Submitted:
15 March 2025
Posted:
17 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- The proposed dual-stream augmentation framework utilizes a single generator with dual perturbations to enhance realism and diversity by effectively capturing both local and global variations in medical images.
- A rigorous mathematical formulation is developed, incorporating a CLP module to preserve semantic integrity and enhance model generalization in image augmentation tasks.
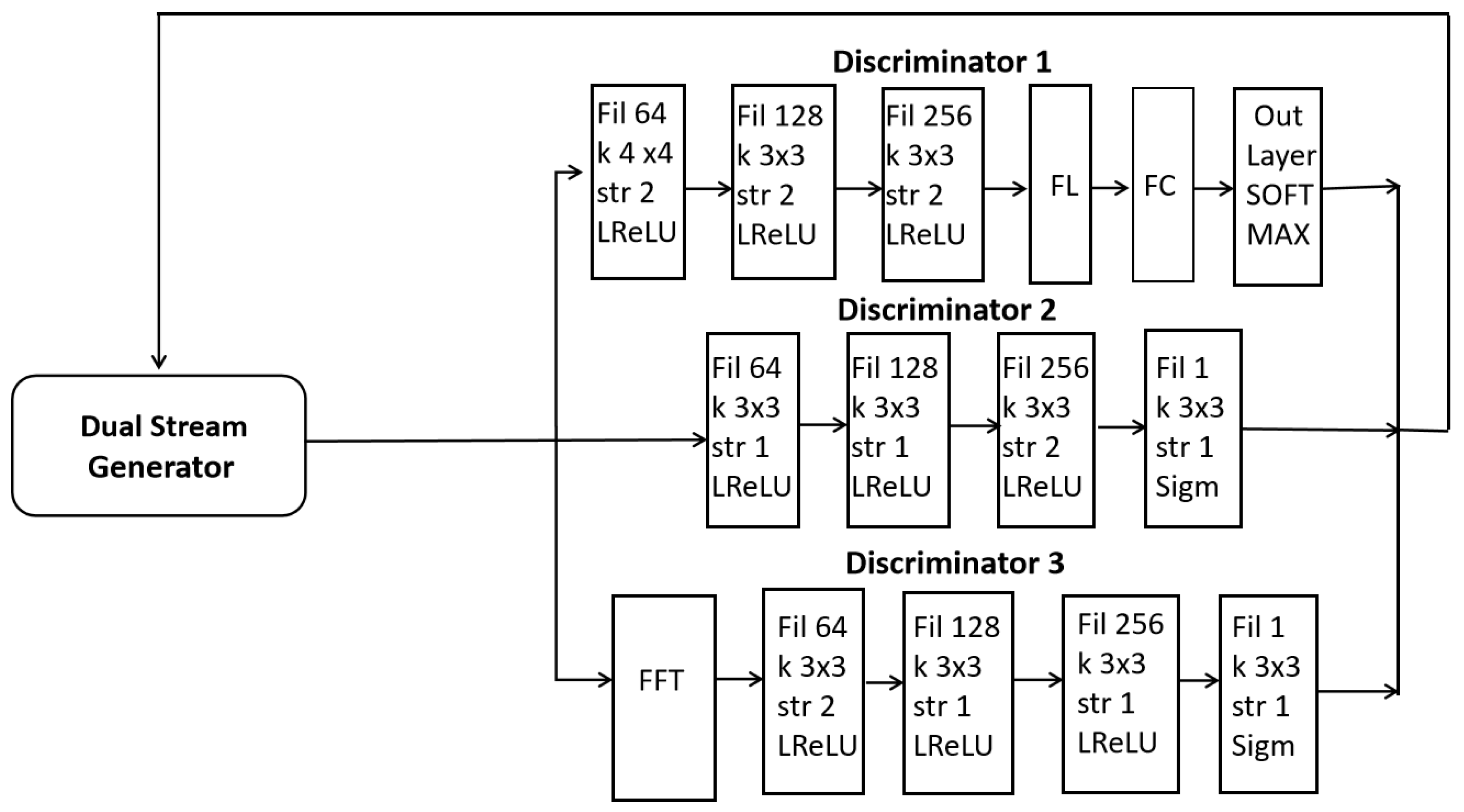
- A three-discriminator architecture is introduced, operating in parallel to assess image quality, diversity, and