Submitted:
28 February 2025
Posted:
03 March 2025
You are already at the latest version
Abstract
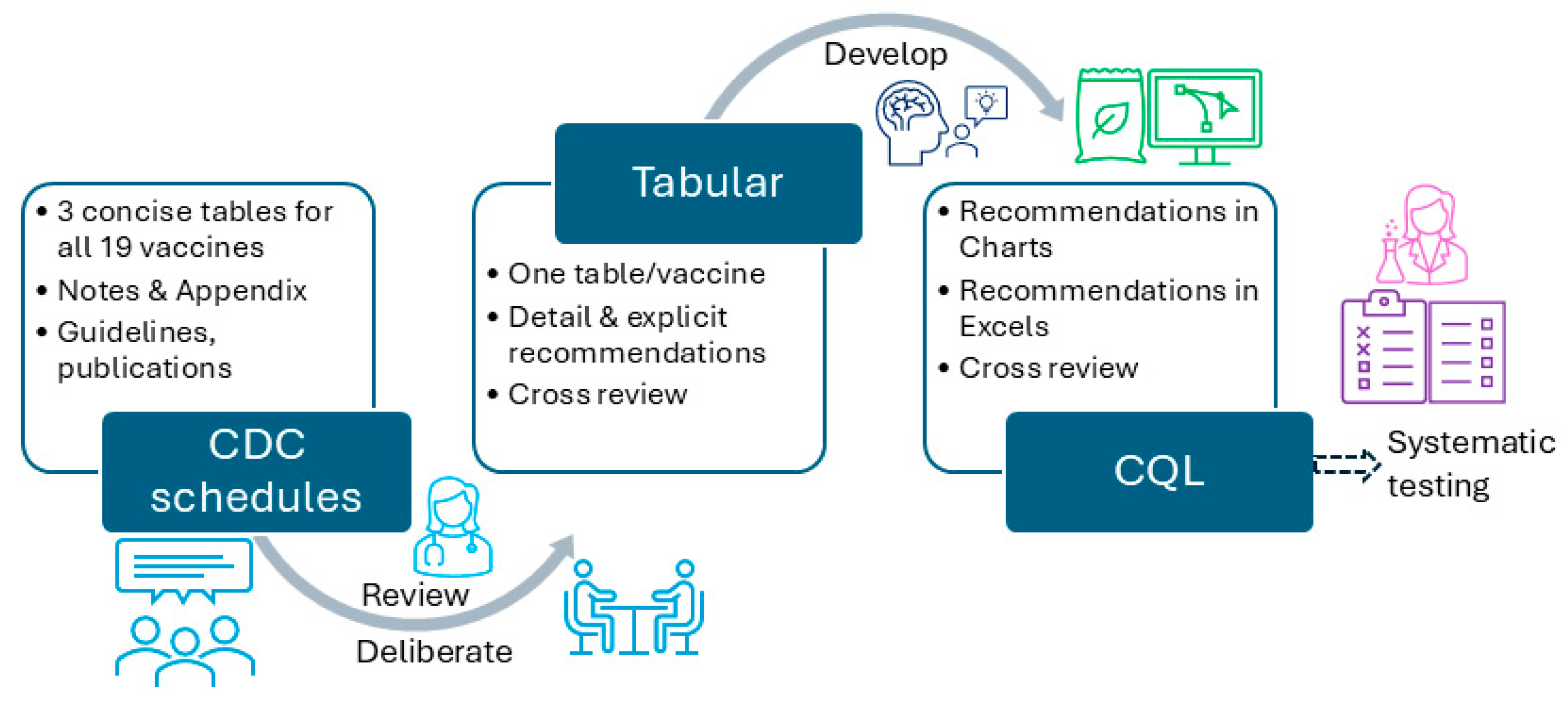
Reusable machine-processable clinical decision support system (CDSS) rules have not been widely achieved in the medical informatics field. This study introduces the process, results, challenges faced, and lessons learned while converting Centers for Disease Control and Prevention (CDC)-recommended immunization schedules (2022) to machine-processable CDSS rules. The study presents our experience in converting the vaccination schedules into tabular, charts, MS Excel, and clinical quality language (CQL) formats. CQL format can be automatically converted to machine-processable format using existing tools. Therefore, it was regarded as a machine-processable format. We have developed 465 rules for 19 vaccines in 13 categories, and we have shared the rules via GitHub to make them publicly available. We used cross-checking to validate CDSS rules in tabular and chart formats. CQL files were tested for syntax and logic with hypothetical patient HL7 FHIR resources. Our rules can be reused and shared by the health IT industry, CDSS developers, medical informatics educators, or clinical care institutions. These CDSS rules can be an important contribution to informatics communities, reducing redundant efforts, which is particularly significant in resource-limited settings. Despite the maturity and concise presentation of the CDC recommendations, careful attention and multiple layers of verification and review are necessary to ensure accurate conversion.
