E.; Brewinska, A.; Guiard, B.; Chacinska, A. Disulfide Bond Formation: Sulfhydryl Oxidase ALR Controls Mitochondrial Biogenesis of Human MIA40. Traffic 2013, 14, 309–320. [Google Scholar] [CrossRef]
- Erdogan, A.J.; Ali, M.; Habich, M.; Salscheider, S.L.; Schu, L.; Petrungaro, C.; Thomas, L.W.; Ashcroft, M.; Leichert, L.I.; Roma, L.P.; et al. The Mitochondrial Oxidoreductase CHCHD4 Is Present in a Semi-Oxidized State in Vivo. Redox Biol. 2018, 17, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Horn, S.; Belkacemi, A.; Kojer, K.; Petrungaro, C.; Habich, M.; Ali, M.; Küttner, V.; Bien, M.; Kauff, F.; et al. Protein Import and Oxidative Folding in the Mitochondrial Intermembrane Space of Intact Mammalian Cells. Mol. Biol. Cell 2013, 24, 2160–2170. [Google Scholar] [CrossRef] [PubMed]
- Habich, M.; Salscheider, S.L.; Riemer, J. Cysteine Residues in Mitochondrial Intermembrane Space Proteins: More than Just Import. Br. J. Pharmacol. 2019, 176, 514–531. [Google Scholar] [CrossRef]
- Mesecke, N.; Terziyska, N.; Kozany, C.; Baumann, F.; Neupert, W.; Hell, K.; Herrmann, J.M. A Disulfide Relay System in the Intermembrane Space of Mitochondria That Mediates Protein Import. Cell 2005, 121, 1059–1069. [Google Scholar] [CrossRef]
- Backes, S.; Herrmann, J.M. Protein Translocation into the Intermembrane Space and Matrix of Mitochondria: Mechanisms and Driving Forces. Front. Mol. Biosci. 2017, 4, 83. [Google Scholar] [CrossRef]
- Herrmann, J.M.; Riemer, J. Mitochondrial Disulfide Relay: Redox-Regulated Protein Import into the Intermembrane Space. J. Biol. Chem. 2012, 287, 4426–4433. [Google Scholar] [CrossRef]
- Koehler, C.M.; Tienson, H.L. Redox Regulation of Protein Folding in the Mitochondrial Intermembrane Space. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2009, 1793, 139–145. [Google Scholar] [CrossRef]
- Busch, J.D.; Fielden, L.F.; Pfanner, N.; Wiedemann, N. Mitochondrial Protein Transport: Versatility of Translocases and Mechanisms. Mol. Cell 2023, 83, 890–910. [Google Scholar] [CrossRef]
- Thiriveedi, V.R.; Mattam, U.; Pattabhi, P.; Bisoyi, V.; Talari, N.K.; Krishnamoorthy, T.; Sepuri, N.B.V. Glutathionylated and Fe–S Cluster Containing hMIA40 (CHCHD4) Regulates ROS and Mitochondrial Complex III and IV Activities of the Electron Transport Chain. Redox Biol. 2020, 37, 101725. [Google Scholar] [CrossRef] [PubMed]
- Zarges, C.; Riemer, J. Oxidative Protein Folding in the Intermembrane Space of Human Mitochondria. FEBS Open Bio 2024, 14, 1610–1626. [Google Scholar] [CrossRef]
- Kawano, S.; Yamano, K.; Naoé, M.; Momose, T.; Terao, K.; Nishikawa, S.; Watanabe, N.; Endo, T. Structural Basis of Yeast Tim40/Mia40 as an Oxidative Translocator in the Mitochondrial Intermembrane Space. Proc. Natl. Acad. Sci. 2009, 106, 14403–14407. [Google Scholar] [CrossRef] [PubMed]
- Sideris, D.P.; Tokatlidis, K. Oxidative Protein Folding in the Mitochondrial Intermembrane Space. Antioxid. Redox Signal. 2010, 13, 1189–1204. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.; Balabanidou, V.; Sideris, D.P.; Lisowsky, T.; Tokatlidis, K. Erv1 Mediates the Mia40-Dependent Protein Import Pathway and Provides a Functional Link to the Respiratory Chain by Shuttling Electrons to Cytochrome c. J. Mol. Biol. 2005, 353, 937–944. [Google Scholar] [CrossRef]
- Milenkovic, D.; Ramming, T.; Müller, J.M.; Wenz, L.-S.; Gebert, N.; Schulze-Specking, A.; Stojanovski, D.; Rospert, S.; Chacinska, A. Identification of the Signal Directing Tim9 and Tim10 into the Intermembrane Space of Mitochondria. Mol. Biol. Cell 2009, 20, 2530–2539. [Google Scholar] [CrossRef]
- Koch, J.R.; Schmid, F.X. Mia40 Targets Cysteines in a Hydrophobic Environment to Direct Oxidative Protein Folding in the Mitochondria. Nat. Commun. 2014, 5, 3041. [Google Scholar] [CrossRef]
