Submitted:
28 June 2024
Posted:
29 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.3. Data Extraction
2.4. Quality Assessment
2.5. Statistical Analysis
3. Results
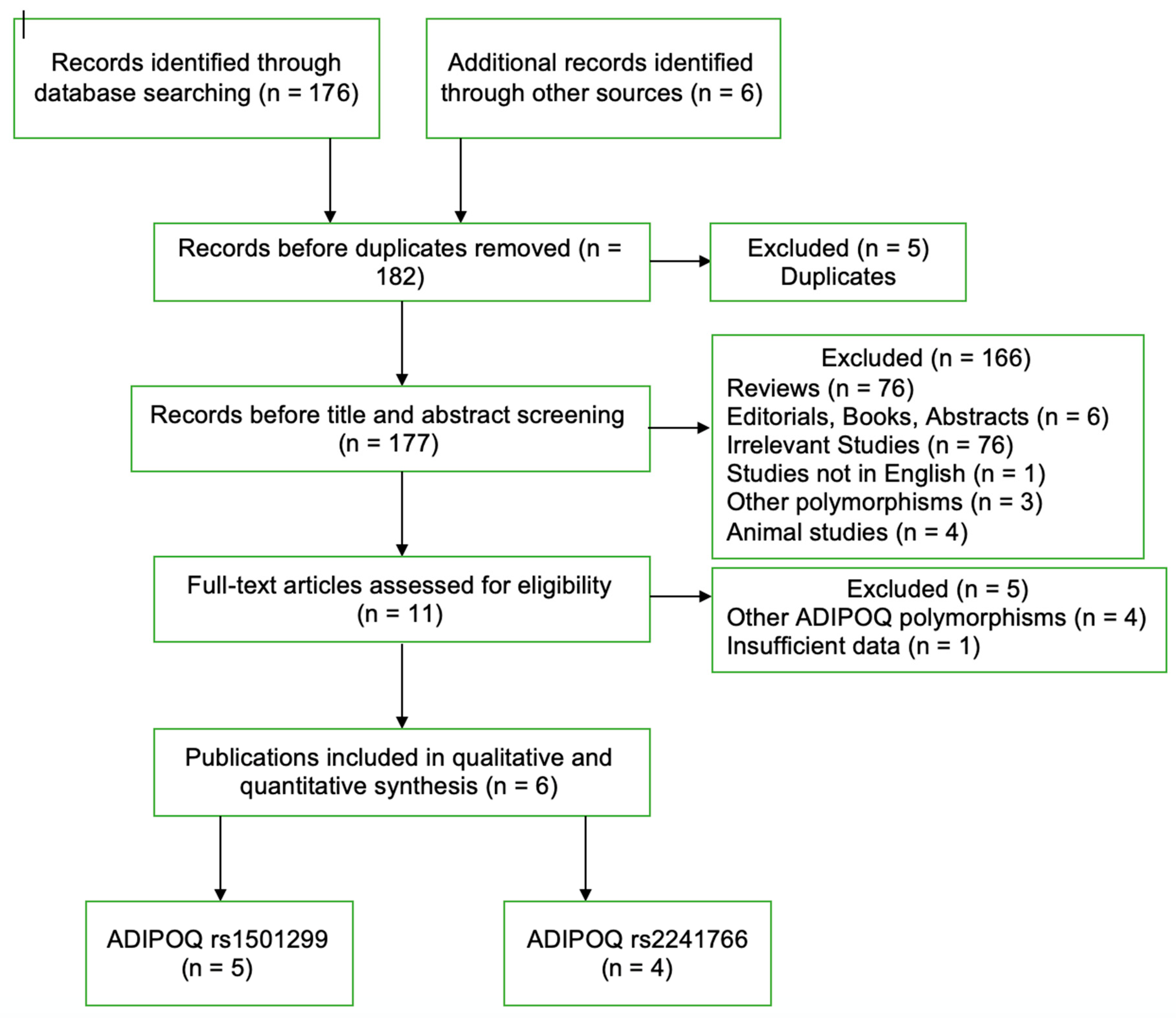
3.1. Literature Search
3.2. Study Characteristics and Summary Statistics
| First Author | Year of Publication | Country/Ethnicity | Study Design | Genotyping Method | Control Type | Type of DM | DR grade | Case | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | Sex (M/F) | Age (years) | ||||||||
| Aioanei | 2021 | Romania/Caucasian | Case–Control | PCR-RFLP | HC | T2DM | NPDR | 198 | 105/93 | 68.72 ± 11.58 |
| Choe | 2013 | Korea/Asian | Cohort Study | PCR-RFLP | NDR | T2DM | Any DR | 231 | N/A | N/A |
| Gouliopoulos | 2022 | Greece/Caucasian | Case–Control | PCR-RFLP | NDR | T2DM | Any DR | 109 | 74/35 | 67.00 ± 8.00 |
| Li | 2015 | China/Asian | Case–Control | PCR-RFLP | NDR | T2DM | Any DR | 372 | 146/226 | 63.39 ± 10.60 |
| Yoshioka | 2004 | Japan/Asian | Case–Control | PCR-RFLP | HC+NDR | T2DM | Any DR | 104 | 55/49 | 62.05 ± 9.20 |
| Choe | 2013 | Korea/Asian | Cohort Study | PCR-RFLP | NDR | T2DM | Any DR | 225 | N/A | N/A |
| Gouliopoulos | 2022 | Greece/Caucasian | Case–Control | PCR-RFLP | NDR | T2DM | Any DR | 109 | 74/35 | 67.00 ± 8.00 |
| Li | 2015 | China/Asian | Case–Control | PCR-RFLP | NDR | T2DM | Any DR | 372 | 146/226 | 63.39 ± 10.60 |
| Sikka | 2014 | India/Asian | Case–Control | PCR-RFLP | HC+NDR | T2DM | Any DR | 169 | N/A | 58.35 ± 9.01 |
| First Author | Control | Genotype distribution | HWE p-value |
MAF | NOS (Stars) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | Sex (M/F) | Age (years) | Case | Control | Control | Case | Control | ||||||
| rs1501299 G/T | |||||||||||||
| GG | GT | TT | GG | GT | TT | ||||||||
| Aioanei | 200 | 143/57 | 58.10 ± 9.00 | 93 | 79 | 26 | 92 | 88 | 20 | 0.876 | 0.33 | 0.32 | 7 |
| Choe | 440 | N/A | N/A | 109 | 102 | 20 | 222 | 178 | 40 | 0.616 | 0.31 | 0.29 | 7 |
| Gouliopoulos | 109 | 75/34 | 66.00 ± 9.00 | 40 | 58 | 11 | 59 | 37 | 13 | 0.069 | 0.37 | 0.29 | 7 |
| Li | 145 | 49/96 | 62.34 ± 10.75 | 164 | 169 | 39 | 82 | 55 | 8 | 0.756 | 0.33 | 0.24 | 8 |
| Yoshioka | 340 | 219/121 | 59.70 ± 10.10 | 50 | 42 | 12 | 163 | 147 | 30 | 0.699 | 0.32 | 0.30 | 7 |
| rs2241766 T/G | |||||||||||||
| TT | TG | GG | TT | TG | GG | ||||||||
| Choe | 442 | N/A | N/A | 111 | 96 | 18 | 213 | 194 | 35 | 0.315 | 0.29 | 0.30 | 7 |
| Gouliopoulos | 109 | 75/34 | 66.00 ± 9.00 | 84 | 23 | 2 | 74 | 32 | 3 | 0.836 | 0.12 | 0.17 | 7 |