Keywords:
1. Introduction
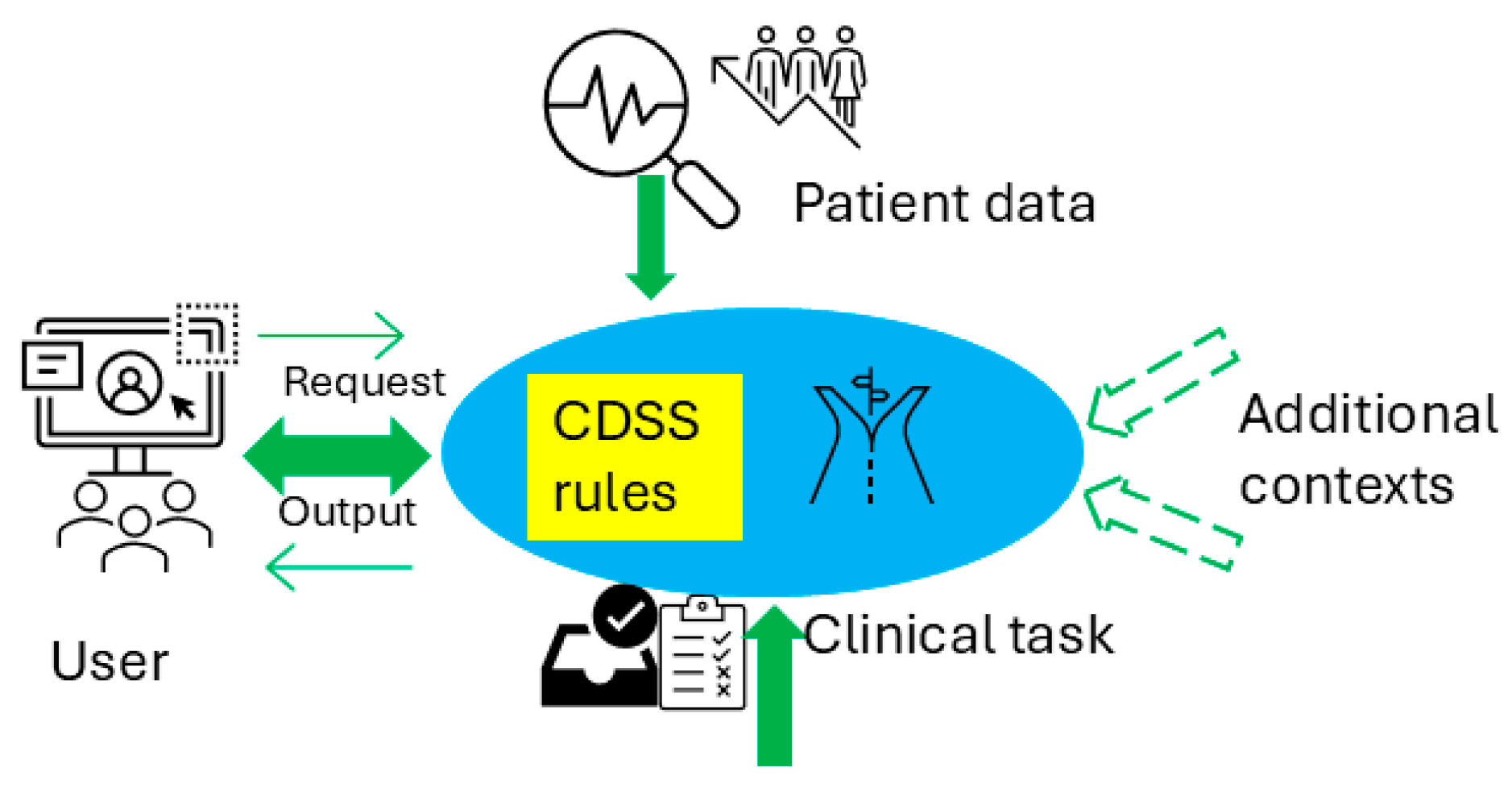
2. Methods
2.1. Vaccination Schedule Rules Converting Workflow
2.2. Specifications Related to Each Format
2.3. Quality Control Strategies
3. Results
4. Discussion
4.1. Significance of the Work
4.2. Challenges and Limitations of the Work
4.3. Lessons Learned Through the Process
4.4. Path to Conversion
4.5. Future Work
5. Conclusions
Acknowledgments
References
- Greenes, R. and G.D. Fiol, Clinical Decision Support and Beyond: Progress and Opportunities in Knowledge-Enhanced Health and Healthcare. 3rd Edition ed. 2023, Cambridge, MA: Elsevier: Academic Press.
- Shortliffe, E.H. J. Cimino, and M.F. Chiang, Biomedical Informatics: Computer Applications in Health Care and Biomedicine. 5th ed. 2021: Springer International Publishing.
- Lobach, D., et al. Enabling Health Care Decisionmaking Through Clinical Decision Support and Knowledge Management. Evidence Report No. 203. (Prepared by the Duke Evidence-based Practice Center under Contract No. 290-2007-10066-I.) AHRQ Publication No. 12-E001-EF. 2012. Rockville, MD.
- Berner, E.S., Clinical Decision Support Systems. Health Informatics. 2016: Springer International Publishing Switzerland.
- Jing, X. Himawan, and T. Law, Availability and usage of clinical decision support systems (CDSSs) in office-based primary care settings in the USA. BMJ Health Care Inform, 2019. 26(1).
- Miller, P.L., et al., Combining tabular, rule-based, and procedural knowledge in computer-based guidelines for childhood immunization. Comput Biomed Res, 1997. 30(3): p. 211-31. [CrossRef]
- Miller, P.L., Domain-constrained generation of clinical condition sets to help test computer-based clinical guidelines. J Am Med Inform Assoc, 2001. 8(2): p. 131-45.
- Miller, P.L., et al., IMM/Serve: a rule-based program for childhood immunization. Proc AMIA Annu Fall Symp, 1996: p. 184-8.