- Sideris, D.P.; Tokatlidis, K. Oxidative Folding of Small Tims Is Mediated by Site-specific Docking onto Mia40 in the Mitochondrial Intermembrane Space. Mol. Microbiol. 2007, 65, 1360–1373. [Google Scholar] [CrossRef]
- Milenkovic, D.; Gabriel, K.; Guiard, B.; Schulze-Specking, A.; Pfanner, N.; Chacinska, A. Biogenesis of the Essential Tim9–Tim10 Chaperone Complex of Mitochondria. J. Biol. Chem. 2007, 282, 22472–22480. [Google Scholar] [CrossRef]
- Lionaki, E.; Aivaliotis, M.; Pozidis, C.; Tokatlidis, K. The N-Terminal Shuttle Domain of Erv1 Determines the Affinity for Mia40 and Mediates Electron Transfer to the Catalytic Erv1 Core in Yeast Mitochondria. Antioxid. Redox Signal. 2010, 13, 1327–1339. [Google Scholar] [CrossRef]
- Bihlmaier, K.; Mesecke, N.; Terziyska, N.; Bien, M.; Hell, K.; Herrmann, J.M. The Disulfide Relay System of Mitochondria Is Connected to the Respiratory Chain. J. Cell Biol. 2007, 179, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Dabir, D.V.; Leverich, E.P.; Kim, S.-K.; Tsai, F.D.; Hirasawa, M.; Knaff, D.B.; Koehler, C.M. A Role for Cytochrome c and Cytochrome c Peroxidase in Electron Shuttling from Erv1. EMBO J. 2007, 26, 4801–4811. [Google Scholar] [CrossRef] [PubMed]
- Rissler, M.; Wiedemann, N.; Pfannschmidt, S.; Gabriel, K.; Guiard, B.; Pfanner, N.; Chacinska, A. The Essential Mitochondrial Protein Erv1 Cooperates with Mia40 in Biogenesis of Intermembrane Space Proteins. J. Mol. Biol. 2005, 353, 485–492. [Google Scholar] [CrossRef]
- Neal, S.E.; Dabir, D.V.; Wijaya, J.; Boon, C.; Koehler, C.M. Osm1 Facilitates the Transfer of Electrons from Erv1 to Fumarate in the Redox-Regulated Import Pathway in the Mitochondrial Intermembrane Space. Mol. Biol. Cell 2017, 28, 2773–2785. [Google Scholar] [CrossRef] [PubMed]
- Murari, A.; Thiriveedi, V.R.; Mohammad, F.; Vengaldas, V.; Gorla, M.; Tammineni, P.; Krishnamoorthy, T.; Sepuri, N.B.V. Human Mitochondrial MIA40 (CHCHD4) Is a Component of the Fe–S Cluster Export Machinery. Biochem. J. 2015, 471, 231–241. [Google Scholar] [CrossRef]
- Spiller, M.P.; Ang, S.K.; Ceh-Pavia, E.; Fisher, K.; Wang, Q.; Rigby, S.E.J.; Lu, H. Identification and Characterization of Mitochondrial Mia40 as an Iron–Sulfur Protein. Biochem. J. 2013, 455, 27–35. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Boscaro, F.; Chatzi, A.; Mikolajczyk, M.; Tokatlidis, K.; Winkelmann, J. Anamorsin Is a [2Fe-2S] Cluster-Containing Substrate of the Mia40-Dependent Mitochondrial Protein Trapping Machinery. Chem. Biol. 2011, 18, 794–804. [Google Scholar] [CrossRef]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. STRUCTURE, FUNCTION, AND FORMATION OF BIOLOGICAL IRON-SULFUR CLUSTERS. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef]
- Thomas, L.W.; Stephen, J.M.; Esposito, C.; Hoer, S.; Antrobus, R.; Ahmed, A.; Al-Habib, H.; Ashcroft, M. CHCHD4 Confers Metabolic Vulnerabilities to Tumour Cells through Its Control of the Mitochondrial Respiratory Chain. Cancer Metab. 2019, 7, 2. [Google Scholar] [CrossRef]
- Thomas, L.W.; Esposito, C.; Stephen, J.M.; Costa, A.S.H.; Frezza, C.; Blacker, T.S.; Szabadkai, G.; Ashcroft, M. CHCHD4 Regulates Tumour Proliferation and EMT-Related Phenotypes, through Respiratory Chain-Mediated Metabolism. Cancer Metab. 2019, 7, 7. [Google Scholar] [CrossRef]
- Longen, S.; Bien, M.; Bihlmaier, K.; Kloeppel, C.; Kauff, F.; Hammermeister, M.; Westermann, B.; Herrmann, J.M.; Riemer, J. Systematic Analysis of the Twin Cx9C Protein Family. J. Mol. Biol. 2009, 393, 356–368. [Google Scholar] [CrossRef]
- Cavallaro, G. Genome-Wide Analysis of Eukaryotic Twin CX9C Proteins. Mol. Biosyst. 2010, 6, 2459. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Jaiswal, D.; Neri, S.; Peruzzini, R.; Winkelmann, J. Structural Characterization of CHCHD5 and CHCHD7: Two Atypical Human Twin CX9C Proteins. J. Struct. Biol. 2012, 180, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Riemer, J. The Mitochondrial Disulfide Relay System: Roles in Oxidative Protein Folding and Beyond. Int. J. Cell Biol. 2013, 2013, 1–12. [Google Scholar] [CrossRef]
