Submitted:
20 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Development of a “Non-Fasting” Gene Signature
Genomic Data Collection
Statistical Analysis
3. Results
3.1. The Non-Fasting Genomic Signature is More Prevalent in Primary Ovarian Tumors Compared with Normal Tissue
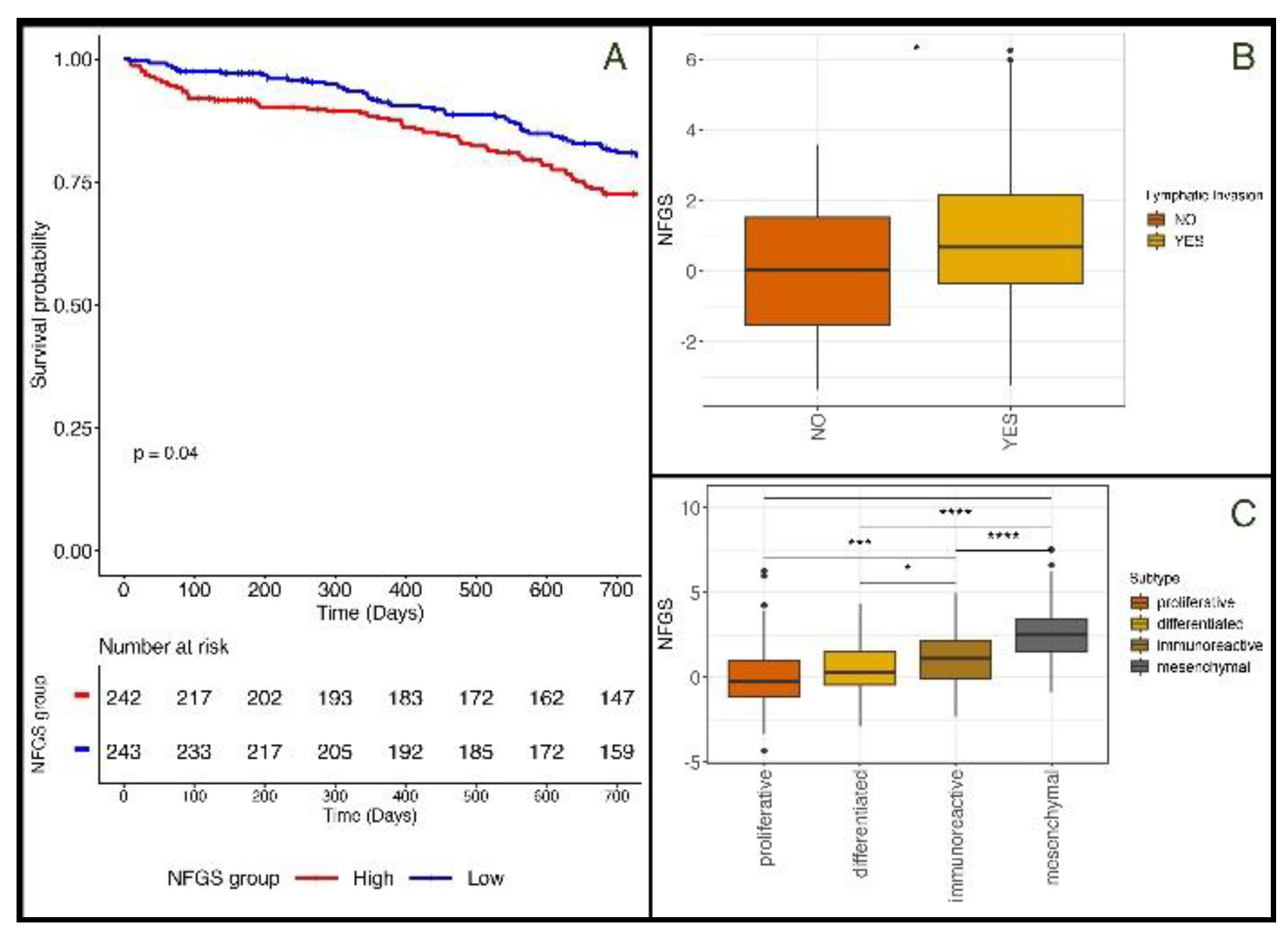
3.2. The Non-Fasting Genomic Signature is Associated with Different Cliniopathological Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Webb, P.M.; Leader, G.; Jordan, S.J.; Head, T.; Causes, C. Best Practice & Research Clinical Obstetrics and Gynaecology Epidemiology of Epithelial Ovarian Cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef] [PubMed]
- AA, W.; K, B.; DK, A.; MA, B.; WA, C.; RL, C.; DS, D.; JJ, K.; LA, M.; KN, M.; et al. Neoadjuvant Chemotherapy for Newly Diagnosed, Advanced Ovarian Cancer: Society of Gynecologic Oncology and American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 3460–3473. [Google Scholar] [CrossRef]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020. JNCCN J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef] [PubMed]
- Bois, A. Du; Reuss, A.; Pujade-Lauraine, E.; Harter, P.; Ray-Coquard, I.; Pfisterer, J. Role of Surgical Outcome as Prognostic Factor in Advanced Epithelial Ovarian Cancer: A Combined Exploratory Analysis of 3 Prospectively Randomized Phase 3 Multicenter Trials: By the Arbeitsgemeinschaft Gynaekologische Onkologie Studiengruppe Ovarialkarzin. Cancer 2009, 115, 1234–1244. [Google Scholar] [CrossRef] [PubMed]
- Cornelison, R.; Llaneza, D.C.; Landen, C.N. Emerging Therapeutics to Overcome Chemoresistance in Epithelial Ovarian Cancer: A Mini-Review. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Han, Y.; Kim, S.I.; Kim, H.S.; Kim, S.J.; Song, Y.S. Tumor Evolution and Chemoresistance in Ovarian Cancer. npj Precis. Oncol. 2018 21 2018, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wadapurkar, R.M.; Sivaram, A.; Vyas, R. RNA-Seq Analysis of Clinical Samples from TCGA Reveal Molecular Signatures for Ovarian Cancer. https://doi-org.wolfson.idm.oclc.org/10.1080/07357907.2023.2182123. 2023. [CrossRef]