- Jing, X., et al., Ontologies Applied in Clinical Decision Support System Rules: Systematic Review. JMIR Med Inform, 2023. 11: p. e43053. [CrossRef]
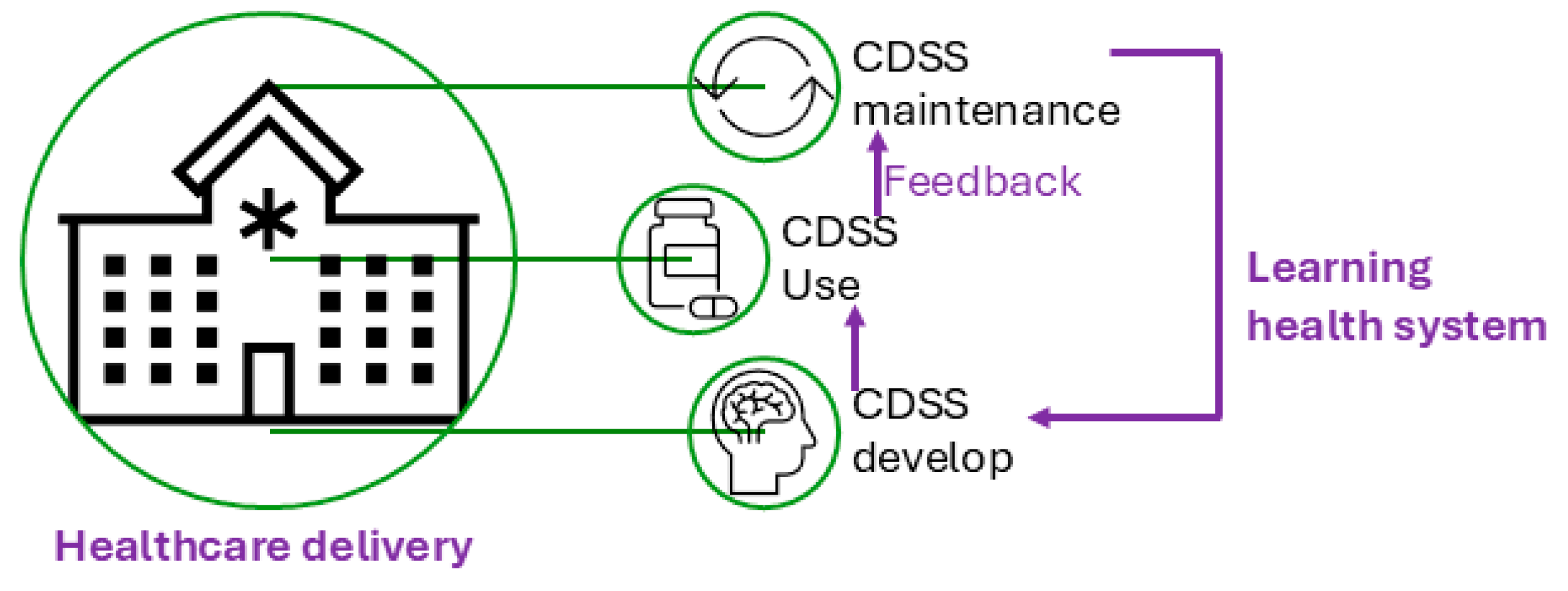
- Friedman, C., et al., Toward a science of learning systems: a research agenda for the high-functioning Learning Health System. J Am Med Inform Assoc, 2015. 22(1): p. 43-50.
- AHRQ. AHRQ CDS Connect. 2024; Available from: https://cds.ahrq.gov/cdsconnect.
- Gruber, T., What is an ontology? Knowledge Acquisition, 1993. 5: p. 199-220.
- Sowa, J.F., Building, sharing and merging ontologies. 2005. 2006.
- Davies, J. Fensel, and F.V. Harmelen, Towards the semantic web: Ontology-driven knowledge management. 2003, Chichester: John Wiley & Sons.
- Rector, A., Foundations of the Semantic Web: Ontology Engineering. 2005. 2006.
- Gruber, T., Ontology, in Encyclopedia of Database Systems, L. Liu and M. Tamer Ozsu, Editors. 2008, Springer-Verlag.
- Jackson R, et al., OBO Foundry in 2021: operationalizing open data principles to evaluate ontologies. Database (Oxford), 2021: p. baab069.
- Whetzel, P., et al., BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res, 2011. 39: p. W541-545. [CrossRef]
- Jing, X., et al., A clinical decision support system (CDSS) ontology to facilitate portable vaccination CDSS rules: preliminary results, in AMIA 2021. 2021: San Diego, CA. p. 1695.
- Jing X, et al., Using Semantic Web Technology to leverage interoperable clinical decision support system rules: a pathway to interoperable patient records. BMC Proceedings, 2023. 17: p. O13.
- Seitz, M.W. Listl, and P. Knaup, Development of an HL7 FHIR Architecture for Implementation of a Knowledge-based Interdisciplinary EHR. Stud Health Technol Inform, 2019. 262: p. 256-259.
- FHIR, H. HL7 FHIR: Release 5. 2025 [cited 2025 Jan 21]; Available from: https://www.hl7.org/fhir/overview.html.
- HealthIT.gov. Clinical Quality Language (CQL). 2024 [cited 2025 Jan 21]; Available from: https://ecqi.healthit.gov/cql?qt-tabs_cql=about.
- HL7 Clinical Decision Support Working Group. Clincial Quality Language (CQL). 2023 [cited 2025 Jan 21]; Available from: https://cql.hl7.org/.
