Submitted:
21 March 2025
Posted:
21 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Related Work
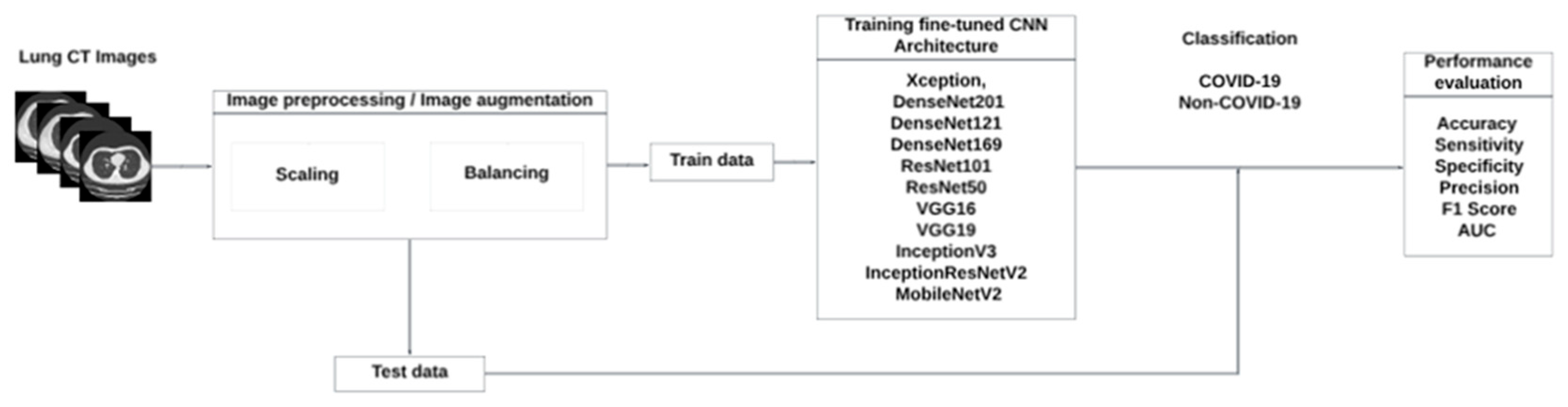
2. Materials and Methods
2.1. Proposed Approach
2.2. Datasets
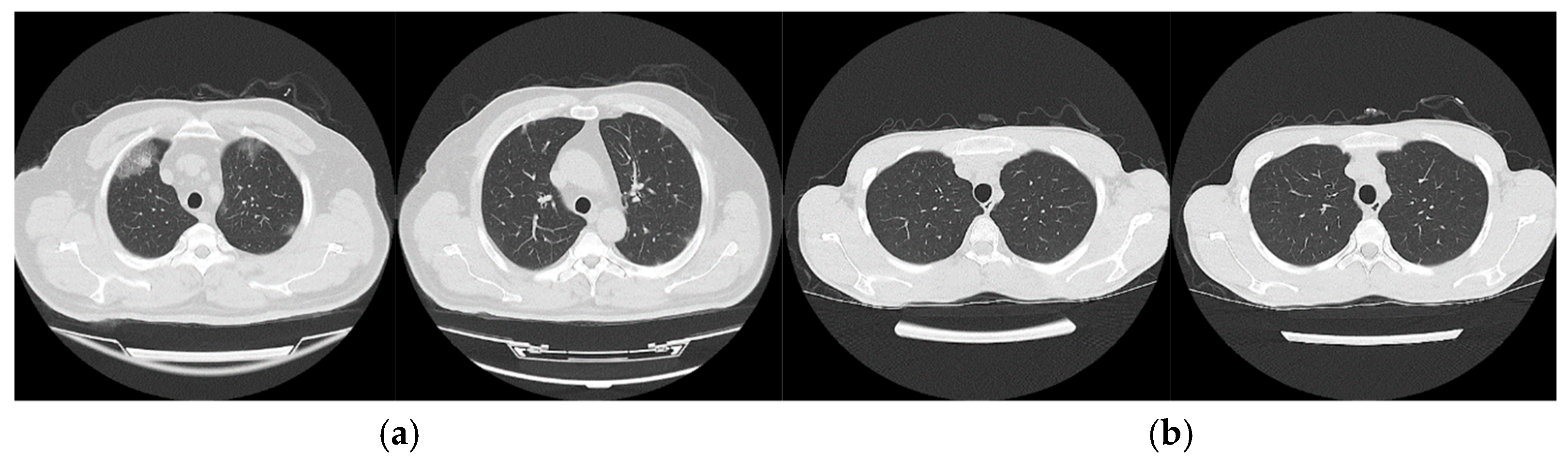
2.2.1. SARS-COV-2 Ct-Scan Dataset
2.2.2. Study Dataset
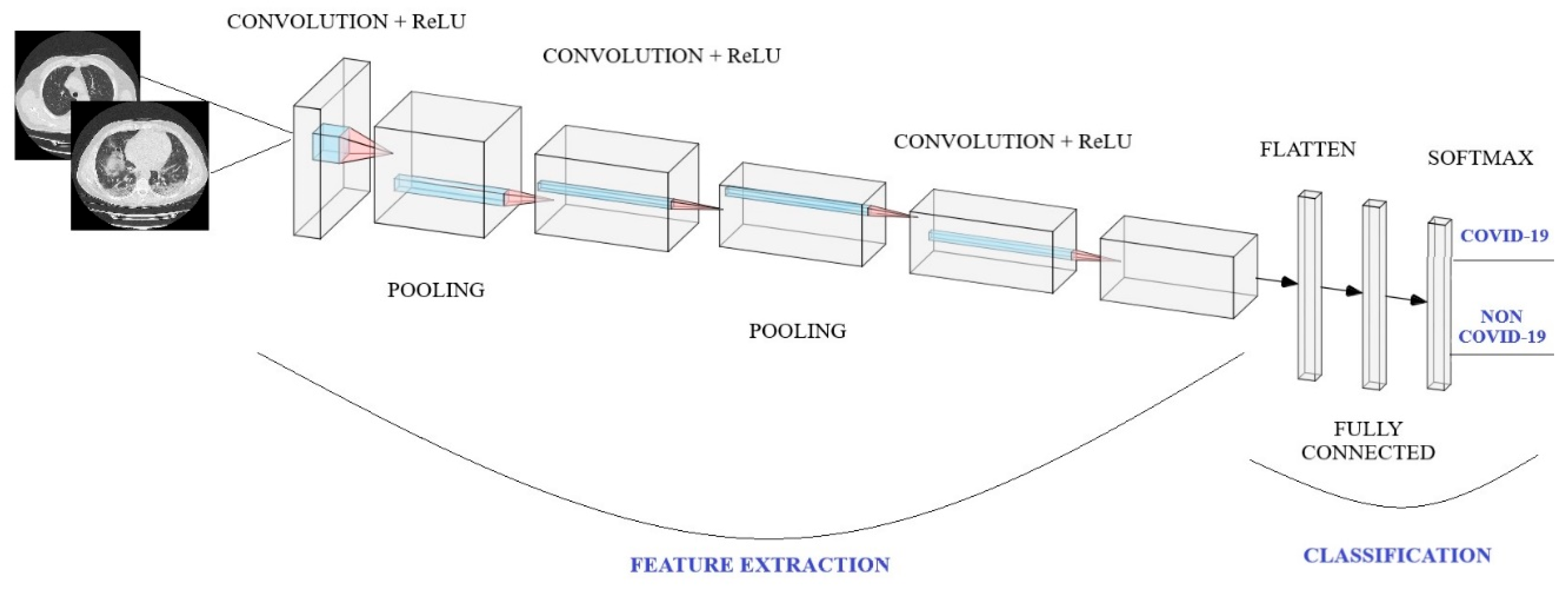
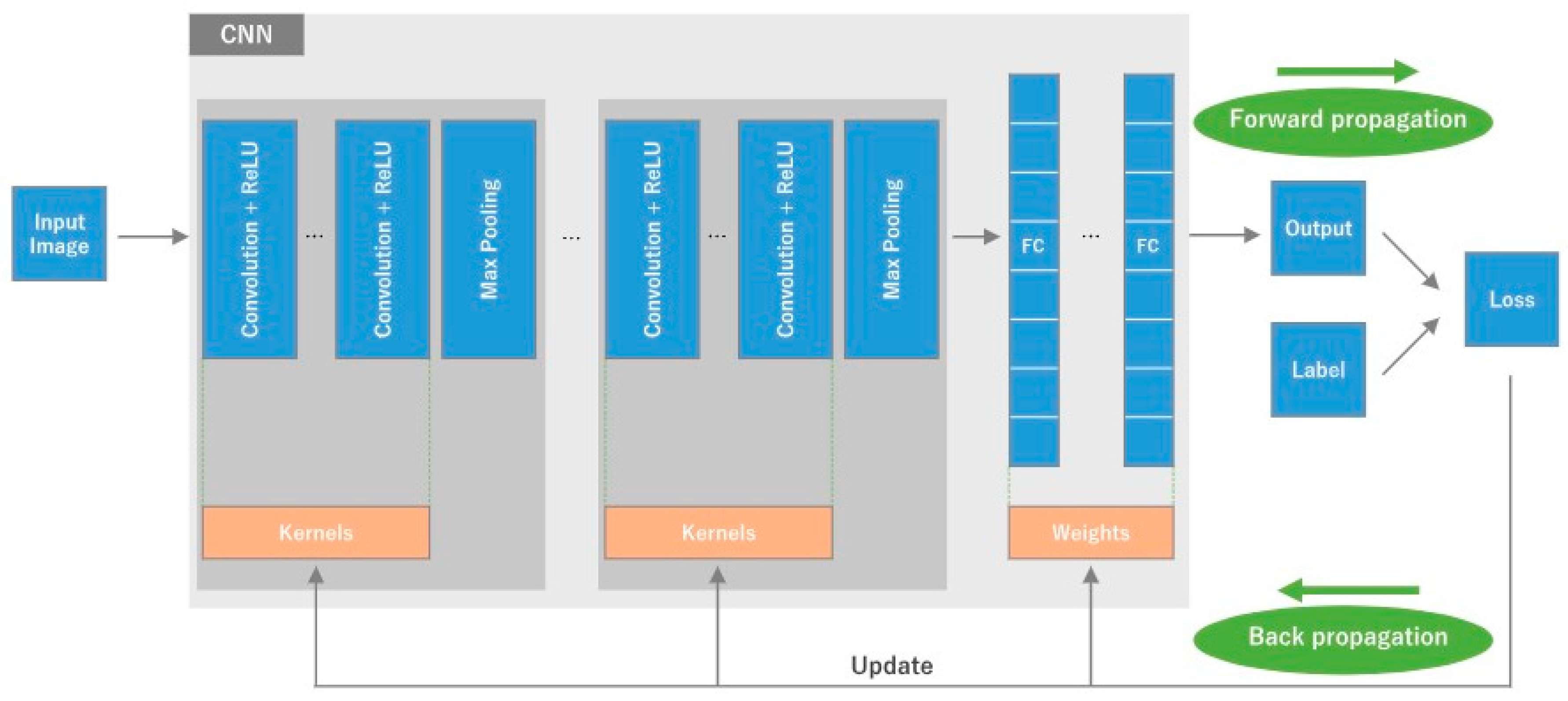
2.3. Convolutional Neural Networks
2.4. Transfer Learning
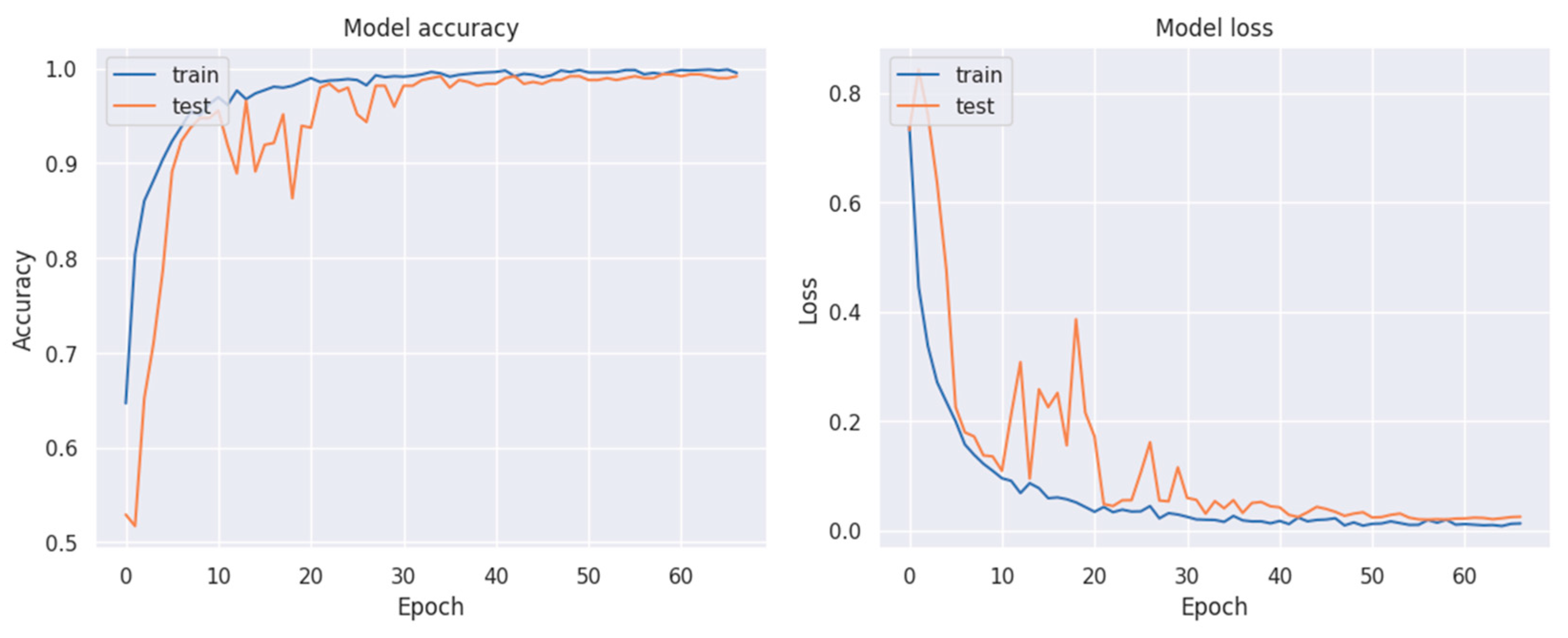
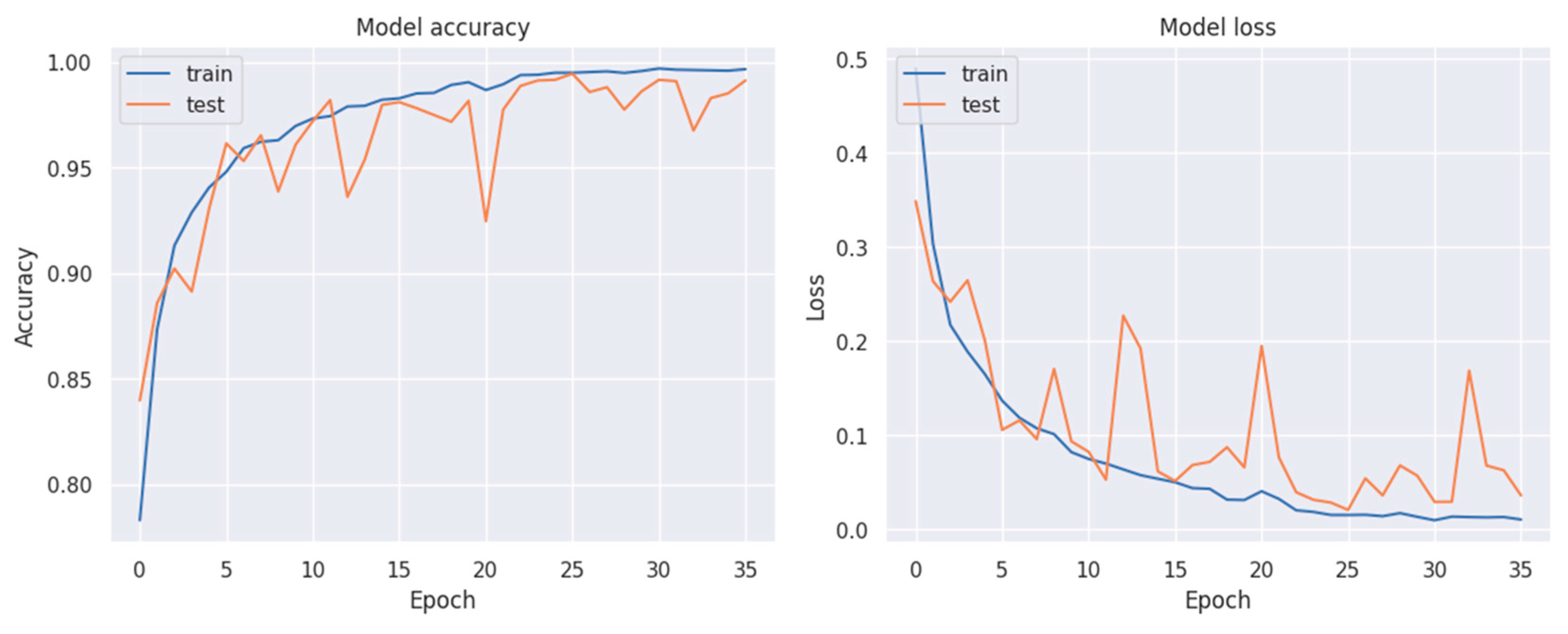
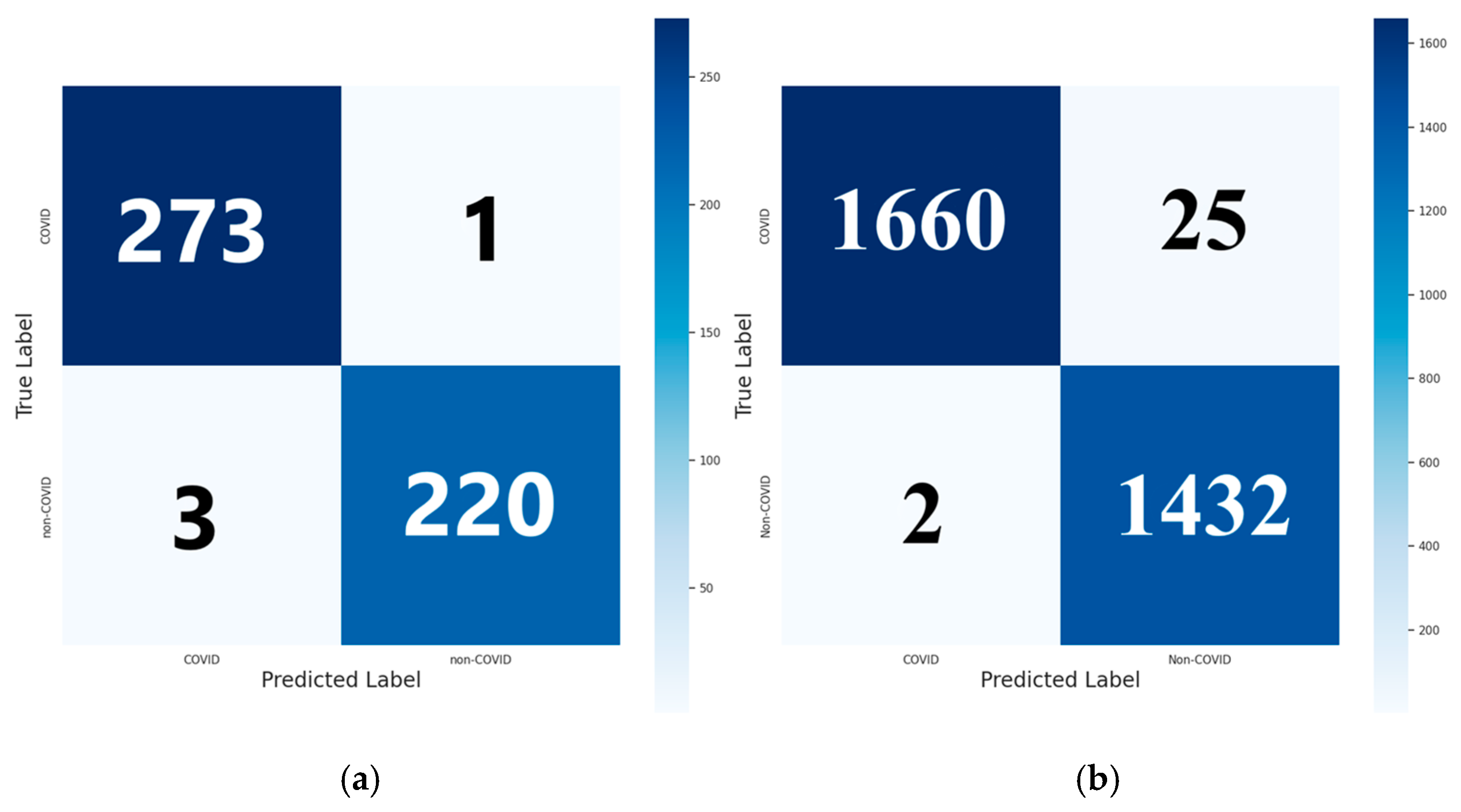
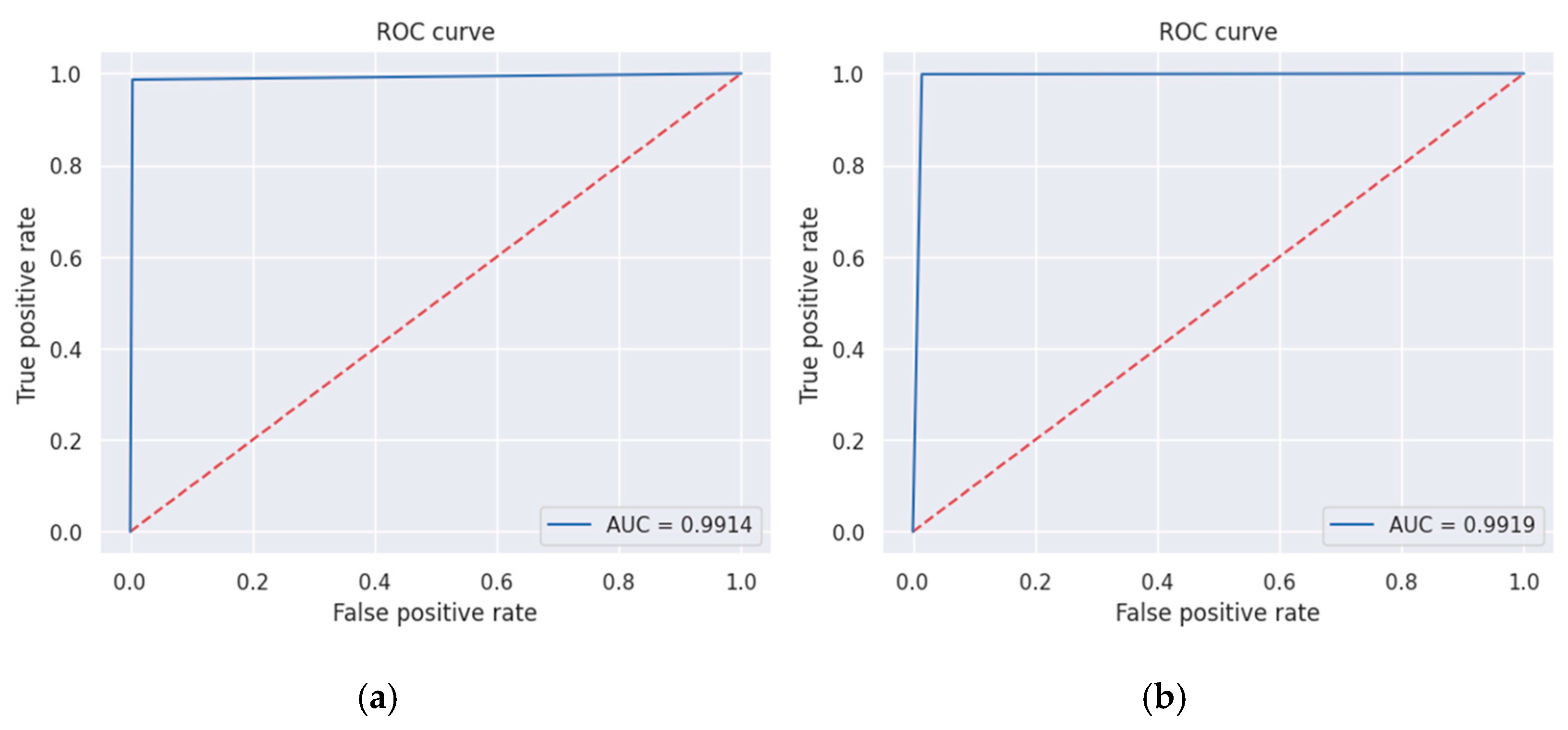
