1. Introduction
Forests are an essential part of global vegetation, widely distributed across various climatic and soil conditions. They play an irreplaceable role in maintaining ecological balance, preserving biodiversity, regulating climate, and providing resources for humanity [
1,
2,
3]. As a key component of Earth’s life-support system and sustainable human development, forests are increasingly challenged by rapid population growth, urbanization, and modernization. Balancing forest ecological protection with technological advancements has become a pressing issue. Strengthening forestry research, advancing forestry science and technology, and enhancing forest monitoring and assessment are crucial research topics in forestry and environmental protection. Efficient vegetation information statistics are a key aspect of this process. They are vital for understanding forest vegetation growth, accurately assessing forest ecological quality, scientifically formulating conservation strategies, and optimizing forestry production layouts [
4]. Vegetation indices serve as an essential tool for measuring vegetation conditions, directly reflecting growth status, coverage, and ecological functions. They provide a simple and effective empirical measure of surface vegetation conditions and are widely applied in land cover analysis, vegetation classification, and environmental change monitoring. By analyzing vegetation time-series data, researchers can assess ecosystem health and stability, indirectly reflecting biodiversity changes. This is significant for natural resource management and ecological balance maintenance [
5,
6]. Additionally, as a crucial tool in ecological research, vegetation indices support scientific studies by offering data and insights, promoting further ecological research development.
The concept and development of vegetation indices date back to the late 1960s to the early 1990s. In 1969, Jordan proposed the first vegetation index—Ratio Vegetation Index (RVI)—which calculates the reflectance ratio between the near-infrared and red bands [
7]. In 1972, Pearson et al. introduced the Vegetation Index Number (VIN), furthering vegetation index research [
8]. In 1973, Rouse et al. applied a nonlinear normalization process to RVI, resulting in the Normalized Difference Vegetation Index (NDVI) [
9], which later became widely used in vegetation remote sensing [
10]. To date, over 40 vegetation indices have been defined globally, covering parameters such as vegetation coverage, chlorophyll content, and canopy structure [
11,
12].
The Leaf Area Index (LAI) is one of the fundamental parameters describing canopy structure. It is defined as half the total leaf surface area per unit ground area [
13]. LAI reflects leaf quantity, distribution, and canopy density and is crucial for studying plant growth, photosynthesis, transpiration, and crop yield [
14,
15]. It has broad applications and research value in forestry and agriculture [
16]. Accurate measurement and analysis of LAI enable a better understanding of plant health and growth, providing a scientific basis for forestry and agricultural management [
17,
18,
19,
20].
The measurement methods for LAI primarily include direct and indirect approaches [
21]. The direct measurement method involves collecting plant leaf samples, measuring their area, and calculating the total leaf area per unit of land to determine LAI. Although this method is accurate, it is labor-intensive, and the collection of leaf samples can cause some damage to the plants. It is typically suitable for small-scale or specific-area measurements and is difficult to apply over large areas [
22]. In contrast, the indirect measurement method estimates LAI by measuring parameters such as canopy reflectance and transmittance using remote sensing technology and optical instruments, combined with mathematical models [
23,
24]. This approach offers advantages such as fast measurement speed, wide coverage, and the ability to conduct large-scale measurements quickly without damaging trees, saving both labor and resources. As a result, it is widely used for LAI measurement [
25].
Measurement methods based on radiative transfer models estimate LAI indirectly by simulating light transmission characteristics within plant canopies. Common instruments include TRAC, AccuPAR, and the LAI-2000 series plant canopy analyzers [
26,
27,
28]. These instruments provide high measurement accuracy and are suitable for various types of vegetation, geographical regions, and ecosystems. However, radiative transfer models often rely on certain assumptions (e.g., uniform canopy structure, single leaf morphology), which may reduce their applicability in complex environments, thereby affecting the ac-curacy of LAI estimation. Additionally, radiative transfer models are highly sensitive to environmental factors such as light conditions, weather, and atmospheric conditions. They require high-precision optical sensors to capture the optical properties of plant canopies, which often come with high costs and technical requirements, increasing the barrier to their use [
29].
In recent years, the hemispherical photography-based LAI measurement method has gained significant attention among researchers, driven by advancements in image processing technology [
29]. This approach involves capturing canopy images from beneath the vegetation using a fisheye lens-equipped camera. By applying image segmentation techniques, the method distinguishes leaf areas (target pixels) from gaps (sky pixels) and estimates LAI using radiative transfer models or empirical models. Compared to traditional radiative transfer model-based methods, hemispherical photography offers the advantage of permanently documenting vegetation dynamics, providing valuable visual data for long-term research and enabling the tracking of canopy changes over time [
30,
31,
32]. Additionally, it is more cost-effective and easier to operate than high-precision optical sensors, making it a practical choice for large-scale ecological monitoring.
The integration of artificial intelligence and automated image analysis software has further enhanced the efficiency of this method [
33], enabling rapid processing of large datasets and extraction of canopy structural information for LAI estimation. However, challenges remain in practical applications. Image segmentation accuracy can be compromised by variable lighting conditions and image quality, particularly in dense forests or complex ecosystems, where distinguishing between leaf and sky pixels becomes difficult [
34,
35,
36]. These limitations highlight the need for continued refinement of the technique to improve robustness and reliability in diverse environmental conditions.
The accuracy of vegetation image segmentation algorithms significantly impacts the precision of hemispherical photography-based LAI measurements. Currently, commonly used image segmentation algorithms primarily include color-based segmentation algorithms and deep learning-based segmentation algorithms. Color-based segmentation algorithms typically rely on grayscale features of images, with common methods including threshold segmentation and edge detection [
37,
38]. These algorithms are relatively simple in structure and computationally efficient, but they are sensitive to factors such as image noise and lighting variations, resulting in limited adaptability to complex scenes and segmentation accuracy [
39,
40]. Additionally, these algorithms struggle to effectively distinguish between leaf areas and trunk areas, often leading to overestimation of the leaf area index. In contrast, deep learning-based segmentation algorithms utilize convolutional neural networks for feature extraction and classification, enabling them to handle complex image segmentation tasks with higher accuracy and processing efficiency. However, these algorithms require substantial computational resources during both training and inference, necessitating high-performance computing hardware. Moreover, the performance of these models heavily depends on the quality and quantity of annotated datasets. Yet, there is currently no publicly available standardized annotated dataset, which limits the further application and development of these algorithms to some extent. As a result, deep learning-based methods remain largely in the experimental research phase and are not yet practical for rapid field measurements [
41,
42].
This study introduces a wavelet-based segmentation paradigm addressing these methodological gaps [
43,
44,
45]. Through multi-scale 9/7 wavelet decomposition, the algorithm simultaneously analyzes grayscale textures and frequency-domain features of canopy components, effectively handling complex forest textures while suppressing noise artifacts. A novel dynamic optimization suppression method improves segmentation precision by adaptively adjusting coefficient thresholds, overcoming the rigidity of conventional fixed-threshold approaches. Experimental validation against LAI-2200C benchmark measurements demonstrates enhanced accuracy across diverse ecosystems, with comparative analysis revealing significant improvements over traditional color-based segmentation in handling mixed-pixel scenarios and structural interference.
The proposed methodology advances LAI quantification by synergizing multi-resolution image analysis with adaptive threshold optimization, particularly effective in heterogeneous canopies where conventional methods struggle [
46]. While maintaining computational practicality for field applications, this approach ad-dresses critical limitations in current segmentation techniques through its inherent noise resilience and multi-scale feature discrimination capabilities. The integration of wavelet transforms with dynamic suppression mechanisms establishes a new framework for vegetation analysis, bridging the gap between computationally intensive deep learning methods and oversimplified traditional algorithms.
2. Materials and Methods
2.1. Study Area and Data Source
2.1.1. Study Area
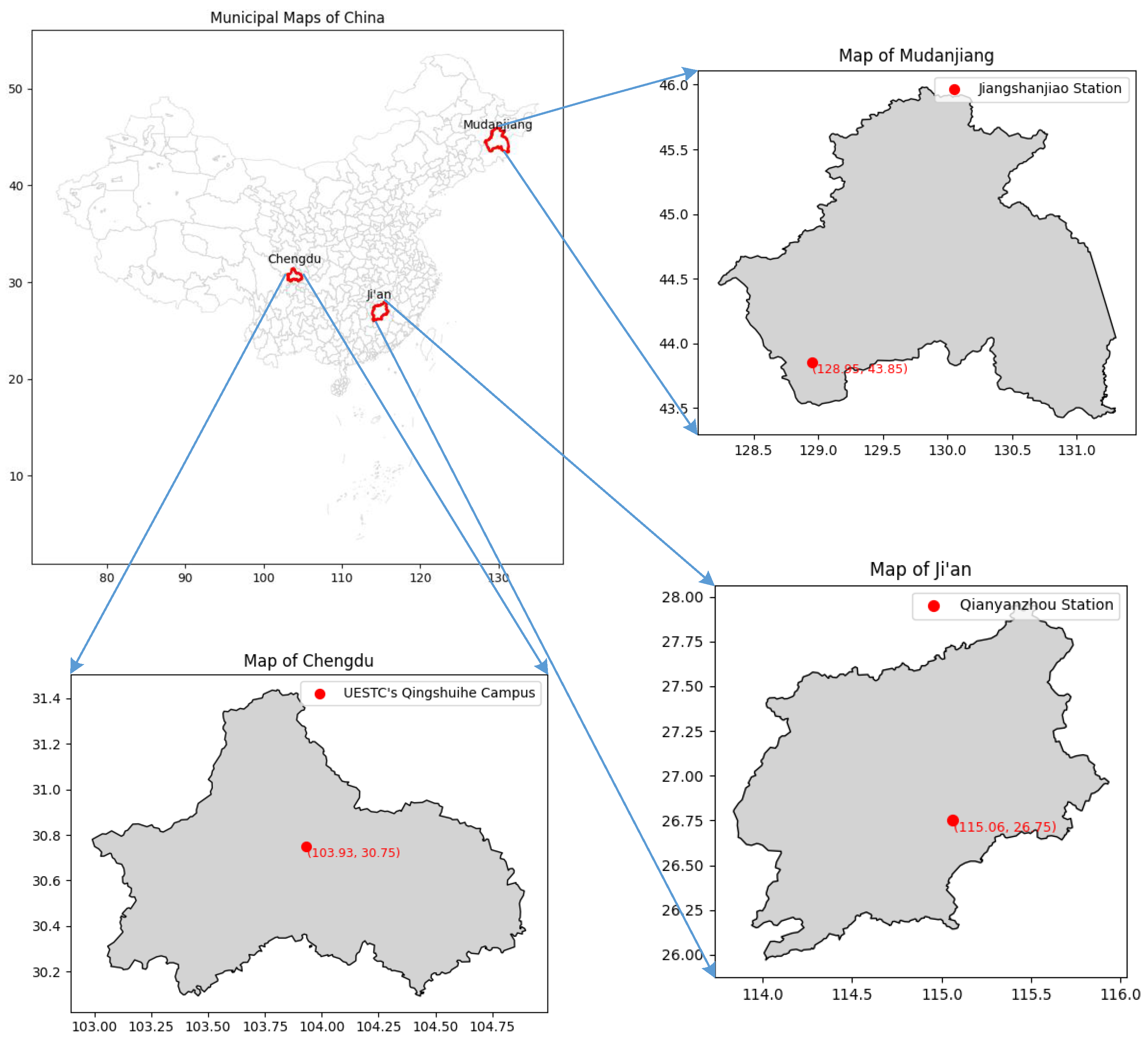
The study selects three forest ecosystems from different geographic regions within China as study areas: Jiangshanjiao Experimental Forest Farm, Forestry Academy of Heilongjiang Province (43°85’N, 128°95’E), Qianyanzhou Subtropical Forest Ecosystem Observation Station, Chinese Academy of Sciences (26°75’N, 115°06’E), and the Qingshui River campus of the University of Electronic Science and Technology of China in Chengdu, Sichuan Province (30°75’N, 103°93’E). The study area map is shown in
Figure 1. The Jiangshanjiao observation station mainly includes temperate mixed forests of larch, red pine, white birch, and mountain ash, while the Qianyanzhou station mainly consists of subtropical evergreen broadleaf forests of oil tea, citrus, and fir trees. The observation site at the University of Electronic Science and Technology includes subtropical evergreen broadleaf forests with camphor, nanmu, and Osmanthus trees.
