Submitted:
15 July 2024
Posted:
15 July 2024
You are already at the latest version
Abstract
Keywords:
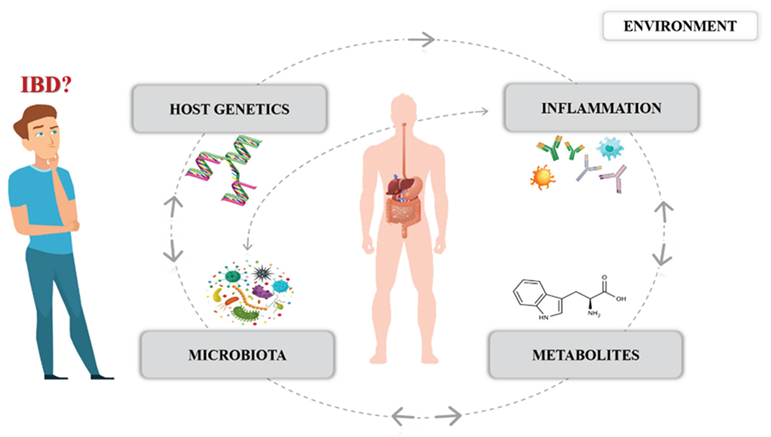
1. Introduction
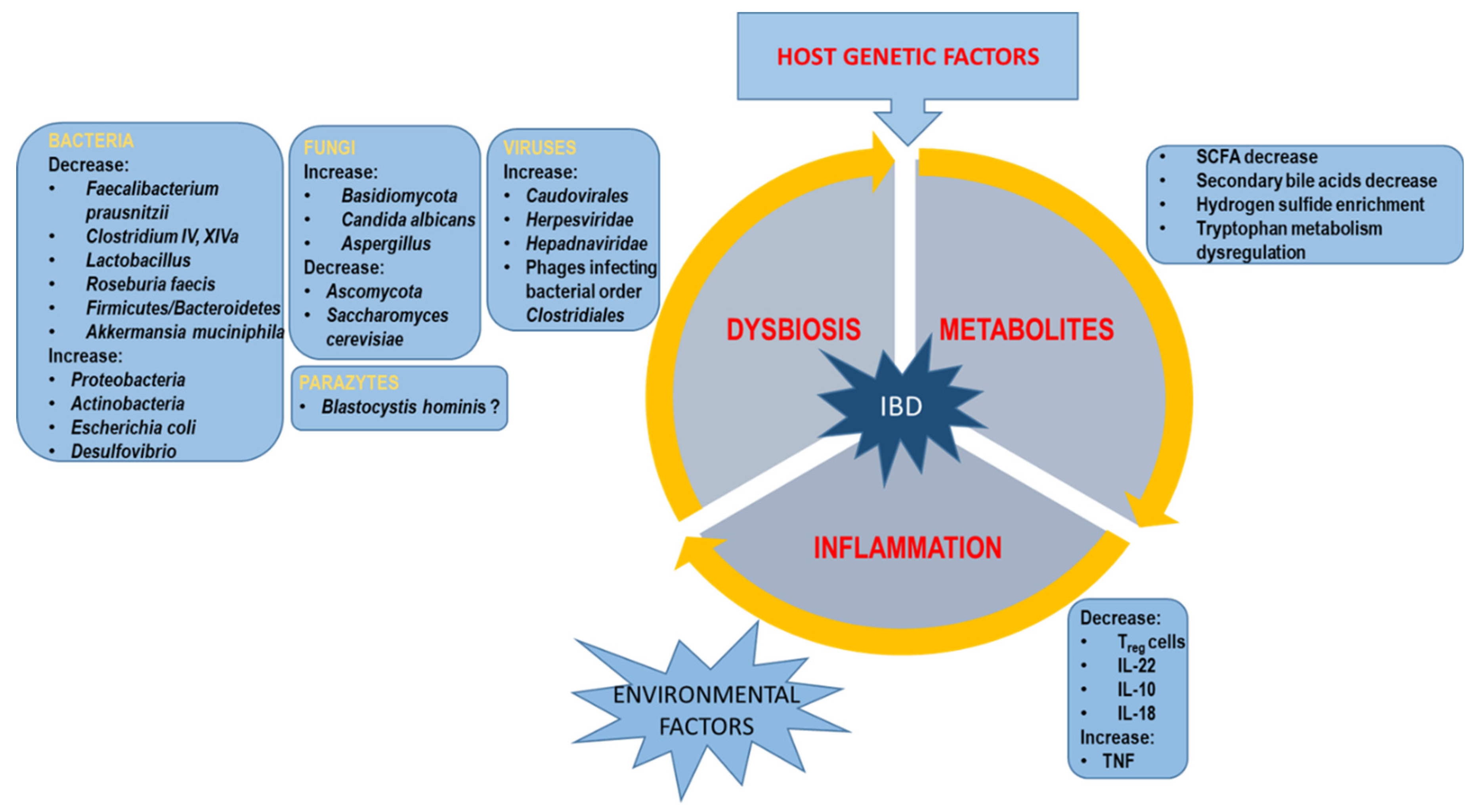
2. Gut Microbiota and IBD
2.1. Bacterial Dysbiosis
2.2. Virome Dysbiosis
2.3. Mycobiome Dysbiosis
2.4. Parasites
3. Metabolomic Changes of Intestinal Microbiota
3.1. SCFAs
3.2. Bile Acid
3.3. Hydrogen Sulfide
3.4. Tryptophan
3.5. Succinate
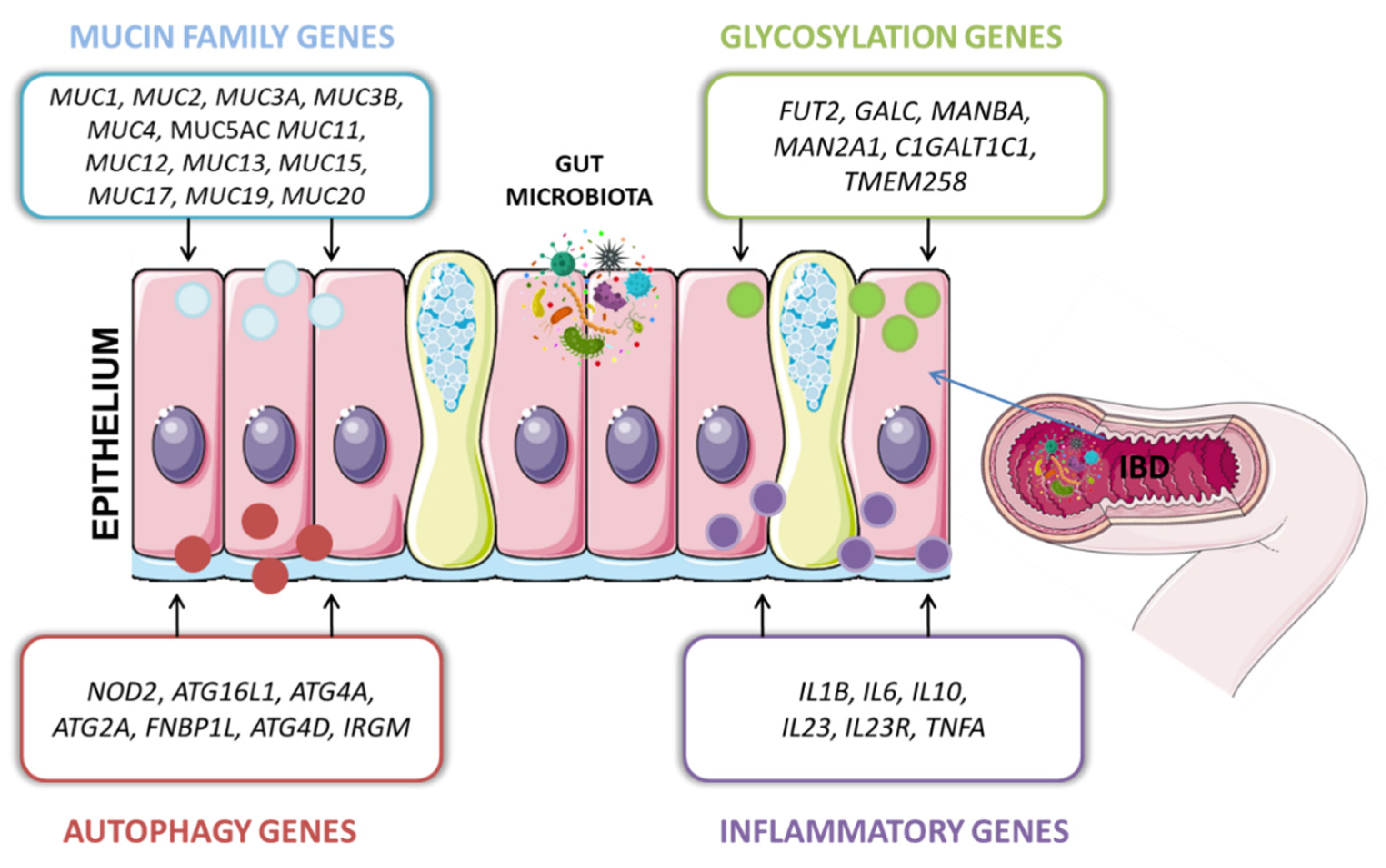
4. Interactions between Host Genes and Microbiota
4.1. Glycosylation Genes
4.2. Autophagy Genes
4.3. Mucins
4.4. Cytokines
| Disease | Gene, full name |
Genetic variant rs number, minor allel, consequence |
Location |
MAF (%) |
Function/ Impact on the host microbiome |
References |
|---|---|---|---|---|---|---|
| Crohn’s Disease (CD) |
ATG16L1 autophagy related 16 like 1 |
rs12994997 (A) intron variant |
2q14.1 | 39 |
↑The risk allele (A) increases abundance of pathogenic symbionts in the intestinal mucosa: Enterobacteriaceae, Bacteroidaceae, and Fusobacteriaceae. ↑ The protective allele (G) increases the number of commensal bacteria Lachnospiraceae |
[95] |
| rs10210302 (T) 2kb upstream variant |
2q14.2 | 39 | Variant C/C is significantly associated with the protection of IBD patients in the Indian population OR = 0.89 (0.71–1.13) | [96] | ||
| rs2241880 (G) missense variant p. Thr216Ala |
2q14.1 | 40 | A significant difference in the incidence of Listeria monocytogenes and Yersinia enterocolitica pathobionts in patients with CD compared to the control group (p<0.05); ↑ The variant T300A leads to impaired autophagy and increased pro-inflammatory cytokine production. ↑ Patients homozygous for variant T300A in the ATG16L1 gene exhibit an increased abundance of pathobionts such as E. coli, Bacteroidesfragilis, Fusobacteriaceae. Muscipirillum schaedleri, ↓Reduction of Bacteroidetes and Firmicutes |
[97,98] |
||
| rs6754677 (A) intron variant |
2q14.2 | 37 | The homozygous genotype A/A showed a risk of developing CD, is associated with terminal ileitis as well, and is related to autophagy. | [99] | ||
|
NOD2 nucleotide-binding oligomerization domain containing 2 |
rs2066844 missense variant p.Arg702Trp |
16q11.2 | 1 | ↑ Increased level of Enterobacteriaceae family and Helicobacter pylori - a risk factor for colon cancer in CD patients. |
[74] |
|
| rs2066845 missense variant p.Gly908Arg p.Gly908Cys |
16q11.2 | 1 | ↓Reduced of Bacteroidetes and Firmicutes, ↑Additionally, the C allele was significantly associated with an increase in relative abundance in the fecal bacterial family Erysipelotrichaceae. |
[74,76] |
||
| rs2066847 (CC/CCCC) frameshift variant p.Leu1007Pro(fs) |
16q12.1 | 1 | Impaired synthesis of pro-inflammatory cytokines (IL1B) and dendritic cells, leading to deregulation of the host’s antimicrobial defense; ↑ Increased abundance of pathobionts and permeability gut. |
[75] |
||
| - | - | - | ↑Increased pathogenic taxa: Yersinia, Campylobacter, Citrobacter, Escherichia coli, Helicobacter, Listeria, Mycobacteria, Pseudomonas, or Staphylococcus | [100] | ||
|
IRGM immunity related GTPase M |
rs11741861 (G) intron variant |
5q14.3 | 16 | ↓The risk variant reduces the abundance of anaerobic bacteria, butyrate-producing - Roseburia in patients with IBD. The protective barrier mucosa of the colon is compromised and, as a result, inflammation is triggered. |
[77] |
|
| rs13361189 (C) intergenic variant |
5q14.3 | 30 | Alteration in the intensity of inflammation of the intestinal mucosa as a result of the implication of an accelerated immune response | [101] | ||
| rs10065172 (T) missense variant p=Leu105= |
