1. Introduction
In the era of information abundance, Large Language Models (LLMs) have achieved significant breakthroughs in general knowledge reasoning tasks, primarily due to extensive pre-training on diverse, publicly accessible datasets [
1]. The widespread adoption of such pre-training paradigms has laid a robust foundation for deploying these models across a broad spectrum of application domains [
2].
Despite their remarkable versatility, LLMs still face crucial limitations. Specifically, in high-stakes fields such as medicine and law, LLM-generated hallucinations pose substantial risks. These risks underscore the importance of reading comprehension, especially in evidence-based question-answering (QA), where models are required to provide answers grounded in specific knowledge sources and accurately attribute them [
3]. Indeed, standalone LLMs often struggle with incomplete or inaccurate internal knowledge, leading to hallucinations and other errors when confronted with complex, domain-specific problems [
4,
5].
To address these shortcomings, Retrieval-Augmented Generation (RAG) has emerged as a promising framework. By integrating LLMs with extensive external knowledge bases, RAG enhances both the factual reliability and traceability of generated responses, thereby offering a more robust solution for specialized domains [
6]. Although RAG significantly mitigates common pitfalls such as hallucinations and incomplete knowledge, it also introduces new obstacles for models with relatively small parameter scales. When presented with multiple disjoint retrieval segments, these models frequently exhibit a decline in reading comprehension, limiting their ability to accurately pinpoint pertinent information and produce coherent, domain-appropriate analyses.
In the medical domain, LLMs are increasingly integral to both research and clinical practice. Models such as Huatuo [
7] and Bianque [
8] illustrate ongoing efforts to adapt general LLMs through fine-tuning on open-source medical data. However, their performance in answering medical questions often remains less than optimal, largely due to the inconsistent quality of these data sources. Moreover, the lack of traceability in model outputs, combined with potential human errors in real or synthetic dialogue-based datasets [
9], presents a further challenge to the professionalism and accuracy required in a medical context.
In response, we propose a more rigorous data construction paradigm, augmented by robust filtering and verification mechanisms, to improve the reliability of training data. By continuously refining the RAG methodology and integrating high-quality, curated medical content, we introduce a novel approach: full-parameter fine-tuning of LLMs within the RAG framework specifically for geriatric medicine. This strategy not only improves the model’s capacity to identify relevant retrieval segments with high precision but also enhances its applicability in clinical settings, resulting in outputs that are both more reliable and more professional.
Our main contributions are summarized as follows:
We develop an automated workflow for generating diverse, high-quality RAG datasets tailored to geriatric medicine, leveraging authoritative Chinese medical encyclopedias to create a specialized Chinese medical RAG QA dataset.
We apply a full-parameter fine-tuning paradigm within the RAG framework for geriatric medicine, significantly boosting the model’s ability to retrieve and utilize the correct evidence segments from external knowledge sources.
We design targeted evaluation metrics to assess the model’s professionalism and accuracy. Empirical results show that, compared to both a “general LLM+RAG” baseline and a “domain-finetuned LLM+RAG” approach, our method achieves substantial performance gains in geriatric medical QA while sustaining robust general-domain competence.
2. Related Work
2.1. Domain Adaptation Strategies for LLM
Large Language Models (LLMs) have demonstrated exceptional performance on general-domain tasks; however, their effectiveness in specialized domains such as geriatric medicine often remains suboptimal [
10]. The core issue lies in the insufficient domain-specific knowledge encoded within these models. To address this gap, researchers have proposed various strategies. One approach involves pre-training on specialized medical corpora to enrich the model’s domain vocabulary and yield more precise knowledge representations. Alternatively, researchers have explored directly infusing medical knowledge into LLMs to augment their comprehension and application of domain-specific concepts [
11]. Additionally, further fine-tuning with synthetic medical dialogues or real clinical interactions has proven effective in enhancing the adaptability of larger models to clinical scenarios [
12].
Despite these advancements, instruction-tuned LLMs encounter notable constraints in medical contexts. They may retain outdated or incomplete internal knowledge, thus failing to reliably address current research findings or rare clinical cases. Moreover, even post-fine-tuning, these models can struggle with complex or specialized queries. This is particularly risky in high-stakes fields like medicine, where inaccuracies carry severe real-world implications.
With the goal of mitigating such limitations, Retrieval-Augmented Generation (RAG) has been proposed as a promising alternative. By combining LLMs with real-time retrieval from external knowledge bases, RAG enables dynamic incorporation of relevant information to enhance both the accuracy and timeliness of responses. Consequently, RAG holds significant potential for improving the reliability and professionalism of LLMs in specialized settings, including geriatric healthcare.
2.2. Enhancing Domain QA with RAG
Retrieval-Augmented Generation (RAG) merges retrieval techniques with generative models, allowing for on-demand integration of external data that can improve the factual grounding of generated text [
13]. In evidence-based QA, RAG first retrieves pertinent documents from large-scale knowledge bases and then generates answers substantiated by specific citations, thereby maintaining both coherence and factual accuracy [
14].
Within the medical field, RAG-based solutions have demonstrated considerable promise in augmenting information retrieval and clinical decision support [
15]. For instance, BioReader [
16] leverages a PubMed-derived corpus of 60 million articles to refine model inputs, delivering more accurate predictions across diverse clinical tasks. BEEP integrates patient data with contextually relevant scientific literature to improve outcome forecasting, such as in-hospital mortality [
17]. Similarly, Almanac leverages up-to-date medical guidelines to help clinicians make real-time, evidence-based decisions [
18].
However, the success of RAG hinges on the availability of high-quality, evidence-based QA datasets, whose creation is typically resource-intensive and thus limited in supply. Even widely utilized datasets such as MedMCQA [
19] and PubMedQA [
20] may contain noisy or inconsistent samples, affecting the overall reliability of RAG systems. Overcoming these challenges is crucial for unlocking the full potential of RAG in healthcare applications.
2.3. Optimizing Fine-Tuning and Data Quality for Domain-Specific Models
Fine-tuning LLMs for specialized tasks ordinarily requires substantial human annotations, a hurdle that becomes particularly acute in domains where experts are scarce or annotation costs are high. To alleviate this data bottleneck, researchers have investigated various strategies, including knowledge distillation [
21,
22,
23], data augmentation [
24,
25], module replacement [
26], semi-supervised learning [
27], and data synthesis [
28].
Data availability poses unique complications in medicine, where “data silos” across institutions often impede the acquisition of large, unified datasets. Moreover, open-source Chinese medical QA corpora frequently exhibit uniform or simplistic question patterns, limiting their efficacy for real-world applications. While existing approaches primarily focus on enhancing LLMs via synthetic or real medical dialogues [
29], these methodologies are prone to human error, making it challenging to train models that consistently deliver accurate and hallucination-free answers.
RAG models, which integrate retrieval and generation, offer a promising avenue to circumvent these limitations. However, the performance of such models strongly depends on the caliber of training data. Domain-specific RAG datasets remain scarce, significantly hindering the deployment of robust applications in professional settings. Thus, further investment in constructing and annotating high-quality datasets, along with the development of novel data enhancement strategies, is imperative for advancing RAG-based solutions in specialized fields like geriatric medicine.