Each study area has significant differences in climate, soil types, etc., with diverse vegetation types, densities, and heights, aiming to comprehensively evaluate the stability, adaptability, and accuracy of the inversion method proposed in this study when dealing with different vegetation types under varying ecological conditions. An overview of the study areas is shown in
Table 1.
2.1.2. Data Collection
This experiment employs a self-developed vegetation image acquisition system, synchronized with the LAI-2200C instrument, to capture high-resolution canopy images, as illustrated in
Figure 2. The vegetation image acquisition device is an embedded device that integrates image capture and storage functions. It is equipped with an STM32F767 microprocessor, a CMOS image sensor OV2640, and a 160° ultra-wide-angle fisheye lens (in comparison, the LAI-2200C has a field of view of 148°), capable of capturing images with up to 2 million pixels. The device is designed to be compact and lightweight, supporting both independent and synchronized collection with the LAI-2200C. During synchronized collection, the device is secured to the optical probe of the LAI-2200C using Velcro straps to ensure the alignment of the fisheye lens center with the center of the optical sensor of the LAI-2200C.
When measuring, the vegetation image acquisition device and LAI-2200C are connected via a cable, and each time the LAI-2200C takes a measurement, the vegetation image device synchronously captures a hemispherical canopy image, which is stored on the built-in TF card of the device. This method ensures that measurements are taken at the same time and location for the same target, effectively reducing the influence of environmental factors on the results and enhancing data accuracy and reliability.
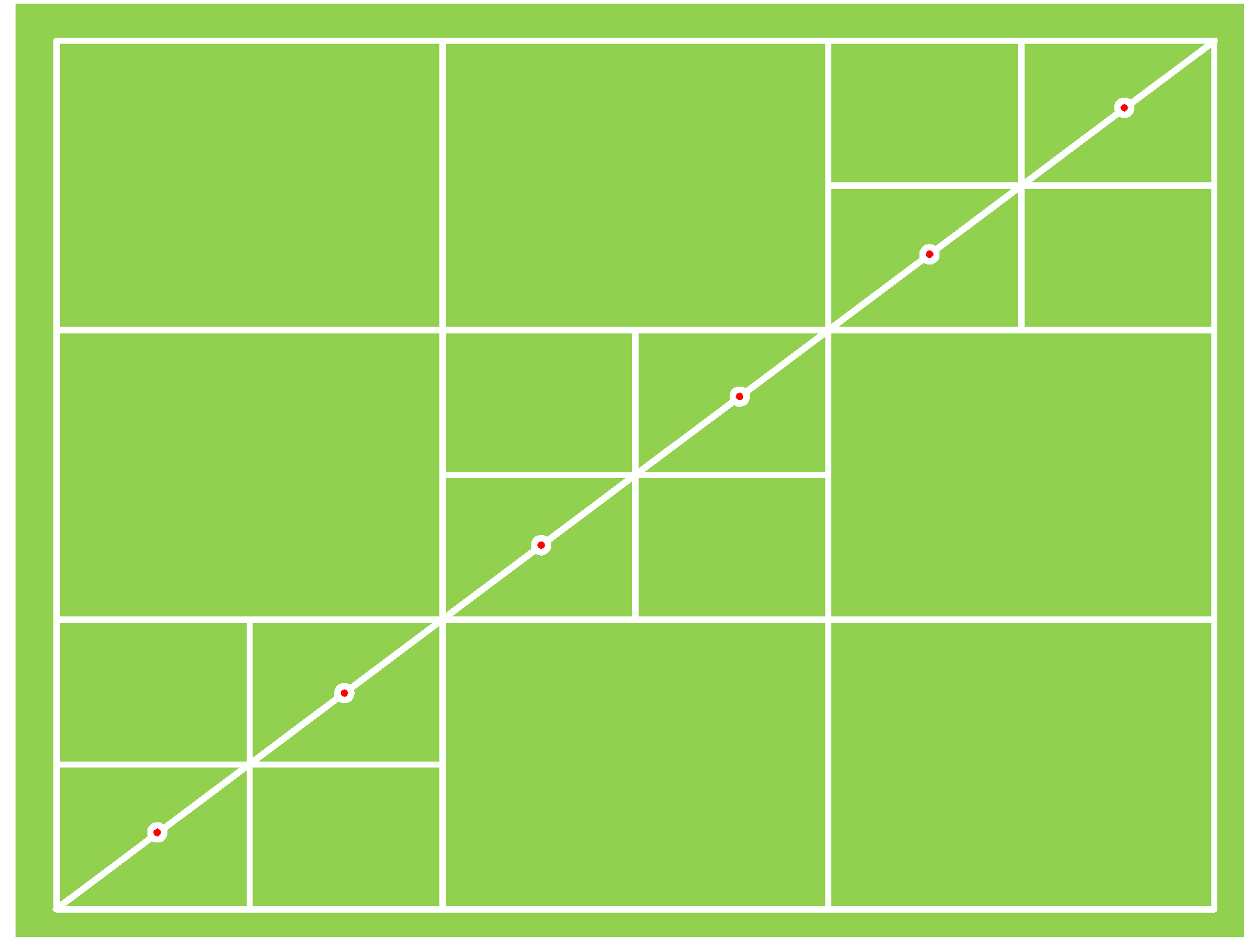
Measurements were conducted in flat areas within the experimental zone using a 6*3 grid sampling method. Specifically, six sample points were collected along the diagonal direction of each sampling area at equal intervals, with a minimum spacing of 10 meters between sampling points. As shown in
Figure 3, the green background represents the sampling area, and the red dots indicate the sample collection points. To minimize single-measurement errors and ensure the stability and accuracy of the data, each sample point was measured three times. Due to the unique characteristics of forest vegetation, trees within the measured area grow non-uniformly, potentially creating irregular gaps of varying sizes, which appear as irregular blank regions in the canopy images.
2.1.3. Dataset
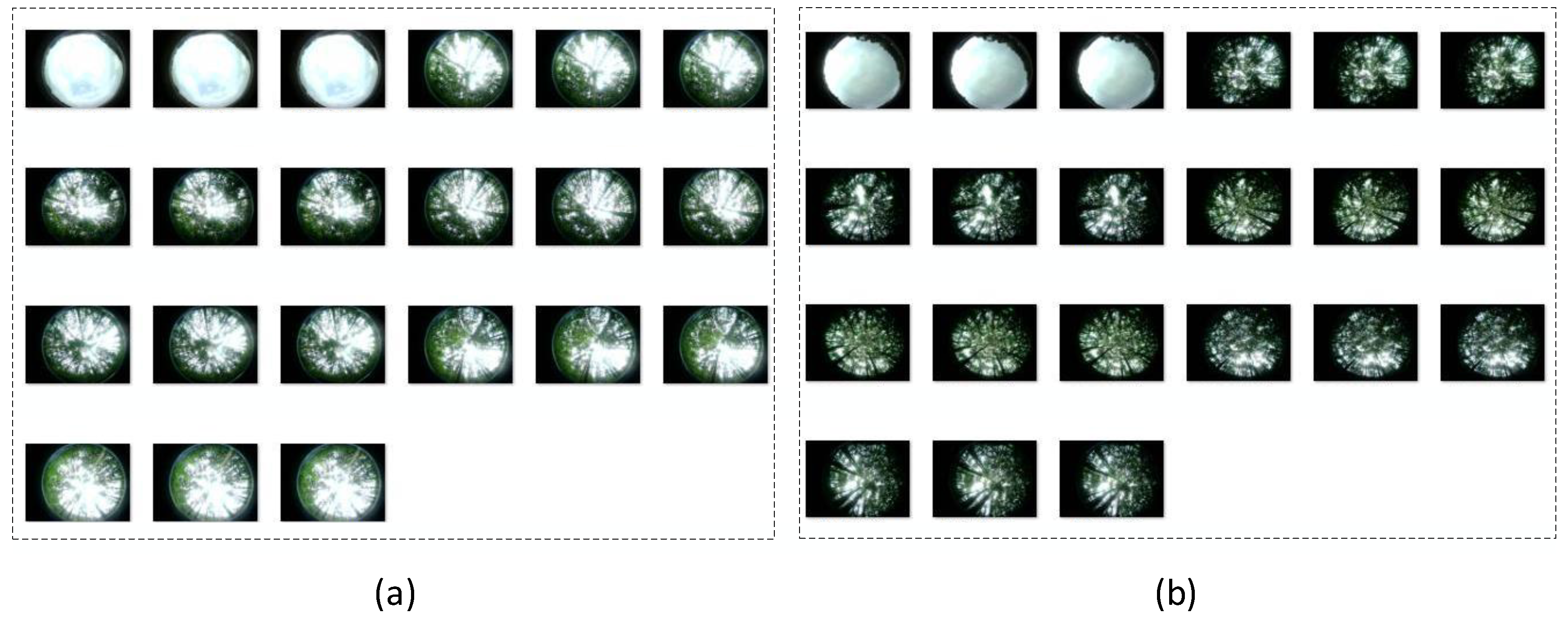
This study collected 60 sets of data, with 20 sets from each study area. Each data set includes one LAI-2200C measurement file and 21 vegetation canopy hemispherical images (3 images are of the outside vegetation, and 18 are of the vegetation canopy). The sample data is shown in
Figure 4. The LAI-2200C measurement file contains information such as firmware version, file creation time, GPS log, and LAI calculation results [
47]. It is important to note that during the measurement process, the LAI-2200C computes the overall LAI of the area by averaging the results of each sample point within a specific sampling region, and generates a measurement file based on this data. In this study, the arithmetic mean of the LAI calculated from the 18 vegetation canopy images was used to represent the measurement results of the inversion method proposed in this paper. This approach ensured a comprehensive consideration of all sample data, effectively reflecting the overall variation in the leaf area index across the sampling area and providing an objective benchmark for evaluating the performance of the inversion method.
2.2. Principles of Wavelet Image Decomposition and the 9/7 Wavelet
Wavelet transform is a very useful technique in image processing. Through wavelet transform, images can be decomposed into different scales and frequency components, allowing information extraction in both spatial and frequency domains. This characteristic makes wavelet transform excellent in many image processing tasks, especially in areas such as image compression, denoising, feature extraction, and segmentation. Applying wavelet decomposition to an image allows for the extraction of information at different frequencies, as shown in
Figure 5.
