Submitted:
19 March 2025
Posted:
20 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing and Analysis of Genomic DNA Integrity
2.3. Diagnostic qPCR
2.4. PCR of Pol Region
2.5. Phylogenetic and Timescale Analyses
2.6. Statistical Analyses
3. Results
3.1. Study Population
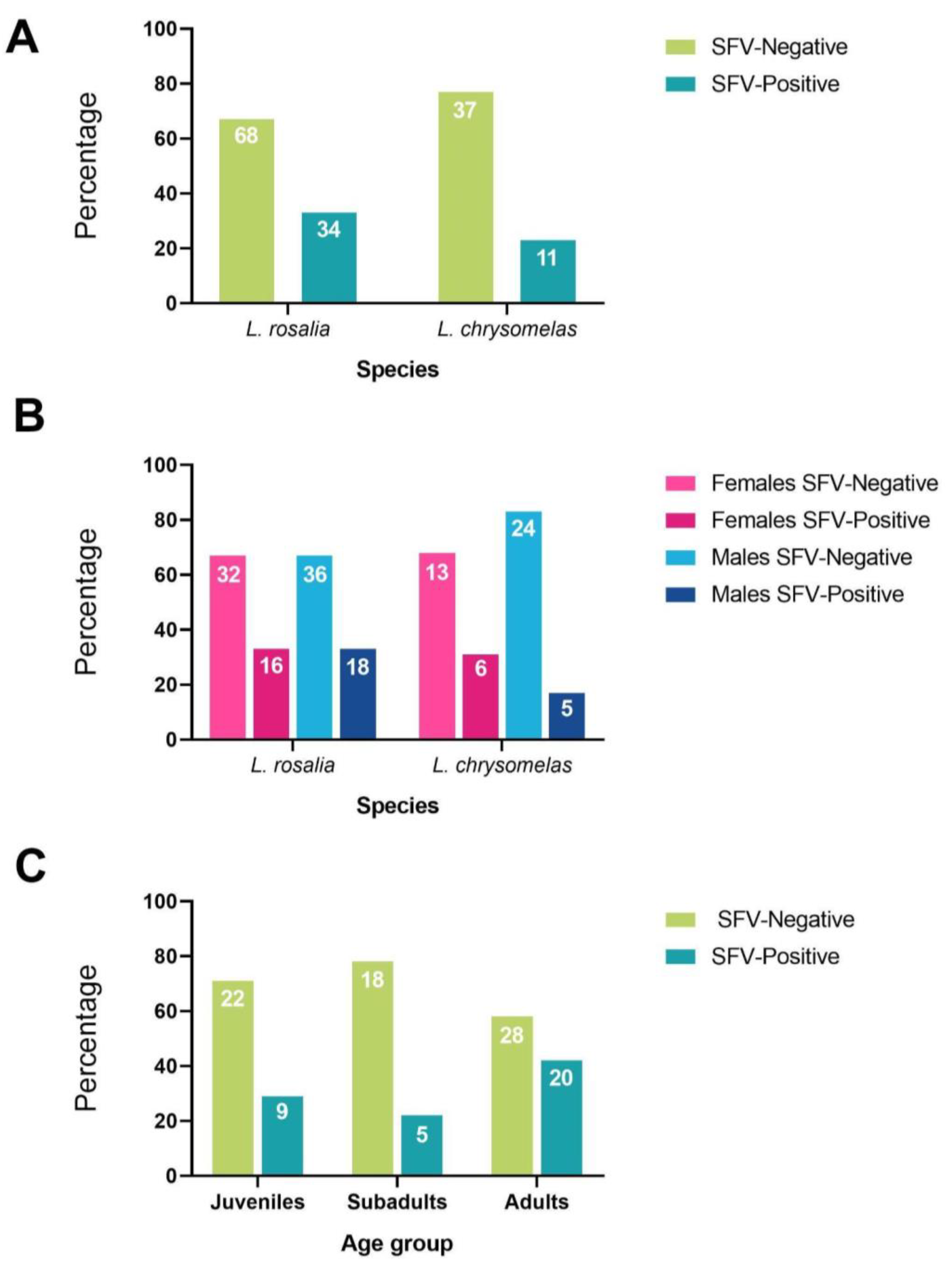
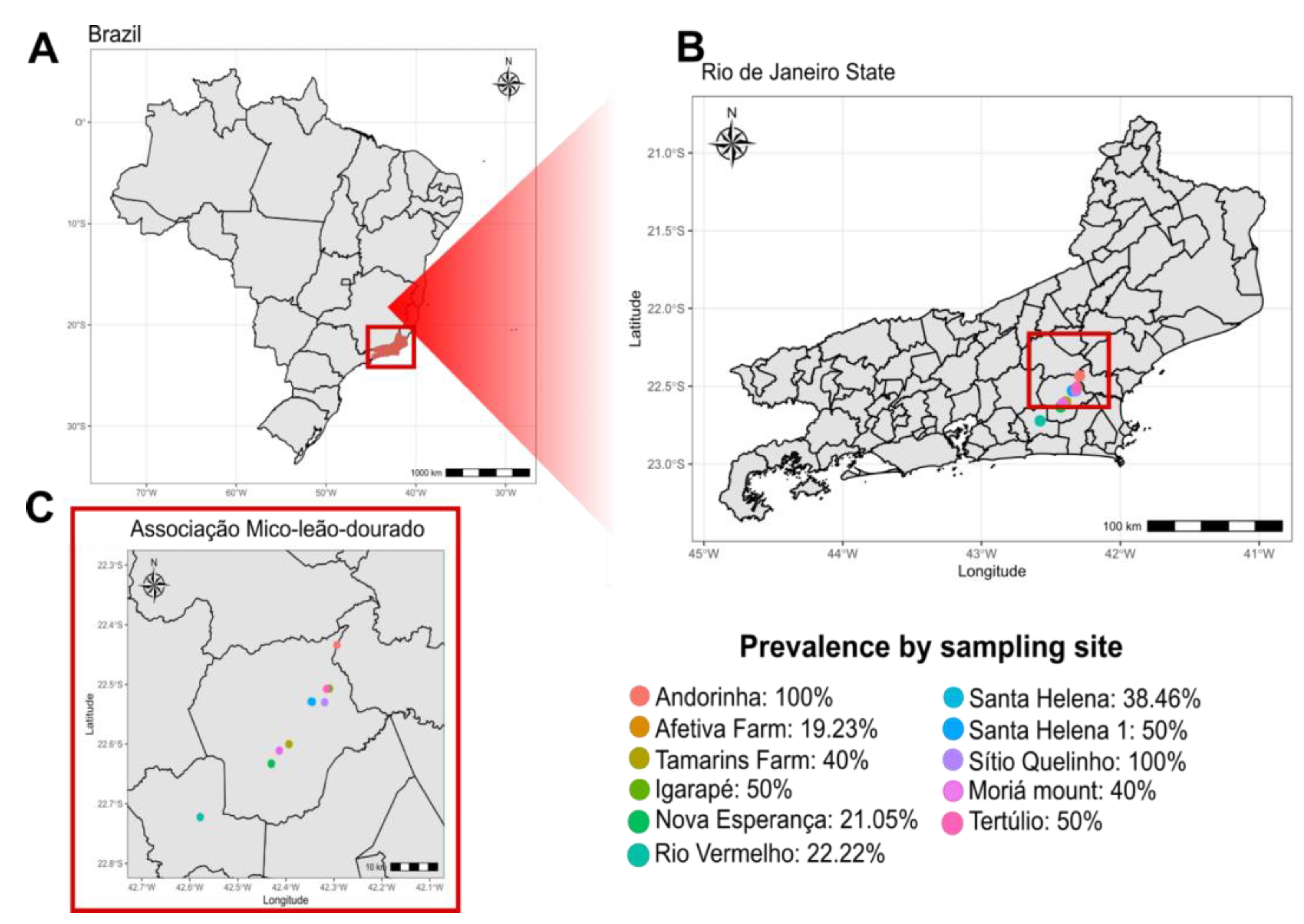
3.2. SFV Prevalence
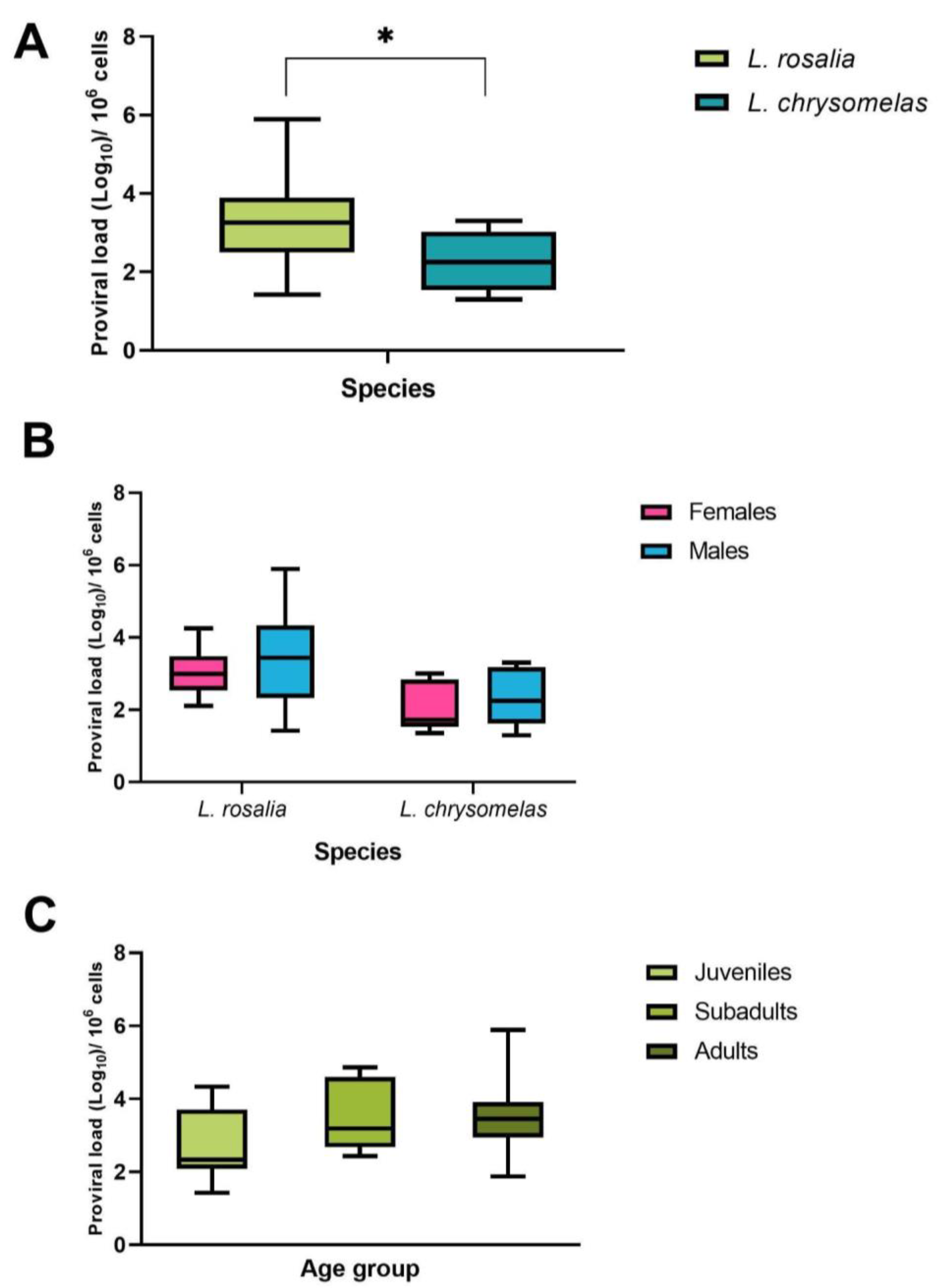
3.3. SFV Proviral Load
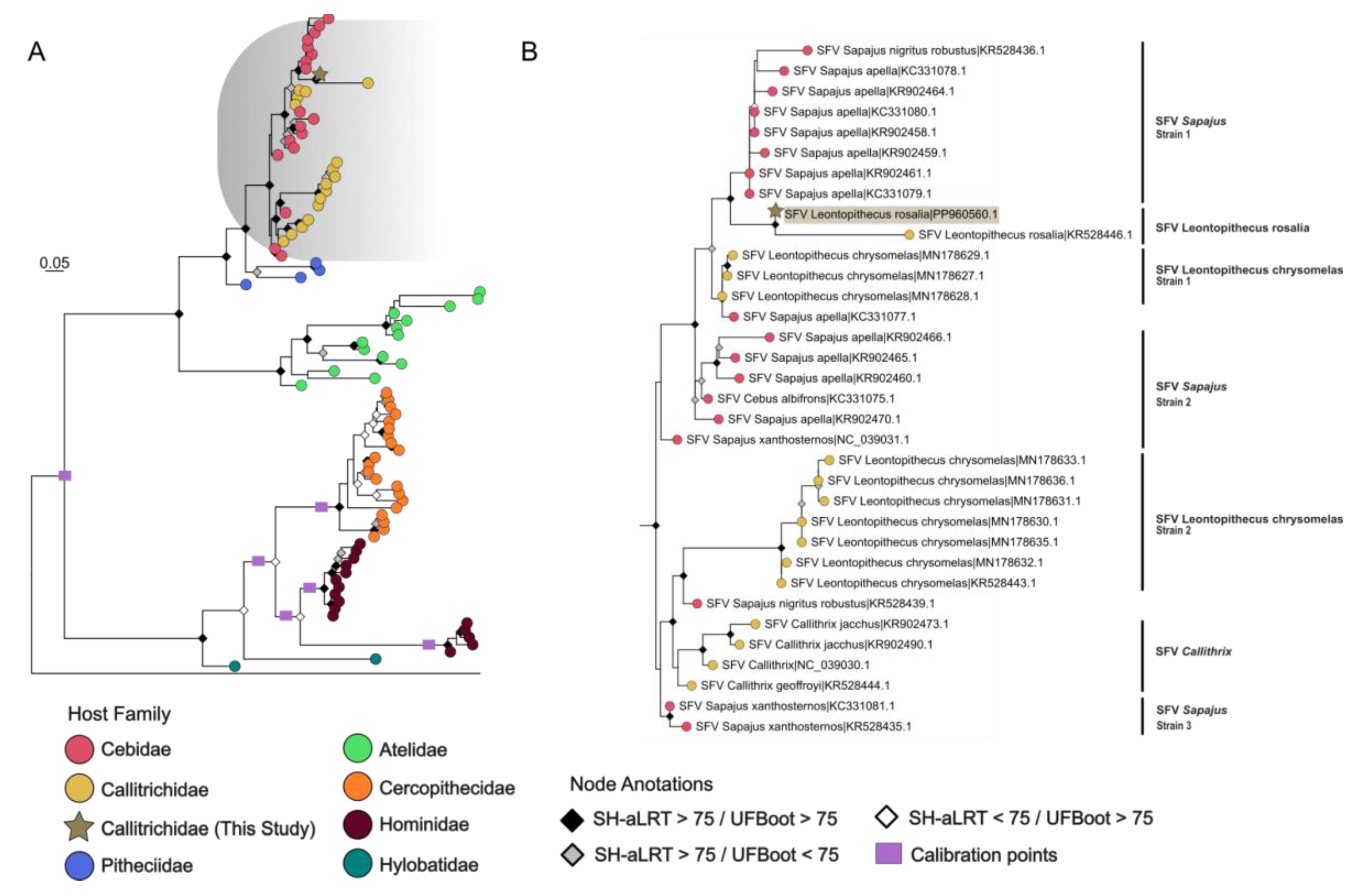
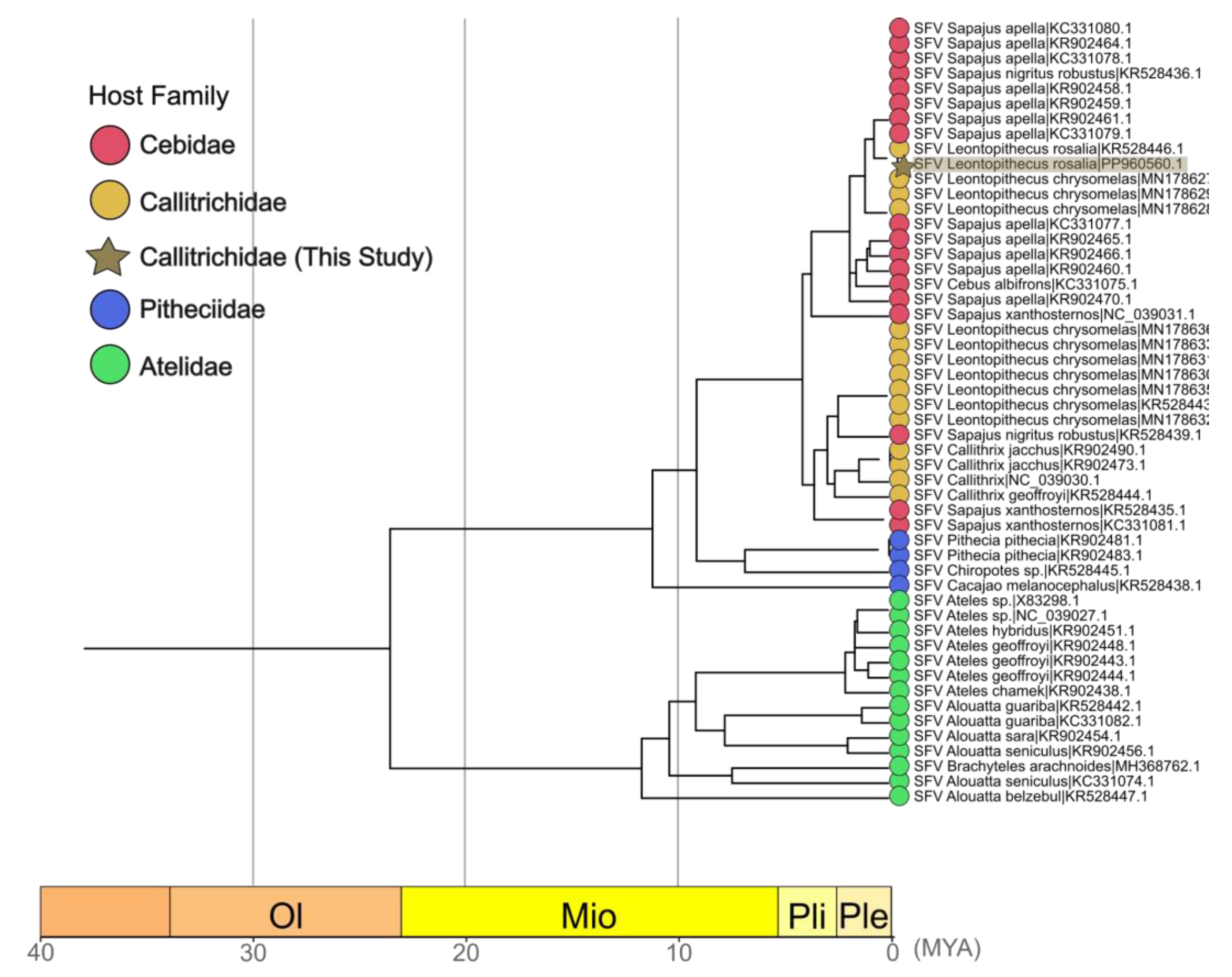
3.4. Phylogenetic and Timescale Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kehl, T.; Tan, J.; Materniak, M. Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/interspecies Transmission. Viruses 2013, 5, 2169–2209. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Bodem, J.; Buseyne, F.; Gessain, A.; Johnson, W.; Kuhn, J.H.; Kuzmak, J.; Lindemann, D.; Linial, M.L.; Löchelt, M.; et al. Spumaretroviruses: Updated Taxonomy and Nomenclature. Virology 2018, 516, 158–164. [Google Scholar] [CrossRef]
- Enders, J.F.; Peebles, T.C. Propagation in Tissue Cultures of Cytopathogenic Agents from Patients with Measles. Proc. Soc. Exp. Biol. Med. 1954, 86, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Rustigian, R.; Johnston, P.; Reihart, H. Infection of Monkey Kidney Tissue Cultures with Virus-like Agents. Proc. Soc. Exp. Biol. Med. 1955, 88, 8–16. [Google Scholar] [CrossRef]