frequency consistency. Additionally, D1 performs classification, eliminating the need for a separate brain tumor (BT) classifier network.
2. Materials & Methods
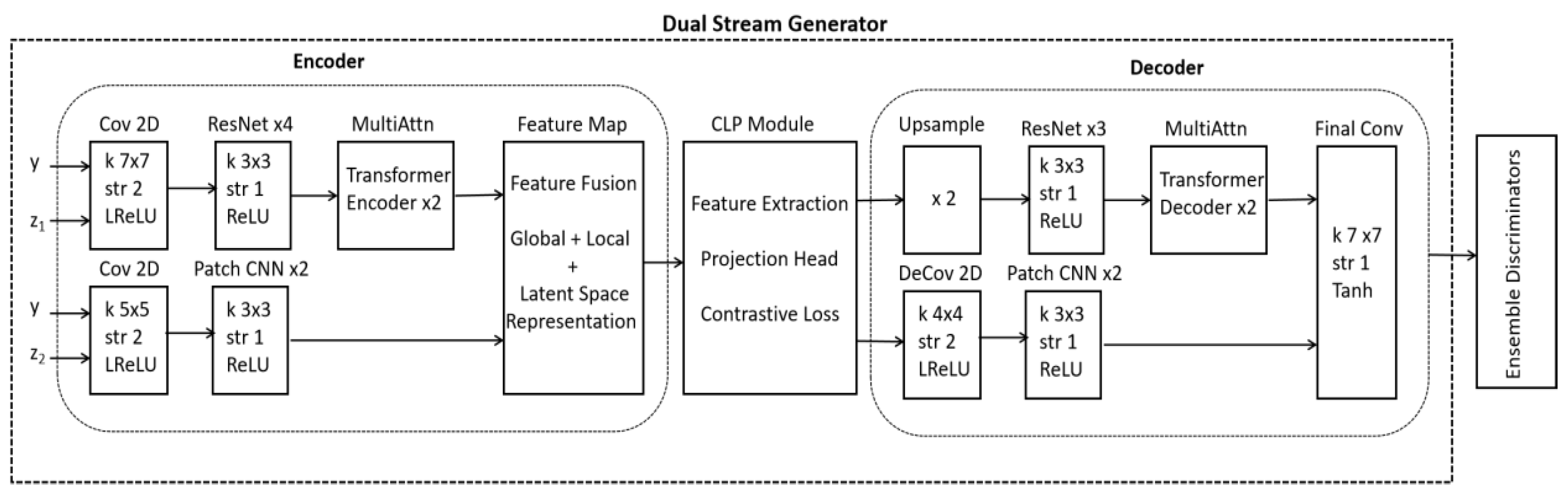
2.1. Dual Stream Generator of Our Proposed Model (DSCLPGAN)
2.2. Complete Architecture of Our Proposed Model (DSCLPGAN)
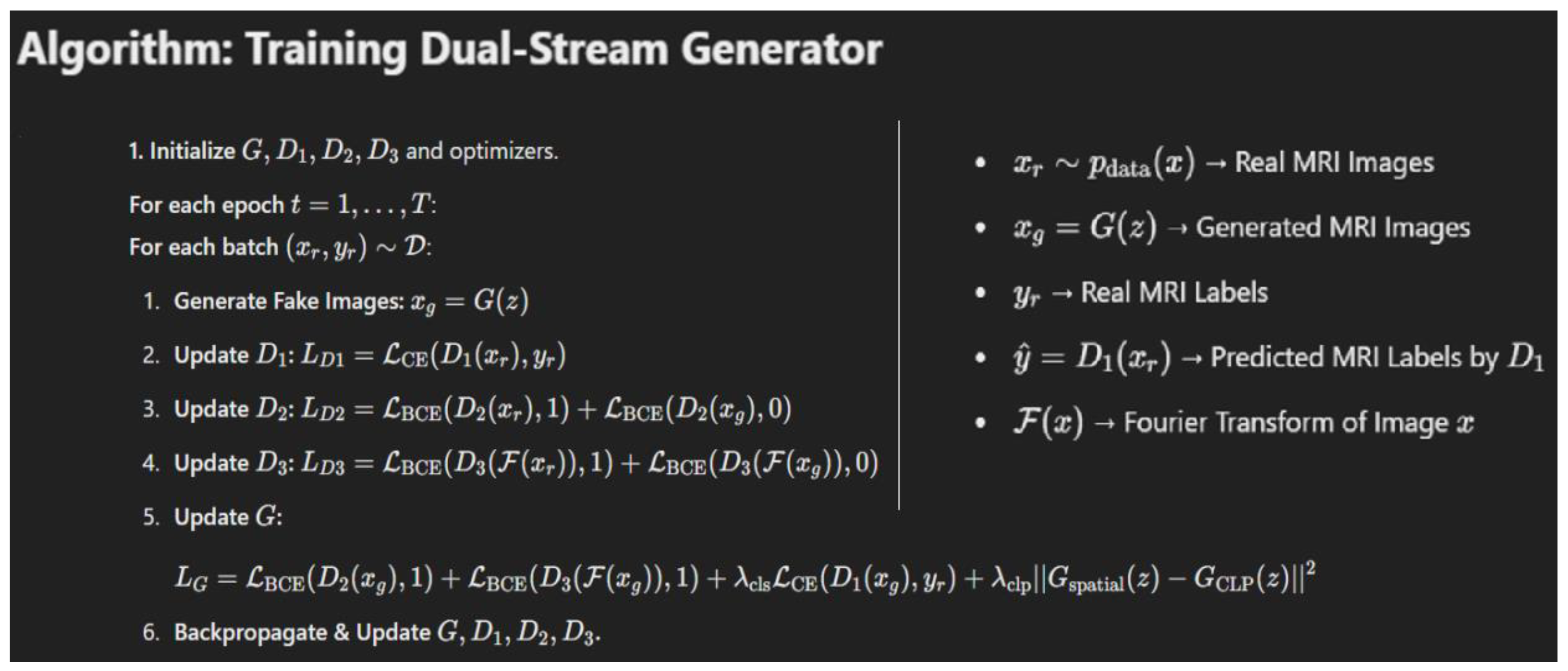
2.3. Mathematical Formulation
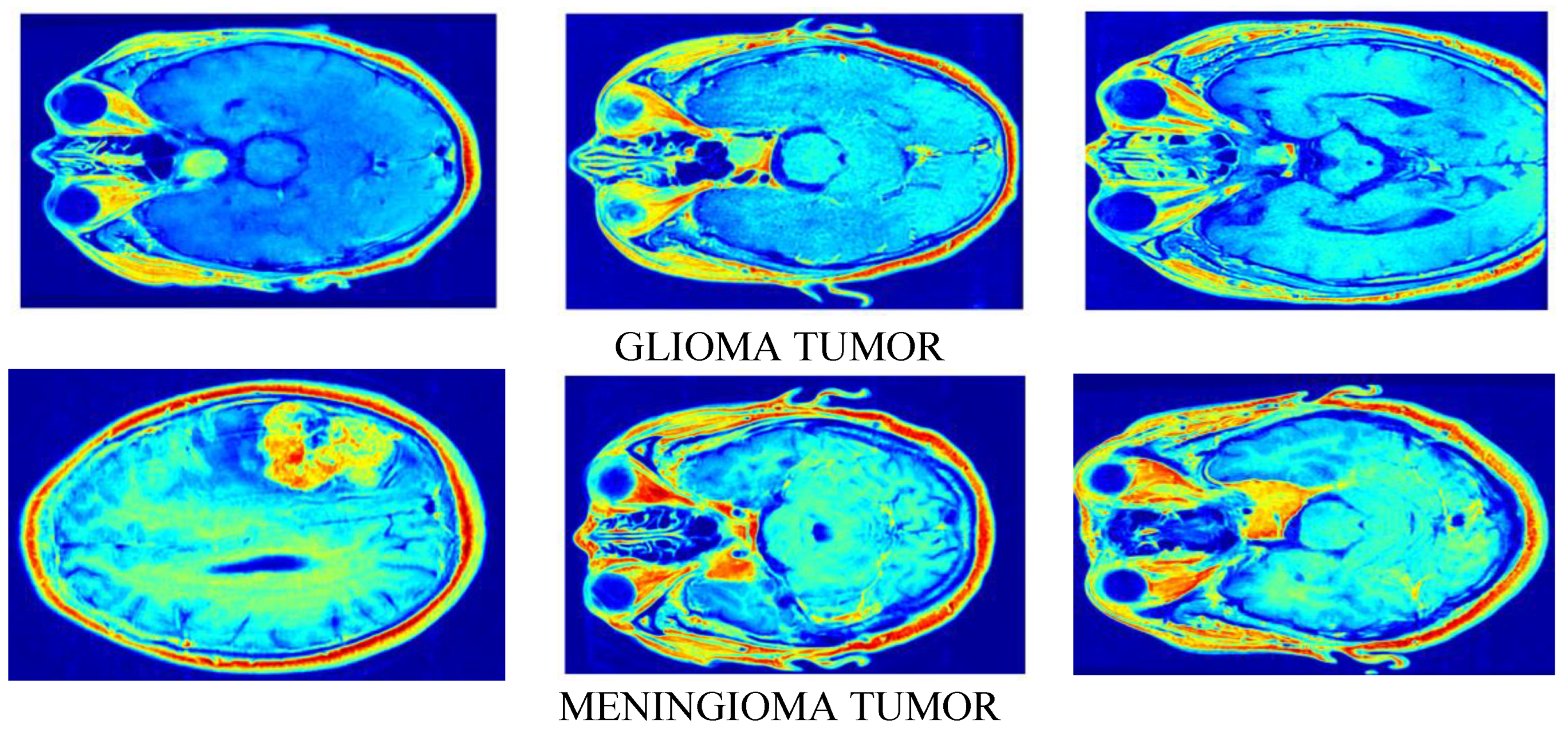
3. Results & Analysis
4. Conclusions
References
- Kaifi, R. A Review of Recent Advances in Brain Tumor Diagnosis Based on AI-Based Classification. Diagnostics (Basel). 2023 Sep 20;13(18):3007.
