Submitted:
07 December 2024
Posted:
09 December 2024
You are already at the latest version
Abstract
Keywords:
| Enzyme | Enzyme Name |
|---|---|
| AADAC | Arylacetamide deacetylase |
| ADH | Alcohol dehydrogenase |
| AKR | Aldo-keto reductase |
| AKR1A1 | Aldo-keto reductase 1A1 |
| AKR1B1 | Aldo-keto reductase 1B1 |
| AKR1B10 | Aldo-keto reductase 1B10 |
| AKR1C1 | Aldo-keto reductase 1C1 |
| AKR1C2 | Aldo-keto reductase 1C2 |
| AKR1C3 | Aldo-keto reductase 1C3 |
| AKR1C4 | Aldo-keto reductase 1C4 |
| AOX1 | Aldehyde oxidase 1 |
| CES1A | Carboxylesterase 1A |
| CES2 | Carboxylesterase 2 |
| COX | Cyclooxygenase |
| COX-1 | Cyclooxygenase 1 |
| COX-2 | Cyclooxygenase 2 |
| EH | Epoxide hydrolase |
| FMO1 | Flavin-containing monooxygenase 1 |
| FMO2 | Flavin-containing monooxygenase 2 |
| FMO3 | Flavin-containing monooxygenase 3 |
| Hb | Hemoglobin |
| LPO | Lactoperoxidase |
| MAO A | Monoamine oxidase A |
| MPO | Myeloperoxidase |
| NAT | N-acetyltransferase |
| NAT1 | N-acetyltransferase 1 |
| NAT2 | N-acetyltransferase 2 |
| NAR | Nitrate reductase |
| NQO | NAD(P)H quinone oxidoreductase |
| NQO1 | NAD(P)H quinone oxidoreductase 1 |
| NPR, POR | NAD(P)H-P450 reductase |
| NR | Nitrate reductase |
| P450 | Cytochrome P450 |
| P450 1A1 | Cytochrome P450 1A1 |
| P450 1A2 | Cytochrome P450 1A2 |
| P450 1B1 | Cytochrome P450 1B1 |
| P450 2A6 | Cytochrome P450 2A6 |
| P450 2B6 | Cytochrome P450 2B6 |
| P450 2C10 | Cytochrome P450 2C10 |
| P450 2C18 | Cytochrome P450 2C18 |
| P450 2C19 | Cytochrome P450 2C19 |
| P450 2C8 | Cytochrome P450 2C8 |
| P450 2C9 | Cytochrome P450 2C9 |
| P450 2C9.1 | Cytochrome P450 2C9.1 |
| P450 2C9.2 | Cytochrome P450 2C9.2 |
| P450 2C9.3 | Cytochrome P450 2C9.3 |
| P450 2D6 | Cytochrome P450 2D6 |
| P450 2E1 | Cytochrome P450 2E1 |
| P450 2F1 | Cytochrome P450 2F1 |
| P450 2J2 | Cytochrome P450 2J2 |
| P450 3A4 | Cytochrome P450 3A4 |
| P450 3A5 | Cytochrome P450 3A5 |
| P450 3A7 | Cytochrome P450 3A7 |
| P450 4A11 | Cytochrome P450 4A11 |
| P450 4B1 | Cytochrome P450 4B1 |
| P450 2W1 | Cytochrome P450 2W1 |
| PGHS | Prostaglandin H synthase |
| PO | Peroxidase |
| SULT | Sulfotransferases |
| SULT1A1 | Sulfotransferase 1A1 |
| SULT1A2 | Sulfotransferase 1A2 |
| SULT1A3 | Sulfotransferase 1A3 |
| SULT1B1 | Sulfotransferase 1B1 |
| SULT1C1 | Sulfotransferase 1C1 |
| SULT1C2 | Sulfotransferase 1C2 |
| SULT1C3 | Sulfotransferase 1C3 |
| SULT1E1 | Sulfotransferase 1E1 |
| SULT2A1 | Sulfotransferase 2A1 |
| SULT2E1 | Sulfotransferase 2E1 |
| XOR | Xanthine oxidoreductase |
| Compound or Metabolite | Compound Category/Source /Metabolite/Toxic Effects | Enzyme | Reactions and Reactive /Toxic Product(s) Formation | References |
|---|---|---|---|---|
| (-)-1-Hydroxyethylpyrene | Metabolite of ethylpyrene, research chemical | SULT1A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [14,15] |
| (-)-1-Hydroxyethylpyrene | As above | SULT1A2 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [15] |
| (-)-1-Hydroxyethylpyrene | As above | SULT1C1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [15] |
| (-)-1-Hydroxyethylpyrene | As above | SULT1C2 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [15] |
| (-)-R,R and (+)-S,S-Benzo[g]chrysene-11,12-diol | Metabolite of B[g]C, fossil fuels, and organic materials combustion product | AKR1B1 | Oxidation, o-quinone | [16] |
| (−)-R,R- and (+)-S,S-Benzo[g]chrysene-11,12-diol | As above | AKR1B10 | Oxidation, o-quinone | [16] |
| (+)-Benz[a]anthracene-3S,4S-diol | Metabolite of B[a]A, fossil fuels, and organic materials combustion products tobacco, smoke constituent | AKR1B1 | Oxidation, o-quinone | [16] |
| (+)-Benz[a]anthracene-3S,4S-diol | As above | AKR1B10 | Oxidation, o-quinone | [16] |
| (+)-Benzo[a]pyrene-7S,8S-dihydrodiol | Metabolite of B[a]P | AKR1B1 | Oxidation, o-quinone | [16] |
| (+)-Benzo[a]pyrene-7S,8S-dihydrodiol | As above | AKR1B10 | Oxidation, o-quinone | [16] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | AKR1C1 | Oxidation, o-quinone, and reactive oxygen species (ROS) | [16,17,18,19,20] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | AKR1C3 | Oxidation, o-quinone, and reactive oxygen species (ROS) | [16,17,19,20] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | AKR1C4 | Oxidation, o-quinone, and reactive oxygen species (ROS) | [16,17,19,20] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | AKR1C2 | Oxidation, o-quinone, and reactive oxygen species (ROS) | [16,17,19,20] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | MAO 2 | Oxidation, peroxyl radicals | [21] |
| (±)- and (-)-Benzo[a]pyrene-7,8-dihydrodiol ((±)- and (-)-B[a]P-7,8-diol) | As above | COX-1 | Oxidation, peroxyl radicals | [21] |
| (±)-, (-)-, and (+)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-, (-)-, and (+)-B[a]P-7,8-diol) | As above | P450 1A1 | trans-(anti)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro- formation, trans-diolepoxide, oxidation * | [1,3,22,23,24,25,27,28,29,30,31,32,34,35,38,39,127] |
| (±)-, (-)-, and (+)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-, (-)-, and (+)-B[a]P-7,8-diol) | As above | P450 1A2 | trans-(anti)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro- formation, trans-diol epoxide, oxidation | [1,22,23,24,38,40,41,42] |
| (±)-, (-)-, and (+)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-, (-)-, and (+)-B[a]P-7,8-diol) | As above | P450 1B1 | trans-(anti)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro- formation, trans-diol epoxide (low Km, high activity, high efficiency), oxidation * | [1,24,25,27,31,36,37,38,41,43,44,45] |
| (±)-, (+)- and (-)-1-Hydroxyethylpyrene | Metabolite of ethylpyrene, research chemicals | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [14,15,46] |
| (±)-, (+)- and (-)-1-Hydroxyethylpyrene | As above | SULT1C3 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [46] |
| (±)-, (+)- and (-)-1-Hydroxyethylpyrene | As above | SULT1E1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * | [14,15,46] |
| (±)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-B[a]P-7,8-diol) | Metabolite of B[a]P | AKR1A1 | Oxidation, o-quinone formation, preferential for (-)-7R,8R-oxidation * | [16,17,31,36,37,47,48] |
| (±)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-B[a]P-7,8-diol) | As above | AKR1C4 | Oxidation, o-quinone formation | [16] |
| (±)-Benzo[a]pyrene-7,8-dihydrodiol ((±)-B[a]P-7,8-diol) | As above | P450 2W1 | Oxidation, diolepoxide formation | [43] |
| 1,10-Diazachrysene [1,10-DAC) | Chrysene derivative | P450 1A2 | Oxidation, enamine epoxide formation | [11,12] |
| 1,2-Dihydro-1,2-dihydroxy-6-nitrochrysene (trans) | Metabolite of 6-nitrochrysene, nitroarene | P450 3A4 | Oxidation | [50] |
| 1,6-Dinitropyrene (1,6-DNP) | Environmental pollutants, diesel engine combustion by-products, nitroarene, pyrene derivative | P450 3A4 | Nitroreduction, aminopyrene, 4-hydroxylamine, formation | [13] |
| 1,6-Dinitropyrene (1,6-DNP) | As above | P450 1B1 (co-expressed with NPR) | 1-Aminopyrene formation, nitroreduction/O-acetylation, at low concentrations, electrophilic nitrenium ion formation | [9] |
| 1,6-Dinitropyrene (1,6-DNP) | As above | NPR | Reduction to 1-Nitro-6-nitrosopyrene, reactive oxygen species formation | [51] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | NPR | Reduction to 1-Nitro-8-nitrosopyrene, reactive oxygen species formation | [51] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | NPR | 1-Aminopyrene formation, nitroreduction/O-acetylation, at low concentrations, electrophilic nitrenium ion formation * | [9] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | P450 3A4 | Epoxidation C4,5-, oxidation, minor reaction | [10,13,52] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | NAT1 | O-Acetylation after nitroreduction, electrophilic nitrenium ion formation | [53] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | NAT2 | O-Acetylation after nitroreduction, electrophilic nitrenium ion formation * | [53] |
| 1,8-Dinitropyrene (1,8-DNP) | As above | P450 1A1 (co-expressed with NPR) | 1-Aminopyrene formation, nitroreduction/O-acetylation, at low concentrations, electrophilic nitrenium ion formation * | [9] |