- Hashimoto-Gotoh, A.; Yoshikawa, R.; Nakagawa, S.; Okamoto, M.; Miyazawa, T. Phylogenetic Analyses Reveal That Simian Foamy Virus Isolated from Japanese Yakushima Macaques (Macaca Fuscata Yakui) Is Distinct from Most of Japanese Hondo Macaques (Macaca Fuscata Fuscata). Gene 2020, 734. [Google Scholar] [CrossRef]
- Shankar, A.; Sibley, S.D.; Goldberg, T.L.; Switzer, W.M. Molecular Analysis of the Complete Genome of a Simian Foamy Virus Infecting Hylobates Pileatus (Pileated Gibbon) Reveals Ancient Co-Evolution with Lesser Apes. Viruses 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Switzer, W.M.; Salemi, M.; Shanmugam, V.; Gao, F.; Cong, M.E.; Kuiken, C.; Bhullar, V.; Beer, B.E.; Vallet, D.; Gautier-Hion, A.; et al. Ancient Co-Speciation of Simian Foamy Viruses and Primates. Nature 2005, 434, 376–380. [Google Scholar] [CrossRef]
- Muniz, C.P.; Troncoso, L.L.; Moreira, M.A.; Soares, E.A.; Pissinatti, A.; Bonvicino, C.R.; Seuánez, H.N.; Sharma, B.; Jia, H.; Shankar, A.; et al. Identification and Characterization of Highly Divergent Simian Foamy Viruses in a Wide Range of New World Primates from Brazil. PLoS One 2013, 8, e67568. [Google Scholar] [CrossRef]
