Submitted:
08 September 2024
Posted:
09 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Selection and Preparation of the Receptors Associated with Infection-Related CVDs
2.2. Selection and Preparation of Antimicrobial Peptides (AMPs)
| Antimicrobial Peptide | PDB ID | Active Site (Number of Residues) | Ref |
|---|---|---|---|
| Aurein | 1VM5 | 1, 2, 5 | [35] |
| Beta-defensin 2 | 1FD4 | 6, 7, 9, 10, 11, 12 | [36] |
| Bombinin | 2AP7 | 1, 3, 4, 6, 7 | [37] |
| Cathelicidin | 2K6O | 1, 4, 5, 8 | [38] |
| Cecropin | 1D9J | 7, 9, 10, 11, 12 | [39] |
| Chim2 | 8EB1 | 10, 11, 14, 15 | [40] |
| Dermcidin | 2NDK | 18, 19, 22, 25, 26, 29 | [41] |
| Esculentin | 5XDJ | 2, 3, 6 | [42] |
| Exendin-4 | 3C59 | 26, 27, 28, 29, 32, 33 | [43] |
| Hepcidin | 3H0T | 13, 14, 16, 18, 19, 20, 21, 22 | [44] |
| Hs05 | 6VLA | 5, 8, 9 | [45] |
| Indolicidin | 1HR1 | 9, 10, 11, 12, 13 | [46] |
| Lactoferrin | 1LFC | 1, 23, 25 | [47] |
| Lavracin | 2N8D | 1, 3, 4, 6 | [48] |
| Magainin | 2MAG | 1, 2, 6, 9, 17, 21 | [49] |
| Melittin | 2MLT | 13, 16, 17, 20 | [50] |
| Microcin J25 | 4CU4 | 9, 10, 19, 20, 21 | [51] |
| Nisin | 1WCO | 9, 12, 17, 19, 20, 21 | [52] |
| Pardaxin | 2KNS | 2, 3, 5, 6, 9, 15, 22, 23, 26, 27, 29, 30, 33 | [53] |
| Piscidin | 6PEZ | 3, 4, 7 | [54] |
| Pleurocidin | 2LS9 | 10, 13, 14, 17, 20, 23, 24 | [55] |
| Polyphemusin I | 1RKK | 7, 9, 12, 14 | [56] |
| Protegrin-1 | 1PG1 | 5, 6, 7, 14, 15, 16 | [57] |
| PvHCt | 2N1C | 14, 15, 16, 17, 18, 19, 20, 22, 23 | [58] |
| Subtilisin A | 1PXQ | 1, 4, 5, 7, 9, 10, 24, 25, 29, 30, 33 | [59] |
| Tachyplesin-1 | 2RTV | 1, 2, 3, 16, 17 | [60] |
| Tachystatin | 1CIX | 5, 11, 12, 15, 18, 22, 23, 29 | [61] |
| Temporin-L | 6GS5 | 4, 7 | [62] |
| Thanatin | 8TFV | 11, 13, 16, 17, 18 | [63] |
| Thermolysin | 6FHP | 258, 263, 267, 305, 306, 309, 310 | [64] |
2.3. Molecular Docking Simulations
2.4. Molecular Dynamics (MD) Simulations
2.5. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
3. Results
3.1. Molecular Docking Simulations of AMPs and Receptors Implicated in Infection-Related CVDs
- ICs: Number of intermolecular contacts
- NIS: Non-interacting surface
3.2. Molecular Dynamics (MD) Simulations
3.3. Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA) Calculations
4. Discussion
4.1. Key Findings from Molecular Simulations and Their Correlation with Other Research
4.2. Clinical Implications and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng J, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204-18. Epub 20171214. PubMed PMID: 29467962; PubMed Central PMCID: PMC5805548. [CrossRef]
- Yang T-H, Gao W-C, Ma X, Liu Q, Pang P-P, Zheng Y-T, et al. A Review on The Pathogenesis of Cardiovascular Disease of Flaviviridea Viruses Infection. Viruses. 2024;16(3):365. PubMed PMID:. [CrossRef]
- Liu C, Waters DD. Chlamydia pneumoniae and atherosclerosis: from Koch postulates to clinical trials. Prog Cardiovasc Dis. 2005;47(4):230-9. PubMed PMID: 15991152; PubMed Central PMCID: PMC7118749. [CrossRef]
- Jung SH, Lee KT. Atherosclerosis by Virus Infection-A Short Review. Biomedicines. 2022;10(10). Epub 20221019. PubMed PMID: 36289895; PubMed Central PMCID: PMC9599298. [CrossRef]
- Wolf D, Ley K. Immunity and Inflammation in Atherosclerosis. Circ Res. 2019;124(2):315-27. PubMed PMID: 30653442; PubMed Central PMCID: PMC6342482. [CrossRef]
- Laera N, Malerba P, Vacanti G, Nardin S, Pagnesi M, Nardin M. Impact of Immunity on Coronary Artery Disease: An Updated Pathogenic Interplay and Potential Therapeutic Strategies. Life (Basel). 2023;13(11). Epub 20231027. PubMed PMID: 38004268; PubMed Central PMCID: PMC10672143. [CrossRef]
- Jaén RI, Val-Blasco A, Prieto P, Gil-Fernández M, Smani T, López-Sendón JL, et al. Innate Immune Receptors, Key Actors in Cardiovascular Diseases. JACC: Basic to Translational Science. 2020;5(7):735-49. [CrossRef]
- Kircheis R, Planz O. The Role of Toll-like Receptors (TLRs) and Their Related Signaling Pathways in Viral Infection and Inflammation. Int J Mol Sci. 2023;24(7). Epub 20230404. PubMed PMID: 37047674; PubMed Central PMCID: PMC10095430. [CrossRef]
- Goulopoulou S, McCarthy CG, Webb RC. Toll-like Receptors in the Vascular System: Sensing the Dangers Within. Pharmacol Rev. 2016;68(1):142-67. PubMed PMID: 26721702; PubMed Central PMCID: PMC4709508. [CrossRef]
- Tanase DM, Valasciuc E, Gosav EM, Ouatu A, Buliga-Finis ON, Floria M, et al. Portrayal of NLRP3 Inflammasome in Atherosclerosis: Current Knowledge and Therapeutic Targets. Int J Mol Sci. 2023;24(9). Epub 20230503. PubMed PMID: 37175869; PubMed Central PMCID: PMC10179095. [CrossRef]
- Karasawa T, Takahashi M. Role of NLRP3 Inflammasomes in Atherosclerosis. J Atheroscler Thromb. 2017;24(5):443-51. Epub 20170304. PubMed PMID: 28260724; PubMed Central PMCID: PMC5429158. [CrossRef]
- Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J Pathol. 2020;251(3):228-48. Epub 20200610. PubMed PMID: 32418199; PubMed Central PMCID: PMC7276767. [CrossRef]
- Liu LP, Zhang XL, Li J. New perspectives on angiotensin-converting enzyme 2 and its related diseases. World J Diabetes. 2021;12(6):839-54. PubMed PMID: 34168732; PubMed Central PMCID: PMC8192247. [CrossRef]
- Cao Q, Lei H, Yang M, Wei L, Dong Y, Xu J, et al. Impact of Cardiovascular Diseases on COVID-19: A Systematic Review. Med Sci Monit. 2021;27:e930032. Epub 20210406. PubMed PMID: 33820904; PubMed Central PMCID: PMC8035813. [CrossRef]
- Olejarz W, Łacheta D, Kubiak-Tomaszewska G. Matrix Metalloproteinases as Biomarkers of Atherosclerotic Plaque Instability. Int J Mol Sci. 2020;21(11). Epub 20200531. PubMed PMID: 32486345; PubMed Central PMCID: PMC7313469. [CrossRef]
- Liu P, Sun M, Sader S. Matrix metalloproteinases in cardiovascular disease. Can J Cardiol. 2006;22 Suppl B(Suppl B):25b-30b. PubMed PMID: 16498509; PubMed Central PMCID: PMC2780831. [CrossRef]
- Porritt RA, Crother TR. Chlamydia pneumoniae Infection and Inflammatory Diseases. For Immunopathol Dis Therap. 2016;7(3-4):237-54. PubMed PMID: 30687565; PubMed Central PMCID: PMC6345537. [CrossRef]
- Muhlestein JB, Anderson JL, Hammond EH, Zhao L, Trehan S, Schwobe EP, Carlquist JF. Infection With Chlamydia pneumoniae Accelerates the Development of Atherosclerosis and Treatment With Azithromycin Prevents It in a Rabbit Model. Circulation. 1998;97(7):633-6. [CrossRef]
- Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229-41. PubMed PMID: 25436105; PubMed Central PMCID: PMC4232501. [CrossRef]