| 10-Azabenzo[a]pyrene | Environmental pollutants, gasoline exhaust, and cooking emissions compounds, aza-aromatic | P450 1A2 | Oxidation at pyridine moiety ** | [54] |
| 10-Azabenzo[a]pyrene | As above | P450 1A1 | Oxidation, minor enzyme * | [54] |
| 10-Hydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene | Metabolite of B[a]P | SULT1E1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [55] |
| 12-Methylbenz[a]anthracene-3,4-diol | Metabolite of 12-methylbenz[a]anthracene | AKR1A1 | Oxidation, o-quinone formation | [48] |
| 1-Acetylpyrene | Industrial chemicals, carbonyl-pyrene | SULT2E1 (in the presence of NADPH-fortified human liver cytosol) | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * (after reduction) | [56] |
| 1-Aminopyrene | Metabolite of 1-nitropyrene, industrial chemicals, arylamine | P450 1A1 | Oxidation, N-hydroxylation, and nitrenium ion formation via O-acetylation, electrophilic nitrenium ion formation | [10] |
| 1-Aminopyrene | As above | P450 1A2 | Oxidation, N-hydroxylation, and nitrenium ion formation via O-acetylation, electrophilic nitrenium ion formation * | [10,57,58,59] |
| 1-Aminopyrene | As above | P450 1B1 | Oxidation, N-hydroxylation, and nitrenium ion formation via O-acetylation, electrophilic nitrenium ion formation * | [10] |
| 1-Aminopyrene | As above | P450 3A4 | Oxidation, N-hydroxylation | [10] |
| 1-Formylpyrene | Fluorescent dye, carbonyl-pyrene | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation (after reduction) | [56] |
| 1-Hydroxy-3-methylcholanthrene (1-OH-MC) | Metabolite of 3-MC | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation, electrophilic nitrenium ion formation | [56] |
| 1-Hydroxymethylpyrene (1-HMP) | Metabolite of 1-MP | SULT1A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * | [14,15,46,60,63] |
| 1-Hydroxymethylpyrene (1-HMP) | As above | SULT1A2 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [15,56,63] |
| 1-Hydroxymethylpyrene (1-HMP) | As above | SULT1A3 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [14,15,60] |
| 1-Hydroxymethylpyrene (1-HMP) | As above | SULT2E1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * | [56] |
| 1-Hydroxymethylpyrene (1-HMP) | As above | SULT1C2 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [56] |
| 1-Hydroxymethylpyrene (1-HMP) | As above | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation, electrophilic nitrenium ion formation * | [56] |
| 1-Methylpyrene (1-MP) | Wood, diesel oil, and gasoline fuels incomplete combustion products, pyrene derivatives. | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * (after hydroxylation) | [14,15,60] |
| 1-Nitro-6-nitrosopyrene | Metabolite of 1,6-dinitropyrene, nitroarene, pyrene derivative | POR | Nitroreduction, reactive oxygen species formation | [51] |
| 1-Nitro-8-nitrosopyrene | Metabolite of 1,8-dinitropyrene, nitroarene | POR | Nitroreduction, reactive oxygen species formation | [51] |
| 1-Nitropyrene (1-NP) | Environmental pollutants, diesel engine combustion by-products, nitroarene, pyrene derivative | P450 1B1 (co-expressed with NPR) | 1-Aminopyrene formation, nitroreduction, and O-acetylation, at low concentrations, electrophilic nitrenium ion formation, epoxidation at high concentrations* | [10] |
| 1-Nitropyrene (1-NP) | As above | P450 1A1 | Oxidation, ring oxidation * | [10,13,64] |
| 1-Nitropyrene (1-NP) | As above | P450 1B1 (co-expressed with NPR) | Oxidation, nitroreduction, epoxidation, ring oxidation *,** | [10,64] |
| 1-Nitropyrene (1-NP) | As above | P450 3A4 | Oxidation, epoxidation, ring oxidation * | [10] |
| 2,3-Dihydroxy-2,3-dihydrofluoranthene | Metabolite of fluoranthene | P450 1B1 | Oxidation | [3] |
| 2-Acetylaminofluorene (2-AAF) | Metabolite of aminofluorene, arylamine | P450 1A2 | N-Hydroxylation, oxidation * | [24,40,52,65,67,68,69] |
| 2-Acetylaminofluorene (2-AAF) | As above | NAT1 | O-Acetylation after N-hydroxylation, electrophilic nitrenium ion formation, electrophilic nitrenium ion formation * | [69] |
| 2-Acetylaminofluorene (2-AAF) | As above | P450 1A1 | N-Hydroxylation, oxidation | [1,24,67] |
| 2-Aminoanthracene (2AA) | Research chemicals, arylamine | P450 1A1 | N-Hydroxylation, oxidation * | [1,3,24,70] |
| 2-Aminoanthracene (2AA) | As above | P450 1A2 | N-Hydroxylation, oxidation (high activity) *, ** | [10,24,40,57,58,59,65,70,71,72] |
| 2-Aminoanthracene (2AA) | As above | P450 1B1 | N-Hydroxylation, oxidation (high activity) * | [1,24,43,70] |
| 2-Aminoanthracene (2AA) | As above | P450 2E1 | N-Hydroxylation, oxidation | [70] |
| 2-Aminoanthracene (2AA) | As above | P450 2W1 | Oxidation | [43] |
| 2-Aminodipyrido[1,2-a:3,2′-d]-imidazole (Glu-P-2) | Cooked meat and fish compounds, a component of tobacco smoke, heterocyclic amine | P450 1A2 | Oxidation | [40,52,65] |
| 2-Aminofluorene (2-AF) | Research chemicals, fluorene derivative, arylamine | P450 1A1 | N-Hydroxylation, oxidation | [1,24,25,70] |
| 2-Aminofluorene (2-AF) | As above | P450 1B1 | N-Hydroxylation, oxidation * | [1,24,25,43,70] |
| 2-Aminofluorene (2-AF) | As above | P450 2E1 | N-Hydroxylation, oxidation * | [70] |
| 2-Aminofluorene (2-AF) | As above | P450 2W1 | Oxidation, diolepoxide formation | [43] |
| 2-Aminofluorene (2-AF) | As above | P450 3A4 | Oxidation, ring oxidation | [70,73] |
| 2-Aminofluorene (2-AF) | As above | P450 3A7 | Oxidation, ring oxidation | [73] |
| 2-Aminofluorene (2-AF) | As above | P450 4B1 | N-Hydroxylation, oxidation | [74,75] |
| 2-Aminofluorene (2-AF) | As above | NAT1 | O-Acetylation after N-hydroxylation, electrophilic nitrenium ion formation | [76,77] |
| 2-Hydroxy-3-methylcholanthrene 2-OH-MC) | Metabolite of 3-MC | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [56] |
| 2-Hydroxymethylpyrene, 2-pyrenemethanol | Metabolite of methylpyrene, research chemical | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [55] |
| 2-Naphthylamine (β-NA) | Industrial chemicals, used in the production of azo dyes, tobacco smoke compounds, arylamine | P450 1A2 | N-Hydroxylation, oxidation | [40,52,65,69,70,82] |
| 2-Naphthylamine (β-NA) | As above | NAT1 | O-Acetylation after N-Hydroxylation, electrophilic nitrenium ion formation | [69] |
| 2-Nitroanisole | Environmental pollutants, industrial chemicals, nitroarene | XOR | Nitroreduction to hydroxylamine | [83] |
| 2-Nitrobenzanthrone (2-NBA) | Ambient air pollutants, nitroarene | NAT2 | O-Acetylation (after nitroreduction to hydroxylamine), electrophilic nitrenium ion formation | [84] |
| 2-Nitrobenzanthrone (2-NBA) | As above | SULT1A1 | O-Sulfation, sulfo-conjugate (after nitroreduction to hydroxylamine), electrophilic nitrenium ion formation | [84] |
| 2-Nitrofluoranthene (2-NF) | As above | P450 1B1 (co-expressed with NPR) | 1-Aminopyrene formation, nitroreduction/O-acetylation, at low concentrations | [9] |
| 2-Nitrofluorene (2-NF) | As above | NAT1 | O-Acetylation after nitroreduction, electrophilic nitrenium ion formation | [77] |
| 2-Nitronaphthalene | Industrial chemicals, nitroarene | P450 1A1 | Oxidation | [78] |
| 2-Nitropyrene (2-NP) | Environmental pollutants, diesel engine combustion by-products, nitroarene | P450 1A1 | Oxidation, ring oxidations | [24,64] |
| 2-Nitropyrene (2-NP) | As above | P450 1B1 | Oxidation, ring oxidations * | [1,24,64] |
| 3,6-Dinitrobenzo[e]pyrene (DNBeP) | Environmental pollutants, surface soil, and airborne particle contaminants, nitroarene | P450 1A1, NPR, OAT2 | Nitroreduction and O-acetylation by NAT enzymes, electrophilic nitrenium ion formation | [79] |
| 3,6-Dinitrobenzo[e]pyrene (DNBeP) | As above | P450 1A2, NPR, OAT2 | Nitroreduction and O-acetylation by NAT enzymes, electrophilic nitrenium ion formation | [79] |
| 3,6-Dinitrobenzo[e]pyrene (DNBeP) | As above | P450 3A4, NPR, OAT2 | Nitroreduction and O-acetylation by NAT enzymes, electrophilic nitrenium ion formation | [79] |
| 3,6-Dinitrobenzo[e]pyrene (DNBeP) | As above | POR | Nitroreduction and O-acetylation by NAT enzymes, electrophilic nitrenium ion formation | [79,80] |
| 3,9-Dinitrofluoranthene | Environmental pollutants, combustion fossil fuels (e.g., diesel engine) products, and research chemicals, fluoranthene derivative, nitroarene | SULT1A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation, electrophilic nitrenium ion formation * (after nitroreduction to hydroxylamine) | [81] |
