1. Introduction
The most recent guidelines of the International Working Group on the Diabetic Foot (IWGDF) and the Infectious Diseases Society of America (IDSA) [
2] recommend that the initial (empirical) antibiotic regimen for diabetic foot infections (DFI) should be based on the results of optimally obtained wound cultures (deep tissue, if available) and the clinical severity of infection. In geographical regions with limited laboratory or surgical facilities [
3,
4] and a high prevalence of antibiotic-resistant bacteria (often gram-negative rods, such as
Pseudomonas spp.) [
5], it is wise to prescribe an initial broad-spectrum regimen to avoid failing to adequately cover pathogens in a rapidly evolving infection. Having a point of care test that could rapidly help predict the likely culture results could help guide (and often constrain) the selection of antibiotic agents.
One such test could be the Gram-stained smear of wound specimens. This could, at a minimum, help to tailor the antibiotics to those active against just gram-positive pathogens, or to add those active against Gram-negative pathogens as well. The Gram-stained smear is an easily performed and inexpensive test that has been used for this purpose for over 100 years and in various infectious scenarios [
6]. It can detect unusual pathogens, help distinguish between contaminated and well-obtained specimens and might partially replace other more expensive microbiological diagnostic procedure that take several days to provide results [
6]. These advantages can make using the Gram-stained smear an important part of antibiotic stewardship programs [
4,
5], especially by reducing unnecessary, costly and often intravenous antibiotic coverage in settings already marked by a high prevalence of antibiotic-resistant microorganisms and limited resources.
Published data on the value of the Gram-stained smear for soft tissue wounds, including DFI are limited. In a study in Tanzania, Abbas et al. probed deep at the base of infected diabetic foot ulcers and aseptically collected two deep tissue specimens with a sterile scalpel [
3,
4]. They then performed Gram-stained smears and standard bacterial cultures on the specimens. Among 128 specimens, 118 (92%) yielded bacterial growth, among which there was 96.4% congruency between the Gram-stain and microbiological culture results [
3]. Of note is that the Gram-stains correctly predicted the presence of Gram-negative organisms in 82% of cases. Similarly, a study in Japan by Taniguchi et al., demonstrated that among 208 patients Gram-stains smears of respiratory, urinary, or skin and soft tissues specimens performed by healthcare professionals directly contributed fewer broad-spectrum antibiotics being prescribed, with 90% of the Gram-stain-based treatments judged as effective [
7,
8,
9].
Based on the results of a narrative review, the authors of a recent opinion paper [
4] called for efforts to evaluate the potential value of the Gram-stain smear as part of antibiotic stewardship programs for managing DFIs, with special emphasis on less developed, limited resources areas. Resource rich areas are facing an ever-increasing prevalence of antibiotic resistance among Gram-negative pathogen groups [
4,
5,
10,
11]. The conditions are, however, still less dire than in less-developed countries such as Tanzania, where access to microbiological cultures is less available, and the prevalence of naturally-resistant Gram-negative rods is higher. In Central and Northern Europe, as in Northern America, the prevalence of gram-negative rods, including
P. aeruginosa, as pathogens in community-acquired DFIs oscillates ranges about 7% to 12% [
13]. In contrast to the developing world, at presentation for surgical assessment for a DFI many patients are already receiving empirical antimicrobial therapy prescribed by their general practitioner [
14,
15,
16], which reduces the likelihood of detecting pathogens in on Gram-stained smears.
Of note is that the time window for the Gram-stain smear providing information about the presence of gram-negative pathogens that can enhance antimicrobial stewarding is quite limited. In patients with a moderately severe DFI undergoing surgical debridement clinicians generally do not need to provide antibiotic coverage for all potential bacteria. In the absence of sepsis, the therapeutic effect of surgery is so profound that even an apparently incorrect initial empirical antimicrobial regimen does not alter the outcomes [
16]. In these resource-rich settings, however, results of a Gram-stain smear could reduce the use of unnecessary broad-spectrum empirical therapy in non-severe DFIs [
9], while supporting its use in severe DFIs with sepsis [
17]. The key issue regarding antibiotic selection is principally about covering antibiotic-resistant gram-negative rods, as virulent gram-positive pathogens (e.g., beta-hemolytic streptococci or
Staphylococcus aureus) are generally covered by the standard empirical narrow-spectrum agents [
1,
2], with the exception of methicillin-resistant
S. aureus (MRSA), the prevalence of which is currently declining globally [
4,
5,
10,
11].