- Habich, M.; Salscheider, S.L.; Murschall, L.M.; Hoehne, M.N.; Fischer, M.; Schorn, F.; Petrungaro, C.; Ali, M.; Erdogan, A.J.; Abou-Eid, S.; et al. Vectorial Import via a Metastable Disulfide-Linked Complex Allows for a Quality Control Step and Import by the Mitochondrial Disulfide Relay. Cell Rep. 2019, 26, 759–774.e5. [Google Scholar] [CrossRef]
- Barchiesi, A.; Wasilewski, M.; Chacinska, A.; Tell, G.; Vascotto, C. Mitochondrial Translocation of APE1 Relies on the MIA Pathway. Nucleic Acids Res. 2015, 43, 5451–5464. [Google Scholar] [CrossRef] [PubMed]
- Reddehase, S.; Grumbt, B.; Neupert, W.; Hell, K. The Disulfide Relay System of Mitochondria Is Required for the Biogenesis of Mitochondrial Ccs1 and Sod1. J. Mol. Biol. 2009, 385, 331–338. [Google Scholar] [CrossRef]
- Klöppel, C.; Suzuki, Y.; Kojer, K.; Petrungaro, C.; Longen, S.; Fiedler, S.; Keller, S.; Riemer, J. Mia40-Dependent Oxidation of Cysteines in Domain I of Ccs1 Controls Its Distribution between Mitochondria and the Cytosol. Mol. Biol. Cell 2011, 22, 3749–3757. [Google Scholar] [CrossRef]
- Groß, D.P.; Burgard, C.A.; Reddehase, S.; Leitch, J.M.; Culotta, V.C.; Hell, K. Mitochondrial Ccs1 Contains a Structural Disulfide Bond Crucial for the Import of This Unconventional Substrate by the Disulfide Relay System. Mol. Biol. Cell 2011, 22, 3758–3767. [Google Scholar] [CrossRef]
- Friederich, M.W.; Erdogan, A.J.; Coughlin, C.R.; Elos, M.T.; Jiang, H.; O’Rourke, C.P.; Lovell, M.A.; Wartchow, E.; Gowan, K.; Chatfield, K.C.; et al. Mutations in the Accessory Subunit NDUFB10 Result in Isolated Complex I Deficiency and Illustrate the Critical Role of Intermembrane Space Import for Complex I Holoenzyme Assembly. Hum. Mol. Genet. 2016, ddw431. [Google Scholar] [CrossRef]
- Finger, Y.; Habich, M.; Gerlich, S.; Urbanczyk, S.; Van De Logt, E.; Koch, J.; Schu, L.; Lapacz, K.J.; Ali, M.; Petrungaro, C.; et al. Proteasomal Degradation Induced by DPP9-mediated Processing Competes with Mitochondrial Protein Import. EMBO J. 2020, 39, e103889. [Google Scholar] [CrossRef]
- Peleh, V.; Riemer, J.; Dancis, A.; Herrmann, J. . Protein Oxidation in the Intermembrane Space of Mitochondria Is Substrate-Specific Rather than General. Microb. Cell 2014, 1, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, L.; Gornicka, A.; Czerwik, T.; Bragoszewski, P.; Loniewska-Lwowska, A.; Schulze-Specking, A.; Truscott, K.N.; Guiard, B.; Milenkovic, D.; Chacinska, A. In Vivo Evidence for Cooperation of Mia40 and Erv1 in the Oxidation of Mitochondrial Proteins. Mol. Biol. Cell 2012, 23, 3957–3969. [Google Scholar] [CrossRef] [PubMed]
- Bourens, M.; Dabir, D.V.; Tienson, H.L.; Sorokina, I.; Koehler, C.M.; Barrientos, A. Role of Twin Cys-Xaa9-Cys Motif Cysteines in Mitochondrial Import of the Cytochrome c Oxidase Biogenesis Factor Cmc1. J. Biol. Chem. 2012, 287, 31258–31269. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Calderone, V.; Cefaro, C.; Ciofi-Baffoni, S.; Gallo, A.; Tokatlidis, K. An Electron-Transfer Path through an Extended Disulfide Relay System: The Case of the Redox Protein ALR. J. Am. Chem. Soc. 2012, 134, 1442–1445. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, E.; Andreadaki, M.; Kritsiligkou, P.; Katrakili, N.; Pozidis, C.; Tokatlidis, K.; Banci, L.; Bertini, I.; Cefaro, C.; Ciofi-Baffoni, S.; et al. Targeting and Maturation of Erv1/ALR in the Mitochondrial Intermembrane Space. ACS Chem. Biol. 2012, 7, 707–714. [Google Scholar] [CrossRef]
- Weckbecker, D.; Longen, S.; Riemer, J.; Herrmann, J.M. Atp23 Biogenesis Reveals a Chaperone-like Folding Activity of Mia40 in the IMS of Mitochondria: Chaperone-like Activity of Mia40 in Atp23 Folding. EMBO J. 2012, 31, 4348–4358. [Google Scholar] [CrossRef]
