Submitted:
02 October 2024
Posted:
07 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Current Applications of TEM and Cryo-EM in Viral Analysis
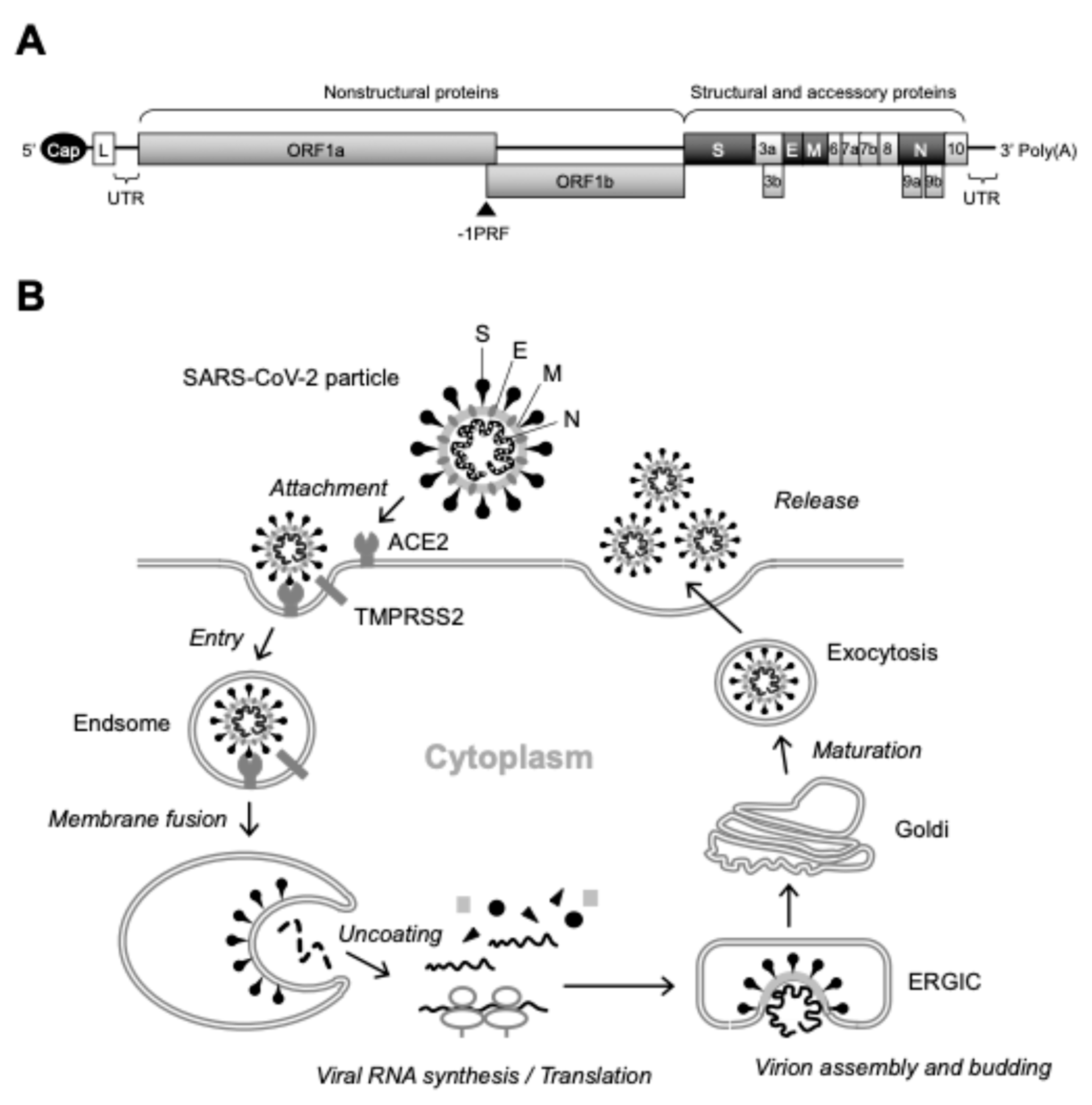
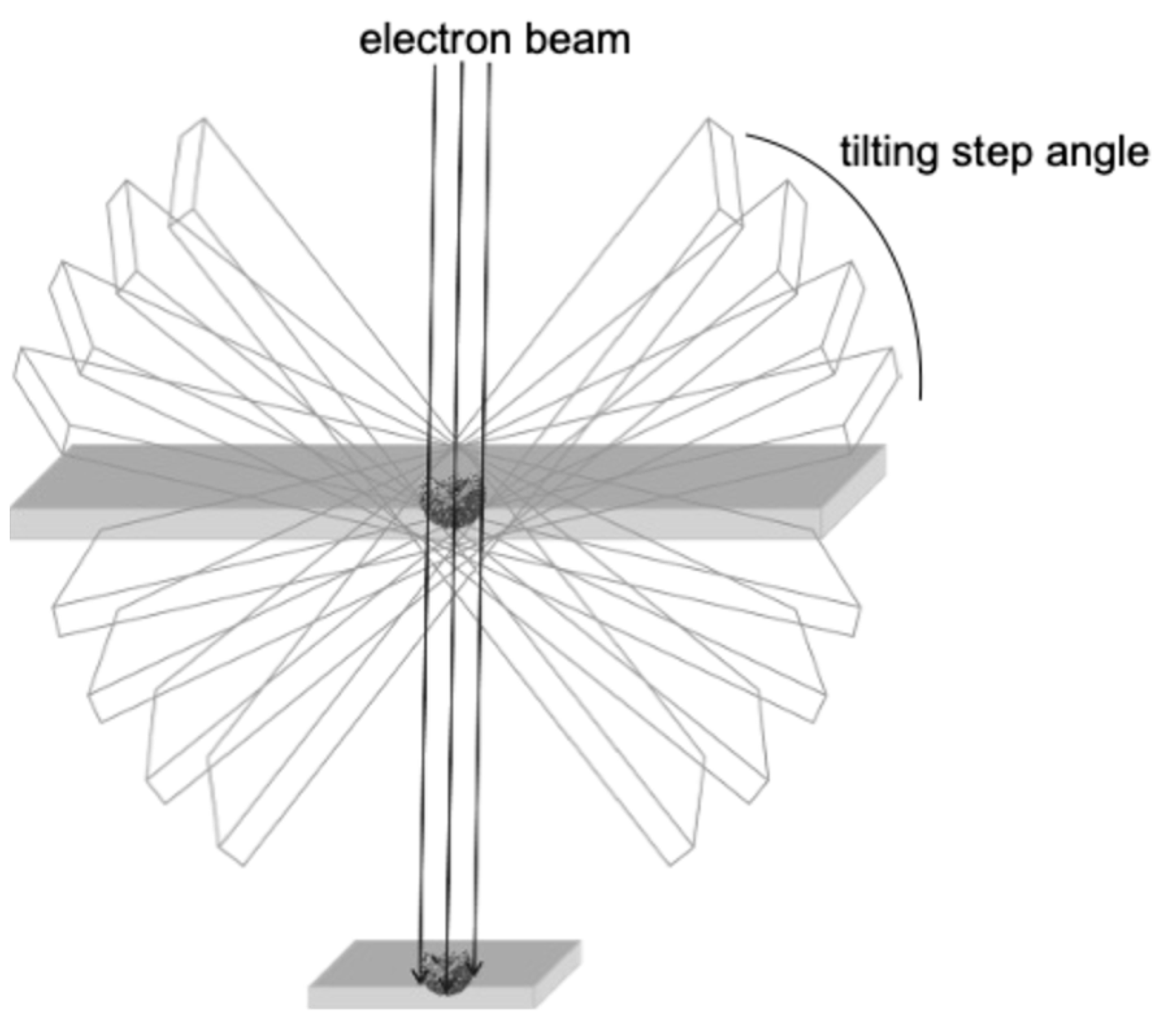
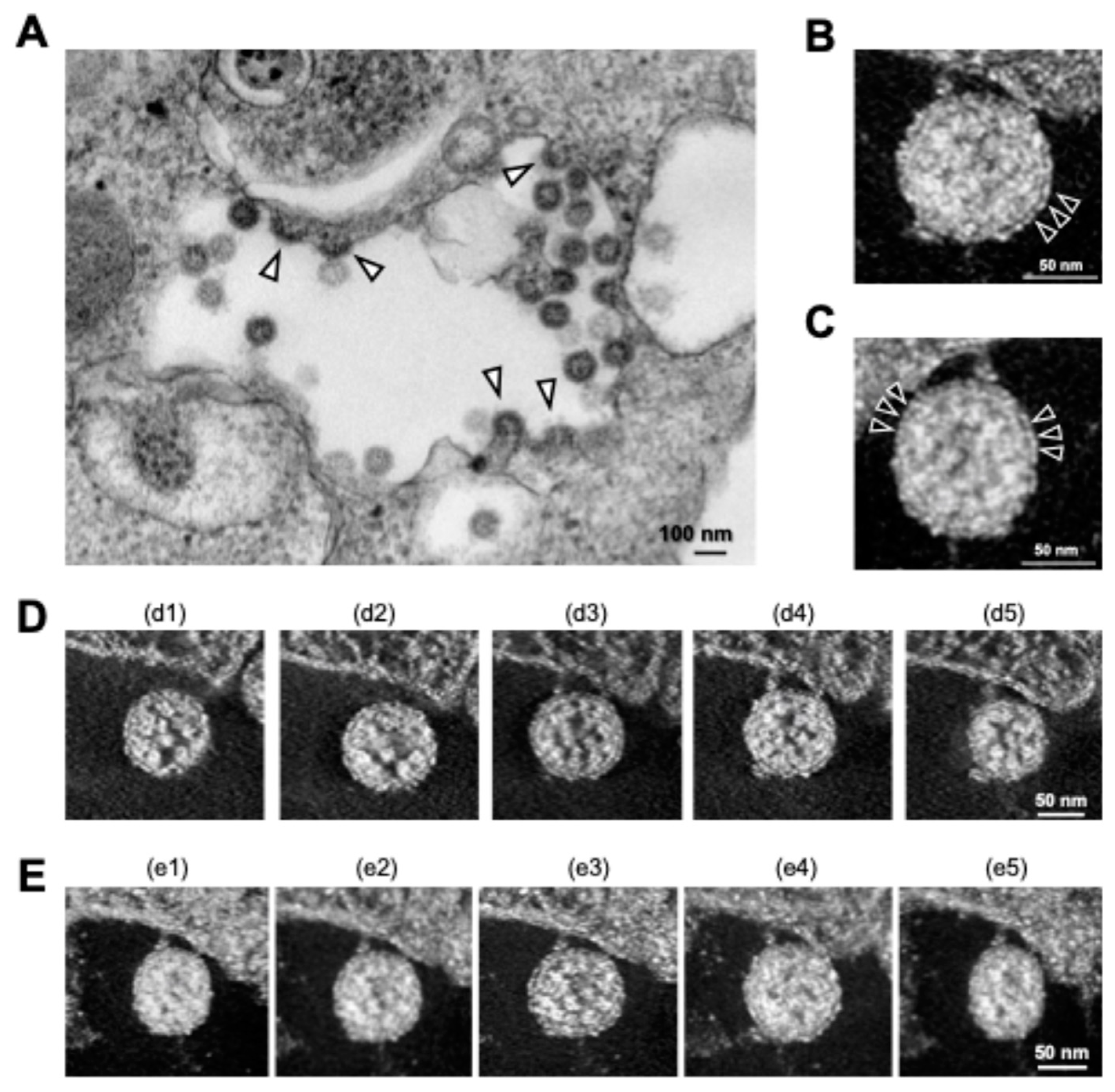
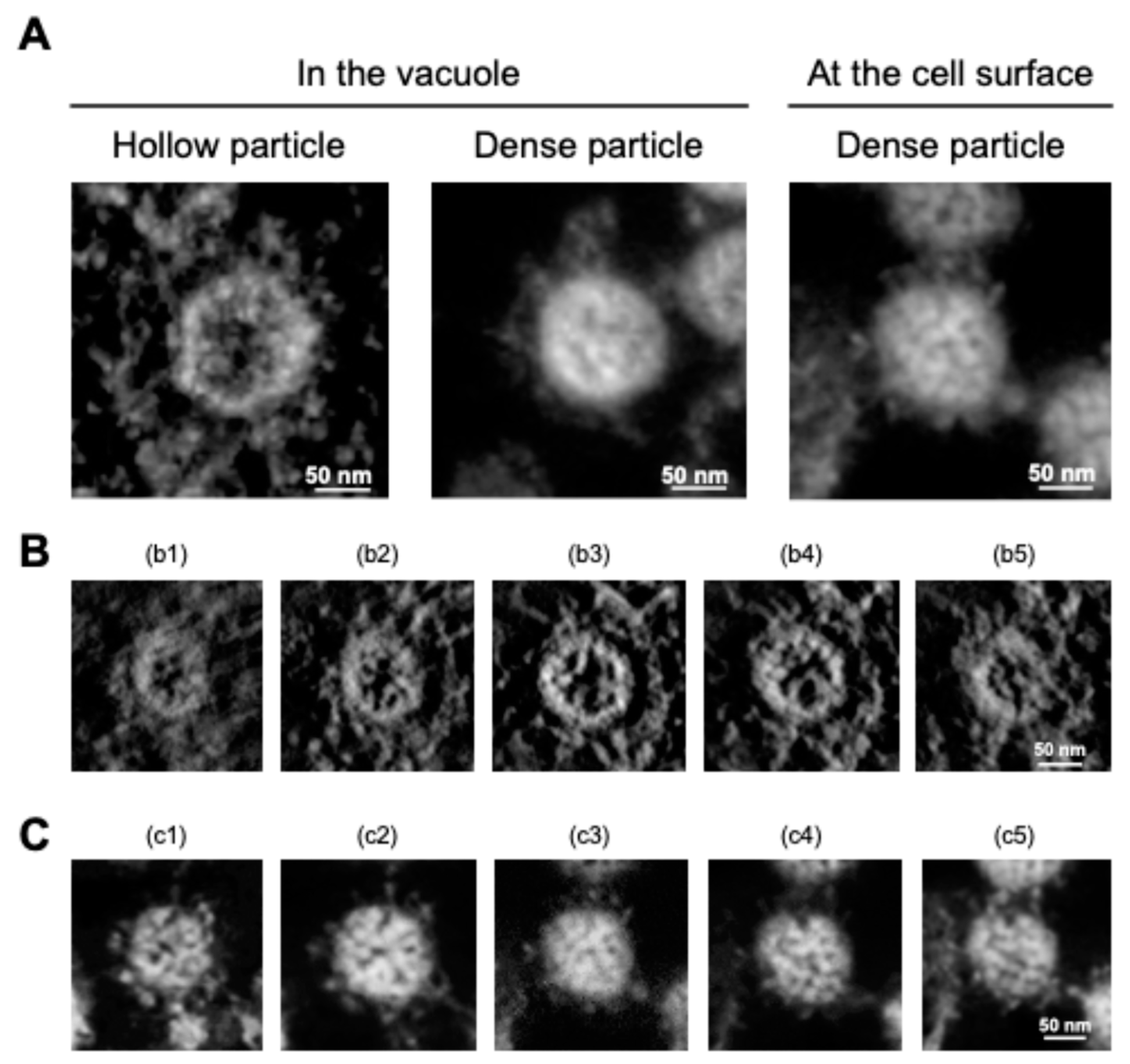
3. New Insights into SARS-CoV-2 Particle Formation Obtained by TEM and ET
4. EM as a Tool to Study the Morphological Integrity of Genetically Engineered Viruses
5. Conclusion
Author Contributions
Conflicts of Interest
References
- Colantuoni, A.; Martini, R.; Caprari, P.; Ballestri, M.; Capecchi, P.L.; Gnasso, A.; Presti, R.L.; Marcoccia, A.; Rossi, M.; Caimi, G. COVID-19 Sepsis and Microcirculation Dysfunction. Front. Physiol. 2020, 11, 747. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, X.; Cai, Y.; Xia, J.; Zhou, X.; Xu, S.; Huang, H.; Zhang, L.; Zhou, X.; Du, C.; et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 2020, 180, 934–943. [Google Scholar] [CrossRef] [PubMed]
- WHO Coronavirus Disease (COVID-19) Dashboard. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 1 August 2024).