- Ghersi, B.M.; Jia, H.; Aiewsakun, P.; Katzourakis, A.; Mendoza, P.; Bausch, D.G.; Kasper, M.R.; Montgomery, J.M.; Switzer, W.M. Wide Distribution and Ancient Evolutionary History of Simian Foamy Viruses in New World Primates. Retrovirology 2015, 12. [Google Scholar] [CrossRef]
- Santos, A.F.; Cavalcante, L.T.F.; Muniz, C.P.; Switzer, W.M.; Soares, M.A. Simian Foamy Viruses in Central and South America: A New World of Discovery. Viruses 2019, 11. [Google Scholar] [CrossRef]
- Miranda, T.S.; Muniz, C.P.; Moreira, S.B.; Bueno, M.G.; Kierulff, M.C.M.; Molina, C.V.; Catão-Dias, J.L.; Pissinatti, A.; Soares, M.A.; Santos, A.F. Eco-Epidemiological Profile and Molecular Characterization of Simian Foamy Virus in a Recently-Captured Invasive Population of Leontopithecus Chrysomelas (golden-Headed Lion Tamarin) in Rio de Janeiro, Brazil. Viruses 2019, 11. [Google Scholar] [CrossRef]
- Hooks, J.J.; Gibbs, C.J., Jr; Chou, S.; Howk, R.; Lewis, M.; Gajdusek, D.C. Isolation of a New Simian Foamy Virus from a Spider Monkey Brain Culture. Infect. Immun. 1973, 8, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Troncoso, L.L.; Muniz, C.P.; Siqueira, J.D.; Curty, G.; Schrago, C.G.; Augusto, A.; Fedullo, L.; Soares, M.A.; Santos, A.F. Characterization and Comparative Analysis of a Simian Foamy Virus Complete Genome Isolated from Brazilian Capuchin Monkeys. Virus Res. 2015, 208, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Muniz, C.P.; Cavalcante, L.T.F.; Dudley, D.M.; Pissinatti, A.; O’Connor, D.H.; Santos, A.F.; Soares, M.A. First Complete Genome Sequence of a Simian Foamy Virus Infecting the Neotropical Primate Brachyteles Arachnoides. Microbiol Resour Announc 2018, 7. [Google Scholar] [CrossRef]
- Thümer, L.; Rethwilm, A.; Holmes, E.C.; Bodem, J. The Complete Nucleotide Sequence of a New World Simian Foamy Virus. Virology 2007, 369, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Schrago, C.G.; Menezes, A.N.; Furtado, C.; Bonvicino, C.R.; Seuanez, H.N. Multispecies Coalescent Analysis of the Early Diversification of Neotropical Primates: Phylogenetic Inference under Strong Gene Trees/species Tree Conflict. Genome Biol. Evol. 2014, 6, 3105–3114. [Google Scholar] [CrossRef]
- Primates-SG - Home Available online:. Available online: http://www.primate-sg.org/ (accessed on 12 August 2024).
- Pacheco, B.; Finzi, A.; McGee-Estrada, K.; Sodroski, J. Species-Specific Inhibition of Foamy Viruses from South American Monkeys by New World Monkey TRIM5{alpha} Proteins. J. Virol. 2010, 84, 4095–4099. [Google Scholar] [CrossRef]
- Marczynska, B.; Jones, C.J.; Wolfe, L.G. Syncytium-Forming Virus of Common Marmosets (Callithrix Jacchus Jacchus). Infect. Immun. 1981, 31, 1261–1269. [Google Scholar] [CrossRef]