| Li | 145 | 49/96 | 62.34 ± 10.75 | 206 | 140 | 25 | 82 | 53 | 10 | 0.720 | 0.26 | 0.25 | 8 |
| Sikka | 355 | N/A | 53.16 ±12.15 | 158 | 9 | 2 | 292 | 58 | 5 | 0.285 | 0.04 | 0.09 | 6 |
3.3. Quantitative and Subgroup Analyses
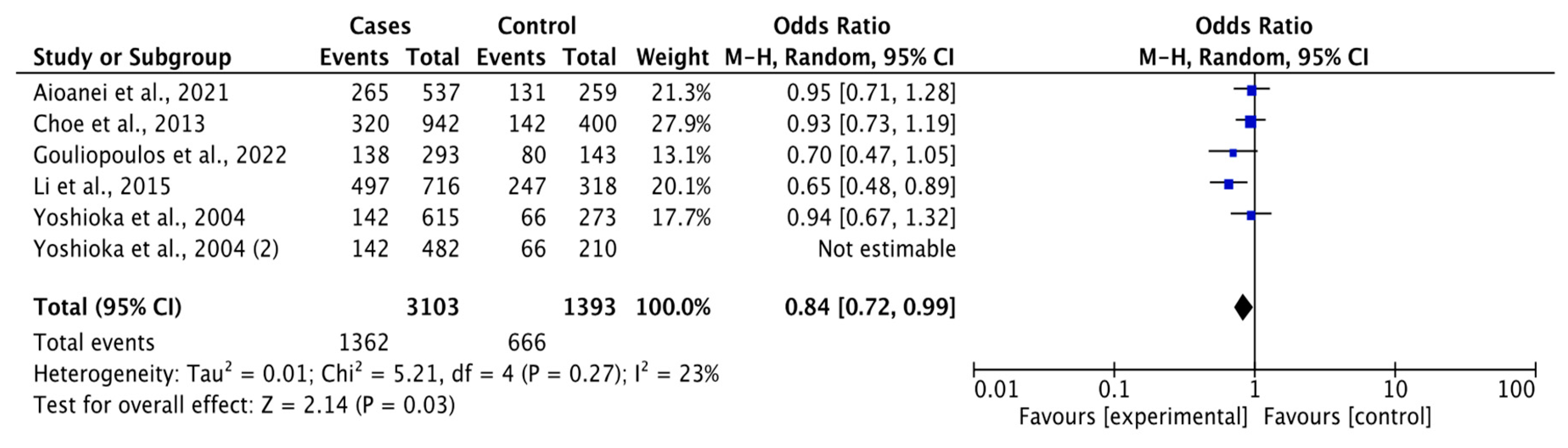
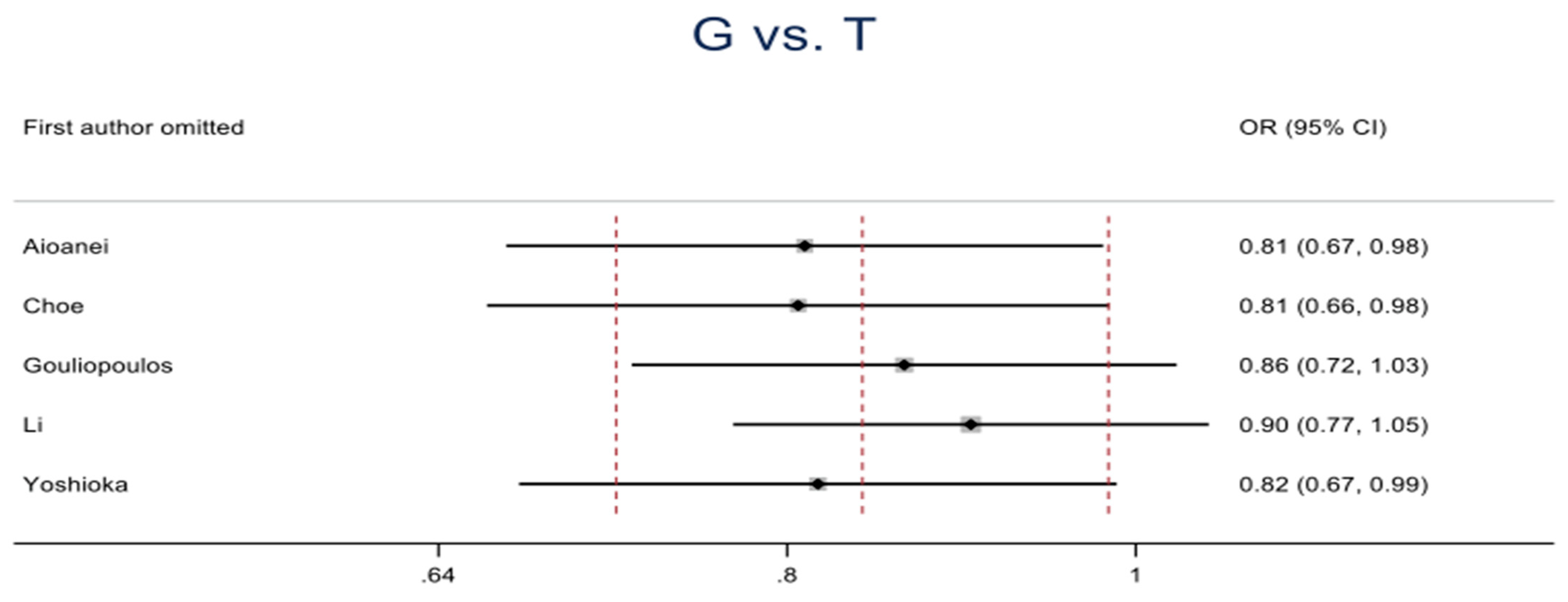
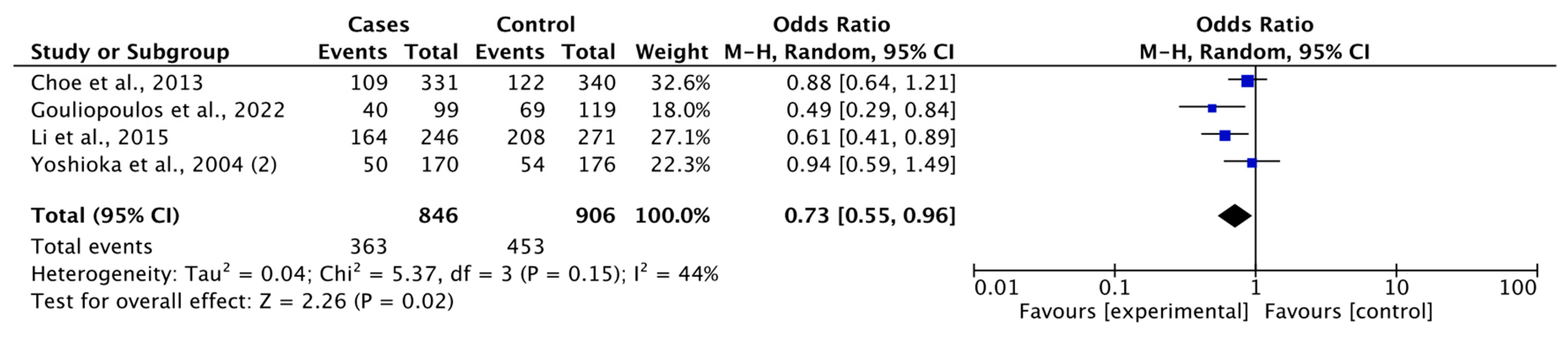
3.3.1. Association between rs1501299 Polymorphism and DR Risk
3.3.2. Association between rs2241766 Polymorphism and DR Risk
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cheung, N.; Mitchell, P.; Wong, T. Y. Diabetic Retinopathy. Lancet (London, England) 2010, 376 (9735), 124–136. [CrossRef]
- Sabanayagam, C.; Banu, R.; Chee, M. L.; Lee, R.; Wang, Y. X.; Tan, G.; Jonas, J. B.; Lamoureux, E. L.; Cheng, C.-Y.; Klein, B. E. K.; Mitchell, P.; Klein, R.; Cheung, C. M. G.; Wong, T. Y. Incidence and Progression of Diabetic Retinopathy: A Systematic Review. lancet. Diabetes Endocrinol. 2019, 7 (2), 140–149. [CrossRef]
- Heng, L. Z.; Comyn, O.; Peto, T.; Tadros, C.; Ng, E.; Sivaprasad, S.; Hykin, P. G. Diabetic Retinopathy: Pathogenesis, Clinical Grading, Management and Future Developments. Diabet. Med. 2013, 30 (6), 640–650. [CrossRef]
- Vujosevic, S.; Aldington, S. J.; Silva, P.; Hernández, C.; Scanlon, P.; Peto, T.; Simó, R. Screening for Diabetic Retinopathy: New Perspectives and Challenges. Lancet Diabetes Endocrinol. 2020, 8 (4), 337–347. [CrossRef]
- Capitão, M.; Soares, R. Angiogenesis and Inflammation Crosstalk in Diabetic Retinopathy. J. Cell. Biochem. 2016, No. April 2016, 2443–2453. [CrossRef]
- Ghamdi, A. H. Al. Clinical Predictors of Diabetic Retinopathy Progression; A Systematic Review. Curr. Diabetes Rev. 2019, 16 (3), 242–247. [CrossRef]
- Simó-Servat, O.; Hernández, C.; Simó, R. Diabetic Retinopathy in the Context of Patients with Diabetes. Ophthalmic Res. 2019, 62 (4), 211–217. [CrossRef]
- Lin, K.-Y.; Hsih, W.-H.; Lin, Y.-B.; Wen, C.-Y.; Chang, T.-J. Update in the Epidemiology, Risk Factors, Screening, and Treatment of Diabetic Retinopathy. J. Diabetes Investig. 2021, 12 (8), 1322–1325. [CrossRef]
- Fung, T. H.; Patel, B.; Wilmot, E. G.; Amoaku, W. M. Diabetic Retinopathy for the Non-Ophthalmologist. Clin. Med. 2022, 22 (2), 112–116. [CrossRef]
- Sharma, A.; Valle, M. L.; Beveridge, C.; Liu, Y.; Sharma, S. Unraveling the Role of Genetics in the Pathogenesis of Diabetic Retinopathy. Eye 2019, 33 (4), 534–541. [CrossRef]
- Xian, L.; He, W.; Pang, F.; Hu, Y. ADIPOQ Gene Polymorphisms and Susceptibility to Polycystic Ovary Syndrome: A HuGE Survey and Meta-Analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012, 161 (2), 117–124. [CrossRef]