3. Methodology
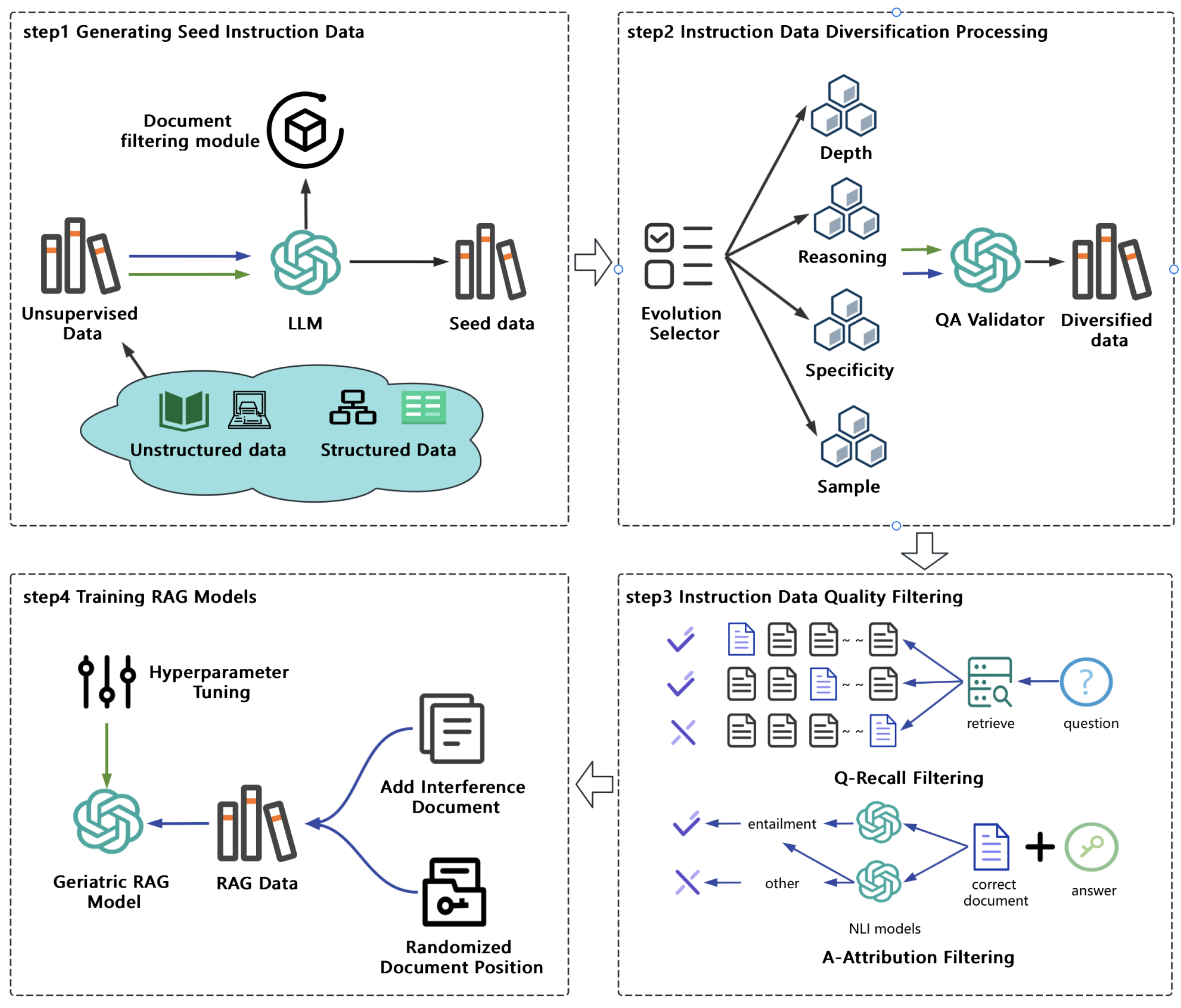
This section outlines our approach for building a robust geriatric medical question-answering system under the Retrieval-Augmented Generation (RAG) framework. First, we describe the collection and preprocessing of unsupervised medical data, focusing on authoritative geriatric resources. We then illustrate how we convert this knowledge into high-quality training data specialized for RAG tasks. Finally, we present our full-parameter fine-tuning strategy for a large language model (LLM), which incorporates domain-relevant medical information retrieved from an external knowledge base. The overall workflow is depicted in
Figure 1.
3.1. Generating Seed Instruction Data
To establish a high-quality geriatric medical dataset, we collected authoritative, up-to-date medical information from reputable websites specializing in geriatric care. Leveraging the advanced generative capabilities of large language models (LLMs), we designed a guiding mechanism that produces medical instruction data closely aligned with real-world clinical contexts. This automated approach significantly alleviates the labor burden of manual annotation and substantially enhances both the breadth and reliability of the resulting dataset.
3.1.1. Unsupervised Knowledge Acquisition
Medical knowledge can manifest in both structured formats—such as knowledge graphs and databases—and unstructured formats—such as medical guidelines and scholarly articles. In this study, we primarily focused on disease encyclopedia entries pertinent to geriatric medicine from a highly regarded Chinese medical website. This data source incorporates both structured metadata (e.g., disease taxonomy) and unstructured descriptions (e.g., symptom explanations, treatment notes) relevant to elderly healthcare.
We first converted the structured metadata into free-text form, creating a unified corpus of unstructured text. Subsequently, we applied a series of domain-specific filtering rules to remove spurious or incomplete information. This rigorous preprocessing ensures that the remaining text is accurate, comprehensive, and well-suited for downstream geriatric QA tasks.
3.1.2. Geriatric Medical Seed Instruction Data Generation
Drawing on this unsupervised repository, we utilized a powerful LLM to generate seed instruction data targeted to geriatric use cases. A cornerstone of this process was our systematic prompting strategy, which explicitly framed the model as a “physician.” By embedding professional medical contexts and clear task objectives into the prompts, the model is guided to produce instruction data that adheres to clinical standards.
To further amplify the logical depth and coherence of these instructions, we integrated the Chain-of-Thought (COT) mechanism [
30] into the prompting pipeline. This incremental reasoning approach compels the model to articulate its thought steps in a sequential manner, thereby maintaining clarity, rigor, and scientific accuracy even when confronting multifaceted geriatric cases. Moreover, by decomposing the instruction generation into smaller reasoning steps, COT enhances the model’s consistency and reliability—an attribute that is especially pertinent for tasks involving complex diagnoses, treatment plans, or extended decision-making scenarios.
3.2. Instruction Data Diversification Processing
A crucial factor influencing model performance is the extent to which synthetic data approximates real-world distributions. Synthetic datasets often exhibit a mismatch with real-world scenarios, thereby inflating training accuracy but yielding suboptimal performance upon deployment [
31]. In contrast, datasets curated via manual annotation typically produce smaller discrepancies between training and testing accuracy. To bridge this gap, we leverage the previously generated seed instruction data as a foundation and further diversify it to ensure more faithful alignment with human-like queries.
In real-world clinical settings, patients typically pose intricate, context-specific, and multifaceted questions rather than uniform or overly simplistic inquiries. Generating such richly varied instructions by hand is labor-intensive and time-consuming, particularly when aiming for highly nuanced content. To address this challenge, we developed a system that employs the generative capabilities of large language models (LLMs) to automatically produce diversified instruction data with varying levels of complexity. Specifically, we designed four targeted “evolutionary strategies” to enhance the depth, specificity, and realism of the instruction dataset:
Depth Evolution Strategy: Increases the complexity of questions by introducing detailed clinical scenarios or requiring multi-step reasoning in the responses.
Reasoning Evolution Strategy: Emphasizes logical progression and causal relationships, enabling the generation of questions that demand comprehensive inferential reasoning.
Specificity Evolution Strategy: Focuses on highly specific questions tailored to unique patient conditions, thereby departing from general or template-based inquiries.
Sample Evolution Strategy: Diversifies the dataset by varying patient demographics, symptom descriptions, or situational contexts, ultimately mirroring a broad spectrum of real-world medical encounters.
Through iterative application of these strategies, we significantly expand the complexity and variability of our instruction dataset, achieving greater fidelity to real clinical interactions. Additionally, we incorporate dynamically switching prompt templates that randomly alternate among the aforementioned strategies. By integrating diverse question-diversification techniques within each prompt, our method introduces stochasticity and adaptability into the data-generation pipeline, ultimately capturing a more representative set of patient inquiries.
Figure 2 illustrates one such prompt template used to generate these diversified instructions, highlighting its capacity for strategic variation and responsiveness to different medical scenarios.
3.3. Instruction Data Quality Filtering
Ensuring the fidelity and consistency of instructional data is paramount for high-quality Retrieval-Augmented Generation (RAG). To this end, we establish two automated evaluation metrics—question recall rate and answer attributability—designed to identify and filter out subpar data instances. By focusing on these two pivotal aspects, we enhance both the reliability and downstream utility of our dataset.