- Muniz, C.P.; Jia, H.; Shankar, A.; Troncoso, L.L.; Augusto, A.M.; Farias, E.; Pissinatti, A.; Fedullo, L.P.; Santos, A.F.; Soares, M.A.; et al. An Expanded Search for Simian Foamy Viruses (SFV) in Brazilian New World Primates Identifies Novel SFV Lineages and Host Age-Related Infections. Retrovirology 2015, 12, 94. [Google Scholar] [CrossRef]
- Kuderna, L.F.K.; Gao, H.; Janiak, M.C.; Kuhlwilm, M.; Orkin, J.D.; Bataillon, T.; Manu, S.; Valenzuela, A.; Bergman, J.; Rousselle, M.; et al. A Global Catalog of Whole-Genome Diversity from 233 Primate Species. Science 2023, 380, 906–913. [Google Scholar] [CrossRef]
- Katzourakis, A.; Aiewsakun, P.; Jia, H.; Wolfe, N.D.; LeBreton, M.; Yoder, A.D.; Switzer, W.M. Discovery of Prosimian and Afrotherian Foamy Viruses and Potential Cross Species Transmissions amidst Stable and Ancient Mammalian Co-Evolution. Retrovirology 2014, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.-Y.; Wei, X.; Cui, J. Multiple Infiltration and Cross-Species Transmission of Foamy Viruses across the Paleozoic to the Cenozoic Era. J. Virol. 2021, 95, e0048421. [Google Scholar] [CrossRef] [PubMed]
- Primates-SG - Primate Diversity by Region Available online:. Available online: http://www.primate-sg.org/primate_diversity_by_region/ (accessed on 12 August 2024).
