Submitted:
09 September 2024
Posted:
10 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Differential Expressed Enzymes in Tumors as Compared to Normal Adjacent Liver Tissues from Hepatoblastoma Human Samples
2.2. Activated Metabolic Transcriptional Program in Hepatoblastoma Tumors
2.3. Activated Metabolic Network in Hepatoblastoma Tumors
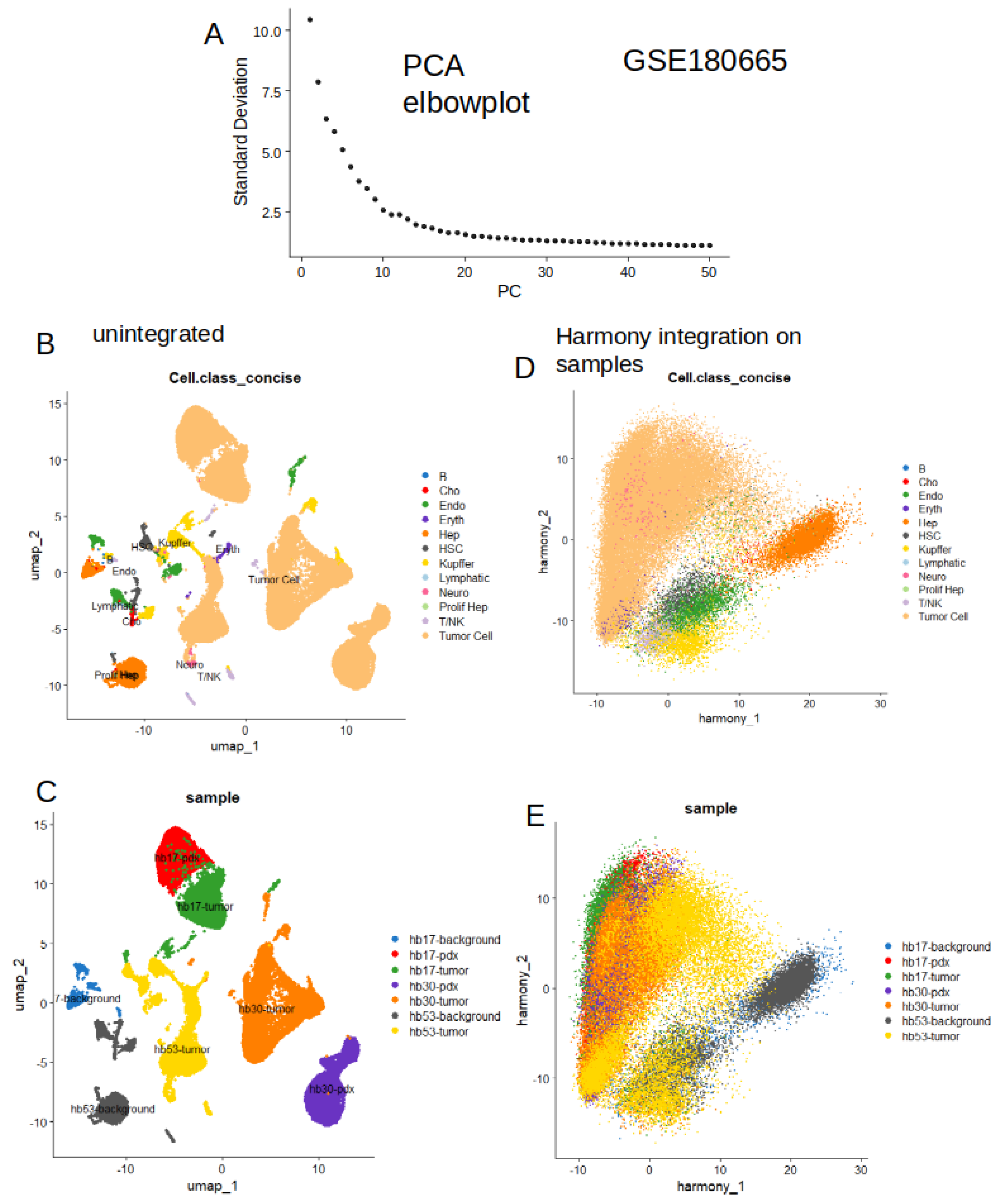
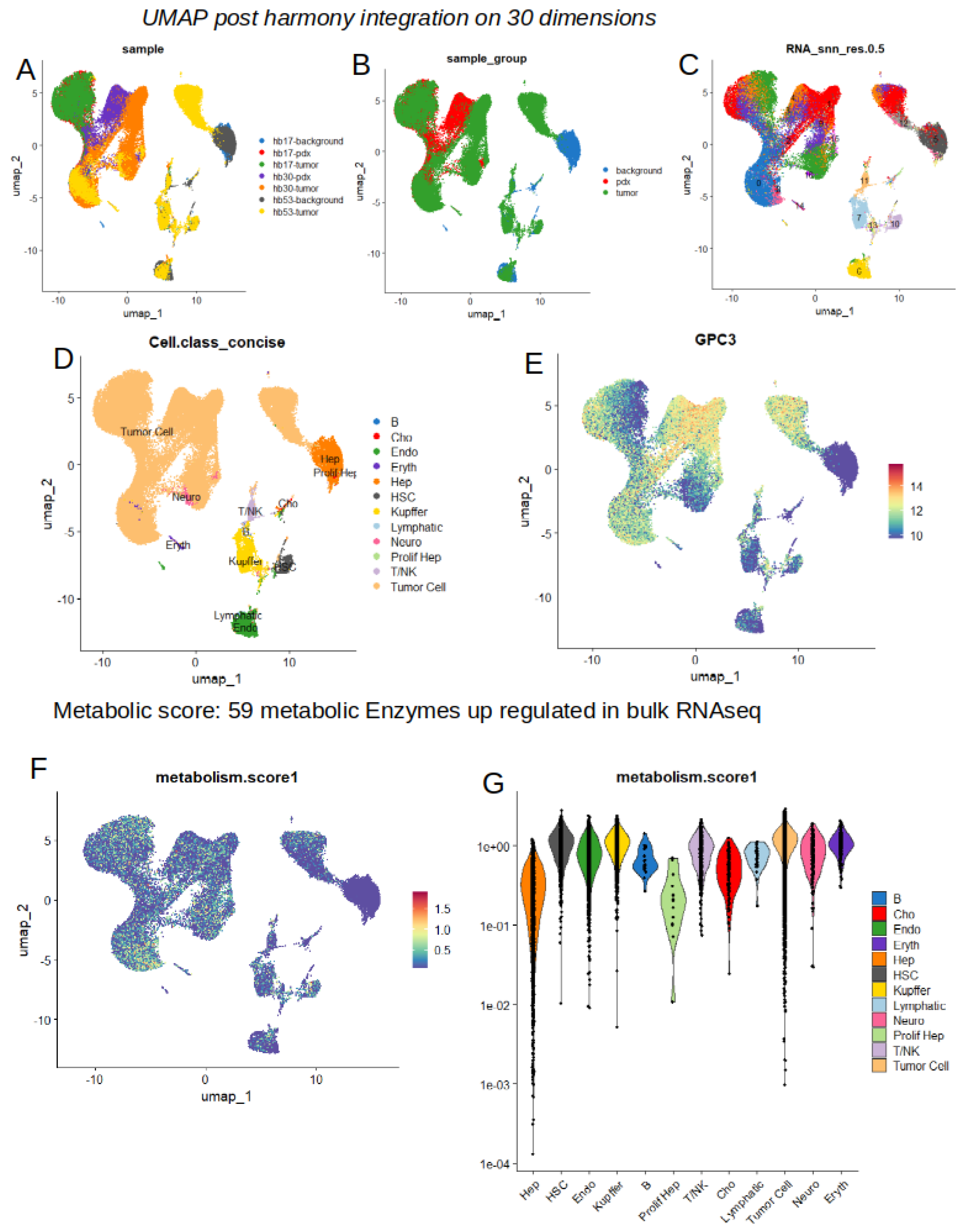
2.4. High Level of Metabolic Score at Single Cell Level in Tumor Cells from Hepatoblastoma
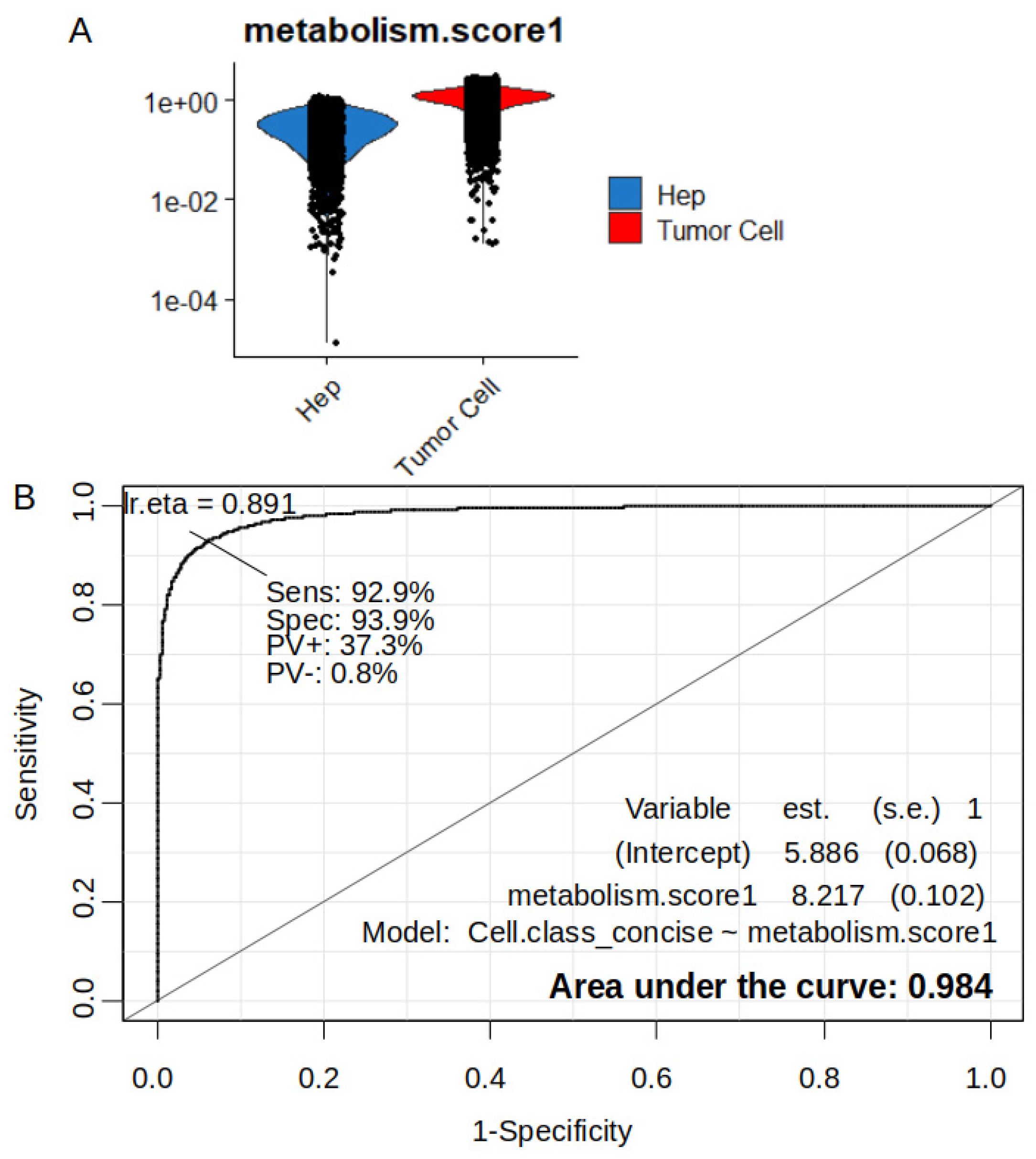
2.5. Increase of Metabolic Score is Highly Predictive of Tumor Cell Status in Single Cell Transcriptome from Hepatoblastoma
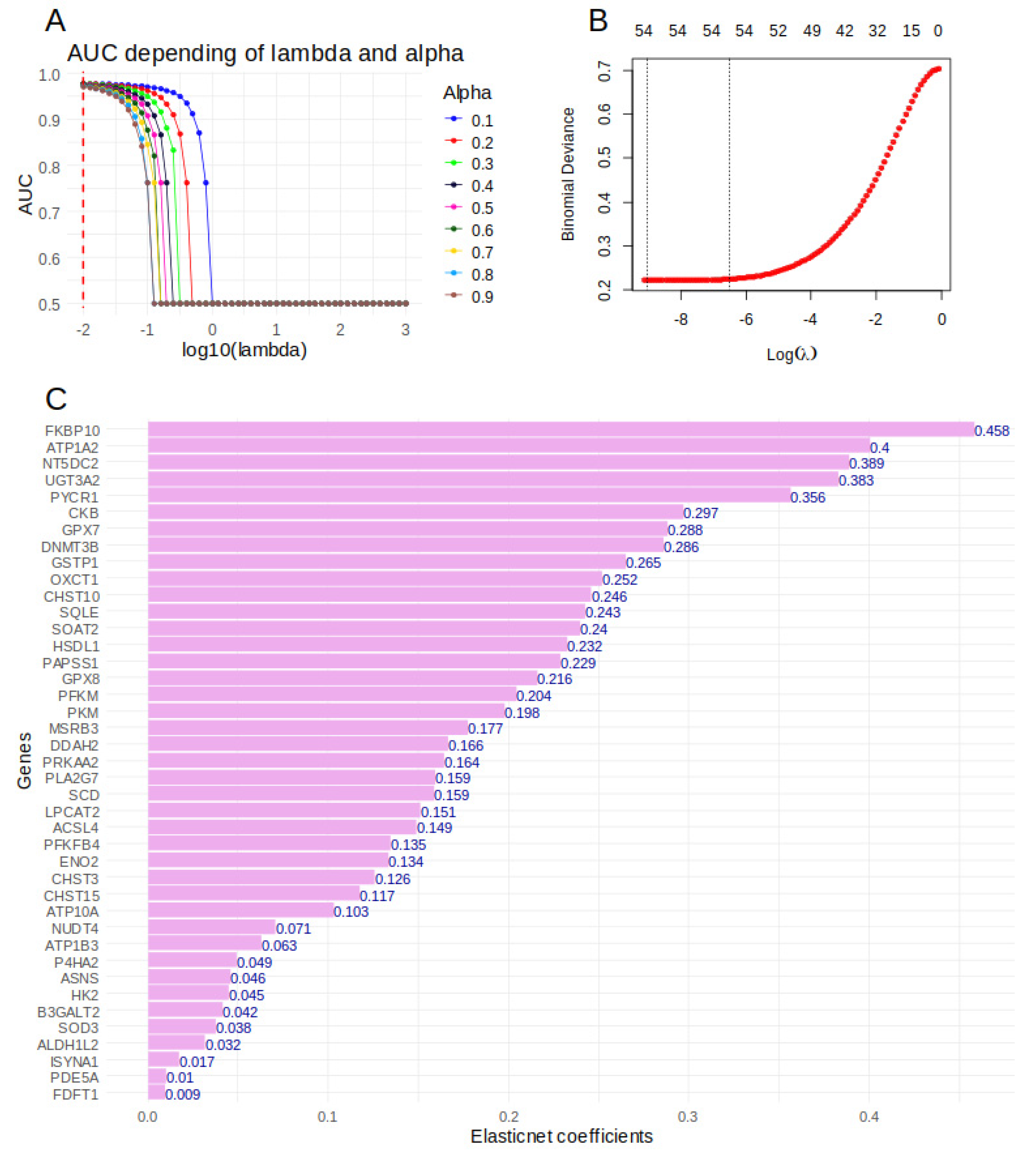
2.6. Importance of Over Expressed Metabolic Markers to Predict Tumor Cell Status in sc-RNAseq of Hepatoblastoma Samples
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Public Dataset of RNA-Sequencing Performed on Human Hepatoblastoma Tissues
5.2. Public Dataset of Single Cell RNA_Sequencing Performed on Hepatoblastoma Samples
5.3. Mammaliam Metabolic Transcriptional Program
5.4. RNA-Sequencing Analyses
5.5. Single Cell RNA-Sequencing Metabolic Score Quantification
5.6. Machine Learning Elasticnet Model on Metabolic Markers
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spector, L.G.; Birch, J. The Epidemiology of Hepatoblastoma. Pediatric Blood & Cancer 2012, 59, 776–779. [Google Scholar] [CrossRef]
- Hager, J.; Sergi, C.M. Hepatoblastoma. In Liver Cancer; Departments of Pediatrics, Laboratory Medicine and Pathology, Stollery Children’s Hospital, University of Alberta, Edmonton, AB, Canada, Sergi, C.M., Eds.; Exon Publications, 2021; pp. 145–164 ISBN 978-0-645-00172-3.
- Yang, T.; Whitlock, R.S.; Vasudevan, S.A. Surgical Management of Hepatoblastoma and Recent Advances. Cancers 2019, 11, 1944. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wu, S.; Tang, H. An Update on Diagnosis and Treatment of Hepatoblastoma. BST 2023, 17, 445–457. [Google Scholar] [CrossRef]
- Hasegawa, M.; Sugiyama, M.; Terashita, Y.; Cho, Y.; Manabe, A. Hepatoblastoma with Bone/Bone Marrow Metastasis in Li-Fraumeni Syndrome Patient. Pediatrics International 2022, 64, e15135. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; H. Feusner, J. Cerebral Metastasis of Hepatoblastoma: A Review. Journal of Pediatric Hematology/Oncology 2016, 38, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Current Biology 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Bao, M.H.-R.; Wong, C.C.-L. Hypoxia, Metabolic Reprogramming, and Drug Resistance in Liver Cancer. Cells 2021, 10, 1715. [Google Scholar] [CrossRef]
- Yang, F.; Hilakivi-Clarke, L.; Shaha, A.; Wang, Y.; Wang, X.; Deng, Y.; Lai, J.; Kang, N. Metabolic Reprogramming and Its Clinical Implication for Liver Cancer. Hepatology 2023, 78, 1602–1624. [Google Scholar] [CrossRef]