5q14.3 | 30 | ↑Increased susceptibility to CD in individuals of European patients (p = 0.008); Haplotype T/T influenced the binding site of a specific microRNA, causing the deregulation of IRGM-dependent xenophagy bacteria in patients with CD; ↑In addition, the T/T genotype is also associated with an increased level of expression of the cytokine TNFα in the peripheral blood, influencing inflammation. |
[72,102] |
||
| Ulcerative colitis |
MUC13 Mucin 13 |
rs1127233 (G) missense variant Arg503Ser |
3q21.2 | 23 |
Variant correlated with UC p = 0.0003; Disturbed MUC13 gene expression correlated with the NFkB pathway can lead to a loss of membrane integrity and thus permeability. |
[103] |
| Inflammatory Bowel Disease (IBD) |
MUC1 Mucin 1 |
- | - | - | MUC1 codes for the main mucus component which is the physical barrier that protects the intestinal epithelium from intestinal bacteria. MUC1 overexpression and hypoglycosylation have been reported in Muc1-knockout IBD mice showing increased damage to the small intestine following infection with C. jejuni. | [104] |
| rs4072037 (C) synonymous variant p.Thr22= |
1q22 | 37 | ↑Increase in the abundance of Ochrobactrum | [105] | ||
|
MUC2 Mucin 2 |
rs2856111 (T) missense variant Leu58Pro |
11p15.5 | 27 | Reduced gene expression is associated with a thinner mucus layer in UC patients - particularly at the site of inflammation due to the reduction of goblet cells. | [106] | |
| rs11825977(A) missense variant p.Val116Met |
11p15.5 | 12 | Decreased MUC2 mRNA expression increases the risk of inflammation and intestinal dysbiosis. | [103] | ||
|
MUC3A Mucin 3A |
- | 7q22.1 | - | Rare alleles change the conformation of the proteins produced. The conformation affects the glycosylation process, which increases the sensitivity to bacterial proteases, and thus breaks the continuity of the protective gel barrier. |
[88] |
|
|
MUC5AC Mucin 5AC |
rs35783651 (G) missense variant p.Ser221Arg |
11p15.5 | 10 | Protective role by participating in the healing of mucosal epithelial wounds and regulating MGL; H. pylori-infected patients indicated a significant decrease in MUC5AC expression level |
[107] |
|
|
MUC19 Mucin 19 |
rs11564245 (C) missense variant p.Asp803His |
12q12 | 5 | Increased susceptibility to CD in the group of patients. | [3,85] |
|
| rs4768261 (T) missense variant p.Ser1226Phe |
12q12 | 5 | ||||
|
CARD9 Caspase Recruitment Domain Family Member 9 |
rs4077515 (T) missense variant p.Ser12Asn/Ile |
9q21.3 | 37 | Innate immune response to peptidoglycan, a macromolecule in the bacterial cell wall ; Aberrant activation of NF-κB and inflammatory factors in response to Aspergillus fumigates, contributing to intestinal inflammation. |
[108] |
|
| rs10781499 (A) synonymous variant p.Pro42= |
9q34.3 | 37 | ↓Decrease butyrate acetate converting bacteria - Roseburia spp | [77] | ||
| rs10870077 (G) intron variant |
9q34.3 | 37 | ↑Increased risk of UC development by modulating the signaling pathway affecting the inflammatory response. |
[109] |
||
|
FUT2 Fucosyltransferase 2 |
Fut2- | 19q13.33 | - | ↓Reduced beneficial bacteria from the Ruminococcaceae and Muribaculaceae, while the pathogenic microorganisms, such as ↑Bilophila, Escherichia, Enterorhabdus, Alistipe, Phascolarctobacterium were increased in the cohort. |
[67,68] |
|
|
C1GALT1C Core 1 Synthase, Glycoprotein-N-Acetylgalactosamine 3-Beta-Galactosyltransferase 1 |
- | 7p22.1-p21.3 | - | Studies demonstrated an 11-fold reduction of Bacteroides and a 3-fold increase of pathogenic Helicobacter microbes in the IBD cohort. | [70] | |
|
IL1B Interleukin 1- Beta |
- | 2q14.1 | - | Increased ILIB level by attack and colonization of Klebsiella pneumoniae, Streptococcus mitis, Streptococcus oralis, and Streptococcus pneumoniae. | [92] |
|
|
IL6 Interleukin 6 |
- | 1q21.3 | The deficiency of IL6 contributes to the dysbiosis of the gut microbiota and increases the abundance of Gram-negative bacteria. | [110] | ||
|
IL10 Interleukin 10 |
rs1800896 (A) 2kb upstream variant |