- Meyer, A.L.S.; Pie, M.R.; Passos, F.C. Assessing the Exposure of Lion Tamarins (Leontopithecus Spp.) to Future Climate Change. Am. J. Primatol. 2014, 76, 551–562. [Google Scholar] [CrossRef]
- Kierulff, M.C.M.; Ruiz-Miranda, C.R.; de Oliveira, P.P.; Beck, B.B.; Martins, A.; Dietz, J.M.; Rambaldi, D.M.; Baker, A.J. The Golden Lion Tamarin Leontopithecus Rosalia: A Conservation Success Story. Int. Zoo Yearbook 2012, 46, 36–45. [Google Scholar] [CrossRef]
- Primates-SG - Home Available online:. Available online: http://www.primate-sg.org/ (accessed on 12 August 2024).
- Associação Mico-Leão-Dourado – Conectando Florestas para salvar a espécie Available online:. Available online: https://micoleao.org.br/ (accessed on 12 August 2024).
- Publicações – Associação Mico-Leão-Dourado Available online:. Available online: https://micoleao.org.br/publicacoes-2/ (accessed on 12 August 2024).
- Ruiz-Miranda, C.R.; Affonso, A.G.; Morais, M.M. de; Verona, C.E.; Martins, A.; Beck, B.B. Behavioral and Ecological Interactions between Reintroduced Golden Lion Tamarins (Leontopithecus Rosalia Linnaeus, 1766) and Introduced Marmosets (Callithrix Spp, Linnaeus, 1758) in Brazil’s Atlantic Coast Forest Fragments. Braz. Arch. Biol. Technol. 2006, 49, 99–109. [Google Scholar] [CrossRef]
- Muniz, C.P.; Zheng, H.Q.; Jia, H.; Cavalcante, L.T.F.; Augusto, A.M.; Fedullo, L.P.; Pissinatti, A.; Soares, M.A.; Switzer, W.M.; Santos, A.F. A Non-Invasive Specimen Collection Method and a Novel Simian Foamy Virus (SFV) DNA Quantification Assay in New World Primates Reveal Aspects of Tissue Tropism and Improved SFV Detection. PLoS One 2017, 12. [Google Scholar] [CrossRef]
- Miranda, T.D.S.; Schiffler, F.B.; D’arc, M.; Moreira, F.R.R.; Cosentino, M.A.C.; Coimbra, A.; Mouta, R.; Medeiros, G.; Girardi, D.L.; Wanderkoke, V.; et al. Metagenomic Analysis Reveals Novel Dietary-Related Viruses in the Gut Virome of Marmosets Hybrids (Callithrix Jacchus X Callithrix Penicillata), Brazil. Virus Res. 2023, 325, 199017. [Google Scholar] [CrossRef]
- Ruiz-Miranda, C.R.; Kleiman, D.G.; Dietz, J.M.; Moraes, E.; Grativol, A.D.; Baker, A.J.; Beck, B.B. Food Transfers in Wild and Reintroduced Golden Lion Tamarins, Leontopithecus rosalia. Am. J. Primatol. 1999, 48, 305–320. [Google Scholar] [CrossRef]
- Dietz, J.M.; Baker, A.J.; Miglioretti, D. Seasonal Variation in Reproduction, Juvenile Growth, and Adult Body Mass in Golden Lion Tamarins (Leontopithecus rosalia). Am. J. Primatol. 1994, 34, 115–132. [Google Scholar] [CrossRef]
- Rylands, A.B. Marmosets and Tamarins: Systematics, Behaviour, and Ecology; Oxford University Press, 1993; ISBN 9780198540229.