| 3-Acetylaminobenzanthrone (3-Ac-ABA) | Metabolite of 3-nitrobenzanthrone (3-NBA), arylamine | P450 1A2 | N-Hydroxylation after deacetylation concentration-dependent), oxidation | [85,87] |
| 3-Acetylaminobenzanthrone (3-Ac-ABA) | As above | NAT1 | O-Acetylation after deacetylation and N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation * | [85] |
| 3-Acetylaminobenzanthrone (3-Ac-ABA) | As above | NAT2 | O-Acetylation after deacetylation and N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation * | [85] |
| 3-Acetylaminobenzanthrone (3-Ac-ABA) | As above | SULT1A1 | O-Sulfation, sulfo-conjugate, after deacetylation and N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation * | [85] |
| 3-Acetylaminobenzanthrone (3-Ac-ABA) | As above | SULT1A2 | O-Sulfation, sulfo-conjugate, after deacetylation and N-hydroxylation, at higher concentrations * | [56,88] |
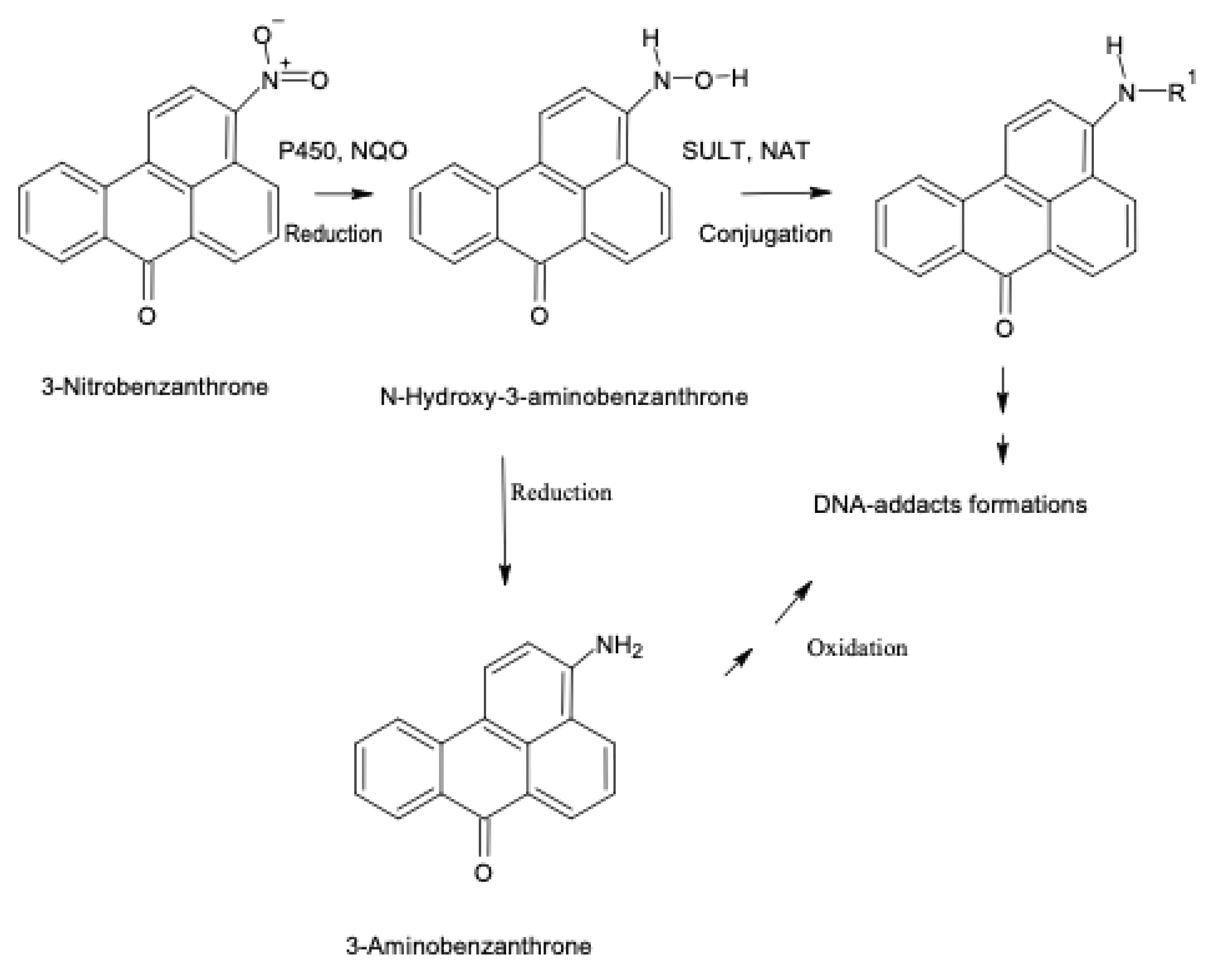
| 3-Aminobenzanthrone (3-ABA) | Metabolite of 3-nitrobenzanthrone (3-NBA) found in diesel fuel exhaust, benzanthrone derivative, arylamine | P450 1A1 | N-Hydroxylation, oxidation ** | [87,88,89] |
| 3-Aminobenzanthrone (3-ABA) | As above | P450 1A2 | N-Hydroxylation, oxidation, concentration-dependent ** | [87,88] |
| 3-Aminobenzanthrone (3-ABA) | As above | P450 1B1 | N-Hydroxylation, oxidation | [89] |
| 3-Aminobenzanthrone (3-ABA) | As above | P450 2A6 | N-Hydroxylation, oxidation | [89] |
| 3-Aminobenzanthrone (3-ABA) | As above | P450 2B6 | N-Hydroxylation, oxidation | [89] |
| 3-Aminobenzanthrone (3-ABA) | As above | LPO | N-Oxidation | [87,88] |
| 3-Aminobenzanthrone (3-ABA) | As above | MPO | N-Oxidation | [87,88] |
| 3-Aminobenzanthrone (3-ABA) | As above | NAT1 | O-Acetylation after N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation * | [85] |
| 3-Aminobenzanthrone (3-ABA) | As above | NAT2 | O-Acetylation after N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation | [85] |
| 3-Aminobenzanthrone (3-ABA) | As above | PGHS | N-Oxidation | [87,88] |
| 3-Aminobenzanthrone (3-ABA) | As above | SULT1A1 | O-Sulfation, sulfo-conjugate, after N-hydroxylation, at higher concentrations * | [85] |
| 3-Aminobenzanthrone (3-ABA) | As above | SULT1A2 | O-Sulfation, sulfo-conjugate, after N-hydroxylation, at higher concentrations, electrophilic nitrenium ion formation * | [85] |
| 3-Methylcholanthrene (3-MC) | Environmental pollutants, incomplete burning organic compounds products coal tar, heavy-end petroleum compounds, cigarette smoke compounds, and research chemical | P450 1A1 | Oxidation, micronucleus frequency increased in CHL-A1 cells | [90] |
| 3-Methylcholanthrene-11,12-diol (3-MC-11,12-diol) | Metabolite of 3-MC | P450 1A1 | Oxidation | [1] |
| 3-Nitrobenzanthrone (3-NBA) | Environmental pollutants, found in diesel fuel exhaust, urban air pollutants, nitroarene | P450 1A1 | Nitroreduction to hydroxylamine | [86,87] |
| 3-Nitrobenzanthrone (3-NBA) | As above | P450 1A2 | Nitroreduction to hydroxylamine | [85,86,87] |
| 3-Nitrobenzanthrone (3-NBA) | As above | P450 2B6 | Nitroreduction to hydroxylamine * | [86] |
| 3-Nitrobenzanthrone (3-NBA) | As above | P450 2D6 | Nitroreduction to hydroxylamine * | [86] |
| 3-Nitrobenzanthrone (3-NBA) | As above | POR | Nitroreduction to hydroxylamine | [86] |
| 3-Nitrobenzanthrone (3-NBA) | As above | XOR | Nitroreduction to hydroxylamine | [85] |
| 3-Nitrobenzanthrone (3-NBA) | As above | NAT1 | O-Acetylation after nitro-reduction to hydroxylamine, at higher concentrations, electrophilic nitrenium ion formation * | [85,87,91] |
| 3-Nitrobenzanthrone (3-NBA) | As above | NAT2 | O-Acetylation after nitro-reduction to hydroxylamine, at higher concentrations, electrophilic nitrenium ion formation * | [84,85,87,91] |
| 3-Nitrobenzanthrone (3-NBA) | As above | NQO | Nitroreduction to hydroxylamine *, ** | [86,87] |
| 3-Nitrobenzanthrone (3-NBA) | As above | SULT1A1 | O-Sulfation, sulfo-conjugate, after nitroreduction to hydroxylamine, electrophilic nitrenium ion formation* | [81,84,85,87,91] |
| 3-Nitrobenzanthrone (3-NBA) | As above | SULT1A2 | O-Sulfation, sulfo-conjugate, after nitroreduction to hydroxylamine, electrophilic nitrenium ion formation | [85,87] |
| 3-Nitrofluoranthene (3-NF) | Constituent of particulate matter in diesel-engine exhaust, urban air pollutants, nitroarene | P450 1B1 (co-expressed with NPR) | 1-Aminopyrene formation, nitroreduction/O-acetylation, at low concentrations, electrophilic nitrenium ion formation | [9] |
| 4,10-Diazachrysene (4,10-DAC) | Chrysene derivative | P450 1A2 | Oxidation, enamine epoxide formation | [11,12] |
| 4,10-Diazachrysene (4,10-DAC) | Chrysenederivative | P450 2A6 | Oxidation, enamine epoxide formation | [11,13] |
| 4-Hydroxycyclopenta[def]chrysene | Metabolite of cyclopenta[def]chrysene, automobile exhaust, and cigarette smoke compound | SULT1B1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * | [14,15] |
| 4-Hydroxycyclopenta[def]chrysene | As above | SULT1E1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [14,15,56] |
| 4-Nitropyrene (4-NP) | As above | P450 3A4 | Nitroreduction, aminopyrene, 4-hydroxylamine, formation ** | [13] |
| 4-Nitropyrene (4-NP) | As above | P450 3A4 | Oxidation, ring oxidations * | [13] |
| 5,6-Dimethylchrysene-1,2-diol | Metabolite of 5,6-dimethylchrysene | P450 1A1 | Oxidation, diolepoxide formation | [1,25,33,38] |
| 5,6-Dimethylchrysene-1,2-diol | As above | P450 1A2 | Oxidation, diolepoxide formation | [1,33,38] |
| 5,6-Dimethylchrysene-1,2-diol | As above | P450 1B1 | Oxidation, diolepoxide formation | [1,3,24,25,33,38,43] |
| 5-Methylchrysene | Environmental pollutants, vehicle emissions, and tobacco smoke compound | P450 1A1 | 1,2-Dihydrodiol formation (medium Km, high activity, high efficiency), oxidation * | [1,33,49,66,92] |
| 5-Methylchrysene | As above | P450 1A2 | 1,2-Dihydrodiol formation, oxidation | [1,33,92] |
| 5-Methylchrysene | As above | P450 1B1 | Oxidation, ring oxidation | [43] |
| 5-Methylchrysene | As above | P450 3A4 | Oxidation, ring oxidations | [92] |
| 5-Methylchrysene | As above | P450 2C10 | 1,2-Dihydrodiol formation, oxidation | [92] |
| 5-Methylchrysene-1,2-diol | Metabolite of 5-methylchrysene | AKR1A1 | Oxidation, o-quinone formation (medium Km, high activity, high efficiency) | [38,47,48] |
| 5-Methylchrysene-1,2-diol | As above | P450 1A1 | Oxidation, o-quinone formation | [1,24,25,33,38,66] |
| 5-Methylchrysene-1,2-diol | As above | P450 1A2 | Oxidation, o-quinone formation | [1,24,33,38] |
| 5-Methylchrysene-1,2-diol | As above | P450 1B1 | Oxidation (medium Km, high activity, high efficiency, o-quinone formation * | [1,3,24,25,33,38,41,43,66] |
| 5-Methylchrysene-1,2-diol | As above | P450 2W1 | Oxidation, o-quinone formation | [43] |
| 5-Methylchrysene-7,8-diol | As above | AKR1C1 | Oxidation, o-quinone formation | [18,19] |
| 5-Methylchrysene-7,8-diol | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| 5-Methylchrysene-7,8-diol | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| 5-Methylchrysene-7,8-diol | As above | AKR1C4 | Oxidation, o-quinone formation** | [19] |
| 5-Nitroacenaphthene | Environmental pollutants, industrial and research chemicals, acenaphthene derivative, nitroarene | SULT1A1 | O-Sulfation, sulfo-conjugation, electrophilic nitrenium ion formation * (after nitroreduction) | [81] |
| 6-Aminochrysene (6-AC) | Metabolite of 6-nitrochrysene, arylamine | P450 1A1 | Oxidation (high activity) * | [1,3,24] |
| 6-Aminochrysene (6-AC) | As above | P450 1A2 | Oxidation | [3,24,57,58,59,93,94] |
| 6-Aminochrysene (6-AC) | As above | P450 1B1 | Oxidation * | [1,24] |
| 6-Aminochrysene (6-AC) | As above | P450 2A6 | Oxidation | [95] |
| 6-Aminochrysene (6-AC) | As above | P450 2B6 | Oxidation | [93,96] |
| 6-Aminochrysene (6-AC) | As above | P450 3A4 | Oxidation, ring oxidation ** | [52,73,89,93,94] |
| 6-Aminochrysene (6-AC) | As above | NAT2 | O-Acetylation after N-hydroxylation, electrophilic nitrenium ion formation | [53] |
| 6-Aminochrysene (6-AC) | As above | P450 3A7 | Oxidation, ring oxidation | [73] |
| 6-Aminochrysene-1,2-diol | As above | P450 1A1 | Diol epoxide formation, oxidation | [24,93,94] |