1q32.1 | 27 | ↓Loss of IL10 receptor function - induction of inflammation in severe course of UC; Allele A was associated with UC p=0.011 in in Mexican cohort. ↑ Increased susceptibility to fungal infections with Candida albicans ↓Decreased IL10 expression is associated with ↑ increased Bacteroides, Prevotella, and Rikenella |
[91] |
|
|
IL23R Interleukin 23 |
rs1004819 (A) intron variant |
1p.31.3 | 40 | Early age of onset of the disease in the Polish population. | [111] | |
| rs76418789 (A) missense variant p.Gly149Arg |
1p.31.3 | 1 | SNP associated with IBD in the Korean population (p = 0.0096) | [112] | ||
| rs11209026 (A) missense variant p.Arg381Gln |
1p.31.3 | 2 | Protective effect in CD but related to UC; ↑ Increased abundance of Christensenellaceae, Bacteroides caccae, and a ↓ decrease in the commensal bacteria Faecalibacterium prausnitzii |
[113] |
||
| rs2201841 (G) intron variant |
1q11 | 40 | Significant association between polymorphisms and UC, especially in Caucasians. | [113] | ||
|
IFNG Interferon Gamma |
- | 12q15 | - | ↑ Increased level of taxa: Dorea, Streptococcus parasanguinis and Streptococcus australis | [94] |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dahlhamer, J.M.; Zammitti, E.P.; Ward, B.W.; Wheaton, A.G.; Croft, J.B. Prevalence of Inflammatory Bowel Disease Among Adults Aged ≥18 Years - United States, 2015, MMWR Morb Mortal Wkly Rep 2016, 65, 1166–1169. [CrossRef]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; Kaplan, G.G. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review, Gastroenterology 2012, 142, 46–54.e42; quiz e30. [CrossRef]
- Kumar, S.; Kumar, A. Microbial pathogenesis in inflammatory bowel diseases, Microbial Pathogenesis 2022, 163, 105383. [CrossRef]
- Li, J.; Butcher, J.; Mack, D.; Stintzi, A. Functional Impacts of the Intestinal Microbiome in the Pathogenesis of Inflammatory Bowel Disease, Inflammatory Bowel Diseases 2015, 21, 139–153. [CrossRef]
- Kudelka, M.R.; Stowell, S.R.; Cummings, R.D.; Neish, A.S. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD, Nat Rev Gastroenterol Hepatol 2020, 17, 597–617. [CrossRef]
- Buttó, L.F.; Haller, D. Dysbiosis in intestinal inflammation: Cause or consequence, International Journal of Medical Microbiology 2016, 306, 302–309. [CrossRef]
- Littman, D.R.; Rudensky, A.Y. Th17 and Regulatory T Cells in Mediating and Restraining Inflammation, Cell 2010, 140, 845–858. [CrossRef]
- Ayabe, T.; Satchell, D.P.; Pesendorfer, P.; Tanabe, H.; Wilson, C.L.; Hagen, S.J.; Ouellette, A.J. Activation of Paneth Cell α-Defensins in Mouse Small Intestine, Journal of Biological Chemistry 2002, 277, 5219–5228. [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; Bleich, A.; Haller, D. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence, Gut 2016, 65, 225–237. [CrossRef]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; Gevers, D.; McGovern, D.P.B.; Singh, N.; Braun, J.; Jacobs, J.P.; Clemente, J.C.; Grinspan, A.; Sands, B.E.; Colombel, J.-F.; Dubinsky, M.C.; Faith, J.J. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice, Immunity 2019, 50, 212–224.e4. [CrossRef]
- Webb, C.R.; den Bakker, H.; Koboziev, I.; Jones-Hall, Y.; Kottapalli, K.R.; Ostanin, D.; Furr, K.L.; Mu, Q.; Luo, X.M.; Grisham, M.B. Differential Susceptibility to T Cell-Induced Colitis in Mice: Role of the Intestinal Microbiota, Inflammatory Bowel Diseases 2018, 24, 361–379. [CrossRef]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. ; Nutrition, IBD and Gut Microbiota: A Review, Nutrients 2020, 12, 944. [CrossRef]