- Muniz, C.P.; Cavalcante, L.T.F.; Jia, H.; Zheng, H.; Tang, S.; Augusto, A.M.; Pissinatti, A.; Fedullo, L.P.; Santos, A.F.; Soares, M.A.; et al. Zoonotic Infection of Brazilian Primate Workers with New World Simian Foamy Virus. PLoS One 2017, 12, e0184502. [Google Scholar] [CrossRef]
- Tandon, R.; Cattori, V.; Gomes-Keller, M.A.; Meli, M.L.; Golder, M.C.; Lutz, H.; Hofmann-Lehmann, R. Quantitation of Feline Leukaemia Virus Viral and Proviral Loads by TaqMan Real-Time Polymerase Chain Reaction. J. Virol. Methods 2005, 130, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Iq-Tree, R.L. 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era., 2020, 37. DOI: https://doi. org/10. 1093/molbev/msaa015 2020, 1530–1534. [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating Divergence Times in Large Molecular Phylogenies. Proc Natl Acad Sci U S A 2012, 109, 19333–19338. [Google Scholar] [CrossRef]
- Tamura, K.; Tao, Q.; Kumar, S. Theoretical Foundation of the RelTime Method for Estimating Divergence Times from Variable Evolutionary Rates. Mol Biol Evol 2018, 35, 1770–1782. [Google Scholar] [CrossRef]
- Mello, B.; Tao, Q.; Tamura, K.; Kumar, S. Fast and Accurate Estimates of Divergence Times from Big Data. Mol Biol Evol 2017, 34, 45–50. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Ripley, B.D. The R Project in Statistical Computing. MSOR Connect. 2001, 1, 23–25. [Google Scholar] [CrossRef]
- Home - GraphPad Available online:. Available online: https://www.graphpad.com/ (accessed on 12 August 2024).
- Pearson, K. X. On the Criterion That a given System of Deviations from the Probable in the Case of a Correlated System of Variables Is Such That It Can Be Reasonably Supposed to Have Arisen from Random Sampling. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science 1900, 50, 157–175. [Google Scholar] [CrossRef]
- Estrada, A.; Garber, P.A.; Rylands, A.B.; Roos, C.; Fernandez-Duque, E.; Di Fiore, A.; Nekaris, K.A.-I.; Nijman, V.; Heymann, E.W.; Lambert, J.E.; et al. Impending Extinction Crisis of the World’s Primates: Why Primates Matter. Sci Adv 2017, 3, e1600946. [Google Scholar] [CrossRef] [PubMed]
- Hood, S.; Mitchell, J.L.; Sethi, M.; Almond, N.M.; Cutler, K.L.; Rose, N.J. Horizontal Acquisition and a Broad Biodistribution Typify Simian Foamy Virus Infection in a Cohort of Macaca Fascicularis. Virol. J. 2013, 10. [Google Scholar] [CrossRef]
- Falcone, V.; Schweizer M; Toniolo, A. ; Neumann-Haefelin, D.; Meyerhans, A. Gamma Interferon Is a Major Suppressive Factor Produced by Activated Human Peripheral Blood Lymphocytes That Is Able to Inhibit Foamy Virus-Induced Cytopathic Effects. Journal of virology 1999, 73. [Google Scholar] [CrossRef]
- Falcone, V.; Leupold, J.; Clotten, J.; Urbanyi, E.; Herchenröder, O.; Spatz, W.; Volk, B.; Böhm, N.; Toniolo, A.; Neumann-Haefelin, D.; et al. Sites of Simian Foamy Virus Persistence in Naturally Infected African Green Monkeys: Latent Provirus Is Ubiquitous, Whereas Viral Replication Is Restricted to the Oral Mucosa. Virology 1999, 257, 7–14. [Google Scholar] [CrossRef]
- Mouinga-Ondémé, A.; Kazanji, M. Simian Foamy Virus in Non-Human Primates and Cross-Species Transmission to Humans in Gabon: An Emerging Zoonotic Disease in Central Africa? Viruses 2013, 5, 1536–1552. [Google Scholar] [CrossRef]
- Blasse, A.; Calvignac-Spencer, S.; Merkel, K.; Goffe, A.S.; Boesch, C.; Mundry, R.; Leendertz, F.H. Mother-Offspring Transmission and Age-Dependent Accumulation of Simian Foamy Virus in Wild Chimpanzees. J. Virol. 2013, 87, 5193–5204. [Google Scholar] [CrossRef]
- Ruiz-Miranda, C.R.; de Morais, M.M., Jr; Dietz, L.A.; Rocha Alexandre, B.; Martins, A.F.; Ferraz, L.P.; Mickelberg, J.; Hankerson, S.J.; Dietz, J.M. Estimating Population Sizes to Evaluate Progress in Conservation of Endangered Golden Lion Tamarins (Leontopithecus rosalia). PLoS One 2019, 14, e0216664. [Google Scholar] [CrossRef]
- Lucas, P. da S.; Alves-Eigenheer, M.; Francisco, T.M.; Dietz, J.M.; Ruiz-Miranda, C.R. Spatial Response to Linear Infrastructures by the Endangered Golden Lion Tamarin. Diversity 2019, 11, 100. [Google Scholar] [CrossRef]