- Fan, Y. , Zhang, X., Gao, C. et al. Burden and trends of brain and central nervous system cancer from 1990 to 2019 at the global, regional, and country levels. Arch Public Health 80, 209 (2022).
- Arnaout, M.M. , Hoz, S., Lee, A. et al. Management of patients with multiple brain metastases. Egypt J Neurosurg 39, 64 (2024).
- Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021 Aug 2;23(8):1231-1251.
- https://www.hopkinsmedicine.org/health/conditions-and-diseases/brain-tumor.
- Louis, D.N., Perry, A., Reifenberger, G. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131, 803–820 (2016).
- Delgado-López, P.D. , Corrales-García, E.M. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol 18, 1062–1071 (2016).
- Abdusalomov, A.B.; Mukhiddinov, M.; Whangbo, T.K. Brain Tumor Detection Based on Deep Learning Approaches and Magnetic Resonance Imaging. Cancers 2023, 15, 4172. [Google Scholar] [CrossRef] [PubMed]
- Lazli, L.; Boukadoum, M.; Mohamed, O.A. A Survey on Computer-Aided Diagnosis of Brain Disorders through MRI Based on Machine Learning and Data Mining Methodologies with an Emphasis on Alzheimer Disease Diagnosis and the Contribution of the Multimodal Fusion. Appl. Sci. 2020, 10, 1894. [Google Scholar] [CrossRef]
- Virupakshappa, Amarapur, B. Computer-aided diagnosis applied to MRI images of brain tumor using cognition based modified level set and optimized ANN classifier. Multimed Tools Appl 79, 3571–3599 (2020).
- Das, S., Goswami, R.S. Advancements in brain tumor analysis: a comprehensive review of machine learning, hybrid deep learning, and transfer learning approaches for MRI-based classification and segmentation. Multimed Tools Appl (2024).
- Md. Naim Islam, Md. Shafiul Azam, Md. Samiul Islam, Muntasir Hasan Kanchan, A.H.M. Shahariar Parvez, Md. Monirul Islam, An improved deep learning-based hybrid model with ensemble techniques for brain tumor detection from MRI image, Informatics in Medicine, Volume 47, 2024, 101483.
- Amran, G.A.; Alsharam, M.S.; Blajam, A.O.A.; Hasan, A.A.; Alfaifi, M.Y.; Amran, M.H.; Gumaei, A.; Eldin, S.M. Brain Tumor Classification and Detection Using Hybrid Deep Tumor Network. Electronics 2022, 11, 3457. [Google Scholar] [CrossRef]
- S. Karim et al., "Developments in Brain Tumor Segmentation Using MRI: Deep Learning Insights and Future Perspectives," in IEEE Access, vol. 12, pp. 26875-26896, 2024.
- Sajjanar, R. , Dixit, U.D. & Vagga, V.K. Advancements in hybrid approaches for brain tumor segmentation in MRI: a comprehensive review of machine learning and deep learning techniques. Multimed Tools Appl 83, 30505–30539 (2024).
- Prakash, R. M. , Kumari, R. S. S., Valarmathi, K., & Ramalakshmi, K. (2022). Classification of brain tumours from MR images with an enhanced deep learning approach using densely connected convolutional network. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 11(2), 266–277.
- https://onlinelibrary.wiley.com/doi/abs/10.1002/ima.22975.
- Mathivanan, S.K., Sonaimuthu, S., Murugesan, S. et al. Employing deep learning and transfer learning for accurate brain tumor detection. Sci Rep 14, 7232 (2024).
- S. Katkam, V. Prema Tulasi, B. Dhanalaxmi and J. Harikiran, "Multi-Class Diagnosis of Neurodegenerative Diseases Using Effective Deep Learning Models With Modified DenseNet-169 and Enhanced DeepLabV 3+," in IEEE Access, vol. 13, pp. 29060-29080, 2025.
- S. Ahmad and P. K. Choudhury, "On the Performance of Deep Transfer Learning Networks for Brain Tumor Detection Using MR Images," in IEEE Access, vol. 10, pp. 59099-59114, 2022.
- Mok, T.C.W. , Chung, A.C.S. (2019). Learning Data Augmentation for Brain Tumor Segmentation with Coarse-to-Fine Generative Adversarial Networks. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science(), vol 11383. Springer, Cham.
- https://onlinelibrary.wiley.com/doi/abs/10.1002/cpe.7031.
- Han, C.; et al. (2020). Infinite Brain MR Images: PGGAN-Based Data Augmentation for Tumor Detection. In: Esposito, A., Faundez-Zanuy, M., Morabito, F., Pasero, E. (eds) Neural Approaches to Dynamics of Signal Exchanges. Smart Innovation, Systems and Technologies, vol 151. Springer, Singapore.
- Goceri, E. Medical image data augmentation: techniques, comparisons and interpretations. Artif Intell Rev 56, 12561–12605 (2023).