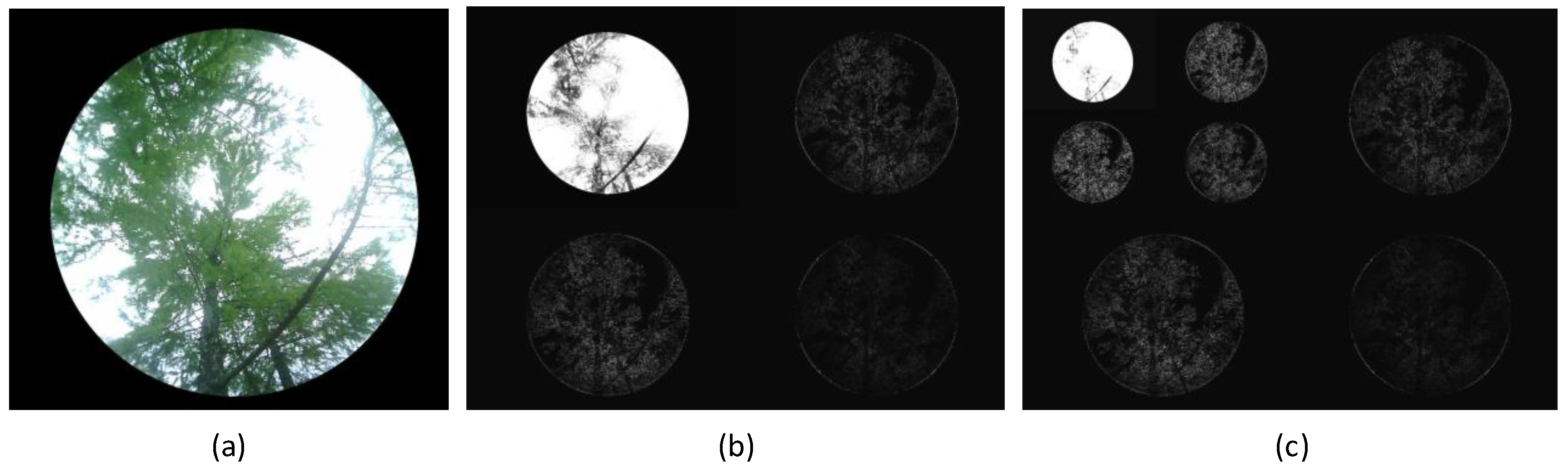
A first wavelet transform of the image results in four subbands: low-low (), low-high (), high-low (), and high-high (). These subbands represent the features of the image at different frequencies and directions. The subband contains the main low-frequency information of the image, reflecting the main structure and background, representing the primary features of the image. The , , and subbands contain high-frequency information in the horizontal, vertical, and diagonal directions, representing details and edges in the image. To extract finer features and improve segmentation accuracy, the subband undergoes further wavelet transformation, resulting in four additional subbands that provide a deeper decomposition.
In wavelet image decomposition, selecting an appropriate transformation method and basis function is crucial for effective image processing. The type of wavelet basis and the level of decomposition depend on the application needs and objectives. In LAI measurement, it is important to emphasize the high-frequency features of leaf areas. Therefore, wavelet decomposition is often carried out at deeper levels to better extract detailed information and suppress low-frequency background interference. The 9/7 wavelet transform is a high-precision, energy-concentrated wavelet transform method with relatively low computational complexity and excellent compression performance. It effectively retains image details and has good edge preservation capability, making it highly suitable for complex background and edge-blurring image segmentation tasks.
Additionally, the 9/7 wavelet transform can be efficiently implemented using only basic addition, subtraction, and bit-shift operations, significantly simplifying the computational process and minimizing storage requirements. This computational efficiency makes the 9/7 wavelet transform particularly well-suited for deployment on resource-constrained microprocessor platforms, enabling high-performance, real-time image processing even in low-power environments.
2.3. Imaging Region Extraction
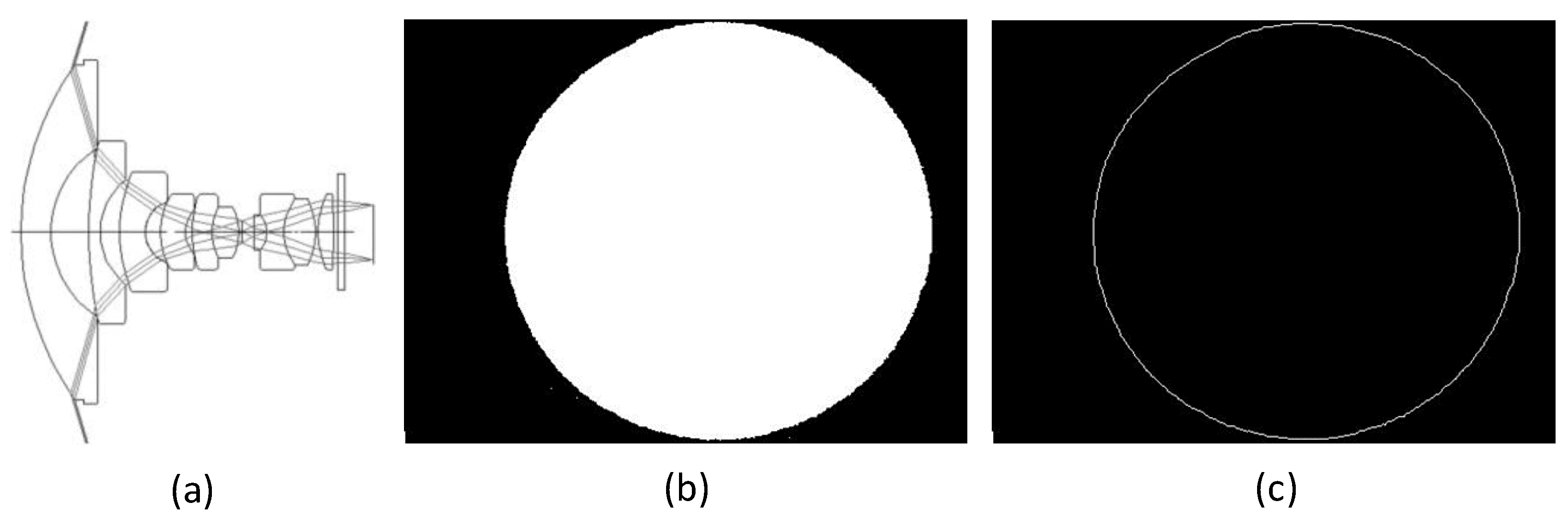
Fisheye lenses typically consist of a group of very short focal length convex lenses. When light passes through them, it refracts at different angles. The refracted light is projected onto the image in a circular field of view, as shown in
Figure 6(a). Due to misalignment during installation, the circular field of view is not exactly centered on the photo. The position of the circular region’s center and its radius must be determined. A white background image is captured using the vegetation image acquisition device, and this fisheye image is then binarized, as shown in
Figure 6(b). Next, morphological operations are performed to smooth the boundaries and extract the boundary points, yielding the circle’s center (840, 634) and radius
, as shown in
Figure 6(c).
According to the equidistant projection model of the fisheye lens, a Cartesian coordinate system is established with the center of the circular imaging area as the origin. The zenith angle
of any pixel
on the image is calculated a using the following formula:
2.4. Analysis of Vegetation Canopy Image Feature
The hemispherical photography method involves capturing canopy images from be-low the plant canopy using a camera equipped with a fisheye lens, with a viewing angle range from 180° to 200°, where the foreground is vegetation and the background is the sky, as shown in
Figure 7(a). The canopy image consists of three regions: the leaf area, the trunk area, and the sky area, which exhibit significant differences in grayscale and frequency. The leaf area has a higher reflectance, finer texture, and is primarily concentrated in high-frequency bands after wavelet transformation. The trunk area is typically darker, with the lowest grayscale and weaker reflection, mainly concentrated in low-frequency bands. The sky area is well-lit, with the highest grayscale, generally appearing in the canopy gaps, and corresponds to the image’s low-frequency information. By performing wavelet transformation, the vegetation image is decomposed into low-frequency and high-frequency subbands. The high-frequency components retain the leaf area details, while the low-frequency components capture the main structure of the trunk and sky. Therefore, by utilizing the different characteristics of these three regions, 9/7 wavelet transformation is employed for multi-scale decomposition of the vegetation image, enhancing or suppressing certain frequency components to increase the grayscale difference between the regions. Finally, threshold segmentation is used to reconstruct the image, achieving segmentation of specific vegetation regions.
The wavelet decomposition results of the vegetation image in
Figure 7(a) are shown in
Figure 7(b) and
Figure 7(c). After the first wavelet decomposition, the
component retains much of the structural information, with the leaf, trunk, and sky regions still clearly visible. This demonstrates that the first wavelet transformation effectively preserves the image’s low-frequency information and presents the main structure of the image. The
and
components contain significant boundary information for the leaf and trunk areas, with sparse leaf information. After a second wavelet decomposition, the high-frequency information is increasingly decomposed, and the leaf area information in the
component decreases, while trunk and sky information is largely preserved. A substantial amount of leaf information is decomposed into
,
, and
parts, displaying high brightness, while the trunk and sky areas appear in black, with significant grayscale differences between adjacent regions.
The wavelet decomposition results of the vegetation image show that the frequency information differs significantly across the regions. The leaf area typically exhibits strong high-frequency features, while the trunk and sky areas are concentrated in low-frequency components. In the hemispherical photography method for measuring LAI, attention is focused on the proportion of the leaf area in the entire image. Therefore, by suppressing the low-frequency components and enhancing the grayscale value of the leaf area, the leaf region becomes more prominent in the image, achieving segmentation of the leaf area. This method not only improves segmentation efficiency but also adapts well to images under varying lighting conditions, making it highly adaptable. This frequency-based segmentation algorithm provides a new and effective technical route for LAI measurement using hemispherical photography.
2.5. LAI Extraction
The LAI inversion method based on hemispherical photography extracts vegetation parameter information by analyzing canopy images. This process includes two key steps: image segmentation and LAI extraction. Image segmentation refers to distinguishing different regions (leaf area, trunk area, and sky area) within the image. The segmented leaf area provides the data foundation for subsequent LAI calculation. LAI extraction is achieved by estimating the proportion of the leaf area in the image.
2.5.1. Image Segmentation
As analyzed previously, vegetation images exhibit distinct frequency characteristics: leaf areas are typically associated with high-frequency components, while trunk and sky regions are dominated by low-frequency components. Accurate segmentation of leaf areas is critical for precise LAI measurement, as it provides the foundational data required for reliable calculations. To achieve this, wavelet decomposition is applied to the vegetation image, with a focus on suppressing low-frequency components. This suppression minimizes the influence of background elements (e.g., trunks and sky) on leaf area segmentation, thereby enhancing the grayscale contrast between leaf regions and other parts of the image. The improved contrast facilitates the effective application of image segmentation algorithms, enabling more accurate extraction of leaf areas and ultimately contributing to higher precision in LAI estimation.
2.5.1.1 Fixed Coefficient Suppression
The vegetation image is decomposed using two layers of 9/7 wavelet transform. The pixel values within the effective imaging area of the
subband from the second wavelet decomposition are normalized:
where
is the original grayscale pixel value of the image, ranging from
, and
is the normalized pixel value, ranging from
. A coefficient
I is applied to globally suppress the grayscale values of the normalized
subband:
The purpose of global grayscale suppression is to enhance the high-frequency components by reducing the low-frequency components, thereby highlighting the edges and details of the leaf area. During this process, the grayscale values of the low-frequency components (trunk and sky areas) are lowered, making the leaf area more distinct in the subsequent image segmentation process. After suppression, the subband is reconstructed with the other subbands using the inverse 9/7 wavelet transform. Finally, Otsu’s method is used to perform automatic threshold segmentation, yielding clear image segmentation results.