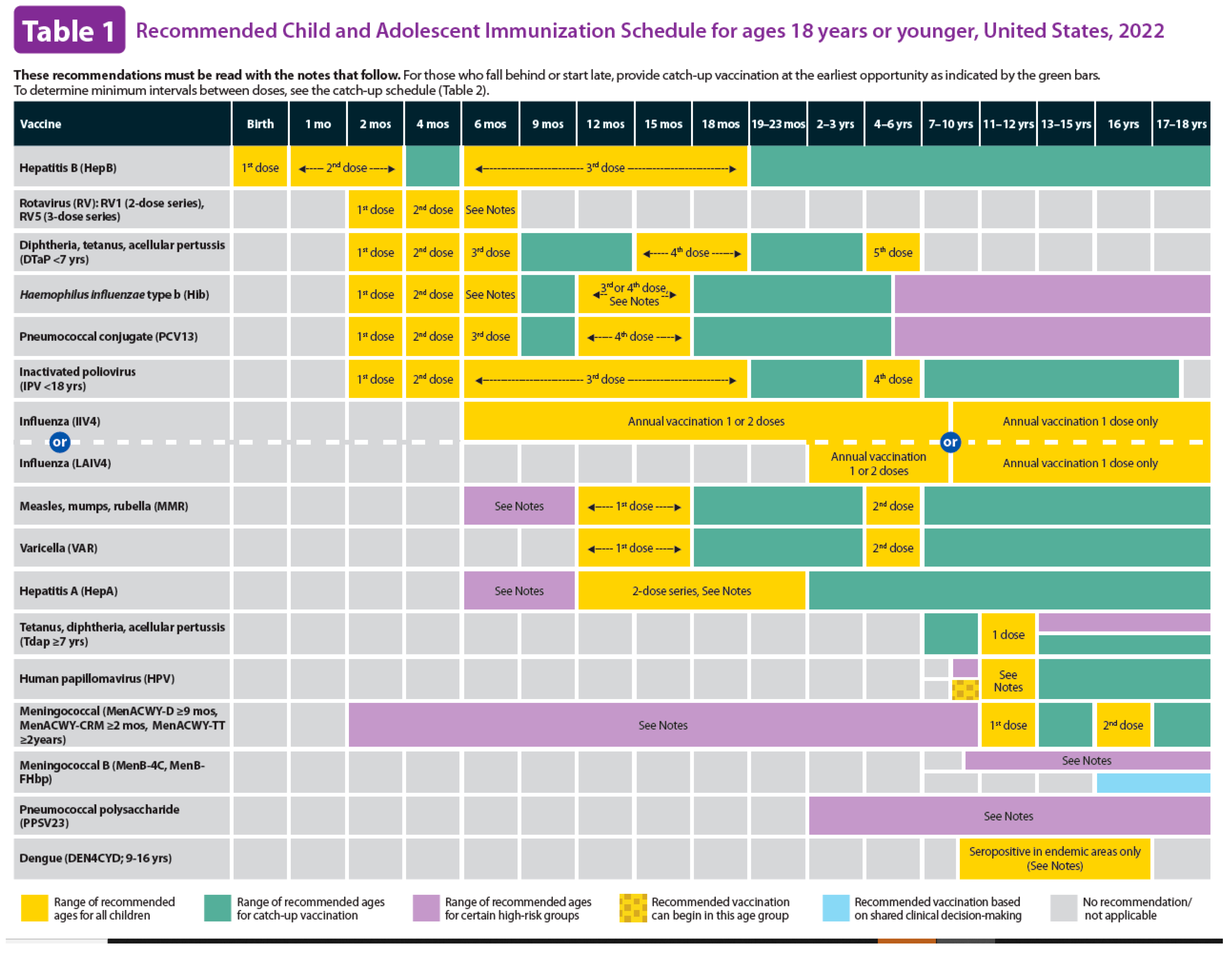
- CDC. Immunization Schedules. 2023 [cited 2024 Sept 12]; Available from: https://www.cdc.gov/vaccines/imz-schedules/index.html.
- Drawio. Diagram.drawio. 2025 [cited 2025 Feb 25]; Available from: https://app.diagrams.net/.
- cqframework. Clinical Quality Language. 2025 [cited 2025 Feb 25]; Available from: https://github.com/cqframework/clinical_quality_language.
- Orlioglu, S. CQLfiles for CDSS testing. 2025 [cited 2025 Feb 28]; Available from: https://github.com/sorliog/cdss-testing-harness.
- mcode. mcode/cql-testing-harness. 2025 [cited 2025 Feb 25]; Available from: https://github.com/mcode/cql-testing-harness.
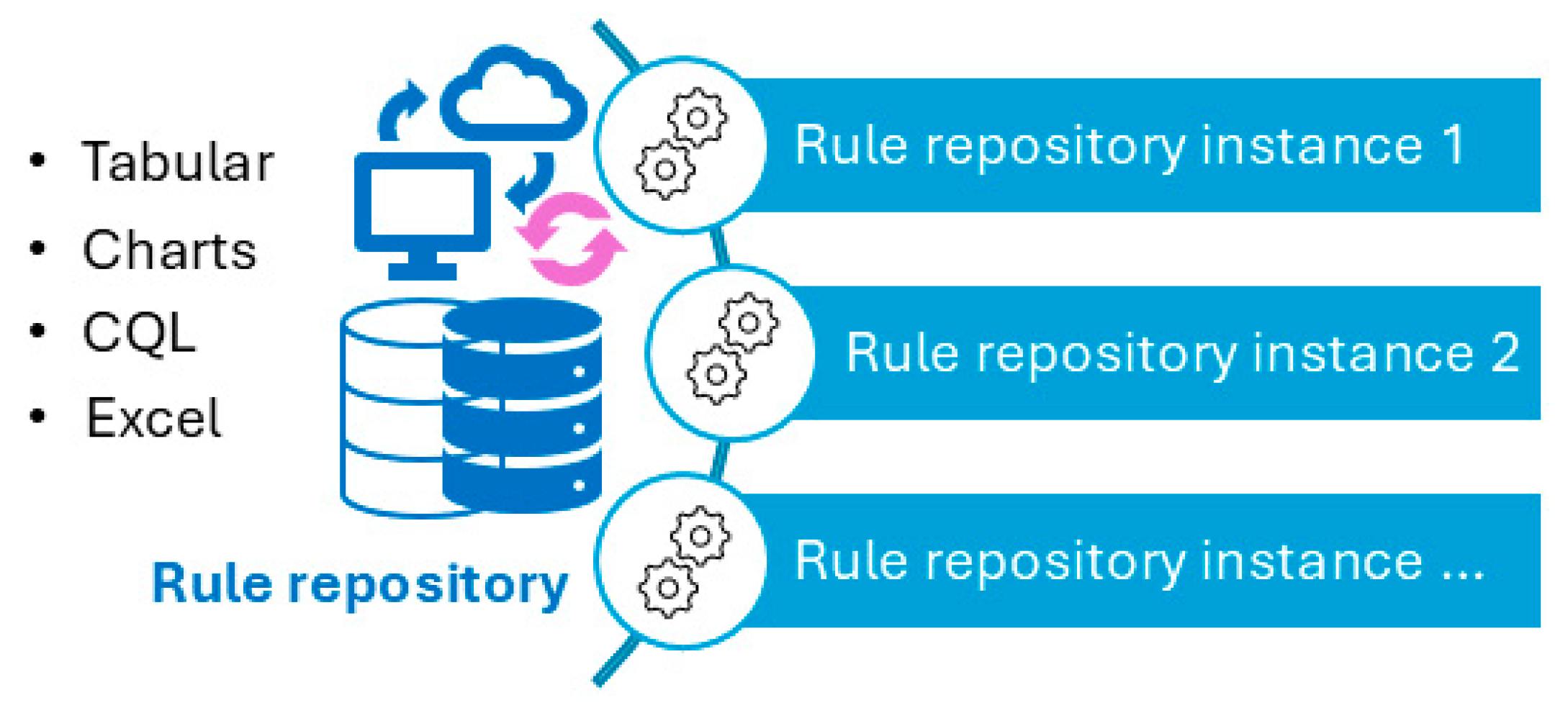
- group, C.P. CDSS Rule Repository. 2025 [cited 2025 Feb 28]; Available from: https://github.com/CDSS4PCP/CDSS_Rules_Repository.