3.3.1. Question Recall Rate
To ensure that the questions generated in the question-answer pairs are relevant to the document content, we follow these steps for quality control of the questions:
First, we convert all document content into vector representations by using an embedding model. Suppose there are N documents, where each document is vectorized as . Similarly, a question is vectorized as . The process for calculating the question recall rate for each document is as follows:
For the set of questions
Q generated from
, we use the vector representation of the question
to retrieve the most relevant documents from the vector library. The cosine similarity between the question
and each document vector
is computed using the following equation:
The top 10 documents with the highest similarity are selected as the retrieval results, denoted as
. Next, we check the position of the correct document
within the retrieved results:
If the correct document appears in the top 3 positions, i.e., , then the question is retained. Otherwise, the question is filtered out.
3.3.2. Answer Attributability
Using an NLI (Natural Language Inference) model for quality control is an effective method, as NLI models validate the logical relationship between an answer and its corresponding document content, thereby assessing the attributability of the answer [
33]. The scoring for answer attributability is defined as follows:
where:
: Total number of sentences in the answer.
: Subset of sentences in the answer identified as entailing the document content.
: Subset of sentences in the answer identified as contradicting the document content.
: Subset of sentences in the answer identified as neutral to the document content.
To achieve high-precision entailment detection, we utilize the top-performing attributability prediction models. If both models predict a sentence as “entailment,” the sentence is considered to be fact-supported and is included in . We set an attributability threshold to filter out synthetic data and retain only high-quality RAG data. This approach ensures that the generated answers are logically consistent with the document content, improving the reliability of the dataset.
3.4. Training RAG Models
To effectively train a high-performing RAG model, traditional reading comprehension datasets, while improving a model’s ability to answer questions based on single documents, are insufficient for the demands of RAG tasks. RAG typically requires retrieving the top 50 or more documents, making it essential to construct multi-document reading comprehension datasets, or RAG-specific datasets. These datasets train the model to first identify the correct document, then perform reading comprehension on it, and finally generate an accurate answer.
Several key considerations must be addressed when constructing RAG datasets. First, since retrieved documents are ranked based on their similarity to the query, correct-answer documents are often placed in the top positions. This creates a misleading assumption that answers are always found in the leading documents, limiting the model’s performance. LLMs naturally prioritize highly ranked documents and may overlook answers located in middle or lower-ranked documents. However, in real-world scenarios, answers can appear randomly within the retrieved documents. Therefore, constructing multi-document datasets must deliberately randomize the placement of correct-answer documents to better reflect practical situations.Second, regarding the selection of distractor documents, a common approach is to use documents entirely unrelated to the correct-answer document to minimize interference and improve the model’s ability to identify the correct document. However, this method is not aligned with the requirements of RAG tasks, where retrieved documents are typically highly relevant to the query. Instead, distractor documents should be selected to be closely related to the correct-answer document but without containing the correct answer. This strategy enhances the model’s ability to distinguish the correct document within highly similar contexts, while also exposing it to domain-relevant distractors, thereby deepening the model’s understanding of the knowledge field.
In summary, constructing multi-document reading comprehension datasets for RAG training requires adherence to the following principles:
Randomized placement of correct-answer documents to prevent the model from developing position-based biases;
Selection of relevant distractor documents to simulate realistic retrieval scenarios, improve discrimination in challenging contexts, and enhance the model’s domain knowledge.
3.4.1. Set Relevant Distractor Documents
The selection of distractor documents is based on a retrieval database of geriatric medical documents. Using an embedding model, documents are randomly selected from a preprocessed and cleaned corpus, ensuring semantic similarity scores between 0.5 and 0.9 relative to the correct-answer document. These distractor documents must exhibit significant content differences from the correct-answer document to increase the difficulty of the retrieval task, thereby effectively enhancing the model’s retrieval accuracy. Additionally, distractor documents should avoid duplication and maintain high quality to prevent introducing noise that could disrupt model training. The primary goal of this step is to incorporate negative samples into the dataset, creating a contrastive learning scenario. This enables the model to accurately identify and retrieve relevant information from semantically similar but content-wise unrelated documents, improving its robustness and precision in complex retrieval tasks.
The semantic similarity between texts, denoted as , is calculated for document pairs and . Using an embedding model, the vector representations of documents, and , are computed. For the correct-answer document , candidate distractor documents are selected from the document corpus based on the following conditions: . Here, represents documents within the corpus that serve as potential distractor documents. For the filtered distractor document set , content redundancy is further eliminated by removing document pairs where the similarity exceeds: . Finally, the correct-answer document is combined with the selected distractor documents to construct the RAG dataset, enabling effective training for retrieval-augmented generation tasks.
3.4.2. Randomly Place Correct Documents
To enhance the model’s ability to perceive the position of correct documents in RAG data, we designed a targeted distribution strategy for correct document placement. Given that correct documents are typically retrieved in the first position in most cases, we allocated 50% of the correct documents to the top position. To address potential decreases in retrieval accuracy, the remaining 50% of the correct documents were distributed as follows: 40% were randomly placed within the top 10 retrieved documents, while the remaining 10% were randomly distributed across all retrieved documents. This strategy aims to balance the model’s adaptability to both high-accuracy and low-accuracy retrieval scenarios, enhancing its robustness and overall performance in practical applications.
3.5. Evaluation Metrics
3.5.1. Domain Metric
In the medical domain, to evaluate the effectiveness of various models in responding to user queries, we propose a comprehensive evaluation metric that incorporates both Answer Correctness and Semantic Similarity. This metric is designed to provide a holistic assessment of the quality of model-generated responses, ensuring that they meet the necessary standards of accuracy and contextual relevance.
The metric integrates
Answer Correctness, which evaluates the classification accuracy of the model, and
Semantic Similarity, which assesses the degree of alignment in linguistic expression and content coverage between the generated answers and standard reference answers. Specifically, Answer Correctness is evaluated using GPT-4, which provides a robust assessment of factual accuracy by leveraging its advanced reasoning and comprehension capabilities. Semantic Similarity, on the other hand, is evaluated using the pre-trained embedding model bge-large-zh-v1.5 [
34], which calculates the degree of alignment between generated and reference answers by analyzing linguistic expression and content representation.
The calculation of the Answer Correctness is defined as follows:
where:
: Number of true positives.
: Number of false positives.
: Number of false negatives.
The overall metric is calculated as a weighted sum of the Answer Correctness and semantic similarity:
where:
: Weight assigned to the Answer Correctness, with a default value of 0.75.
: Weight assigned to Semantic Similarity, with a default value of 0.25.
This comprehensive metric ensures that the evaluation framework is both robust and practical. It measures the ability of the model to deliver accurate responses while maintaining a high degree of semantic alignment, thereby reflecting the model’s overall effectiveness in real-world applications. This comprehensive framework is particularly suited for the medical domain, where the reliability and contextual appropriateness of responses are critical. By employing this metric, we provide a systematic and reliable tool for assessing and improving the quality of model-generated answers in medical settings.
3.5.2. General Metric
To evaluate the general capabilities of the model in the general domain, we utilized the Chinese tasks from the Longbench [
32] benchmarking suite. Longbench offers a diverse collection of tasks and datasets, making it an ideal framework for assessing the multifaceted competencies of language models across various dimensions.
We primarily utilized the following tasks from Longbench for our evaluation:
LSHT: A Chinese classification task that involves categorizing news articles into 24 distinct categories;
DuReader: A task requiring the answering of relevant Chinese questions based on multiple retrieved documents;
MultiFieldQA_ZH: A question-answering task based on a single document, where the documents span diverse domains;
VCSum: A summarization task that entails generating concise summaries of Chinese meeting transcripts;
Passage_Retrieval_ZH: A retrieval task where, given several Chinese passages from the C4 dataset, the model must identify which passage corresponds to a given summary.
4. Results
4.1. Data Diversity
To validate the effectiveness of our approach in generating diversified instructional data, we introduced two metrics for diversity assessment:
Verb Usage Frequency: A higher number of verbs exceeding a predefined frequency threshold indicates greater diversity;
ROUGE-L: A lower average ROUGE-L score within the same dataset signifies higher diversity.