- Longen, S.; Woellhaf, M.W.; Petrungaro, C.; Riemer, J.; Herrmann, J.M. The Disulfide Relay of the Intermembrane Space Oxidizes the Ribosomal Subunit Mrp10 on Its Transit into the Mitochondrial Matrix. Dev. Cell 2014, 28, 30–42. [Google Scholar] [CrossRef]
- Vascotto, C.; Bisetto, E.; Li, M.; Zeef, L.A.H.; D’Ambrosio, C.; Domenis, R.; Comelli, M.; Delneri, D.; Scaloni, A.; Altieri, F.; et al. Knock-in Reconstitution Studies Reveal an Unexpected Role of Cys-65 in Regulating APE1/Ref-1 Subcellular Trafficking and Function. Mol. Biol. Cell 2011, 22, 3887–3901. [Google Scholar] [CrossRef]
- Petrungaro, C.; Zimmermann, K.M.; Küttner, V.; Fischer, M.; Dengjel, J.; Bogeski, I.; Riemer, J. The Ca2+-Dependent Release of the Mia40-Induced MICU1-MICU2 Dimer from MCU Regulates Mitochondrial Ca2+ Uptake. Cell Metab. 2015, 22, 721–733. [Google Scholar] [CrossRef]
- Wrobel, L.; Trojanowska, A.; Sztolsztener, M.E.; Chacinska, A. Mitochondrial Protein Import: Mia40 Facilitates Tim22 Translocation into the Inner Membrane of Mitochondria. Mol. Biol. Cell 2013, 24, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Aras, S.; Bai, M.; Lee, I.; Springett, R.; Hüttemann, M.; Grossman, L.I. MNRR1 (Formerly CHCHD2) Is a Bi-Organellar Regulator of Mitochondrial Metabolism. Mitochondrion 2015, 20, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Darshi, M.; Trinh, K.N.; Murphy, A.N.; Taylor, S.S. Targeting and Import Mechanism of Coiled-Coil Helix Coiled-Coil Helix Domain-Containing Protein 3 (ChChd3) into the Mitochondrial Intermembrane Space. J. Biol. Chem. 2012, 287, 39480–39491. [Google Scholar] [CrossRef] [PubMed]
- Sakowska, P.; Jans, D.C.; Mohanraj, K.; Riedel, D.; Jakobs, S.; Chacinska, A. The Oxidation Status of Mic19 Regulates MICOS Assembly. Mol. Cell. Biol. 2015, 35, 4222–4237. [Google Scholar] [CrossRef]
- Wang, T.; Liu, H.; Itoh, K.; Oh, S.; Zhao, L.; Murata, D.; Sesaki, H.; Hartung, T.; Na, C.H.; Wang, J. C9orf72 Regulates Energy Homeostasis by Stabilizing Mitochondrial Complex I Assembly. Cell Metab. 2021, 33, 531–546.e9. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, T.; Ji, Y.J.; Johnson, K.; Liu, H.; Johnson, K.; Bailey, S.; Suk, Y.; Lu, Y.-N.; Liu, M.; et al. A C9orf72–CARM1 Axis Regulates Lipid Metabolism under Glucose Starvation-Induced Nutrient Stress. Genes Dev. 2018, 32, 1380–1397. [Google Scholar] [CrossRef]
- Lehmer, C.; Schludi, M.H.; Ransom, L.; Greiling, J.; Junghänel, M.; Exner, N.; Riemenschneider, H.; Van Der Zee, J.; Van Broeckhoven, C.; Weydt, P.; et al. A Novel CHCHD10 Mutation Implicates a Mia40-dependent Mitochondrial Import Deficit in ALS. EMBO Mol. Med. 2018, 10, e8558. [Google Scholar] [CrossRef]
- Mohanraj, K.; Wasilewski, M.; Benincá, C.; Cysewski, D.; Poznanski, J.; Sakowska, P.; Bugajska, Z.; Deckers, M.; Dennerlein, S.; Fernandez-Vizarra, E.; et al. Inhibition of Proteasome Rescues a Pathogenic Variant of Respiratory Chain Assembly Factor COA7. EMBO Mol. Med. 2019, 11, e9561. [Google Scholar] [CrossRef]
- Harada, H.; Moriya, K.; Kobuchi, H.; Ishihara, N.; Utsumi, T. Protein N-Myristoylation Plays a Critical Role in the Mitochondrial Localization of Human Mitochondrial Complex I Accessory Subunit NDUFB7. Sci. Rep. 2023, 13, 22991. [Google Scholar] [CrossRef]
- Zhuang, J.; Kamp, W.M.; Li, J.; Liu, C.; Kang, J.-G.; Wang, P.; Hwang, P.M. Forkhead Box O3A (FOXO3) and the Mitochondrial Disulfide Relay Carrier (CHCHD4) Regulate P53 Protein Nuclear Activity in Response to Exercise. J. Biol. Chem. 2016, 291, 24819–24827. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, Y.; Hou, X.; Tao, Z.; Ren, H.; Wang, G. Dependence of PINK1 Accumulation on Mitochondrial Redox System. Aging Cell 2020, 19, e13211. [Google Scholar] [CrossRef] [PubMed]