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RT-PCR | Reverse transcription-polymerase chain reaction |
| CT | Conputed tomography |
| CNN | Convolutional neural network |
| 2D | Two dimensional |
References
- Chetoui, M.; Akhloufi, M. A. Explainable vision transformers and radiomics for covid-19 detection in chest x-rays. Journal of Clinical Medicine 2022, 11(11), 3013. [CrossRef]
- Meem, A. T.; Khan, M. M.; Masud, M.; Aljahdali, S. Prediction of Covid-19 Based on Chest X-Ray Images Using Deep Learning with CNN. Computer Systems Science and Engineering 2022, 1223-1240.
- Lee, M. H.; Shomanov, A.; Kudaibergenova, M.; Viderman, D. (2023). Deep learning methods for interpretation of pulmonary CT and X-ray images in patients with COVID-19-related lung involvement: a systematic review. Journal of Clinical Medicine 2023, 12(10), 3446. [CrossRef]
- Qian, X.; Wang, X.; et al. M3Lung-Sys: A deep learning system for multi-class lung pneumonia screening from CT imaging. IEEE Journal of Biomedical and Health Informatics 2020, 24(12), 3539-3550. [CrossRef]
- Saurabh, N.; Shetty, J. A review of intelligent medical imaging diagnosis for the COVID-19 infection. Intelligent Decision Technologies 2022, 16(1), 127-144.