- Clavería-Cabello, A.; Herranz, J.M.; Latasa, M.U.; Arechederra, M.; Uriarte, I.; Pineda-Lucena, A.; Prosper, F.; Berraondo, P.; Alonso, C.; Sangro, B.; et al. Identification and Experimental Validation of Druggable Epigenetic Targets in Hepatoblastoma. Journal of Hepatology 2023, 79, 989–1005. [Google Scholar] [CrossRef]
- Wang, H.; Lu, J.; Chen, X.; Schwalbe, M.; Gorka, J.E.; Mandel, J.A.; Wang, J.; Goetzman, E.S.; Ranganathan, S.; Dobrowolski, S.F.; et al. Acquired Deficiency of Peroxisomal Dicarboxylic Acid Catabolism Is a Metabolic Vulnerability in Hepatoblastoma. Journal of Biological Chemistry 2021, 296, 100283. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, F.; Wan, T.; Chen, A.; Wang, H.; Jiang, B.; Zhao, W.; Liao, S.; Wang, S.; Li, G.; et al. ZEB1 Enhances Warburg Effect to Facilitate Tumorigenesis and Metastasis of HCC by Transcriptionally Activating PFKM. Theranostics 2021, 11, 5926–5938. [Google Scholar] [CrossRef] [PubMed]
- Duda, P.; Janczara, J.; McCubrey, J.A.; Gizak, A.; Rakus, D. The Reverse Warburg Effect Is Associated with Fbp2-Dependent Hif1α Regulation in Cancer Cells Stimulated by Fibroblasts. Cells 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Alasadi, A.; Cao, B.; Guo, J.; Tao, H.; Collantes, J.; Tan, V.; Su, X.; Augeri, D.; Jin, S. Mitochondrial Uncoupler MB1-47 Is Efficacious in Treating Hepatic Metastasis of Pancreatic Cancer in Murine Tumor Transplantation Models. Oncogene 2021, 40, 2285–2295. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, E.K.; Adamski, J.; Zahn, G.; Gaumann, A.; Flores-Borja, F.; Ziegler, C.; Mycielska, M.E. Extracellular Citrate and Metabolic Adaptations of Cancer Cells. Cancer Metastasis Rev 2021, 40, 1073–1091. [Google Scholar] [CrossRef]
- Povero, D.; Chen, Y.; Johnson, S.M.; McMahon, C.E.; Pan, M.; Bao, H.; Petterson, X.-M.T.; Blake, E.; Lauer, K.P.; O’Brien, D.R.; et al. HILPDA Promotes NASH-Driven HCC Development by Restraining Intracellular Fatty Acid Flux in Hypoxia. Journal of Hepatology 2023, 79, 378–393. [Google Scholar] [CrossRef]
- Sprinzl, M.F.; Puschnik, A.; Schlitter, A.M.; Schad, A.; Ackermann, K.; Esposito, I.; Lang, H.; Galle, P.R.; Weinmann, A.; Heikenwälder, M.; et al. Sorafenib Inhibits Macrophage-Induced Growth of Hepatoma Cells by Interference with Insulin-like Growth Factor-1 Secretion. Journal of Hepatology 2015, 62, 863–870. [Google Scholar] [CrossRef]
- Buscher, H.-P. Defective Drug Uptake Contributing to Multidrug Resistance in Hepatoma Cells Can Be Evaluated in Vitro. Klin Wochenschr 1990, 68, 443–446. [Google Scholar] [CrossRef]
- Rivera-Aguirre, J.; López-Sánchez, G.N.; Chávez-Tapia, N.C.; Uribe, M.; Nuño-Lámbarri, N. Metabolic-Associated Fatty Liver Disease Regulation through Nutri EpigeneticMethylation. MRMC 2023, 23, 1680–1690. [Google Scholar] [CrossRef]
- El Taghdouini, A.; Van Grunsven, L.A. Epigenetic Regulation of Hepatic Stellate Cell Activation and Liver Fibrosis. Expert Review of Gastroenterology & Hepatology 2016, 10, 1397–1408. [Google Scholar] [CrossRef]