- Brunelli, F.; Valente, E.M.; Arena, G. Mechanisms of Neurodegeneration in Parkinson’s Disease: Keep Neurons in the PINK1. Mech. Ageing Dev. 2020, 189, 111277. [Google Scholar] [CrossRef]
- Pujols, J.; Fornt-Suñé, M.; Gil-García, M.; Bartolomé-Nafría, A.; Canals, F.; Cerofolini, L.; Teilum, K.; Banci, L.; Esperante, S.A.; Ventura, S. MIA40 Circumvents the Folding Constraints Imposed by TRIAP1 Function. J. Biol. Chem. 2025, 301, 108268. [Google Scholar] [CrossRef] [PubMed]
- Hasson, S.A.; Damoiseaux, R.; Glavin, J.D.; Dabir, D.V.; Walker, S.S.; Koehler, C.M. Substrate Specificity of the TIM22 Mitochondrial Import Pathway Revealed with Small Molecule Inhibitor of Protein Translocation. Proc. Natl. Acad. Sci. 2010, 107, 9578–9583. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cefaro, C.; Cenacchi, L.; Ciofi-Baffoni, S.; Felli, I.C.; Gallo, A.; Gonnelli, L.; Luchinat, E.; Sideris, D.; et al. Molecular Chaperone Function of Mia40 Triggers Consecutive Induced Folding Steps of the Substrate in Mitochondrial Protein Import. Proc. Natl. Acad. Sci. 2010, 107, 20190–20195. [Google Scholar] [CrossRef]
- Fraga, H.; Bech-Serra, J.-J.; Canals, F.; Ortega, G.; Millet, O.; Ventura, S. The Mitochondrial Intermembrane Space Oxireductase Mia40 Funnels the Oxidative Folding Pathway of the Cytochrome c Oxidase Assembly Protein Cox19. J. Biol. Chem. 2014, 289, 9852–9864. [Google Scholar] [CrossRef]
- Bien, M.; Longen, S.; Wagener, N.; Chwalla, I.; Herrmann, J.M.; Riemer, J. Mitochondrial Disulfide Bond Formation Is Driven by Intersubunit Electron Transfer in Erv1 and Proofread by Glutathione. Mol. Cell 2010, 37, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Koch, J.R.; Schmid, F.X. Mia40 Combines Thiol Oxidase and Disulfide Isomerase Activity to Efficiently Catalyze Oxidative Folding in Mitochondria. J. Mol. Biol. 2014, 426, 4087–4098. [Google Scholar] [CrossRef]
- Fraga, H.; Pujols, J.; Gil-Garcia, M.; Roque, A.; Bernardo-Seisdedos, G.; Santambrogio, C.; Bech-Serra, J.-J.; Canals, F.; Bernadó, P.; Grandori, R.; et al. Disulfide Driven Folding for a Conditionally Disordered Protein. Sci. Rep. 2017, 7, 16994. [Google Scholar] [CrossRef]
- Thangamani, S.; Maland, M.; Mohammad, H.; Pascuzzi, P.E.; Avramova, L.; Koehler, C.M.; Hazbun, T.R.; Seleem, M.N. Repurposing Approach Identifies Auranofin with Broad Spectrum Antifungal Activity That Targets Mia40-Erv1 Pathway. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Dabir, D.V.; Hasson, S.A.; Setoguchi, K.; Johnson, M.E.; Wongkongkathep, P.; Douglas, C.J.; Zimmerman, J.; Damoiseaux, R.; Teitell, M.A.; Koehler, C.M. A Small Molecule Inhibitor of Redox-Regulated Protein Translocation into Mitochondria. Dev. Cell 2013, 25, 81–92. [Google Scholar] [CrossRef]
- Pacheu-Grau, D.; Wasilewski, M.; Oeljeklaus, S.; Gibhardt, C.S.; Aich, A.; Chudenkova, M.; Dennerlein, S.; Deckers, M.; Bogeski, I.; Warscheid, B.; et al. COA6 Facilitates Cytochrome c Oxidase Biogenesis as Thiol-Reductase for Copper Metallochaperones in Mitochondria. J. Mol. Biol. 2020, 432, 2067–2079. [Google Scholar] [CrossRef]
- Stroud, D.A.; Maher, M.J.; Lindau, C.; Vögtle, F.-N.; Frazier, A.E.; Surgenor, E.; Mountford, H.; Singh, A.P.; Bonas, M.; Oeljeklaus, S.; et al. COA6 Is a Mitochondrial Complex IV Assembly Factor Critical for Biogenesis of mtDNA-Encoded COX2. Hum. Mol. Genet. 2015, 24, 5404–5415. [Google Scholar] [CrossRef]