Global grayscale suppression was applied to the vegetation image shown in
Figure 8(a) using coefficients
, and
. The segmentation results are displayed in
Figure 8, where the trunk and sky regions are marked in white, and the leaf areas are marked in black. It is important to note that smaller suppression coefficients result in stronger suppression effects on the image.
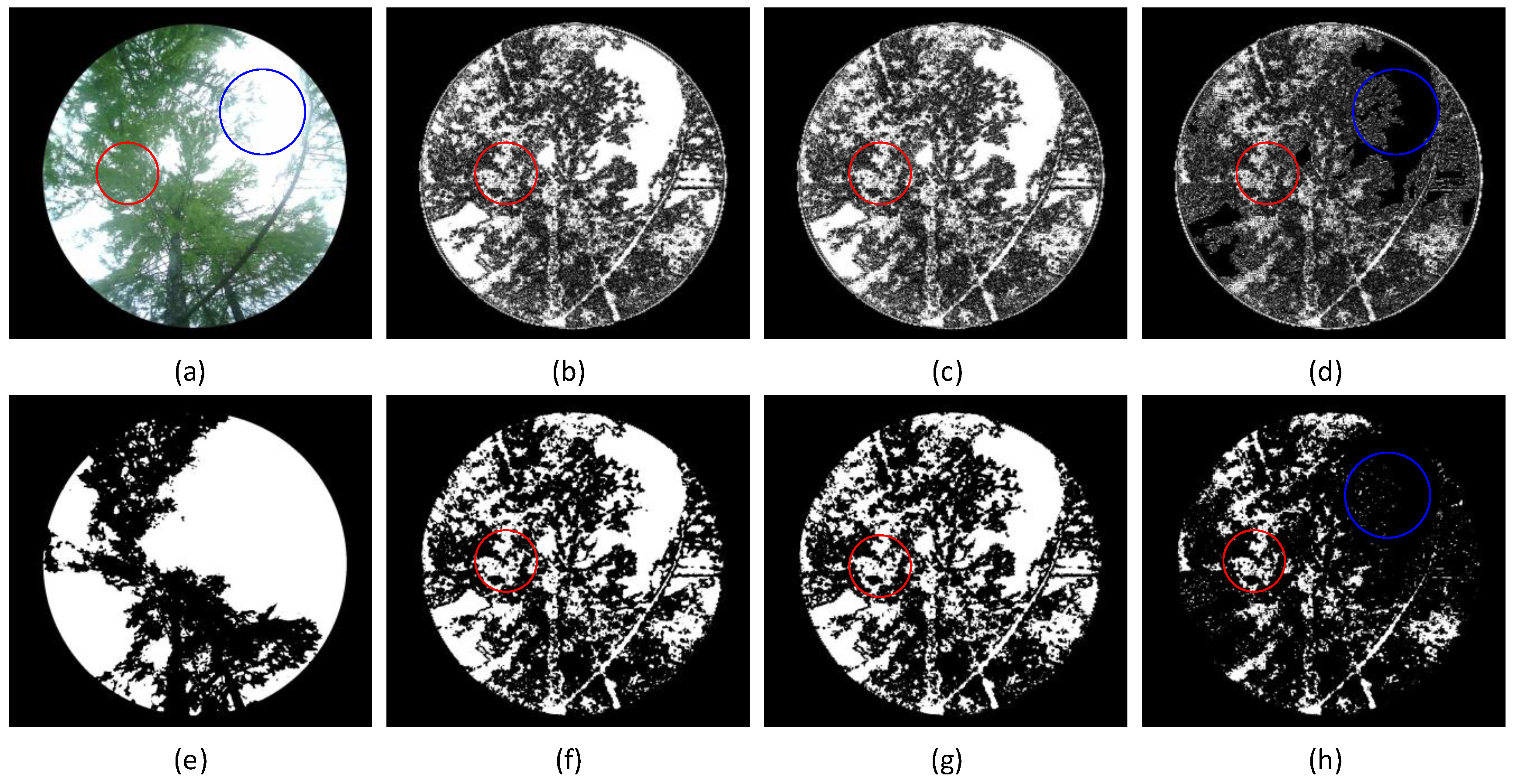
By comparing the segmentation results, the following five observations can be made:
- (1)
When the vegetation image is directly processed using Otsu’s method, some of the brighter leaf areas are misclassified as the sky, and some darker trunk areas are misclassified as leaf areas due to insufficient grayscale contrast.
- (2)
After the wavelet transformation of the image, Otsu’s method accurately segments the trunk area.
- (3)
When the suppression coefficient is too small (
), some leaf areas with subtle grayscale variations are misclassified as sky regions, as shown in the red-circled areas in
Figure 8(b),
Figure 8(c), and
Figure 8(d). This occurs because these leaf areas, located in the low-frequency components, experience excessive suppression, reducing their grayscale values to levels similar to other low-frequency areas, resulting in misclassification.
- (4)
As the suppression coefficient increases, the grayscale suppression of low-frequency components weakens, improving the contrast between the leaf area and the sky area, thereby enhancing the leaf area segmentation.
- (5)
When the suppression coefficient is excessively large
, the sky region is erroneously classified as part of the leaf area, as illustrated in the blue-circled region of
Figure 9(d). This misclassification arises because the high grayscale values of the sky are inadequately suppressed, diminishing the grayscale contrast between the sky and the leaf area and resulting in segmentation errors.
When collecting vegetation images in natural environments, factors such as weather conditions, leaf density, and distribution differences result in significant global grayscale variations across different vegetation images. The low-frequency components of the image may include the leaf area, sky area, and trunk area. Due to these variations in grayscale distribution, the same suppression coefficient may produce inconsistent results across different images, leading to under-suppression or over-suppression in some images, affecting the accuracy of segmentation results. Therefore, when applying grayscale suppression, it is beneficial to consider a suppression scheme based on the image’s grayscale mean to improve segmentation accuracy.
2.5.1.2 Grayscale Mean-Based Suppression
For vegetation images, after performing the two-layer 9/7 wavelet decomposition, all pixel values in the effective imaging area of the
subband are normalized as
. By iterating through all pixel points, the arithmetic mean of the normalized grayscale image is calculated as follows:
where
N represents the number of pixel points within the effective imaging area. Global grayscale suppression is applied to the normalized
subband using the coefficient
I as follows:
The suppressed subband is then reconstructed with the other subbands using the inverse 9/7 wavelet transform. Finally, Otsu’s method is used to segment the reconstructed image automatically.
Subsequently, the suppressed subband, denoted as , along with the other subbands, is reconstructed into an image using the inverse 9/7 wavelet transform. Finally, Otsu’s method is employed to perform automatic threshold segmentation on the reconstructed image, yielding clear and precise image segmentation results.
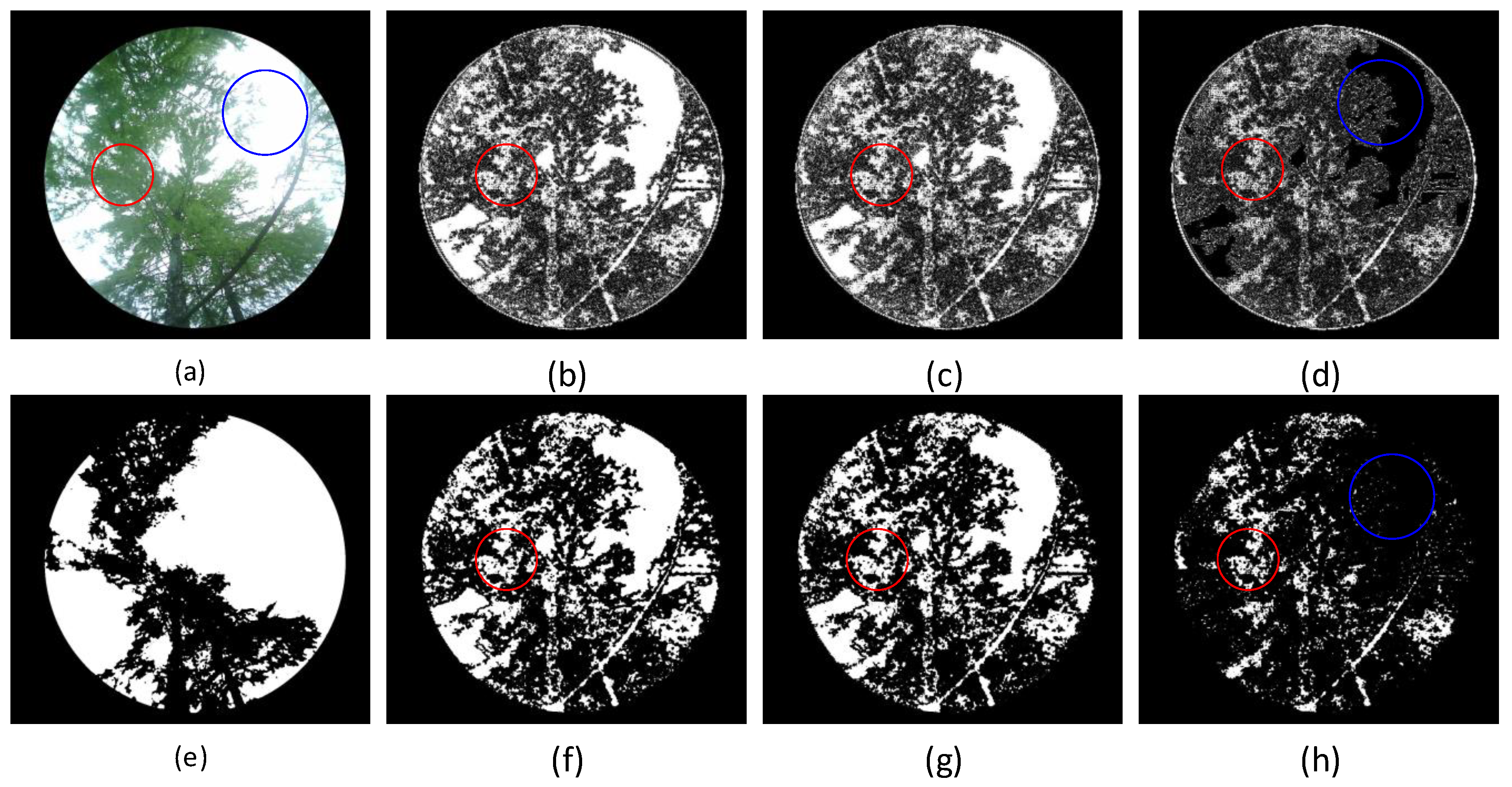
For the vegetation image shown in
Figure 9(a), global grayscale suppression is applied with coefficients
, with the segmentation results shown in the
Figure 9. The trunk and sky areas are marked in white and the leaf area in black. It should be noted that the smaller the suppression coefficient, the stronger the suppression effect on the image.
Segmentation results show that the grayscale mean-based suppression algorithm still provides good segmentation results. However, some leaf areas with smooth grayscale transitions are still misclassified as sky regions. Compared with the fixed coefficient global suppression algorithm, the following two differences were observed:
- (1)
When the suppression coefficient does not exceed , the sky region is correctly segmented without misclassification.
- (2)
When the suppression coefficient increases to , the sky region is misclassified as part of the leaf area.
The analysis of the two suppression algorithms yields the following conclusions:
- (1)
The trunk area in vegetation images typically has the lowest grayscale values, and even with a high suppression coefficient, it is correctly identified without misclassification.