- Muteeb G, Rehman MT, Shahwan M, Aatif M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals (Basel). 2023;16(11). Epub 20231115. PubMed PMID: 38004480; PubMed Central PMCID: PMC10675245. [CrossRef]
- Ahmed SK, Hussein S, Qurbani K, Ibrahim RH, Fareeq A, Mahmood KA, Mohamed MG. Antimicrobial resistance: Impacts, challenges, and future prospects. Journal of Medicine, Surgery, and Public Health. 2024;2:100081. [CrossRef]
- Heianza Y, Zheng Y, Ma W, Rimm EB, Albert CM, Hu FB, et al. Duration and life-stage of antibiotic use and risk of cardiovascular events in women. Eur Heart J. 2019;40(47):3838-45. PubMed PMID: 31216010; PubMed Central PMCID: PMC6911167. [CrossRef]
- Luong HX, Thanh TT, Tran TH. Antimicrobial peptides - Advances in development of therapeutic applications. Life Sci. 2020;260:118407. Epub 20200912. PubMed PMID: 32931796; PubMed Central PMCID: PMC7486823. [CrossRef]
- Xuan J, Feng W, Wang J, Wang R, Zhang B, Bo L, et al. Antimicrobial peptides for combating drug-resistant bacterial infections. Drug Resistance Updates. 2023;68:100954. [CrossRef]
- Benfield AH, Henriques ST. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front Med Technol. 2020;2:610997. Epub 20201211. PubMed PMID: 35047892; PubMed Central PMCID: PMC8757789. [CrossRef]
- Gong H, Hu X, Zhang L, Fa K, Liao M, Liu H, et al. How do antimicrobial peptides disrupt the lipopolysaccharide membrane leaflet of Gram-negative bacteria? Journal of Colloid and Interface Science. 2023;637:182-92. [CrossRef]
- Zhang Q-Y, Yan Z-B, Meng Y-M, Hong X-Y, Shao G, Ma J-J, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Military Medical Research. 2021;8(1):48. [CrossRef]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714-23. PubMed PMID: 9504803. [CrossRef]
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(1):363-7. [CrossRef]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215-20. [CrossRef]
- Thompson D, Pepys MB, Wood SP. The physiological structure of human C-reactive protein and its complex with phosphocholine. Structure. 1999;7(2):169-77. [CrossRef]
- Rowsell S, Hawtin P, Minshull CA, Jepson H, Brockbank SMV, Barratt DG, et al. Crystal Structure of Human MMP9 in Complex with a Reverse Hydroxamate Inhibitor. Journal of Molecular Biology. 2002;319(1):173-81. [CrossRef]
- Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570(7761):338-43. [CrossRef]
- Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 2009;458(7242):1191-5. [CrossRef]
- Wang G, Li Y, Li X. Correlation of Three-dimensional Structures with the Antibacterial Activity of a Group of Peptides Designed Based on a Nontoxic Bacterial Membrane Anchor *. Journal of Biological Chemistry. 2005;280(7):5803-11. [CrossRef]
- Hoover DM, Rajashankar KR, Blumenthal R, Puri A, Oppenheim JJ, Chertov O, Lubkowski J. The Structure of Human β-Defensin-2 Shows Evidence of Higher Order Oligomerization *. Journal of Biological Chemistry. 2000;275(42):32911-8. [CrossRef]
- Zangger K, Gößler R, Khatai L, Lohner K, Jilek A. Structures of the glycine-rich diastereomeric peptides bombinin H2 and H4. Toxicon. 2008;52(2):246-54. [CrossRef]
- Wang G. Structures of Human Host Defense Cathelicidin LL-37 and Its Smallest Antimicrobial Peptide KR-12 in Lipid Micelles *<sup></sup>. Journal of Biological Chemistry. 2008;283(47):32637-43. [CrossRef]
- Oh D, Shin SY, Kang JH, Hahm KS, Kim KL, Kim Y. NMR structural characterization of cecropin A(1-8) - magainin 2(1-12) and cecropin A (1-8) - melittin (1-12) hybrid peptides. J Pept Res. 1999;53(5):578-89. PubMed PMID: 10424354. [CrossRef]
- Viana de Freitas T, Karmakar U, Vasconcelos AG, Santos MA, Oliveira do Vale Lira B, Costa SR, et al. Release of immunomodulatory peptides at bacterial membrane interfaces as a novel strategy to fight microorganisms. Journal of Biological Chemistry. 2023;299(4). [CrossRef]
- Nguyen VS, Tan KW, Ramesh K, Chew FT, Mok YK. Structural basis for the bacterial membrane insertion of dermcidin peptide, DCD-1L. Scientific Reports. 2017;7(1):13923. [CrossRef]
- Loffredo MR, Ghosh A, Harmouche N, Casciaro B, Luca V, Bortolotti A, et al. Membrane perturbing activities and structural properties of the frog-skin derived peptide Esculentin-1a(1-21)NH2 and its Diastereomer Esc(1-21)-1c: Correlation with their antipseudomonal and cytotoxic activity. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2017;1859(12):2327-39. [CrossRef]
- Runge S, Thøgersen H, Madsen K, Lau J, Rudolph R. Crystal Structure of the Ligand-bound Glucagon-like Peptide-1 Receptor Extracellular Domain * . Journal of Biological Chemistry. 2008;283(17):11340-7. [CrossRef]
- Jordan JB, Poppe L, Haniu M, Arvedson T, Syed R, Li V, et al. Hepcidin Revisited, Disulfide Connectivity, Dynamics, and Structure. Journal of Biological Chemistry. 2009;284(36):24155-67. [CrossRef]
- Mariano GH, Gomes de Sá LG, Carmo da Silva EM, Santos MA, Cardozo Fh JL, Lira BOV, et al. Characterization of novel human intragenic antimicrobial peptides, incorporation and release studies from ureasil-polyether hybrid matrix. Materials Science and Engineering: C. 2021;119:111581. [CrossRef]
- Friedrich CL, Rozek A, Patrzykat A, Hancock REW. Structure and Mechanism of Action of an Indolicidin Peptide Derivative with Improved Activity against Gram-positive Bacteria *. Journal of Biological Chemistry. 2001;276(26):24015-22. [CrossRef]
- Hwang PM, Zhou N, Shan X, Arrowsmith CH, Vogel HJ. Three-Dimensional Solution Structure of Lactoferricin B, an Antimicrobial Peptide Derived from Bovine Lactoferrin. Biochemistry. 1998;37(12):4288-98. [CrossRef]
- Pillong M, Hiss JA, Schneider P, Lin Y-C, Posselt G, Pfeiffer B, et al. Rational Design of Membrane-Pore-Forming Peptides. Small. 2017;13(40):1701316. [CrossRef]
- Gesell J, Zasloff M, Opella SJ. Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an α-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution. Journal of Biomolecular NMR. 1997;9(2):127-35. [CrossRef]
- Terwilliger TC, Weissman L, Eisenberg D. The structure of melittin in the form I crystals and its implication for melittin’s lytic and surface activities. Biophys J. 1982;37(1):353-61. PubMed PMID: 7055627; PubMed Central PMCID: PMC1329151. [CrossRef]
- Mathavan I, Zirah S, Mehmood S, Choudhury HG, Goulard C, Li Y, et al. Structural basis for hijacking siderophore receptors by antimicrobial lasso peptides. Nature Chemical Biology. 2014;10(5):340-2. [CrossRef]
- Hsu S-TD, Breukink E, Tischenko E, Lutters MAG, de Kruijff B, Kaptein R, et al. The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nature Structural & Molecular Biology. 2004;11(10):963-7. [CrossRef]
