Submitted:
20 November 2024
Posted:
21 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
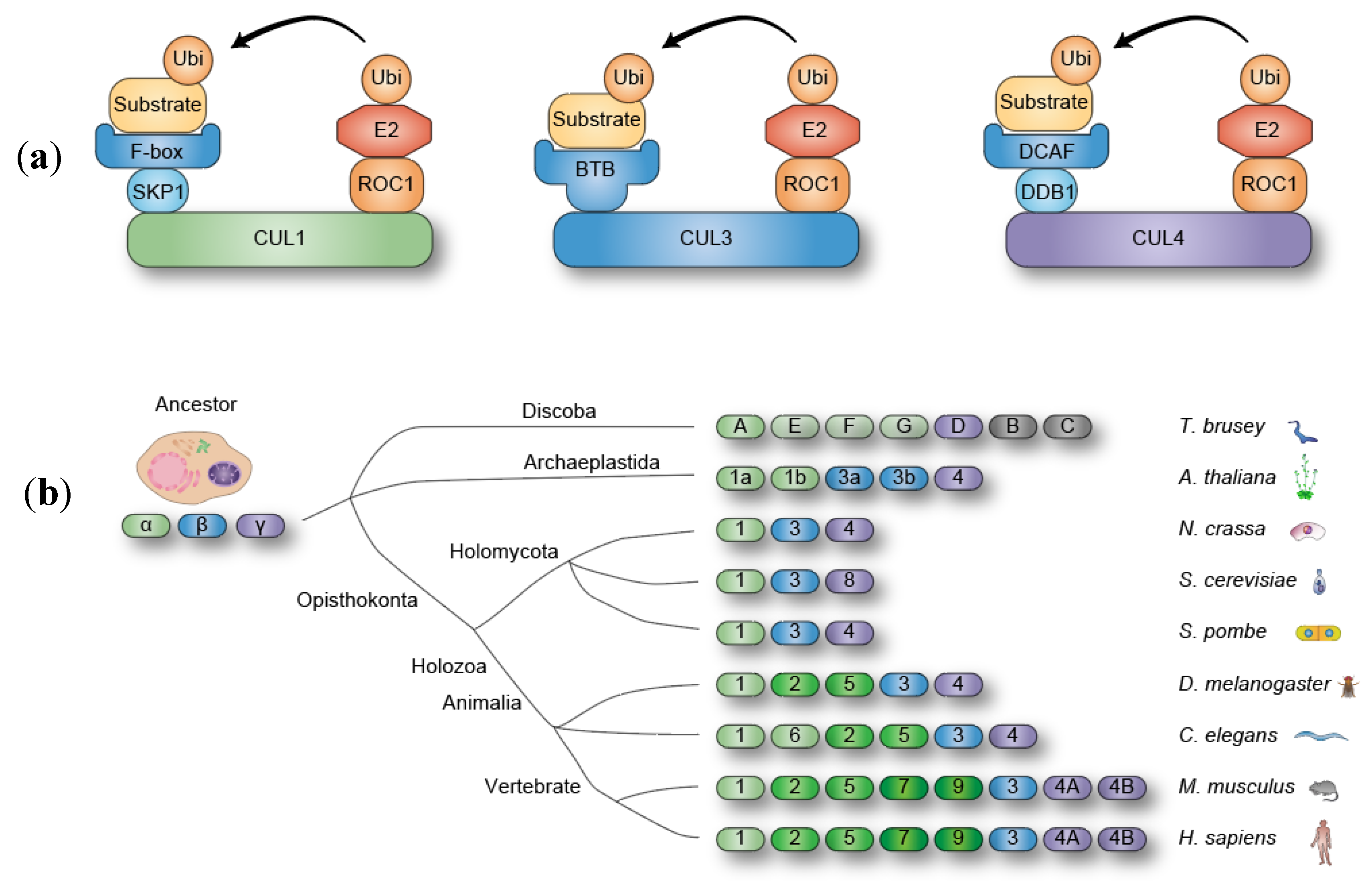
2. CRL4 Structure
3. Organismal Phenotypes of CUL4 Mutation
4. CRL4 Functions in Trypanosoma
5. CRL4 Functions in Plants
6. CRL4 Functions in Fungi
7. CRL4 Functions in Worms
8. CRL4 Functions in Flies
9. CRL4 Functions in Mammals
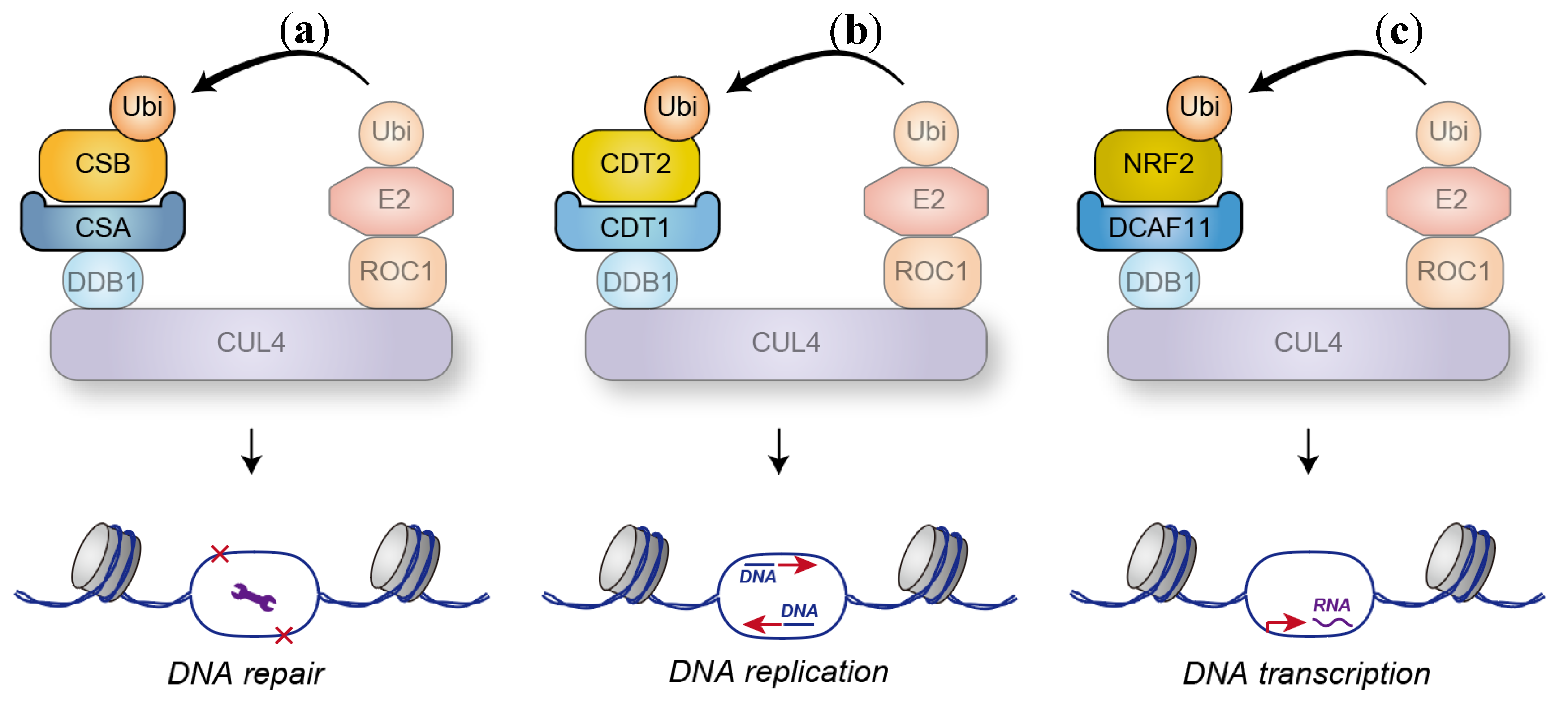
9.1. DNA Modification-Related Proteins
9.2. Histone Modification-Related Enzymes
9.3. Other Substrates Targeted by Nuclear DCAFs
10. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Labbadia, J.; Morimoto, R.I. The biology of proteostasis in aging and disease. Annu Rev Biochem 2015, 84, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; P. Kasturi,; Hartl, F.U. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Bio 2019, 20, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Verma, K.; et al. Recent advances in understanding the role of proteostasis. Fac Rev 2021, 10, 72. [Google Scholar] [CrossRef]
- Liu, F.; Walters, K.J. Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci 2010, 35, 352–60. [Google Scholar] [CrossRef]
- Fulda, S.; Rajalingam, K.; Dikic, I. Ubiquitylation in immune disorders and cancer: from molecular mechanisms to therapeutic implications. EMBO Mol Med 2012, 4, 545–56. [Google Scholar] [CrossRef]
- Iwai, K.; Fujita, H.; Sasaki, Y. Linear ubiquitin chains: NF-κB signalling, cell death and beyond. Nat Rev Mol Cell Bio 2014, 15, 503–508. [Google Scholar] [CrossRef]
- Nakagawa, T.; Nakayama, K. Protein monoubiquitylation: targets and diverse functions. Genes Cells 2015, 20, 543–562. [Google Scholar] [CrossRef]
- Choudhary, C.; Mann, M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 2010, 11, 427–439. [Google Scholar] [CrossRef]
- Steger, M.; Karayel, Ö.; Demichev, V. Ubiquitinomics: History, methods, and applications in basic research and drug discovery. Proteomics 2022, 22, e2200074. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Liao, S.; et al. The Protein Neddylation Pathway in Trypanosoma brucei: FUNCTIONAL CHARACTERIZATION AND SUBSTRATE IDENTIFICATION. J Biol Chem 2017, 292, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Marín, I. , Diversification of the cullin family. BMC Evol Biol 2009, 9, 267. [Google Scholar] [CrossRef]
- Sarikas, A.; Hartmann, T.; Pan, Z.Q. The cullin protein family. Genome Biol, 2011. 12(4): p. 220.
- Del Pino R.C. et al. Evolution of cullin E3 ubiquitin ligases and function in trypanosomes. bioRxiv [Preprint] 2023, 2023.07.24.550360.
- Jackson, S.; Xiong, Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 2009, 34, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Nag, A. CUL4A ubiquitin ligase: a promising drug target for cancer and other human diseases. Open Biol 2014, 4, 130217. [Google Scholar] [CrossRef]
- Hannah, J.; Zhou, P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene 2015, 573, 33–45. [Google Scholar] [CrossRef]
- Zou, Y.; et al. Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem 2009, 284, 33320–32. [Google Scholar] [CrossRef]
- Nakagawa, T.; Xiong, Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol Cell 2011, 43, 381–91. [Google Scholar] [CrossRef]
- Kapetanaki, M.G.; et al. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A 2006, 103, 2588–2593. [Google Scholar] [CrossRef]
- Guerrero-Santoro, J.; et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res 2008, 68, 5014–5022. [Google Scholar] [CrossRef]
- Chen, H.; et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 2006, 18, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; et al. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.Q.; et al. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 2000, 5, 169–190. [Google Scholar] [CrossRef]
- Matsuyama, A.; et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 2006, 24, 841–847. [Google Scholar] [CrossRef]
- Swenson, J.M.; et al. The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic. Elife, 2016, 5, e16096. [Google Scholar] [CrossRef]
- Alleva, B.; et al. CRL4 regulates recombination and synaptonemal complex aggregation in the Caenorhabditis elegans germline. PLoS Genet 2019, 15, e1008486. [Google Scholar] [CrossRef]
- Lee, J.; Zhou, P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 2007, 26, 775–780. [Google Scholar] [CrossRef]
- Pan, Z.Q.; et al. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 2004, 23, 1985–1997. [Google Scholar] [CrossRef]
- Schulze-Niemand, E.; Naumann, M. The COP9 signalosome: A versatile regulatory hub of Cullin-RING ligases. Trends Biochem Sci 2023, 48, 82–95. [Google Scholar] [CrossRef]
- Angers, S.; et al. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef]
- Fischer, E.S.; et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 2011, 147, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, W.I.; et al. , The CRL4(DCAF1) cullin-RING ubiquitin ligase is activated following a switch in oligomerization state. EMBO J 2021, 40, e108008. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; et al. , Structural basis of human transcription-DNA repair coupling. Nature 2021, 598, 368–372. [Google Scholar] [CrossRef]