In this pilot study, we aimed to retrospectively assess the theoretical potential of the employing routine Gram-stained smears in tailoring a “semi-empirical” antibiotic prescribing in an effort to improve antibiotic stewardship [
18]. We sought to determine how accurate the Gram-stain smear was in predicting the main pathogen groups in monomicrobial and operated polymicrobial DFI’s, including for
P. aeruginosa, requiring immediate antibiotic treatment. We concentrated our evaluation on moderate and severe DFIs, as these are most likely to require urgent and appropriate antibiotic selections.
2. Materials and Methods
2.1. Setting, Database, Inclusion and Exclusion Criteria
The Balgrist University Hospital is a referral orthopedic center for diabetic foot problems. From January 1, 2000 until December 11, 2018, we included all adult DFI patients with (partial) lower extremity amputations or surgical debridement in our cohort. Exclusion criteria were: follow-up time less than six months; treatment with major amputation; or, orthopedic implant-related DFIs [
16]. For this analysis, the first author (D.A) re-assessed our DFI cohort and focused on Gram-stained smears. Microbiologically, we used Gram-stain and culture results done on intraoperative (or exceptionally deep tissue) samples [
16]. In our hospital we attempt to treat these infections with oral (as opposed to parenteral) antibiotic agents as soon as possible and to target only the “prevalent” or “virulent” pathogen groups in polymicrobial culture results of soft tissue infections.
2.2. Statistical Analyses
We analyzed only DFI episodes for which there were visible bacteria on a wound Gram-stained smear. We elected not to include a non-contributive Gram-stain attempt within the entire DFI cohort, because the lack of bacterial vizualization clinically depends on numerous clinical and laboratory factors that could considerably bias our performance testing on germ identification [
6]. For each culture, we used descriptive statistics to compared the Gram-stain results with the microbiological culture reports. Specifically, we computed sensitivity, specificity, positive and negative predictive values of the Gram-stain regarding microbiological cultures [
19]. Of note, while sensitivity and specificity are inherent to the performance of the Gram-stain, independently of the epidemiological circumstances, the predictive values strongly depend on the local prevalence of a bacteria group in the patient population [
19].
3. Results
3.1. Study Population and General Antibiotic Treatment
Among patients in our cohort we analyzed 1,235 moderate or severe DFI cases among adult patients. Overall, 86% of DFIs involved the forefoot, 8% the midfoot, and 3% the hindfoot (among these, 2% were in the calcaneus). Diabetes was type II in 68% and the patients and 54% of all operated cases underwent lower extremity revascularization. Empirically selected preoperative antibiotic therapy was prescribed by the patients’ general practitioner in 73.9% of operated episodes, for a median duration of thirteen days preoperatively, using 59 different regimens and dosing schemas [
15]. Our initial choice of post-surgical antibiotic regimen did not cover all causative pathogens in 32% of DFI episodes [
16]. Routine operative wound care was performed by nurses on the ward for inpatients and by professional wound nurses in the outpatient setting. All patients had appropriate wound pressure off-loaded, but we have no direct information on patient compliance with this in the outpatient setting.
3.2. Gram-Staining and Microbiological Cultures
We recorded 99 different bacterial culture constellations according to criteria of Clinical Laboratory Standards Institute (CLSI [previously]) and European Committee on Antimicrobial Susceptibility Testing (EUCAST [new]) [
20], while the Gram-staining procedures remained unchanged throughout the study protocol. All Gram-staining was performed by professional microbiologists (and their laboratory assistants) in the University of Zürich’s IMM (Institut für Medizinische Mikrobiologie [IMM]). Results of wound cultures were available 1-3 days after sampling, at least for antibiotic-naïve DFIs. The microbiological laboratory did not report Gram-stain results in 74% of cases (914/1235). Among the 321 (26%) for which Gram-stain results were reported, the microbiologists did not visualize bacteria in 46% of episodes (149/321). Thus, useful Gram-staining results were available in only 54% of the cases.