- (2)
When the suppression coefficient is too small, the suppression effect on low-frequency components is too strong, causing some gently varying leaf areas to be overly suppressed, making them indistinguishable from the sky, leading to misclassification.
- (3)
When the suppression coefficient is too large, the sky area is not effectively suppressed, reducing the contrast between the sky and leaf areas and causing misclassification of the sky as the leaf area.
- (4)
An appropriate suppression coefficient creates a clear grayscale contrast between the leaf area, sky, and trunk, improving segmentation accuracy.
Thus, there is an optimal suppression coefficient. If the coefficient is too low, it can effectively identify the sky area; however, if it is too high, the ability to recognize the sky decreases, though misclassification of the leaf area improves. Based on this analysis, a dynamic optimal suppression coefficient algorithm is proposed. This algorithm adapts to complex vegetation image segmentation needs, ensuring accurate segmentation results under various conditions.
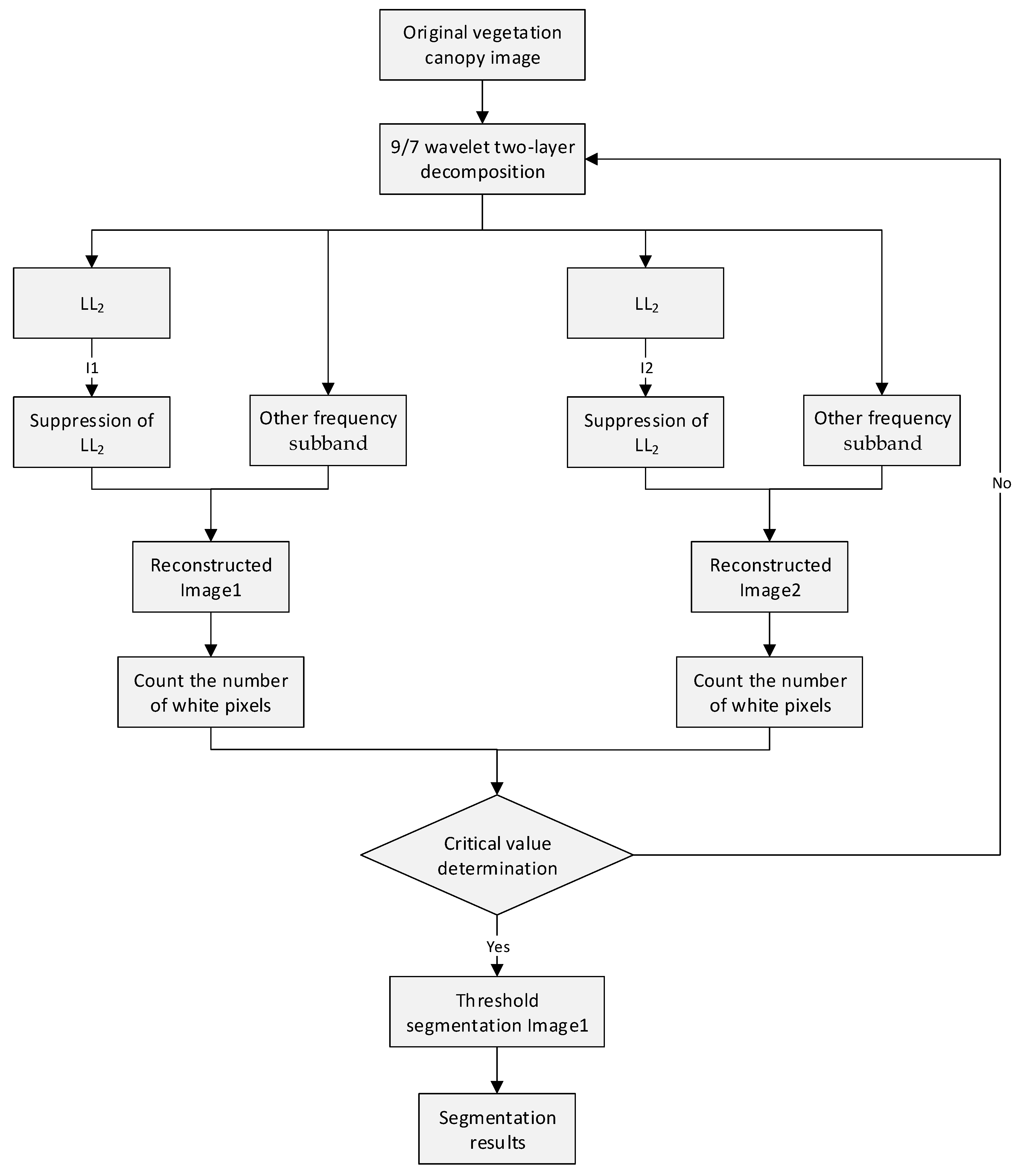
2.5.1.3 Dynamic Optimization of Suppression
Dynamic optimization of suppression is an algorithm that dynamically adjusts the suppression coefficient. Through multiple dynamic iterations, the algorithm gradually approaches the optimal suppression coefficient to adapt to the grayscale characteristics of different vegetation images, enabling more accurate image segmentation. This provides precise data for subsequent LAI extraction. The algorithm process is shown in
Figure 10.
The vegetation image undergoes a two-level 9/7 wavelet decomposition, and all pixel values within the effective imaging region of the
subband from the second-level decomposition are normalized as
.The arithmetic mean of the normalized grayscale images calculated as
. Suppression coefficients
and
are defined such that:
where
C is a positive increment. These coefficients are applied to globally suppress the normalized
and
subband:
The suppressed subbands and are then reconstructed with other subbands using the 9/7 inverse wavelet transform, yielding reconstructed grayscale images and . Finally, Otsu’s method is applied for automatic threshold segmentation to obtain clear segmentation results.
White pixels (sky and trunk regions) are labeled as 1, and black pixels (leaf regions) as 0. The number of white pixels in
and
, denoted
and
are counted. The difference between the two images is calculated as:
During execution, the algorithm repeats or terminates based on the value of
. If iteration is required,
increases by a step size
:
when
is small, the segmentation results of
and
are similar (
). As
increases,
may misclassify sky regions as leaf regions earlier than
, causing
to decrease sharply while
remains stable. A sudden increase in
indicates optimal suppression coefficients have been reached, terminating the optimization.
To address potential misclassification of leaf regions as sky in
(due to small
),
is used to correct
. A pure sky image (vegetation-free) is normalized:
where
is the original grayscale (range:
).
represents the normalized pixel value, scaled to the range
. By iterating through all pixel points, the arithmetic mean of the normalized grayscale values for the vegetation-free image is calculated as follows:
For each white pixel in , if the corresponding pixel in is black (non-trunk) and the original pixel’s grayscale is less than , the pixel is corrected to black (leaf region).
4. Discussion
4.1. Discussion on Wavelet Transform-Based Segmentation Algorithms
The fixed coefficient suppression algorithm, when processing images, produces consistent suppression effects on all images due to the fixed suppression coefficient, leading to over-segmentation or misclassification in some images.
The grayscale mean-based suppression algorithm dynamically adjusts based on grayscale mean, improving the image segmentation effect. However, its flexibility is limited, and it has limitations when dealing with complex backgrounds or detailed regions.
The dynamic optimal suppression algorithm dynamically adjusts the suppression coefficient based on image characteristics and performs well for different types of vegetation images. Additionally, using the reconstruction results to repair misclassification can more accurately separate the leaf area, avoid over-suppression issues, and significantly improve the segmentation accuracy and stability. Future work could continue to explore how to further optimize the algorithm to ensure its adaptability and reliability in large-scale or different environments.
4.2. Suppression Coefficient Selection
This paper compares the segmentation results of different suppression coefficients and selects the suppression coefficient parameters based on experience, especially the initial values
and
, and increment
for the dynamic optimal suppression algorithm, which is not universally applicable. In setting the suppression coefficients
and
,
, meaning that the increment of
compared to
is the same as the increment for
itself:
This parameter design borrows from the iterative computing idea, simplifying the computational load. In actual execution, the segmentation result of suppression coefficient is used as the segmentation result for in the next iteration, thus only recalculating the new suppression coefficient ’s result, which reduces computational load by half compared to when .
The advantage of the dynamic optimal suppression algorithm is its ability to adjust the suppression coefficient based on image characteristics, but this process relies on understanding and prior knowledge of image properties. To enhance the algorithm’s adaptability, future research could use automated parameter optimization, machine learning, and multi-scale analysis methods to automatically determine the suppression coefficient parameters, further improving the algorithm’s applicability and reliability.
4.3. Instrument Selection for Comparison
Currently, commonly used indirect measurement instruments are divided into two categories: one category measures the light radiation above or below the vegetation using specific optical sensors, and then estimates the leaf area index based on the light attenuation, with representative instruments including the LAI-2200 series, TRAC, and DEMON; the other category uses optical cameras to obtain hemispherical canopy images and then analyzes the images to calculate the LAI, with representative instruments including the CI-100. Among these instruments, the LAI-2200 series, developed by the U.S. company LI-COR, occupies a crucial position in plant physiology and ecological research due to its efficiency, non-destructive nature, and wide application [
29].
The LAI-2200C uses a fisheye optical sensor (vertical field of view of 148°) to measure projected light at 5 zenith angles (7°, 23°, 38°, 53°, 68°) on the canopy, using a vegetation canopy radiation propagation model to calculate the leaf area index. As a high-precision leaf area index measuring instrument, the LAI-2200C has been widely used worldwide and is the instrument of choice for researchers in plant physiology and ecology. Its measurement results are widely recognized within the industry for their high reliability and accuracy and are regarded as the industry standard.
4.4. Result Analysis and Discussion
The experimental results show that the leaf area index extracted by the inversion method designed in this paper is overall smaller than the LAI-2200C measurement results, which is similar to other studies. This discrepancy occurs because the LAI-2200C assumes that tree leaves are optical black bodies, absorbing all light and neglecting reflection and refraction effects between leaves. In reality, tree leaves have certain reflective and transmissive properties, which affect the light propagation within the canopy and consequently the gap rate measurement. This leads to a smaller gap rate and an overestimation of the LAI in the LAI-2200C measurements. Moreover, the LAI-2200C is sensitive to all shading objects in the field of view, including leaves, stems, and branches [
52], which are all included in the measurements, causing an overestimation of the LAI.
In this paper’s designed inversion method, the trunk area is treated as a non-leaf area. During actual measurements, the trunk may block some leaf areas, resulting in an under-estimation of the leaf area, and thus the extracted leaf area index is smaller. Therefore, in practical applications, the LAI-2200C measurement results can be combined with the image-based inversion method through weighted averaging or data fusion techniques to obtain more accurate LAI values or verified using destructive sampling or other independent methods (e.g., ground LiDAR) [
52,
53,
54].