- Jandhyala, S.M. Role of the normal gut microbiota, WJG 2015, 21, 8787. [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; Morgan, X.C.; Kostic, A.D.; Luo, C.; González, A.; McDonald, D.; Haberman, Y.; Walters, T.; Baker, S.; Rosh, J.; Stephens, M.; Heyman, M.; Markowitz, J.; Baldassano, R.; Griffiths, A.; Sylvester, F.; Mack, D.; Kim, S.; Crandall, W.; Hyams, J.; Huttenhower, C.; Knight, R.; Xavier, R.J. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease, Cell Host & Microbe 2014, 15, 382–392. [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; Ferrante, M.; Verhaegen, J.; Rutgeerts, P.; Vermeire, S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis, Gut 2014, 63, 1275–1283. [CrossRef]
- Zhang, M.; Qiu, X.; Zhang, H.; Yang, X.; Hong, N.; Yang, Y.; Chen, H.; Yu, C. Faecalibacterium prausnitzii Inhibits Interleukin-17 to Ameliorate Colorectal Colitis in Rats, PLoS ONE 2014, 9, e109146. [CrossRef]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; Myridakis, A.; Delzenne, N.M.; Klievink, J.; Bhattacharjee, A.; van der Ark, K.C.H.; Aalvink, S.; Martinez, L.O.; Dumas, M.-E.; Maiter, D.; Loumaye, A.; Hermans, M.P.; Thissen, J.-P.; Belzer, C.; de Vos, W.M.; Cani, P.D. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice, Nat Med 2017, 23, 107–113. [CrossRef]
- Zakerska-Banaszak, O.; Tomczak, H.; Gabryel, M.; Baturo, A.; Wolko, L.; Michalak, M.; Malinska, N.; Mankowska-Wierzbicka, D.; Eder, P.; Dobrowolska, A.; Slomski, R.; Skrzypczak-Zielinska, M. Dysbiosis of gut microbiota in Polish patients with ulcerative colitis: a pilot study, Sci Rep 2021, 11, 2166. [CrossRef]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; Guerin, E.; Velayudhan, V.; Ross, R.P.; Hill, C. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific, Cell Host Microbe 2019, 26, 527–541.e5. [CrossRef]
- Clooney, A.G.; Sutton, T.D.S.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’Regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; Hill, C. Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflammatory Bowel Disease, Cell Host & Microbe 2019, 26, 764–778.e5. [CrossRef]
- Fernandes, M.A.; Verstraete, S.G.; Phan, T.G.; Deng, X.; Stekol, E.; LaMere, B.; Lynch, S.V.; Heyman, M.B.; Delwart, E. Enteric Virome and Bacterial Microbiota in Children With Ulcerative Colitis and Crohn Disease, Journal of Pediatric Gastroenterology & Nutrition 2019, 68, 30–36. 68. [CrossRef]
- Zuo, T.; Lu, X.-J.; Zhang, Y.; Cheung, C.P.; Lam, S.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Zhao, R.; Chan, P.K.S.; Sung, J.J.Y.; Yu, J.; Chan, F.K.L.; Cao, Q.; Sheng, J.-Q.; Ng, S.C. Gut mucosal virome alterations in ulcerative colitis, Gut 2019, 68, 1169–1179. [CrossRef]
- Ungaro, F.; Massimino, L.; D’Alessio, S.; Danese, S. The gut virome in inflammatory bowel disease pathogenesis: From metagenomics to novel therapeutic approaches, United European Gastroenterol. J. 2019, 7, 999–1007. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; Porter, N.; Martens, E.; O’Connell, R.; Jacob, V.; Scherl, E.; Crawford, C.; Stephens, W.Z.; Casjens, S.R.; Longman, R.S.; Round, J.L. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis, Cell Host & Microbe 2019, 25, 285–299.e8. [CrossRef]
- Iliev, I.D.; Leonardi, I. Fungal dysbiosis: immunity and interactions at mucosal barriers, Nat Rev Immunol 2017, 17, 635–646. [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; Cosnes, J.; Seksik, P.; Langella, P.; Skurnik, D.; Richard, M.L.; Beaugerie, L. Fungal microbiota dysbiosis in IBD, Gut 2017, 66, 1039–1048. [CrossRef]
- Knox, N.C.; Forbes, J.D.; Peterson, C.-L.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiome in Inflammatory Bowel Disease: Lessons Learned From Other Immune-Mediated Inflammatory Diseases, Am J Gastroenterol 2019, 114, 1051–1070. [CrossRef]
- Zwolinska-Wcislo, M.; Brzozowski, T.; Budak, A.; Kwiecien, S.; Sliwowski, Z.; Drozdowicz, D.; Trojanowska, D.; Rudnicka-Sosin, L.; Mach, T.; Konturek, S.J.; Pawlik, W.W. Effect of Candida colonization on human ulcerative colitis and the healing of inflammatory changes of the colon in the experimental model of colitis ulcerosa, J Physiol Pharmacol 2009, 60, 107–118.