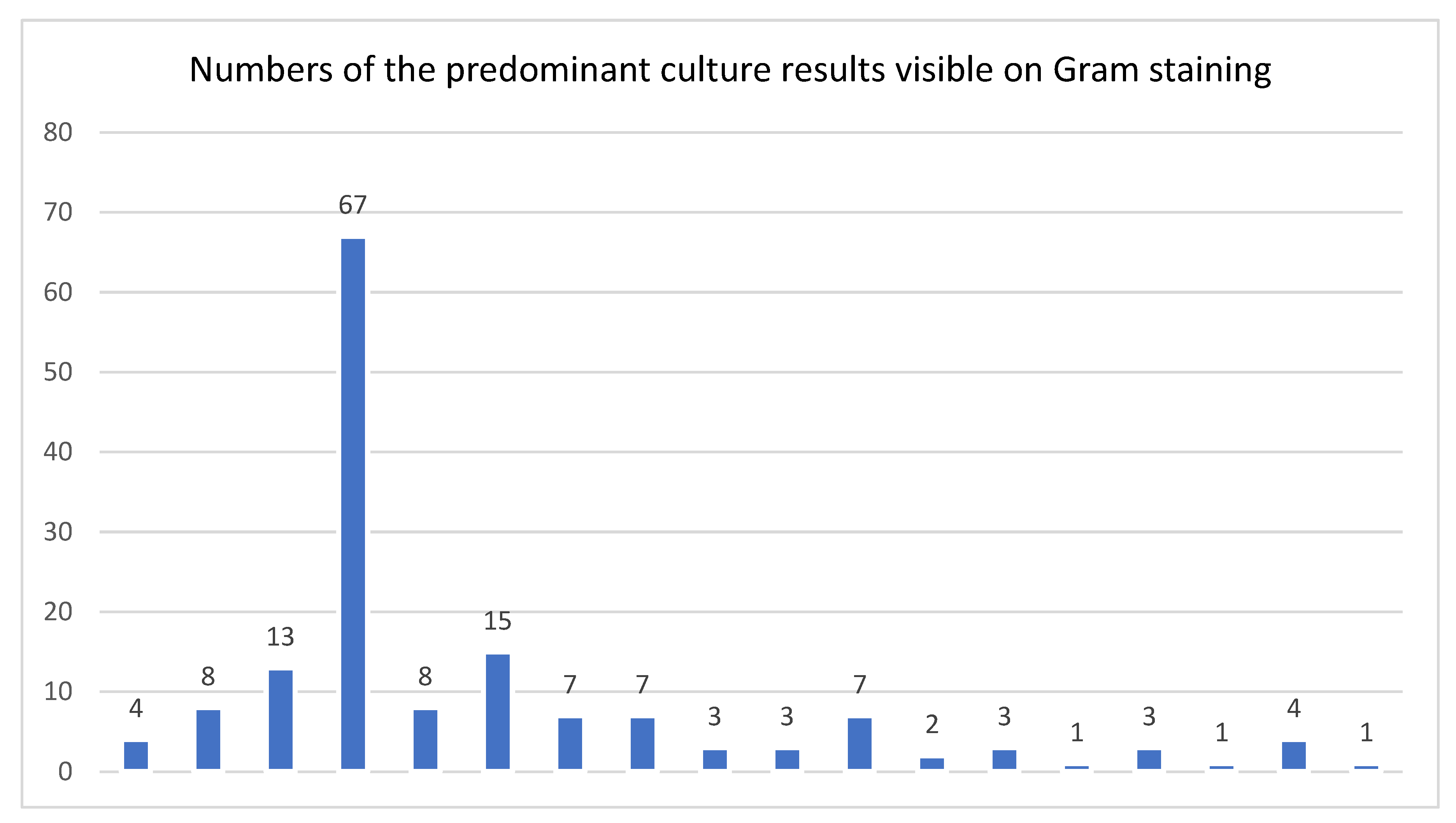
Overall, 35% of all DFI cultures were polymicrobial. Among isolated pathogens, 25% were Gram-negative bacteria. The eight most frequently-cultured pathogenic organisms or groups from all cases were: coagulase-negative staphylococci (n=258), Staphylococcus aureus (224), enterococci (60), Pseudomonas spp. (52); streptococci (33); Escherichia coli (31); Proteus spp. (23); and, obligate anaerobes (17). Among the cases in which organisms were visible on the Gram-stained smear the main pathogen groups were quite different: S. aureus (n=77); enterococci (33); coagulase-negative staphylococci (32); obligate anaerobes (24); Pseudomonas spp. (13); Proteus spp. (13); streptococci (12); and. E. coli (12). Specifically, coagulase-negative staphylococci were less frequently noted on the Gram-stain while S. aureus was the best detected. We don’t know if they were less often seen, or less often reported (as technicians may have considered them non-pathogens). For cultures that grew P. aeruginosa, Gram-negative organisms were only noted on the Gram-stained smears in one-quarter cases.