- Stacey, M., C. McGregor, and M. Tracy, An architecture for multi-dimensional temporal abstraction and its application to support neonatal intensive care. Annu Int Conf IEEE Eng Med Biol Soc, 2007. 2007: p. 3752-6.
- Hripcsak, G. , Writing Arden Syntax medical logic modules. Computers in Biology and Medicine, 1994. 24(5): p. 331-363.
- Samwald, M. , et al., Towards an interoperable information infrastructure providing decision support for genomic medicine. Stud Health Technol Inform, 2011. 169: p. 165-9.
- Brandt, C.A. , et al., Visualizing the logic of a clinical guideline: a case study in childhood immunization. Methods Inf Med, 1997. 36(3): p. 179-83.
- Miller, P.L. , Tools for immunization guideline knowledge maintenance. I. Automated generation of the logic "kernel" for immunization forecasting. Comput Biomed Res, 1998. 31(3): p. 172-89.
- Miller, P.L., S. J. Frawley, and F.G. Sayward, Issues in accommodating national changes and local variation in a computer-based guideline for childhood immunization and in related knowledge maintenance tools. Proc AMIA Symp, 1998: p. 563-7.
- Geissbuhler, A. and R. Miller, distributing knowledge maintenance for clinical decision support systems, in Proc AMIA Symp. 1999. p. 770774.
- Miller, P.L., S. J. Frawley, and F.G. Sayward, Exploring three approaches for handling incomplete patient histories in a computer-based guideline for childhood immunization. Proc AMIA Symp, 1999: p. 878-82.
- Miller, P.L., Opportunities at the intersection of bioinformatics and health informatics: a case study. J Am Med Inform Assoc, 2000: p. 431-438. [CrossRef]
- Miller, P.L., S. J. Frawley, and F.G. Sayward, Maintaining and incrementally revalidating a computer-based clinical guideline: a case study. J Biomed Inform, 2001. 34(2): p. 99-111.