| 6-Aminochrysene-1,2-diol | As above | P450 1A2 | Diol epoxide formation, oxidation | [24,93,94] |
| 6-Aminochrysene-1,2-diol | As above | P450 1B1 | Diol epoxide formation, oxidation * | [24,93,94] |
| 6-Aminochrysene-1,2-diol | As above | P450 3A4 | Diol epoxide formation, oxidation | [93,94] |
| 6-Hydroxymethylanthracene | Metabolite of methylanthracene, research chemicals, benzylic alcohol | SULT1C3 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [46] |
| 6-Hydroxymethylbenzo[a]pyrene | Metabolite of methylbenzo[a]pyrene, research chemicals, PAH derivative | SULT1B1 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [15] |
| 6-Hydroxymethylbenzo[a]pyrene | As above | SULT1C3 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [46] |
| 6-Hydroxymethylbenzo[a]pyrene | As above | SULT2A1 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [14,15] |
| 6-Methylchrysene | Environmental pollutants, tobacco smoke constituent | P450 1A1 | 1,2-Dihydrodiol formation, oxidation ** | [92] |
| 6-Methylchrysene | As above | P450 1A2 | 1,2-Dihydrodiol formation, oxidation | [92] |
| 6-Methylchrysene | As above | P450 2C10 | 1,2-Dihydrodiol formation, oxidation | [92] |
| 6-Methylchrysene | As above | P450 1A2 | 6-Methylhydroxylation, oxidation | [92] |
| 6-Methylchrysene | As above | P450 3A4 | 6-Methylhydroxylation, oxidation | [92] |
| 6-Nitrochrysene | Environmental pollutants, research chemicals, nitroarene | P450 1A2 | Oxidation, trans-1,2-dihydro-1,2-dihydroxy-6-nitrochrysene formation * | [1,50] |
| 6-Nitrochrysene | As above | P450 1A1 | Oxidation, trans-1,2-dihydro-1,2-dihydroxy-6-nitrochrysene formation * | [1,50] |
| 6-Nitrochrysene | As above | P450 3A4 | Nitroreduction, 6-amino chrysene formation * | [1,50] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | Product of incomplete combustion product of gasoline and coal | P450 1A1 | Oxidation (low Km, high activity and efficiency) * | [1,25,33,97,98] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 1A2 | Oxidation | [1,33,98] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 1B1 | Oxidation (low Km, high activity, and efficiency) | [1,25,33,43,98] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 2C9 | Oxidation | [1,33,97] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 2D6 | Oxidation | [97] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | Metabolite of 7,12-DMBA | AKR1A1 | Oxidation, o-quinone formation | [47,48] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | AKR1B10 | Oxidation, o-quinone formation | [16] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | AKR1C1 | Oxidation, o-quinone formation, minor enzyme | [18,19] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | AKR1C4 | Oxidation, o-quinone formation ** | [19] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 1A1 | 3,4-Dihydrodiol-1,2-epoxide formation (medium Km, high activity, high efficiency), oxidation*, also, micronucleus frequency increased in CHL-A1 cells | [1,3,25,33,38,90] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 1A2 | Oxidation | [1,33,38] |
| 7,12-Dimethylbenz[a]anthracene (7,12-DMBA) | As above | P450 1B1 | 3,4-Dihydrodiol-1,2-epoxide formation (medium Km, high activity, high efficiency), oxidation * | [1,3,24,25,33,38] |
| 7-Hydroxy-12-methylbenz[a]anthracene | Metabolite of DMBA | SULT2A1 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [55] |
| 7-Hydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene | Metabolite of B[a]P | SULT1A1 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation* | [55,56] |
| 7-Hydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene | As above | SULT1E1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation * | [56] |
| 7-Methylbenz[a]anthracene-3,4-diol | Metabolite of 7-methylbenz[a]anthracene | AKR1A1 | Oxidation, o-quinone formation, preferential for (-)-3S,4S-oxidation | [47,48] |
| 7-Methylbenz[a]anthracene-3,4-diol | As above | AKR1C1 | Oxidation, o-quinone formation, minor enzyme | [19] |
| 7-Methylbenz[a]anthracene-3,4-diol | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| 7-Methylbenz[a]anthracene-3,4-diol | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| 7-Methylbenz[a]anthracene-3,4-diol | As above | AKR1C4 | Oxidation, o-quinone formation | [19] |
| 7-Methylbenz[c]acridine (7MBAC) | Research chemicals, aza-aromatic | P450 1A2 | 3,4-Dihydrodiol formation, oxidation | [100] |
| 7-Methylbenz[c]acridine (7MBAC) | As above | P450 1A1 | K-region oxide formation, oxidation * | [100] |
| 7-Methylbenz[c]acridine (7MBAC) | As above | P450 1A2 | K-region oxide formation, oxidation | [100] |
| 7-Methylbenz[c]acridine (7MBAC) | As above | P450 3A4 | K-region oxide formation, oxidation | [100] |
| 9-Hydroxybenzo[a]pyrene | Metabolite of B[a]P | P450 1A1 | Oxidation | [1] |
| 9-Hydroxybenzo[a]pyrene | As above | P450 1B1 | Oxidation | [1] |
| 9-Hydroxymethyl-10-methylanthracene | Industrial and research chemicals, used in the synthesis of fluorescent dyes and pigments | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [56] |
| 9-Hydroxymethylanthracene | Research chemical | SULT2A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [56] |
| Benz[a]anthracene | Incomplete combustion products of organic matter, found in gasoline and diesel fuel exhaust, tobacco smoke compound | P450 1A1 | Oxidation | [1,33] |
| Benz[a]anthracene-1,2-diol | Metabolite of benz[a]anthracene | P450 1A1 | Oxidation, micronucleus frequency increased in CHL-A1 cells | [1,38,90,101] |
| Benz[a]anthracene-3,4-diol | As above | AKR1A1 | Oxidation, o-quinone formation | [47,48] |
| Benz[a]anthracene-3,4-diol | As above | AKR1C1 | Oxidation, o-quinone formation | [19] |
| Benz[a]anthracene-3,4-diol | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| Benz[a]anthracene-3,4-diol | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| Benz[a]anthracene-3,4-diol | As above | AKR1C4 | Oxidation, o-quinone formation | [19] |
| Benz[a]anthracene-3,4-diol | As above | P450 1A1 | Oxidation | [1,33] |
| Benz[a]anthracene-3,4-diol | As above | P450 1A2 | Oxidation | [1,33] |
| Benz[a]anthracene-5,6-diol | As above | P450 1A1 | Oxidation | [1,38] |
| Benzo[a]perylene | Incomplete combustion products present in automobile exhaust, tobacco smoke, grilled meat, edible oil compound | P450 1A1 | Oxidation | [102] |
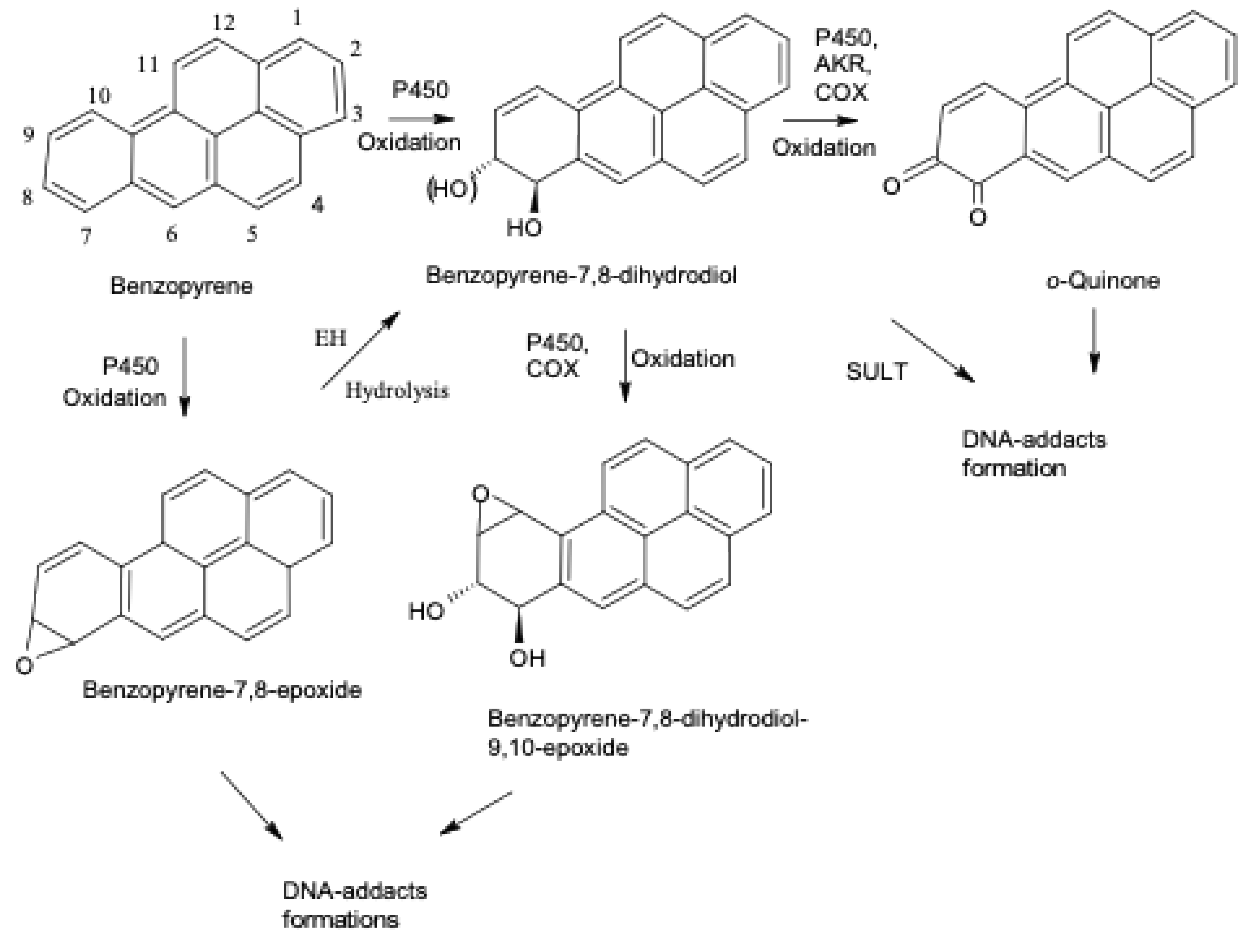
| Benzo[a]pyrene (B[a]P) | Incomplete combustion product of organic matter, coal tar, tobacco smoke, and many foods (e.g., grilled meat) compound | P450 1A1 | trans-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro- formation (low activity, medium activity, or high activity, high efficiency), 1,6-,3,6-,6,12-dione (quinone formation, low activity), oxidation * | [1,24,27,28,30,31,32,33,52,67,103,105,106,107,108] |
| Benzo[a]pyrene (B[a]P) | As above | P450 1B1 | trans-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro- formation (medium Km, high activity, high efficiency), 1,6-,3,6-Dione (quinone formation, low activity), oxidation ** | [1,24,25,26,27,33,38,43,44,70,102,104,109,129] |
| Benzo[a]pyrene-7,8-oxide (B[a]P-7,8-oxide) | Metabolite of B[a]P | Epoxide hydrolase, EH | Hydrolysis to B[a]P-7,8-diol, participation in B[a]P toxicity | [1,27] |
| Benzo[b]fluoroanthene-9,10-diol (B[b]F-11,12-diol) | Metabolite of B[b]F | P450 1A1 | Oxidation * | [1,3,24,25,33,38] |
| Benzo[b]fluoroanthene-9,10-diol (B[b]F-11,12-diol) | As above | P450 1A2 | Oxidation | [1,24,33,38] |
| Benzo[b]fluoroanthene-9,10-diol (B[b]F-11,12-diol) | As above | P450 1B1 | Oxidation | [1,24,25,33,38] |
| Benzo[c]phenanthrene (B[c]P) | Wood and fossil fuel exhaust compound | P450 1A1 | Dihydrodiol 3,4-, 1,2-epoxide formation, oxidation ** | [106] |
| Benzo[c]phenanthrene (B[c]P) | As above | P450 1B1 | Dihydrodiol 3,4-, 1,2-epoxide formation, oxidation ** | [1,25,33,110,111,112] |
| Benzo[c]phenanthrene (B[c]P) | As above | P450 2C9 | Oxidation | [33] |
| Benzo[c]phenanthrene-3,4-dio (B[c]P-3,4-diol) | Metabolite of B[c]P | AKR1C2 | Oxidation, o-quinone formation | [19] |
| Benzo[c]phenanthrene-3,4-dio (B[c]P-3,4-diol) | As above | AKR1C4 | Oxidation, o-quinone formation | [19] |
| Benzo[c]phenanthrene-3,4-dio (B[c]P-3,4-diol) | As above | P450 1A2 | Oxidation | [1] |