- Kokic, G.; et al. , Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair. Nat Struct Mol Biol 2024, 31, 536–547. [Google Scholar] [CrossRef]
- Osaka, F.; et al. , Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J 2000, 19, 3475–3484. [Google Scholar] [CrossRef]
- Michel, J.J.; McCarville, J.F.; Xiong, Y. A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J Biol Chem 2003, 278, 22828–22837. [Google Scholar] [CrossRef]
- Holmberg, C.; et al. , Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 2005, 19, 853–862. [Google Scholar] [CrossRef]
- Mimura, S.; et al. , Cul8/Rtt101 forms a variety of protein complexes that regulate DNA damage response and transcriptional silencing. J Biol Chem 2010, 285, 9858–9867. [Google Scholar] [CrossRef]
- Zhao, Y.; et al. , Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 2010, 285, 4355–4365. [Google Scholar] [CrossRef]
- Lewis, Z.A.; et al. , DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet 2010, 6, e1001196. [Google Scholar] [CrossRef]
- Gabilly, S.T.; et al. , Regulation of photoprotection gene expression in Chlamydomonas by a putative E3 ubiquitin ligase complex and a homolog of CONSTANS. Proc Natl Acad Sci U S A 2019, 116, 17556–17562. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, A.; et al. , CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J 2006, 47, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. , et al. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 2008, 20, 1437–1455. [Google Scholar] [CrossRef] [PubMed]
- Dumbliauskas, E.; et al. , The Arabidopsis CUL4-DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J 2011, 30, 731–743. [Google Scholar] [CrossRef]
- Pazhouhandeh, M.; et al. , MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci U S A 2011, 108, 3430–3435. [Google Scholar] [CrossRef]
- Jahns, M.T.; et al. Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biol 2014, 12, e1001930. [Google Scholar] [CrossRef]
- Zhong, W.; et al. CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 2003, 423, 885–889. [Google Scholar] [CrossRef]
- Hu, J.; et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev 2008, 22, 866–871. [Google Scholar] [CrossRef]
- Lin, H.C.; et al. Cul4 and DDB1 regulate Orc2 localization, BrdU incorporation and Dup stability during gene amplification in Drosophila follicle cells. J Cell Sci 2009, 122, 2393–2401. [Google Scholar] [CrossRef]
- Tare, M.; et al. An E3 ubiquitin ligase, cullin-4 regulates retinal differentiation in Drosophila eye. Genesis 2020, 58, e23395. [Google Scholar] [CrossRef]
- Zhao, X.; et al. Zebrafish cul4a, but not cul4b, modulates cardiac and forelimb development by upregulating tbx5a expression. Hum Mol Genet 2015, 24, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; et al. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell 2009, 34, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Zha, Z.; et al. A Non-Canonical Function of Gβ as a Subunit of E3 Ligase in Targeting GRK2 Ubiquitylation. Mol Cell 2015, 58, 794–803. [Google Scholar] [CrossRef] [PubMed]
- Kopanja, D.; et al. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene 2009, 28, 2456–2465. [Google Scholar] [CrossRef]
- Yin, Y.; et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev Biol 2011, 356, 51–62. [Google Scholar] [CrossRef]
- Jiang, B.; et al. Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PLoS One 2012, 7, e37070. [Google Scholar] [CrossRef]
- Liu, L.; et al. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res 2012, 22, 1258–1269. [Google Scholar] [CrossRef]
- Tarpey, P.S.; et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet 2007, 80, 345–352. [Google Scholar] [CrossRef]
- Zou, Y.; et al. Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am J Hum Genet 2007, 80, 561–566. [Google Scholar] [CrossRef]
- Tarpey, P.S.; et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet 2009, 41, 535–543. [Google Scholar] [CrossRef]
- Ruban, A.V.; Wilson, S. The Mechanism of Non-Photochemical Quenching in Plants: Localization and Driving Forces. Plant Cell Physiol 2021, 62, 1063–1072. [Google Scholar] [CrossRef]
- Li, B.; Ruiz, J.C.; Chun, K.T. CUL-4A is critical for early embryonic development. Mol Cell Biol 2002, 22, 4997–5005. [Google Scholar] [CrossRef]
- Kopanja, D.; et al. Cul4A is essential for spermatogenesis and male fertility. Dev Biol 2011, 352, 278–287. [Google Scholar] [CrossRef]
- Chen, L.C.; et al. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res 1998, 58, 3677–3683. [Google Scholar]
- Melchor, L.; et al. Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res 2009, 11, R86. [Google Scholar] [CrossRef]
- Schindl, M.; et al. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res 2007, 27, 949–952. [Google Scholar]
- Yasui, K.; et al. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology 2002, 35, 1476–1484. [Google Scholar] [CrossRef]
- Shinomiya, T.; et al. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer, 1999, 24, 337–344. [Google Scholar] [CrossRef]
- Dohna, M.; et al. Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer 2000, 28, 145–152. [Google Scholar] [CrossRef]