- Marco-Ruiz, L., et al. Interoperability Mechanisms of Clinical Decision Support Systems: A Systematic Review. in 14th Scandinavian Conference on Health Informatics. 2016. Gothenburg, Sweden.
- Samwald, M. , et al., The Arden Syntax standard for clinical decision support: Experiences and directions. J Biomed Infor, 2012. 45(4): p. 711-718.
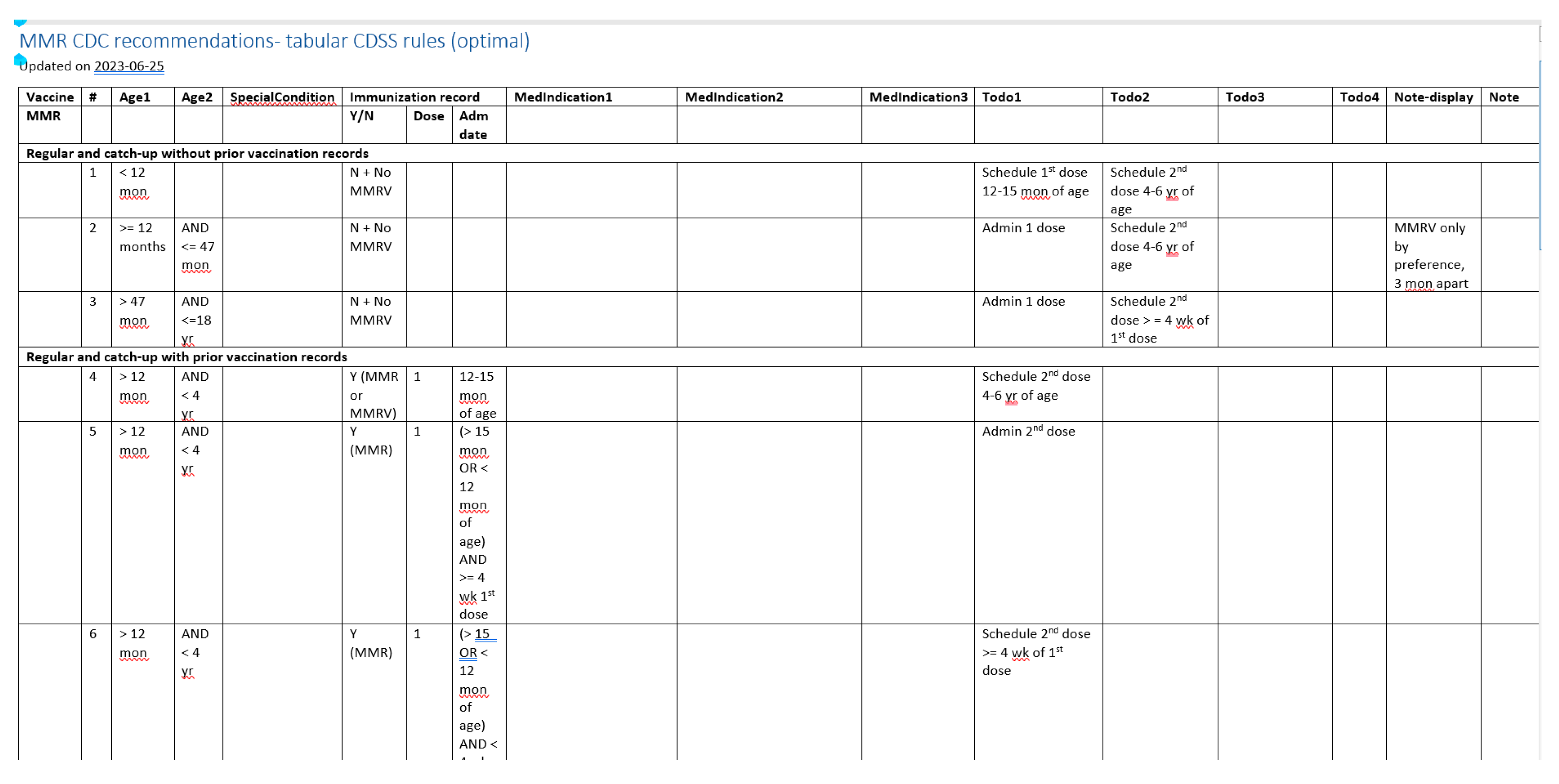
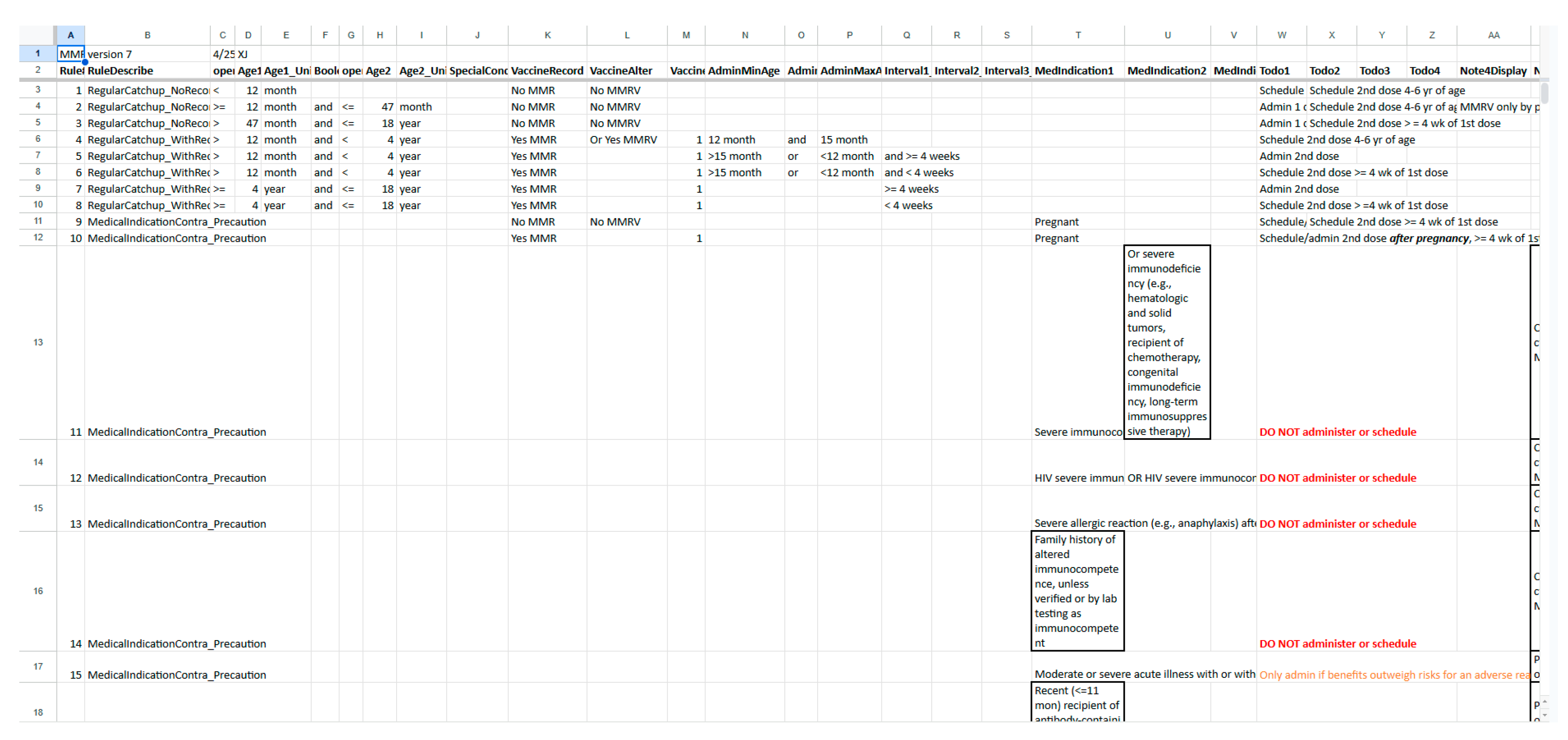
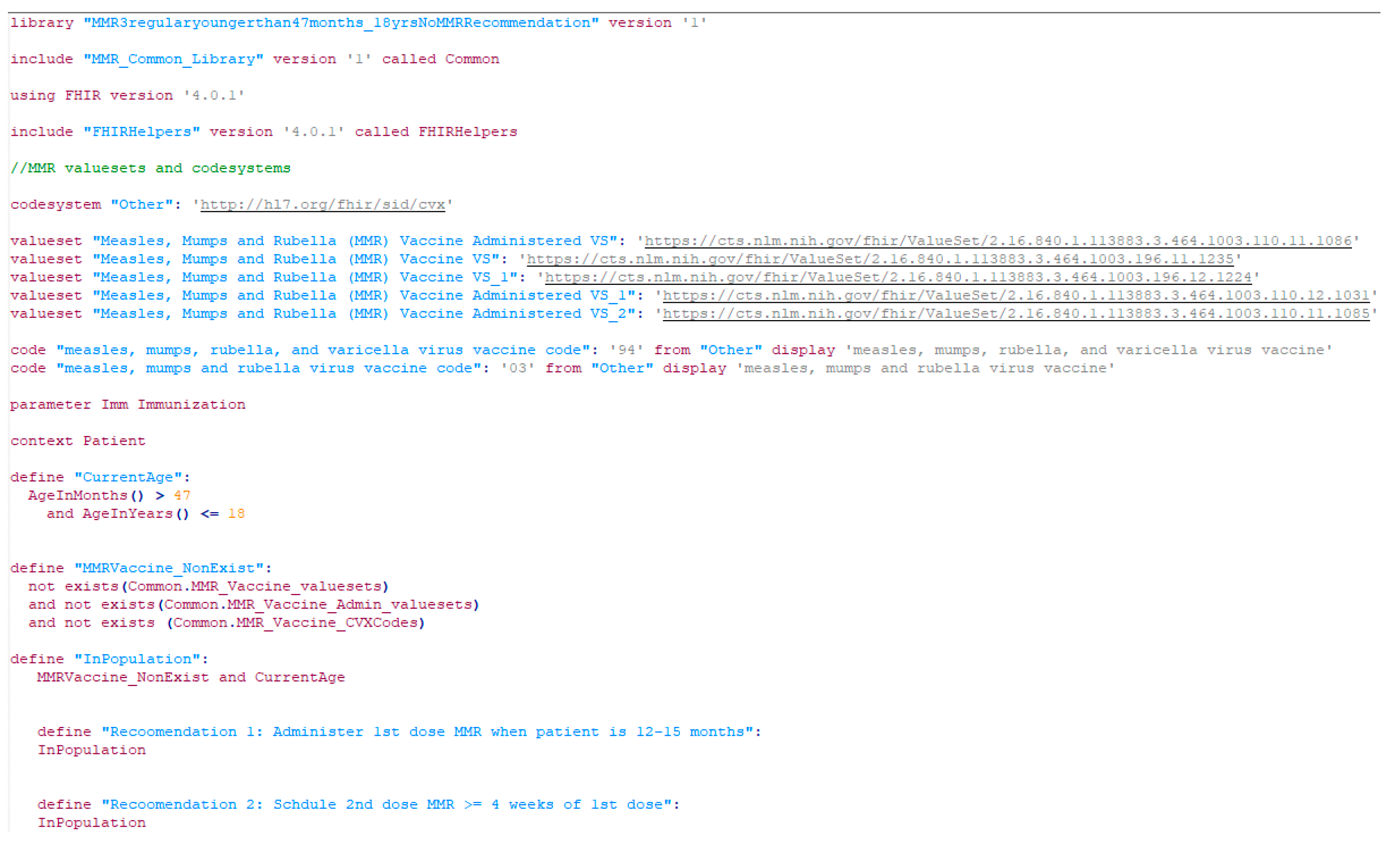
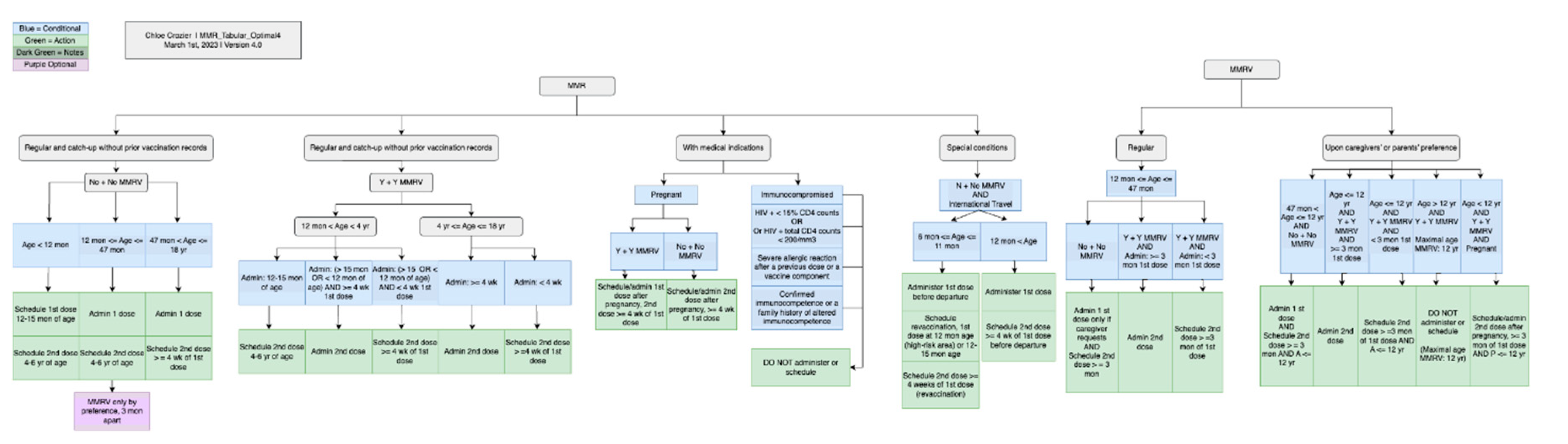
| Vaccine category | Abbreviation | Regular or catch up | Medical conditions /special situations | Contraindication /precaution | Total |
|---|---|---|---|---|---|
| Dengue | DEN4CYD | 2 | - | 7 | 9 |
| Diphtheria, tetanus, and acellular pertussis | DTaP, DT | 16 | 14 | 7 | 37 |
| Haemophilus influenzae type b | Hib | 16 | 20 | 4 | 40 |