Among the Gram stain smear on which organisms were detected, in 40% (69/172) of cases the organisms were typically resistant to our standard empirical antibiotic choice of amoxicillin/clavulanic acid, 40 of which were non-fermenting Gram-negative rods.
Figure 1 displays the distribution of pathogen groups, which were visible on Gram-stain, in graphical form.
A pathogen was seen on an intraoperative Gram-stain smear in 172 of 1235 cases (14%), 101 (59%) of which were only Gram-positive, 32 (19%) only Gram-negative, and 39 (33%) had both.
Table 1 summarizes the concordance between the culture results and a prior Gram-stained smear (interpreted by an infectious diseases physician (I.U).
3.3. Prediction Performances of Cultured Pathogen Groups by the Gram-Stain
Compared to the culture results, the statistical performance characteristics of the Gram-stained smear for gram-positive organisms were: sensitivity 56%; specificity 93%; positive predictive value 97%; and negative predictive value 38%. The corresponding values for Gram-negative organisms were: 61%; 97%; 50%; and, 82%. When stratified for episodes under current (pre-operative) antibiotic treatment, the combined values were 67%, 96%, 97%, and 49%. When stratified for just monomicrobial infections, these performances were 51%, 92%, 93%, and 48% for Gram-positive cultures, and 52%, 98%, 88%, and 90% Gram-negative cultures.
Table 2 shows the links of the Gram-stain results to the cultures yielding non-fermenting Gram-negative rods.
4. Discussion
In our setting of a large Swiss city hospital, the results of routine Gram-stain smears of intraoperative DFI samples revealed a relatively low sensitivity throughout all investigated strata in predicting the pathogens found on cultured specimens. Overall, the Gram-stained smears reported pathogens only in half of the specimens from patients with moderate or severe DFI. Of note, many of the patients had been treated empirically with antibiotics before surgery. The pathogen most often visualized on the Gram-stained smears, as well found growing in the corresponding cultures, was
S. aureus. The overall negative predictive values of the Gram-stained smear were moderate at best, and only reasonably clinically useful for mono-bacterial Gram-negative rods. The positive and negative predictive values of the Gram-stained smear largely depended on the local epidemiology of DFI pathogens, which might certainly be different in other centers [
19]. When bacteria are observed in high numbers on the Gram stain, the positive predictive value can be quite high. On the other hand, the negative predictive calue is lower, which means that a negative Gram stain result does not definitively exclude a bacterial infection. Negative Gram stain results do not exclude the possibility of infection, and culture methods remain essential for identifying specific pathogens and determining their antibiotic susceptibilities. This discrepancy is especially important in cases where the bacterial load is low or the infection is in its early stages. Clinicians should interpret Gram stain results in conjunction with clinical findings and consider specimen quality to make informed decisions about patient management.
The published literature on the clinical value of Gram-stained smears is sparse and dominated by three publications: the Abbas trial performed in Dar es Salaam, Tanzania [
3]; the Taniguchi study from Okinawa, Japan [
7]; and, the Chisman study from Birmingham, England [
14]. Data from all three publications favor using the Gram-stain smear in the initial management of DFI and other infections [
4]. While the potential use of Gram-stained smear for assessing the likely causative pathogens in DFI is mentioned in some other articles [e.g., 21-25], athey are limited in assessing its potential. The Abbas paper is the most favorable for the use of the Gram-stain, at least for a resource-limited setting in the less-developed world where availability of microbiological cultures is often limited. Both the available literature and our own experience suggest that using the results of a Gram-stained smear is better than just available clinical (and other laboratory) evidence for tailoring the empirical antibiotic choice. In particular, it is likely to reduce unnecessary prescribing of anti-Gram-negative coverage in regions already dealing with a high prevalence of multidrug resistance among these frequent pathogens [
4,
5,
11,
12].