- Han, Q.; Geng, W.; Zhang, D.; Cai, G.; Zhu, H. ADIPOQ Rs2241766 Gene Polymorphism and Predisposition to Diabetic Kidney Disease. J. Diabetes Res. 2020, 2020, 5158497. [CrossRef]
- Fang, H.; Judd, R. L. Adiponectin Regulation and Function. Compr. Physiol. 2018, 8 (3), 1031–1063. [CrossRef]
- Al-Nbaheen, M. S. Effect of Genetic Variations in the ADIPOQ Gene on Susceptibility to Type 2 Diabetes Mellitus. Diabetes. Metab. Syndr. Obes. 2022, 15, 2753–2761. [CrossRef]
- Alfaqih, M. A.; Al-Hawamdeh, A.; Amarin, Z. O.; Khader, Y. S.; Mhedat, K.; Allouh, M. Z. Single Nucleotide Polymorphism in the ADIPOQ Gene Modifies Adiponectin Levels and Glycemic Control in Type Two Diabetes Mellitus Patients. Biomed Res. Int. 2022, 2022. [CrossRef]
- Chung, H. F.; Long, K. Z.; Hsu, C. C.; Mamun, A. Al; Chiu, Y. F.; Tu, H. P.; Chen, P. S.; Jhang, H. R.; Hwang, S. J.; Huang, M. C. Adiponectin Gene (ADIPOQ) Polymorphisms Correlate with the Progression of Nephropathy in Taiwanese Male Patients with Type 2 Diabetes. Diabetes Res. Clin. Pract. 2014, 105 (2), 261–270. [CrossRef]
- Isakova, J.; Talaibekova, E.; Vinnikov, D.; Saadanov, I.; Aldasheva, N. ADIPOQ, KCNJ11 and TCF7L2 Polymorphisms in Type 2 Diabetes in Kyrgyz Population: A Case-Control Study. J. Cell. Mol. Med. 2019, 23 (2), 1628–1631. [CrossRef]
- Aioanei, C. S.; Ilies, R. F.; Bala, C.; Petrisor, M. F.; Porojan, M. D.; Popp, R. A.; Catana, A. The Role of Adiponectin and Toll-like Receptor 4 Gene Polymorphisms on Non-Proliferative Retinopathy in Type 2 Diabetes Mellitus Patients. A Casecontrol Study in Romanian Caucasians Patients. Acta Endocrinol. (Copenh). 2019, 15 (1), 32–38. [CrossRef]
- Gouliopoulos, N.; Siasos, G.; Bouratzis, N.; Oikonomou, E.; Kollia, C.; Konsola, T.; Oikonomou, D.; Rouvas, A.; Kassi, E.; Tousoulis, D.; Moschos, M. M. Polymorphism Analysis of ADIPOQ Gene in Greek Patients with Diabetic Retinopathy. Ophthalmic Genet. 2022, 43 (3), 326–331. [CrossRef]
- Sikka, R.; Raina, P.; Matharoo, K.; Bandesh, K.; Bhatia, R.; Chakrabarti, S.; Bhanwer, A. J. S. TNF-α (g.-308 G > A) and ADIPOQ (g. + 45 T > G) Gene Polymorphisms in Type 2 Diabetes and Microvascular Complications in the Region of Punjab (North-West India). Curr. Eye Res. 2014, 39 (10), 1042–1051. [CrossRef]
- Choe, E. Y.; Wang, H. J.; Kwon, O.; Kim, K. J.; Kim, B. S.; Lee, B. W.; Ahn, C. W.; Cha, B. S.; Lee, H. C.; Kang, E. S.; Mantzoros, C. S. Variants of the Adiponectin Gene and Diabetic Microvascular Complications in Patients with Type 2 Diabetes. Metabolism. 2013, 62 (5), 677–685. [CrossRef]