- Mandrekar, P. Epigenetic Regulation in Alcoholic Liver Disease. WJG 2011, 17, 2456. [Google Scholar] [CrossRef]
- Wang, S.; Zha, L.; Cui, X.; Yeh, Y.; Liu, R.; Jing, J.; Shi, H.; Chen, W.; Hanover, J.; Yin, J.; et al. Epigenetic Regulation of Hepatic Lipid Metabolism by DNA Methylation. Advanced Science 2023, 10, 2206068. [Google Scholar] [CrossRef] [PubMed]
- Moran-Salvador, E.; Mann, J. Epigenetics and Liver Fibrosis. Cellular and Molecular Gastroenterology and Hepatology 2017, 4, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Jung, Y. DNA Methylation in Nonalcoholic Fatty Liver Disease. IJMS 2020, 21, 8138. [Google Scholar] [CrossRef] [PubMed]
- Raggi, C.; Invernizzi, P. Methylation and Liver Cancer. Clinics and Research in Hepatology and Gastroenterology 2013, 37, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Meza, G.; Von Felden, J.; Gonzalez-Kozlova, E.E.; Garcia-Lezana, T.; Peix, J.; Portela, A.; Craig, A.J.; Sayols, S.; Schwartz, M.; Losic, B.; et al. DNA Methylation Profiling of Human Hepatocarcinogenesis. Hepatology 2021, 74, 183–199. [Google Scholar] [CrossRef]
- Wang, Q.; Liang, N.; Yang, T.; Li, Y.; Li, J.; Huang, Q.; Wu, C.; Sun, L.; Zhou, X.; Cheng, X.; et al. DNMT1-Mediated Methylation of BEX1 Regulates Stemness and Tumorigenicity in Liver Cancer. Journal of Hepatology 2021, 75, 1142–1153. [Google Scholar] [CrossRef]
- Rivas, M.; Aguiar, T.; Fernandes, G.; Lemes, R.; Caires-Júnior, L.; Goulart, E.; Telles-Silva, K.; Maschietto, M.; Cypriano, M.; De Toledo, S.; et al. DNA Methylation as a Key Epigenetic Player for Hepatoblastoma Characterization. Clinics and Research in Hepatology and Gastroenterology 2021, 45, 101684. [Google Scholar] [CrossRef]
- Cui, X.; Liu, B.; Zheng, S.; Dong, K.; Dong, R. Genome-Wide Analysis of DNA Methylation in Hepatoblastoma Tissues. Oncology Letters 2016, 12, 1529–1534. [Google Scholar] [CrossRef]
- O’Malley, J.; Kumar, R.; Inigo, J.; Yadava, N.; Chandra, D. Mitochondrial Stress Response and Cancer. Trends in Cancer 2020, 6, 688–701. [Google Scholar] [CrossRef]
- Fromenty, B.; Roden, M. Mitochondrial Alterations in Fatty Liver Diseases. Journal of Hepatology 2023, 78, 415–429. [Google Scholar] [CrossRef]
- Lee, H.-Y.; Nga, H.T.; Tian, J.; Yi, H.-S. Mitochondrial Metabolic Signatures in Hepatocellular Carcinoma. Cells 2021, 10, 1901. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, H.; Waseem, M.; Parvez, S.; Qureshi, M.I. Oxaliplatin-Induced Oxidative Stress Provokes Toxicity in Isolated Rat Liver Mitochondria. Archives of Medical Research 2015, 46, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Gong, Y.-X.; Xie, D.-P.; Jeong, H.; Seo, H.; Kim, J.; Park, Y.H.; Sun, H.-N.; Kwon, T. Anticancer Effect of ERM210 on Liver Cancer Cells Through ROS/Mitochondria-Dependent Apoptosis Signaling Pathways. In Vivo 2021, 35, 2599–2608. [Google Scholar] [CrossRef] [PubMed]