The 2017 Chisman paper is the most similar to our study [
14]. The authors investigated the role of microscopy in the management of DFI [
14]. Among 114 tissue samples included, in 50% on Gram-stain smear there were either no organisms or no predominant organism seen. Gram-stain results were in complete agreement with the final culture results only in 46%. Similar to our findings, 72% of the patients were already receiving systemic antibiotic agents at the time of sampling. Also, as in our study, the overall sensitivity and specificity of the Gram-stains were relatively low (75% and 70%, respectively), but the specificity for Gram-negative pathogens was almost 99%. The authors concluded that while the Gram-stain smear was a poor predictor of culture results, because it was quick and inexpensive it had a role in investigating the microbiology of DFI. Furthermore, they speculated that the presence of Gram-negative rods was specific enough to allow the early broadening of the empirical antibiotic regimen [
14], but did not provide evidence for this in their treatment outcomes. In that sense, we confirm all findings of the 2017 Chisman paper. The difference is that this paper might consider using the Gram-stain for an anti-Gram-negative antibiotic broadening, whereas we investigate if it could reduce the recommended spectrum.
While there are certainly potential benefits to obtaining a Gram-stained stain smear to help in selection of empiric therapy, there remain some problems. These include the possibility of missing non-predominant or poorly visualized co-pathogens in a polymicrobial DFI, and the lack of being able to confidently identify the particular species of pathogen by just its Gram-stain appearance [
4]. For example, a Gram-negative rod could be a susceptible
Proteus spp., or an
Enterobacter spp. naturally resistant to many first-line β-lactam antibiotics, which would require different antibiotic treatments. For this reason, and to determine the antibiotic susceptibilities of visualized organisms, cultures of specimens are still required. Another problem with the Gram-stained smear is that it requires at least a certain amount of inoculum, with an adequate load of bacteria, to see the microorganisms; and very probably a antibiotic-naïve sampling. This is often not the case in paucibacillary mild DFI with minimal drainage, or in patients already on antibiotic therapy. Additionally, inter-observer variabilities in reading Gram-stained smears can be substantial, especially when comparing experienced clinicians or microbiologists versus junior houseofficers or medical students, who are now less often training in this skill than in the past. With fewer hospital clinical microbiology laboratories now performing their own Gram-stained smears, there are fewer opportunities for clinicians or microbiologists to attain or maintain proficiency.
Regarding selecting antibiotic regimens, in mild and moderate DFI, clinicians do not usually need to attempt to cover all pathogens in soft-tissue DFI from the start, especially if the patient undergoes surgical treatment [
4,
16]. For example, many moderate and ischemic DFI harbor obligately anaerobic organisms that may be secondary pathogens, but we often leave these to open air after debridement without administering specific anti-anaerobe antibiotics [
4]. In accordance with international guidance [
1], we usually concentrate on covering the one or two most virulent pathogens, assuming that additional isolates found on culture are either non-virulent colonizers or contaminants. This approach, however, requires having adequate clinical and microbiological experience, and careful assessment of the clinical response to the empiric therapy. After initiating empiric antibiotic therapy, the choice of the definitive regimen must consider the patient’s clinical response, as well as the local epidemiology and antibiotic susceptibilities of pathogens [
4]. The Gram-stain smear also shares an important shortcoming of cultures. When more than one type of organism is seen or grown, especially from macerated or necrotic areas, neither can definitively distinguish between true pathogens and colonization/contamination. For example, in a similar setting of superficial burn infections, studies have demonstrated only a fair degree of microbiological correlation between Gram-stain and the pathogens of clinical infection (kappa statistic 0.32) [
26].