4.5. Image Segmentation Method Comparison and Discussion
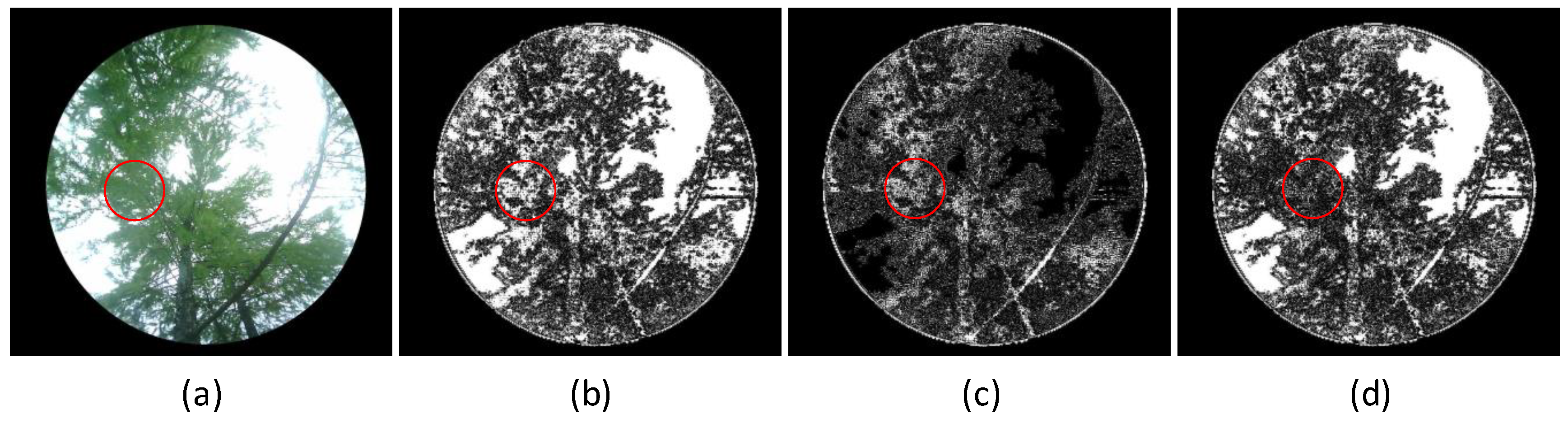
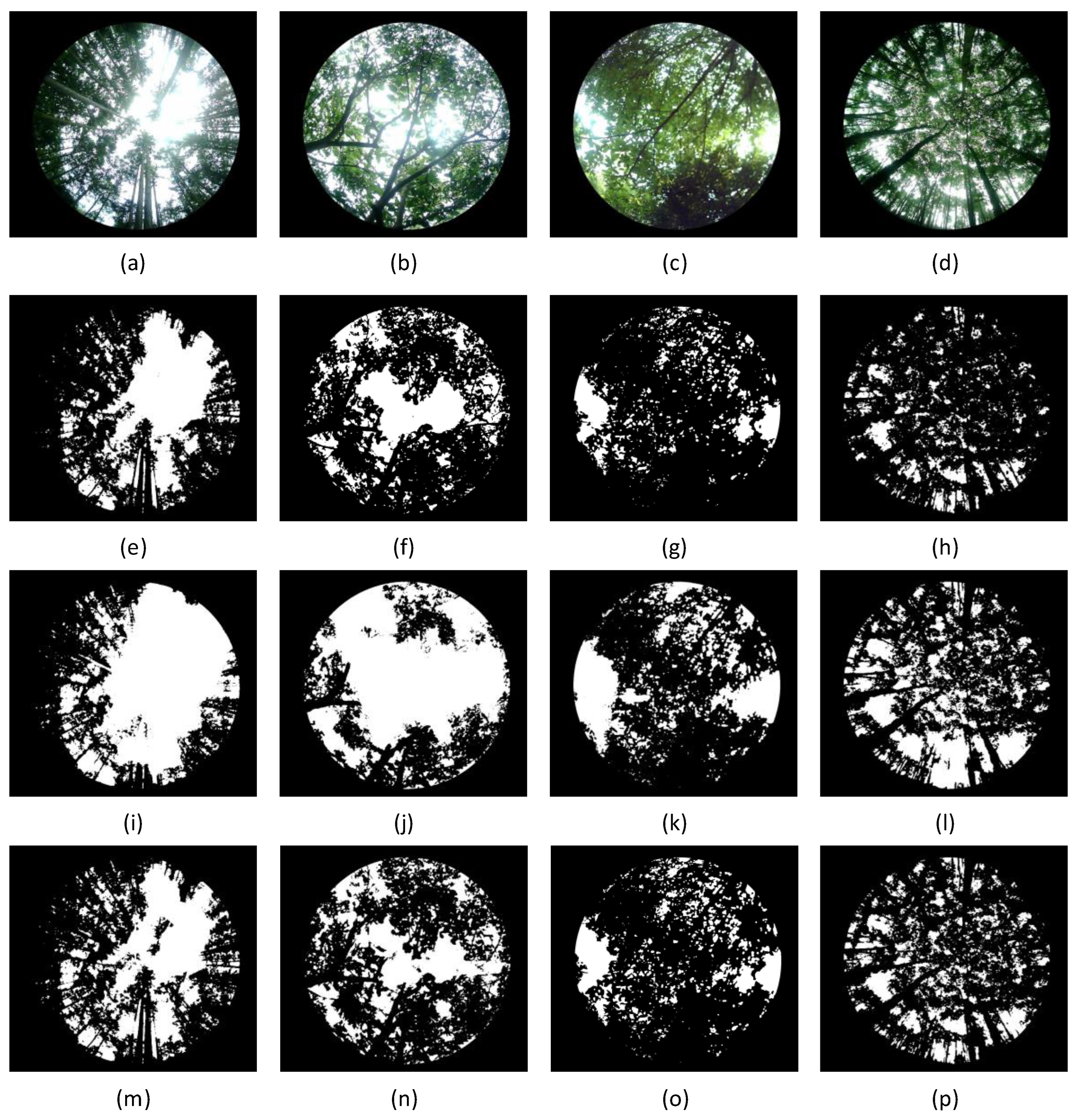
The hemispherical photography method captures hemispherical canopy images to calculate the leaf area index. Gap rate extraction is one of the key steps in measuring LAI, and its accuracy directly affects the LAI measurement results. Currently, commonly used image processing algorithms include the traditional Otsu’s method and the improved Otsu’s method [
56]. Different algorithms exhibit differences and limitations in segmenting leaf and sky pixels, leading to inaccurate gap rate extraction. Four typical forest vegetation canopy images were selected, and the segmentation results using fixed threshold methods (
), traditional Otsu’s method, and improved Otsu’s method are shown in
Figure 15. The results show that all segmentation algorithms fail to effectively segment the trunk area, and significant differences are present in the segmentation results. Among these methods, the improved Otsu’s method performs the best.
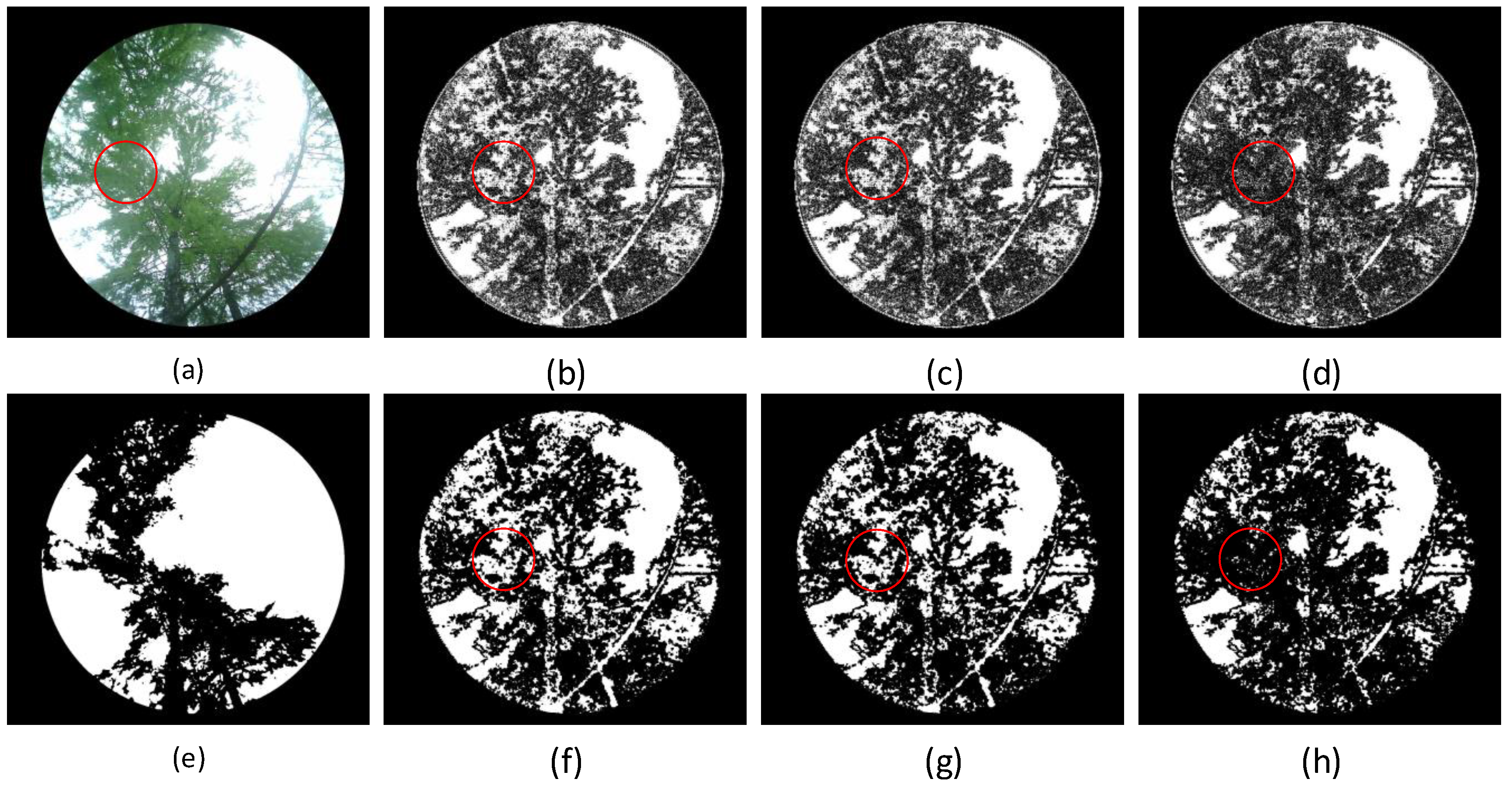
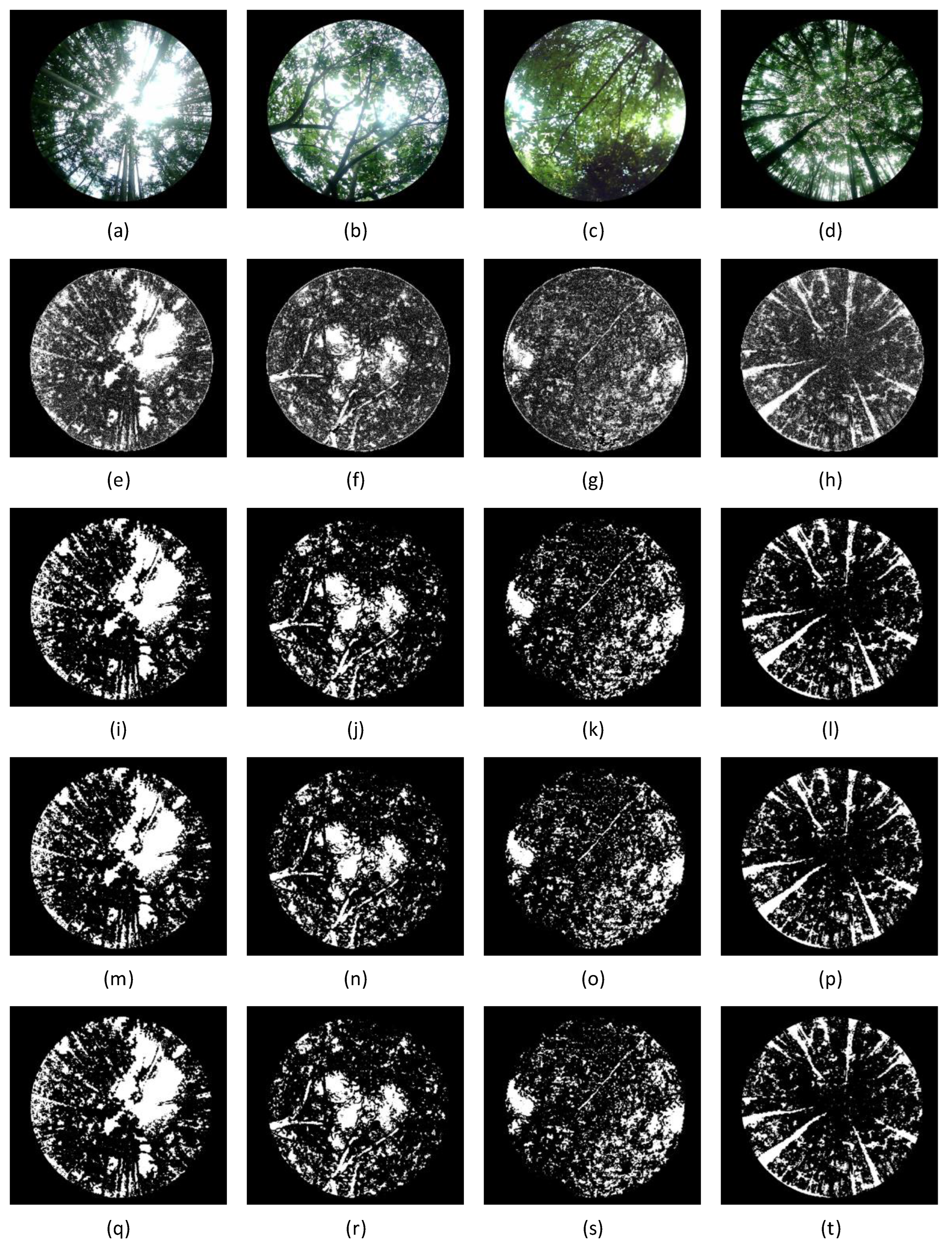
By applying the dynamic optimal suppression algorithm to decompose, reconstruct, and repair these four typical forest vegetation canopy images, and then segmenting them using the fixed threshold method (
), traditional Otsu’s method, and improved Otsu’s method[
56], the results are shown in
Figure 16. The wavelet transform significantly improves the contrast of the target areas, allowing the segmentation algorithms to more accurately separate the leaf area from the background area. Moreover, after wavelet transformation, the segmentation results are more consistent and show significantly better accuracy than those directly processed with these algorithms on the original images. Therefore, combining wavelet transformation with image segmentation algorithms can significantly reduce the complexity of segmentation and improve both efficiency and accuracy.