- Li, Y.; Wu, Q. H.; Jiao, M. L.; Fan, X. H.; Hu, Q.; Hao, Y. H.; Liu, R. H.; Zhang, W.; Cui, Y.; Han, L. Y. Gene-Environment Interaction between Adiponectin Gene Polymorphisms and Environmental Factors on the Risk of Diabetic Retinopathy. J. Diabetes Investig. 2015, 6 (1), 56–66. [CrossRef]
- Yoshioka, K.; Yoshida, T.; Takakura, Y.; Umekawa, T.; Kogure, A.; Toda, H.; Yoshikawa, T. Adiponectin Gene Polymorphism (G276T) and Diabetic Retinopathy in Japanese Patients with Type 2 Diabetes. Diabetic medicine : a journal of the British Diabetic Association. England October 2004, pp 1158–1159. [CrossRef]
- Page, M. J.; McKenzie, J. E.; Bossuyt, P. M.; Boutron, I.; Hoffmann, T. C.; Mulrow, C. D.; Shamseer, L.; Tetzlaff, J. M.; Akl, E. A.; Brennan, S. E.; Chou, R.; Glanville, J.; Grimshaw, J. M.; Hróbjartsson, A.; Lalu, M. M.; Li, T.; Loder, E. W.; Mayo-Wilson, E.; McDonald, S.; McGuinness, L. A.; Stewart, L. A.; Thomas, J.; Tricco, A. C.; Welch, V. A.; Whiting, P.; Moher, D. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372. [CrossRef]
- Ding, Y.; Hu, Z.; Yuan, S.; Xie, P.; Liu, Q. Association between Transcription Factor 7-like 2 Rs7903146 Polymorphism and Diabetic Retinopathy in Type 2 Diabetes Mellitus: A Meta-Analysis. Diabetes Vasc. Dis. Res. 2015, 12 (6), 436–444. [CrossRef]
- Jiao, J.; Li, Y.; Xu, S.; Wu, J.; Yue, S.; Liu, L. Association of FokI, TaqI, BsmI and ApaI Polymorphisms with Diabetic Retinopathy: A Pooled Analysis of Case-Control Studies. Afr. Health Sci. 2018, 18 (4), 891–899. [CrossRef]
- Luo, S.; Wang, F.; Shi, C.; Wu, Z. A Meta-Analysis of Association between Methylenetetrahydrofolate Reductase Gene (MTHFR) 677C/T Polymorphism and Diabetic Retinopathy. Int. J. Environ. Res. Public Health 2016, 13 (8). [CrossRef]
- Horita, N.; Kaneko, T. Genetic Model Selection for a Case-Control Study and a Meta-Analysis. Meta Gene 2015, 5, 1–8. [CrossRef]
- DerSimonian, R.; Laird, N. Meta-Analysis in Clinical Trials. Control. Clin. Trials 1986, 7 (3), 177–188. [CrossRef]
- Li, X. F.; Jiang, G. Bin; Cheng, S. Y.; Song, Y. F.; Deng, C.; Niu, Y. M.; Cai, J. W. Association between PPAR-Γ2 Gene Polymorphisms and Diabetic Retinopathy Risk: A Meta-Analysis. Aging (Albany. NY). 2021, 13 (4), 5136–5149. [CrossRef]
- Montesanto, A.; Bonfigli, A. R.; Crocco, P.; Garagnani, P.; De Luca, M.; Boemi, M.; Marasco, E.; Pirazzini, C.; Giuliani, C.; Franceschi, C.; Passarino, G.; Testa, R.; Olivieri, F.; Rose, G. Genes Associated with Type 2 Diabetes and Vascular Complications. Aging (Albany. NY). 2018, 10 (2), 178–196. [CrossRef]