- Qiu, X.; Ma, J.; Jiao, C.; Mao, X.; Zhao, X.; Lu, M.; Wang, K.; Zhang, H. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis, Oncotarget 2017, 8, 107577–107588. [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; Murciano, C.; Blagojevic, M.; Thavaraj, S.; Förster, T.M.; Hebecker, B.; Kasper, L.; Vizcay, G.; Iancu, S.I.; Kichik, N.; Häder, A.; Kurzai, O.; Luo, T.; Krüger, T.; Kniemeyer, O.; Cota, E.; Bader, O.; Wheeler, R.T.; Gutsmann, T.; Hube, B.; Naglik, J.R. Candidalysin is a fungal peptide toxin critical for mucosal infection, Nature 2016, 532, 64–68. [CrossRef]
- Kasper, L.; König, A.; Koenig, P.-A.; Gresnigt, M.S.; Westman, J.; Drummond, R.A.; Lionakis, M.S.; Groß, O.; Ruland, J.; Naglik, J.R.; Hube, B. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes, Nat Commun 2018, 9, 4260. [CrossRef]
- The International IBD Genetics Consortium (IIBDGC). Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease, Nature 2012, 491, 119–124. [CrossRef]
- Taylor, P.R.; Brown, G.D.; Reid, D.M.; Willment, J.A.; Martinez-Pomares, L.; Gordon, S.; Wong, S.Y.C.; Receptor, G. ; Dectin-1, Is Predominantly Expressed on the Surface of Cells of the Monocyte/Macrophage and Neutrophil Lineages, The Journal of Immunology 2002, 169, 3876–3882. [CrossRef]
- Miyoshi, J.; Sofia, M.A.; Pierre, J.F. The evidence for fungus in Crohn’s disease pathogenesis, Clin J Gastroenterol 2018, 11, 449–456. [CrossRef]
- Eichenberger, R.M.; Ryan, S.; Jones, L.; Buitrago, G.; Polster, R.; de Oca, M.M.; Zuvelek, J.; Giacomin, P.R.; Dent, L.A.; Engwerda, C.R.; Field, M.A.; Sotillo, J.; Loukas, A. Hookworm Secreted Extracellular Vesicles Interact With Host Cells and Prevent Inducible Colitis in Mice, Front. Immunol. 2018, 9, 850. [Google Scholar] [CrossRef]
- Hamad, I.; Raoult, D.; Bittar, F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: taxonomy and detection methods, Parasite Immunol 2016, 38, 12–36. [CrossRef]
- Coskun, A.; Malatyali, E.; Ertabaklar, H.; Yasar, M.B.; Karaoglu, A.O.; Ertug, S. Blastocystis in ulcerative colitis patients: Genetic diversity and analysis of laboratory findings, Asian Pacific Journal of Tropical Medicine 2016, 9, 916–919. [CrossRef]
- The Blastocystis Investigation Group, C. Audebert, G. Even, A. Cian, A. Loywick, S. Merlin, E. Viscogliosi, M. Chabé, Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota, Sci Rep 2016, 6, 25255. [Google Scholar] [CrossRef]
- Tai, W.-P.; Hu, P.-J.; Wu, J.; Lin, X.-C. Six ulcerative colitis patients with refractory symptoms co-infective with Blastocystis hominis in China, Parasitol Res 2011, 108, 1207–1210. [CrossRef]
- Verstockt, B.; Vermeire, S.; Van Assche, G.; Ferrante, M. When IBD is not IBD, Scandinavian Journal of Gastroenterology 2018, 53, 1085–1088. [CrossRef]
- Vadlamudi, N.; Maclin, J.; Dimmitt, R.A.; Thame, K.A. Cryptosporidial infection in children with inflammatory bowel disease, Journal of Crohn’s and Colitis 2013, 7, e337–e343. [CrossRef]
- Arai, T.; Lopes, F. Potential of human helminth therapy for resolution of inflammatory bowel disease: The future ahead, Experimental Parasitology 2022, 232, 108189. [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: causation or correlation? , Nat Rev Gastroenterol Hepatol 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Tefas, C.; Ciobanu, L.; Tanțău, M.; Moraru, C.; Socaciu, C. The potential of metabolic and lipid profiling in inflammatory bowel diseases: a pilot study, Bosn J of Basic Med Sci 2019. [CrossRef]
- Gallagher, K.; Catesson, A.; Griffin, J.L.; Holmes, E.; Williams, H.R.T. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review, Journal of Crohn’s and Colitis 2021, 15, 813–826. [CrossRef]
- Paramsothy, S.; Nielsen, S.; Kamm, M.A.; Deshpande, N.P.; Faith, J.J.; Clemente, J.C.; Paramsothy, R.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; Leong, R.W.L.; Connor, S.; Ng, W.; Lin, E.; Borody, T.J.; Wilkins, M.R.; Colombel, J.-F.; Mitchell, H.M.; Kaakoush, N.O. Specific Bacteria and Metabolites Associated With Response to Fecal Microbiota Transplantation in Patients With Ulcerative Colitis, Gastroenterology 2019, 156, 1440–1454.e2. [CrossRef]
- Magnusson, M.K.; Isaksson, S.; Öhman, L. The Anti-inflammatory Immune Regulation Induced by Butyrate Is Impaired in Inflamed Intestinal Mucosa from Patients with Ulcerative Colitis, Inflammation 2020, 43, 507–517. [CrossRef]
- Fiorucci, S.; Carino, A.; Baldoni, M.; Santucci, L.; Costanzi, E.; Graziosi, L.; Distrutti, E.; Biagioli, M. Bile Acid Signaling in Inflammatory Bowel Diseases, Dig Dis Sci 2021, 66, 674–693. [CrossRef]
- Bares, M.; Cantero, J.M.B.; Flores, E.I.; Alcalde, B.G.; Ortega, E.M.; Muret, F.R.M.; Asenjo, E.C.; Sánchez, V.G.; Casas, J.A.V. Bile acid malabsorption in patients with chronic diarrhea and Crohn�s disease, Rev Esp Enferm Dig 2018, 111,. [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; Bridonneau, C.; Dumetz, F.; Grill, J.-P.; Masliah, J.; Beaugerie, L.; Cosnes, J.; Chazouillères, O.; Poupon, R.; Wolf, C.; Mallet, J.-M.; Langella, P.; Trugnan, G.; Sokol, H.; Seksik, P. ; Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases, Gut 2013, 62, 531–539. [CrossRef]
- Washio, J.; Sato, T.; Koseki, T.; Takahashi, N. Hydrogen sulfide-producing bacteria in tongue biofilm and their relationship with oral malodour, Journal of Medical Microbiology 2005, 54, 889–895. [CrossRef]