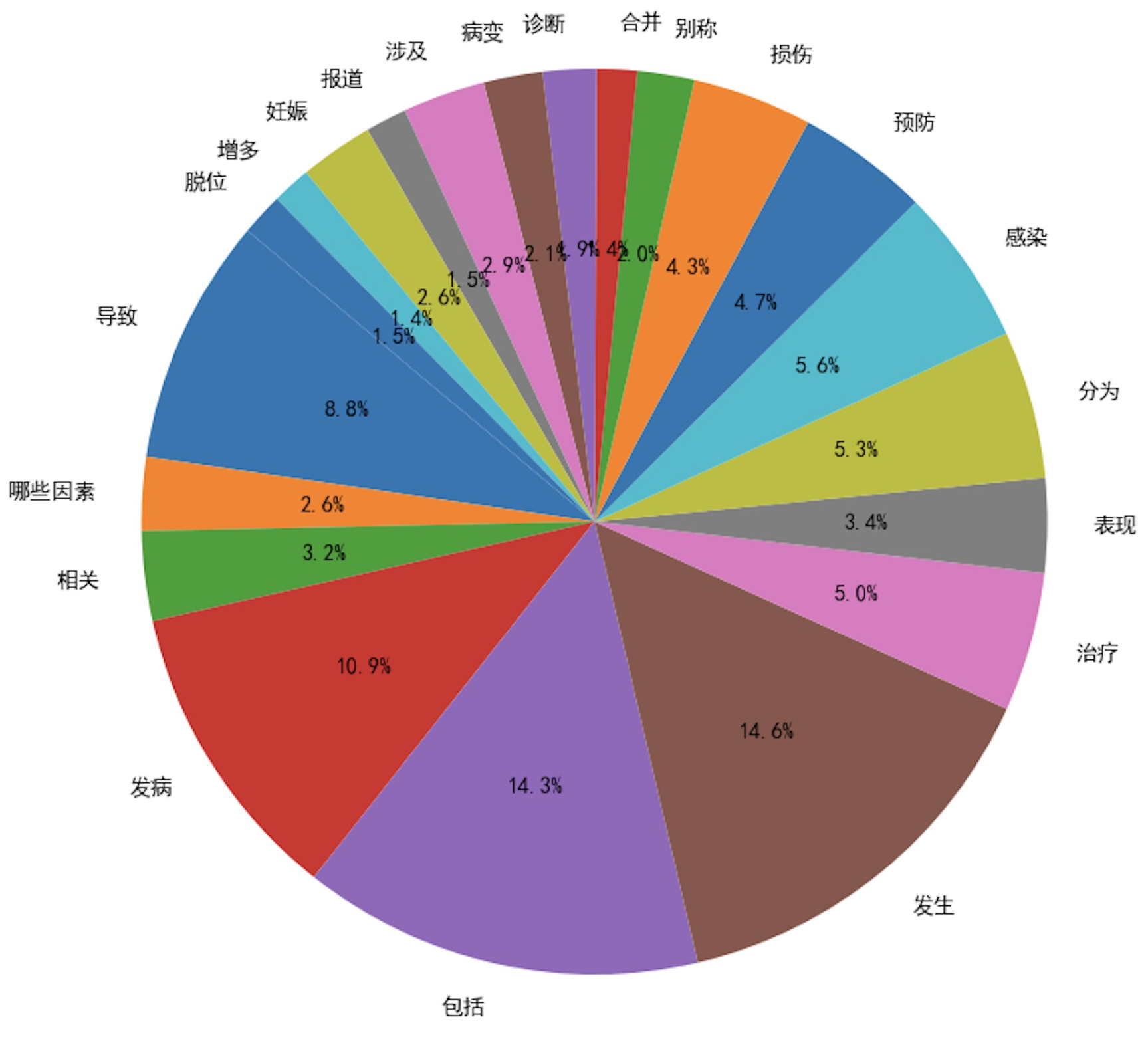
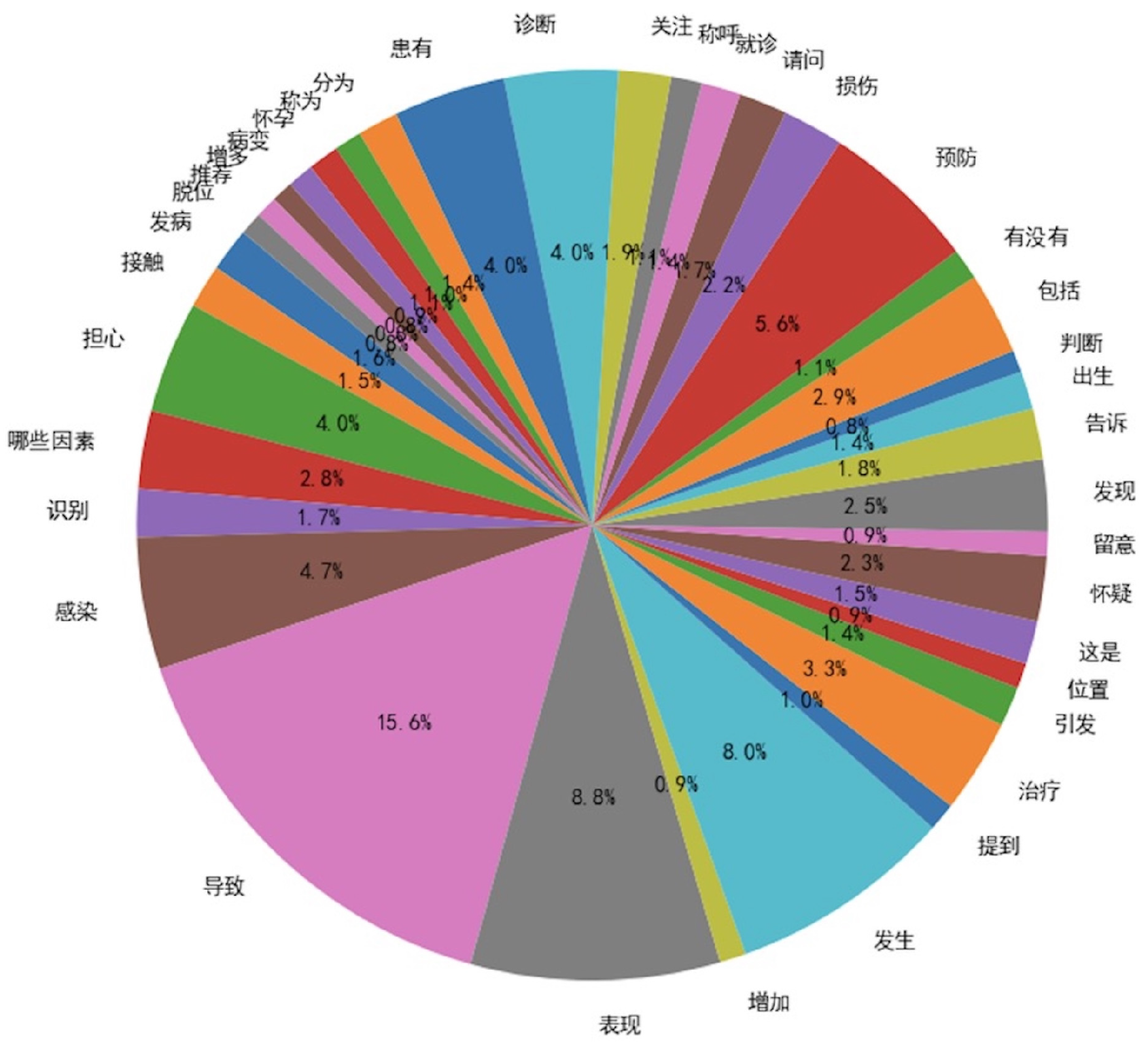
In the evaluation process, we analyzed and compared the verb usage frequency in the seed instruction data and the diversified instruction data, using a frequency threshold of 50. As illustrated in
Figure 3 and
Figure 4, the diversified instruction data incorporates a significantly greater variety of verbs compared to the seed instruction data. Furthermore, we examined the ROUGE-L score distributions of the two datasets. As depicted in
Figure 5, the average ROUGE-L score for the diversified instruction data is notably lower than that of the seed instruction data, reinforcing the conclusion that our method successfully enhances diversity.