| Benzo[c]phenanthrene-3,4-dio (B[c]P-3,4-diol) | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | Metabolite of B[g]C, fossil fuels and organic materials incomplete combustion product | P450 1A1 | Oxidation | [1,25,33,38] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | AKR1C1 | Oxidation, o-quinone formation | [18,19] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | AKR1C4 | Oxidation, o-quinone formation ** | [19] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | P450 1A2 | Oxidation | [1,33,38] |
| Benzo[g]chrysene-11,12-diol (B[g]C-11,12-diol) | As above | P450 1B1 | Oxidation * | [1,3,24,25,33,38] |
| Chrysene-1,2-diol | As above | AKR1C2 | Oxidation, o-quinone formation | [19] |
| Chrysene-1,2-diol | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| Chrysene-1,2-diol | As above | AKR1C4 | Oxidation, o-quinone formation | [19] |
| Chrysene-1,2-diol | As above | P450 1A1 | Oxidation * | [1,25,33] |
| Chrysene-1,2-diol | As above | P450 1A2 | Oxidation | [1,33,38] |
| Chrysene-1,2-diol | As above | P450 1B1 | Oxidation, diolepoxide formation * | [1,3,24,25,33,38,43] |
| Chrysene-1,2-diol | As above | P450 2W1 | Oxidation, diolepoxide formation | [43] |
| Cyclopenta[c,d]pyrene | As above | P450 1B1 | Oxidation | [113] |
| Cyclopenta[c,d]pyrene | Incomplete combustion product of organic matter, gasoline engine exhaust compound | P450 1A1 | Oxidation | [106] |
| Dibenz[a,h]acridine | Incomplete combustion product of organic substances, primarily found in gasoline exhaust, petroleum refinery incinerator emissions, coal combustion emissions, cigarette smoke, and coal tar pitch | P450 1A1 | 10,11-Diol formation, oxidation * | [114] |
| Dibenz[a,h]acridine | As above | P450 1B1 | 10,11-Diol formation, oxidation | [114] |
| Dibenz[a,h]anthracene | Incomplete combustion product of organic substances found in air, soil, or sediment, and on pyrolysis of tobacco | P450 1A1 | Oxidation | [1] |
| Dibenz[a,h]anthracene | As above | P450 1A2 | 1,2-Dihydrodiol formation, oxidation ** | [115] |
| Dibenz[a,h]anthracene | As above | P450 1A2 | trans-3,4-Dihydrodiol formation, oxidation | [115] |
| Dibenz[a,h]anthracene | As above | P450 2B6 | trans-3,4-Dihydrodiol formation, oxidation | [115] |
| Dibenz[a,h]anthracene | As above | P450 2C9 | trans-3,4-Dihydrodiol formation, oxidation ** | [115] |
| Dibenz[a,j]acridine (DBJAC) | Automobile exhaust, coal burning, incinerator waste streams, cigarette smoke compound, heteroarene | P450 3A4 | 3,4-Dihydrodiol formation, oxidation * | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A1 | K-region oxide formation, oxidation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A2 | K-region oxide formation, oxidation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A1 | K-region dihydrodiol formation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A1 | 3,4-Dihydrodiol formation, oxidation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A2 | 3,4-Dihydrodiol formation, oxidation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 3A5 | 3,4-Dihydrodiol formation, oxidation | [100] |
| Dibenz[a,j]acridine (DBJAC) | As above | P450 1A2 | K-region dihydrodiol formation, oxidation | [100] |
| Dibenzo[a,e]fluoranthene | Research chemical | P450 1A1 | Oxidation, mutagenicity | [102] |
| Dibenzo[a,e]pyrene (DB[a,e]P) | Tobacco smoke compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Dibenzo[a,f]fluoranthene | Incomplete combustion of organic materials, such as fossil fuels, wood, and tobacco smoke compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Dibenzo[a,h]pyrene (DB[a,h]P) | Tobacco smoke compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Dibenzo[a,k]fluoranthene | Incomplete burning of coal, oil, gas, wood, garbage, and other organic substances compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Dibenzo[a,l]pyrene (DB[a,l]P) | Combustion of wood and coal, gasoline and diesel exhaust, and tobacco smoke compound | P450 1A1 | (-)-syn- and (-)-anti-11,12-Dihydrodiol-13,14-epoxide formation (medium Km, high activity, high efficiency, oxidation * | [1,28,33,102,116,117,118,119,120,121,122,123] |
| Dibenzo[a,l]pyrene (DB[a,l]P) | As above | P450 1A2 | (-)-anti-11,12-Dihydrodiol-13,14-epoxide formation, oxidation | [33,119,120] |
| Dibenzo[a,l]pyrene (DB[a,l]P) | As above | P450 1B1 | (-)-anti-11,12-Dihydrodiol-13,14-epoxide formation (medium Km, high activity, high efficiency), oxidation * | [1,28,33,43,44,116,117,118,119,120,121,122,123,128] |
| Dibenzo[a,l]pyrene-11,12-diol (DB[a,l] -11,12-diol) | Metabolite of DB[a,l]P | P450 1A1 | 11,12-Dihydrodiol-13,14-epoxide formation (medium Km, high activity, high efficiency, oxidation * | [1,25,28,33,38,41,104,116,117,118,123] |
| Dibenzo[a,l]pyrene-11,12-diol (DB[a,l] -11,12-diol) | As above | P450 1A2 | Oxidation | [31,38,104] |
| Dibenzo[a,l]pyrene-11,12-diol (DB[a,l] -11,12-diol) | As above | P450 1B1 | 11,12-Dihydrodiol-13,14-epoxide formation (medium Km, high activity, high efficiency), oxidation * | [1,24,25,28,33,38,41,43,44,104,116,117,118,123] |
| Dibenzo[a,l]pyrene-11,12-diol (DB[a,l] -11,12-diol) | As above | P450 2W1 | Oxidation, diolepoxide formation | [43] |
| Dibenzo[b,k]fluoranthene | Environmental pollutants, diesel fuel particulate compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Fluoranthene-2,3-diol | Metabolite of fluoranthene | P450 1A1 | Oxidation, diolepoxide formation | [1,38] |
| Naphthalene | As above | P450 2F1 | Oxidation | [124] |
| Naphthalene 1,2-diol | Metabolite of naphthalene | AKR1C1 | Oxidation, o-quinone formation ** | [18,19] |
| Naphthalene 1,2-diol | As above | AKR1C2 | Oxidation, o-quinone formation ** | [19] |
| Naphthalene 1,2-diol | As above | AKR1C3 | Oxidation, o-quinone formation | [19] |
| Naphthalene 1,2-diol | As above | AKR1C4 | Oxidation, o-quinone formation | [19] |
| Naphtho[1,2-k]fluoranthene | Environmental pollutants, incomplete combustion of organic matter compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Naphtho[2,3-a]pyrene | Air pollutants, applied in biological and electronic fields, incomplete combustion processes, and tobacco smoke compound | P450 1A1 | Oxidation, mutagenicity | [102] |
| Naphtho[2,3-e]pyrene | As above | P450 1A1 | Oxidation, mutagenicity | [102] |
| N-Hydroxy-2-acetylaminofluorene | Metabolite of 2-acetylaminofluorene, hydroxamic acid, heterocyclic amine | SULT1A1 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation | [14,15] |
| N-Hydroxy-2-acetylaminofluorene | As above | SULT1A2 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [14,15,63] |
| N-Hydroxy-2-aminofluorene | Metabolite of 2-aminofluorene, hydroxamic acid, heterocyclic amine | NAT1 | O-Acetylation, electrophilic nitrenium ion formation | [130] |
| N-Hydroxy-2-aminofluorene | As above | NAT2 | O-Acetylation, electrophilic nitrenium ion formation | [130] |
| N-Hydroxy-2-aminofluorene | As above | SULT1A1 | O-Sulfation, sulfo-conjugate, electrophilic nitrenium ion formation | [14,125] |
| N-Hydroxy-2-aminofluorene | As above | SULT1A2 | O-Sulfation, sulfo-conjugate formation, electrophilic nitrenium ion formation * | [81] |
| Phenanthrene | Environmental contaminants, industrial chemicals, tobacco smoke compound | P450 1A1 | Oxidation to 1,2- (major reaction), 9,10- and 3,4-dihydrodiols (minor reactions) and phenols, at high concentration | [126] |
| Phenanthrene | As above | P450 1A2 | Oxidation to 1,2- (major reaction), 3,4-, 9,10-dihydrodiols and phenols | [126] |
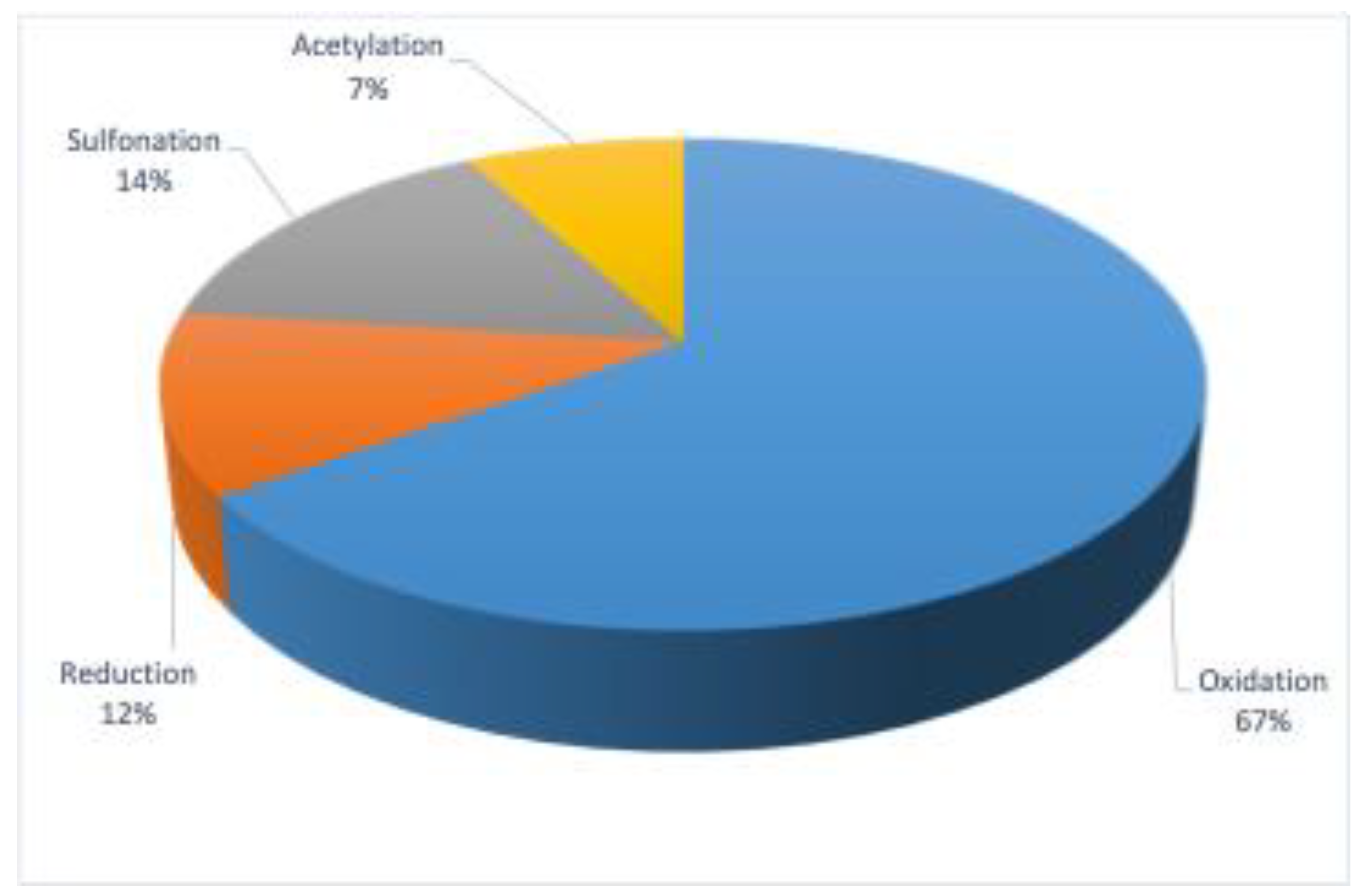
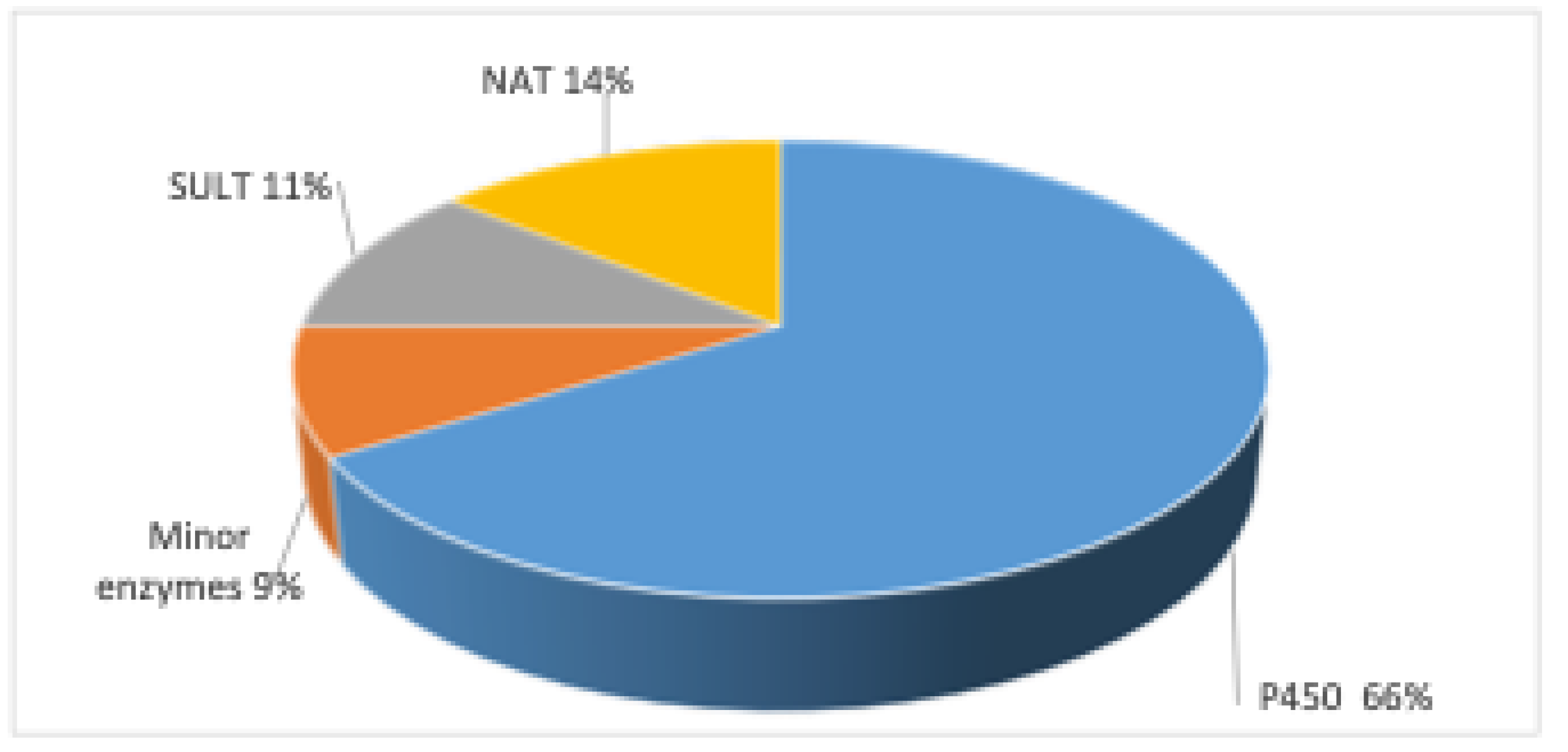
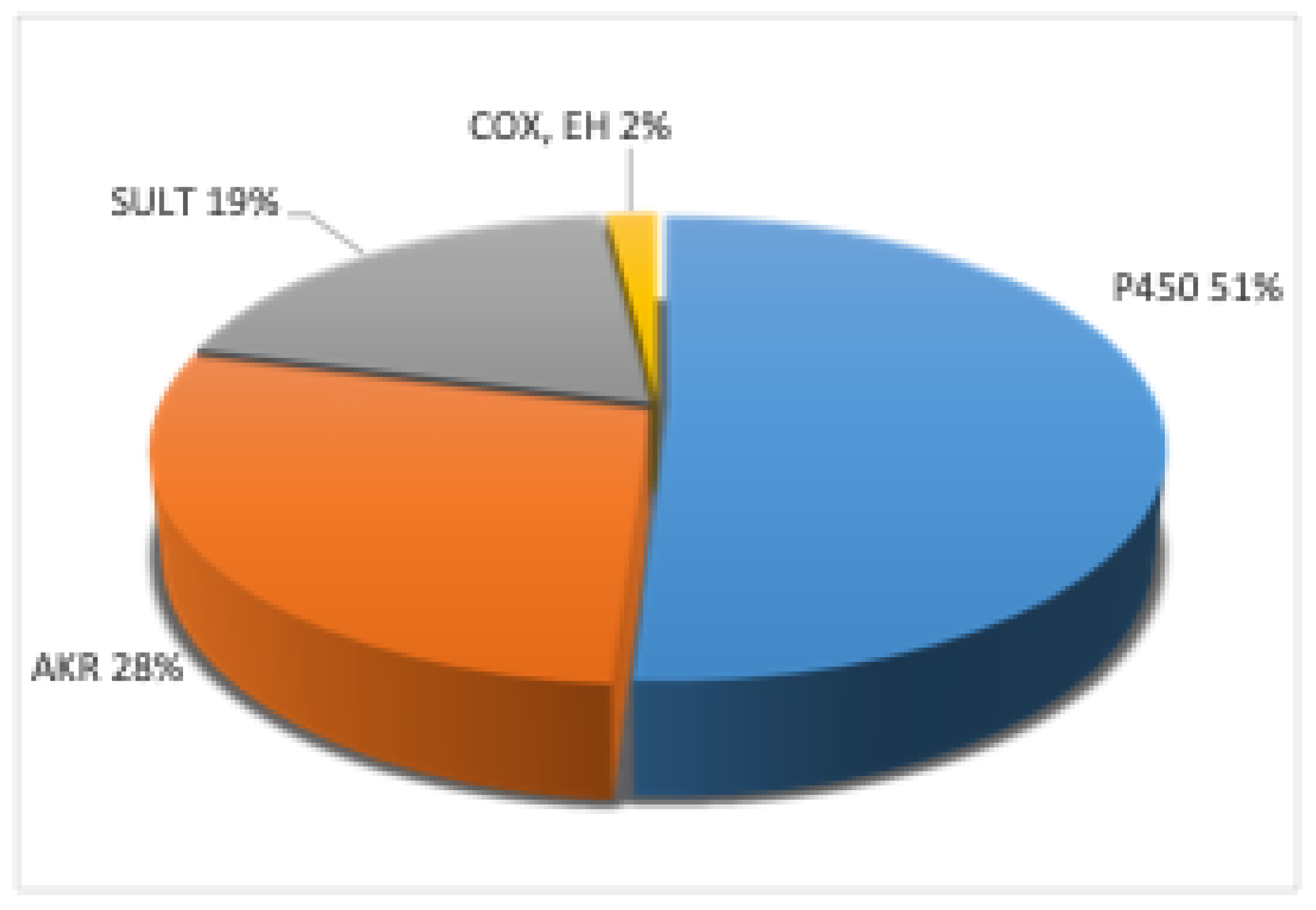
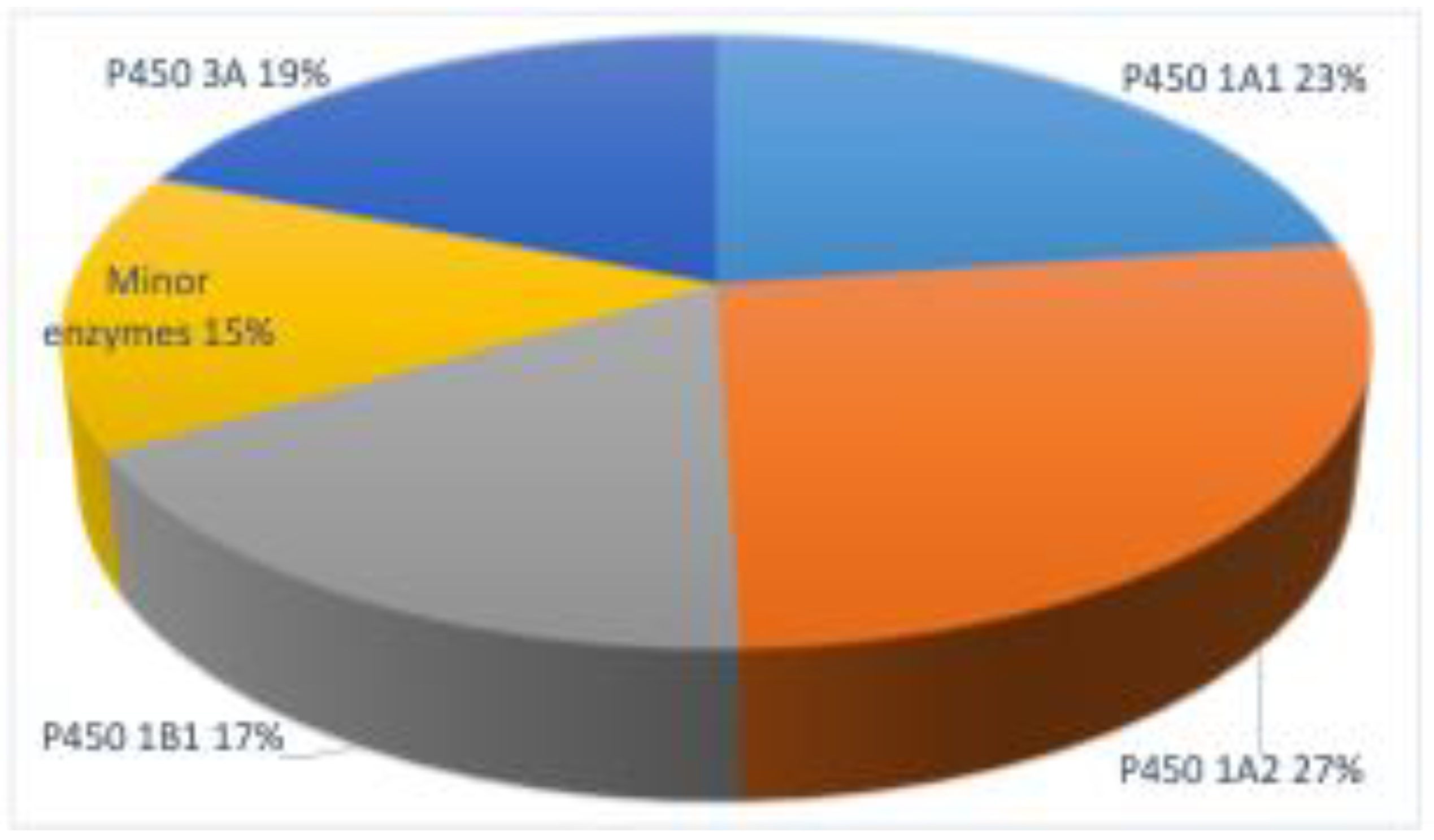
2. Metabolic Toxication of PAHs
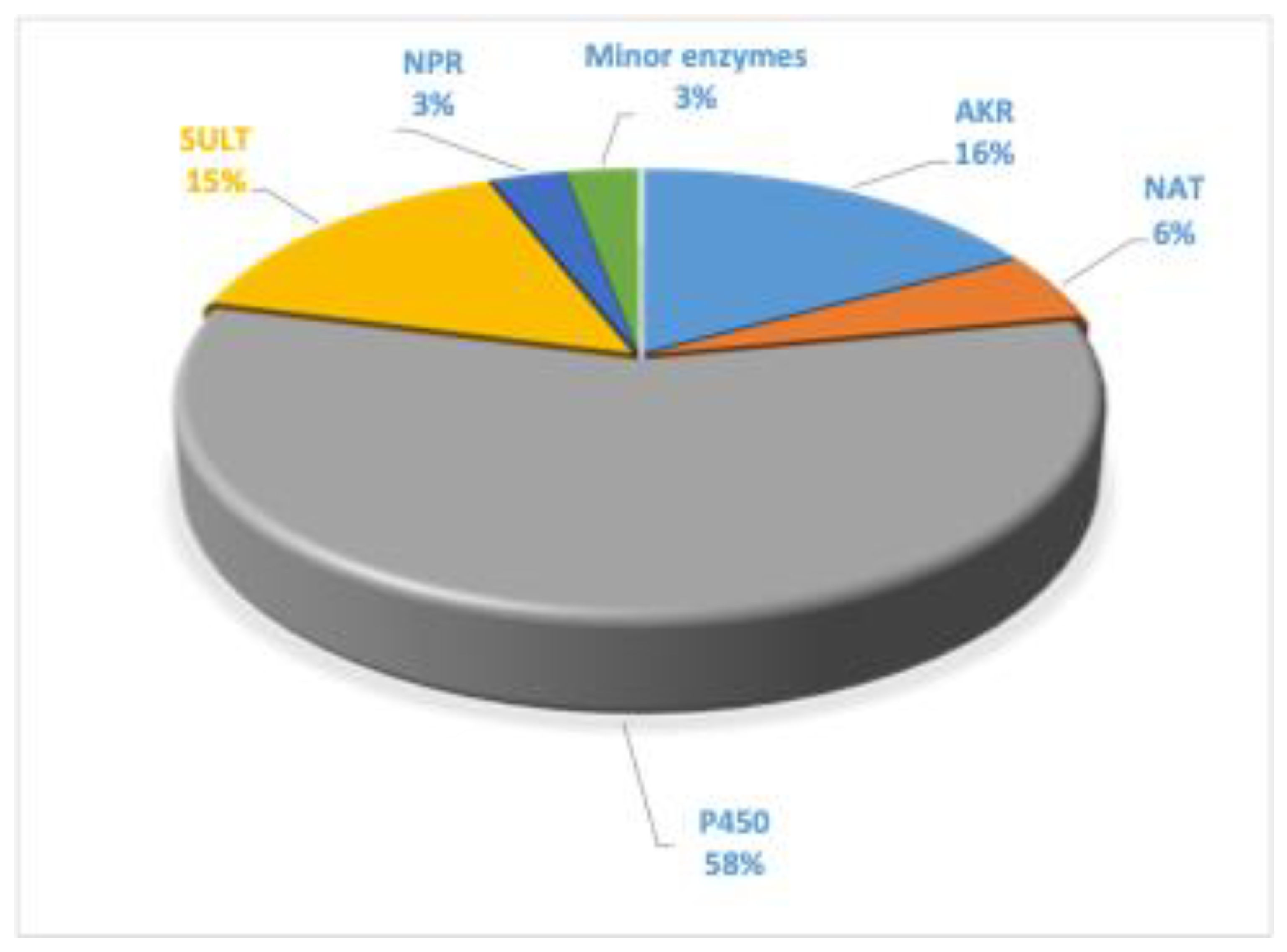
3. Enzymes
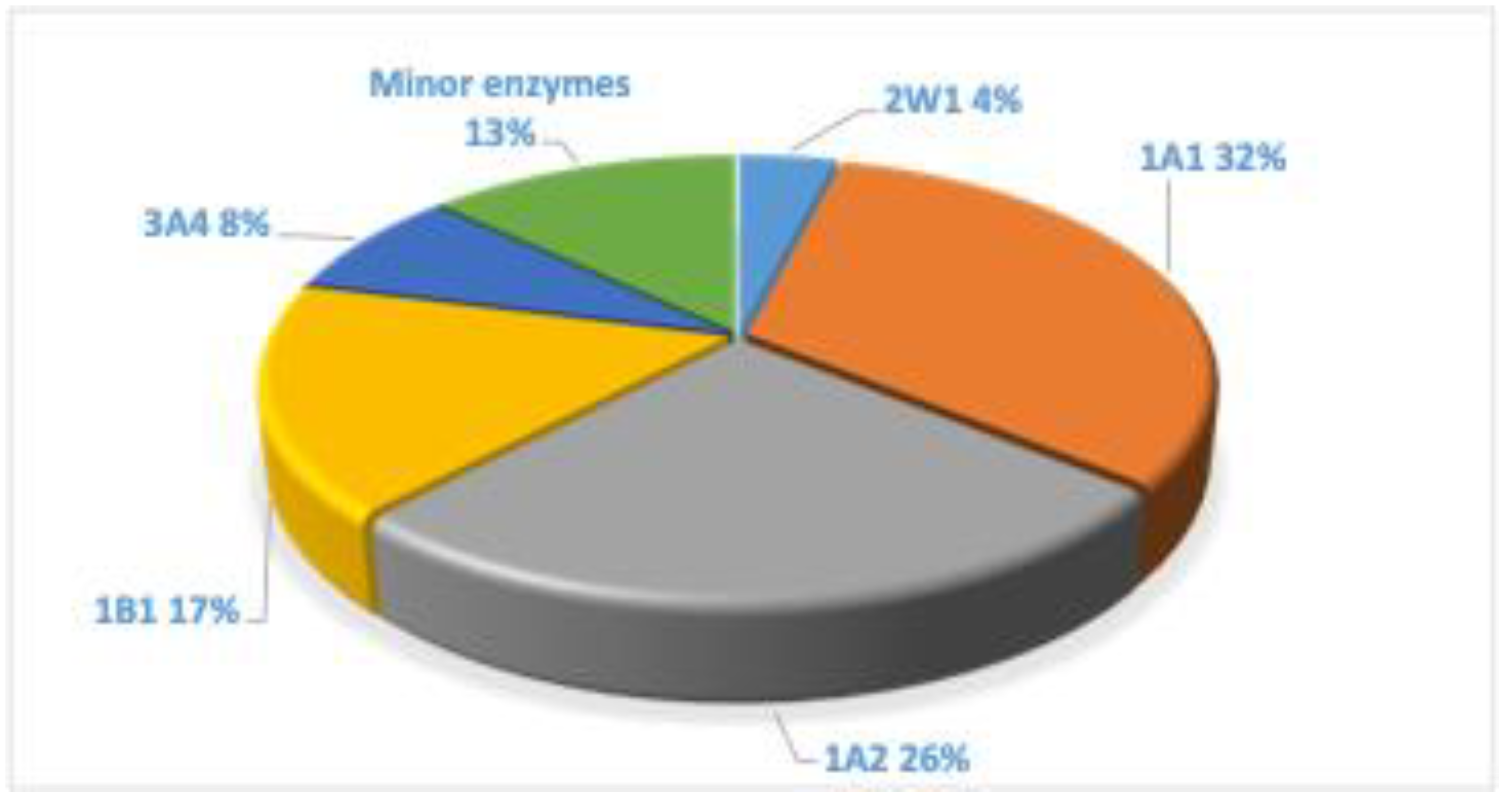
4. P450 Enzymes
5. Effect of the Structure of PAHs on the Toxication Reactions
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shimada, T.; Oda, Y.; Gillam, E.M.; Guengerich, F.P.; Inoue, K. Metabolic Activation of Polycyclic Aromatic Hydrocarbons and Other Procarcinogens by Cytochromes P450 1A1 and P450 1B1 Allelic Variants and Other Human Cytochromes P450 in Salmonella Typhimurium NM2009. Drug Metab Dispos 2001, 29, 1176–1182, PMID: 11502724.