- Wang, Y.; et al. CUL4A overexpression enhances lung tumor growth and sensitizes lung cancer cells to erlotinib via transcriptional regulation of EGFR. Mol Cancer 2014, 13, 252. [Google Scholar] [CrossRef]
- Li, X.; et al. CUL4A expression in pediatric osteosarcoma tissues and its effect on cell growth in osteosarcoma cells. Tumour Biol 2016, 37, 8139–8144. [Google Scholar] [CrossRef]
- Hung, M.S.; et al. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med 2011, 15, 350–358. [Google Scholar] [CrossRef]
- Michiels, E.M.; et al. Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J Pediatr Hematol Oncol 2002, 24, 205–210. [Google Scholar] [CrossRef]
- Birner, P.; et al. Human homologue for Caenorhabditis elegans CUL-4 protein overexpression is associated with malignant potential of epithelial ovarian tumours and poor outcome in carcinoma. J Clin Pathol 2012, 65, 507–511. [Google Scholar] [CrossRef]
- Ren, S.; et al. Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J Mol Med (Berl) 2012, 90, 1121–1132. [Google Scholar] [CrossRef]
- Saucedo-Cuevas, L.P.; et al. CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response. Oncotarget 2014, 5, 2330–2343. [Google Scholar] [CrossRef]
- Wang, Y.; et al. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res 2014, 74, 520–531. [Google Scholar] [CrossRef]
- Pan, Y.; et al. CUL4A facilitates hepatocarcinogenesis by promoting cell cycle progression and epithelial-mesenchymal transition. Sci Rep 2015, 5, 17006. [Google Scholar] [CrossRef]
- Wang, Y.; et al. Analysis of lung tumor initiation and progression in transgenic mice for Cre-inducible overexpression of Cul4A gene. Thorac Cancer 2015, 6, 480–487. [Google Scholar] [CrossRef]
- Jiang, T.; et al. Cullin 4B is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med Oncol 2013, 30, 534. [Google Scholar] [CrossRef]
- Hu, H.; et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 2012, 22, 781–95. [Google Scholar] [CrossRef]
- Yuan, J.; et al. Accelerated hepatocellular carcinoma development in CUL4B transgenic mice. Oncotarget 2015, 6, 15209–15221. [Google Scholar] [CrossRef]
- Aihara, Y.; et al. Algal photoprotection is regulated by the E3 ligase CUL4-DDB1(DET1). Nat Plants 2019, 5, 34–40. [Google Scholar] [CrossRef]
- Tokutsu, R.; et al. The CONSTANS flowering complex controls the protective response of photosynthesis in the green alga Chlamydomonas. Nat Commun 2019, 10, 4099. [Google Scholar] [CrossRef]
- Jang, S.; et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 2008, 27, 1277–1288. [Google Scholar] [CrossRef]
- Liu, L.J.; et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef]
- Liu, Q.; et al. The Blue Light-Dependent Polyubiquitination and Degradation of Arabidopsis Cryptochrome2 Requires Multiple E3 Ubiquitin Ligases. Plant Cell Physiol 2016, 57, 2175–2186. [Google Scholar] [CrossRef]
- Zhu, L.; et al. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat Commun 2015, 6, 7245. [Google Scholar] [CrossRef]
- Yanagawa, Y.; et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 2004, 18, 2172–2181. [Google Scholar] [CrossRef]
- Castells, E.; et al. The conserved factor DE-ETIOLATED 1 cooperates with CUL4-DDB1DDB2 to maintain genome integrity upon UV stress. EMBO J 2011, 30, 1162–1172. [Google Scholar] [CrossRef]
- Lee, J.H.; et al. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732. [Google Scholar] [CrossRef]
- Seo, K.I.; et al. ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 2014, 26, 695–711. [Google Scholar] [CrossRef]
- Irigoyen, M.L.; et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728. [Google Scholar] [CrossRef]
- Murchie, E.H.; Ruban, A.V. Dynamic non-photochemical quenching in plants: from molecular mechanism to productivity. Plant J 2020, 101, 885–896. [Google Scholar] [CrossRef]
- Xu, X.; et al. Illuminating Progress in Phytochrome-Mediated Light Signaling Pathways. Trends Plant Sci 2015, 20, 641–650. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, C. Mechanisms of Cryptochrome-Mediated Photoresponses in Plants. Annu Rev Plant Biol 2020, 71, 103–129. [Google Scholar] [CrossRef]
- Cheng, M.C.; et al. Phytochrome Signaling Networks. Annu Rev Plant Biol 2021, 72, 217–244. [Google Scholar] [CrossRef]
- Zuo, Z.; et al. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 2011, 21, 841–847. [Google Scholar] [CrossRef]
- Kciuk, M.; et al. Focus on UV-Induced DNA Damage and Repair-Disease Relevance and Protective Strategies. Int J Mol Sci 2020, 21, 7264. [Google Scholar] [CrossRef]
- Zhang, C.; et al. Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 2010, 22, 2353–2369. [Google Scholar] [CrossRef]
- Lim, J.; Lim, C.W.; Lee, S.C. Core Components of Abscisic Acid Signaling and Their Post-translational Modification. Front Plant Sci 2022, 13, 895698. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin-Proteasome System in ABA Signaling: From Perception to Action. Mol Plant 2016, 9, 21–33. [Google Scholar] [CrossRef]
- Zaidi, I.W.; et al. Rtt101 and Mms1 in budding yeast form a CUL4(DDB1)-like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 2008, 9, 1034–1040. [Google Scholar] [CrossRef]