- Formosa, L.E.; Maghool, S.; Sharpe, A.J.; Reljic, B.; Muellner-Wong, L.; Stroud, D.A.; Ryan, M.T.; Maher, M.J. Mitochondrial COA7 Is a Heme-Binding Protein with Disulfide Reductase Activity, Which Acts in the Early Stages of Complex IV Assembly. Proc. Natl. Acad. Sci. 2022, 119, e2110357119. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, K.; Milenkovic, D.; Chacinska, A.; Müller, J.; Guiard, B.; Pfanner, N.; Meisinger, C. Novel Mitochondrial Intermembrane Space Proteins as Substrates of the MIA Import Pathway. J. Mol. Biol. 2007, 365, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Gladyck, S.; Aras, S.; Hüttemann, M.; Grossman, L.I. Regulation of COX Assembly and Function by Twin CX9C Proteins—Implications for Human Disease. Cells 2021, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Vellanki, R.N.; Coyaud, É.; Ignatchenko, V.; Li, L.; Krieger, J.R.; Taylor, P.; Tong, J.; Pham, N.-A.; Liu, G.; et al. CHCHD2 Is Coamplified with EGFR in NSCLC and Regulates Mitochondrial Function and Cell Migration. Mol. Cancer Res. 2015, 13, 1119–1129. [Google Scholar] [CrossRef]
- Dzeja, P.; Terzic, A. Adenylate Kinase and AMP Signaling Networks: Metabolic Monitoring, Signal Communication and Body Energy Sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef]
- Yoshida, Y.; Izumi, H.; Torigoe, T.; Ishiguchi, H.; Itoh, H.; Kang, D.; Kohno, K. P53 Physically Interacts with Mitochondrial Transcription Factor A and Differentially Regulates Binding to Damaged DNA. Cancer Res. 2003, 63, 3729–3734. [Google Scholar]
- Singh, K.; Kulawiec, M.; Ayyasamy, V. P53 Regulates mtDNA Copy Number and Mitocheckpoint Pathway. J. Carcinog. 2009, 8, 8. [Google Scholar] [CrossRef]
- Hainaut, P.; Mann, K. Zinc Binding and Redox Control of P53 Structure and Function. Antioxid. Redox Signal. 2001, 3, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Zavileyskiy, L.; Bunik, V. Regulation of P53 Function by Formation of Non-Nuclear Heterologous Protein Complexes. Biomolecules 2022, 12, 327. [Google Scholar] [CrossRef]
- Miliara, X.; Tatsuta, T.; Berry, J.-L.; Rouse, S.L.; Solak, K.; Chorev, D.S.; Wu, D.; Robinson, C.V.; Matthews, S.; Langer, T. Structural Determinants of Lipid Specificity within Ups/PRELI Lipid Transfer Proteins. Nat. Commun. 2019, 10, 1130. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, M.J.; Friedman, J.R.; Osman, C.; Salin, B.; Di Rago, J.-P.; Nunnari, J.; Langer, T.; Tatsuta, T. MICOS and Phospholipid Transfer by Ups2–Mdm35 Organize Membrane Lipid Synthesis in Mitochondria. J. Cell Biol. 2016, 213, 525–534. [Google Scholar] [CrossRef]
- Claypool, S.M.; Koehler, C.M. The Complexity of Cardiolipin in Health and Disease. Trends Biochem. Sci. 2012, 37, 32–41. [Google Scholar] [CrossRef]
- Dudek, J. Role of Cardiolipin in Mitochondrial Signaling Pathways. Front. Cell Dev. Biol. 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, T.; Langer, T. Intramitochondrial Phospholipid Trafficking. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids 2017, 1862, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Nedara, K.; Reinhardt, C.; Lebraud, E.; Arena, G.; Gracia, C.; Buard, V.; Pioche-Durieu, C.; Castelli, F.; Colsch, B.; Bénit, P.; et al. Relevance of the TRIAP1/P53 Axis in Colon Cancer Cell Proliferation and Adaptation to Glutamine Deprivation. Front. Oncol. 2022, 12, 958155. [Google Scholar] [CrossRef]
- Park, W.-R.; Nakamura, Y. p53CSV, a Novel P53-Inducible Gene Involved in the P53-Dependent Cell-Survival Pathway. Cancer Res. 2005, 65, 1197–1206. [Google Scholar] [CrossRef]
- Potting, C.; Tatsuta, T.; König, T.; Haag, M.; Wai, T.; Aaltonen, M.J.; Langer, T. TRIAP1/PRELI Complexes Prevent Apoptosis by Mediating Intramitochondrial Transport of Phosphatidic Acid. Cell Metab. 2013, 18, 287–295. [Google Scholar] [CrossRef]
- Miliara, X.; Garnett, J.A.; Tatsuta, T.; Abid Ali, F.; Baldie, H.; Pérez-Dorado, I.; Simpson, P.; Yague, E.; Langer, T.; Matthews, S. Structural Insight into the TRIAP 1/ PRELI -like Domain Family of Mitochondrial Phospholipid Transfer Complexes. EMBO Rep. 2015, 16, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, P.; Zhuang, J.; Son, A.Y.; Karius, A.K.; Syed, A.M.; Nishi, M.; Wu, Z.; Mori, M.P.; Kim, Y.-C.; et al. CHCHD4-TRIAP1 Regulation of Innate Immune Signaling Mediates Skeletal Muscle Adaptation to Exercise. Cell Rep. 2024, 43, 113626. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; M. Wilson Iii, D. Overview of Base Excision Repair Biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef]