- Bhunia A, Domadia PN, Torres J, Hallock KJ, Ramamoorthy A, Bhattacharjya S. NMR Structure of Pardaxin, a Pore-forming Antimicrobial Peptide, in Lipopolysaccharide Micelles: MECHANISM OF OUTER MEMBRANE PERMEABILIZATION 2. Journal of Biological Chemistry. 2010;285(6):3883-95. [CrossRef]
- Comert F, Greenwood A, Maramba J, Acevedo R, Lucas L, Kulasinghe T, et al. The host-defense peptide piscidin P1 reorganizes lipid domains in membranes and decreases activation energies in mechanosensitive ion channels. Journal of Biological Chemistry. 2019;294(49):18557-70. [CrossRef]
- Amos ST, Vermeer LS, Ferguson PM, Kozlowska J, Davy M, Bui TT, et al. Antimicrobial Peptide Potency is Facilitated by Greater Conformational Flexibility when Binding to Gram-negative Bacterial Inner Membranes. Sci Rep. 2016;6:37639. Epub 20161122. PubMed PMID: 27874065; PubMed Central PMCID: PMC5118786. [CrossRef]
- Powers J-PS, Rozek A, Hancock REW. Structure–activity relationships for the β-hairpin cationic antimicrobial peptide polyphemusin I. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2004;1698(2):239-50. [CrossRef]
- 57. Fahrner RL, Dieckmann T, Harwig SS, Lehrer RI, Eisenberg D, Feigon J. Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem Biol. 1996;3(7):543-50. PubMed PMID: 8807886. [CrossRef]
- Petit VW, Rolland J-L, Blond A, Cazevieille C, Djediat C, Peduzzi J, et al. A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes. Biochimica et Biophysica Acta (BBA) - General Subjects. 2016;1860(3):557-68. [CrossRef]
- Kawulka KE, Sprules T, Diaper CM, Whittal RM, McKay RT, Mercier P, et al. Structure of Subtilosin A, a Cyclic Antimicrobial Peptide from Bacillus subtilis with Unusual Sulfur to α-Carbon Cross-Links: Formation and Reduction of α-Thio-α-Amino Acid Derivatives. Biochemistry. 2004;43(12):3385-95. [CrossRef]
- Kushibiki T, Kamiya M, Aizawa T, Kumaki Y, Kikukawa T, Mizuguchi M, et al. Interaction between tachyplesin I, an antimicrobial peptide derived from horseshoe crab, and lipopolysaccharide. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2014;1844(3):527-34. [CrossRef]
- Fujitani N, Kawabata S-i, Osaki T, Kumaki Y, Demura M, Nitta K, Kawano K. Structure of the Antimicrobial Peptide Tachystatin A *. Journal of Biological Chemistry. 2002;277(26):23651-7. [CrossRef]
- Manzo G, Ferguson PM, Hind CK, Clifford M, Gustilo VB, Ali H, et al. Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities. Scientific Reports. 2019;9(1):10934. [CrossRef]
- Mandard N, Sodano P, Labbe H, Bonmatin J-M, Bulet P, Hetru C, et al. Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data. European Journal of Biochemistry. 1998;256(2):404-10. [CrossRef]
- Fiebig D, Storka J, Roeder M, Meyners C, Schmelz S, Blankenfeldt W, et al. Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487. The FEBS Journal. 2018;285(22):4246-64. [CrossRef]
- Mehdi SF, Pusapati S, Anwar MS, Lohana D, Kumar P, Nandula SA, et al. Glucagon-like peptide-1: a multi-faceted anti-inflammatory agent. Front Immunol. 2023;14:1148209. Epub 20230517. PubMed PMID: 37266425; PubMed Central PMCID: PMC10230051. [CrossRef]
- Amezcua-Castillo E, González-Pacheco H, Sáenz-San Martín A, Méndez-Ocampo P, Gutierrez-Moctezuma I, Massó F, et al. C-Reactive Protein: The Quintessential Marker of Systemic Inflammation in Coronary Artery Disease-Advancing toward Precision Medicine. Biomedicines. 2023;11(9). Epub 20230902. PubMed PMID: 37760885; PubMed Central PMCID: PMC10525787. [CrossRef]
- Sortino O, Hullsiek KH, Richards E, Rupert A, Schminke A, Tetekpor N, et al. The Effects of Recombinant Human Lactoferrin on Immune Activation and the Intestinal Microbiome Among Persons Living with Human Immunodeficiency Virus and Receiving Antiretroviral Therapy. J Infect Dis. 2019;219(12):1963-8. PubMed PMID: 30721997; PubMed Central PMCID: PMC6784498. [CrossRef]
- Agier J, Efenberger M, Brzezińska-Błaszczyk E. Cathelicidin impact on inflammatory cells. Cent Eur J Immunol. 2015;40(2):225-35. Epub 20150803. PubMed PMID: 26557038; PubMed Central PMCID: PMC4637384. [CrossRef]
- Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. Journal of the American Chemical Society. 2003;125(7):1731-7. [CrossRef]
- Huang L, Sexton DJ, Skogerson K, Devlin M, Smith R, Sanyal I, et al. Novel peptide inhibitors of angiotensin-converting enzyme 2. J Biol Chem. 2003;278(18):15532-40. Epub 20030226. PubMed PMID: 12606557. [CrossRef]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-9. [CrossRef]
- Vangone A, Bonvin A. PRODIGY: A Contact-based Predictor of Binding Affinity in Protein-protein Complexes. BIO-PROTOCOL. 2017;7. [CrossRef]
- Grassmann G, Miotto M, Desantis F, Di Rienzo L, Tartaglia GG, Pastore A, et al. Computational Approaches to Predict Protein–Protein Interactions in Crowded Cellular Environments. Chemical Reviews. 2024;124(7):3932-77. [CrossRef]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845-54. [CrossRef]
- Robertson MJ, Tirado-Rives J, Jorgensen WL. Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J Chem Theory Comput. 2015;11(7):3499-509. Epub 2015/07/21. PubMed PMID: 26190950; PubMed Central PMCID: PMC4504185. [CrossRef]
- Yuet P, Blankschtein D. Molecular Dynamics Simulation Study of Water Surfaces: Comparison of Flexible Water Models. The journal of physical chemistry B. 2010;114:13786-95. [CrossRef]
- Dermawan D, Sumirtanurdin R, Dewantisari D. Simulasi dinamika molekular reseptor estrogen alfa dengan andrografolid sebagai anti kanker payudara. Indones J Pharm Sci Technol. 2019;6(2):65-76.
- Saini RS, Binduhayyim RIH, Gurumurthy V, Alshadidi AAF, Aldosari LIN, Okshah A, et al. Dental biomaterials redefined: molecular docking and dynamics-driven dental resin composite optimization. BMC Oral Health. 2024;24(1):557. [CrossRef]
- Schrödinger. The PyMOL Molecular Graphics System. 2.4 ed2020.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-12. PubMed PMID: 15264254. [CrossRef]
- Tian S, Sun H, Pan P, Li D, Zhen X, Li Y, Hou T. Assessing an ensemble docking-based virtual screening strategy for kinase targets by considering protein flexibility. J Chem Inf Model. 2014;54(10):2664-79. Epub 20140929. PubMed PMID: 25233367. [CrossRef]
- Yuan Z, Chen X, Fan S, Chang L, Chu L, Zhang Y, et al. Binding Free Energy Calculation Based on the Fragment Molecular Orbital Method and Its Application in Designing Novel SHP-2 Allosteric Inhibitors. Int J Mol Sci [Internet]. 2024; 25(1):[1-24 pp.].