- Lockman, K.A.; Baren, J.P.; Pemberton, C.J.; Baghdadi, H.; Burgess, K.E.; Plevris-Papaioannou, N.; Lee, P.; Howie, F.; Beckett, G.; Pryde, A.; et al. Oxidative Stress Rather than Triglyceride Accumulation Is a Determinant of Mitochondrial Dysfunction in in Vitro Models of Hepatic Cellular Steatosis. Liver International 2012, 32, 1079–1092. [Google Scholar] [CrossRef]
- Esmaeili, M.A.; Farimani, M.M.; Kiaei, M. Anticancer Effect of Calycopterin via PI3K/Akt and MAPK Signaling Pathways, ROS-Mediated Pathway and Mitochondrial Dysfunction in Hepatoblastoma Cancer (HepG2) Cells. Mol Cell Biochem 2014, 397, 17–31. [Google Scholar] [CrossRef]
- Yumnam, S.; Hong, G.E.; Raha, S.; Saralamma, V.V.G.; Lee, H.J.; Lee, W.; Kim, E.; Kim, G.S. Mitochondrial Dysfunction and Ca 2+ Overload Contributes to Hesperidin Induced Paraptosis in Hepatoblastoma Cells, HepG2. Journal Cellular Physiology 2016, 231, 1261–1268. [Google Scholar] [CrossRef]
- Hooks, K.B.; Audoux, J.; Fazli, H.; Lesjean, S.; Ernault, T.; Dugot-Senant, N.; Leste-Lasserre, T.; Hagedorn, M.; Rousseau, B.; Danet, C.; et al. New Insights into Diagnosis and Therapeutic Options for Proliferative Hepatoblastoma. Hepatology 2018, 68, 89–102. [Google Scholar] [CrossRef]
- Bondoc, A.; Glaser, K.; Jin, K.; Lake, C.; Cairo, S.; Geller, J.; Tiao, G.; Aronow, B. Identification of Distinct Tumor Cell Populations and Key Genetic Mechanisms through Single Cell Sequencing in Hepatoblastoma. Commun Biol 2021, 4, 1049. [Google Scholar] [CrossRef]
- Zhou, S.; O’Gorman, M.R.G.; Yang, F.; Andresen, K.; Wang, L. Glypican 3 as a Serum Marker for Hepatoblastoma. Sci Rep 2017, 7, 45932. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Ma, W.K.; Voss, D.M.; Scharner, J.; Costa, A.S.H.; Lin, K.-T.; Jeon, H.Y.; Wilkinson, J.E.; Jackson, M.; Rigo, F.; Bennett, C.F.; et al. ASO-Based PKM Splice-Switching Therapy Inhibits Hepatocellular Carcinoma Growth. Cancer Res 2022, 82, 900–915. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Armento, A.; Bussolati, O.; Chiu, M.; Ellerkamp, V.; Scharpf, M.O.; Sander, P.; Schmid, E.; Warmann, S.W.; Fuchs, J. Hepatoblastoma: Glutamine Depletion Hinders Cell Viability in the Embryonal Subtype but High GLUL Expression Is Associated with Better Overall Survival. J Cancer Res Clin Oncol 2021, 147, 3169–3181. [Google Scholar] [CrossRef]
- Camarota, L.M.; Woollett, L.A.; Howles, P.N. Reverse Cholesterol Transport Is Elevated in Carboxyl Ester Lipase-Knockout Mice. FASEB J 2011, 25, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Fjeld, K.; Beer, S.; Johnstone, M.; Zimmer, C.; Mössner, J.; Ruffert, C.; Krehan, M.; Zapf, C.; Njølstad, P.R.; Johansson, S.; et al. Length of Variable Numbers of Tandem Repeats in the Carboxyl Ester Lipase (CEL) Gene May Confer Susceptibility to Alcoholic Liver Cirrhosis but Not Alcoholic Chronic Pancreatitis. PLoS One 2016, 11, e0165567. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Zhou, J.; Cheng, Z.; Lu, P. Squalene Epoxidase (SQLE) Promotes the Growth and Migration of the Hepatocellular Carcinoma Cells. Tumour Biol 2015, 36, 6173–6179. [Google Scholar] [CrossRef]