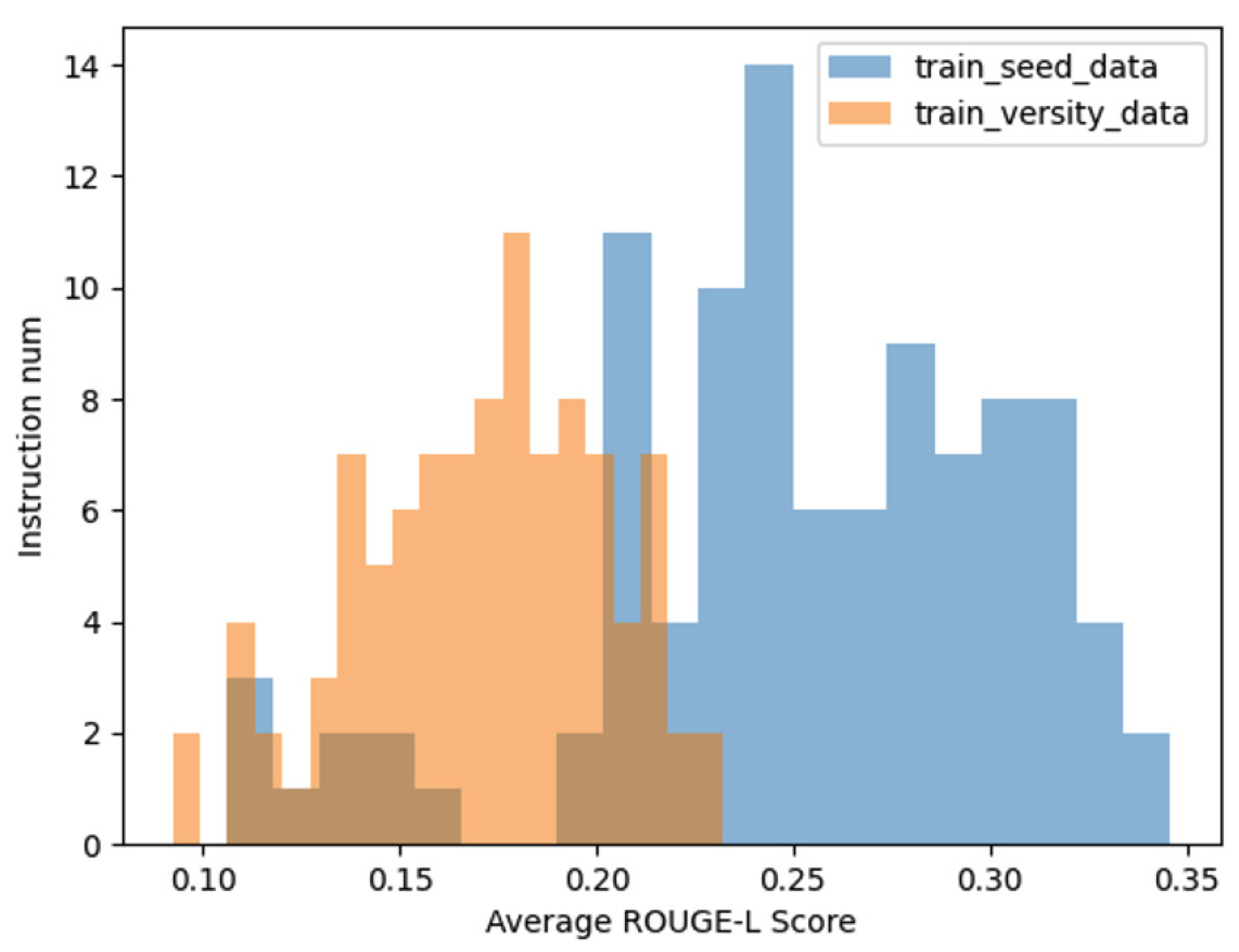
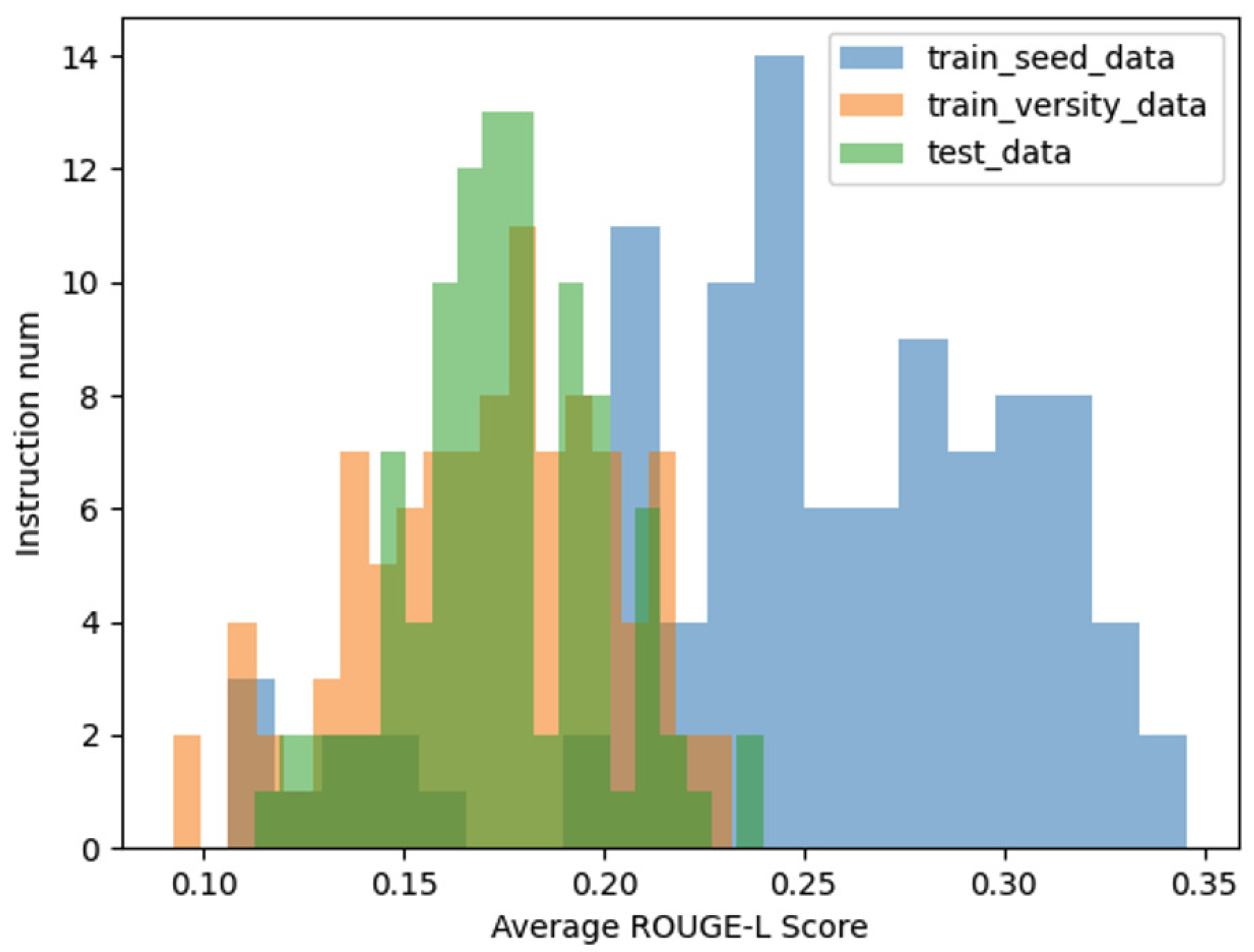
In addition to the aforementioned analyses, we further compared the average ROUGE-L score distributions among three datasets: the seed instruction data, the diversified instruction data, and a set of 100 manually curated instruction datasets, which serve as a benchmark for real-world data. As illustrated in
Figure 7, the distribution of the ROUGE-L scores for the diversified instruction data exhibits a closer alignment with the distribution observed in the manually curated data.
This comparison underscores the effectiveness of our method in approximating the characteristics of real-world instructional data. By achieving a ROUGE-L score distribution that closely mirrors that of human-generated data, the diversified instruction data demonstrates not only increased variety but also enhanced representational fidelity to real-world scenarios. This alignment further validates the practical utility of our approach in generating high-quality, diverse instructional datasets.