- Li, X.; Mi, Z.; Liu, Z.; Rong, P. SARS-CoV-2: Pathogenesis, Therapeutics, Variants, and Vaccines. Front. Microbiol. 2024, 15, 1334152. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Zhang, J.; Lan, Y.; Sanyal, S. Membrane Heist: Coronavirus Host Membrane Remodeling during Replication. Biochimie 2020, 179, 229–236. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 Structure and Replication Characterized by in Situ Cryo-Electron Tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef] [PubMed]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; Jong, A.W.M. de; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A Molecular Pore Spans the Double Membrane of the Coronavirus Replication Organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.; Lee, J.-Y.; Cerikan, B.; Neufeldt, C.J.; Oorschot, V.M.J.; Köhrer, S.; Hennies, J.; Schieber, N.L.; Ronchi, P.; Mizzon, G.; et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe. 2020, 28, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535. [Google Scholar] [CrossRef]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S.; et al. Olfactory Transmucosal SARS-CoV-2 Invasion as a Port of Central Nervous System Entry in Individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef]
- Martines, R.B.; Ritter, J.M.; Matkovic, E.; Gary, J.; Bollweg, B.C.; Bullock, H.; Goldsmith, C.S.; Silva-Flannery, L.; Seixas, J.N.; Reagan-Steiner, S.; et al. Pathology and Pathogenesis of SARS-CoV-2 Associated with Fatal Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 2005–2015. [Google Scholar] [CrossRef]
- Dittmayer, C.; Meinhardt, J.; Radbruch, H.; Radke, J.; Heppner, B.I.; Heppner, F.L.; Stenzel, W.; Holland, G.; Laue, M. Why Misinterpretation of Electron Micrographs in SARS-CoV-2-Infected Tissue Goes Viral. Lancet 2020, 396, e64–e65. [Google Scholar] [CrossRef]
- G. , K.T.; Dean, E.; S., G.C.; R., Z.S.; Teresa, P.; Shannon, E.; Suxiang, T.; Carlo, U.; A., C.J.; Wilina, L.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef]
- Frank, J. “Just in Time”: The Role of Cryo-Electron Microscopy in Combating Recent Pandemics. Biochemistry 2021, 60, 3449–3451. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and Distributions of SARS-CoV-2 Spike Proteins on Intact Virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Turoňová, B.; Sikora, M.; Schürmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; Bülow, S. von; Gecht, M.; Bagola, K.; Hörner, C.; et al. In Situ Structural Analysis of SARS-CoV-2 Spike Reveals Flexibility Mediated by Three Hinges. Science 2020, 370, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, L.; Howe, A.; Gilchrist, J.B.; Sheng, Y.; Sun, D.; Knight, M.L.; Zanetti-Domingues, L.C.; Bateman, B.; Krebs, A.-S.; Chen, L.; et al. Correlative Multi-Scale Cryo-Imaging Unveils SARS-CoV-2 Assembly and Egress. Nat. Commun. 2021, 12, 4629. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Fujioka, Y.; Sakaguchi, S.; Suzuki, Y.; Nakano, T. Three-Dimensional Reconstruction by Electron Tomography for the Application to Ultrastructural Analysis of SARS-CoV-2 Particles. Med Mol Morphol 2021, 55, 1–8. [Google Scholar] [CrossRef]
- Wu, H.; Fujioka, Y.; Sakaguchi, S.; Suzuki, Y.; Nakano, T. Morphological Analysis for Two Types of Viral Particles in Vacuoles of SARS-CoV-2-Infected Cells. Med. Mol. Morphol. 2024, 1–12. [Google Scholar] [CrossRef]