- Huang, Y. C.; Chang, Y. W.; Cheng, C. W.; Wu, C. M.; Liao, W. L.; Tsai, F. J. Causal Relationship between Adiponectin and Diabetic Retinopathy: A Mendelian Randomization Study in an Asian Population. Genes (Basel). 2021, 12 (1), 1–11. [CrossRef]
- Rudofsky, G.; Schlimme, M.; Schlotterer, A.; von Eynatten, M.; Reismann, P.; Tafel, J.; Grafe, I.; Morcos, M.; Nawroth, P.; Bierhaus, A.; Hamann, A. No Association of the 94T/G Polymorphism in the Adiponectin Gene with Diabetic Complications. Diabetes, Obes. Metab. 2005, 7 (4), 455–459. [CrossRef]
- Zietz, B.; Buechler, C.; Kobouch, K.; Neumeier, M.; Schölmerich, J.; Scäffler, A. Serum Levels of Adiponectin Are Associated with Diabetic Retinopathy and with Adiponectin Gene Mutations in Caucasian Patients with Diabetes Mellitus Type 2. Exp. Clin. Endocrinol. Diabetes 2008, 116 (9), 532–536. [CrossRef]
- Liao, W. L.; Chen, Y. H.; Chen, C. C.; Huang, Y. C.; Lin, H. J.; Chen, Y. T.; Ban, B.; Wu, C. M.; Chang, Y. W.; Hsieh, A. R.; Tsai, F. J. Effect of Adiponectin Level and Genetic Variation of Its Receptors on Diabetic Retinopathy A Case-Control Study. Med. (United States) 2019, 98 (11), 1–7. [CrossRef]
- Tan, T. E.; Wong, T. Y. Diabetic Retinopathy: Looking Forward to 2030. Front. Endocrinol. (Lausanne). 2023, 13 (January), 1–8. [CrossRef]
- Lightman, S.; Towler, H. M. A. Diabetic Retinopathy. Clin. Cornerstone 2003, 5 (2), 12–21. [CrossRef]
- Al-Shabrawey, M.; Zhang, W.; McDonald, D. Diabetic Retinopathy: Mechanism, Diagnosis, Prevention, and Treatment. Biomed Res. Int. 2015, 2015. [CrossRef]
- Stitt, A. W.; Lois, N.; Medina, R. J.; Adamson, P.; Curtis, T. M. Advances in Our Understanding of Diabetic Retinopathy. Clin. Sci. 2013, 125 (1), 1–17. [CrossRef]
- Alimi, M.; Goodarzi, M. T.; Nekoei, M. Association of ADIPOQ Rs266729 and Rs1501299 Gene Polymorphisms and Circulating Adiponectin Level with the Risk of Type 2 Diabetes in a Population of Iran: A Case-Control Study. J. Diabetes Metab. Disord. 2021, 20 (1), 87–93. [CrossRef]
- Zhao, N.; Li, N.; Zhang, S.; Ma, Q.; Ma, C.; Yang, X.; Yin, J.; Zhang, R.; Li, J.; Yang, X.; Cui, T. Associations between Two Common Single Nucleotide Polymorphisms (Rs2241766 and Rs1501299) of ADIPOQ Gene and Coronary Artery Disease in Type 2 Diabetic Patients: A Systematic Review and Meta-Analysis. Oncotarget 2017, 8 (31), 51994–52005. [CrossRef]