| Hepatitis A | HepA, Twinrix | 6 | 2 | 4 | 12 |
| Hepatitis B | HepB | 8 | 14 | 3 | 25 |
| Human papillomavirus | HPV | 15 | 18 | 2 | 35 |
| Influenza (inactivated and live and attenuated) | IIV4, LAIV4 | 13 | - | 33 | 46 |
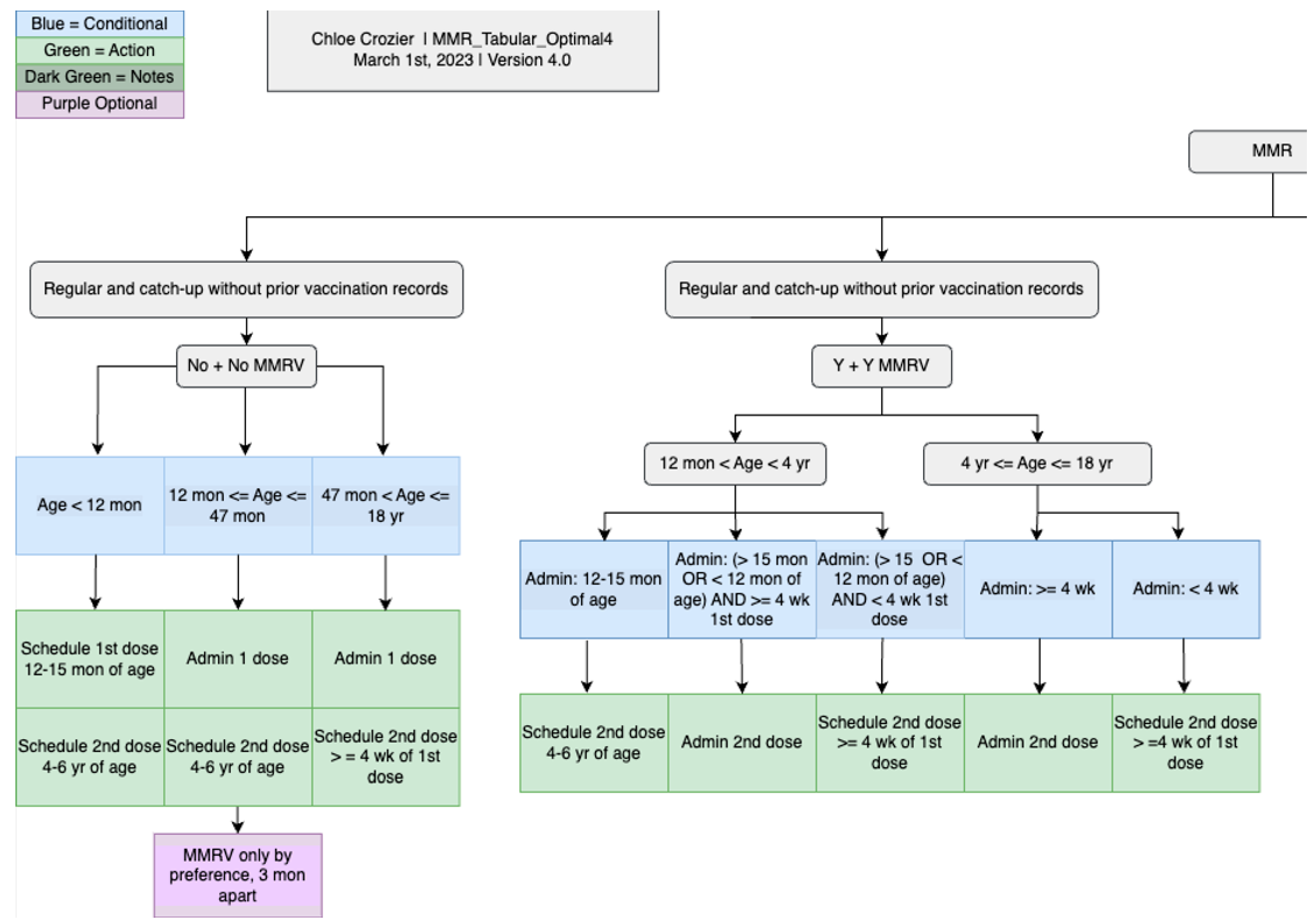
| Measles, mumps, and rubella | MMR, MMRV | 16 | 2 | 10 | 28 |
| Meningococcal serogroups A, C, W, Y | MenACWY-D, MenACWY-CRM, MenACWY-TT | 12 | 34 | 5 | 51 |
| Meningococcal serogroup B | MenB-4C, MenB-FHbp | 8 | 8 | 4 | 20 |
| Pneumococcal 13-valent conjugate, 23-valent polysaccharide | PCV13, PPSV23 | 20 | 30 | 4 | 54 |
| Poliovirus (inactivated) | IPV, tOPV | 33 | 6 | 3 | 42 |
| Rotavirus | RV1, RV5 | 11 | - | 8 | 19 |
| Tetanus, diphtheria, and acellular pertussis | Tdap, Td | 12 | 9 | 6 | 27 |
| Varicella | VAR, MMRV | 12 | - | 8 | 20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