- Metwaly, A.; Dunkel, A.; Waldschmitt, N.; Raj, A.C.D.; Lagkouvardos, I.; Corraliza, A.M.; Mayorgas, A.; Martinez-Medina, M.; Reiter, S.; Schloter, M.; Hofmann, T.; Allez, M.; Panes, J.; Salas, A.; Haller, D. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism, Nat Commun 2020, 11, 4322. [CrossRef]
- Leschelle, X.; Goubern, M.; Andriamihaja, M.; Blottière, H.M.; Couplan, E.; Gonzalez-Barroso, M.-M.; Petit, C.; Pagniez, A.; Chaumontet, C.; Mignotte, B.; Bouillaud, F.; Blachier, F. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide, Biochimica et Biophysica Acta (BBA) - General Subjects 2005, 1725, 201–212. [CrossRef]
- Smith, F.M.; Coffey, J.C.; Kell, M.R.; O’Sullivan, M.; Redmond, H.P.; Kirwan, W.O. A characterization of anaerobic colonization and associated mucosal adaptations in the undiseased ileal pouch, Colorect Dis 2005, 7, 563–570. [CrossRef]
- Li, X.; Zhang, Z.; Zabed, H.M.; Yun, J.; Zhang, G.; Qi, X. An Insight into the Roles of Dietary Tryptophan and Its Metabolites in Intestinal Inflammation and Inflammatory Bowel Disease, Mol. Nutr. Food Res. 2021, 65, 2000461. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous Bacteria from the Gut Microbiota Regulate Host Serotonin Biosynthesis, Cell 2015, 161, 264–276. [CrossRef]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; Colgan, S.P. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor, The American Journal of Pathology 2018, 188, 1183–1194. [CrossRef]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; Corr, S.C.; McManus, G.; Ryan, D.; Jacobs, H.T.; Szibor, M.; Xavier, R.J.; Braun, T.; Frezza, C.; Murphy, M.P.; O’Neill, L.A. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages, Cell 2016, 167, 457–470.e13. [CrossRef]
- Macias-Ceja, D.C.; Ortiz-Masiá, D.; Salvador, P.; Gisbert-Ferrándiz, L.; Hernández, C.; Hausmann, M.; Rogler, G.; Esplugues, J.V.; Hinojosa, J.; Alós, R.; Navarro, F.; Cosin-Roger, J.; Calatayud, S.; Barrachina, M.D. Succinate receptor mediates intestinal inflammation and fibrosis, Mucosal Immunol 2019, 12, 178–187. [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; Bousvaros, A.; Korzenik, J.; Sands, B.E.; Xavier, R.J.; Huttenhower, C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment, Genome Biol 2012, 13, R79. [CrossRef]
- Zhang, Y.; Bhosle, A.; Bae, S.; McIver, L.J.; Pishchany, G.; Accorsi, E.K.; Thompson, K.N.; Arze, C.; Wang, Y.; Subramanian, A.; Kearney, S.M.; Pawluk, A.; Plichta, D.R.; Rahnavard, A.; Shafquat, A.; Xavier, R.J.; Vlamakis, H.; Garrett, W.S.; Krueger, A.; Huttenhower, C.; Franzosa, E.A. Discovery of bioactive microbial gene products in inflammatory bowel disease, Nature 2022, 606, 754–760. [CrossRef]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; Binder, V.; Finkel, Y.; Cortot, A.; Modigliani, R.; Laurent-Puig, P.; Gower-Rousseau, C.; Macry, J.; Colombel, J.F.; Sahbatou, M.; Thomas, G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease, Nature 2001, 411, 599–603. [CrossRef]
- Krela-Kaźmierczak, I.; Zakerska-Banaszak, O.; Skrzypczak-Zielińska, M.; Łykowska-Szuber, L.; Szymczak-Tomczak, A.; Zawada, A.; Rychter, A.M.; Ratajczak, A.E.; Skoracka, K.; Skrzypczak, D.; Marcinkowska, E.; Słomski, R.; Dobrowolska, A. Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota-A Narrative Review, Nutrients 2022, 14, 2520. [Google Scholar] [CrossRef]
- Arabyan, N.; Park, D.; Foutouhi, S.; Weis, A.M.; Huang, B.C.; Williams, C.C.; Desai, P.; Shah, J.; Jeannotte, R.; Kong, N.; Lebrilla, C.B.; Weimer, B.C. Salmonella Degrades the Host Glycocalyx Leading to Altered Infection and Glycan Remodeling, Sci Rep 2016, 6, 29525. [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD, Autophagy 2020, 16, 38–51. [CrossRef]
- Pickard, J.M.; Chervonsky, A.V. Intestinal fucose as a mediator of host-microbe symbiosis, J Immunol 2015, 194, 5588–5593. [CrossRef]
- Tang, X.; Wang, W.; Hong, G.; Duan, C.; Zhu, S.; Tian, Y.; Han, C.; Qian, W.; Lin, R.; Hou, X. Gut microbiota-mediated lysophosphatidylcholine generation promotes colitis in intestinal epithelium-specific Fut2 deficiency, J Biomed Sci 2021, 28, 20. 28. [CrossRef]
- Cheng, S.; Hu, J.; Wu, X.; Pan, J.-A.; Jiao, N.; Li, Y.; Huang, Y.; Lin, X.; Zou, Y.; Chen, Y.; Zhu, L.; Zhi, M.; Lan, P. Altered gut microbiome in FUT2 loss-of-function mutants in support of personalized medicine for inflammatory bowel diseases, J Genet Genomics 2021, 48, 771–780. [CrossRef]
- Philpott, D.J.; Sorbara, M.T.; Robertson, S.J.; Croitoru, K.; Girardin, S.E. NOD proteins: regulators of inflammation in health and disease, Nat Rev Immunol 2014, 14, 9–23. [CrossRef]
- Kudelka, M.R.; Hinrichs, B.H.; Darby, T.; Moreno, C.S.; Nishio, H.; Cutler, C.E.; Wang, J.; Wu, H.; Zeng, J.; Wang, Y.; Ju, T.; Stowell, S.R.; Nusrat, A.; Jones, R.M.; Neish, A.S.; Cummings, R.D. Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk, Proc Natl Acad Sci U S A 2016, 113, 14787–14792. [CrossRef]
- Nighot, P.K.; Hu, C.-A.A.; Ma, T.Y. Autophagy Enhances Intestinal Epithelial Tight Junction Barrier Function by Targeting Claudin-2 Protein Degradation, Journal of Biological Chemistry 2015, 290, 7234–7246. [CrossRef]