- De Luis, D. A.; Izaola, O.; De La Fuente, B.; Primo, D.; Ovalle, H. F.; Romero, E. Rs1501299 Polymorphism in the Adiponectin Gene and Their Association with Total Adiponectin Levels, Insulin Resistance and Metabolic Syndrome in Obese Subjects. Ann. Nutr. Metab. 2017, 69 (3–4), 226–231. [CrossRef]
- Cai, X.; Gan, Y.; Fan, Y.; Hu, J.; Jin, Y.; Chen, F.; Chen, T.; Sun, Y.; Wang, J.; Qin, W.; Tu, H. The Adiponectin Gene Single-Nucleotide Polymorphism Rs1501299 Is Associated with Hepatocellular Carcinoma Risk. Clin. Transl. Oncol. 2014, 16 (2), 166–172. [CrossRef]
- Bieńkiewicz, J.; Smolarz, B.; Malinowski, A. Association Between Single Nucleotide Polymorphism +276G > T (Rs1501299) in ADIPOQ and Endometrial Cancer. Pathol. Oncol. Res. 2016, 22 (1), 135–138. [CrossRef]
- Tu, Y.; Yu, Q.; Fan, G.; Yang, P.; Lai, Q.; Yang, F.; Zhang, S.; Wang, W.; Wang, D.; Yu, X.; Wang, C. Y. Assessment of Type 2 Diabetes Risk Conferred by SNPs Rs2241766 and Rs1501299 in the ADIPOQ Gene, a Case/Control Study Combined with Meta-Analyses. Mol. Cell. Endocrinol. 2014, 396 (1–2), 1–9. [CrossRef]
- Leon-Cachon, R. B. R.; Salinas-Santander, M. A.; Aguilar-Tamez, D. A.; Marianavaldez-Ortiz, P.; Rios-Ibarra, C. P.; Cepeda-Nieto, A. C.; Suarez-Valencia, V. de J.; Morlett-Chavez, J. A. ADIPOQ-Rs2241766 Polymorphism Is Associated with Changes in Cholesterol Levels of Mexican Adolescents. J. Appl. Biomed. 2022, 20 (4), 146–153. [CrossRef]
- Liu, Y.; Kanu, J. S.; Qiu, S.; Cheng, Y.; Li, R.; Kou, C.; Gu, Y.; Bai, Y.; Shi, J.; Li, Y.; Liu, Y.; Yu, Y. Associations between Three Common Single Nucleotide Polymorphisms (Rs266729, Rs2241766, and Rs1501299) of ADIPOQ and Cardiovascular Disease: A Meta-Analysis. Lipids Health Dis. 2018, 17 (1), 1–21. [CrossRef]
- ZHOU, J. M.; ZHANG, M.; WANG, S.; WANG, B. Y.; HAN, C. Y.; REN, Y. C.; ZHANG, L.; ZHANG, H. Y.; YANG, X. Y.; ZHAO, Y.; HU, D. S. Association of the ADIPOQ Rs2241766 and Rs266729 Polymorphisms with Metabolic Syndrome in the Chinese Population: A Meta-Analysis. Biomed. Environ. Sci. 2016, 29 (7), 505–515. [CrossRef]
- Wu, J.; Liu, Z.; Meng, K.; Zhang, L. Association of Adiponectin Gene (ADIPOQ) Rs2241766 Polymorphism with Obesity in Adults: A Meta-Analysis. PLoS One 2014, 9 (4). [CrossRef]
| rs1501299 | Study | Sample size | Studies (n) | Test of association | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | Z | p-value | χ2 | p-value | I2 (%) | T2 | |||
| G vs. T | Overall | 3103 | 1393 | 5 | 0.84 (0.72-0.99) | 2.14 | 0.03 | 5.21 | 0.27 | 23 | 0.01 |
| Asian | 2273 | 991 | 3 | 0.84 (0.66-1.05) | 1.51 | 0.13 | 3.76 | 0.15 | 47 | 0.02 | |
| Caucasian | 830 | 402 | 2 | 0.84 (0.63-1.13) | 1.14 | 0.25 | 1.44 | 0.23 | 31 | 0.01 | |