- Sanaat, A. , Shiri, I., Ferdowsi, S. et al. Robust-Deep: A Method for Increasing Brain Imaging Datasets to Improve Deep Learning Models’ Performance and Robustness. J Digit Imaging 35, 469–481 (2022).
- Gab Allah, A.M.; Sarhan, A.M.; Elshennawy, N.M. Classification of Brain MRI Tumor Images Based on Deep Learning PGGAN Augmentation. Diagnostics 2021, 11, 2343. [Google Scholar] [CrossRef] [PubMed]
- https://arxiv.org/abs/2411.00875.
- Asiri, A.A.; Shaf, A.; Ali, T.; Aamir, M.; Irfan, M.; Alqahtani, S.; Mehdar, K.M.; Halawani, H.T.; Alghamdi, A.H.; Alshamrani, A.F.A.; et al. Brain Tumor Detection and Classification Using Fine-Tuned CNN with ResNet50 and U-Net Model: A Study on TCGA-LGG and TCIA Dataset for MRI Applications. Life 2023, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Andrés Anaya-Isaza, Leonel Mera-Jiménez, Lucía Verdugo-Alejo, Luis Sarasti, Optimizing MRI-based brain tumor classification and detection using AI: A comparative analysis of neural networks, transfer learning, data augmentation, and the cross-transformer network, European Journal of Radiology Open, Volume 10, 2023, 100484.
- A.M. Kocharekar, S. Datta, Padmanaban and R. R., "Comparative Analysis of Vision Transformers and CNN-based Models for Enhanced Brain Tumor Diagnosis," 2024 3rd International Conference on Automation, Computing and Renewable Systems (ICACRS), Pudukkottai, India, 2024, pp. 1217-1223.
- Zakariah, Mohammed et al, Dual vision Transformer-DSUNET with feature fusion for brain tumor segmentation Heliyon, Volume 10, Issue 18, e37804, 2024.
- L. ZongRen, W. Silamu, W. Yuzhen and W. Zhe, "DenseTrans: Multimodal Brain Tumor Segmentation Using Swin Transformer," in IEEE Access, vol. 11, pp. 42895-42908, 2023. [CrossRef]
- Swetha, A. V. S. , Bala, M., & Sharma, K. (2024). A Linear Time Shrinking-SL(t)-ViT Approach for Brain Tumor Identification and Categorization. IETE Journal of Research, 70(11), 8300–8322.
- Sadafossadat Tabatabaei, Khosro Rezaee, Min Zhu, Attention transformer mechanism and fusion-based deep learning architecture for MRI brain tumor classification system, Biomedical Signal Processing and Control, Volume 86, Part A, 2023, 105119,ISSN 1746-8094.
- Pacal, I. A novel Swin transformer approach utilizing residual multi-layer perceptron for diagnosing brain tumors in MRI images. Int. J. Mach. Learn. & Cyber. 15, 3579–3597 (2024).
- Tehsin, S.; Nasir, I.M.; Damaševičius, R. GATransformer: A Graph Attention Network-Based Transformer Model to Generate Explainable Attentions for Brain Tumor Detection. Algorithms 2025, 18, 89. [Google Scholar] [CrossRef]
- Usman Akbar, M. , Larsson, M., Blystad, I. et al. Brain tumor segmentation using synthetic MR images - A comparison of GANs and diffusion models. Sci Data 11, 259 (2024).
- A. Saeed, K. Shehzad, S. S. Bhatti, S. Ahmed and A. T. Azar, "GGLA-NeXtE2NET: A Dual-Branch Ensemble Network With Gated Global-Local Attention for Enhanced Brain Tumor Recognition," in IEEE Access, vol. 13, pp. 7234-7257, 2025.
- Karpakam, S. , Kumareshan, N. Enhanced brain tumor detection and classification using a deep image recognition generative adversarial network (DIR-GAN): a comparative study on MRI, X-ray, and FigShare datasets. Neural Comput & Applic (2025). [CrossRef]
- L. Desalegn and W. Jifara, "HARA-GAN: Hybrid Attention and Relative Average Discriminator Based Generative Adversarial Network for MR Image Reconstruction," in IEEE Access, vol. 12, pp. 23240-23251, 2024.
- Lyu Y, Tian X. MWG-UNet++: Hybrid Transformer U-Net Model for Brain Tumor Segmentation in MRI Scans. Bioengineering (Basel). 2025 Jan 31;12(2):140. [CrossRef] [PubMed] [PubMed Central]
- Ahmed, S. , Feng, J., Ferzund, J. et al. FAME: A Federated Adversarial Learning Framework for Privacy-Preserving MRI Reconstruction. Appl Magn Reson (2025).
- Yang Yang, Xianjin Fang, Xiang Li, Yuxi Han, Zekuan Yu, CDSG-SAM: A cross-domain self-generating prompt few-shot brain tumor segmentation pipeline based on SAM, Biomedical Signal Processing and Control, Volume 100, Part B, 2025, 106936, ISSN 1746-8094.
- J. Donahue and K. Simonyan, “Large scale adversarial representation learning,” Advances in neural information processing systems, vol. 32, 2019.
- T. Li, H. Chang, S. K. Mishra, H. Zhang, D. Katabi, and D. Krishnan, “Mage: Masked generative encoder to unify representation learning and image synthesis,” 2022.
- https://arxiv.org/abs/2102.07074.
- https://arxiv.org/abs/2312.01999.
- Xu M, Cui J, Ma X, Zou Z, Xin Z, Bilal M. Image enhancement with art design: a visual feature approach with a CNN-transformer fusion model. PeerJ Comput Sci. 2024 Nov 13;10:e2417.
- Yurtsever MME, Atay Y, Arslan B, Sagiroglu S. Development of brain tumor radiogenomic classification using GAN-based augmentation of MRI slices in the newly released gazi brains dataset. BMC Med Inform Decis Mak. 2024 Oct 4;24(1):285.
- https://arxiv.org/abs/2501.07055v1.
- Zhou M, Wagner MW, Tabori U, Hawkins C, Ertl-Wagner BB, Khalvati F. Generating 3D brain tumor regions in MRI using vector-quantization Generative Adversarial Networks. Comput Biol Med. 2025 Feb;185:109502.
- https://arxiv.org/abs/2412.11849.