4.6. Improvement Measures Discussion
The image processing algorithm proposed in this paper is currently implemented on a PC. If this algorithm is to be migrated to a microprocessor platform (such as embedded systems or Raspberry Pi), the primary task is to evaluate the algorithm’s operating speed and resource usage, including processing time, memory usage, and other key indicators. To improve the running efficiency on microprocessors, optimization of both the code (e.g., reducing unnecessary computations, using more efficient algorithms) and hardware (e.g., utilizing specific hardware acceleration features, optimizing memory access patterns) will be necessary.
The image processing algorithm proposed in this paper performs well with the data from the three selected forest experimental areas. However, due to the limited dataset, it is not possible to determine if the algorithm will still show good segmentation results when processing more diverse tree species datasets. Therefore, it is necessary to collect image datasets containing multiple tree species and evaluate the segmentation results, studying the algorithm’s adaptability to different tree species.
Lastly, by comparing the differences between the leaf area index extracted from different tree species and the actual values, the applicability of the inversion method pro-posed in this paper can be evaluated. Furthermore, analyzing possible influencing factors, such as tree structure features, leaf morphology, and distribution, will help better under-stand how these factors affect the leaf area index inversion results. Once these influencing factors are fully understood, the inversion method can be further optimized and improved to enhance its accuracy and robustness.
5. Conclusions
To address the demand for fast and accurate measurement of LAI in forest ecosystems, this paper proposes a LAI inversion method based on 9/7 wavelet transform. This method focuses on the core issue of accurate segmentation of leaf elements and background elements in vegetation canopy images. By utilizing differences in grayscale and frequency features between the leaf area, trunk area, and sky area, the 9/7 wavelet transform is used to decompose, process, and reconstruct vegetation images, achieving efficient and precise segmentation of the leaf area. By extracting the canopy gap rate, rapid LAI measurement is achieved. Since the wavelet transform enhances the grayscale difference between the leaf area and the background areas (trunk area, sky area), the image segmentation algorithms can more easily identify and separate the leaf area, significantly simplifying the segmentation process. Compared to directly using image segmentation algorithms on raw vegetation images, the wavelet transform-based image segmentation algorithm proposed in this paper shows significant advantages, especially in complex backgrounds and different lighting conditions, achieving higher segmentation accuracy.
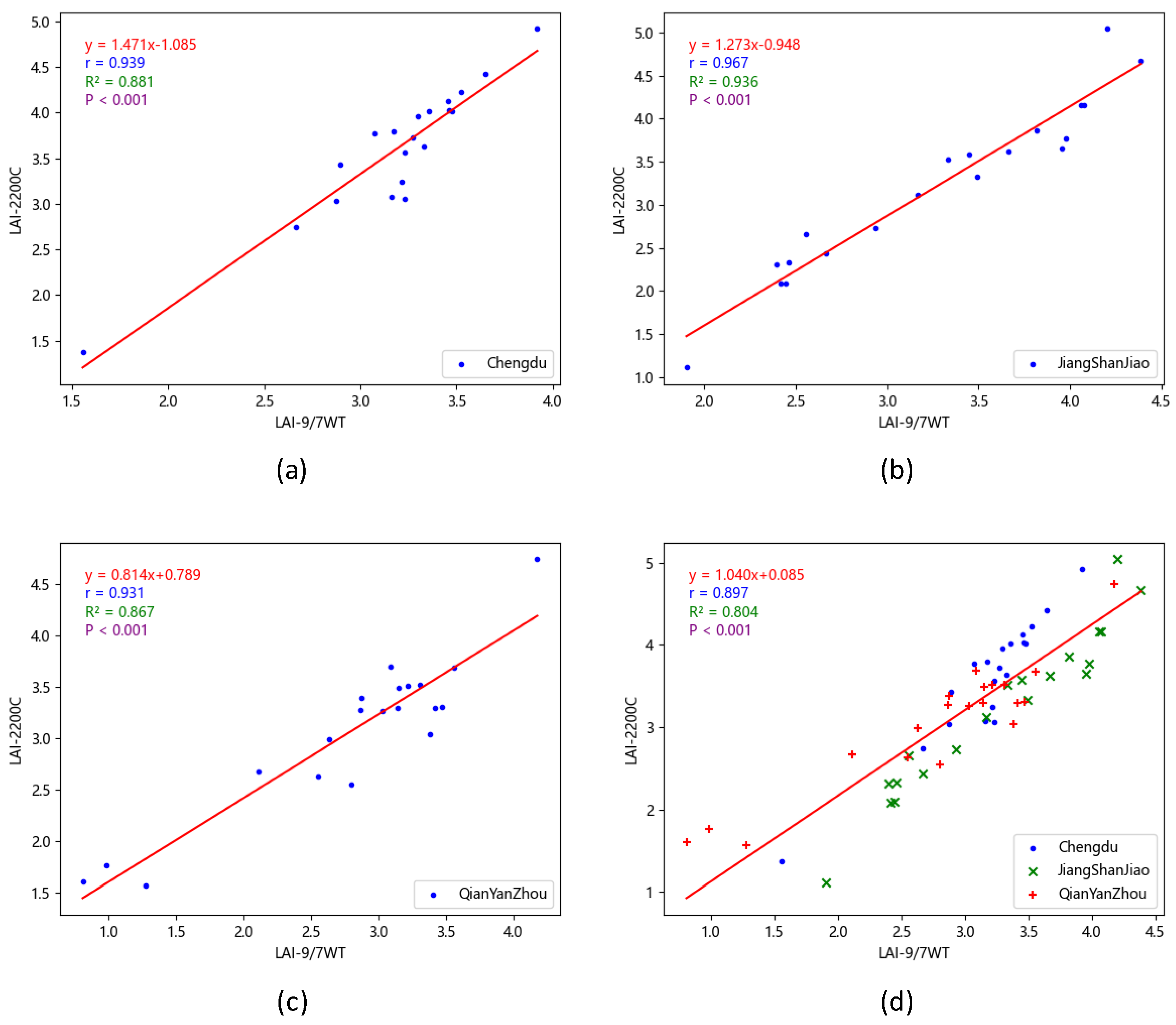
To validate the effectiveness of the proposed inversion method, three forest experimental areas from different geographic regions were selected, and vegetation canopy images were synchronized with the LAI-2200C. The LAI values for 60 sampling areas were calculated using the proposed method and compared with the LAI-2200C measurements. The results show that the proposed method is highly correlated with the LAI-2200C measurements (, ), indicating that it has high accuracy under different environmental conditions and is a reliable LAI measurement method.
This paper uses an enhanced algorithm to implement wavelet transforms, significantly simplifying the 9/7 wavelet transform’s computation process, making it suitable for real-time applications. The dynamic optimal suppression algorithm’s parameter setup reduces intermediate data during the calculation process, lowering memory and storage requirements, and providing the potential for application on microprocessor platforms. Combining wavelet transforms makes segmentation results consistent across different segmentation algorithms, showcasing the algorithm’s adaptability to complex scenarios. Therefore, the research results not only apply to efficient and high-precision image processing tasks on PC but also provide strong support for developing portable and high-precision LAI measurement devices. As the technology continues to mature and expand, it is expected to have a profound impact on sustainable agriculture, ecological protection, and addressing climate change.
Figure 1.
Image of the study area location.
Figure 1.
Image of the study area location.
Figure 2.
Experimental data collection. (a)LAI-2200C; (b) LAI-2200C measurement principle; (c) Experimental equipment connection diagram; (d) Experimental site map.
Figure 2.
Experimental data collection. (a)LAI-2200C; (b) LAI-2200C measurement principle; (c) Experimental equipment connection diagram; (d) Experimental site map.
Figure 3.
Schematic diagram of sample point collection.
Figure 3.
Schematic diagram of sample point collection.
Figure 4.
Sample data example. (a) Sample 1; (b) Sample 2.
Figure 4.
Sample data example. (a) Sample 1; (b) Sample 2.
Figure 5.
Schematic diagram of image wavelet decomposition.
Figure 5.
Schematic diagram of image wavelet decomposition.
Figure 6.
Image area extraction. (a) Schematic diagram of fisheye lens imaging; (b) Binary image; (c) Imaging area boundary.
Figure 6.
Image area extraction. (a) Schematic diagram of fisheye lens imaging; (b) Binary image; (c) Imaging area boundary.
Figure 7.
Wavelet decomposition results. (a) Vegetation canopy image; (b) One layer decomposition result; (c) Second level decomposition results.
Figure 7.
Wavelet decomposition results. (a) Vegetation canopy image; (b) One layer decomposition result; (c) Second level decomposition results.
Figure 8.
Fixed coefficient suppression algorithm segmentation results. (a) Original image; (b) reconstructed image; (c) reconstructed image; (d) reconstructed image; Figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 8.