- Jalehi, M. K.; Albaker, B. M. Highly accurate multiclass classification of respiratory system diseases from chest radiography images using deep transfer learning technique. Biomedical Signal Processing and Control 2023, 84, 104745. [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W. et al. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology 2020, 296(2), E32-E40. [CrossRef]
- Dong, D.; Tang, Z.; Wang, S.; Hui, H.; Gong, L.; Lu, Y. et al. The role of imaging in the detection and management of COVID-19: a review. IEEE reviews in biomedical engineering 2020, 14, 16-29. [CrossRef]
- Islam, M. M.; Karray, F.; Alhajj, R.; Zeng, J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). Ieee Access 2021, 9, 30551-30572. [CrossRef]
- Clark, S.; Kamalinejad, E.; Magpantay, C.; Sahota, S.; Zhong, J.; Hu, Y. A Review of CNN on Medical Imaging to Diagnose COVID-19 Infections. In Proceedings of ISCA 34th International Conference on (Vol. 79, pp. 91-98) (November, 2021).
- Liu, S.; Cai, T.; Tang, X.; Wang, C. MRL-Net: Multi-Scale Representation Learning Network for COVID-19 Lung CT Image Segmentation. IEEE Journal of Biomedical and Health Informatics 2023, 27(9), 4317-4328. [CrossRef]
- Fallahpoor, M.; Chakraborty, S.; Heshejin, M. T.; Chegeni, H.; Horry, M. J.; Pradhan, B. Generalizability assessment of COVID-19 3D CT data for deep learning-based disease detection. Computers in Biology and Medicine 2022, 145, 105464. [CrossRef]
- Ardakani, A. A.; Kanafi, A. R.; Acharya, U. R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Computers in biology and medicine 2020, 121, 103795. [CrossRef]
- Apostolopoulos, I. D.; Mpesiana, T. A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and engineering sciences in medicine 2020, 43(2), 635-640. [CrossRef]
- Horry, M. J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. X-ray image based COVID-19 detection using pre-trained deep learning models, 2020, https://doi.org/10.31224/osf.io/wx89s.