- Oya, E.; et al. H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep 2019, 20, e48111. [Google Scholar] [CrossRef]
- Liu, H.; et al. The Cross-Regulation Between Set1, Clr4, and Lsd1/2 in Schizosaccharomyces pombe. PLoS Genet 2024, 20, e1011107. [Google Scholar] [CrossRef]
- Braun, S.; et al. The Cul4-Ddb1(Cdt)² ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 2011, 144, 41–54. [Google Scholar] [CrossRef]
- Bondar, T.; Ponomarev, A.; Raychaudhuri, P. Ddb1 is required for the proteolysis of the Schizosaccharomyces pombe replication inhibitor Spd1 during S phase and after DNA damage. J Biol Chem 2004, 279, 9937–9943. [Google Scholar] [CrossRef]
- Liu, C.; et al. Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4-Ddb1-CSN ubiquitin ligase. EMBO J 2005, 24, 3940–3951. [Google Scholar] [CrossRef]
- Kim, Y.; Kipreos, E.T. Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div 2007, 2, 18. [Google Scholar] [CrossRef]
- Han, J.; et al. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 2013, 155, 817–829. [Google Scholar] [CrossRef]
- Han, J.; et al. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev 2010, 24, 1485–1490. [Google Scholar] [CrossRef]
- Xu, H.; et al. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 2010, 6, e1001132. [Google Scholar] [CrossRef]
- Adhvaryu, K.K.; et al. The cullin-4 complex DCDC does not require E3 ubiquitin ligase elements to control heterochromatin in Neurospora crassa. Eukaryot Cell 2015, 14, 25–28. [Google Scholar] [CrossRef]
- Jia, S.; Kobayashi, R.; Grewal, S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 2005, 7, 1007–1013. [Google Scholar] [CrossRef]
- Horn, P.J.; Bastie, J.N.; Peterson, C.L. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 2005, 19, 1705–1714. [Google Scholar] [CrossRef]
- Ralph, E.; Boye, E.; Kearsey, S.E. DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep 2006, 7, 1134–1139. [Google Scholar] [CrossRef]
- Scholes, D.T.; et al. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 2001, 159, 1449–1465. [Google Scholar] [CrossRef]
- Collins, S.R.; et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 2007, 446, 806–810. [Google Scholar] [CrossRef]
- Roberts, T.M.; et al. Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase. Mol Biol Cell 2008, 19, 171–180. [Google Scholar] [CrossRef]
- Young, T.J.; et al. CAF-1 and Rtt101p function within the replication-coupled chromatin assembly network to promote H4 K16ac, preventing ectopic silencing. PLoS Genet 2020, 16, e1009226. [Google Scholar] [CrossRef]
- Noireterre, A.; et al. The cullin Rtt101 promotes ubiquitin-dependent DNA-protein crosslink repair across the cell cycle. Nucleic Acids Res 2024, 52, 9654–9670. [Google Scholar] [CrossRef] [PubMed]
- Buser, R. , et al. The Replisome-Coupled E3 Ubiquitin Ligase Rtt101Mms22 Counteracts Mrc1 Function to Tolerate Genotoxic Stress. PLoS Genet 2016, 12, e1005843. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, P.K.; et al. , Molecular cloning and characterization of Saccharomyces cerevisiae RAD28, the yeast homolog of the human Cockayne syndrome A (CSA) gene. J Bacteriol 1996, 178, 5977–5988. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, Y.; Dohmae, N.; Hanaoka, F. Schizosaccharomyces pombe Ddb1 recruits substrate-specific adaptor proteins through a novel protein motif, the DDB-box. Mol Cell Biol 2008, 28, 6746–6756. [Google Scholar] [CrossRef]
- Kim, S.H.; Michael, W.M. Regulated proteolysis of DNA polymerase eta during the DNA-damage response in C. elegans. Mol Cell 2008, 32, 757–766. [Google Scholar] [CrossRef]
- Choe, K.P.; Przybysz, A.J.; Strange, K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol 2009, 29, 2704–2715. [Google Scholar] [CrossRef]
- Shibutani, S.T.; et al. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev Cell 2008, 15, 890–900. [Google Scholar] [CrossRef]
- Higa, L.A.; et al. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 2006, 5, 71–77. [Google Scholar] [CrossRef]
- Ozturk, N.; et al. Ramshackle (Brwd3) promotes light-induced ubiquitylation of Drosophila Cryptochrome by DDB1-CUL4-ROC1 E3 ligase complex. Proc Natl Acad Sci U S A 2013, 110, 4980–4985. [Google Scholar] [CrossRef]
- Kumar, A.; Baker, N.E. The CRL4 E3 ligase Mahjong/DCAF1 controls cell competition through the transcription factor Xrp1, independently of polarity genes. Development 2022, 149, dev200795. [Google Scholar] [CrossRef]
- Cho, Y.S.; et al. CDK7 regulates organ size and tumor growth by safeguarding the Hippo pathway effector Yki/Yap/Taz in the nucleus. Genes Dev 2020, 34, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.S.; Takata, K.; Wood, R.D. DNA polymerases and cancer. Nat Rev Cancer 2011, 11, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, T.K.; et al. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 2015, 88, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol Cell Biol 2020, 40, e00099–20. [Google Scholar] [CrossRef]
- Lo, J.Y.; Spatola, B.N.; Curran, S.P. WDR23 regulates NRF2 independently of KEAP1. PLoS Genet 2017, 13, e1006762. [Google Scholar] [CrossRef]
- Lui, D.Y.; Colaiácovo, M.P. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol 2013, 757, 133–170. [Google Scholar]