- Chang, N.-Y.; Chan, Y.-J.; Ding, S.-T.; Lee, Y.-H.; HuangFu, W.-C.; Liu, I.-H. Sterol O-Acyltransferase 2 Contributes to the Yolk Cholesterol Trafficking during Zebrafish Embryogenesis. PLoS One 2016, 11, e0167644. [Google Scholar] [CrossRef]
- Rivas, M.P.; Aguiar, T.F.M.; Fernandes, G.R.; Caires-Júnior, L.C.; Goulart, E.; Telles-Silva, K.A.; Cypriano, M.; De Toledo, S.R.C.; Rosenberg, C.; Carraro, D.M.; et al. TET Upregulation Leads to 5-Hydroxymethylation Enrichment in Hepatoblastoma. Front. Genet. 2019, 10, 553. [Google Scholar] [CrossRef]
- Antwi, S.O.; Heckman, M.; White, L.; Yan, I.; Sarangi, V.; Lauer, K.P.; Reddy, J.; Ahmed, F.; Veliginti, S.; Mejías Febres, E.D.; et al. Metabolic Liver Cancer: Associations of Rare and Common Germline Variants in One-Carbon Metabolism and DNA Methylation Genes. Hum Mol Genet 2023, 32, 2646–2655. [Google Scholar] [CrossRef]
- Ashkavand, Z.; O’Flanagan, C.; Hennig, M.; Du, X.; Hursting, S.D.; Krupenko, S.A. Metabolic Reprogramming by Folate Restriction Leads to a Less Aggressive Cancer Phenotype. Mol Cancer Res 2017, 15, 189–200. [Google Scholar] [CrossRef]
- Sun, X.; Han, L.; Seth, P.; Bian, S.; Li, L.; Csizmadia, E.; Junger, W.G.; Schmelzle, M.; Usheva, A.; Tapper, E.B.; et al. Disordered Purinergic Signaling and Abnormal Cellular Metabolism Are Associated with Development of Liver Cancer in Cd39/ENTPD1 Null Mice. Hepatology 2013, 57, 205–216. [Google Scholar] [CrossRef]
- Staller, D.W.; Panigrahi, S.S.; Jayasinghe, Y.P.; Dong, Y.; Mahto, S.; Kumar, V.; Ronning, D.R.; Mahato, R.I. A Novel Phosphodiesterase Inhibitor for the Treatment of Chronic Liver Injury and Metabolic Diseases. Hepatology 2024. [Google Scholar] [CrossRef] [PubMed]
- Barrett, K.G.; Fang, H.; Gargano, M.D.; Markovich, D.; Kocarek, T.A.; Runge-Morris, M. Regulation of Murine Hepatic Hydroxysteroid Sulfotransferase Expression in Hyposulfatemic Mice and in a Cell Model of 3’-Phosphoadenosine-5’-Phosphosulfate Deficiency. Drug Metab Dispos 2013, 41, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Pike, S.T.; Rajendra, R.; Artzt, K.; Appling, D.R. Mitochondrial C1-Tetrahydrofolate Synthase (MTHFD1L) Supports the Flow of Mitochondrial One-Carbon Units into the Methyl Cycle in Embryos. J Biol Chem 2010, 285, 4612–4620. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Xu, I.M.-J.; Chiu, D.K.-C.; Lai, R.K.-H.; Tse, A.P.-W.; Lan Li, L.; Law, C.-T.; Tsang, F.H.-C.; Wei, L.L.; Chan, C.Y.-K.; et al. Folate Cycle Enzyme MTHFD1L Confers Metabolic Advantages in Hepatocellular Carcinoma. J Clin Invest 2017, 127, 1856–1872. [Google Scholar] [CrossRef] [PubMed]