- Shimada, T.; Murayama, N.; Yamazaki, H,; Tanaka, K.; Takenaka, S.; Komori, M.; Kim, D.; Guengerich, F.P. Metabolic Activation of Polycyclic Aromatic Hydrocarbons and Aryl and Heterocyclic Amines by Human Cytochromes P450 2A13 and 2A6. Chem Res Toxicol 2013, 26, 529–537. [CrossRef]
- Guengerich, F.P.; Shimada, T. Activation of Procarcinogens by Human Cytochrome P450 Enzymes. Mutat Res 1998, 400, 201–213. [CrossRef]
- Rendic, S.; Guengerich, F.P. Contributions of Human Enzymes in Carcinogen Metabolism. Chem Res Toxicol 2012, 25, 1316–1383. [CrossRef]
- Rendic, S.; Guengerich, F.P. Survey of Human Oxidoreductases and Cytochrome P450 Enzymes Involved in the Metabolism of Xenobiotic and Natural Chemicals. Chem Res Toxicol 2015, 28, 38–42. [CrossRef]
- Rendic, S.; Guengerich, F.P. Development and Uses of Offline and Web-Searchable Metabolism Databases – The Case of Benzo[a]pyrene. Curr Drug Metab 2018, 19, 3–46. [CrossRef]
- Rendic, S.; Guengerich, F.P. Human Family 1–4 cytochrome P450 enzymes involved in the metabolic activation of xenobiotic and physiological chemicals: An update. Arch Toxicol 2021, 95, 95–472. [CrossRef]
- Rendic, S.; Guengerich, F.P. Formation of Potentially Toxic Metabolites of Drugs in Reactions Catalyzed by Human Drug-Metabolizing Enzymes. Arch Toxicol 2024, 98, 1581–1628. [CrossRef]
- Yamazaki, H.; Hatanaka, N.; Kizu, R.; Hayakawa, K.; Shimada, N.; Guengerich, F.P.; Nakajima, M.; Yokoi, T. Bioactivation of Diesel Exhaust Particle Extracts and Their Major Nitrated Polycyclic Aromatic Hydrocarbon Components, 1-Nitropyrene and Dinitropyrenes, by Human Cytochromes P450 1A1, 1A2, and 1B1. Mutat Res 2000, 472, 129–138. [CrossRef]
- Hatanaka, N.; Yamazaki, H.; Oda, Y.; Guengerich, F.P.; Nakajima, M.; Yokoi, T. Metabolic Activation of Carcinogenic 1-Nitropyrene by Human Cytochrome P450 1B1 in Salmonella Typhimurium Strain Expressing an O-Acetyltransferase in SOS/Umu Assay. Mutat Res 2001, 497, 223–233. [CrossRef]
- Yamada, K.; Suzuki, T.; Kohara, A.; Kato, T.A.; Hayashi, M.; Mizutani, T.; Saeki, K. (2005). Nitrogen-substitution effect on in vivo mutagenicity of chrysene. Mutat Res 2005, 586, 1–17. [CrossRef]
- Yamada, K.; Hakura, A.; Kato, T.-A.; Mizutani, T.; Saeki, K.-I. Nitrogen-Substitution Effects on the Mutagenicity and Cytochrome P450 Isoform-Selectivity of Chrysene Analogs. Mutat Res 2005, 586, 87–95. [CrossRef]
- Chae, Y.H.; Thomas, T.; Guengerich, F.P.; Fu, P.P.; El-Bayoumy, K. Comparative Metabolism of 1-, 2-, and 4-Nitropyrene by Human Hepatic and Pulmonary Microsomes. Cancer Res 1999, 59, 1473–1480.
- Glatt, H.; Engelke, C.E.; Pabel, U.; Teubner, W.; Jones, A.L.; Coughtrie, M.W.; Andrae, U.; Falany, C.N.; Meinl, W. Sulfotransferases: Genetics and Role in Toxicology. Toxicol Lett 2000, 112–113, 341–348. [CrossRef]
- Glatt, H.; Boeing, H.; Engelke, C.E.; Ma, L.; Kuhlow, A.; Pabel, U.; Pomplun, D.; Teubner, W.; Meinl, W. Human Cytosolic Sulphotransferases: Genetics, Characteristics, Toxicological Aspects. Mutat Res 2001, 482, 27–40. [CrossRef]
- Quinn, A.M.; Harvey, R.G.; Penning, T.M. Oxidation of PAH Trans-Dihydrodiols by Human Aldo-Keto Reductase AKR1B10. Chem Res Toxicol 2008, 21, 2207–2215. [CrossRef]
- Burczynski, M.E.; Lin, H.K.; Penning, T.M. Isoform-Specific Induction of a Human Aldo-Keto Reductase by Polycyclic Aromatic Hydrocarbons (PAHs), Electrophiles, and Oxidative Stress: Implications for the Alternative Pathway of PAH Activation Catalyzed by Human Dihydrodiol Dehydrogenase. Cancer Res 1999, 59, 607–614.
- Burczynski, M.E.; Sridhar, G.R.; Palackal, N.T.; Penning, T.M. The Reactive Oxygen Species--and Michael Acceptor-Inducible Human Aldo-Keto Reductase AKR1C1 Reduces the Alpha, Beta-Unsaturated Aldehyde 4-Hydroxy-2-Nonenal to 1,4-Dihydroxy-2-Nonene. J Biol Chem 2001, 276, 2890–2897. [CrossRef]
- Palackal, N.T.; Lee, S.H.; Harvey, R.G.; Blair, I.A.; Penning, T.M. Activation of Polycyclic Aromatic Hydrocarbon Trans-Dihydrodiol Proximate Carcinogens by Human Aldo-Keto Reductase (AKR1C) Enzymes and Their Functional Overexpression in Human Lung Carcinoma (A549) Cells. J Biol Chem 2002, 277, 24799–24808. [CrossRef]
- Penning, T.M. Human Aldo-Keto Reductases and the Metabolic Activation of Polycyclic Aromatic Hydrocarbons. Chem Res Toxicol 2014, 27, 1901–1917. [CrossRef]
- Wiese, F.W.; Thompson, P.A.; Kadlubar, F.F. Carcinogen Substrate Specificity of Human COX-1 and COX-2. Carcinogenesis 2001, 22, 5–10. [CrossRef]
- Shimada, T.; Martin, M.V.; Pruess-Schwartz, D.; Marnett, L.J.; Guengerich, F.P. Roles of Individual Human Cytochrome P-450 Enzymes in the Bioactivation of Benzo(a)pyrene, 7,8-Dihydroxy-7,8-Dihydrobenzo(a)Pyrene, and Other Dihydrodiol Derivatives of Polycyclic Aromatic Hydrocarbons. Cancer Res 1989, 49, 6304–6312, PMID: 2509067.
- Shimada, T.; Gillam, E.M.; Sandhu, P.; Guo, Z.; Tukey, R.H.; Guengerich, F.P. Activation of Procarcinogens by Human Cytochrome P450 Enzymes Expressed in Escherichia Coli. Simplified Bacterial Systems for Genotoxicity Assays. Carcinogenesis 1994, 15, 2523–2529. [CrossRef]
- Shimada, T.; Hayes, C.L.; Yamazaki, H.; Amin, S.; Hecht, S.S.; Guengerich, F.P.; Sutter, T.R. Activation of Chemically Diverse Procarcinogens by Human Cytochrome P-450 1B1. Cancer Res 1996, 56, 2979–2984, PMID: 8674051.
- Shimada, T.; Watanabe, J.; Kawajiri, K.; Sutter, T.R.; Guengerich, F.P.; Gillam, E.M.; Inoue, K. Catalytic Properties of Polymorphic Human Cytochrome P450 1B1 Variants. Carcinogenesis 1999, 20, 1607–1613. [CrossRef]
- Shimada, T.; Watanabe, J.; Inoue, K.; Guengerich, F.P.; Gillam, E.M. Specificity of 17beta-Oestradiol and Benzo[a]Pyrene Oxidation by Polymorphic Human Cytochrome P4501B1 Variants Substituted at Residues 48, 119 and 432. Xenobiotica 2001, 31, 163–176. [CrossRef]
- Gelhaus, S.L.; Harvey, R.G.; Penning, T.M.; Blair, I.A. Regulation of Benzo[a]Pyrene-Mediated DNA- and Glutathione-Adduct Formation by 2,3,7,8-Tetrachlorodibenzo-p-Dioxin in Human Lung Cells. Chem Res Toxicol 2011, 24, 89–98. [CrossRef]
- Kushman, M.E.; Kabler, S.L.; Ahmad, S.; Doehmer, J.; Morrow, C.S.; Townsend, A.J. Cytotoxicity and Mutagenicity of Dibenzo[a,l]Pyrene and (+/-)-Dibenzo[a,l]Pyrene-11,12-Dihydrodiol in V79MZ Cells Co-Expressing Either hCYP1A1 or hCYP1B1 Together with Human Glutathione-S-Transferase A1. Mutat Res 2007, 624, 80–87. [CrossRef]
- Kushman, M.E.; Kabler, S.L.; Ahmad, S.; Doehmer, J.; Morrow, C.S.; Townsend, A.J. Protective Efficacy of hGSTM1-1 against B[a]P and (+)- or (-)-B[a]P-7,8-Dihydrodiol Cytotoxicity, Mutagenicity, and Macromolecular Adducts in V79 Cells Coexpressing hCYP1A1. Toxicol Sci 2007, 99, 51–57. [CrossRef]
- Kushman, M.E.; Kabler, S.L.; Fleming, M.H.; Ravoori, S.; Gupta, R.C.; Doehmer, J.; Morrow, C.S.; Townsend, A.J. Expression of Human Glutathione S-Transferase P1 Confers Resistance to Benzo[a]Pyrene or Benzo[a]Pyrene-7,8-Dihydrodiol Mutagenesis, Macromolecular Alkylation and Formation of Stable N2-Gua-BPDE Adducts in Stably Transfected V79MZ Cells Co-Expressing hCYP1A1. Carcinogenesis 2007, 28, 207–214. [CrossRef]
- Ruan, Q.; Gelhaus, S.L.; Penning, T.M.; Harvey, R.G.; Blair, I.A. Aldo-Keto Reductase- and Cytochrome P450-Dependent Formation of Benzo[a]Pyrene-Derived DNA Adducts in Human Bronchoalveolar Cells. Chem Res Toxicol 2007, 20, 424–431. [CrossRef]
- Schwarz, D.; Kisselev, P.; Cascorbi, I.; Schunck, W.H.; Roots, I. Differential Metabolism of Benzo[a]Pyrene and Benzo[a]Pyrene-7,8-Dihydrodiol by Human CYP1A1 Variants. Carcinogenesis 2001, 22, 453–459. [CrossRef]
- Schwarz, D.; Kisselev, P.; Honeck, H.; Cascorbi, I.; Schunck, W.H.; Roots, I. Co-Expression of Human Cytochrome P4501A1 (CYP1A1) Variants and Human NADPH-Cytochrome P450 Reductase in the Baculovirus/Insect Cell System. Xenobiotica 2001, 31, 345–356. [CrossRef]
- Shimada, T.; Fujii-Kuriyama, Y. Metabolic Activation of Polycyclic Aromatic Hydrocarbons to Carcinogens by Cytochromes P450 1A1 and 1B1. Cancer Sci 2004, 95, 1–6. [CrossRef]
- Shou, M.; Korzekwa, K.R.; Crespi, C.L.; Gonzalez, F.J.; Gelboin, H.V. The Role of 12 cDNA-Expressed Human, Rodent, and Rabbit Cytochromes P450 in the Metabolism of Benzo[a]Pyrene and Benzo[a]Pyrene Trans-7,8-Dihydrodiol. Mol Carcinog 1994, 10, 159–168. [CrossRef]
- Doehmer, J.; Holtkamp, D.; Soballa, V.; Raab, G.; Schmalix, W.; Seidel, A.; Greim, H.; Jacob, J. Cytochrome P450 Mediated Reactions Studied in Genetically Engineered V79 Chinese Hamster Cells. Pharmacogenetics 1995, 5 Spec No, S91-96. [CrossRef]
- Jiang, H.; Shen, Y.-M.; Quinn, A.M.; Penning, T.M. Competing Roles of Cytochrome P450 1A1/1B1 and Aldo-Keto Reductase 1A1 in the Metabolic Activation of (+/-)-7,8-Dihydroxy-7,8-Dihydro-Benzo[a]Pyrene in Human Bronchoalveolar Cell Extracts. Chem Res Toxicol 2005, 18, 365–374. [CrossRef]
- Jiang, H.; Vudathala, D.K.; Blair, I.A.; Penning, T.M. Competing Roles of Aldo-Keto Reductase 1A1 and Cytochrome P4501B1 in Benzo[a]Pyrene-7,8-Diol Activation in Human Bronchoalveolar H358 Cells: Role of AKRs in P4501B1 Induction. Chem Res Toxicol 2006, 19, 68–78. [CrossRef]
- Shimada, T. Xenobiotic-Metabolizing Enzymes Involved in Activation and Detoxification of Carcinogenic Polycyclic Aromatic Hydrocarbons. Drug Metab Pharmacokinet 2006, 21, 257–276. [CrossRef]
- Schmalix, W.A.; Mäser, H.; Kiefer, F.; Reen, R.; Wiebel, F.J.; Gonzalez, F.; Seidel, A.; Glatt, H.; Greim, H.; Doehmer, J. Stable Expression of Human Cytochrome P450 1A1 cDNA in V79 Chinese Hamster Cells and Metabolic Activation of Benzo[a]Pyrene. Eur J Pharmacol 1993, 248, 251–261. [CrossRef]
- Aoyama, T.; Gonzalez, F.J.; Gelboin, H.V. Human cDNA-Expressed Cytochrome P450 IA2: Mutagen Activation and Substrate Specificity. Mol Carcinog 1989, 2, 192–198. [CrossRef]
- Shimada, T.; Guengerich, F.P. Inhibition of Human Cytochrome P450 1A1-, 1A2-, and 1B1-Mediated Activation of Procarcinogens to Genotoxic Metabolites by Polycyclic Aromatic Hydrocarbons. Chem Res Toxicol 2006, 19, 288–294. [CrossRef]
- Gautier, J.C.; Lecoeur, S.; Cosme, J.; Perret, A.; Urban, P.; Beaune, P.; Pompon, D. Contribution of Human Cytochrome P450 to Benzo[a]Pyrene and Benzo[a]Pyrene-7,8-Dihydrodiol Metabolism, as Predicted from Heterologous Expression in Yeast. Pharmacogenetics 1996, 6, 489–499. [CrossRef]
- Wu, Z.-L.; Sohl, C.D.; Shimada, T.; Guengerich, F.P. Recombinant Enzymes Overexpressed in Bacteria Show Broad Catalytic Specificity of Human Cytochrome P450 2W1 and Limited Activity of Human Cytochrome P450 2S1. Mol Pharmacol 2006, 69, 2007–2014. [CrossRef]
- Guengerich, F.P.; Chun, Y.-J.; Kim, D.; Gillam, E.M.J.; Shimada, T. Cytochrome P450 1B1: A Target for Inhibition in Anticarcinogenesis Strategies. Mutat Res 2003, 523–524, 173–182. [CrossRef]
- Mammen, J.S.; Pittman, G.S.; Li, Y.; Abou-Zahr, F.; Bejjani, B.A.; Bell, D.A.; Strickland, P.T.; Sutter, T.R. Single Amino Acid Mutations, but Not Common Polymorphisms, Decrease the Activity of CYP1B1 against (-)Benzo[a]Pyrene-7R-Trans-7,8-Dihydrodiol. Carcinogenesis 2003, 24, 1247–1255. [CrossRef]
- Meinl, W.; Donath, C.; Schneider, H.; Sommer, Y.; Glatt, H. SULT1C3, an Orphan Sequence of the Human Genome, Encodes an Enzyme Activating Various Promutagens. Food Chem Toxicol 2008, 46, 1249–1256. [CrossRef]
- Palackal, N.T.; Burczynski, M.E.; Harvey, R.G.; Penning, T.M. Metabolic Activation of Polycyclic Aromatic Hydrocarbon Trans-Dihydrodiols by Ubiquitously Expressed Aldehyde Reductase (AKR1A1). Chem Biol Interact 2001, 130–132, 815–824. [CrossRef]
- Palackal, N.T.; Burczynski, M.E.; Harvey, R.G.; Penning, T.M. The Ubiquitous Aldehyde Reductase (AKR1A1) Oxidizes Proximate Carcinogen Trans-Dihydrodiols to o-Quinones: Potential Role in Polycyclic Aromatic Hydrocarbon Activation. Biochemistry 2001, 40, 10901–10910. [CrossRef]
- Cheung, Y. L.; Gray, T. J.; Ioannides, C. Mutagenicity of chrysene, its methyl and benzo derivatives, and their interactions with cytochromes P-450 and the Ah-receptor; relevance to their carcinogenic potency. Toxicology 1993, 81, 69–86. [CrossRef]
- Chae, Y.H.; Yun, C.H.; Guengerich, F.P.; Kadlubar, F.F.; el-Bayoumy, K. Roles of Human Hepatic and Pulmonary Cytochrome P450 Enzymes in the Metabolism of the Environmental Carcinogen 6-Nitrochrysene. Cancer Res 1993, 53, 2028–2034, PMID: 8481905.