- Rifai EA, Ferrario V, Pleiss J, Geerke DP. Combined Linear Interaction Energy and Alchemical Solvation Free-Energy Approach for Protein-Binding Affinity Computation. J Chem Theory Comput. 2020;16(2):1300-10. [CrossRef]
- Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. Journal of Chemical Theory and Computation. 2021;17(10):6281-91. [CrossRef]
- Miller BR, 3rd, McGee TD, Jr., Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. Journal of chemical theory and computation. 2012;8(9):3314-21. [CrossRef]
- Panday SK, Alexov E. Protein-Protein Binding Free Energy Predictions with the MM/PBSA Approach Complemented with the Gaussian-Based Method for Entropy Estimation. ACS Omega. 2022;7(13):11057-67. Epub 2022/04/14. PubMed PMID: 35415339; PubMed Central PMCID: PMC8991903. [CrossRef]
- Nnyigide OS, Lee SG, Hyun K. In Silico Characterization of the Binding Modes of Surfactants with Bovine Serum Albumin. Sci Rep. 2019;9(1):10643. Epub 20190723. PubMed PMID: 31337814; PubMed Central PMCID: PMC6650617. [CrossRef]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389-95. [CrossRef]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3(9):710-20. PubMed PMID: 12949495. [CrossRef]
- Festa M, Sansone C, Brunet C, Crocetta F, Di Paola L, Lombardo M, et al. Cardiovascular Active Peptides of Marine Origin with ACE Inhibitory Activities: Potential Role as Anti-Hypertensive Drugs and in Prevention of SARS-CoV-2 Infection. Int J Mol Sci. 2020;21(21). Epub 20201107. PubMed PMID: 33171852; PubMed Central PMCID: PMC7664667. [CrossRef]
- Lee RT. Matrix metalloproteinase inhibition and the prevention of heart failure. Trends Cardiovasc Med. 2001;11(5):202-5. PubMed PMID: 11597832. [CrossRef]
- Ndinguri MW, Bhowmick M, Tokmina-Roszyk D, Robichaud TK, Fields GB. Peptide-based selective inhibitors of matrix metalloproteinase-mediated activities. Molecules. 2012;17(12):14230-48. Epub 20121130. PubMed PMID: 23201642; PubMed Central PMCID: PMC3678729. [CrossRef]
- Al Musaimi O, Lombardi L, Williams DR, Albericio F. Strategies for Improving Peptide Stability and Delivery. Pharmaceuticals (Basel). 2022;15(10). Epub 20221019. PubMed PMID: 36297395; PubMed Central PMCID: PMC9610364. [CrossRef]
- Hossain MS, Shovon MTI, Hasan MR, Hakim FT, Hasan MM, Esha SA, et al. Therapeutic Potential of Antiviral Peptides against the NS2B/NS3 Protease of Zika Virus. ACS Omega. 2023;8(38):35207-18. Epub 20230913. PubMed PMID: 37779969; PubMed Central PMCID: PMC10536883. [CrossRef]
- Kastritis PL, Bonvin AM. On the binding affinity of macromolecular interactions: daring to ask why proteins interact. J R Soc Interface. 2013;10(79):20120835. Epub 20121212. PubMed PMID: 23235262; PubMed Central PMCID: PMC3565702. [CrossRef]
- Lin D, Sun L-C, Huo W-S, Zhang L-J, Chen Y-L, Miao S, Cao M-J. Improved functionality and safety of peptides by the formation of peptide-polyphenol complexes. Trends in Food Science & Technology. 2023;141:104193. [CrossRef]
- Fan Z, Fu M, Xu Z, Zhang B, Li Z, Li H, et al. Sustained Release of a Peptide-Based Matrix Metalloproteinase-2 Inhibitor to Attenuate Adverse Cardiac Remodeling and Improve Cardiac Function Following Myocardial Infarction. Biomacromolecules. 2017;18(9):2820-9. [CrossRef]
- Evans BJ, King AT, Katsifis A, Matesic L, Jamie JF. Methods to Enhance the Metabolic Stability of Peptide-Based PET Radiopharmaceuticals. Molecules. 2020;25(10). Epub 20200514. PubMed PMID: 32423178; PubMed Central PMCID: PMC7287708. [CrossRef]
- Böttger R, Hoffmann R, Knappe D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS One. 2017;12(6):e0178943. Epub 20170602. PubMed PMID: 28575099; PubMed Central PMCID: PMC5456363. [CrossRef]