- Barchiesi, A.; Bazzani, V.; Jabczynska, A.; Borowski, L.S.; Oeljeklaus, S.; Warscheid, B.; Chacinska, A.; Szczesny, R.J.; Vascotto, C. DNA Repair Protein APE1 Degrades Dysfunctional Abasic mRNA in Mitochondria Affecting Oxidative Phosphorylation. J. Mol. Biol. 2021, 433, 167125. [Google Scholar] [CrossRef] [PubMed]
- Arena, G.; Modjtahedi, N.; Kruger, R. Exploring the Contribution of the Mitochondrial Disulfide Relay System to Parkinson’s Disease: The PINK1/CHCHD4 Interplay. Neural Regen. Res. 2021, 16, 2222. [Google Scholar] [CrossRef]
- Sekine, S.; Youle, R.J. PINK1 Import Regulation; a Fine System to Convey Mitochondrial Stress to the Cytosol. BMC Biol. 2018, 16, 2. [Google Scholar] [CrossRef]
- Sekine, S.; Wang, C.; Sideris, D.P.; Bunker, E.; Zhang, Z.; Youle, R.J. Reciprocal Roles of Tom7 and OMA1 during Mitochondrial Import and Activation of PINK1. Mol. Cell 2019, 73, 1028–1043.e5. [Google Scholar] [CrossRef]
- Antony, A.N.; Paillard, M.; Moffat, C.; Juskeviciute, E.; Correnti, J.; Bolon, B.; Rubin, E.; Csordás, G.; Seifert, E.L.; Hoek, J.B.; et al. MICU1 Regulation of Mitochondrial Ca2+ Uptake Dictates Survival and Tissue Regeneration. Nat. Commun. 2016, 7, 10955. [Google Scholar] [CrossRef]
- Terziyska, N.; Grumbt, B.; Bien, M.; Neupert, W.; Herrmann, J.M.; Hell, K. The Sulfhydryl Oxidase Erv1 Is a Substrate of the Mia40-dependent Protein Translocation Pathway. FEBS Lett. 2007, 581, 1098–1102. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cefaro, C.; Ciofi-Baffoni, S.; Gajda, K.; Felli, I.C.; Gallo, A.; Pavelkova, A.; Kallergi, E.; Andreadaki, M.; et al. An Intrinsically Disordered Domain Has a Dual Function Coupled to Compartment-Dependent Redox Control. J. Mol. Biol. 2013, 425, 594–608. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Cefaro, C.; Cenacchi, L.; Ciofi-Baffoni, S.; Felli, I.C.; Gallo, A.; Gonnelli, L.; Luchinat, E.; Sideris, D.; et al. Molecular Chaperone Function of Mia40 Triggers Consecutive Induced Folding Steps of the Substrate in Mitochondrial Protein Import. Proc. Natl. Acad. Sci. 2010, 107, 20190–20195. [Google Scholar] [CrossRef] [PubMed]
- Fagnani, E.; Cocomazzi, P.; Pellegrino, S.; Tedeschi, G.; Scalvini, F.G.; Cossu, F.; Da Vela, S.; Aliverti, A.; Mastrangelo, E.; Milani, M. CHCHD4 Binding Affects the Active Site of Apoptosis Inducing Factor (AIF): Structural Determinants for Allosteric Regulation. Structure 2024, 32, 594–602.e4. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.T.; Gorman, M.A.; Lazarou, M.; Ryan, M.T.; Gulbis, J.M. Crystal Structure of the Mitochondrial Chaperone TIM9.10 Reveals a Six-Bladed Alpha-Propeller. Mol. Cell 2006, 21, 123–133. [Google Scholar] [CrossRef]
- Zong, S.; Wu, M.; Gu, J.; Liu, T.; Guo, R.; Yang, M. Structure of the Intact 14-Subunit Human Cytochrome c Oxidase. Cell Res. 2018, 28, 1026–1034. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Janicka, A.; Martinelli, M.; Kozlowski, H.; Palumaa, P. A Structural-Dynamical Characterization of Human Cox17. J. Biol. Chem. 2008, 283, 7912–7920. [Google Scholar] [CrossRef] [PubMed]
- Barthe, P.; Yang, Y.S.; Chiche, L.; Hoh, F.; Strub, M.P.; Guignard, L.; Soulier, J.; Stern, M.H.; van Tilbeurgh, H.; Lhoste, J.M.; et al. Solution Structure of Human p8MTCP1, a Cysteine-Rich Protein Encoded by the MTCP1 Oncogene, Reveals a New Alpha-Helical Assembly Motif. J. Mol. Biol. 1997, 274, 801–815. [Google Scholar] [CrossRef]
- Maghool, S.; Cooray, N.D.G.; Stroud, D.A.; Aragão, D.; Ryan, M.T.; Maher, M.J. Structural and Functional Characterization of the Mitochondrial Complex IV Assembly Factor Coa6. Life Sci. Alliance 2019, 2, e201900458. [Google Scholar] [CrossRef]