- Menezo, Y.; Elder, K.; Clement, A.; Clement, P. Folic Acid, Folinic Acid, 5 Methyl TetraHydroFolate Supplementation for Mutations That Affect Epigenesis through the Folate and One-Carbon Cycles. Biomolecules 2022, 12, 197. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Ma, X.; Chang, W.; Zhang, X.; Liu, Y.; Shen, H.; Hu, X.; Ren, A.-J. Subcellular Expression Patterns of FKBP Prolyl Isomerase 10 (FKBP10) in Colorectal Cancer and Its Clinical Significance. Int J Mol Sci 2023, 24, 11415. [Google Scholar] [CrossRef]
- Ramadori, G.; Ioris, R.M.; Villanyi, Z.; Firnkes, R.; Panasenko, O.O.; Allen, G.; Konstantinidou, G.; Aras, E.; Brenachot, X.; Biscotti, T.; et al. FKBP10 Regulates Protein Translation to Sustain Lung Cancer Growth. Cell Rep 2020, 30, 3851–3863. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, X.; Bai, G.; Huang, X.; Hu, S.; Mao, H.; Liu, P. Identification of Three Potential Prognostic Genes in Platinum-Resistant Ovarian Cancer via Integrated Bioinformatics Analysis. Cancer Manag Res 2021, 13, 8629–8646. [Google Scholar] [CrossRef]
- Li, K.-S.; Zhu, X.-D.; Liu, H.-D.; Zhang, S.-Z.; Li, X.-L.; Xiao, N.; Liu, X.-F.; Xu, B.; Lei, M.; Zhang, Y.-Y.; et al. NT5DC2 Promotes Tumor Cell Proliferation by Stabilizing EGFR in Hepatocellular Carcinoma. Cell Death Dis 2020, 11, 335. [Google Scholar] [CrossRef]
- Vergara, A.G.; Watson, C.J.W.; Watson, J.M.; Chen, G.; Lazarus, P. Altered Metabolism of Polycyclic Aromatic Hydrocarbons by UDP-Glycosyltransferase 3A2 Missense Variants. Chem Res Toxicol 2020, 33, 2854–2862. [Google Scholar] [CrossRef]
- Ding, Z.; Ericksen, R.E.; Lee, Q.Y.; Han, W. Reprogramming of Mitochondrial Proline Metabolism Promotes Liver Tumorigenesis. Amino Acids 2021, 53, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Yan, M.; Liu, T.; Wang, Z.; Duan, Y.; Xia, Y.; Ji, G.; Shen, Y.; Wang, L.; Li, L.; et al. Creatine Kinase B Suppresses Ferroptosis by Phosphorylating GPX4 through a Moonlighting Function. Nat Cell Biol 2023, 25, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, E.; Capone, F.; Accardo, M.; Sorice, A.; Costantini, M.; Colonna, G.; Castello, G.; Costantini, S. GPX4 and GPX7 Over-Expression in Human Hepatocellular Carcinoma Tissues. Eur J Histochem 2015, 59, 2540. [Google Scholar] [CrossRef] [PubMed]
- Gurioli, G.; Martignano, F.; Salvi, S.; Costantini, M.; Gunelli, R.; Casadio, V. GSTP1 Methylation in Cancer: A Liquid Biopsy Biomarker? Clin Chem Lab Med 2018, 56, 702–717. [Google Scholar] [CrossRef]
- Huang, D.; Li, T.; Wang, L.; Zhang, L.; Yan, R.; Li, K.; Xing, S.; Wu, G.; Hu, L.; Jia, W.; et al. Hepatocellular Carcinoma Redirects to Ketolysis for Progression under Nutrition Deprivation Stress. Cell Res 2016, 26, 1112–1130. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets--Update. Nucleic Acids Res. 2013, 41, D991–995. [Google Scholar] [CrossRef]
- Zappia, L.; Lun, A.; Kamm, J.; Cannoodt, R. Zellkonverter: Conversion Between scRNA-Seq Objects 2024.