- Otegui, M.S.; Pennington, J.G. Electron Tomography in Plant Cell Biology. Microscopy 2019, 68, 69–79. [Google Scholar] [CrossRef]
- Hardenbrook, N.J.; Zhang, P. A Structural View of the SARS-CoV-2 Virus and Its Assembly. Curr. Opin. Virol. 2022, 52, 123–134. [Google Scholar] [CrossRef]
- Redondo, N.; Zaldívar-López, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264. [Google Scholar] [CrossRef]
- Riegger, R.J.; Caliskan, N. Thinking Outside the Frame: Impacting Genomes Capacity by Programmed Ribosomal Frameshifting. Front. Mol. Biosci. 2022, 9, 842261. [Google Scholar] [CrossRef]
- Saibil, H.R. Cryo-EM in Molecular and Cellular Biology. Mol. Cell 2022, 82, 274–284. [Google Scholar] [CrossRef]
- Parthasarathy, M. V. Transmission electron microscopy: Chemical fixation, freezing methods, and inmunolocalization. In The Maize Handbook. Springer, New York, 1994; pp. 118–134.
- Carter, M. , Essner, R., Goldstein, N., Iyer, M. Microscopy. In Guide to Research Techniques in Neuroscience, 3rd ed.; Academic Press, Massachusetts, 2022; pp. 115–143.