- Guo, R.; Zong, S.; Wu, M.; Gu, J.; Yang, M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170, 1247–1257.e12. [Google Scholar] [CrossRef]
- Fantini, D.; Vascotto, C.; Marasco, D.; D’Ambrosio, C.; Romanello, M.; Vitagliano, L.; Pedone, C.; Poletto, M.; Cesaratto, L.; Quadrifoglio, F.; et al. Critical Lysine Residues within the Overlooked N-Terminal Domain of Human APE1 Regulate Its Biological Functions. Nucleic Acids Res. 2010, 38, 8239–8256. [Google Scholar] [CrossRef]
- Wu, W.; Shen, Q.; Zhang, R.; Qiu, Z.; Wang, Y.; Zheng, J.; Jia, Z. The Structure of the MICU 1- MICU 2 Complex Unveils the Regulation of the Mitochondrial Calcium Uniporter. EMBO J. 2020, 39, e104285. [Google Scholar] [CrossRef]
- Esposito, L.; Balasco, N.; Smaldone, G.; Berisio, R.; Ruggiero, A.; Vitagliano, L. AlphaFold-Predicted Structures of KCTD Proteins Unravel Previously Undetected Relationships among the Members of the Family. Biomolecules 2021, 11, 1862. [Google Scholar] [CrossRef] [PubMed]
- Esposito, L.; Balasco, N.; Vitagliano, L. Alphafold Predictions Provide Insights into the Structural Features of the Functional Oligomers of All Members of the KCTD Family. Int. J. Mol. Sci. 2022, 23, 13346. [Google Scholar] [CrossRef] [PubMed]
| PUBMED QUERY | TOTAL PAPERS |
| CHCHD4 SUBSTRATES | 25 |
| CHCHD4 OXIDOREDUCTASE | 53 |
| CHCHD4 REDOX | 50 |
| MIA40 SUBSTRATES | 89 |
| MIA40 OXIDOREDUCTASE | 118 |
| MIA40 REDOX | 113 |
| Symbol | UniprotKB | Yeast homolog |
Length (aa) |
The recognized cysteine motif (CXnC)2 |
Docked Cysteine residue on the substrates | Reference |
| GROUP I | ||||||
| ALR | P55789 | Erv2 | 205 | CX2C-X15-CX16C | C71 or C74 | [31] |
| COX17 | Q14061 | Cox17 | 67 | CX9C-X8-CX9C | C45 | [30] |
| TIMM9 | Q9Y5J7 | Tim9 | 89 | CX3C-X15-CX3C |
*C35 **C28 |
[30] |
| TIMM10 | P62072 | Tim10 | 90 | CX3C-X16-CX3C |
*C35 **C29 |
[30] |
| GROUP IIa | ||||||
| AK2 | P54819 | - | 239 | - | C42 | [91] |
| APE1 | P27695 | - | 318 | - | C93 | [86,99] |
| CHCHD2 (MNRR1) |
Q9Y6H1 | Mix17 | 151 | CX9C-X9-CX9C | C144 | [48,102] |
| CHCHD3 (MIC19) |
Q9NX63 | Mic19 | 227 | CX9C-X10-CX9C | C193 | [48,85,103,104] |
| CMC1 | Q7Z7K0 | Cmc1 | 106 | CX9C-X11-CX9C |
*C42 **C31 |
[85,94] |
| COX19 | Q49B96 | Cox19 | 90 | CX9C-X10-CX9C | C51 | [22] |
| MICU1 | Q9BPX6 | - | 476 | - |
*C463 **C463 |
[100] |
| NDUFB10 | O96000 | - | 172 | CX6C-X29CX11C | C107 | [90] |
| GROUP IIb | ||||||
| Anamorsin | Q6FI81 | Dre2 | 312 | - | - | [77] |
| C9orf72 | Q96LT7 | - | 481 | CX2C-X29-CX3C | - | [105,106] |
| CHCHD1 | Q96BP2 | Mrp10 | 118 | CX9C-X10-CX9C | - | [98] |
| CHCHD5 | Q9BSY4 | Mic14 | 110 | CX9C-X11-CX9C-X13-CX9C-X10-CX9C | - | [83,85] |
| CHCHD7 (COX23) | Q9BUK0 | Cox23 | 85 | CX9C-X10-CX9C | - | [83] |
| CHCHD10 | Q8WYQ3 | Mic17 | 142 | CX9C-X9-CX9C | - | [107] |
| CMC2 | Q9NRP2 | Cmc2 | 79 | CX9C-X12-CX9C | - | [85] |
| CMC4 | P56277 | Cmc4 | 68 | CX9C-X9-CX9C-CX10C | - | [85] |
| COA4 (CMC3) |
Q9NYJ1 | Coa4 | 87 | CX9C-X9-CX9C | - | [48,54,85] |
| COA5 (PET191, C2orf64) | Q86WW8 | Pet191 | 74 | CX9C-X22-CX9C CX9C-X5-CX9C-X5-CX9C |
- | [85] |
| COA6 (C1orf31) | Q5JTJ3 | Coa6 | 125 | CX9C-X10-CX10C | - | [85] |
| COA7 | Q96BR5 | - | 231 | - | - | [108] |
| Cox6B1 | P14854 | Cox12 | 86 | CX9C-X13-CX10C | - | [85] |
| NDUFA8 | P51970 | - | 172 | CX9C-X9-CX9C-X11-CX9C-X10-CX9C | - | [54] |
| NDUFB7 | P17568 | - | 137 | CX9C-X10-CX9C | - | [46,85] |
| NDUFS5 | O43920 | - | 106 | CX9C-X12-CX9C | - | [48,85] |
| NDUFS8 | O00217 | - | 210 | CX9C-X28-CX9C | - | [85] |
| p53 | P04637 | - | 393 | CX5C-X134-CX2C | - | [110] |
| PINK1 | Q9BXM7 | - | 581 | - | - | [111,112] |
| TIMM8A | O60220 | Tim8 | 97 | CX3C-X14-CX3C | - | [34] |
| TIMM13 | Q9Y5L4 | Tim13 | 95 | CX3C-X15-CX3C | - | [85] |
| TRIAP1 (MDM35) | O43715 | Mdm35 | 76 | CX9C-X18-CX9C | - | [113] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