| DR vs DM | 2433 | 1071 | 4 | 0.81 (0.67-0.97) | 2.28 | 0.02 | 4.13 | 0.25 | 27 | 0.01 | |
| DR vs HC | 812 | 388 | 2 | 0.97 (0.76-1.24) | 0.21 | 0.83 | 0.07 | 0.79 | 0 | 0.00 | |
| GG vs. TT | Overall | 1074 | 219 | 5 | 0.76 (0.55-1.04) | 1.73 | 0.08 | 3.02 | 0.55 | 0 | 0.00 |
| Asian | 790 | 149 | 3 | 0.72 (0.44-1.18) | 1.32 | 0.19 | 2.99 | 0.22 | 33 | 0.06 | |
| Caucasian | 284 | 70 | 2 | 0.79 (0.46-1.33) | 0.90 | 0.37 | 0.00 | 0.96 | 0 | 0.00 | |
| DR vs DM | 846 | 165 | 4 | 0.75 (0.52-1.08) | 1.55 | 0.12 | 3.01 | 0.39 | 0 | 0.00 | |
| DR vs HC | 278 | 66 | 2 | 0.78 (0.45-1.34) | 0.91 | 0.36 | 0.00 | 1.00 | 0 | 0.00 | |
| GT vs. TT | Overall | 955 | 219 | 5 | 0.90 (0.63-1.29) | 0.56 | 0.57 | 4.82 | 0.31 | 17 | 0.03 |
| Asian | 693 | 149 | 3 | 0.87 (0.58-1.30) | 0.70 | 0.48 | 1.70 | 0.43 | 0 | 0.00 | |
| Caucasian | 262 | 70 | 2 | 1.08 (0.41-2.81) | 0.15 | 0.88 | 3.00 | 0.08 | 67 | 0.32 | |
| DR vs DM | 741 | 165 | 4 | 1.00 (0.66-1.52) | 0.01 | 0.99 | 3.64 | 0.30 | 18 | 0.03 | |
| DR vs HC | 256 | 66 | 2 | 0.66 (0.38-1.14) | 1.49 | 0.14 | 0.06 | 0.81 | 0 | 0.00 | |
| GG vs. GT | Overall | 1074 | 955 | 5 | 0.80 (0.60-1.08) | 1.44 | 0.15 | 9.55 | 0.05 | 58 | 0.07 |
| Asian | 790 | 693 | 3 | 0.83 (0.64-1.08) | 1.40 | 0.16 | 2.60 | 0.27 | 23 | 0.01 | |
| Caucasian | 284 | 262 | 2 | 0.71 (0.28-1.82) | 0.71 | 0.48 | 6.94 | 0.008 | 86 | 0.39 | |
| DR vs DM | 846 | 741 | 4 | 0.72 (0.53-0.98) | 2.06 | 0.04 | 5.85 | 0.12 | 49 | 0.05 | |
| DR vs HC | 278 | 256 | 2 | 1.18 (0.84-1.66) | 0.97 | 0.33 | 0.16 | 0.69 | 0 | 0.00 | |
| GG vs. GT+TT | Overall | 1074 | 1174 | 5 | 0.79 (0.61-1.03) | 1.76 | 0.08 | 8.15 | 0.09 | 51 | 0.04 |
| Asian | 790 | 842 | 3 | 0.81 (0.61-1.07) | 1.48 | 0.14 | 3.34 | 0.19 | 40 | 0.02 | |
| Caucasian | 284 | 332 | 2 | 0.73 (0.35-1.52) | 0.83 | 0.40 | 4.81 | 0.03 | 79 | 0.22 | |
| DR vs DM | 846 | 906 | 4 | 0.73 (0.55-0.96) | 2.26 | 0.02 | 5.37 | 0.15 | 44 | 0.04 | |
| DR vs HC | 278 | 322 | 2 | 1.09 (0.79-1.50) | 0.50 | 0.61 | 0.14 | 0.71 | 0 | 0.00 | |
| GG+GT vs.TT | Overall | 2031 | 219 | 5 | 0.82 (0.61-1.12) | 1.26 | 0.21 | 3.31 | 0.51 | 0 | 0.00 |
| Asian | 1485 | 149 | 3 | 0.78 (0.51-1.19) | 1.14 | 0.26 | 2.38 | 0.30 | 16 | 0.02 | |
| Caucasian | 546 | 70 | 2 | 0.87 (0.53-1.44) | 0.53 | 0.59 | 0.85 | 0.36 | 0 | 0.00 | |
| DR vs DM | 1587 | 165 | 4 | 0.86 (0.60-1.23) | 0.81 | 0.42 | 3.06 | 0.38 | 2 | 0.00 | |
| DR vs HC | 534 | 66 | 2 | 0.72 (0.43-1.21) | 1.25 | 0.21 | 0.02 | 0.90 | 0 | 0.00 | |
| GG+TT vs. GT | Overall | 1293 | 955 | 5 | 0.85 (0.63-1.13) | 1.13 | 0.26 | 9.80 | 0.04 | 59 | 0.06 |