- Cavallaro PA, De Santo M, Belsito EL, Longobucco C, Curcio M, Morelli C, et al. Peptides Targeting HER2-Positive Breast Cancer Cells and Applications in Tumor Imaging and Delivery of Chemotherapeutics. Nanomaterials (Basel). 2023;13(17). Epub 20230901. PubMed PMID: 37686984; PubMed Central PMCID: PMC10490457. [CrossRef]
- Ahn WY, Busemeyer JR. Challenges and promises for translating computational tools into clinical practice. Curr Opin Behav Sci. 2016;11:1-7. PubMed PMID: 27104211; PubMed Central PMCID: PMC4834893. [CrossRef]
- Mahadik R, Kiptoo P, Tolbert T, Siahaan TJ. Immune Modulation by Antigenic Peptides and Antigenic Peptide Conjugates for Treatment of Multiple Sclerosis. Med Res Arch. 2022;10(5). Epub 20220601. PubMed PMID: 36381196; PubMed Central PMCID: PMC9648198. [CrossRef]
- Jawa V, Terry F, Gokemeijer J, Mitra-Kaushik S, Roberts BJ, Tourdot S, De Groot AS. T-Cell Dependent Immunogenicity of Protein Therapeutics Pre-clinical Assessment and Mitigation-Updated Consensus and Review 2020. Front Immunol. 2020;11:1301. Epub 20200630. PubMed PMID: 32695107; PubMed Central PMCID: PMC7338774. [CrossRef]
- Rossino G, Marchese E, Galli G, Verde F, Finizio M, Serra M, et al. Peptides as Therapeutic Agents: Challenges and Opportunities in the Green Transition Era. Molecules. 2023;28(20). Epub 20231019. PubMed PMID: 37894644; PubMed Central PMCID: PMC10609221. [CrossRef]
- Wang L, Wang N, Zhang W, Cheng X, Yan Z, Shao G, et al. Therapeutic peptides: current applications and future directions. Signal Transduction and Targeted Therapy. 2022;7(1):48. [CrossRef]
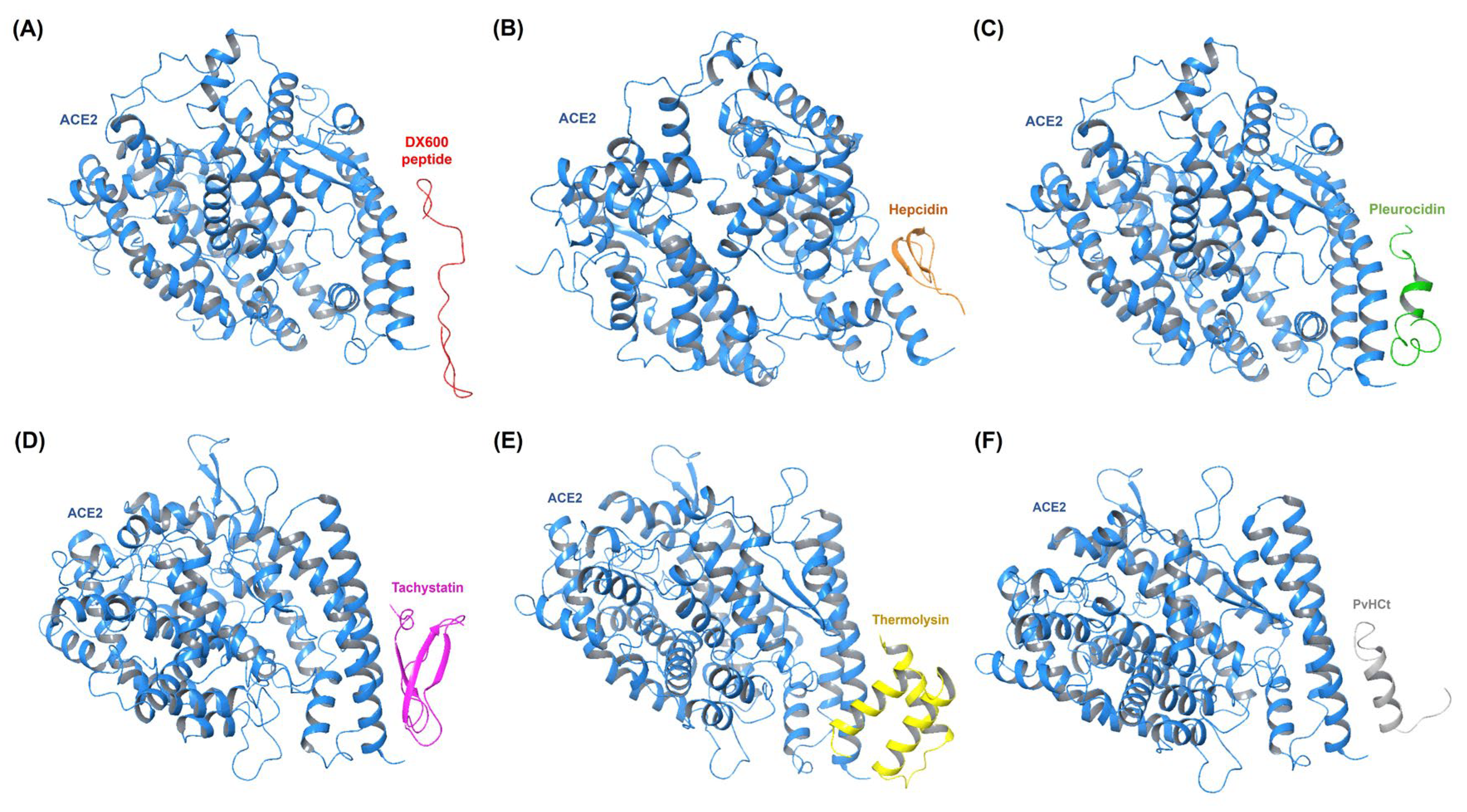
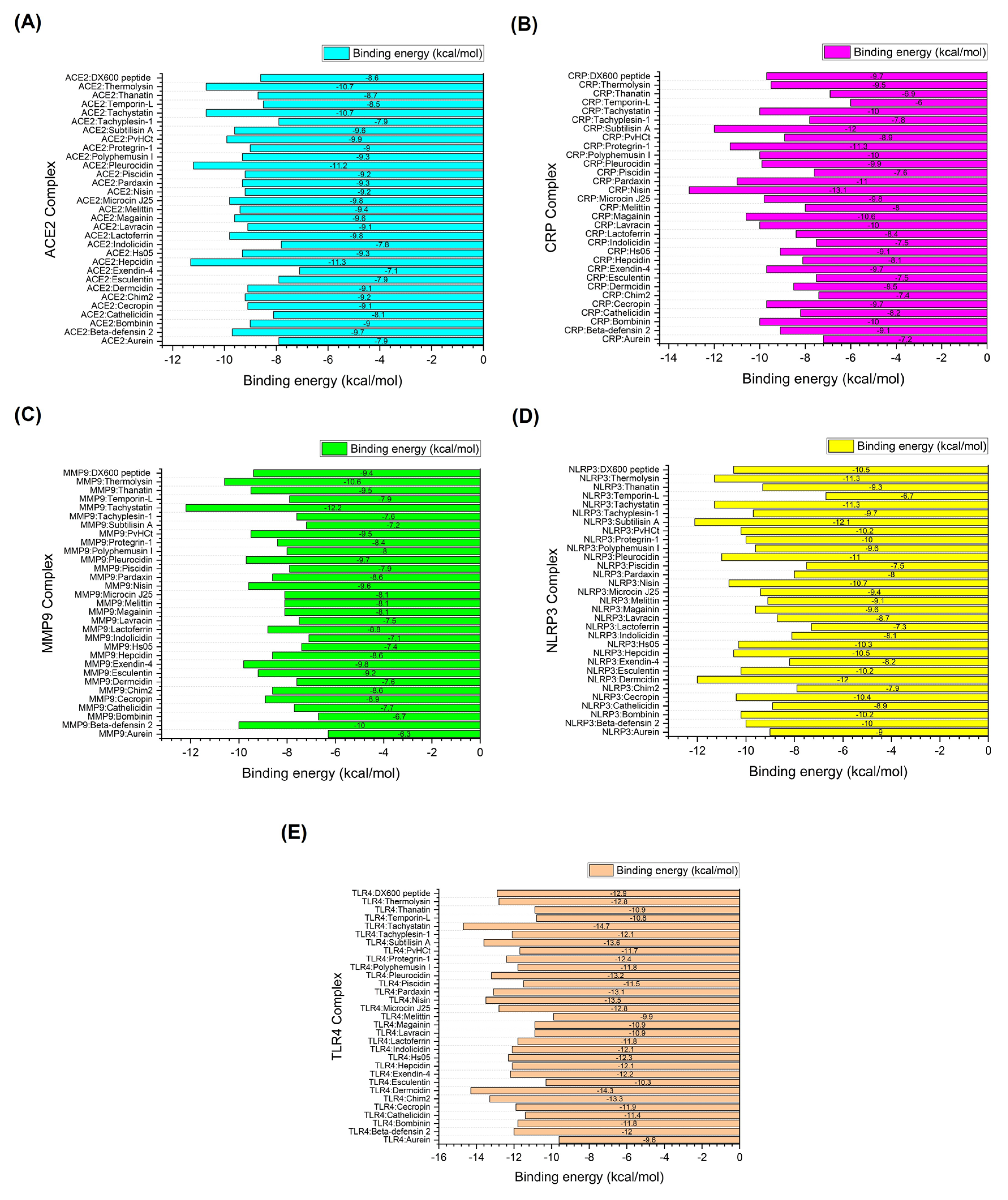
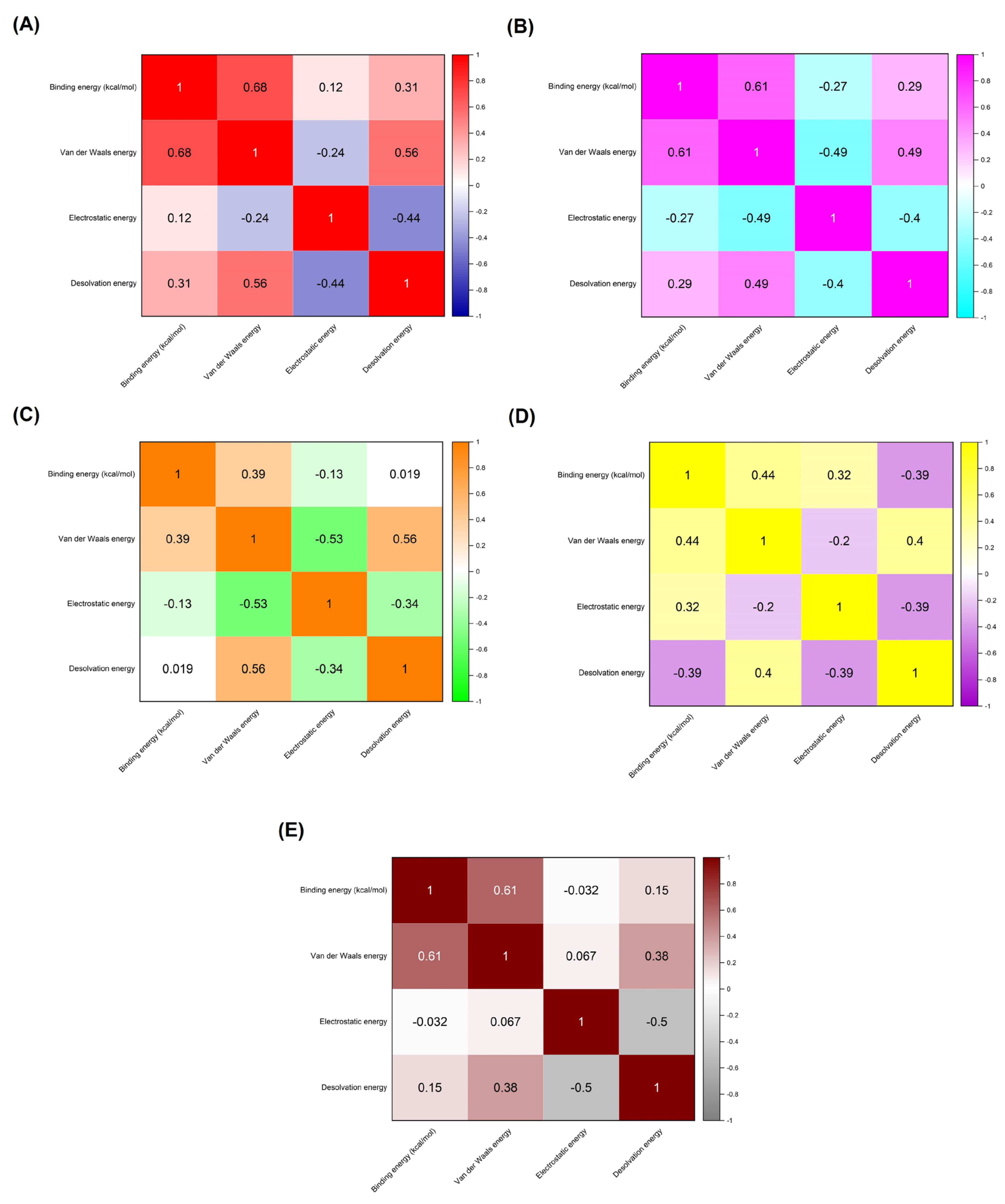

| Receptors Associated with Infection-related CVDs | PDB ID | Active Site (Number of Residues) |
Ref |
|---|---|---|---|
| Angiotensin-Converting Enzyme 2 (ACE2) | 6M0J | 24, 30, 35, 38, 41, 42, 83, 353 | [30] |
| C-reactive protein (CRP) | 1B09 | 58, 60, 61, 66, 74, 81, 138, 139, 140, 147, 150 | [31] |
| Matrix Metalloproteinase-9 (MMP9) | 1GKC | 131, 149, 165, 175, 177, 182, 183, 185, 186, 187, 188, 189, 190, 197, 199, 201, 203, 205, 206, 208, 211, 212, 213, 214, 215, 393, 401, 402, 405, 421, 422, 423 | [32] |
| NLRP3 Inflammasome | 6NPY | 165, 166, 167, 229, 230, 231, 232, 379, 411, 414, 520 | [33] |
| Toll-Like Receptor-4 (TLR4) | 3FXI | 363, 364, 365, 386, 409, 410, 411, 433, 458, 507, 533 | [34] |
| Complex | HADDOCK score (a.u.) | Binding energy (kcal/mol) | Van der Waals energy | Electrostatic energy | Desolvation energy | RMSD |
|---|---|---|---|---|---|---|
| ACE2 Complexes | ||||||
| ACE2:DX600 peptide (standard inhibitor) | -76.8 +/- 0.9 | -8.6 | -38.7 +/- 2.9 | -79.0 +/- 28.3 | -26.1 +/- 4.2 | 1.4 +/- 0.2 |
| ACE2:Hepcidin | -96.3 +/- 7.2 | -11.3 | -44.7 +/- 1.2 | -250.1 +/- 24.0 | -3.1 +/- 4.7 | 0.5 +/- 0.3 |
| ACE2:Pleurocidin | -104.8 +/- 1.9 | -11.2 | -46.6 +/- 6.0 | -220.6 +/- 23.5 | -18.0 +/- 2.2 | 1.5 +/- 0.1 |
| ACE2:Tachystatin | -102.0 +/- 3.7 | -10.7 | -48.4 +/- 4.4 | -220.9 +/- 32.4 | -20.0 +/- 4.3 | 1.5 +/- 0.0 |
| ACE2:Thermolysin | -91.4 +/- 2.4 | -10.7 | -49.2 +/- 2.5 | -131.8 +/- 20.5 | -19.0 +/- 1.1 | 0.7 +/- 0.5 |
| ACE2:PvHCt | -94.0 +/- 2.8 | -9.9 | -44.8 +/- 2.2 | -117.5 +/- 15.6 | -28.6 +/- 1.0 | 0.6 +/- 0.4 |
| CRP Complexes | ||||||
| CRP:DX600 peptide (standard inhibitor) | -90.7 +/- 8.8 | -9.7 | -40.8 +/- 4.6 | -171.9 +/- 13.0 | -15.7 +/- 2.1 | 0.8 +/- 0.5 |
| CRP:Nisin | -79.3 +/- 4.1 | -13.1 | -43.0 +/- 4.4 | -198.5 +/- 14.5 | 3.1 +/- 1.3 | 2.6 +/- 0.6 |
| CRP:Subtilisin A | -128.1 +/- 8.3 | -12.0 | -46.5 +/- 6.1 | -306.9 +/- 57.8 | -22.6 +/- 7.6 | 0.6 +/- 0.5 |
| CRP:Protegrin-1 | -92.7 +/- 4.0 | -11.3 | -37.5 +/- 3.9 | -330.3 +/- 21.8 | 9.7 +/- 2.4 | 1.3 +/- 0.1 |