While we think there is practical value in obtaining Gram-stained smears for selecting empiric antibiotics for treating a DFI, especially in resource poor settings, their usefulness (especially in resource-rich settings) are still mostly hypothetical. We lack well-designed prospective trials assessing the effectiveness of Gram-stained smears in improving antibiotic therapy and clinical outcomes. The conclusions of the Chisman study [
14] suggested using the Gram-stained smear could allow early broadening of antibiotic treatment (if Gram-negative rods are seen), rather than the narrowing aimed at by antimicrobial stewardship. A prospective trial aimed at assessing the value of narrowing the antibiotic spectrum would be difficult because of ethical concerns about failing to adequately cover potential pathogens. In resource-rich settings, one potential design might be investigating the potential usefulness of the Gram-stained smear in a study limiting the empiric antibiotic spectrum for mild and moderate infections, while administering broad-spectrum agents for severe infections, as recommended in the IWGDF/IDSA guidelines [
1,
2]. For individual DFI cases this benefit might be small, but from a large scale societal viewpoint, even a short time period of avoidance of a broad-spectrum therapy could still be beneficiary. When performing trials, the quality of the tissue sampling is key in all trial considerations. We cannot emphasize enough that the Gram smear quality grading will always largely depend on the sampling approach [
27].
5. Conclusions
While Gram-stained smears offer immediate insights and can guide initial antimicrobial therapy in soft-tissue DFI, they should not be solely relied upon for definitive diagnosis. The Gram-stain is very probably useful in the management of DFI and regarding antibiotic stewardship in many resource-poor settings with limited access to microbiology, a high prevalence of Gram-negative pathogens, and difficulties for surgery. Based on an evaluation of data from our urban Swiss setting, and the sparse literature in temperate developed regions [
4,
7,
14], the results of Gram-stain smear from deep intraoperative DFI tissue samples provided limited usefulness for the prediction of the cultured pathogens in major clinical strata investigated. The overall negative predictive values for the Gram-stained smears were unsatisfactory, and only reasonably acceptable for the subgroup of cases caused by gram-negative rods. Prospective trials are needed to determine the value of the Gram-stained smear in supporting antibiotic stewardship in resource-rich settings such as in Switzerland. Such trials would be pragmatically difficult, and their results likely to provide benefit that would be more epidemiological than clinically helpful to patients.
Supplementary Materials
No supplementary materials.
Author Contributions
Conceptualization: I.U, F.W.A.W, B.A.L, and D.A; Methodology: F.W.A.W, D.A, and I.U. Validation: F.W.A.W, M.S, D.A, and I.U. Investigation: D.A, L.L, F.S, M.S, I.U, F.N, and F.W.A.W. Resources: D.A, M.S, F.W.A.W, J.L, and I.U; Data curation: D.A, F.W.A.W, I.U, and M.S. Data collection: D.A, F.W.A.W, M.S, and F.N. Data analysis: I.U; Analysis verification: I.U. Writing: original draft preparation, J.L, B.A.L, and I.U. Writing-review and editing, B.A.L, F.W.A.W, M.S, and I.U; Visual, I.U.; Supervision: I.U, F.W.A.W, and M.S. Project administration: D.A, I.U, and F.W.A.W. All authors have agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the local Ethics Committee of Zurich Canton.
Informed Consent Statement
All the included patients have signed a generic informed consent form, in which they authorized us to use their data in an anonymous form and in accordance with the regulations. The project was approved by the local Ethics Committee’ of Zurich Canton, Switzerland (BASEC-Number 2019-01994).
Data Availability Statement
Key data are available in an anonymous form upon reasonable scientific request to the corresponding author. They are not publicly available.
Acknowledgments
We are indebted to Ms. Nathalie Kühne, research assistant of the former Unit for Clinical and Applied Research (UCAR), for her help.
Conflicts of Interest
The authors declare no conflict of interest.
Congress workshop
Parts of this manuscript has been presented twice as different posters at the 84th Swiss National Annual Congress for Orthopedic Surgery and Traumatology (swissorthopaedics), 26-28 June 2024, in Lausanne, Switzerland, and at the Joint Annual Meeting for Infectious Diseases, 28-30 August 2024, in Berne, Switzerland (Poster Nr 0.095).