- Zielke, N.; et al. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 2011, 480, 123–127. [Google Scholar] [CrossRef]
- Cermakian, N.; Sassone-Corsi, P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol 2000, 1, 59–67. [Google Scholar] [CrossRef]
- Han, D.; et al. BRWD3 promotes KDM5 degradation to maintain H3K4 methylation levels. Proc Natl Acad Sci U S A 2023, 120, e2305092120. [Google Scholar] [CrossRef]
- Tamori, Y.; et al. Involvement of Lgl and Mahjong/VprBP in cell competition. PLoS Biol 2010, 8, e1000422. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes Dev 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nita, A.; Moroishi, T. Hippo pathway in cell-cell communication: emerging roles in development and regeneration. Inflamm Regen 2024, 44, 18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Kipreos, E.T.; et al. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 1996, 85, 829–839. [Google Scholar] [CrossRef]
- Nakagawa, T.; et al. SPT16 ubiquitylation by DCAF14-CRL4 regulates FACT binding to histones. Cell Rep 2022, 38, 110541. [Google Scholar] [CrossRef]
- Raisch, J.; et al. Pulse-SILAC and Interactomics Reveal Distinct DDB1-CUL4-Associated Factors, Cellular Functions, and Protein Substrates. Mol Cell Proteomics 2023, 22, 100644. [Google Scholar] [CrossRef]
- Huang, Y.H.; et al. Systematic Profiling of DNMT3A Variants Reveals Protein Instability Mediated by the DCAF8 E3 Ubiquitin Ligase Adaptor. Cancer Discov 2022, 12, 220–235. [Google Scholar] [CrossRef]
- Leng, F.; et al. Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4(DCAF5) ubiquitin ligase. Nat Commun 2018, 9, 1641. [Google Scholar] [CrossRef]
- Huang, D.; et al. CRL4(DCAF8) dependent opposing stability control over the chromatin remodeler LSH orchestrates epigenetic dynamics in ferroptosis. Cell Death Differ 2021, 28, 1593–1609. [Google Scholar] [CrossRef]
- Ren, P.; et al. CRL4(DCAF13) E3 ubiquitin ligase targets MeCP2 for degradation to prevent DNA hypermethylation and ensure normal transcription in growing oocytes. Cell Mol Life Sci 2024, 81, 165. [Google Scholar] [CrossRef]
- Nakagawa, T.; et al. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell 2015, 57, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Shibata, E.; Dar, A.; Dutta, A. CRL4Cdt2 E3 ubiquitin ligase and proliferating cell nuclear antigen (PCNA) cooperate to degrade thymine DNA glycosylase in S phase. J Biol Chem 2014, 289, 23056–23064. [Google Scholar] [CrossRef] [PubMed]
- Slenn, T.J.; et al. Thymine DNA glycosylase is a CRL4Cdt2 substrate. J Biol Chem 2014, 289, 23043–23055. [Google Scholar] [CrossRef] [PubMed]
- Le, R.; et al. Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell Stem Cell 2021, 28, 732–747.e9. [Google Scholar] [CrossRef]
- Wang, H.; et al. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 2006, 22, 383–394. [Google Scholar] [CrossRef]
- Biterge, B.; et al. Methylation of histone H4 at aspartate 24 by protein L-isoaspartate O-methyltransferase (PCMT1) links histone modifications with protein homeostasis. Sci Rep 2014, 4, 6674. [Google Scholar] [CrossRef]
- Niikura, Y.; et al. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev Cell 2015, 32, 589–603. [Google Scholar] [CrossRef]
- Mouysset, J.; et al. CRL4(RBBP7) is required for efficient CENP-A deposition at centromeres. J Cell Sci 2015, 128, 1732–1745. [Google Scholar]
- Djakbarova, U.; Marzluff, W.F.; Köseoğlu, M.M. DDB1 and CUL4 associated factor 11 (DCAF11) mediates degradation of Stem-loop binding protein at the end of S phase. Cell Cycle 2016, 15, 1986–1996. [Google Scholar] [CrossRef]
- Matsunuma, R.; et al. UV Damage-Induced Phosphorylation of HBO1 Triggers CRL4DDB2-Mediated Degradation To Regulate Cell Proliferation. Mol Cell Biol 2016, 36, 394–406. [Google Scholar] [CrossRef]
- Abbas, T.; et al. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 2010, 40, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Centore, R.C.; et al. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 2010, 40, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Oda, H.; et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 2010, 40, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 2010, 12, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.; et al. SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J Cell Biol 2011, 192, 43–54. [Google Scholar] [CrossRef]
- Zhang, Y.L.; et al. DCAF13 promotes pluripotency by negatively regulating SUV39H1 stability during early embryonic development. EMBO J 2018, 37, e98981. [Google Scholar] [CrossRef]
- Luo, D.; et al. CUL4B-DDB1-COP1-mediated UTX downregulation promotes colorectal cancer progression. Exp Hematol Oncol 2023, 12, 77. [Google Scholar] [CrossRef]
- Xu, J.; et al. MEN1 Degradation Induced by Neddylation and the CUL4B-DCAF7 Axis Promotes Pancreatic Neuroendocrine Tumor Progression. Cancer Res 2023, 83, 2226–2247. [Google Scholar] [CrossRef]
- Vulto-van Silfhout, A.T.; et al. Variants in CUL4B are associated with cerebral malformations. Hum Mutat 2015, 36, 106–117. [Google Scholar] [CrossRef]