| CRP:Pardaxin | -102.4 +/- 2.9 | -11.0 | -50.1 +/- 0.3 | -202.1 +/- 37.0 | -16.5 +/- 5.7 | 1.7 +/- 0.2 |
| CRP:Magainin | -108.2 +/- 5.6 | -10.6 | -25.2 +/- 4.2 | -352.4 +/- 45.0 | -14.6 +/- 1.7 | 1.0 +/- 0.2 |
| MMP9 Complexes | ||||||
| MMP9:DX600 peptide (standard inhibitor) | -98.3 +/- 2.5 | -9.4 | -55.5 +/- 3.8 | -205.3 +/- 34.7 | -22.0 +/- 2.3 | 1.4 +/- 0.3 |
| MMP9:Tachystatin | -136.4 +/- 3.4 | -12.2 | -85.2 +/- 7.8 | -235.8 +/- 14.4 | -28.6 +/- 3.1 | 1.6 +/- 0.0 |
| MMP9:Thermolysin | -114.4 +/- 3.8 | -10.6 | -70.3 +/- 6.6 | -173.2 +/- 46.2 | -27.6 +/- 3.6 | 1.2 +/- 0.2 |
| MMP9:Beta-defensin 2 | -93.6 +/- 4.7 | -10.0 | -55.2 +/- 3.8 | -285.5 +/- 37.5 | -3.9 +/- 1.5 | 0.5 +/- 0.3 |
| MMP9:Exendin-4 | -48.2 +/- 3.2 | -9.8 | -47.3 +/- 5.7 | -55.4 +/- 20.3 | -14.5 +/- 1.5 | 0.9 +/- 0.5 |
| MMP9:Pleurocidin | -143.7 +/- 4.0 | -9.7 | -62.7 +/- 6.7 | -406.5 +/- 40.9 | -27.8 +/- 2.8 | 1.2 +/- 0.1 |
| NLRP3 Complexes | ||||||
| NLRP3:DX600 peptide (standard inhibitor) | -87.9 +/- 2.7 | -10.5 | -50.1 +/- 6.8 | -237.3 +/- 44.7 | -5.9 +/- 4.6 | 0.9 +/- 0.5 |
| NLRP3:Subtilisin A | -138.7 +/- 2.8 | -12.1 | -75.8 +/- 4.3 | -271.9 +/- 59.6 | -31.2 +/- 6.0 | 0.7 +/- 0.4 |
| NLRP3:Dermcidin | -98.6 +/- 14.7 | -12.0 | -45.9 +/- 4.1 | -374.3 +/- 87.9 | 11.9 +/- 4.2 | 1.0 +/- 0.6 |
| NLRP3:Tachystatin | -100.1 +/- 8.7 | -11.3 | -50.4 +/- 9.3 | -281.3 +/- 36.1 | -0.2 +/- 5.0 | 1.4 +/- 0.2 |
| NLRP3:Thermolysin | -82.3 +/- 4.9 | -11.3 | -51.1 +/- 4.7 | -193.6 +/- 19.4 | 0.3 +/- 2.8 | 1.2 +/- 0.9 |
| NLRP3:Pleurocidin | -110.5 +/- 9.2 | -11.0 | -63.0 +/- 6.9 | -253.8 +/- 21.7 | -15.0 +/- 6.3 | 1.1 +/- 0.7 |
| TLR4 Complexes | ||||||
| TLR4:DX600 peptide (standard inhibitor) | -78.4 +/- 4.4 | -12.9 | -41.0 +/- 7.0 | -111.4 +/- 34.1 | -22.9 +/- 5.1 | 1.3 +/- 0.2 |
| TLR4:Tachystatin | -125.7 +/- 2.9 | -14.7 | -71.8 +/- 3.2 | -172.7 +/- 15.0 | -23.1 +/- 2.0 | 0.4 +/- 0.2 |
| TLR4:Dermcidin | -91.1 +/- 8.6 | -14.3 | -57.5 +/- 3.3 | -137.5 +/- 37.3 | -10.2 +/- 1.9 | 1.0 +/- 0.6 |
| TLR4:Subtilisin A | -108.7 +/- 3.4 | -13.6 | -59.9 +/- 2.4 | -70.5 +/- 21.6 | -40.4 +/- 3.5 | 1.3 +/- 0.1 |
| TLR4:Nisin | -94.6 +/- 6.3 | -13.5 | -55.8 +/- 5.0 | -153.0 +/- 28.9 | -13.6 +/- 3.1 | 1.3 +/- 0.1 |
| TLR4:Chim2 | -93.3 +/- 4.4 | -13.3 | -47.7 +/- 5.0 | -119.0 +/- 23.8 | -27.3 +/- 5.7 | 0.4 +/- 0.3 |
| Complex | ICs charged-charged | ICs charged-polar | ICs charged-apolar | ICs polar-polar | ICs polar-apolar | ICs apolar-apolar | NIS charged | NIS apolar |
|---|---|---|---|---|---|---|---|---|
| ACE2 Complexes | ||||||||
| ACE2:DX600 peptide (standard inhibitor) | 3 | 3 | 14 | 0 | 5 | 6 | 27.88 | 33.63 |
| ACE2:Hepcidin | 12 | 3 | 28 | 0 | 7 | 7 | 27.54 | 33.86 |
| ACE2:Pleurocidin | 8 | 5 | 23 | 0 | 11 | 4 | 27.87 | 34.61 |
| ACE2:Tachystatin | 5 | 9 | 20 | 2 | 12 | 8 | 26.88 | 33.76 |
| ACE2:Thermolysin | 4 | 4 | 29 | 2 | 9 | 4 | 27.33 | 34.11 |
| ACE2:PvHCt | 14 | 3 | 16 | 2 | 7 | 2 | 27.35 | 34.30 |
| CRP Complexes | ||||||||
| CRP:DX600 peptide (standard inhibitor) | 4 | 7 | 16 | 2 | 15 | 7 | 26.83 | 40.24 |
| CRP:Nisin | 1 | 3 | 27 | 1 | 26 | 19 | 24.71 | 42.35 |
| CRP:Subtilisin A | 5 | 4 | 29 | 4 | 23 | 14 | 25.9 | 43.98 |
| CRP:Protegrin-1 | 9 | 11 | 19 | 0 | 18 | 11 | 29.03 | 40.00 |
| CRP:Pardaxin | 2 | 10 | 24 | 3 | 21 | 18 | 25.29 | 44.12 |
| CRP:Magainin | 8 | 5 | 20 | 2 | 18 | 10 | 28.12 | 42.50 |
| MMP9 Complexes | ||||||||
| MMP9:DX600 peptide (standard inhibitor) | 6 | 2 | 26 | 0 | 7 | 30 | 23.08 | 43.36 |
| MMP9:Tachystatin | 4 | 10 | 22 | 0 | 20 | 28 | 22.29 | 41.40 |
| MMP9:Thermolysin | 6 | 3 | 29 | 0 | 11 | 33 | 24.36 | 42.31 |
| MMP9:Beta-defensin 2 | 11 | 3 | 28 | 0 | 10 | 24 | 23.33 | 47.33 |
| MMP9:Exendin-4 | 1 | 4 | 19 | 0 | 16 | 22 | 26.06 | 43.66 |
| MMP9:Pleurocidin | 12 | 0 | 27 | 1 | 8 | 34 | 22.3 | 46.04 |