- Corcoran, C.C.; Grady, C.R.; Pisitkun, T.; Parulekar, J.; Knepper, M.A. From 20th Century Metabolic Wall Charts to 21st Century Systems Biology: Database of Mammalian Metabolic Enzymes. American Journal of Physiology-Renal Physiology 2017, 312, F533–F542. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Research 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision Weights Unlock Linear Model Analysis Tools for RNA-Seq Read Counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wong, L.; Goh, W.W.B. How to Do Quantile Normalization Correctly for Gene Expression Data Analyses. Sci Rep 2020, 10, 15534. [Google Scholar] [CrossRef] [PubMed]
- Desterke, C.; Xiang, Y.; Elhage, R.; Duruel, C.; Chang, Y.; Hamaï, A. Ferroptosis Inducers Upregulate PD-L1 in Recurrent Triple-Negative Breast Cancer. Cancers 2023, 16, 155. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. The Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of Biological Networks and Gene Expression Data Using Cytoscape. Nat Protoc 2007, 2, 2366–2382. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinformatics 2011, 12, 77. [Google Scholar] [CrossRef]
- Kuhn, M. Building Predictive Models in R Using the Caret Package. J. Stat. Soft. 2008, 28. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Soft. 2023, 106. [Google Scholar] [CrossRef]