| Asian | 939 | 693 | 3 | 0.87 (0.70-1.08) | 1.23 | 0.22 | 1.99 | 0.37 | 0 | 0.00 | |
| Caucasian | 354 | 262 | 2 | 0.75 (0.29-1.91) | 0.61 | 0.54 | 7.78 | 0.005 | 87 | 0.40 | |
| DR vs DM | 1011 | 741 | 4 | 0.76 (0.57-1.02) | 1.82 | 0.13 | 5.72 | 0.13 | 48 | 0.04 | |
| DR vs HC | 344 | 256 | 2 | 1.24 (0.90-1.72) | 1.30 | 0.19 | 0.16 | 0.69 | 0 | 0.00 | |
| rs2241766 | Study | Sample size | Studies (n) | Test of association | Test of heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | Z | p-value | χ2 | p-value | I2 (%) | T2 | |||
| T vs. G | Overall | 3045 | 805 | 4 | 1.30 (1.00-2.09) | 1.42 | 0.15 | 10.03 | 0.02 | 70 | 0.09 |
| Asian | 2674 | 740 | 3 | 1.27 (0.82-1.96) | 1.06 | 0.29 | 8.93 | 0.01 | 78 | 0.11 | |
| DR vs DM | 2681 | 763 | 4 | 1.21 (0.90-1.64) | 1.27 | 0.20 | 6.76 | 0.08 | 56 | 0.05 | |
| TT vs. GG | Overall | 1220 | 100 | 4 | 1.34 (0.87-2.07) | 1.31 | 0.19 | 1.74 | 0.63 | 0 | 0.00 |
| Asian | 1062 | 95 | 3 | 1.32 (0.84-2.07) | 1.21 | 0.23 | 1.66 | 0.44 | 0 | 0.00 | |
| DR vs DM | 1056 | 97 | 4 | 1.33 (0.86-2.07) | 1.28 | 0.20 | 1.74 | 0.63 | 0 | 0.00 | |
| TG vs. GG | Overall | 605 | 100 | 4 | 0.94 (0.60-1.49) | 0.26 | 0.79 | 1.06 | 0.79 | 0 | 0.00 |
| Asian | 550 | 95 | 3 | 0.93 (0.58-1.50) | 0.29 | 0.77 | 1.03 | 0.60 | 0 | 0.00 | |
| DR vs DM | 569 | 97 | 4 | 0.96 (0.60-1.52) | 0.18 | 0.86 | 0.70 | 0.87 | 0 | 0.00 | |
| TT vs. TG+GG | Overall | 1220 | 705 | 4 | 1.38 (0.89-2.15) | 1.45 | 0.15 | 10.61 | 0.01 | 72 | 0.14 |
| Asian | 1062 | 645 | 3 | 1.35 (0.78-2.35) | 1.07 | 0.28 | 9.69 | 0.008 | 79 | 0.18 | |
| DR vs DM | 1056 | 666 | 4 | 1.29 (0.89-1.87) | 1.35 | 0.18 | 7.21 | 0.07 | 58 | 0.08 | |
| TT vs. TG | Overall | 1220 | 605 | 4 | 1.41 (0.88-2.25) | 1.43 | 0.15 | 10.92 | 0.01 | 73 | 0.16 |
| Asian | 1062 | 550 | 3 | 1.39 (0.76-2.53) | 1.08 | 0.28 | 10.13 | 0.006 | 80 | 0.22 | |
| DR vs DM | 1056 | 569 | 4 | 1.31 (0.88-1.95) | 1.33 | 0.18 | 7.54 | 0.06 | 60 | 0.09 | |
| TT+TG vs. GG | Overall | 1845 | 100 | 4 | 0.99 (0.64-1.54) | 0.03 | 0.97 | 0.33 | 0.95 | 0 | 0.00 |
| Asian | 1632 | 95 | 3 | 0.97 (0.62-1.52) | 0.14 | 0.88 | 0.11 | 0.95 | 0 | 0.00 | |
| DR vs DM | 1645 | 97 | 4 | 0.99 (0.63-1.53) | 0.06 | 0.95 | 0.30 | 0.96 | 0 | 0.00 | |
| TT+GG vs. TG | Overall | 1320 | 605 | 4 | 1.40 (0.88-2.21) | 1.42 | 0.16 | 10.91 | 0.01 | 72 | 0.15 |
| Asian | 1157 | 550 | 3 | 1.38 (0.77-2.49) | 1.08 | 0.28 | 10.14 | 0.006 | 80 | 0.21 | |
| DR vs DM | 1153 | 569 | 4 | 1.30 (0.88-1.92) | 1.31 | 0.19 | 7.51 | 0.06 | 60 | 0.09 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