References
- Senneville, É.; Albalawi, Z.; Van Asten, S.A.; Abbas, Z.G.; Allison, G.; Aragón-Sánchez, J.; Embil, J.M.; Lavery, L.A.; Alhasan, M.; Öz, O.; Uçkay, I.; Urbančič-Rovan, V.; Xu, Z.-R.; Peters, E.J.G. IWGDF/IDSA guidelines on the diagnosis and treatment of diabetes-related foot infections (IWGDF/IDSA 2023. Diabetes Metab Res Rev 2024, 40, e3687. [Google Scholar] [CrossRef] [PubMed]
- Lipsky, B.A.; Berendt, A.R.; Cornia, P.B.; Pile, J.C.; Peters, E.J.; Armstrong, D.G.; Deery, H.G.; Embil, J.M.; Joseph, W.S.; Karchmer, A.W.; Pinzur, M.S.; Senneville, E.; Infectious Diseases Society of America. 2012 Infectious Diseases Society of America clinical practice guideline for the diagnosis and treatment of diabetic foot infections. Clin Infect Dis 2012, 54, 132–173. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Z.G.; Lutale, J.K.; Ilondo, M.M.; Archibald, L.K. The utility of Gram stains and culture in the management of limb ulcers in persons with diabetes. Int Wound J 2012, 9, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Abbas, Z.G.; Gangji, R.R.; Uçkay, I. Antibiotic Stewardship in the Management of Infected Diabetic Foot Ulcer Disease in Less Developed Countries. Endocrinol Diabetes Metab 2024, 7, 00503. [Google Scholar] [CrossRef]
- Sürme, S.; Saltoğlu, N.; Kurt, A.F.; Karaali, R.; Balkan, I.I.; Baghaki, S.; et al. Changing Bacterial Etiology and Antimicrobial Resistance Profiles as Prognostic Determinants of Diabetic Foot Infections: A Ten-Year Retrospective Cohort Study. Surg Infect (Larchmt) 2022, 23, 667–674. [Google Scholar] [CrossRef]
- Boyanova, L. Direct Gram staining and its various benefits in the diagnosis of bacterial infections. Postgrad Med 2018, 130, 105–110. [Google Scholar] [CrossRef]
- Taniguchi, T.; Tsuha, S.; Shiiki, S.; Narita, M. Gram-stain-based antimicrobial selection reduces cost and overuse compared with Japanese guidelines. BMC Infect Dis 2015, 15, 458. [Google Scholar] [CrossRef]
- Thairu, Y.; Abdullahi, N.I.; Yahaya, U. Laboratory Perspective of Gram Staining and its Significance in Investigations of Infectious Diseases. Sub-Saharan African J Med 2014, 1, 168–174. [Google Scholar] [CrossRef]
- Yoshimura, J.; Ogura, H.; Oda, J. Can Gram staining be a guiding tool for optimizing initial antimicrobial agents in bacterial infections? Acute Med Surg 2023, 10, 862. [Google Scholar] [CrossRef]
- Benavent, E.; Murillo, O.; Grau, I.; Laporte-Amargos, J.; Gomez-Junyent, J.; Soldevila, L.; Tubau, F.; Ariza, J.; Pallares, R. The Impact of Gram-Negative Bacilli in Bacteremic Skin and Soft Tissue Infections Among Patients With Diabetes. Diabetes Care 2019, 42, 110–112. [Google Scholar] [CrossRef]
- Ertuğrul, M.B.; Uyar-Güleç, G.; Baktıroğlu, S.; Çörekli, E.; Türe, M. The Distribution of Causative Microorganisms in Diabetic Foot Infection: Has There Been Any Alterations? Klimik Dergisi 2017, 30, 27–31. [Google Scholar] [CrossRef]
- Ramakant, P.; Verma, A.K.; Misra, R.; Prasad, K.N.; Chand, G.; Mishra, A.; Agarwal, G.; Agarwal, A.; Mishra, S.K. Changing microbiological profile of pathogenic bacteria in diabetic foot infections: Time for a rethink on which empirical therapy to choose? Diabetologia 2011, 54, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Uçkay, I.; Holy, D.; Schöni, M.; Waibel, F.W.A.; Trache, T.; Burkhard, J.; Böni, T.; Lipsky, B.A.; Berli, M.C. How good are clinicians in predicting the presence of Pseudomonas spp. in diabetic foot infections? A prospective clinical evaluation. Endocrinol Diabetes Metab 2021, 4, e00225. [Google Scholar] [CrossRef] [PubMed]