| NLRP3 Complexes | ||||||||
| NLRP3:DX600 peptide (standard inhibitor) | 5 | 7 | 23 | 2 | 15 | 8 | 23.84 | 42.36 |
| NLRP3:Subtilisin A | 1 | 10 | 37 | 2 | 19 | 23 | 23.74 | 44.13 |
| NLRP3:Dermcidin | 14 | 8 | 28 | 0 | 15 | 14 | 25.24 | 42.86 |
| NLRP3:Tachystatin | 8 | 11 | 22 | 5 | 15 | 11 | 23.99 | 41.14 |
| NLRP3:Thermolysin | 6 | 11 | 31 | 0 | 8 | 4 | 24 | 41.93 |
| NLRP3:Pleurocidin | 13 | 6 | 31 | 1 | 10 | 20 | 23.97 | 42.84 |
| TLR4 Complexes | ||||||||
| TLR4:DX600 peptide (standard inhibitor) | 4 | 6 | 15 | 0 | 23 | 8 | 24.43 | 30.77 |
| TLR4:Tachystatin | 5 | 10 | 24 | 4 | 26 | 10 | 23.90 | 31.58 |
| TLR4:Dermcidin | 10 | 2 | 26 | 1 | 21 | 16 | 25.49 | 33.26 |
| TLR4:Subtilisin A | 4 | 5 | 16 | 2 | 24 | 28 | 23.57 | 32.82 |
| TLR4:Nisin | 6 | 9 | 20 | 7 | 25 | 14 | 23.87 | 31.98 |
| TLR4:Chim2 | 6 | 8 | 16 | 1 | 21 | 14 | 25.34 | 31.51 |
| Complex | Residue (Receptor) | Protein Atom (Receptor) |
Residue (Interacting Peptide) |
Protein Atom (Interacting Peptide) |
Interaction Distance (Å) |
|---|---|---|---|---|---|
| ACE2:Hepcidin | Ser19 | OG | Lys24 | NZ | 2.81 |
| Glu23 | OE2 | Lys24 | NZ | 2.58 | |
| Asp30 | OD1 | Arg16 | NH1 | 2.71 | |
| Asp30 | OD2 | Arg16 | NH2 | 2.59 | |
| Asp38 | OD2 | Lys18 | NZ | 2.61 | |
| CRP:Nisin | Asn61 | OD1 | Cys19 | SG | 2.94 |
| Glu147 | OE1 | Lys22 | NZ | 2.66 | |
| Glu147 | OE2 | Asn20 | ND2 | 2.93 | |
| Gln150 | NE2 | Gly18 | O | 2.97 | |
| Gln150 | NE2 | Cys19 | O | 2.86 | |
| MMP9:Tachystatin | Glu111 | OE2 | Leu6 | N | 2.66 |
| Tyr179 | OH | Arg3 | NE | 2.90 | |
| Pro180 | O | Thr20 | OG1 | 2.64 | |
| Asp182 | O | Arg14 | NH2 | 2.79 | |
| Gly183 | O | Arg14 | NH1 | 2.88 | |
| Asp185 | O | Arg14 | NH1 | 2.81 | |
| Leu188 | N | Tyr38 | OH | 2.85 | |
| Gln199 | OE1 | Arg3 | NH1 | 3.11 | |
| Gln199 | OE1 | Arg3 | NH2 | 2.93 | |
| Tyr393 | OH | Thr37 | OG1 | 2.89 | |
| His411 | O | Arg40 | NH2 | 2.86 | |
| His411 | ND1 | Asn10 | N | 3.27 | |
| Ser412 | O | Arg40 | NH1 | 3.28 | |
| Ser412 | O | Arg40 | NH2 | 2.75 | |
| NLRP3:Subtilisin A | Gln147 | OE1 | Lys2 | NZ | 2.70 |
| Glu150 | OE2 | Ala5 | N | 2.95 | |
| Glu150 | OE2 | Thr6 | N | 3.17 | |
| Glu150 | OE2 | Cys7 | SG | 2.95 | |
| Lys164 | O | Trp34 | NE1 | 2.77 | |
| Glu425 | OE2 | Cys13 | N | 2.68 | |
| Arg452 | NE | Glu23 | OE2 | 2.61 | |
| Arg452 | NH2 | Glu23 | OE1 | 2.61 | |
| Arg502 | NH1 | Thr6 | O | 2.78 | |
| TLR4:Tachystatin | Asn383 | O | Arg30 | NH1 | 3.27 |
| Asn383 | O | Arg30 | NH2 | 2.94 | |
| Ser386 | O | Tyr44 | OH | 2.78 | |
| Lys435 | NZ | Cys23 | O | 2.74 | |
| Lys435 | NZ | Cys24 | O | 2.94 | |
| Lys435 | NZ | Leu27 | O | 2.71 | |
| His458 | NE2 | Val12 | O | 2.69 | |
| Arg460 | NH2 | Gly17 | O | 2.73 |
| Complex | Average RMSD (Å) | Average RMSF (Å) | Average RoG (Å) | Number of Hydrogen Bonds Between the Two Proteins | Potential Energy (kcal/mol) |
|---|---|---|---|---|---|
| ACE2 Complexes | |||||
| ACE2 (apo-protein) | 2.870 | 0.772 | 2.501 | N/A | -440,543.758 |
| ACE2:DX600 peptide (standard inhibitor) | 3.559 | 0.792 | 2.597 | 50 | -583,916.418 |
| ACE2:Hepcidin | 3.245 | 0.822 | 2.548 | 49 | -477,259.112 |
| ACE2:Pleurocidin | 3.218 | 0.791 | 2.572 | 50 | -466,473.033 |
| ACE2:Tachystatin | 3.327 | 0.747 | 2.658 | 53 | -660,317.634 |
| ACE2:Thermolysin | 3.340 | 0.789 | 2.703 | 54 | -572,443.081 |
| ACE2:PvHCt | 3.493 | 0.795 | 2.614 | 51 | -603,748.294 |
| CRP Complexes | |||||
| CRP (apo-protein) | 2.010 | 0.726 | 1.608 | N/A | -136,101.530 |
| CRP:DX600 peptide (standard inhibitor) | 2.402 | 0.958 | 1.735 | 14 | -184,719.286 |
| CRP:Nisin | 2.313 | 0.787 | 1.737 | 16 | -288,567.674 |
| CRP:Subtilisin A | 1.977 | 0.757 | 1.704 | 16 | -166,979.914 |
| CRP:Protegrin-1 | 2.416 | 0.951 | 1.653 | 17 | -179,352.210 |