4.2. Model Training
Due to computational resource constraints, we selected three models as the base models for our experiments: Qwen2.5-7B-Instruct, DeepSeek-V2-Lite-Chat, and GLM-4-9B-Chat. Qwen2.5-7B-Instruct represents the latest iteration of Alibaba’s large language model, incorporating 7 billion parameters and improved instruction-following capabilities. DeepSeek-V2-Lite-Chat is a lightweight, high-performance model optimized for conversational tasks, while GLM-4-9B-Chat is a 9-billion-parameter model designed for advanced generative language modeling.
Leveraging our constructed diversified instruction data, we applied supervised fine-tuning (SFT) and RAG instruction fine-tuning to these base models. This multi-model approach enables us to evaluate the effectiveness of our methods across varying model architectures and parameter scales, ensuring robust and comprehensive performance analysis.
The experimental results presented in
Table 1 underscore significant variations in model performance across different strategies, particularly in terms of correctness and similarity within the test set in the medical domain. The baseline model, while establishing a foundational benchmark, exhibited limitations in both correctness and similarity. However, when integrated with the RAG strategy, a marked improvement was observed, reflecting the ability of RAG to enhance both the accuracy and semantic consistency of the model’s responses. This suggests that incorporating external retrieval mechanisms, as implemented in the RAG strategy, can substantially enrich the model’s understanding and alignment with domain-specific information. Further analysis reveals that models utilizing the SFT strategy demonstrated notable advancements in correctness, surpassing the baseline model. Nevertheless, this gain in correctness was accompanied by a marginal decline in similarity, indicating a potential trade-off between precision in reasoning and semantic alignment. Importantly, when the SFT strategy was combined with RAG, the model achieved significant gains across both metrics, demonstrating the complementary nature of these approaches. This combination effectively balances domain-specific fine-tuning with enhanced contextual retrieval, leading to more robust performance. The final optimized model, which integrates our advanced SFT strategy, achieved the highest scores in both correctness and similarity between all configurations. This result highlights the effectiveness and superiority of our approach in addressing the complex challenges of medical test sets. By leveraging the strengths of SFT and RAG in a unified framework, the model demonstrates its capability to achieve exceptional performance in tasks requiring high accuracy and semantic alignment. These findings not only emphasize the robustness of our methodology but also underscore its potential for broader applications in domains where precision and reliability are paramount.
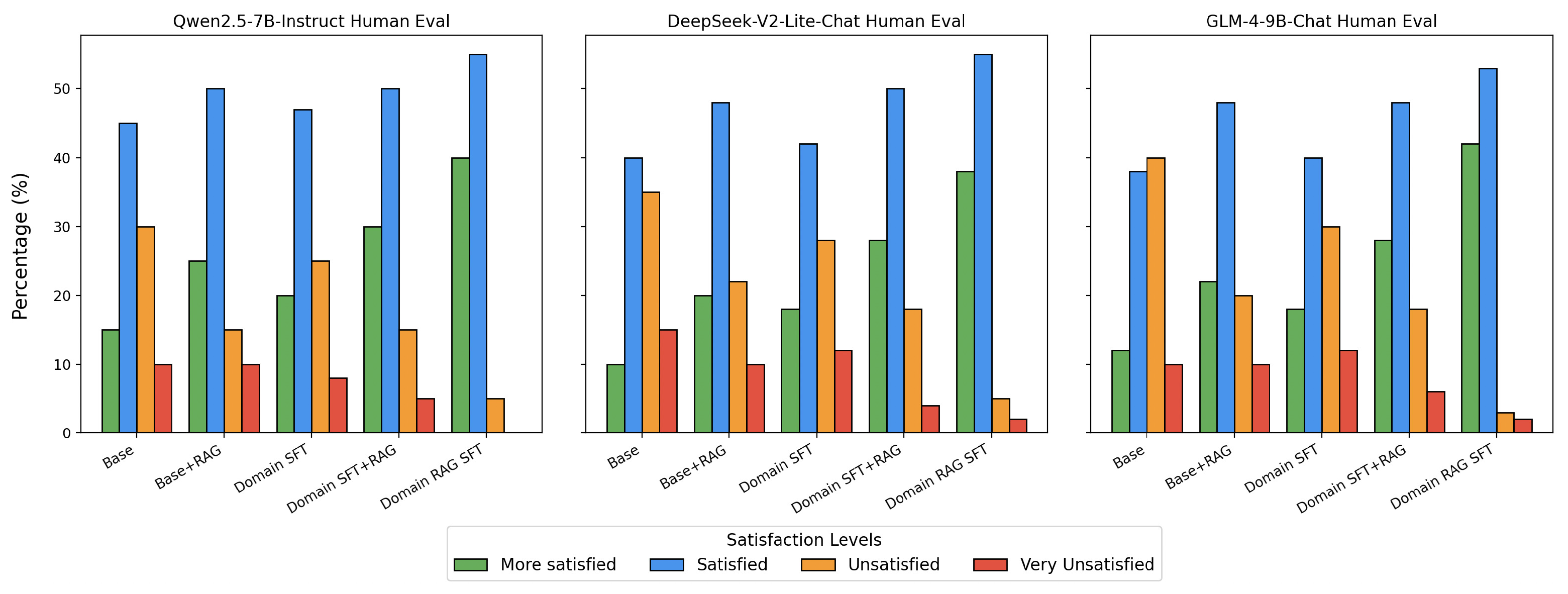
To further validate the effectiveness of our approach beyond automated metrics, we conducted human evaluations with medical domain experts.
Figure 7 illustrates the human evaluation results conducted by five medical students who were tasked with assessing model-generated answers on a predefined medical test set. Each student categorized the responses into four levels of satisfaction: More Satisfied, Satisfied, Unsatisfied, and Very Unsatisfied. The baseline model exhibited moderate levels of satisfaction, indicating foundational performance but leaving room for improvement. The integration of the RAG strategy significantly improved satisfaction scores, as evidenced by a noticeable reduction in dissatisfaction rates. Models incorporating the SFT strategy further demonstrated enhanced correctness and contextual understanding, with higher proportions of responses in the “More Satisfied” and “Satisfied” categories. The combination of SFT and RAG strategies yielded the most notable results, with the final model achieving the highest satisfaction rates among all configurations. These evaluations, conducted by individuals with medical domain expertise, provide robust evidence for the effectiveness of our approach in optimizing model performance and user satisfaction in practical, domain-specific scenarios.
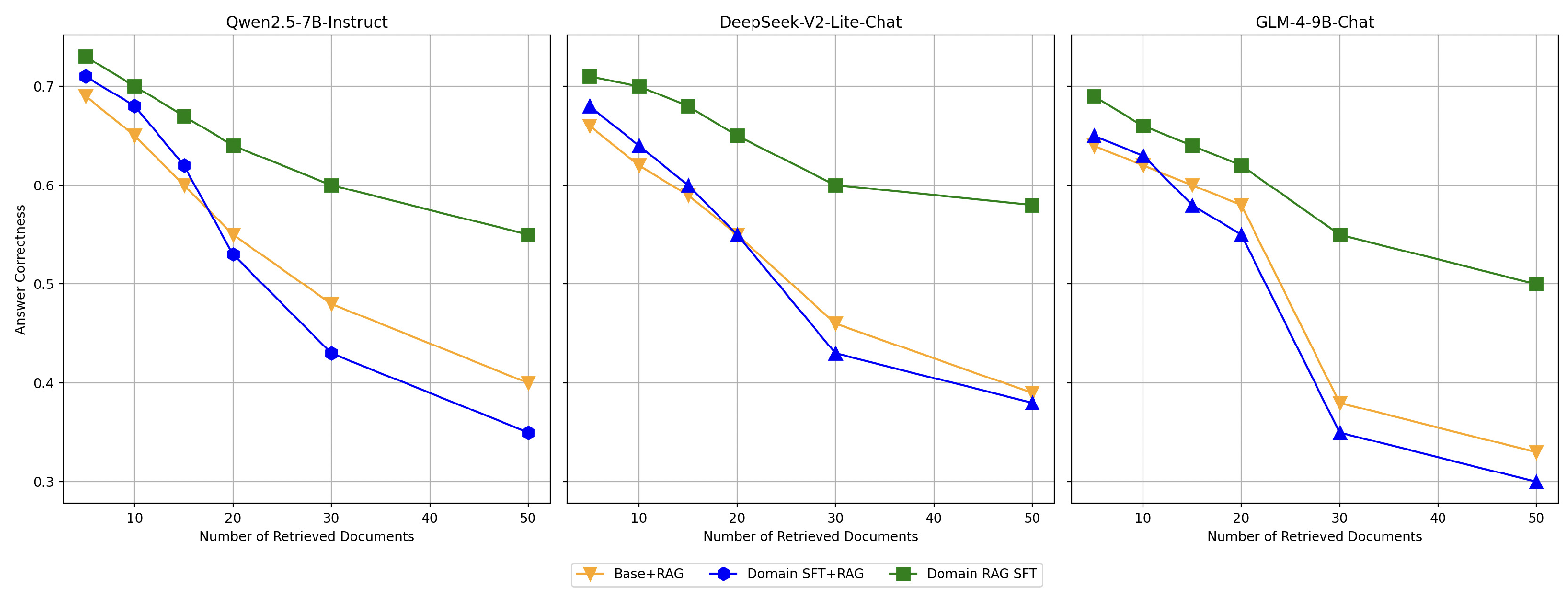
To evaluate the impact of increasing the number of retrieved documents on model performance, we conducted the following experiment: using the same test set, we varied the number of retrieved documents. The results demonstrate that as the number of retrieved documents increases, the interference from irrelevant or low-relevance documents becomes more pronounced, leading to a gradual decline in model performance. This trend highlights the challenges of maintaining correctness in scenarios with abundant retrieved information. The experimental results are shown in
Figure 8.