- Wang, X.; et al. VprBP/DCAF1 Regulates the Degradation and Nonproteolytic Activation of the Cell Cycle Transcription Factor FoxM1. Mol Cell Biol 2017, 37, e00609–16. [Google Scholar] [CrossRef]
- Jung, E.S.; et al. Jmjd2C increases MyoD transcriptional activity through inhibiting G9a-dependent MyoD degradation. Biochim Biophys Acta 2015, 1849, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; et al. DCAF1 controls T-cell function via p53-dependent and -independent mechanisms. Nat Commun 2016, 7, 10307. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; et al. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell 2012, 48, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.F.; et al. Sirtuin 7-dependent deacetylation of DDB1 regulates the expression of nuclear receptor TR4. Biochem Biophys Res Commun 2017, 490, 423–428. [Google Scholar] [CrossRef]
- Kaur, M.; et al. CRL4-DDB1-VPRBP ubiquitin ligase mediates the stress triggered proteolysis of Mcm10. Nucleic Acids Res 2012, 40, 7332–7346. [Google Scholar] [CrossRef]
- Wen, X.; et al. The HIV1 protein Vpr acts to enhance constitutive DCAF1-dependent UNG2 turnover. PLoS One 2012, 7, e30939. [Google Scholar] [CrossRef]
- Kassmeier, M.D.; et al. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J 2012, 31, 945–958. [Google Scholar] [CrossRef]
- Guo, P.; et al. The assembly of mammalian SWI/SNF chromatin remodeling complexes is regulated by lysine-methylation dependent proteolysis. Nat Commun 2022, 13, 6696. [Google Scholar] [CrossRef]
- Zhang, C.; et al. Proteolysis of methylated SOX2 protein is regulated by L3MBTL3 and CRL4(DCAF5) ubiquitin ligase. J Biol Chem 2019, 294, 476–489. [Google Scholar] [CrossRef]
- Peng, Z.; et al. Human DNA Ligase I Interacts with and Is Targeted for Degradation by the DCAF7 Specificity Factor of the Cul4-DDB1 Ubiquitin Ligase Complex. J Biol Chem 2016, 291, 21893–21902. [Google Scholar] [CrossRef]
- Sun, Y.; et al. Targeting neddylation sensitizes colorectal cancer to topoisomerase I inhibitors by inactivating the DCAF13-CRL4 ubiquitin ligase complex. Nat Commun 2023, 14, 3762. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; et al. CRL4A(DTL) degrades DNA-PKcs to modulate NHEJ repair and induce genomic instability and subsequent malignant transformation. Oncogene 2021, 40, 2096–2111. [Google Scholar] [CrossRef] [PubMed]
- Ndoja, A.; et al. Ubiquitin Ligase COP1 Suppresses Neuroinflammation by Degrading c/EBPβ in Microglia. Cell 2020, 182, 1156–1169.e12. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; et al. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004, 303, 1371–1374. [Google Scholar] [CrossRef]
- Lin, H.; et al. IP(6)-assisted CSN-COP1 competition regulates a CRL4-ETV5 proteolytic checkpoint to safeguard glucose-induced insulin secretion. Nat Commun 2021, 12, 2461. [Google Scholar] [CrossRef]
- Lu, G.; et al. Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell 2014, 26, 222–234. [Google Scholar] [CrossRef]
- Dornan, D.; et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef]
- Su, Y.; et al. Glucose-induced CRL4(COP1)-p53 axis amplifies glycometabolism to drive tumorigenesis. Mol Cell 2023, 83, 2316–2331.e7. [Google Scholar] [CrossRef]
- Groisman, R.; et al. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev, 2006, 20, 1429–34. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Shamay, M.; et al. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J Biol Chem 2010, 285, 36377–36386. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, E.; et al. The Chromatin Remodeler HELLS: A New Regulator in DNA Repair, Genome Maintenance, and Cancer. Int J Mol Sci 2022, 23, 9313. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; et al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 2015, 7, 1051–1073. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef]
- Chakravarti, D.; LaBella, K.A.; DePinho, R.A. Telomeres: history, health, and hallmarks of aging. Cell 2021, 184, 306–322. [Google Scholar] [CrossRef]
- Luger, K.; et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Cao, J.; Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol 2012, 2, 26. [Google Scholar] [CrossRef]
- Chen, J.J.; Stermer, D.; Tanny, J.C. Decoding histone ubiquitylation. Front Cell Dev Biol 2022, 10, 968398. [Google Scholar] [CrossRef]
- Yang, Y.; et al. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 2015, 34, 104–118. [Google Scholar] [CrossRef]
- Ekwall, K. Epigenetic control of centromere behavior. Annu Rev Genet 2007, 41, 63–81. [Google Scholar] [CrossRef]
- Beck, D.B.; et al. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev 2012, 26, 2580–2589. [Google Scholar] [CrossRef]
- Jin, Q.; et al. USP7 Cooperates with NOTCH1 to Drive the Oncogenic Transcriptional Program in T-Cell Leukemia. Clin Cancer Res 2019, 25, 222–239. [Google Scholar] [CrossRef]
- Rao, R.C.; Dou, Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 2015, 15, 334–346. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Hoshii, T.; Armstrong, S.A. Mixed-Lineage Leukemia Fusions and Chromatin in Leukemia. Cold Spring Harb Perspect Med 2017, 7, a026658. [Google Scholar] [CrossRef]