- Murata, M.; Ohnishi, S.; Seike, K.; Fukuhara, K.; Miyata, N.; Kawanishi, S. Oxidative DNA Damage Induced by Carcinogenic Dinitropyrenes in the Presence of P450 Reductase. Chem Res Toxicol 2004, 17, 1750–1756. [CrossRef]
- Guengerich, F.P.; Shimada, T.; Raney, K.D.; Yun, C.H.; Meyer, D.J.; Ketterer, B.; Harris, T.M.; Groopman, J.D.; Kadlubar, F.F. Elucidation of Catalytic Specificities of Human Cytochrome P450 and Glutathione S-Transferase Enzymes and Relevance to Molecular Epidemiology. Environ Health Perspect 1992, 98, 75–80. [CrossRef]
- Yamada, K.; Suzuki, T.; Hakura, A.; Mizutani, T.; Saeki, K. Metabolic Activation of 10-Aza-Substituted Benzo[a]Pyrene by Cytochrome P450 1A2 in Human Liver Microsomes. Mutat Res 2004, 557, 159–165. [CrossRef]
- Glatt, H. Sulfotransferases in the Bioactivation of Xenobiotics. Chem Biol Interact 2000, 129, 141–170. [CrossRef]
- Duarte, M.P.; Palma, B.B.; Gilep, A.A.; Laires, A.; Oliveira, J.S.; Usanov, S.A.; Rueff, J.; Kranendonk, M. The Stimulatory Role of Human Cytochrome B5 in the Bioactivation Activities of Human CYP1A2, 2A6 and 2E1: A New Cell Expression System to Study Cytochrome P450 Mediated Biotransformation. Mutagenesis 2005, 20, 93–100. [CrossRef]
- Duarte, M.P.; Palma, B.B.; Laires, A.; Oliveira, J.S.; Rueff, J.; Kranendonk, M. Escherichia Coli BTC, a Human Cytochrome P450 Competent Tester Strain with a High Sensitivity towards Alkylating Agents: Involvement of Alkyltransferases in the Repair of DNA Damage Induced by Aromatic Amines. Mutagenesis 2005, 20, 199–208. [CrossRef]
- Duarte, M.P.; Palma, B.B.; Gilep, A.A.; Laires, A.; Oliveira, J.S.; Usanov, S.A.; Rueff, J.; Kranendonk, M. The Stimulatory Role of Human Cytochrome B5 in the Bioactivation Activities of Human CYP1A2, 2A6 and 2E1: A New Cell Expression System to Study Cytochrome P450-Mediated Biotransformation (a Corrigendum Report on Duarte et al. (2005) Mutagenesis 20, 93-100). Mutagenesis 2007, 22, 75–81. [CrossRef]
- H. Sulfation and Sulfotransferases 4: Bioactivation of Mutagens via Sulfation. FASEB J 1997, 11, 314–321. [CrossRef]
- Kreis, P.; Brandner, S.; Coughtrie, M.W.; Pabel, U.; Meinl, W.; Glatt, H.; Andrae, U. Human Phenol Sulfotransferases hP-PST and hM-PST Activate Propane 2-Nitronate to a Genotoxicant. Carcinogenesis 2000, 21, 295–299. [CrossRef]
- Glatt, H.; Meinl, W. Use of Genetically Manipulated Salmonella Typhimurium Strains to Evaluate the Role of Sulfotransferases and Acetyltransferases in Nitrofen Mutagenicity. Carcinogenesis 2004, 25, 779–786. [CrossRef]
- Meinl, W.; Meerman, J.H.N.; Glatt, H. Differential Activation of Promutagens by Alloenzymes of Human Sulfotransferase 1A2 Expressed in Salmonella Typhimurium. Pharmacogenetics 2002, 12, 677–689. [CrossRef]
- Sun, Y.-W.; Guengerich, F.P.; Sharma, A.K.; Boyiri, T.; Amin, S.; el-Bayoumy, K. Human Cytochromes P450 1A1 and 1B1 Catalyze Ring Oxidation but Not Nitroreduction of Environmental Pollutant Mononitropyrene Isomers in Primary Cultures of Human Breast Cells and Cultured MCF-10A and MCF-7 Cell Lines. Chem Res Toxicol 2004, 17, 1077–1085. [CrossRef]
- Shimada, T.; Iwasaki, M.; Martin, M.V.; Guengerich, F.P. Human Liver Microsomal Cytochrome P-450 Enzymes Involved in the Bioactivation of Procarcinogens Detected by Umu Gene Response in Salmonella Typhimurium TA 1535/pSK1002. Cancer Res 1989, 49, 3218–3228, PMEDID 2655891.
- Ahmad, S.; Kabler, S.L.; Rudd, L.; Amin, S.; Doehmer, J.; Morrow, C.S.; Townsend, A.J. Cytotoxicity and Mutagenicity of 5-Methylchrysene and Its 1,2-Dihydrodiol in V79MZ Cells Modified to Express Human CYP1A1 or CYP1B1, in the Presence or Absence of Human GSTP1 Coexpression. Toxicol Lett 2008, 183, 99–104. [CrossRef]
- Yamazaki, Y.; Fujita, K.-I.; Nakayama, K.; Suzuki, A.; Nakamura, K.; Yamazaki, H.; Kamataki, T. Establishment of Ten Strains of Genetically Engineered Salmonella Typhimurium TA1538 Each Co-Expressing a Form of Human Cytochrome P450 with NADPH-Cytochrome P450 Reductase Sensitive to Various Promutagens. Mutat Res 2004, 562, 151–162. [CrossRef]
- Yueh, M.F.; Nguyen, N.; Famourzadeh, M.; Strassburg, C.P.; Oda, Y.; Guengerich, F.P.; Tukey, R.H. The Contribution of UDP-Glucuronosyltransferase 1A9 on CYP1A2-Mediated Genotoxicity by Aromatic and Heterocyclic Amines. Carcinogenesis 2001, 22, 943–950. [CrossRef]
- Oda, Y. Analysis of the Involvement of Human N-Acetyltransferase 1 in the Genotoxic Activation of Bladder Carcinogenic Arylamines Using a SOS/Umu Assay System. Mutat Res 2004, 554, 399–406. [CrossRef]
- Oda, Y.; Aryal, P.; Terashita, T.; Gillam, E.M.; Guengerich, F.P.; Shimada, T. Metabolic Activation of Heterocyclic Amines and Other Procarcinogens in Salmonella Typhimurium Umu Tester Strains Expressing Human Cytochrome P4501A1, 1A2, 1B1, 2C9, 2D6, 2E1, and 3A4 and Human NADPH-P450 Reductase and Bacterial O-Acetyltransferase. Mutat Res 2001, 492, 81–90. [CrossRef]
- Kranendonk, M.; Fisher, C.W.; Roda, R.; Carreira, F.; Theisen, P.; Laires, A.; Rueff, J.; Vermeulen, N.P.; Estabrook, R.W. Escherichia Coli MTC, a NADPH Cytochrome P450 Reductase Competent Mutagenicity Tester Strain for the Expression of Human Cytochrome P450: Comparison of Three Types of Expression Systems. Mutat Res 1999, 439, 287–300. [CrossRef]
- Josephy, P.D.; Evans, D.H.; Parikh, A.; Guengerich, F.P. Metabolic Activation of Aromatic Amine Mutagens by Simultaneous Expression of Human Cytochrome P450 1A2, NADPH-Cytochrome P450 Reductase, and N-Acetyltransferase in Escherichia Coli. Chem Res Toxicol 1998, 11, 70–74. [CrossRef]
- Gillam, E.M.; Wunsch, R.M.; Ueng, Y.F.; Shimada, T.; Reilly, P.E.; Kamataki, T.; Guengerich, F.P. Expression of Cytochrome P450 3A7 in Escherichia Coli: Effects of 5’ Modification and Catalytic Characterization of Recombinant Enzyme Expressed in Bicistronic Format with NADPH-Cytochrome P450 Reductase. Arch Biochem Biophys 1997, 346, 81–90. [CrossRef]
- Imaoka, S.; Hayashi, K.; Hiroi, T.; Yabusaki, Y.; Kamataki, T.; Funae, Y. A Transgenic Mouse Expressing Human CYP4B1 in the Liver. Biochem Biophys Res Commun 2001, 284, 757–762. [CrossRef]
- Imaoka, S.; Yoneda, Y.; Sugimoto, T.; Hiroi, T.; Yamamoto, K.; Nakatani, T.; Funae, Y. CYP4B1 Is a Possible Risk Factor for Bladder Cancer in Humans. Biochem Biophys Res Commun 2000, 277, 776–780. [CrossRef]
- Grant, D.M.; Josephy, P.D.; Lord, H.L.; Morrison, L.D. Salmonella Typhimurium Strains Expressing Human Arylamine N-Acetyltransferases: Metabolism and Mutagenic Activation of Aromatic Amines. Cancer Res 1992, 52, 3961–3964, PMID: 1617672.
- Oda, Y.; Yamazaki, H.; Shimada, T. Role of Human N-Acetyltransferases, NAT1 or NAT2, in Genotoxicity of Nitroarenes and Aromatic Amines in Salmonella Typhimurium NM6001 and NM6002. Carcinogenesis 1999, 20, 1079–1083. [CrossRef]
- Sasaki, J.C.; Arey, J.; Eastmond, D.A.; Parks, K.K.; Phousongphouang, P.T.; Grosovsky, A.J. Evidence for Oxidative Metabolism in the Genotoxicity of the Atmospheric Reaction Product 2-Nitronaphthalene in Human Lymphoblastoid Cell Lines. Mutat Res 1999, 445, 113–125. [CrossRef]
- Oda, Y.; Hirayama, T.; Watanabe, T. Genotoxic Activation of the Environmental Pollutant 3,6-Dinitrobenzo[e]Pyrene in Salmonella Typhimurium Umu Strains Expressing Human Cytochrome P450 and N-Acetyltransferase. Toxicol Lett 2009, 188, 258–262. [CrossRef]
- Kawanishi, M.; Watanabe, T.; Hagio, S.; Ogo, S.; Shimohara, C.; Jouchi, R.; Takayama, S.; Hasei, T.; Hirayama, T.; Oda, Y.; et al. Genotoxicity of 3,6-Dinitrobenzo[e]Pyrene, a Novel Mutagen in Ambient Air and Surface Soil, in Mammalian Cells in Vitro and in Vivo. Mutagenesis 2009, 24, 279–284. [CrossRef]
- Oda, Y.; Zhang, Y.; Buchinger, S.; Reifferscheid, G.; Yang, M. Roles of Human Sulfotransferases in Genotoxicity of Carcinogens Using Genetically Engineered Umu Test Strains. Environ Mol Mutagen 2012, 53, 152–164. [CrossRef]
- Butler, M.A.; Iwasaki, M.; Guengerich, F.P.; Kadlubar, F.F. Human Cytochrome P-450PA (P-450IA2), the Phenacetin O-Deethylase, Is Primarily Responsible for the Hepatic 3-Demethylation of Caffeine and N-Oxidation of Carcinogenic Arylamines. Proc Natl Acad Sci U S A 1989, 86, 7696–7700. [CrossRef]
- Stiborová, M.; Naiman, K.; Martínková, M.; Martínek, V.; Svobodová, M.; Schmeiser, H.H.; Frei, E. Genotoxic Mechanisms for the Carcinogenicity of the Environmental Pollutants and Carcinogens O-Anisidine and 2-Nitroanisole Follow from Adducts Generated by Their Metabolite N-(2-Methoxyphenyl)-Hydroxylamine with Deoxyguanosine in DNA. Interdiscip Toxicol 2009, 2, 24–27. [CrossRef]
- Arlt, V.M.; Glatt, H.; Gamboa da Costa, G.; Reynisson, J.; Takamura-Enya, T.; Phillips, D.H. Mutagenicity and DNA Adduct Formation by the Urban Air Pollutant 2-Nitrobenzanthrone. Toxicol Sci 2007, 98, 445–457. [CrossRef]
- Arlt, V.M.; Glatt, H.; Muckel, E.; Pabel, U.; Sorg, B.L.; Seidel, A.; Frank, H.; Schmeiser, H.H.; Phillips, D.H. Activation of 3-Nitrobenzanthrone and Its Metabolites by Human Acetyltransferases, Sulfotransferases and Cytochrome P450 Expressed in Chinese Hamster V79 Cells. Int J Cancer 2003, 105, 583–592. [CrossRef]
- Arlt, V.M.; Stiborova, M.; Hewer, A.; Schmeiser, H.H.; Phillips, D.H. Human Enzymes Involved in the Metabolic Activation of the Environmental Contaminant 3-Nitrobenzanthrone: Evidence for Reductive Activation by Human NADPH:Cytochrome P450 Reductase. Cancer Res 2003, 63, 2752–2761, PMID: 12782579.