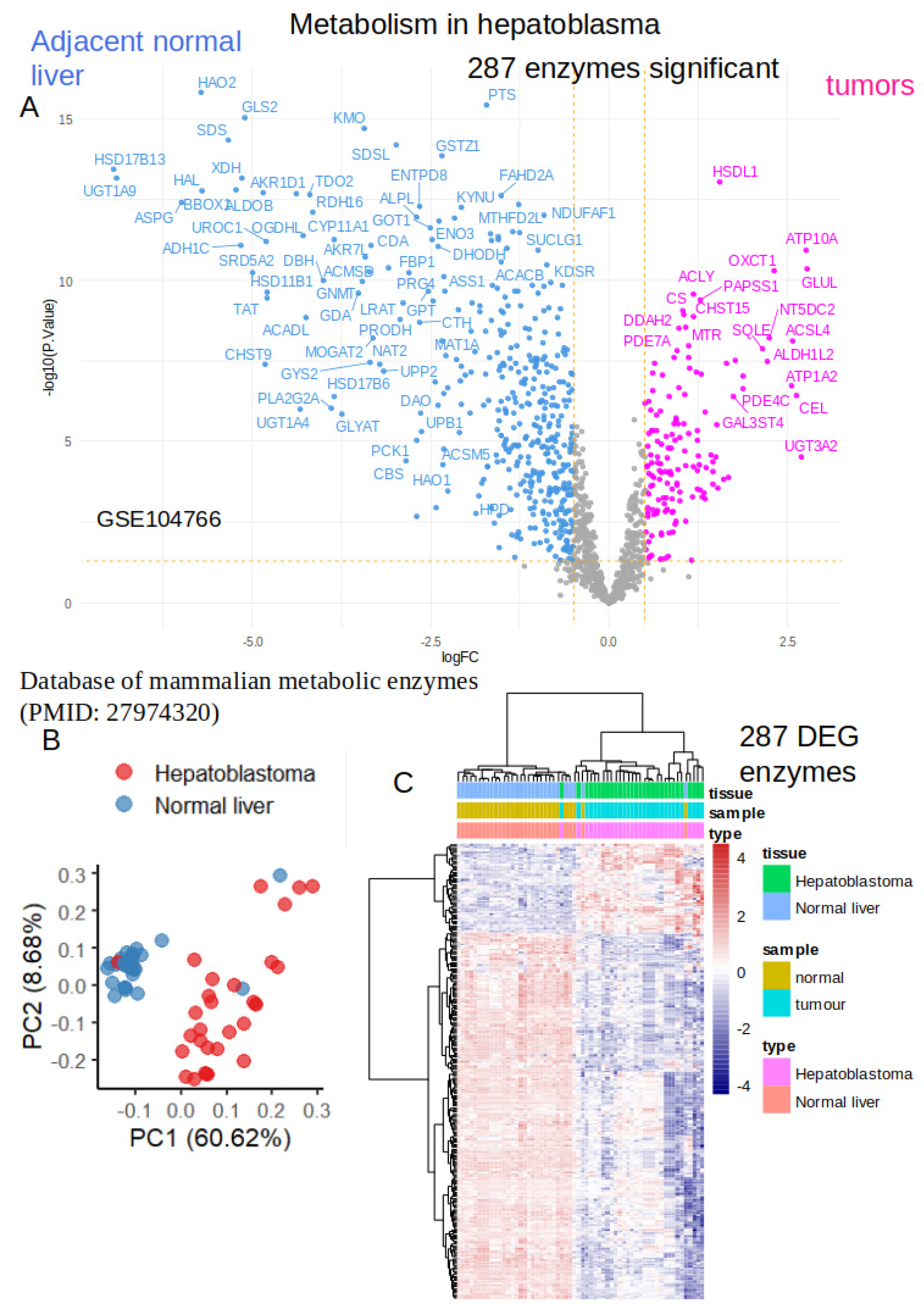
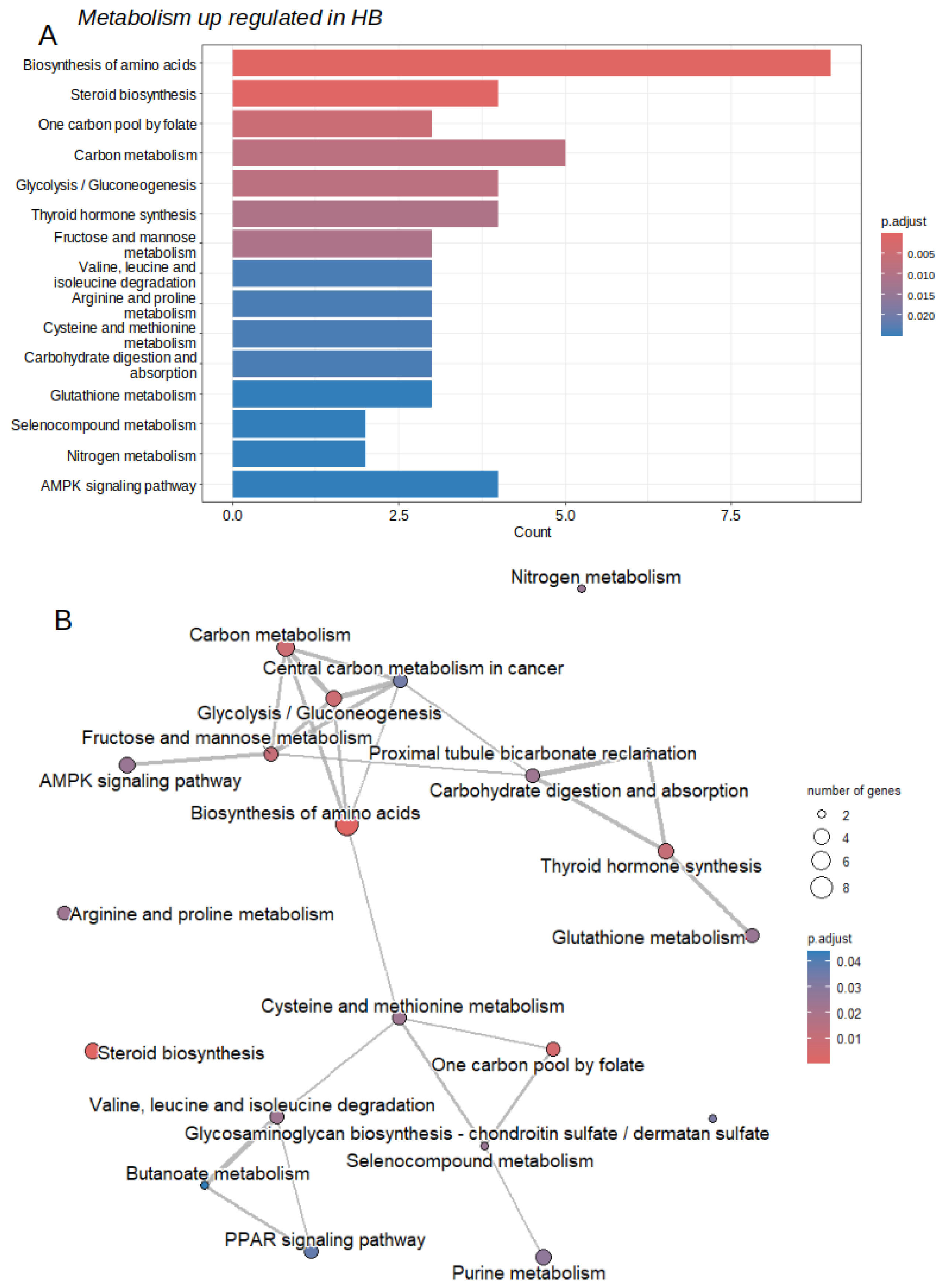
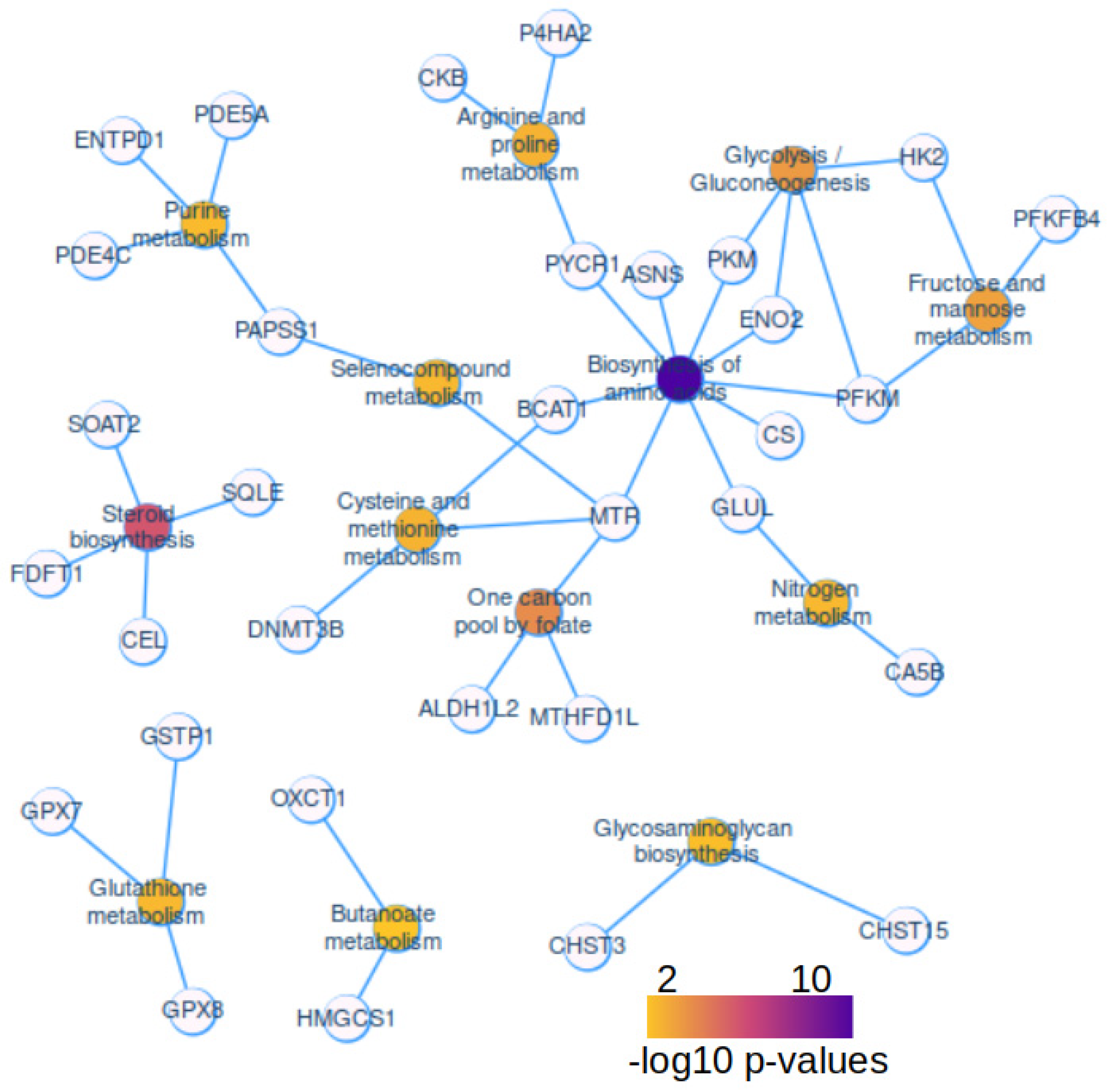
| ID | Description | subcategory | count | qvalue |
|---|---|---|---|---|
| hsa01230 | Biosynthesis of amino acids | Amin acid | 9 | 7.40E-09 |
| hsa00100 | Steroid biosynthesis | Lipid | 4 | 1.03E-04 |
| hsa00670 | One carbon pool by folate | Cofactors and vitamins | 3 | 3.39E-03 |
| hsa01200 | Carbon metabolism | Carbon | 5 | 5.46E-03 |
| hsa00010 | Glycolysis / Gluconeogenesis | Carbohydrate | 4 | 5.46E-03 |
| hsa00051 | Fructose and mannose metabolism | Carbohydrate | 3 | 7.25E-03 |
| hsa00280 | Valine, leucine and isoleucine degradation | Amino acid | 3 | 1.60E-02 |
| hsa00330 | Arginine and proline metabolism | Amino acid | 3 | 1.60E-02 |
| hsa00270 | Cysteine and methionine metabolism | Amino acid | 3 | 1.60E-02 |
| hsa00480 | Glutathione metabolism | Amino acid | 3 | 1.70E-02 |
| hsa00450 | Selenocompound metabolism | Amino acid | 2 | 1.70E-02 |
| hsa00910 | Nitrogen metabolism | Energy | 2 | 1.70E-02 |
| hsa00230 | Purine metabolism | Nucleotide | 4 | 1.90E-02 |
| hsa00532 | Glycosaminoglycan biosynthesis | Glycan | 2 | 2.26E-02 |
| hsa00650 | Butanoate metabolism | Carbohydrate | 2 | 3.00E-02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