- Okada, Y.; et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005, 121, 167–178. [Google Scholar] [CrossRef]
- Gilan, O.; et al. CRISPR-ChIP reveals selective regulation of H3K79me2 by Menin in MLL leukemia. Nat Struct Mol Biol 2023, 30, 1592–1606. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008, 14, 355–368. [Google Scholar] [CrossRef]
- Nakagawa, T.; Mondal, K.; Swanson, P.C. VprBP (DCAF1): a promiscuous substrate recognition subunit that incorporates into both RING-family CRL4 and HECT-family EDD/UBR5 E3 ubiquitin ligases. BMC Mol Biol 2013, 14, 22. [Google Scholar] [CrossRef]
- Schabla, N.M.; Mondal, K.; Swanson, P.C. DCAF1 (VprBP): emerging physiological roles for a unique dual-service E3 ubiquitin ligase substrate receptor. J Mol Cell Biol 2019, 11, 725–735. [Google Scholar] [CrossRef]
- Vanegas-Torres, C.A.; Schindler, M. HIV-1 Vpr Functions in Primary CD4(+) T Cells. Viruses 2024, 16, 420. [Google Scholar] [CrossRef]
- Dobransky, A.; et al. CRL4-DCAF1 Ubiquitin Ligase Dependent Functions of HIV Viral Protein R and Viral Protein X. Viruses 2024, 16, 1313. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, H. Lysine Methylation-Dependent Proteolysis by the Malignant Brain Tumor (MBT) Domain Proteins. Int J Mol Sci 2024, 25, 2248. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; et al. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Perez, G. et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res 2024, Online ahead of print.
- Potter, S.C.; et al. HMMER web server: 2018 update. Nucleic Acids Res 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Cherry, J.M.; et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 2012, 40, D700–D705. [Google Scholar] [CrossRef]
- Rutherford, K.M.; Lera-Ramírez, M.; Wood, V. PomBase: a Global Core Biodata Resource-growth, collaboration, and sustainability. Genetics 2024, 227, iyae007. [Google Scholar] [CrossRef]
- Berardini, T.Z.; et al. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Harris, T.W.; et al. WormBase: a modern Model Organism Information Resource. Nucleic Acids Res 2020, 48, D762–D767. [Google Scholar] [CrossRef]
- Öztürk-Çolak, A.; et al. FlyBase: updates to the Drosophila genes and genomes database. Genetics 2024, 227, iyad211. [Google Scholar] [CrossRef]
- Tang, J.; Chu, G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst) 2002, 1, 601–616. [Google Scholar] [CrossRef]
- Han, X.; Huang, X.; Deng, X.W. The Photomorphogenic Central Repressor COP1: Conservation and Functional Diversification during Evolution. Plant Commun 2020, 1, 100044. [Google Scholar] [CrossRef]
- Ponnu, J.; Hoecker, U. Illuminating the COP1/SPA Ubiquitin Ligase: Fresh Insights Into Its Structure and Functions During Plant Photomorphogenesis. Front Plant Sci 2021, 12, 662793. [Google Scholar] [CrossRef] [PubMed]
- Truong, L.N.; Wu, X. Prevention of DNA re-replication in eukaryotic cells. J Mol Cell Biol 2011, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pozo, P.N.; Cook, J.G. Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes (Basel) 2016, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Dale, B.; et al. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer 2021, 21, 638–654. [Google Scholar] [CrossRef]
- Tsai, J.M.; et al. Targeted protein degradation: from mechanisms to clinic. Nat Rev Mol Cell Biol 2024, 25, 740–757. [Google Scholar] [CrossRef]
- Ito, T.; et al. Identification of a primary target of thalidomide teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef]
| Species | Cul4 gene | Phenotype | Reference |
|---|---|---|---|
| S. cerevisiae$$$S. Pombe$$$N. crassa | Cul8/Cul4 | Hypersensitivity to DNA damage | [37,38,39,40,41,42] |
| Growth retardation | |||
| Mitotic defect | |||
| C. reinhardtii | Cul4 | Induction of photoprotection-related gene expression | [43] |
| A. thaliana | Cul4 | Developmental defects | [23,44,45,46,47,48] |
| Constitutive photomorphogenesis | |||
| C. elegans | Cul-4 | Developmental defects | [28,49] |
| DNA re-replication | |||
| D. melanogaster | Cul-4 | Developmental defects | [50,51,52] |
| D. rerio | Cul4a | Developmental defects | [53] |
| M. musculus | Cul4a | Resistance to skin cancer | [54] |
| Cardiac hypertrophy | [55] | ||
| Male infertility | [56,57] | ||
| Cul4b | Lethality at embryonic stage | [58,59] | |
| H. sapiens | CUL4B | X-linked intellectual disability | [60,61,62] |
| Species | Substrate | Receptor | Function of CRL4 | Reference |
|---|---|---|---|---|
| C. reinhardtii | CONSTANS | SPA/COP1$$$DET1 | Inhibition of light energy dissipation-related gene expression | [43,85,86] |
| A. thaliana | CONSTANS | SPA/COP1 | Inhibition of flowering-related gene expression | [87,88] |
| CRY2 | SPA/COP1 | Inhibition of light response-related gene expression | [89] | |
| PIF1 | SPA/COP1 | Promotion of photomorphogenesis-related gene expression | [90] | |
| HY5 | SPA/COP1$$$DET1 | Inhibition of photomorphogenesis-related gene expression | [23,91] | |
| DDB2 | DET1 | Promotion of DNA repair | [92] | |
| ABI5 | DWA1/2 | Inhibition of ABA signaling-related gene expression | [93] | |
| ABI5 | ABD1 | Inhibition of ABA signaling-related gene expression | [94] | |