- Chisman, R.; Lowry, D.; Saeed, M.A.; Tiwari, A.; David, M.D. Prescribing antibiotics in diabetic foot infection: What is the role of initial microscopy and culture of tissue samples? Int Wound J 2017, 14, 685–690. [Google Scholar] [CrossRef]
- Muri, T.; Schöni, M.; Waibel, F.W.A.; Altmann, D.; Sydler, C.; Furrer, P.R.; Napoli, F.; Uçkay, I. Preoperative Antibiotic Administration Does Not Improve the Outcomes of Operated Diabetic Foot Infections. Antibiotics (Basel) 2024, 13, 1136. [Google Scholar] [CrossRef]
- Nieuwland, A.J.; Waibel, F.W.A.; Flury, A.; Lisy, M.; Berli, M.C.; Lipsky, B.A.; Uçkay, I.; Schöni, M. Initial antibiotic therapy for postoperative moderate or severe diabetic foot infections: Broad versus narrow spectrum, empirical versus targeted. Diabetes Obes Metab 2023, 25, 3290–3297. [Google Scholar] [CrossRef]
- Wuarin, L.; Abbas, M.; Harbarth, S.; Waibel, F.; Holy, D.; Burkhard, J.; Uçkay, I. Changing perioperative prophylaxis during antibiotic therapy and iterative debridement for orthopedic infections? PLoS ONE 2019, 14, 0226674. [Google Scholar] [CrossRef]
- Uçkay, I.; Berli, M.; Sendi, P.; Lipsky, B.A. Principles and practice of antibiotic stewardship in the management of diabetic foot infections. Curr Opin Infect Dis 2019, 32, 95–101. [Google Scholar] [CrossRef]
- Monaghan, T.F.; Rahman, S.N.; Agudelo, C.W.; Wein, A.J.; Lazar, J.M.; Everaert, K.; Dmochowski, R.R. Foundational Statistical Principles in Medical Research: Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value. Medicina (Kaunas) 2021, 57, 503. [Google Scholar] [CrossRef]
- Wolfensberger, A.; Sax, H.; Weber, R.; Zbinden, R.; Kuster, S.P.; Hombach, M. Change of antibiotic susceptibility testing guidelines from CLSI to EUCAST: Influence on cumulative hospital antibiograms. PLoS ONE 2013, 8, e79130. [Google Scholar] [CrossRef]
- Rahman, M.Z.; Uddin, M.J.; Mollah, A.H.; Khan, H.H.; Kabir, A.Z. Common Microorganisms Present in Diabetic Foot Infection and Its Spectrum of Antibiotic Sensitivity. Medicine Today 2022, 34, 65–69. [Google Scholar] [CrossRef]
- Oates, A.; Bowling, F.L.; Boulton, A.J.; Bowler, P.G.; Metcalf, D.G.; McBain, A. The visualization of biofilms in chronic diabetic foot wounds using routine diagnostic microscopy methods. J Diabetes Res 2014, 2014, 153586. [Google Scholar] [CrossRef] [PubMed]
- Barshes, N.R.; Rodriguez-Barradas, M.C.; Bechara, C.F.; Pisimisis, G.; Young, E.J.; Kougias, P. Microbial isolates and their antimicrobial susceptibilities in inframalleolar foot infections. Surg Infect (Larchmt) 2014, 15, 585–591. [Google Scholar] [CrossRef]
- Bouza, E.; Onori, R.; Semiglia-Chong, M.A.; Álvarez-Uría, A.; Alcalá, L.; Burillo, A. Fast track SSTI management program based on a rapid molecular test (GeneXpert® MRSA/SA SSTI) and antimicrobial stewardship. J Microbiol Immunol Infect 2020, 53, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, V.P.; Alves, C.A.S.; Queiroz, A.B.; Barberino, M.G.M.A.; Fidelis, R.J.R.; Fidelis, C.; de Araújo, J.S. Is there concordance between bone and tendon cultures in patients with foot tissue loss? J Vasc Bras 2019, 18, e20190063. [Google Scholar] [CrossRef]
- Elsayed, S.; Gregson, D.B.; Lloyd, T.; Crichton, M.; Church, D.L. Utility of Gram stain for the microbiological analysis of burn wound surfaces. Arch Pathol Lab Med 2003, 127, 1485–1488. [Google Scholar] [CrossRef]
- Clark, S.T.; Forbes, J.D.; Matukas, L.M. Wound swab quality grading is dependent on Gram smear screening approach. Sci Rep 2023, 13, 3160. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).