Among the configurations, the Domain Fine-Tuned model (Domain SFT) exhibits a significant decline in performance as the number of retrieved documents increases. Notably, when a large number of documents are retrieved, its performance even falls below that of the Base+RAG model. This indicates that while domain-specific fine-tuning enhances correctness in scenarios with fewer retrieved documents, it struggles to effectively manage the noise introduced by larger retrieval sets. In contrast, the Domain RAG Fine-Tuned model (Domain RAG SFT) demonstrates remarkable robustness across varying retrieval quantities. By applying domain-specific optimization to the retrieval-augmented generation (RAG) framework, this configuration improves the model’s ability to identify and focus on relevant documents, mitigating the negative impact of irrelevant retrievals. As a result, its performance remains relatively stable, showing only moderate decline even when the number of retrieved documents increases significantly.
These findings validate the effectiveness of the Domain RAG Fine-Tuning strategy. By enhancing the model’s sensitivity to relevant documents, this approach addresses the limitations of traditional fine-tuning methods. It ensures superior performance in retrieval-augmented generation tasks, even under challenging scenarios with a large number of retrieved documents.
To verify whether fine-tuning tailored for RAG would compromise the model’s original general capabilities, we conducted experiments on five Chinese tasks from Longbench. The experiments involved testing the Base model, the Domain SFT model, and the Domain RAG SFT model. The results, as shown in
Table 2, the Domain SFT model, which relies solely on traditional domain-specific fine-tuning, appears to harm the model’s original general capabilities due to its narrow focus. For example, its performance dropped on tasks like multifieldqa_zh and passage_retrieval_zh, suggesting that the singular nature of domain-specific fine-tuning may impair the model’s ability to generalize effectively. In contrast, the Domain RAG SFT model, designed to enhance the model’s sensitivity to relevant documents, not only avoided reducing the model’s general capabilities but also achieved modest improvements in reading comprehension tasks. This demonstrates that the Domain RAG SFT strategy effectively balances the trade-off between domain-specific enhancements and general-purpose performance.
In summary, the results highlight that while traditional domain fine-tuning may risk diminishing a model’s general capabilities, the Domain RAG SFT strategy successfully preserves these capabilities. Moreover, it introduces measurable improvements in tasks such as reading comprehension, proving its effectiveness in augmenting both specialized and general abilities.
5. Discussion
Our experimental findings demonstrate that integrating Retrieval-Augmented Generation (RAG) with a domain-specific fine-tuning paradigm substantially improves performance in geriatric medical question-answering (QA). Compared to general-purpose LLMs and baseline “LLM + RAG” configurations, our proposed approach achieves marked gains in accuracy and domain relevance, as measured by metrics such as Answer Correctness and Answer Similarity. Notably, these enhancements are most pronounced in complex geriatric scenarios, wherein nuanced clinical knowledge is crucial for producing accurate and contextually relevant responses.
A key driver behind these improvements is the domain-specific data pipeline, composed of seed instruction generation, data diversification, and rigorous quality filtering. Our results show that this pipeline effectively increases the quantity and variety of high-quality medical instruction data, reducing both factual drift (hallucinations) and superficial question-answer pairings. By incorporating question recall rate and answer attributability filters, we retained only those QA instances that demonstrate strong alignment between questions and their underlying knowledge sources. In addition, applying Chain-of-Thought (COT) prompting during seed data generation contributed to the logical coherence of multi-step reasoning tasks, a capability that is often critical in geriatrics due to the complexity of elderly patient care.
Another notable outcome is that while our approach is specifically tailored for geriatric healthcare, the resulting model also exhibits strong generalization capacity across broader Chinese QA tasks. This finding indicates that careful domain adaptation—through full-parameter fine-tuning—does not necessarily compromise performance on more general tasks. Instead, retrieval mechanisms help the model selectively incorporate specialized knowledge when required, without diminishing its broader linguistic and reasoning capabilities.
5.1. Practical and Theoretical Implications
From a practical standpoint, this research underscores the feasibility of constructing specialized QA systems that balance domain depth with general-purpose utility. In real-world clinical environments, such hybrid capability can prove invaluable, allowing medical practitioners to consult a single system for both routine informational queries and more complex, domain-specific questions.
On the theoretical side, our study enriches the body of literature on domain adaptation for large language models by illustrating how RAG architectures can be optimized for specialized fields. The results indicate that systematic data curation—involving both generative data augmentation and stringent quality filtering—serves as a cornerstone for successful domain adaptation. Moreover, the ability to integrate fine-grained external knowledge bases points to new directions for exploring multi-modal or multi-domain retrieval, particularly relevant for medical research and practice where diverse data types (e.g., lab results, imaging, guidelines) co-exist.
5.2. Limitations
Despite the improvements demonstrated, several limitations warrant discussion. First, our dataset and evaluation settings, while extensively curated, still rely on publicly available medical information. Proprietary clinical data—involving patient records or more specialized cases—might yield different performance outcomes and additional domain complexities. Second, the RAG approach, though effective in mitigating hallucinations, remains dependent on the quality and recency of external knowledge sources; outdated or inconsistently updated references could impair the system’s reliability. Third, while our method shows robust general-domain performance, it would be worth investigating scenarios where deeper specialization might be necessary (e.g., rare diseases within geriatric medicine) and whether that specialization would compromise broader QA versatility.
5.3. Future Work
Looking ahead, future research could explore dynamic knowledge updating to ensure that the external knowledge base remains aligned with evolving medical guidelines. Integrating multimodal data (such as imaging results or sensor-based patient data) could further enhance the system’s clinical applicability. Additionally, extending this methodology to multi-lingual contexts or other specialized medical subfields (e.g., oncology, pediatrics) would validate the transferability of our approach. Finally, real-world clinical deployment studies, involving user feedback from healthcare providers and patients, are essential for assessing the model’s practical impacts, ethical considerations, and overall acceptance in healthcare workflows.
6. Conclusions
In this study, we investigated the application of advanced Retrieval-Augmented Generation (RAG) methodologies in the domain of geriatric medicine to address the critical need for accurate and reliable question-answering (QA) in clinical contexts. By leveraging publicly accessible medical resources, we constructed an automated pipeline for generating high-quality, geriatric-specific RAG datasets, culminating in the creation of a specialized Chinese medical knowledge QA corpus. This innovation substantially reduces manual efforts associated with data curation while improving the fidelity and breadth of domain coverage.