- Cheng, Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 2015, 161, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Taha, B.A.; Mashhadany, Y.A.; Al-Jumaily, A.H.J.; Zan, M.S.D.B.; Arsad, N. SARS-CoV-2 Morphometry Analysis and Prediction of Real Virus Levels Based on Full Recurrent Neural Network Using TEM Images. Viruses 2022, 14, 2386. [Google Scholar] [CrossRef] [PubMed]
- Pizzato, M.; Baraldi, C.; Sopetto, G.B.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022, 1, 815388. [Google Scholar] [CrossRef]
- Chenavier, F.; Estrozi, L.F.; Teulon, J.-M.; Zarkadas, E.; Freslon, L.-L.; Pellequer, J.-L.; Ruigrok, R.W.H.; Schoehn, G.; Ballandras-Colas, A.; Crépin, T. Cryo-EM Structure of Influenza Helical Nucleocapsid Reveals NP-NP and NP-RNA Interactions as a Model for the Genome Encapsidation. Sci. Adv. 2023, 9, eadj9974. [Google Scholar] [CrossRef]
- Knoops, K.; Kikkert, M.; Worm, S.H.E. van den; Zevenhoven-Dobbe, J.C.; Meer, Y. van der; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum. PLoS Biol. 2008, 6, e226. [Google Scholar] [CrossRef]
- Pinto, A.L.; Rai, R.K.; Brown, J.C.; Griffin, P.; Edgar, J.R.; Shah, A.; Singanayagam, A.; Hogg, C.; Barclay, W.S.; Futter, C.E.; et al. Ultrastructural Insight into SARS-CoV-2 Entry and Budding in Human Airway Epithelium. Nat. Commun. 2022, 13, 1609. [Google Scholar] [CrossRef]
- Barreto-Vieira, D.F.; Silva, M.A.N. da; Almeida, A.L.T. de; Rasinhas, A. da C.; Monteiro, M.E.; Miranda, M.D.; Motta, F.C.; Siqueira, M.M.; Girard-Dias, W.; Archanjo, B.S.; et al. SARS-CoV-2: Ultrastructural Characterization of Morphogenesis in an In Vitro System. Viruses 2022, 14, 201. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, H.; Huang, W.; Zhou, J.; Qiu, M.; Deng, Z.; Chen, L.; Weng, Y.; Cai, L.; Gu, Y.; et al. Cell Morphological Analysis of SARS-CoV-2 Infection by Transmission Electron Microscopy. J. Thorac. Dis. 2020, 12, 4368–4373. [Google Scholar] [CrossRef]
- Calder, L.J.; Calcraft, T.; Hussain, S.; Harvey, R.; Rosenthal, P.B. Electron Cryotomography of SARS-CoV-2 Virions Reveals Cylinder-Shaped Particles with a Double Layer RNP Assembly. Commun. Biol. 2022, 5, 1210. [Google Scholar] [CrossRef]
- Vogt, V. M. Retroviral virions and genomes. In Retroviruses. Cold Spring Harbor Laboratory Press, New York, 1997.
- Takasaki, T.; Kurane, I.; Aihara, H.; Ohkawa, N.; Yamaguchi, J. Electron Microscopic Study of Human Immunodeficiency Virus Type 1 (HIV-1) Core Structure: Two RNA Strands in the Core of Mature and Budding Particles. Arch. Virol. 1997, 142, 375–382. [Google Scholar] [CrossRef]
- Saha, I.; Saffarian, S. Dynamics of the HIV Gag Lattice Detected by Localization Correlation Analysis and Time-Lapse IPALM. Biophys. J. 2020, 119, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Lau, D.; Márquez, C.L.; Parker, M.W.; Böcking, A.T. Negative Staining Transmission Electron Microscopy of HIV Viral Particles Permeabilized with PFO and Capsid Stabilized with IP6. Bio Protoc. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.; Ganser-Pornillos, B.K.; Tivol, W.F.; Sundquist, W.I.; Jensen, G.J. Three-Dimensional Structure of HIV-1 Virus-like Particles by Electron Cryotomography. J. Mol. Biol. 2005, 346, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.G.; Wilk, T.; Welker, R.; Kräusslich, H.; Fuller, S.D. Structural Organization of Authentic, Mature HIV-1 Virions and Cores. EMBO J. 2003, 22, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.M.; Leaman, D.P.; Lovendahl, K.N.; Croft, J.T.; Benhaim, M.A.; Hodge, E.A.; Zwick, M.B.; Lee, K.K. Cryo-ET of Env on Intact HIV Virions Reveals Structural Variation and Positioning on the Gag Lattice. Cell 2022, 185, 641–653. [Google Scholar] [CrossRef]
- Ward, A.E.; Kiessling, V.; Pornillos, O.; White, J.M.; Ganser-Pornillos, B.K.; Tamm, L.K. HIV-Cell Membrane Fusion Intermediates Are Restricted by Serincs as Revealed by Cryo-Electron and TIRF Microscopy. J. Biol. Chem. 2020, 295, 15183–15195. [Google Scholar] [CrossRef]