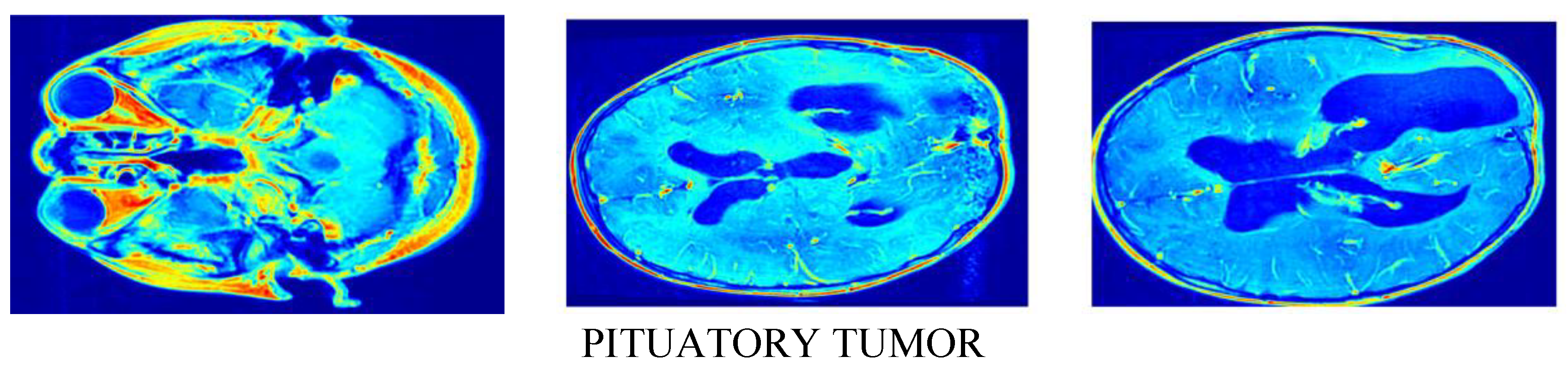
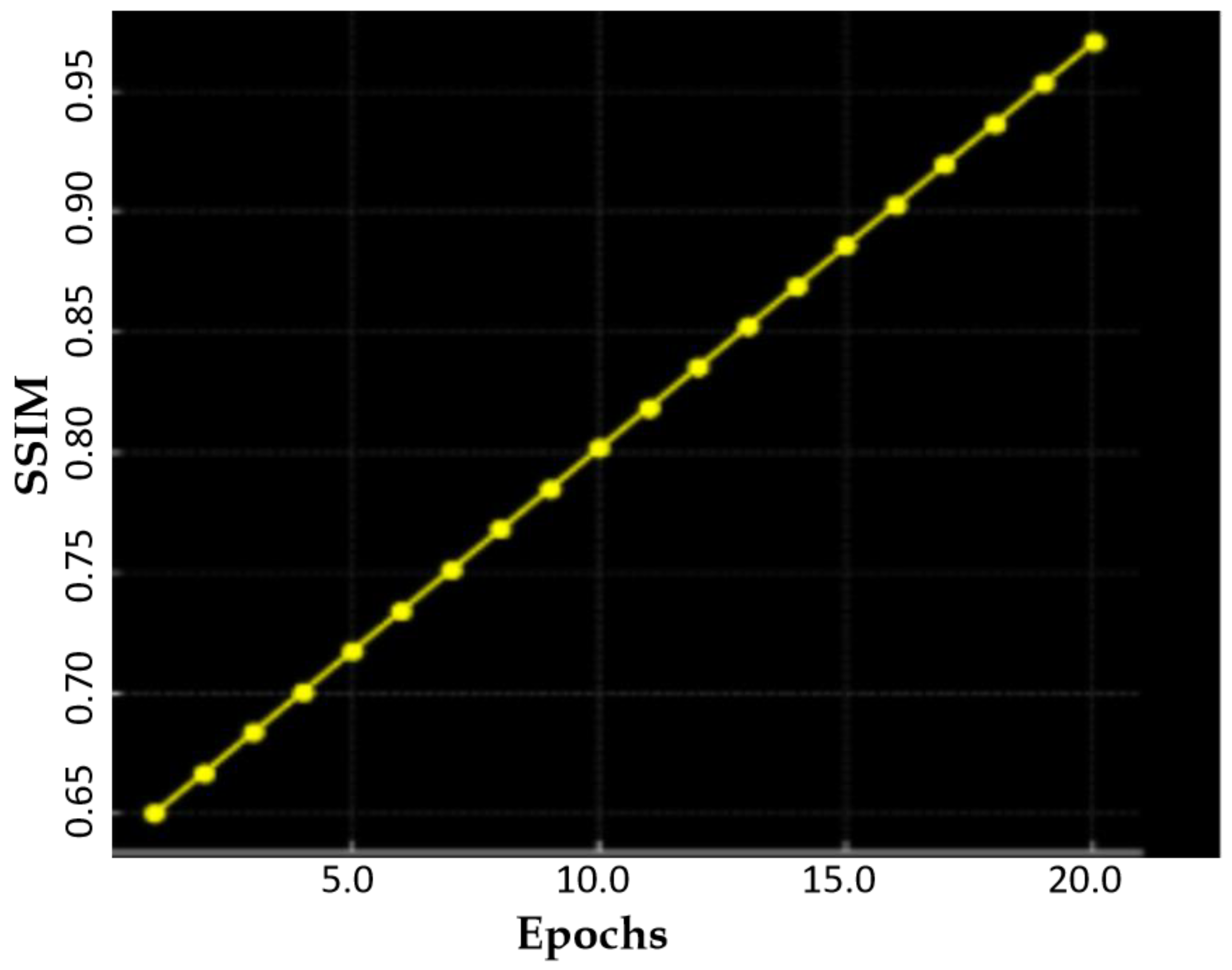
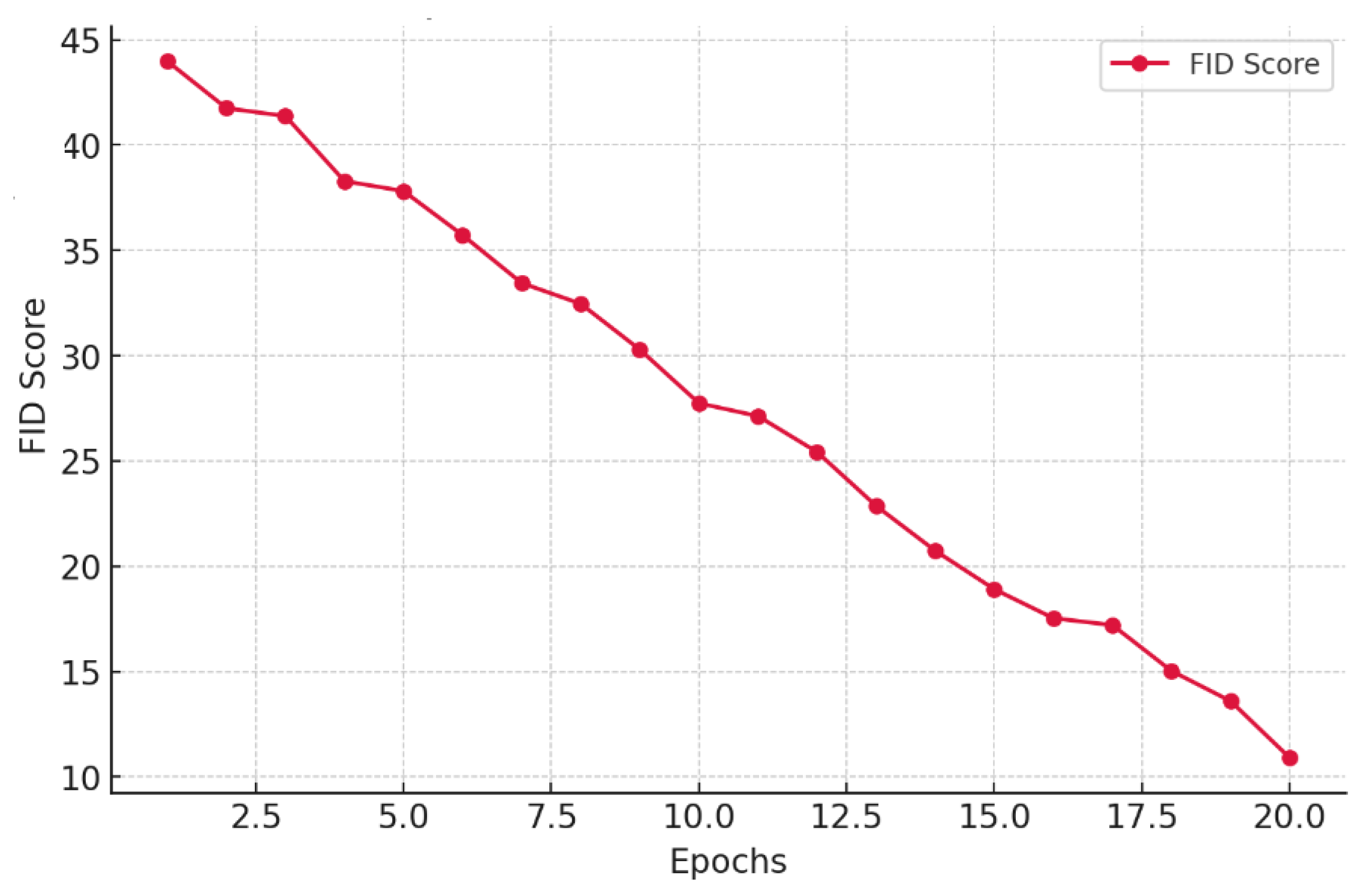
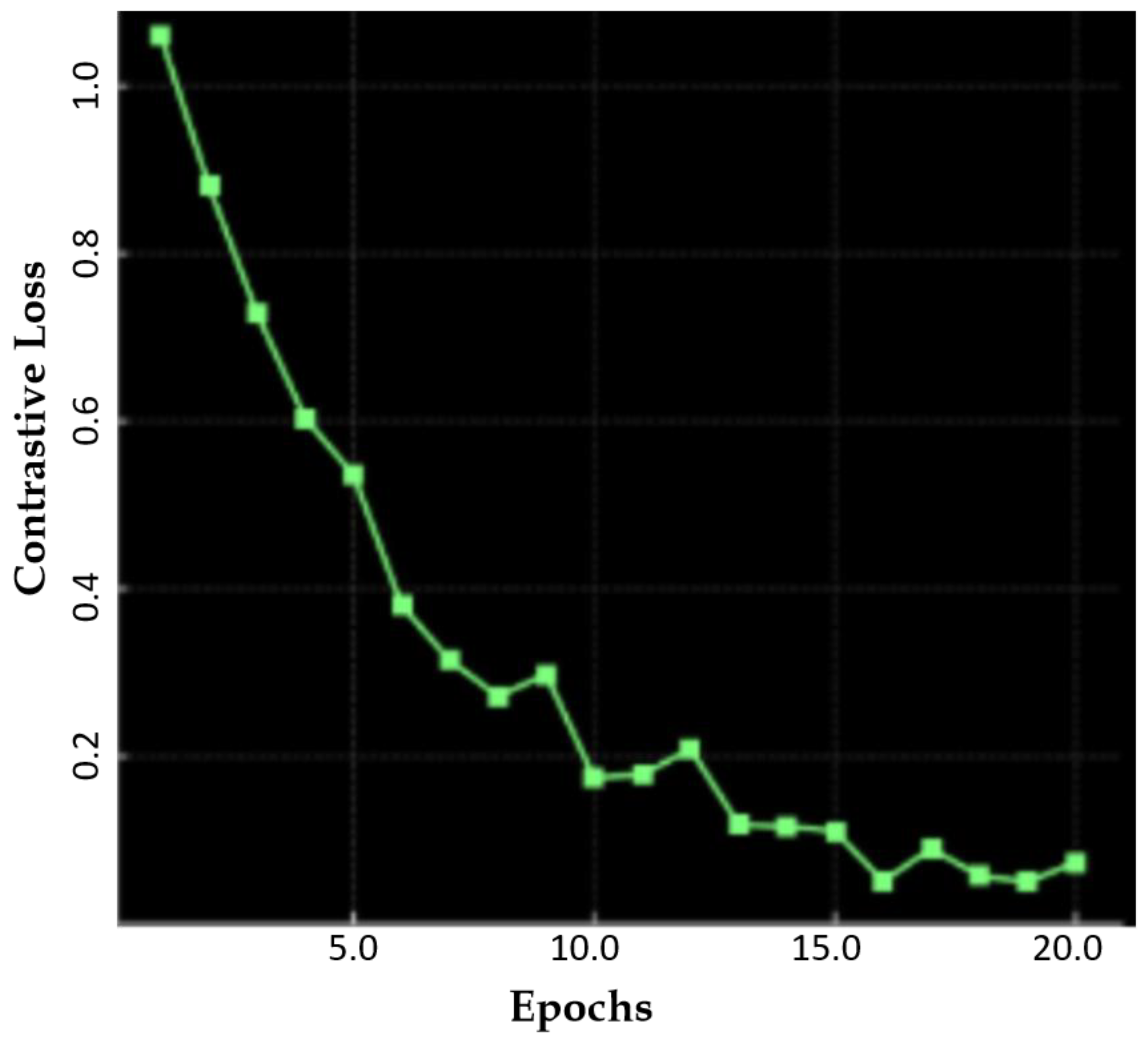
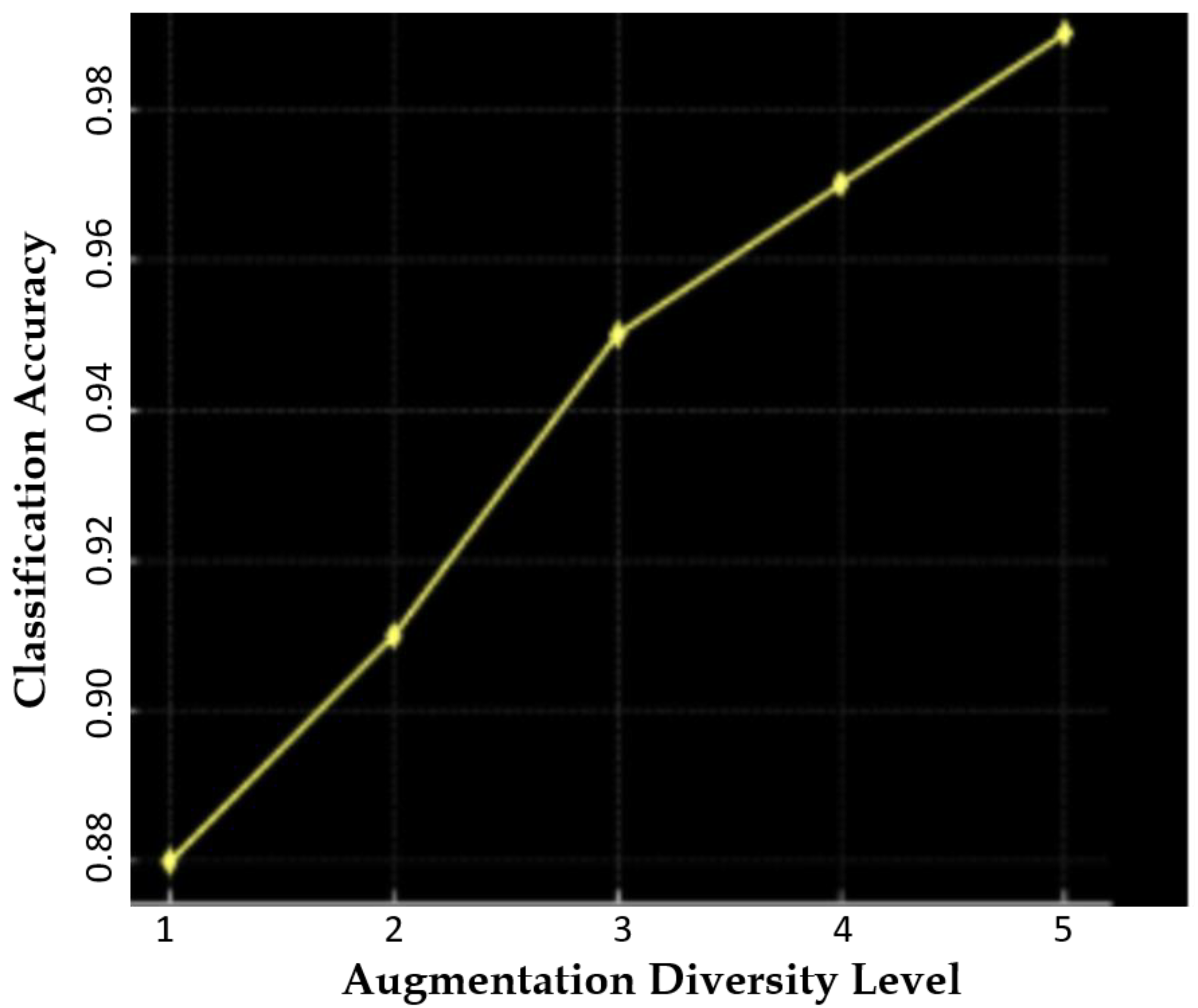