- Punn, N. S.; Agarwal, S. Automated diagnosis of COVID-19 with limited posteroanterior chest X-ray images using fine-tuned deep neural networks. Applied Intelligence 2021, 51(5), 2689-2702. [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Applic 2021, 24, 1207–1220. [CrossRef]
- Asif, S.; Zhao, M.; Tang, F. et al. A deep learning-based framework for detecting COVID-19 patients using chest X-rays. Multimedia Systems 2022, 28, 1495–1513. [CrossRef]
- Keerthana, R.; Gladston, A.; Nehemiah, H. K. Transfer learning-based CNN diagnostic framework for diagnosis of COVID-19 from lung CT images. The Imaging Science Journal 2022, 70(7), 413-438. [CrossRef]
- Huang, M. L.; Liao, Y. C. A lightweight CNN-based network on COVID-19 detection using X-ray and CT images. Computers in Biology and Medicine 2022, 146, 105604. [CrossRef]
- Kaur, J.; Kaur, P. A CNN Transfer Learning-Based Automated Diagnosis of COVID-19 From Lung Computerized Tomography Scan Slices. New Generation Computing 2023, 1-44. [CrossRef]
- Malik, H.; Anees, T.; Al-Shamaylehs, A. S.; Alharthi, S. Z.; Khalil, W.; Akhunzada, A. Deep Learning-Based Classification of Chest Diseases Using X-rays, CT Scans, and Cough Sound Images. Diagnostics 2023, 13(17), 2772. [CrossRef]
- Kathamuthu, N. D.; Subramaniam, S.; Le, Q. H.; Muthusamy, S.; Panchal, H.; Sundararajan, S. C. M. et al. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Advances in Engineering Software 2023, 175, 103317. [CrossRef]
- Hossain, M. M.; Walid, M. A. A.; Galib, S. S.; Azad, M. M.; Rahman, W.; Shafi, A. S. M.; Rahman, M. M. Covid-19 detection from chest ct images using optimized deep features and ensemble classification. Systems and Soft Computing 2024, 6, 200077. [CrossRef]
- Zolya, M. A.; Baltag, C.; Bratu, D. V.; Coman, S.; Moraru, S. A. COVID-19 Detection and Diagnosis Model on CT Scans Based on AI Techniques. Bioengineering 2024, 11(1), 79. [CrossRef]
- Joshi, K. K.; Gupta, K.; Agrawal, J. An efficient transfer learning approach for prediction and classification of SARS–COVID-19. Multimedia Tools and Applications 2024, 83(13), 39435-39457. [CrossRef]
- Yousefpanah, K.; Ebadi, M. J.; Sabzekar, S.; Zakaria, N. H.; Osman, N. A.; Ahmadian, A. An Emerging Network for COVID-19 CT-Scan Classification using an ensemble deep transfer learning model. Acta Tropica 2024, 107277. [CrossRef]
- Özkan, Y. Uygulamalı Derin Öğrenme. Papatya Yayıncılık Eğitim 2021, İstanbul.
- Koresh, H. J. Implementation and Efficient Analysis of Preprocessing Techniques in Deep Learning for Image Classification. Current Medical Imaging 2023, 20, e290823220482.
- Murcia-Gomez, D.; Rojas-Valenzuela, I.; Valenzuela, O. Impact of image preprocessing methods and deep learning models for classifying histopathological breast cancer images. Applied Sciences 2022, 12(22), 11375. [CrossRef]
- Joshi, A. M.; Nayak, D. R. MFL-Net: An Efficient Lightweight Multi-Scale Feature Learning CNN for COVID-19 Diagnosis From CT Images. IEEE Journal of Biomedical and Health Informatics 2022, 26(11), 5355-5363. [CrossRef]
- Shamrat, F. J. M.; Azam, S.; Karim, A.; Ahmed, K.; Bui, F. M.; De Boer, F. High-precision multiclass classification of lung disease through customized MobileNetV2 from chest X-ray images. Computers in Biology and Medicine 2023, 155, 06646. [CrossRef]
- Soares, E.; Angelov, P.; Biaso, S.; Froes, M. H.; Abe, D. K. SARS-CoV-2 CT-scan dataset: A large dataset of real patients CT scans for SARS-CoV-2 identification. MedRxiv 2020, 2020-04. [CrossRef]
- Sharma, A.; Singh, K.; Koundal, D. A novel fusion-based convolutional neural network approach for classification of COVID-19 from chest X-ray images. Biomedical Signal Processing and Control 2022, 77, 103778, 2022. [CrossRef]
- Taye, M. M. Theoretical understanding of convolutional neural network: concepts, architectures, applications, future directions. Computation 2023, 11(3), 52. [CrossRef]
- Kugunavar, S.; Prabhakar, C. J. Convolutional neural networks for the diagnosis and prognosis of the coronavirus disease pandemic. Visual computing for industry, biomedicine, and art 2021, 4(1), 12. [CrossRef]
- Khanday, N. Y.; Sofi, S. A. Deep insight: Convolutional neural network and its applications for COVID-19 prognosis. Biomedical Signal Processing and Control 2021, 69, 102814, 2021. [CrossRef]
- Ergin, T. Convolutional Neural Network (ConvNet yada CNN) nedir, nasıl çalışır. Medium 2018. Available online: https://medium. com/@ tuncerergin/convolutional-neural-network-convnetyada-cnn-nedir-nasil-calisir-97a0f5d34cad (accessed on 17 October 2024).