- Passos, D.O.; Li, M.; Yang, R.; Rebensburg, S.V.; Ghirlando, R.; Jeon, Y.; Shkriabai, N.; Kvaratskhelia, M.; Craigie, R.; Lyumkis, D. Cryo-EM Structures and Atomic Model of the HIV-1 Strand Transfer Complex Intasome. Science 2017, 355, 89–92. [Google Scholar] [CrossRef]
- Philippe, N.; Legendre, M.; Doutre, G.; Couté, Y.; Poirot, O.; Lescot, M.; Arslan, D.; Seltzer, V.; Bertaux, L.; Bruley, C.; et al. Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes. Science 2013, 341, 281–286. [Google Scholar] [CrossRef]
- Burton-Smith, R.N.; Murata, K. Cryo-Electron Microscopy of the Giant Viruses. Microscopy 2021, 70, 477–486. [Google Scholar] [CrossRef]
- Chihara, A.; Burton-Smith, R.N.; Kajimura, N.; Mitsuoka, K.; Okamoto, K.; Song, C.; Murata, K. A Novel Capsid Protein Network Allows the Characteristic Internal Membrane Structure of Marseilleviridae Giant Viruses. Sci. Rep. 2022, 12, 21428. [Google Scholar] [CrossRef]
- Xiao, C.; Fischer, M.G.; Bolotaulo, D.M.; Ulloa-Rondeau, N.; Avila, G.A.; Suttle, C.A. Cryo-EM Reconstruction of the Cafeteria Roenbergensis Virus Capsid Suggests Novel Assembly Pathway for Giant Viruses. Sci. Rep. 2017, 7, 5484. [Google Scholar] [CrossRef] [PubMed]
- Downing, K.H.; Glaeser, R.M. Estimating the Effect of Finite Depth of Field in Single-Particle Cryo-EM. Ultramicroscopy 2018, 184, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced Isolation of SARS-CoV-2 by TMPRSS2-Expressing Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Mastronarde, D.N. Automated Electron Microscope Tomography Using Robust Prediction of Specimen Movements. J. Struct. Biol. 2005, 152, 36–51. [Google Scholar] [CrossRef]
- Gan, L.; Jensen, G.J. Electron Tomography of Cells. Q. Rev. Biophys. 2012, 45, 27–56. [Google Scholar] [CrossRef]
- McEwen, B.F.; Marko, M. The Emergence of Electron Tomography as an Important Tool for Investigating Cellular Ultrastructure. J. Histochem. Cytochem. 2000, 49, 553–563. [Google Scholar] [CrossRef]
- Low, J.G.H.; Lee, L.S.; Ooi, E.E.; Ethirajulu, K.; Yeo, P.; Matter, A.; Connolly, J.E.; Skibinski, D.A.G.; Saudan, P.; Bachmann, M.; et al. Safety and Immunogenicity of a Virus-like Particle Pandemic Influenza A (H1N1) 2009 Vaccine: Results from a Double-Blinded, Randomized Phase I Clinical Trial in Healthy Asian Volunteers. Vaccine 2014, 32, 5041–5048. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Zha, L.; Cabral-Miranda, G.; Bachmann, M.F. Major Findings and Recent Advances in Virus–like Particle (VLP)-Based Vaccines. Semin. Immunol. 2017, 34, 123–132. [Google Scholar] [CrossRef]
- Chen, G.L.; Coates, E.E.; Plummer, S.H.; Carter, C.A.; Berkowitz, N.; Conan-Cibotti, M.; Cox, J.H.; Beck, A.; O’Callahan, M.; Andrews, C.; et al. Effect of a Chikungunya Virus–Like Particle Vaccine on Safety and Tolerability Outcomes. JAMA 2020, 323, 1369–1377. [Google Scholar] [CrossRef]
- Swann, H.; Sharma, A.; Preece, B.; Peterson, A.; Eldredge, C.; Belnap, D.M.; Vershinin, M.; Saffarian, S. Minimal System for Assembly of SARS-CoV-2 Virus like Particles. Sci. Rep. 2020, 10, 21877. [Google Scholar] [CrossRef]
- Syed, A.M.; Taha, T.Y.; Tabata, T.; Chen, I.P.; Ciling, A.; Khalid, M.M.; Sreekumar, B.; Chen, P.-Y.; Hayashi, J.M.; Soczek, K.M.; et al. Rapid Assessment of SARS-CoV-2 Evolved Variants Using Virus-like Particles. Science 2021, 374, 1626–1632. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.M.; Ciling, A.; Taha, T.Y.; Chen, I.P.; Khalid, M.M.; Sreekumar, B.; Chen, P.-Y.; Kumar, G.R.; Suryawanshi, R.; Silva, I.; et al. Omicron Mutations Enhance Infectivity and Reduce Antibody Neutralization of SARS-CoV-2 Virus-like Particles. Proc. Natl. Acad. Sci. USA 2022, 119, e2200592119. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Shi, M.; Li, J.; Song, P.; Li, N. Construction of SARS-CoV-2 Virus-Like Particles by Mammalian Expression System. Front. Bioeng. Biotechnol. 2020, 8, 862. [Google Scholar] [CrossRef]