- Battistini, C.; Ballan, R.; Herkenhoff, M.E.; Saad, S.M.I.; Sun, J. Vitamin D Modulates Intestinal Microbiota in Inflammatory Bowel Diseases, Int J Mol Sci 2020, 22, E362. [CrossRef]
- Sidiq, T.; Yoshihama, S.; Downs, I.; Kobayashi, K.S. Nod2: A Critical Regulator of Ileal Microbiota and Crohn’s Disease, Front. Immunol. 2016, 7. [Google Scholar] [CrossRef]
- Turpin, W.; Bedrani, L.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Smith, M.I.; Garay, J.A.R.; Lee, S.-H.; Guttman, D.S.; Griffiths, A.; Moayyedi, P.; Panaccione, R.; Huynh, H.; Steinhart, H.A.; Aumais, G.; Dieleman, L.A.; Turner, D. CCC IBD GEM Project research team, A. D. Paterson, K. Croitoru, Associations of NOD2 polymorphisms with Erysipelotrichaceae in stool of in healthy first degree relatives of Crohn’s disease subjects, BMC Med Genet 2020, 21, 204. [Google Scholar] [CrossRef]
- Seiderer, J.; Brand, S.; Herrmann, K.A.; Schnitzler, F.; Hatz, R.; Crispin, A.; Pfennig, S.; Schoenberg, S.O.; Göke, B.; Lohse, P.; Ochsenkuhn, T. Predictive value of the CARD15 variant 1007fs for the diagnosis of intestinal stenoses and the need for surgery in Crohn’s disease in clinical practice: results of a prospective study, Inflamm Bowel Dis 2006, 12, 1114–1121. [CrossRef]
- Frank, D.N.; Robertson, C.E.; Hamm, C.M.; Kpadeh, Z.; Zhang, T.; Chen, H.; Zhu, W.; Sartor, R.B.; Boedeker, E.C.; Harpaz, N.; Pace, N.R.; Li, E. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases, Inflamm Bowel Dis 2011, 17, 179–184. [CrossRef]
- Imhann, F.; Vila, A.V.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; Steege, R.W.F.T.; Huttenhower, C.; Dijkstra, G.; Xavier, R.J.; Festen, E.A.M.; Wijmenga, C.; Zhernakova, A.; Weersma, R.K. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease, Gut 2018, 67, 108–119. [CrossRef]
- Liu, H.; Gao, P.; Jia, B.; Lu, N.; Zhu, B.; Zhang, F. IBD-Associated Atg16L1T300A Polymorphism Regulates Commensal Microbiota of the Intestine, Front. Immunol. 2022, 12, 772189. [Google Scholar] [CrossRef]
- Rokhsefat, S.; Lin, A.; Comelli, E.M. Mucin-Microbiota Interaction During Postnatal Maturation of the Intestinal Ecosystem: Clinical Implications, Dig Dis Sci 2016, 61, 1473–1486. [CrossRef]
- Colquhoun, C.; Duncan, M.; Grant, G. Inflammatory Bowel Diseases: Host-Microbial-Environmental Interactions in Dysbiosis, Diseases 2020, 8, 13. 8. [CrossRef]
- Taman, H.; Fenton, C.G.; Hensel, I.V.; Anderssen, E.; Florholmen, J.; Paulssen, R.H. Transcriptomic Landscape of Treatment-Naïve Ulcerative Colitis, J Crohns Colitis 2018, 12, 327–336. [CrossRef]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in Intestinal Mucosal Defense and Inflammation: Learning From Clinical and Experimental Studies, Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef] [PubMed]
- Padra, M.; Adamczyk, B.; Flahou, B.; Erhardsson, M.; Chahal, G.; Smet, A.; Jin, C.; Thorell, A.; Ducatelle, R.; Haesebrouck, F.; Karlsson, N.G.; Lindén, S.K. Helicobacter suis infection alters glycosylation and decreases the pathogen growth inhibiting effect and binding avidity of gastric mucins, Mucosal Immunol 2019, 12, 784–794. [CrossRef]
- Farooq, S.M.; Stillie, R.; Svensson, M.; Svanborg, C.; Strieter, R.M.; Stadnyk, A.W. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulfate-induced colitis, J Pharmacol Exp Ther 2009, 329, 123–129. [CrossRef]
- Melhem, H.; Regan-Komito, D.; Niess, J.H. Mucins Dynamics in Physiological and Pathological Conditions, IJMS 2021, 22, 13642. [CrossRef]
- Johansson, M.E.V.; Larsson, J.M.H.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions, Proc Natl Acad Sci U S A 108 Suppl 2011, 1, 4659–4665. [CrossRef]
- Van der Sluis, M.; De Koning, B.A.E.; De Bruijn, A.C.J.M.; Velcich, A.; Meijerink, J.P.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; Einerhand, A.W.C. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection, Gastroenterology 2006, 131, 117–129. [CrossRef]
- McCole, D.F. IBD candidate genes and intestinal barrier regulation, Inflamm Bowel Dis 2014, 20, 1829–1849. [CrossRef]
- Yamamoto-Furusho, J.K.; Ascaño-Gutiérrez, I.; Furuzawa-Carballeda, J.; Fonseca-Camarillo, G. Differential Expression of MUC12, MUC16, and MUC20 in Patients with Active and Remission Ulcerative Colitis, Mediators of Inflammation 2015, 2015, 1–8. [CrossRef]
- Graham, D.B.; Xavier, R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease, Nature 2020, 578, 527–539. [CrossRef]
- Janney, A.; Powrie, F.; Mann, E.H. Host–microbiota maladaptation in colorectal cancer, Nature 2020, 585, 509–517. [CrossRef]
- Read, E.; Curtis, M.A.; Neves, J.F. The role of oral bacteria in inflammatory bowel disease, Nat Rev Gastroenterol Hepatol 2021, 18, 731–742. [CrossRef]
- Jergens, A.E.; Parvinroo, S.; Kopper, J.; Wannemuehler, M.J. Rules of Engagement: Epithelial-Microbe Interactions and Inflammatory Bowel Disease, Front. Med. 2021, 8, 669913. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Horst, R.T.; Jansen, T.; Jacobs, L.; Bonder, M.J.; Kurilshikov, A.; Fu, J.; Joosten, L.A.B.; Zhernakova, A.; Huttenhower, C.; Wijmenga, C.; Netea, M.G.; Xavier, R.J. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity, Cell 2016, 167, 1125–1136.e8. [CrossRef]
- M. Sadaghian, Interaction between the gut and its microbiota in inflammatory bowel disease. (n.d.).