- Romano, V.; Martins, A.F.; Ruiz-Miranda, C.R. Unraveling the Dispersal Patterns and the Social Drivers of Natal Emigration of a Cooperative Breeding Mammal, the Golden Lion Tamarin. Am. J. Primatol. 2019, 81, e22959. [Google Scholar] [CrossRef] [PubMed]
- Moraes, A.M.; Ruiz-Miranda, C.R.; Galetti, P.M., Jr; Niebuhr, B.B.; Alexandre, B.R.; Muylaert, R.L.; Grativol, A.D.; Ribeiro, J.W.; Ferreira, A.N.; Ribeiro, M.C. Landscape Resistance Influences Effective Dispersal of Endangered Golden Lion Tamarins within the Atlantic Forest. Biol. Conserv. 2018, 224, 178–187. [Google Scholar] [CrossRef]
- Sapajus Apella: Boubli, J.P. , Stevenson, P.R., Palacios, E., de La Torre, S., Ravetta, A.L., Messias, M.R., Carvalho, A.S. & Mittermeier, R.A. IUCN Red List Threat Species 2020. [Google Scholar] [CrossRef]
- D’arc, M.; Moreira, F.R.R.; Dias, C.A.; Souza, A.R.; Seuánez, H.N.; Soares, M.A.; Tavares, M.C.H.; Santos, A.F.A. The Characterization of Two Novel Neotropical Primate Papillomaviruses Supports the Ancient within-Species Diversity Model. Virus Evol. 2020, 6, veaa036. [Google Scholar] [CrossRef]
- Jameson Kiesling, N.M.; Yi, S.V.; Xu, K.; Gianluca Sperone, F.; Wildman, D.E. The Tempo and Mode of New World Monkey Evolution and Biogeography in the Context of Phylogenomic Analysis. Mol. Phylogenet. Evol. 2015, 82, 386–399. [Google Scholar] [CrossRef]
- Schneider, H.; Sampaio, I. The Systematics and Evolution of New World Primates - A Review. Mol. Phylogenet. Evol. 2015, 82 Pt B, 348–357. [Google Scholar] [CrossRef]
| Individuals | L. rosalia | L. chrysomelas |
| All individuals | 102 | 48 |
| Males | 54 (53%) | 29 (60%) |
| Females | 48 (7%) | 19 (40%) |
| Adults | 48 (47%) | N/A* |
| Subadults | 23 (23%) | N/A |
| Juveniles | 31 (30%) | N/A |
| Median weight (grams) | 521 (259 - 754) g | N/A |
| Average knee-heel distance | 83 (65 -97) cm | N/A |
| Median collection per site | 5 (2-26) | N/A |
| Collection point | Animals sampled | Juveniles | Subadults | Adults | Prevalence (%) | Average proviral load* |
| Afetiva Farm | 26 | 12 | 9 | 5 | 19 | 03.04 |
| Tamarins Farm | 5 | 0 | 2 | 3 | 40 | 3.81 |
| Igarapé | 12 | 3 | 3 | 6 | 50 | 3.30 |
| Nova esperança | 19 | 4 | 4 | 11 | 21 | 3.14 |
| Rio Vermelho | 9 | 4 | 0 | 5 | 22 | 2.74 |
| Ribeirão | 2 | 0 | 0 | 2 | 0 | N/A |
| Santa Helena | 13 | 1 | 4 | 8 | 35 | 3.73 |
| Santa Helena I | 4 | 1 | 1 | 2 | 50 | 4.56 |
| Sítio Quelinho | 2 | 1 | 0 | 1 | 100 | 3.91 |
| Tertúlio | 2 | 1 | 0 | 1 | 50 | 3.93 |
| Monte Moriá | 5 | 4 | 0 | 1 | 40 | 3.63 |
| Andorinha | 3 | 0 | 0 | 3 | 100 | 4.22 |
| Collection point | Group | Animals | Prevalence (%) | #break#Average proviral load* |
| Afetiva | Afetiva 1 | 2 | 50%% | 2.05 |
| Afetiva | Afetiva 2/ AF2 | 12 | 25% | 2.19 |
| Afetiva | Afetiva 3/ AF3 | 2 | 0% | N/A* |
| Afetiva | UR | 4 | 0% | N/A |
| Afetiva | FP | 5 | 20% | 2.33 |
| Afetiva | FP3 | 1 | 0% | N/A |
| Andorinha | CH2 | 3 | 100% | 4.22 |
| Tamarins Farm | Sidney 3 | 1 | 100% | 3.49 |
| Tamarins Farm | TM2 | 4 | 25,00% | 4.12 |
| Igarapé | IG | 8 | 62% | 2.81 |
| Igarapé | ph2 | 4 | 25% | 1.88 |
| Moriá Mount | Ronaldo Machado (RM) | 2 | 40% | 6.63 |
| Nova Esperança | GM2 | 3 | 0% | N/A |
| Nova Esperança | GM3 | 7 | 29% | 3.02 |
| Nova Esperança | GM4 | 2 | 100% | 3.05 |
| Nova Esperança | GM5 | 4 | 0% | N/A |
| Nova Esperança | GM7 | 3 | 0% | N/A |
| Rio vermelho | M6 | 1 | 100% | 2.51 |
| Rio vermelho | Mistura fina | 3 | 0% | N/A |
| Rio vermelho | RV | 4 | 25% | 2.96 |
| Rio vermelho | RT | 1 | 0% | N/A |
| Ribeirão | ZN | 2 | 0% | N/A |
| Santa Helena | FN | 2 | 100% | 2.65 |
| Santa Helena | JA | 5 | 20% | 4.34 |
| Santa Helena | JN | 2 | 100% | 4.30 |
| Santa Helena | JR | 4 | 0% | N/A |
| Santa Helena 1 | SH | 1 | 0% | N/A |
| Santa Helena 1 | SS2 | 3 | 67% | 4.56 |
| Sítio Quelinho | q1 | 2 | 100% | 3.91 |
| Tertulio | JD | 2 | 50% | 3.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