- Stiborová, M.; Arlt, V.M.; Henderson, C.J.; Wolf, C.R.; Frei, E.; Schmeiser, H.H.; Phillips, D.H. Molecular Mechanism of Genotoxicity of the Environmental Pollutant 3-Nitrobenzanthrone. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2005, 149, 191–197. [CrossRef]
- Arlt, V.M.; Henderson, C.J.; Wolf, C.R.; Schmeiser, H.H.; Phillips, D.H.; Stiborova, M. Bioactivation of 3-Aminobenzanthrone, a Human Metabolite of the Environmental Pollutant 3-Nitrobenzanthrone: Evidence for DNA Adduct Formation Mediated by Cytochrome P450 Enzymes and Peroxidases. Cancer Lett 2006, 234, 220–231. [CrossRef]
- Arlt, V.M.; Hewer, A.; Sorg, B.L.; Schmeiser, H.H.; Phillips, D.H.; Stiborova, M. 3-Aminobenzanthrone, a Human Metabolite of the Environmental Pollutant 3-hatanakaRat Liver Microsomes: Evidence for Activation by Cytochrome P450 1A1 and P450 1A2. Chem Res Toxicol 2004, 17, 1092–1101. [CrossRef]
- Wu, J.; Dong, H.; Cai, Z.; Yu, Y. Stable Expression of Human Cytochrome CYP2B6 and CYP1A1 in Chinese Hamster CHL Cells: Their Use in Micronucleus Assays. Chin Med Sci J 1997, 12, 148–155, PMID: 11360624.
- Arlt, V.M.; Glatt, H.; Muckel, E.; Pabel, U.; Sorg, B.L.; Schmeiser, H.H.; Phillips, D.H. Metabolic Activation of the Environmental Contaminant 3-Nitrobenzanthrone by Human Acetyltransferases and Sulfotransferase. Carcinogenesis 2002, 23, 1937–1945. [CrossRef]
- Koehl, W.; Amin, S.; Staretz, M.E.; Ueng, Y.F.; Yamazaki, H.; Tateishi, T.; Guengerich, F.P.; Hecht, S.S. Metabolism of 5-Methylchrysene and 6-Methylchrysene by Human Hepatic and Pulmonary Cytochrome P450 Enzymes. Cancer Res 1996, 56, 316–324, PMID: 8542586.
- Yamazaki, H.; Mimura, M.; Oda, Y.; Inui, Y.; Shiraga, T.; Iwasaki, K.; Guengerich, F.P.; Shimada, T. Roles of Different Forms of Cytochrome P450 in the Activation of the Promutagen 6-Aminochrysene to Genotoxic Metabolites in Human Liver Microsomes. Carcinogenesis 1993, 14, 1271–1278. [CrossRef]
- Yamazaki, H.; Mimura, M.; Oda, Y.; Gonzalez, F.J.; el-Bayoumy, K.; Chae, Y.H.; Guengerich, F.P.; Shimada, T. Activation of Trans-1,2-Dihydro-1,2-Dihydroxy-6-Aminochrysene to Genotoxic Metabolites by Rat and Human Cytochromes P450. Carcinogenesis 1994, 15, 465–470. [CrossRef]
- Yun, C.H.; Shimada, T.; Guengerich, F.P. Purification and Characterization of Human Liver Microsomal Cytochrome P-450 2A6. Mol Pharmacol 1991, 40, 679–685, PMID: 1944238.
- Mimura, M.; Baba, T.; Yamazaki, H.; Ohmori, S.; Inui, Y.; Gonzalez, F.J.; Guengerich, F.P.; Shimada, T. Characterization of Cytochrome P-450 2B6 in Human Liver Microsomes. Drug Metab Dispos 1993, 21, 1048–1056. PMID: 7905383.
- Hashizume, T.; Yoshitomi, S.; Asahi, S.; Uematsu, R.; Matsumura, S.; Chatani, F.; Oda, H. Advantages of Human Hepatocyte-Derived Transformants Expressing a Series of Human Cytochrome P450 Isoforms for Genotoxicity Examination. Toxicol Sci 2010, 116, 488–497. [CrossRef]
- Buters, J.; Quintanilla-Martinez, L.; Schober, W.; Soballa, V.J.; Hintermair, J.; Wolff, T.; Gonzalez, F.J.; Greim, H. CYP1B1 Determines Susceptibility to Low Doses of 7,12-Dimethylbenz[a]Anthracene-Induced Ovarian Cancers in Mice: Correlation of CYP1B1-Mediated DNA Adducts with Carcinogenicity. Carcinogenesis 2003, 24, 327–334. [CrossRef]
- Roberts-Thomson, S.J.; McManus, M.E.; Tukey, R.H.; Gonzalez, F.J.; Holder, G.M. Metabolism of Polycyclic Aza-Aromatic Carcinogens Catalyzed by Four Expressed Human Cytochromes P450. Cancer Res 1995, 55, 1052–1059, PMID: 7866988.
- Yoshitomi, S.; Ikemoto, K.; Takahashi, J.; Miki, H.; Namba, M.; Asahi, S. Establishment of the Transformants Expressing Human Cytochrome P450 Subtypes in HepG2, and Their Applications on Drug Metabolism and Toxicology. Toxicol In Vitro 2001, 15, 245–256. [CrossRef]
- Durant, J.L.; Lafleur, A.L.; Busby, W.F.; Donhoffner, L.L.; Penman, B.W.; Crespi, C.L. Mutagenicity of C24H14 PAH in Human Cells Expressing CYP1A1. Mutat Res 1999, 446, 1–14. [CrossRef]
- Kim, J.H.; Stansbury, K.H.; Walker, N.J.; Trush, M.A.; Strickland, P.T.; Sutter, T.R. Metabolism of Benzo[a]Pyrene and Benzo[a]Pyrene-7,8-Diol by Human Cytochrome P450 1B1. Carcinogenesis 1998, 19, 1847–1853. [CrossRef]
- Shimada, T.; Wunsch, R.M.; Hanna, I.H.; Sutter, T.R.; Guengerich, F.P.; Gillam, E.M. Recombinant Human Cytochrome P450 1B1 Expression in Escherichia Coli. Arch Biochem Biophys 1998, 357, 111–120. [CrossRef]
- Guo, Z.; Gillam, E.M.; Ohmori, S.; Tukey, R.H.; Guengerich, F.P. Expression of Modified Human Cytochrome P450 1A1 in Escherichia Coli: Effects of 5’ Substitution, Stabilization, Purification, Spectral Characterization, and Catalytic Properties. Arch Biochem Biophys 1994, 312, 436–446. [CrossRef]
- Penman, B.W.; Chen, L.; Gelboin, H.V.; Gonzalez, F.J.; Crespi, C.L. Development of a Human Lymphoblastoid Cell Line Constitutively Expressing Human CYP1A1 cDNA: Substrate Specificity with Model Substrates and Promutagens. Carcinogenesis 1994, 15, 1931–1937. [CrossRef]
- Sengstag, C.; Eugster, H.P.; Würgler, F.E. High Promutagen Activating Capacity of Yeast Microsomes Containing Human Cytochrome P-450 1A and Human NADPH-Cytochrome P-450 Reductase. Carcinogenesis 1994, 15, 837–843. [CrossRef]
- Hashizume, T.; Yoshitomi, S.; Asahi, S.; Matsumura, S.; Chatani, F.; Oda, H. In Vitro Micronucleus Test in HepG2 Transformants Expressing a Series of Human Cytochrome P450 Isoforms with Chemicals Requiring Metabolic Activation. Mutat Res 2009, 677, 1–7. [CrossRef]
- Aklillu, E.; Øvrebø, S.; Botnen, I.V.; Otter, C.; Ingelman-Sundberg, M. Characterization of Common CYP1B1 Variants with Different Capacity for Benzo[a]Pyrene-7,8-Dihydrodiol Epoxide Formation from Benzo[a]Pyrene. Cancer Res 2005, 65, 5105–5111. [CrossRef]
- Baum, M.; Amin, S.; Guengerich, F.P.; Hecht, S.S.; Köhl, W.; Eisenbrand, G. Metabolic Activation of Benzo[c]Phenanthrene by Cytochrome P450 Enzymes in Human Liver and Lung. Chem Res Toxicol 2001, 14, 686–693. [CrossRef]
- Einolf, H.J.; Story, W.T.; Marcus, C.B.; Larsen, M.C.; Jefcoate, C.R.; Greenlee, W.F.; Yagi, H.; Jerina, D.M.; Amin, S.; Park, S.S.; et al. Role of Cytochrome P450 Enzyme Induction in the Metabolic Activation of Benzo[c]Phenanthrene in Human Cell Lines and Mouse Epidermis. Chem Res Toxicol 1997, 10, 609–617. [CrossRef]
- Seidel, A.; Soballa, V.J.; Raab, G.; Frank, H.; Greim, H.; Grimmer, G.; Jacob, J.; Doehmer, J. Regio- and Stereoselectivity in the Metabolism of Benzo[c]Phenanthrene Mediated by Genetically Engineered V79 Chinese Hamster Cells Expressing Rat and Human Cytochromes P450. Environ Toxicol Pharmacol 1998, 5, 179–196. [CrossRef]
- Crespi, C.L.; Penman, B.W.; Steimel, D.T.; Smith, T.; Yang, C.S.; Sutter, T.R. Development of a Human Lymphoblastoid Cell Line Constitutively Expressing Human CYP1B1 cDNA: Substrate Specificity with Model Substrates and Promutagens. Mutagenesis 1997, 12, 83–89. [CrossRef]
- Yuan, Z.-X.; Kumar, S.; Sikka, H.C. Comparative Metabolism of the Aza Polynuclear Aromatic Hydrocarbon Dibenz[a,h]Acridine by Recombinant Human and Rat Cytochrome P450s. Chem Res Toxicol 2004, 17, 672–678. [CrossRef]
- Shou, M.; Krausz, K.W.; Gonzalez, F.J.; Gelboin, H.V. Metabolic Activation of the Potent Carcinogen Dibenzo[a,h]anthracene by cDNA-Expressed Human Cytochromes P450. Arch Biochem Biophys 1996, 328, 201–207. [CrossRef]
- Luch, A.; Coffing, S.L.; Tang, Y.M.; Schneider, A.; Soballa, V.; Greim, H.; Jefcoate, C.R.; Seidel, A.; Greenlee, W.F.; Baird, W.M.; et al. Stable Expression of Human Cytochrome P450 1B1 in V79 Chinese Hamster Cells and Metabolically Catalyzed DNA Adduct Formation of Dibenzo[a,l]Pyrene. Chem Res Toxicol 1998, 11, 686–695. [CrossRef]
- Luch, A.; Kishiyama, S.; Seidel, A.; Doehmer, J.; Greim, H.; Baird, W.M. The K-Region Trans-8,9-Diol Does Not Significantly Contribute as an Intermediate in the Metabolic Activation of Dibenzo[a,l]Pyrene to DNA-Binding Metabolites by Human Cytochrome P450 1A1 or 1B1. Cancer Res 1999, 59, 4603–4609, PMID: 10493514.
- Luch, A.; Schober, W.; Soballa, V.J.; Raab, G.; Greim, H.; Jacob, J.; Doehmer, J.; Seidel, A. Metabolic Activation of Dibenzo[a,l]Pyrene by Human Cytochrome P450 1A1 and P450 1B1 Expressed in V79 Chinese Hamster Cells. Chem Res Toxicol 1999, 12, 353–364. [CrossRef]
- Melendez-Colon, V.J.; Luch, A.; Seidel, A.; Baird, W.M. Comparison of Cytochrome P450- and Peroxidase-Dependent Metabolic Activation of the Potent Carcinogen Dibenzo[a,l]Pyrene in Human Cell Lines: Formation of Stable DNA Adducts and Absence of a Detectable Increase in Apurinic Sites. Cancer Res 1999, 59, 1412–1416.
- Melendez-Colon, V.J.; Luch, A.; Seidel, A.; Baird, W.M. Formation of Stable DNA Adducts and Apurinic Sites upon Metabolic Activation of Bay and Fjord Region Polycyclic Aromatic Hydrocarbons in Human Cell Cultures. Chem Res Toxicol 2000, 13, 10–17. [CrossRef]
- King, L.C.; Adams, L.; Allison, J.; Kohan, M.J.; Nelson, G.; Desai, D.; Amin, S.; Ross, J.A. A Quantitative Comparison of Dibenzo[a,l]Pyrene-DNA Adduct Formation by Recombinant Human Cytochrome P450 Microsomes. Mol Carcinog 1999, 26, 74–82, PMID: 10506751.
- Shou, M.; Krausz, K.W.; Gonzalez, F.J.; Gelboin, H.V. Metabolic Activation of the Potent Carcinogen Dibenzo[a,l]Pyrene by Human Recombinant Cytochromes P450, Lung and Liver Microsomes. Carcinogenesis 1996, 17, 2429–2433. [CrossRef]
- Schober, W.; Luch, A.; Soballa, V.J.; Raab, G.; Stegeman, J.J.; Doehmer, J.; Jacob, J.; Seidel, A. On the Species-Specific Biotransformation of Dibenzo[a,l]Pyrene. Chem Biol Interact 2006, 161, 37–48. [CrossRef]
- Lanza, D.L.; Code, E.; Crespi, C.L.; Gonzalez, F.J.; Yost, G.S. Specific Dehydrogenation of 3-Methylindole and Epoxidation of Naphthalene by Recombinant Human CYP2F1 Expressed in Lymphoblastoid Cells. Drug Metab Dispos 1999, 27, 798–803, PMID: 10383923.
- Chou, H.C.; Lang, N.P.; Kadlubar, F.F. Metabolic Activation of N-Hydroxy Arylamines and N-Hydroxy Heterocyclic Amines by Human Sulfotransferase(s). Cancer Res 1995, 55, 525–529, PMID: 7834621.
- Schober, W.; Pusch, G.; Oeder, S.; Reindl, H.; Behrendt, H.; Buters, J.T.M. Metabolic Activation of Phenanthrene by Human and Mouse Cytochromes P450 and Pharmacokinetics in CYP1A2 Knockout Mice. Chem Biol Interact 2010, 183, 57–66. [CrossRef]
- Kisselev, P.; Schwarz, D.; Platt, K.-L.; Schunck, W.-H.; Roots, I. Epoxidation of Benzo[a]Pyrene-7,8-Dihydrodiol by Human CYP1A1 in Reconstituted Membranes. Effects of Charge and Nonbilayer Phase Propensity of the Membrane. Eur J Biochem 2002, 269, 1799–1805. [CrossRef]
- Mahadevan, B.; Luch, A.; Atkin, J.; Haynes, M.; Nguyen, T.; Baird, W.M. Inhibition of Human Cytochrome P450 1b1 Further Clarifies Its Role in the Activation of Dibenzo[a,l]Pyrene in Cells in Culture. J Biochem Mol Toxicol 2007, 21, 101–109. [CrossRef]
- Shimada, T.; Gillam, E.M.; Oda, Y.; Tsumura, F.; Sutter, T.R.; Guengerich, F.P.; Inoue, K. Metabolism of Benzo[a]Pyrene to Trans-7,8-Dihydroxy-7, 8-Dihydrobenzo[a]Pyrene by Recombinant Human Cytochrome P450 1B1 and Purified Liver Epoxide Hydrolase. Chem Res Toxicol 1999, 12, 623–629. [CrossRef]
- Hein, D.W.; Doll, M.A.; Rustan, T.D.; Gray, K.; Feng, Y.; Ferguson, R.J.; Grant, D.M. Metabolic Activation and Deactivation of Arylamine Carcinogens by Recombinant Human NAT1 and Polymorphic NAT2 Acetyltransferases. Carcinogenesis 1993, 14, 1633–1638. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