| CRP:Pardaxin | 2.355 | 0.864 | 1.687 | 16 | -143,351.144 |
| CRP:Magainin | 2.192 | 0.814 | 1.688 | 17 | -145,486.514 |
| MMP9 Complexes | |||||
| MMP9 (apo-protein) | 2.078 | 1.094 | 1.495 | N/A | -111,366.323 |
| MMP9:DX600 peptide (standard inhibitor) | 2.555 | 1.267 | 1.610 | 11 | -164,237.839 |
| MMP9:Tachystatin | 2.209 | 1.359 | 1.631 | 13 | -195,363.451 |
| MMP9:Thermolysin | 1.992 | 1.295 | 1.666 | 15 | -131,816.946 |
| MMP9:Beta-defensin 2 | 2.498 | 1.330 | 1.641 | 13 | -163,272.695 |
| MMP9:Exendin-4 | 2.820 | 1.461 | 1.661 | 12 | -191,827.586 |
| MMP9:Pleurocidin | 2.274 | 1.095 | 1.573 | 11 | -119,726.773 |
| NLRP3 Complexes | |||||
| NLRP3 (apo-protein) | 3.176 | 0.924 | 3.614 | N/A | -900,750.476 |
| NLRP3:DX600 peptide (standard inhibitor) | 3.636 | 1.056 | 3.652 | 45 | -1,127,743.577 |
| NLRP3:Subtilisin A | 3.621 | 1.044 | 3.667 | 48 | -1,070,921.883 |
| NLRP3:Dermcidin | 3.501 | 1.048 | 3.728 | 50 | -1,370,475.374 |
| NLRP3:Tachystatin | 3.459 | 0.972 | 3.678 | 49 | -1,467,888.897 |
| NLRP3:Thermolysin | 3.362 | 0.941 | 3.638 | 53 | -1,239,438.186 |
| NLRP3:Pleurocidin | 3.207 | 1.051 | 3.587 | 49 | -977,382.530 |
| TLR4 Complexes | |||||
| TLR4 (apo-protein) | 2.642 | 0.720 | 3.201 | N/A | -718,765.881 |
| TLR4:DX600 peptide (standard inhibitor) | 2.968 | 0.775 | 3.268 | 38 | -755,432.195 |
| TLR4:Tachystatin | 2.914 | 0.878 | 3.221 | 46 | -780,309.925 |
| TLR4:Dermcidin | 2.938 | 0.900 | 3.234 | 43 | -708,531.477 |
| TLR4:Subtilisin A | 3.106 | 0.831 | 3.194 | 45 | -711,005.102 |
| TLR4:Nisin | 2.962 | 0.815 | 3.193 | 44 | -712,561.934 |
| TLR4:Chim2 | 3.143 | 0.918 | 3.220 | 45 | -743,878.601 |
| Complex | MM/PBSA Calculation Results ΔGbinding (kcal/mol) | Average (kcal/mol) | ||
|---|---|---|---|---|
| I | II | III | ||
| ACE2 Complexes | ||||
| ACE2:DX600 peptide (standard inhibitor) | -22.07 | -22.51 | -22.27 | -22.28 |
| ACE2:Hepcidin | -53.81 | -53.35 | -52.56 | -53.24 |
| ACE2:Pleurocidin | -46.94 | -46.17 | -46.64 | -46.58 |
| ACE2:Tachystatin | -62.34 | -60.47 | -61.93 | -61.58 |
| ACE2:Thermolysin | -44.14 | -45.63 | -44.70 | -44.82 |
| ACE2:PvHCt | -31.42 | -31.08 | -31.18 | -31.22 |
| CRP Complexes | ||||
| CRP:DX600 peptide (standard inhibitor) | -28.23 | -27.82 | -27.93 | -27.99 |
| CRP:Nisin | -38.81 | -38.65 | -38.75 | -38.73 |
| CRP:Subtilisin A | -70.24 | -70.99 | -70.92 | -70.71 |
| CRP:Protegrin-1 | -67.52 | -67.86 | -67.31 | -67.56 |
| CRP:Pardaxin | -60.72 | -56.91 | -60.72 | -59.45 |
| CRP:Magainin | -53.41 | -53.47 | -53.35 | -53.41 |
| MMP9 Complexes | ||||
| MMP9:DX600 peptide (standard inhibitor) | -53.14 | -53.07 | -51.63 | -52.61 |
| MMP9:Tachystatin | -96.59 | -96.7 | -96.55 | -96.61 |
| MMP9:Thermolysin | -66.02 | -68.04 | -65.97 | -66.67 |
| MMP9:Beta-defensin 2 | -78.69 | -78.72 | -77.75 | -78.38 |
| MMP9:Exendin-4 | -23.95 | -23.51 | -23.60 | -23.68 |
| MMP9:Pleurocidin | -94.82 | -94.15 | -93.77 | -94.24 |
| NLRP3 Complexes | ||||
| NLRP3:DX600 peptide (standard inhibitor) | -43.87 | -45.63 | -43.57 | -44.35 |
| NLRP3:Subtilisin A | -69.45 | -71.10 | -72.81 | -71.12 |
| NLRP3:Dermcidin | -61.90 | -62.54 | -62.43 | -62.29 |
| NLRP3:Tachystatin | -69.04 | -69.56 | -69.02 | -69.20 |
| NLRP3:Thermolysin | -28.14 | -28.87 | -28.77 | -28.59 |
| NLRP3:Pleurocidin | -60.13 | -55.24 | -60.83 | -58.73 |
| TLR4 Complexes | ||||
| TLR4:DX600 peptide (standard inhibitor) | -33.05 | -32.48 | -32.93 | -32.82 |
| TLR4:Tachystatin | -59.73 | -59.72 | -59.62 | -59.69 |
| TLR4:Dermcidin | -45.92 | -46.23 | -44.51 | -45.55 |
| TLR4:Subtilisin A | -43.62 | -42.51 | -43.61 | -43.24 |
| TLR4:Nisin | -56.90 | -57.13 | -56.90 | -56.97 |
| TLR4:Chim2 | -57.95 | -58.19 | -58.45 | -58.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