Fixed coefficient suppression algorithm segmentation results. (a) Original image; (b) reconstructed image; (c) reconstructed image; (d) reconstructed image; Figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 9.
Grayscale mean-based suppression segmentation results. (a) Original image; (b) reconstructed image; (c) reconstructed image; (d) reconstructed image; Figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 9.
Grayscale mean-based suppression segmentation results. (a) Original image; (b) reconstructed image; (c) reconstructed image; (d) reconstructed image; Figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 10.
Dynamic optimization of suppression algorithm process.
Figure 10.
Dynamic optimization of suppression algorithm process.
Figure 11.
Results of dynamic optimization suppression method. (a) Original image; (b) Reconstruct image (); (c) Reconstruct image (); (d) The final output image.
Figure 11.
Results of dynamic optimization suppression method. (a) Original image; (b) Reconstruct image (); (c) Reconstruct image (); (d) The final output image.
Figure 12.
Comparison of segmentation results. (a) Original vegetation image; (b) Fixed coefficient suppression algorithm (); (c) Grayscale mean-based suppression algorithm (); (d) Dynamic optimization suppression algorithm; figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 12.
Comparison of segmentation results. (a) Original vegetation image; (b) Fixed coefficient suppression algorithm (); (c) Grayscale mean-based suppression algorithm (); (d) Dynamic optimization suppression algorithm; figures (e) to (h) show the Otsu’s method segmentation results of figures (a) to (d).
Figure 13.
Correlation analysis of LAI values. (a)Chengdu;(b)Jiangshanjiao;(c)Qianyanzhou;(d)All.
Figure 13.
Correlation analysis of LAI values. (a)Chengdu;(b)Jiangshanjiao;(c)Qianyanzhou;(d)All.
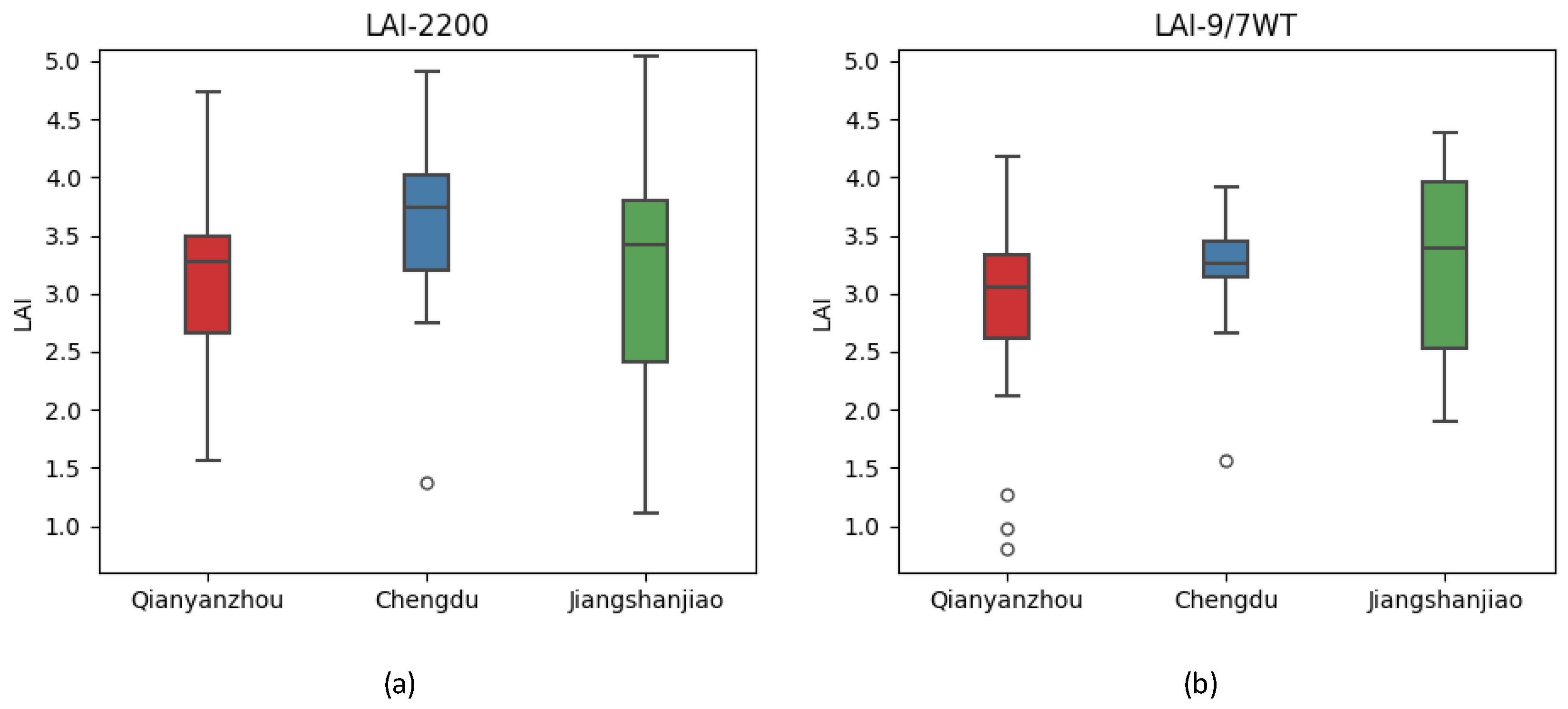
Figure 14.
Boxplot of the LAI value. (a)LAI-2200C;(b)LAI-9/7WT.
Figure 14.
Boxplot of the LAI value. (a)LAI-2200C;(b)LAI-9/7WT.
Figure 15.
Image segmentation results. Figures (a) to (d) show the original images; Figures (e) to (h) show the fixed threshold method segmentation results of figures (a) to (d); Figures (i) to (l) show the traditional Otsu’s method segmentation results of figures (a) to (d); Figures (m) to (p) show the improve Otsu’s method segmentation results of figures (a) to (d).
Figure 15.
Image segmentation results. Figures (a) to (d) show the original images; Figures (e) to (h) show the fixed threshold method segmentation results of figures (a) to (d); Figures (i) to (l) show the traditional Otsu’s method segmentation results of figures (a) to (d); Figures (m) to (p) show the improve Otsu’s method segmentation results of figures (a) to (d).
Figure 16.
Dynamic optimization suppression algorithm segmentation results. Figures (a) to (d) show the original images; Figures (e) to (h) show the reconstructing images with wavelet transform of figures (a) to (d); Figures (i) to (l) show the fixed threshold method segmentation results of figures (e) to (h); Figures (m) to (p) show the traditional Otsu’s method segmentation results of figures (e) to (h); Figures (q) to (t) show the improve Otsu’s method segmentation results of figures (e) to (h).
Figure 16.
Dynamic optimization suppression algorithm segmentation results. Figures (a) to (d) show the original images; Figures (e) to (h) show the reconstructing images with wavelet transform of figures (a) to (d); Figures (i) to (l) show the fixed threshold method segmentation results of figures (e) to (h); Figures (m) to (p) show the traditional Otsu’s method segmentation results of figures (e) to (h); Figures (q) to (t) show the improve Otsu’s method segmentation results of figures (e) to (h).
Table 1.
Overview the study area.
Table 1.
Overview the study area.
| SN |
Area name |
Vegetation type |
Climate type |
Landform type |
| 1 |
Jiangshanjiao Station, Heilongjiang,China |
coniferous and broad- leaved mixed forest |
Temperate monsoon climate |
Low mountain and hilly landform |
| 2 |
Qianyanzhou Station, Jiangxi, China |
evergreen broad-leaved forest |
Subtropical monsoon climate |
Red Soil Hills |
| 3 |
Qingshuihe Campus of UESTC,Sichuan,china |
evergreen broad-leaved forest |
Subtropical monsoon climate |
Plain |
Table 2.
Weight Factors.
|
|
|
|
| 7° |
12.5 |
0.0266 |
0.0427 |
| 23° |
12.5 |
0.0852 |
0.1369 |
| 38° |
12.5 |
0.1343 |
0.2157 |
| 53° |
12.5 |
0.1742 |
0.2798 |
| 68° |
12.5 |
0.2023 |
0.3248 |
Table 3.
Evaluation of Single Sample Reconstruction Results.
Table 3.
Evaluation of Single Sample Reconstruction Results.
| |
Fixed coefficient
suppression algorithm |
Grayscale mean-based
suppression algorithm |
Dynamic optimization
suppression algorithm |
| |
0.01 |
0.02 |
0.01g |
0.02g |
- |
| DIR |
0.6077 |
0.6272 |
0.6487 |
0.6663 |
0.7859 |
| Ui |
0.978 |
0.9774 |
0.9799 |
0.9798 |
0.9835 |
|
1.4856 |
1.5141 |
1.3049 |
1.2796 |
0.9549 |
|
23.6957 |
23.403 |
22.9683 |
22.7328 |
20.9293 |
|
0.5266 |
0.4819 |
0.4625 |
0.4266 |
0.4256 |
|
2.638 |
2.6618 |
2.4393 |
2.4062 |
2.2444 |
Table 4.
Evaluation of Multi-Sample Reconstruction Results.
Table 4.
Evaluation of Multi-Sample Reconstruction Results.
| |
Fixed coefficient
suppression algorithm |
Grayscale mean-based
suppression algorithm |
Dynamic optimization
suppression algorithm |
| |
0.01 |
0.02 |
0.01g |
0.02g |
- |
| DIR |
0.6437 |
0.6537 |
0.6668 |
0.6723 |
0.7481 |
| Ui |
0.9442 |
0.9437 |
0.944 |
0.944 |
0.9589 |
|
2.6149 |
2.6527 |
2.4204 |
2.414 |
2.1597 |
|
35.2194 |
35.135 |
34.2992 |
34.2531 |
33.1133 |
|
1.986 |
1.9812 |
1.9371 |
1.9312 |
1.7034 |
|
2.9535 |
2.9765 |
2.7295 |
2.7167 |
2.4775 |
Table 5.
Boxplot data sheet of the LAI value.
Table 5.
Boxplot data sheet of the LAI value.
| |
Study area |
|
LAI-2200C |
Dynamic optimization suppression algorithm |
| Chengdu |
median |
3.748 |
3.252 |
| 75th percentile and 25th percentile |
75th percentile |
4.016 |
3.456 |
| 25th percentile |
3.199 |
3.140 |
| Difference |
0.817 |
0.316 |
| extremum |
maximum |
4.917 |
3.919 |
| minimum |
1.374 |
1.557 |
| Jiangshanjiao |
median |
3.425 |
3.391 |
| 75th percentile and 25th percentile |
75th percentile |
3.792 |
3.961 |
| 25th percentile |
2.412 |
2.533 |
| Difference |
1.38 |
1.428 |
| extremum |
maximum |
5.040 |
4.387 |
| minimum |
1.110 |
1.904 |
| Qianyanzhou |
median |
3.279 |
3.061 |
| 75th percentile and 25th percentile |
75th percentile |
3.494 |
3.328 |
| 25th percentile |
2.664 |
2.612 |
| Difference |
0.83 |
0.716 |
| extremum |
maximum |
4.738 |
4.177 |
| minimum |
1.566 |
0.807 |
Table 6.
Analysis of LAI value Error
Table 6.
Analysis of LAI value Error
| |
Chengdu |
Jiangshanjiao |
Qianyanzhou |
ALL |
| RMSE |
0.533 |
0.318 |
0.415 |
0.431 |
| MAE |
0.461 |
0.234 |
0.358 |
0.351 |