- Pugazhendhi, S.; Baskaran, K.; Santhanam, S.; Ramakrishna, B.S. Association of ATG16L1 gene haplotype with inflammatory bowel disease in Indians, PLoS One 2017, 12, e0178291. [CrossRef]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of Gut Microbiota in Inflammatory Bowel Disease (IBD): Cause or Consequence? IBD Treatment Targeting the Gut Microbiome, Pathogens 2019, 8, 126. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Dalmasso, G.; Müller, S.; Carrière, J.; Seibold, F.; Darfeuille-Michaud, A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy, Gastroenterology 2014, 146, 508–519. [CrossRef]
- Kee, B.P.; Ng, J.G.; Ng, C.C.; Hilmi, I.; Goh, K.L.; Chua, K.H. Genetic polymorphisms of ATG16L1 and IRGM genes in Malaysian patients with Crohn’s disease, J Dig Dis 2020, 21, 29–37. [CrossRef]
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 inflammation current insights. J Inflamm Res 2018, 11, 49–60. [Google Scholar] [CrossRef]
- Parkes, M.; Barrett, J.C.; Prescott, N.J.; Tremelling, M.; Anderson, C.A.; Fisher, S.A.; Roberts, R.G.; Nimmo, E.R.; Cummings, F.R.; Soars, D.; Drummond, H.; Lees, C.W.; Khawaja, S.A.; Bagnall, R.; Burke, D.A.; Todhunter, C.E.; Ahmad, T.; Onnie, C.M.; McArdle, W.; Strachan, D.; Bethel, G.; Bryan, C.; Lewis, C.M.; Deloukas, P.; Forbes, A.; Sanderson, J.; Jewell, D.P.; Satsangi, J.; Mansfield, J.C. Wellcome Trust Case Control Consortium, L. Cardon, C.G. Mathew, Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility, Nat Genet 2007, 39, 830–832. [Google Scholar] [CrossRef]
- Parkes, M.; Barrett, J.C.; Prescott, N.J.; Tremelling, M.; Anderson, C.A.; Fisher, S.A.; Roberts, R.G.; Nimmo, E.R.; Cummings, F.R.; Soars, D.; Drummond, H.; Lees, C.W.; Khawaja, S.A.; Bagnall, R.; Burke, D.A.; Todhunter, C.E.; Ahmad, T.; Onnie, C.M.; McArdle, W.; Strachan, D.; Bethel, G.; Bryan, C.; Lewis, C.M.; Deloukas, P.; Forbes, A.; Sanderson, J.; Jewell, D.P.; Satsangi, J.; Mansfield, J.C. Wellcome Trust Case Control Consortium, L. Cardon, C.G. Mathew, Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility, Nat Genet 2007, 39, 830–832. [Google Scholar] [CrossRef]
- Moehle, C.; Ackermann, N.; Langmann, T.; Aslanidis, C.; Kel, A.; Kel-Margoulis, O.; Schmitz-Madry, A.; Zahn, A.; Stremmel, W.; Schmitz, G. Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease, J Mol Med 2006, 84, 1055–1066. [CrossRef]
- McAuley, J.L.; Linden, S.K.; Png, C.W.; King, R.M.; Pennington, H.L.; Gendler, S.J.; Florin, T.H.; Hill, G.R.; Korolik, V.; McGuckin, M.A. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection, J. Clin. Invest. 2007, 117, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xin, Y.; Zhou, J.; Tian, Z.; Liu, C.; Yu, X.; Meng, X.; Jiang, W.; Zhao, S.; Dong, Q. Gastric Mucosa-Associated Microbial Signatures of Early Gastric Cancer, Front Microbiol 2020, 11, 1548. [CrossRef]
- Visschedijk, M.C.; Alberts, R.; Mucha, S.; Deelen, P.; de Jong, D.J.; Pierik, M.; Spekhorst, L.M.; Imhann, F.; van der Meulen-de Jong, A.E.; van der Woude, C.J.; van Bodegraven, A.A.; Oldenburg, B.; Löwenberg, M.; Dijkstra, G.; Ellinghaus, D.; Schreiber, S.; Wijmenga, C. The Initiative on Crohn and Colitis, Parelsnoer Institute, M. A. Rivas, A. Franke, C.C. van Diemen, R.K. Weersma, Pooled Resequencing of 122 Ulcerative Colitis Genes in a Large Dutch Cohort Suggests Population-Specific Associations of Rare Variants in MUC2, PLoS ONE 2016, 11, e0159609. [Google Scholar] [CrossRef]
- Padra, M.; Adamczyk, B.; Flahou, B.; Erhardsson, M.; Chahal, G.; Smet, A.; Jin, C.; Thorell, A.; Ducatelle, R.; Haesebrouck, F.; Karlsson, N.G.; Lindén, S.K. Helicobacter suis infection alters glycosylation and decreases the pathogen growth inhibiting effect and binding avidity of gastric mucins, Mucosal Immunol 2019, 12, 784–794. [CrossRef]
- Luo, P.; Yang, Z.; Chen, B.; Zhong, X. The multifaceted role of CARD9 in inflammatory bowel disease, J Cell Mol Med 2020, 24, 34–39. [CrossRef]
- Zhernakova, A.; Festen, E.M.; Franke, L.; Trynka, G.; van Diemen, C.C.; Monsuur, A.J.; Bevova, M.; Nijmeijer, R.M.; van ’t Slot, R.; Heijmans, R.; Boezen, H.M.; van Heel, D.A.; van Bodegraven, A.A.; Stokkers, P.C.F.; Wijmenga, C.; Crusius, J.B.A.; Weersma, R.K. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP, Am J Hum Genet 2008, 82, 1202–1210. [CrossRef]
- Wu, S.; Zhang, Y.; Ma, J.; Liu, Y.; Li, W.; Wang, T.; Xu, X.; Wang, Y.; Cheng, K.; Zhuang, R. Interleukin-6 absence triggers intestinal microbiota dysbiosis and mucosal immunity in mice, Cytokine 2022, 153, 155841. [CrossRef]
- Borecki, K.; Zawada, I.; Salkić, N.N.; Karakiewicz, B.; Adler, G. Relationship between the IL23R SNPs and Crohn’s Disease Susceptibility and Phenotype in the Polish and Bosnian Populations: A Case-Control Study, IJERPH 2019, 16, 1551. [CrossRef]
- Alharbi, R.S.; Shaik, N.A.; Almahdi, H.; ElSokary, H.A.; Jamalalail, B.A.; Mosli, M.H.; Alsufyani, H.A.; Al-Aama, J.Y.; Elango, R.; Saadah, O.I.; Banaganapalli, B. Genetic association study of NOD2 and IL23R amino acid substitution polymorphisms in Saudi Inflammatory Bowel Disease patients, Journal of King Saud University - Science 2022, 34, 101726. [CrossRef]
- Peng, L.-L.; Wang, Y.; Zhu, F.-L.; Xu, W.-D.; Ji, X.-L.; Ni, J. IL-23R mutation is associated with ulcerative colitis: A systemic review and meta-analysis, Oncotarget 2017, 8, 4849–4863. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