| PYL8 | DDA1 | Inhibition of ABA signaling-related gene expression | [95] |
| Species | Substrate | Receptor | Function of CRL4 | Reference |
|---|---|---|---|---|
| S. pombe | Histone H3 | Dos1 | Promotion of H3K9 methylation | [106] |
| Lsd1/2 | Dos1 | Inhibition of H3K9 demethylation | [107] | |
| Set1 | Dos1 | Inhibition of H3K4 methylation | [107] | |
| Epe1 | Cdt1 | Establishment of heterochromatin boundary | [108] | |
| Spd1 | Cdt1 | Promotion of dNTP synthesis | [39,109,110] | |
| Cdt2 | Cdt1 | Inhibition of DNA re-replication | [111] | |
| S. cerevisiae | Histone H3 | Mms22 | Promotion of histone transfer to replicated DNA | [112] |
| Spt16 | Mms22 | Promotion of DNA replication | [113] |
| Species | Substrate | Receptor | Function of CRL4 | Reference |
|---|---|---|---|---|
| C. elegans | CDT-1 | CDT-2 | Inhibition of DNA re-replication | [49] |
| Pol η | CDT-2 | Completion of DNA repair | [127] | |
| SKN-1 | WDR-23 | Inhibition of stress-related gene expression | [128] | |
| D. melanogaste | Dup (Cdt1) | L(2)dtl | Inhibition of DNA re-replication | [51] |
| E2f1 | L(2)dtl | Promote of cell proliferation | [129] | |
| Dap | Unknown | Promote of cell proliferation | [130] | |
| Cyclin E | Unknown | Inhibition of cell proliferation | [130] | |
| Cry | Ramshackle (Brwd3) | Inhibition of circadian gene expression | [131] | |
| Xrp1 | Mahjong$$$(Dcaf1) | Inhibition of cell elimination-related gene expression | [132] | |
| Yki | Dcaf12 | Inhibition of cell proliferation-related gene expression | [133] |
| Category | Substrate | Receptor | Function of CRL4 | Reference |
|---|---|---|---|---|
| DNA modification-related | DNMT3A | DCAF8 | Inhibition of DNA methylation | [149] |
| DNMT1 | DCAF5/L3MBTL3 | Inhibition of DNA methylation | [150] | |
| LSH | DCAF8 | Promotion of DNA demethylation | [151] | |
| MeCP2 | DCAF13 | Promotion of gene expression | [152] | |
| TET1/2/3 | DCAF1 | Promotion of DNA demethylation | [153] | |
| TDG | CDT2 | Inhibition of DNA demethylation | [154,155]4 | |
| KAP1 | DCAF11 | Promotion of gene expression | [156] | |
| Histone modification-related | Histone H2A | RBBP4/7 | Inhibition of gene expression | [83] |
| Histone H2A | DDB2 | Promotion of DNA repair | [21,22,157] | |
| Histone H3 | DDB2 | Promotion of DNA repair | [157] | |
| Histone H4 | DDB2 | Promotion of DNA repair | [157] | |
| Histone H4 | DCAF1 | Promotion of DNA repair | [158] | |
| CENP-A | COPS8 | Promotion of centromere formation | [159] | |
| CENP-A | RBBP4/7 | Promotion of centromere formation | [160] | |
| SLBP | DCAF11 | Inhibition of histone translation | [161] | |
| SPT16 | DCAF14 | Promotion of DNA replication | [147] | |
| HBO1 | DDB2 | Inhibition of DNA replication | [162] | |
| SET8 | CDT2 | Inhibition of DNA re-replication and transcription | [163,164,165,166,167] | |
| SUV39H1 | DCAF13 | Promotion of gene expression | [168] | |
| KDM6A | COP1 | Promotion of gene expression | [169] | |
| Menin | DCAF7 | Inhibition of gene expression | [170] | |
| WDR5 | Not required | Inhibition of gene expression | [20,171] | |
| Others | NRF2 | DCAF11 | Inhibition of gene expression | [137] |
| FoxM1 | DCAF1 | Inhibition of gene expression | [172] | |
| MyoD | DCAF1 | Inhibition of gene expression | [173] | |
| p53 | DCAF1 | Inhibition of gene expression | [174] | |
| RORa | DCAF1 | Inhibition of gene expression | [175] | |
| TR4 | DCAF1 | Inhibition of gene expression | [176] | |
| MCM10 | DCAF1 | Inhibition of DNA replication | [177] | |
| UNG2 | DCAF1 | Inhibition of DNA repair | [178] | |
| SMUG1 | DCAF1 | Inhibition of DNA repair | [178] | |
| RAG1 | DCAF1 | Inhibition of DNA recombination | [179] | |
| E2F1 | DCAF5/L3MBTL3 | Inhibition of gene expression | [150] | |
| DNMT1 | DCAF5/L3MBTL3 | Inhibition of DNA methylation | [150] | |
| SMARCC1 | DCAF5/L3MBTL3 | Inhibition of gene expression | [180] | |
| SMARCC2 | DCAF5/L3MBTL3 | Inhibition of gene expression | [180] | |
| SOX2 | DCAF5/L3MBTL3 | Inhibition of gene expression | [181] | |
| LIG1 | DCAF7 | Inhibition of DNA replication | [182] | |
| TOP1-DPCs | DCAF13 | Promotion of DNA repair | [183] | |
| DNA-PKcs | CDT2 | Inhibition of DNA repair | [184] | |
| C/EBPβ | COP1 | Inhibition of gene expression | [185] | |
| c-JUN | COP1 | Inhibition of gene expression | [186] | |
| ETV5 | COP1 | Inhibition of gene expression | [187] | |
| ETS1 | COP1 | Inhibition of gene expression | [188] | |
| ETS2 | COP1 | Inhibition of gene expression | [188] | |
| p53 | COP1 | Inhibition of gene expression | [189,190] | |
| CSB | CSA | Completion of DNA repair | [191] |
| Mammalian DCAF | Trypanosoma | Yeasts | Plants | Insects |
|---|---|---|---|---|
| DCAF1 | (-) | (-) | ✔︎ | ✔︎ |
| CDT2 (DCAF2) | (-) | ✔︎ | (-) | ✔︎ |
| DCAF5 | (-) | (-) | (-) | (-) |
| DCAF7 | (-) | ✔︎ | (-) | (-) |
| DCAF8 | (-) | (-) | (-) | (-) |
| DCAF11 | (-) | ✔︎ | (-) | ✔︎ |
| DCAF12 | (-) | (-) | (-) | ✔︎ |
| DCAF13 | ✔︎ | ✔︎ | (-) | ✔︎ |
| DCAF14 | (-) | (-) | (-) | ✔︎ |
| COP1 | (-) | ✔︎ | ✔︎ | ✔︎ |
| CSA | (-) | ✔︎ | ✔︎ | ✔︎ |
| DDB2 | (-) | (-) | ✔︎ | (-) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