- Miura, K.; Suzuki, Y.; Ishida, K.; Arakawa, M.; Wu, H.; Fujioka, Y.; Emi, A.; Maeda, K.; Hamajima, R.; Nakano, T.; et al. Distinct Motifs in the E Protein Are Required for SARS-CoV-2 Virus Particle Formation and Lysosomal Deacidification in Host Cells. J. Virol. 2023, e00426–23. [Google Scholar] [CrossRef]
- Racaniello, V.R.; Baltimore, D. Cloned Poliovirus Complementary DNA Is Infectious in Mammalian Cells. Science 1981, 214, 916–919. [Google Scholar] [CrossRef]
- Ávila-Pérez, G.; Nogales, A.; Martín, V.; Almazán, F.; Martínez-Sobrido, L. Reverse Genetic Approaches for the Generation of Recombinant Zika Virus. Viruses 2018, 10, 597. [Google Scholar] [CrossRef]
- Aubry, F.; Nougairède, A.; Gould, E.A.; Lamballerie, X. de Flavivirus Reverse Genetic Systems, Construction Techniques and Applications: A Historical Perspective. Antiviral Res 2015, 114, 67–85. [Google Scholar] [CrossRef]
- Kurhade, C.; Xie, X.; Shi, P.-Y. Reverse Genetic Systems of SARS-CoV-2 for Antiviral Research. Antiviral Res. 2023, 210, 105486. [Google Scholar] [CrossRef]
- Kril, V.; Aïqui-Reboul-Paviet, O.; Briant, L.; Amara, A. New Insights into Chikungunya Virus Infection and Pathogenesis. Annu. Rev. Virol. 2021, 8, 1–21. [Google Scholar] [CrossRef]
- Halstead, S.B. Reappearance of Chikungunya, Formerly Called Dengue, in the Americas. Emerg. Infect. Dis. 2015, 21, 557–561. [Google Scholar] [CrossRef]
- Suzuki, Y.; Tanaka, A.; Maeda, Y.; Emi, A.; Fujioka, Y.; Sakaguchi, S.; Vasudevan, S.G.; Kobayashi, T.; Lim, C.-K.; Takasaki, T.; et al. Construction and Characterization of an Infectious Clone Generated from Chikungunya Virus SL11131 Strain. Virology 2021, 552, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, M.; Nakano, T.; Sakuragi, S.; Shioda, T.; Sano, K.; Sakuragi, J. The Relationship between HIV-1 Genome RNA Dimerization, Virion Maturation and Infectivity. Nucleic Acids Res. 2011, 39, 3404–3417. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P.; Barrett, A.D.T. Pathogenesis and Pathophysiology of Yellow Fever. Adv. Virus Res. 2003, 60, 343–395. [Google Scholar] [CrossRef] [PubMed]
- Monath, T.P. Treatment of Yellow Fever. Antiviral Res. 2008, 78, 116–124. [Google Scholar] [CrossRef]
- Bodakuntla, S.; Kuhn, C.C.; Biertümpfel, C.; Mizuno, N. Cryo-Electron Microscopy in the Fight against COVID-19—Mechanism of Virus Entry. Front. Mol. Biosci. 2023, 10, 1252529. [Google Scholar] [CrossRef]
- Saville, J.W.; Berezuk, A.M.; Srivastava, S.S.; Subramaniam, S. Three-Dimensional Visualization of Viral Structure, Entry, and Replication Underlying the Spread of SARS-CoV-2. Chem. Rev. 2022, 122, 14066–14084. [Google Scholar] [CrossRef]
- Leigh, K.E.; Modis, Y. Imaging and Visualizing SARS-CoV-2 in a New Era for Structural Biology. Interface Focus 2021, 11, 20210019. [Google Scholar] [CrossRef]
- Liu, C.; Mendonça, L.; Yang, Y.; Gao, Y.; Shen, C.; Liu, J.; Ni, T.; Ju, B.; Liu, C.; Tang, X.; et al. The Architecture of Inactivated SARS-CoV-2 with Postfusion Spikes Revealed by Cryo-EM and Cryo-ET. Structure 2020, 28, 1218–1224. [Google Scholar] [CrossRef]
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural Organization of a Filamentous Influenza A Virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar] [CrossRef]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor Binding and Priming of the Spike Protein of SARS-CoV-2 for Membrane Fusion. Nature 2020, 588, 327–330. [Google Scholar] [CrossRef]
- Fujiyoshi, Y.; Kume, N.P.; Sakata, K.; Sato, S.B. Fine Structure of Influenza A Virus Observed by Electron Cryo-microscopy. EMBO J. 1994, 13, 318–326. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