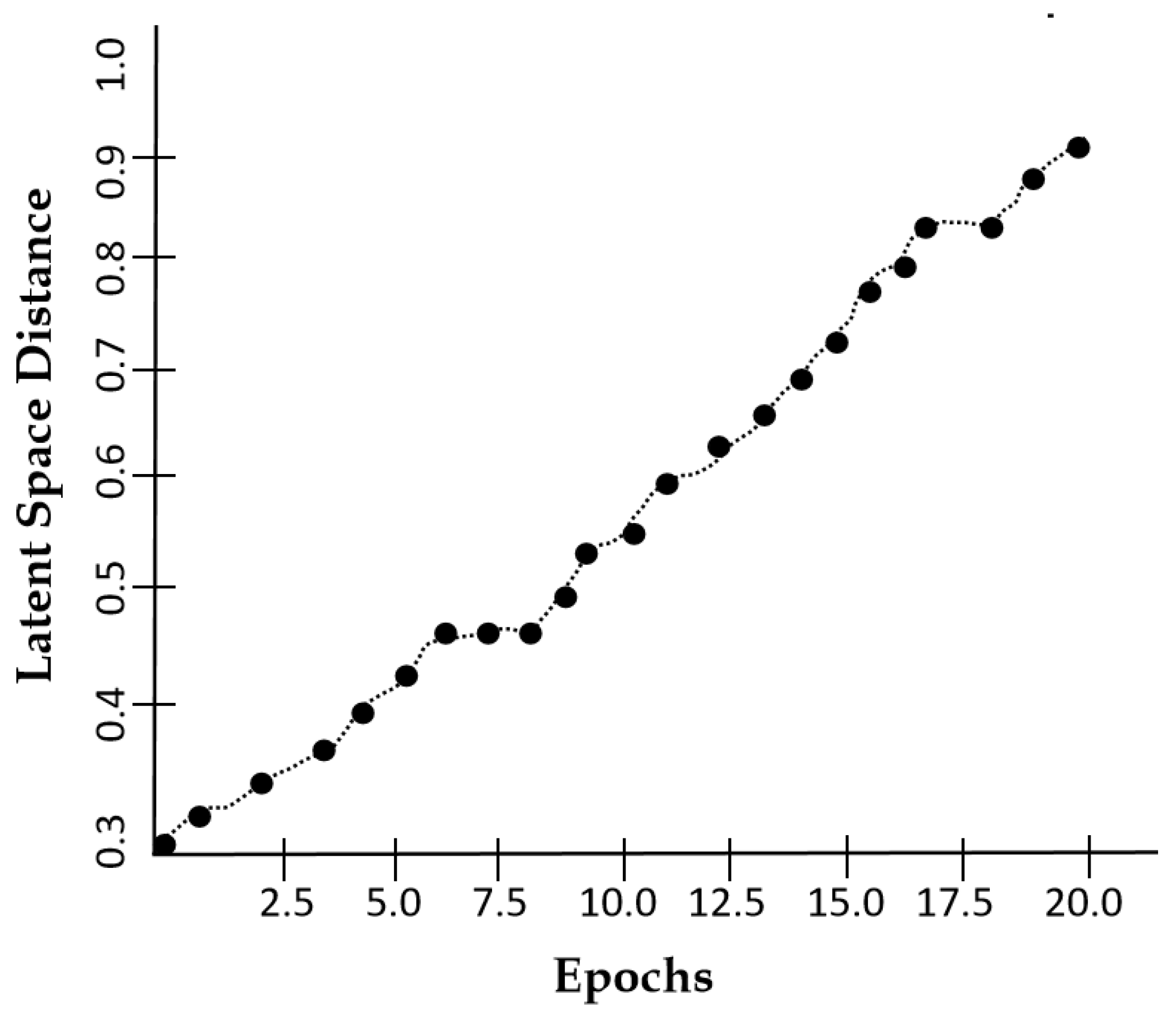
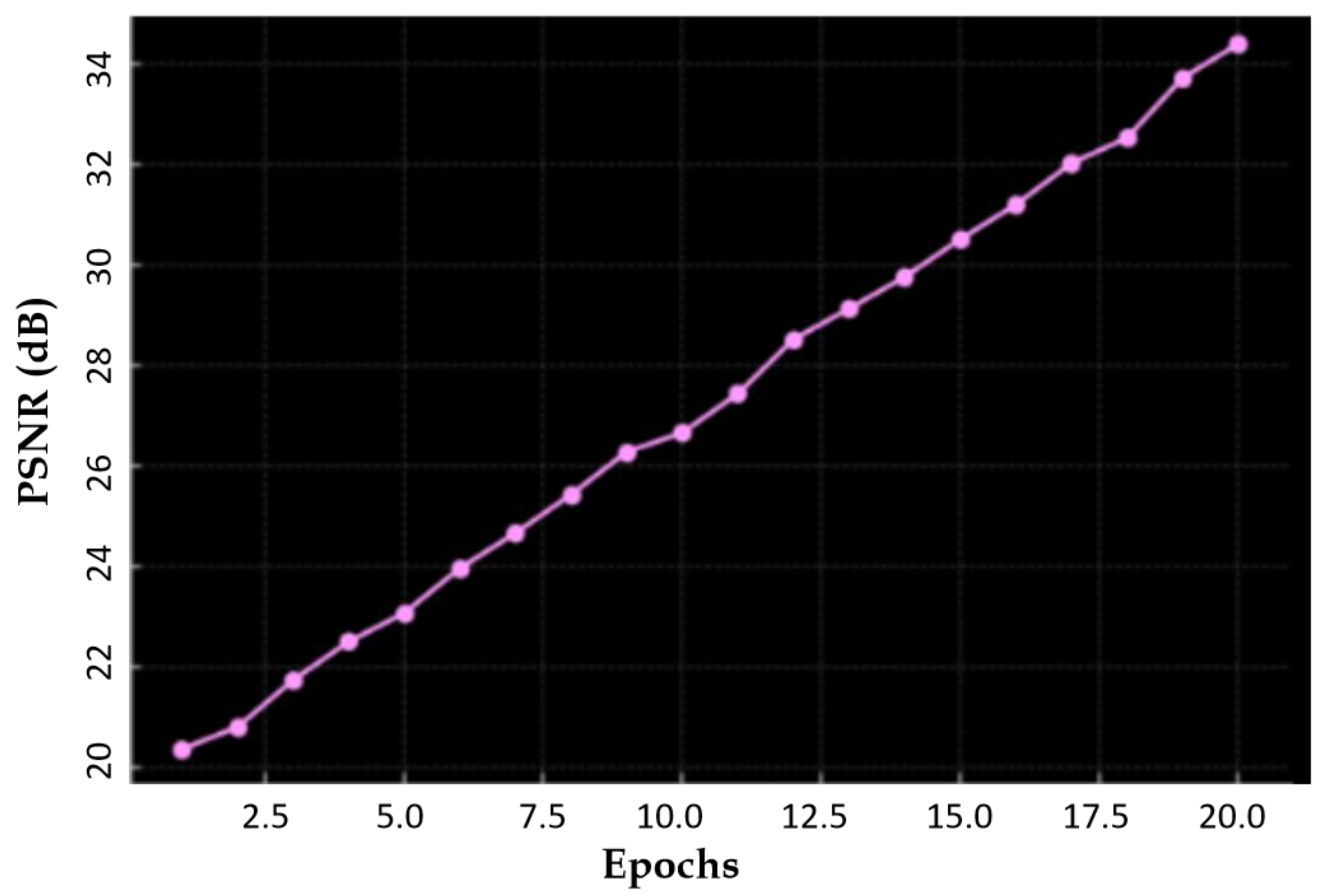
| Method | SSIM | FID | PSNR |
| BIGGAN [44] | 0.7314 | 47.63 | 25.89 |
| MAGE [45] | 0.8220 | 45.62 | 27.28 |
| TransGAN [46] | 0.8376 | 35.45 | 27.66 |
| SR TransGAN [47] | 0.8504 | 31.29 | 30.28 |
| CTGAN[48] | 0.8755 | 29.10 | 26.47 |
| StyleGANv2 [49] | 0.8841 | 32.56 | 29.31 |
| SFCGAN [50] | 0.9077 | 28.04 | 29.14 |
| VQ- GAN [51] | 0.9166 | 26.55 | 31.04 |
| 3D Pix2Pix GAN [52] | 0.9210 | 27.87 | 30.19 |
| Proposed DSCLPGAN | 0.9861 | 12 | 34.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