- Yamashita, R.; Nishio, M.; Do, R. K. G.; Togashi, K. Convolutional neural networks: an overview and application in radiology. Insights into imaging 2018, 9, 611-629. [CrossRef]
- Zhang, H.; Qie, Y. Applying Deep Learning to Medical Imaging: A Review. Applied Sciences 2023, 13(18), 10521. [CrossRef]
- Morid, M. A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Computers in Biology and Medicine 2021, 128, 104115. [CrossRef]
- Ayana, G.; Dese, K.; Choe, S. W. Transfer learning in breast cancer diagnoses via ultrasound imaging. Cancers 2021, 13(4), 738. [CrossRef]
- Wang, J.; Zhu, H.; Wang, S. H.; Zhang, Y. D. A review of deep learning on medical image analysis. Mobile Networks and Applications 2021, 26, 351-380. [CrossRef]

| Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1 Score (%) | AUC (%) |
|
|---|---|---|---|---|---|---|
| Xception | 96.78 | 97.76 | 95.99 | 95.20 | 96.46 | 96.87 |
| DenseNet201 | 99.19 | 98.65 | 99.64 | 99.55 | 99.10 | 99.14 |
| DenseNet121 | 96.58 | 99.10 | 94.53 | 93.64 | 96.30 | 96.81 |
| DenseNet169 | 97.59 | 95.96 | 98.91 | 98.62 | 97.27 | 97.43 |
| ResNet101 | 42.25 | 44.84 | 40.15 | 37.88 | 41.07 | 42.49 |
| ResNet50 | 96.98 | 95.96 | 97.81 | 97.27 | 96.61 | 96.89 |
| VGG16 | 96.78 | 95.52 | 97.81 | 97.26 | 96.38 | 96.66 |
| VGG19 | 96.18 | 96.86 | 95.62 | 94.74 | 95.79 | 96.24 |
| InceptionV3 | 96.98 | 97.31 | 96.72 | 96.02 | 96.66 | 97.01 |
| InceptionResNetV2 | 94.57 | 96.41 | 93.07 | 91.88 | 94.09 | 94.74 |
| MobileNetV2 | 98.19 | 98.21 | 98.18 | 97.77 | 97.99 | 98.19 |
| NasNetLarge | 45.47 | 100.00 | 1.09 | 45.14 | 62.20 | 50.55 |
| NasNetMobile | 59.76 | 99.55 | 27.37 | 52.73 | 68.94 | 63.46 |
| Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1 Score (%) | AUC (%) |
|
|---|---|---|---|---|---|---|
| Xception | 96.31 | 98.95 | 94.07 | 93.41 | 96.10 | 96.51 |
| DenseNet201 | 99.13 | 99.86 | 98.52 | 98.28 | 99.07 | 99.19 |
| DenseNet121 | 97.85 | 98.60 | 97.21 | 96.78 | 97.68 | 97.91 |
| DenseNet169 | 97.47 | 98.88 | 96.27 | 95.74 | 97.29 | 97.57 |
| ResNet101 | 94.58 | 98.60 | 91.17 | 90.45 | 94.35 | 94.89 |
| ResNet50 | 95.38 | 93.72 | 96.80 | 96.13 | 94.91 | 95.26 |
| VGG16 | 97.27 | 95.39 | 98.87 | 98.63 | 96.98 | 97.13 |
| VGG19 | 92.47 | 99.79 | 86.25 | 86.03 | 92.40 | 93.02 |
| InceptionV3 | 96.09 | 96.75 | 95.49 | 95.08 | 95.91 | 96.12 |
| InceptionResNetV2 | 96.86 | 94.62 | 98.76 | 98.47 | 96.51 | 96.69 |
| MobileNetV2 | 96.38 | 98.11 | 94.90 | 94.23 | 96.13 | 96.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