A key contribution of our work lies in the systematic integration of RAG mechanisms with full-parameter fine-tuning of large language models (LLMs). This approach not only leverages external knowledge sources to mitigate hallucinations and enhance factual accuracy but also refines the model’s internal representations to better capture the nuances of geriatric healthcare. Our empirical results underscore the effectiveness of this dual strategy: two tailored evaluation metrics—answer similarity and correctness—demonstrated substantial improvements over baseline models, underscoring the robustness and precision of the proposed framework. Moreover, we validated the generalization capability of our RAG-based system on diverse Chinese QA tasks beyond the medical domain, attesting to its adaptability and broad applicability.
Notably, this research provides new insights into optimizing data construction and quality assurance for specialized QA systems. By introducing advanced filtering methods—such as question recall rate and answer attributability—we systematically enhance data reliability and mitigate the propagation of errors or noisy samples. These methodological advances form a viable blueprint for extending RAG-driven QA to other high-stakes fields where domain specificity and factual consistency are paramount.
Author Contributions
Conceptualization, B.W.; methodology, B.W. and S.L.; software, B.W.; validation, B.W., Z.H. and C.L.; formal analysis, B.W.; investigation, B.W.; resources, S.L.; data curation, B.W. and C.L.; writing—original draft preparation, B.W. , S.L. and Z.H.; writing—review and editing, B.W. , S.L. , Z.H. and C.L.; visualization, B.W.; supervision, S.L.; project administration, B.W. , S.L. and Z.H.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research on Key Technologies and System Development of Global Collaborative Cognitive Computing for Livable Cities grant number 2020YFB2104402.
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| QA |
Question-answering |
| LLM |
Large Language Model |
| RAG |
Retrieval-augmented Generation |
| SFT |
Supervised Fine-Tuning |
| CoT |
Chain-of-Thought |
References
- Brown, T.; Mann, B.; Ryder, N.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Lazaridou, A.; Gribovskaya, E.; Stokowiec, W.; et al. Internet-augmented language models through few-shot prompting for open-domain question answering. arXiv Preprint 2022, arXiv:2203.05115. [Google Scholar]
- Ni, J.; Bingler, J.; Colesanti-Senni, C.; et al. Chatreport: Democratizing sustainability disclosure analysis through LLM-based tools. arXiv Preprint 2023, arXiv:2307.15770. [Google Scholar]
- Ji, Z.; Lee, N.; Frieske, R.; et al. Survey of hallucination in natural language generation. ACM Comput. Surv. 2023, 55, 1–38. [Google Scholar] [CrossRef]
- Hu, X.; Chen, J.; Li, X.; et al. Do large language models know about facts? arXiv Preprint 2023, arXiv:2310.05177. [Google Scholar]
- Gao, Y.; Xiong, Y.; Gao, X.; et al. Retrieval-augmented generation for large language models: A survey. arXiv Preprint 2023, arXiv:2312.10997. [Google Scholar]
- Wang, H.; Liu, C.; Xi, N.; et al. Huatuo: Tuning llama model with Chinese medical knowledge. arXiv Preprint 2023, arXiv:2304.06975. [Google Scholar]
- Chen, Y.; Wang, Z.; Xing, X.; et al. Bianque: Balancing the questioning and suggestion ability of health LLMs with multi-turn health conversations polished by ChatGPT. arXiv Preprint 2023, arXiv:2310.15896. [Google Scholar]
- Li, L.; Wang, P.; Yan, J.; et al. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef] [PubMed]
- Lewis, P.; Ott, M.; Du, J.; et al. Pretrained language models for biomedical and clinical tasks: Understanding and extending the state-of-the-art. In Proceedings of the 3rd Clinical Natural Language Processing Workshop, Online, 19 November 2020; pp. 146–157. [Google Scholar]
- Zhang, T.; Cai, Z.; Wang, C.; et al. SMedBERT: A knowledge-enhanced pre-trained language model with structured semantics for medical text mining. arXiv Preprint 2021, arXiv:2108.08983. [Google Scholar]
- Li, L.; Wang, P.; Yan, J.; et al. Real-world data medical knowledge graph: Construction and applications. Artif. Intell. Med. 2020, 103, 101817. [Google Scholar] [CrossRef]
- Lewis, P.; Perez, E.; Piktus, A.; et al. Retrieval-augmented generation for knowledge-intensive NLP tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Borgeaud, S.; Mensch, A.; Hoffmann, J.; et al. Improving language models by retrieving from trillions of tokens. In Proceedings of the International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2022; PMLR; pp. 2206–2240. [Google Scholar]
- Bora, A.; Cuayáhuitl, H. Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. Machine Learning and Knowledge Extraction 2024, 6, 2355–2374. [Google Scholar] [CrossRef]
- Frisoni, G.; Mizutani, M.; Moro, G.; et al. Bioreader: A retrieval-enhanced text-to-text transformer for biomedical literature. In Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, Online, 7–11 December 2022; pp. 5770–5793. [Google Scholar]
- Naik, A.; Parasa, S.; Feldman, S.; et al. Literature-augmented clinical outcome prediction. arXiv Preprint 2021, arXiv:2111.08374. [Google Scholar]
- Zakka, C.; Shad, R.; Chaurasia, A.; et al. Almanac—retrieval-augmented language models for clinical medicine. NEJM AI 2024, 1, AIoa2300068. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Umapathi, L.K.; Sankarasubbu, M. MedMCQA: A large-scale multi-subject multi-choice dataset for medical domain question answering. In Proceedings of the Conference on Health, Inference, and Learning, Virtual, 20–23 April 2022; PMLR; pp. 248–260. [Google Scholar]
- Jin, Q.; Dhingra, B.; Liu, Z.; et al. PubMedQA: A dataset for biomedical research question answering. arXiv Preprint 2019, arXiv:1909.06146. [Google Scholar]
- Fan, A.; Jernite, Y.; Perez, E.; et al. ELI5: Long form question answering. arXiv Preprint 2019, arXiv:1907.09190. [Google Scholar]
- Hinton, G. Distilling the knowledge in a neural network. arXiv Preprint 2015, arXiv:1503.02531. [Google Scholar]
- Beyer, L.; Zhai, X.; Royer, A.; et al. Knowledge distillation: A good teacher is patient and consistent. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10925–10934. [Google Scholar]
- Hsieh, C.Y.; Li, C.L.; Yeh, C.K.; et al. Distilling step-by-step! Outperforming larger language models with less training data and smaller model sizes. arXiv Preprint 2023, arXiv:2305.02301. [Google Scholar]
- DeVries, T.; Taylor, G.W. Dataset augmentation in feature space. arXiv Preprint 2017, arXiv:1702.05538. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Zhou, W.; Bras, R.L.; Choi, Y. Modular transformers: Compressing transformers into modularized layers for flexible efficient inference. arXiv Preprint 2023, arXiv:2306.02379. [Google Scholar]
- Chen, T.; Kornblith, S.; Swersky, K.; et al. Big self-supervised models are strong semi-supervised learners. Adv. Neural Inf. Process. Syst. 2020, 33, 22243–22255. [Google Scholar]
- Puri, R.; Spring, R.; Patwary, M.; et al. Training question answering models from synthetic data. arXiv Preprint 2020, arXiv:2002.09599. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; et al. Chain-of-thought prompting elicits reasoning in large language models. Advances in Neural Information Processing Systems 2022, 35, 24824–24837. [Google Scholar]
- Xu, C.; Sun, Q.; Zheng, K.; et al. WizardLM: Empowering large language models to follow complex instructions. arXiv Preprint 2023, arXiv:2304.12244. [Google Scholar]
- Bai, Y.; Lv, X.; Zhang, J.; et al. Longbench: A bilingual, multitask benchmark for long context understanding. arXiv Preprint 2023, arXiv:2308.14508. [Google Scholar]
- Chen, J.; Choi, E.; Durrett, G. Can NLI models verify QA systems’ predictions? arXiv preprint 2021, arXiv:2104.08731. [Google Scholar]
- Xiao, S.; Liu, Z.; Zhang, P.; et al. C-pack: Packed resources for general chinese embeddings. In Proceedings of the 47th International ACM SIGIR Conference on Research and Development in Information Retrieval; 2024; pp. 641–649. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).