- Zhang, K.; Erkan, E.P.; Jamalzadeh, S.; Dai, J.; Andersson, N.; Kaipio, K.; Lamminen, T.; Mansuri, N.; Huhtinen, K.; Carpén, O.; et al. Longitudinal Single-Cell RNA-Seq Analysis Reveals Stress-Promoted Chemoresistance in Metastatic Ovarian Cancer. Sci. Adv. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Nencioni, A.; Caffa, I.; Cortellino, S.; Longo, V.D. Fasting and Cancer: Molecular Mechanisms and Clinical Application. Nat. Rev. Cancer 2018, 18, 707–719. [Google Scholar] [CrossRef]
- Mercken, E.M.; Crosby, S.D.; Lamming, D.W.; Jebailey, L.; Krzysik-Walker, S.; Villareal, D.T.; Capri, M.; Franceschi, C.; Zhang, Y.; Becker, K.; et al. Calorie Restriction in Humans Inhibits the PI3K/AKT Pathway and Induces a Younger Transcription Profile. Aging Cell 2013, 12, 645–651. [Google Scholar] [CrossRef]
- Wahl, D.; LaRocca, T.J. Transcriptomic Effects of Healthspan-Promoting Dietary Interventions: Current Evidence and Future Directions. Front. Nutr. 2021, 8, 1–10. [Google Scholar] [CrossRef]
- Komatsu, T.; Park, S.; Hayashi, H.; Mori, R.; Yamaza, H.; Shimokawa, I. Mechanisms of Calorie Restriction: A Review of Genes Required for the Life-Extending and Tumor-Inhibiting Effects of Calorie Restriction. Nutrients 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.N.; Saccon, T.D.; Pradiee, J.; Rincón, J.A.A.; Andrade, K.R.S.; Rovani, M.T.; Mondadori, R.G.; Cruz, L.A.X.; Barros, C.C.; Masternak, M.M.; et al. Effect of Caloric Restriction and Rapamycin on Ovarian Aging in Mice. GeroScience 2019, 41, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Grymula, K.; Piotrowska, K.; Słuczanowska-Gła̧bowska, S.; Mierzejewska, K.; Tarnowski, M.; Tkacz, M.; Poniewierska-Baran, A.; Pȩdziwiatr, D.; Suszyńska, E.; Laszczyńska, M.; et al. Positive Effects of Prolonged Caloric Restriction on the Population of Very Small Embryonic-like Stem Cells - Hematopoietic and Ovarian Implications. J. Ovarian Res. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Zhang, X.M.; Wang, N.; Zhou, X.L.; Fu, Y.C.; Luo, L.L. Calorie Restriction Inhibits Ovarian Follicle Development and Follicle Loss through Activating SIRT1 Signaling in Mice. Eur. J. Med. Res. 2015, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Shen, X.; Liu, H.; Lu, S.; Peng, J.; Kuang, H. Caloric Restriction in Female Reproduction: Is It Beneficial or Detrimental? Reprod. Biol. Endocrinol. 2021, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; Salmon-Divon, M.; Salgado, R.; Dvash, E.; Pondé, N.; Zahavi, T.; Salmon, A.; Loibl, S.; Denkert, C.; Joensuu, H.; et al. Reactive Stroma and Trastuzumab Resistance in HER2-Positive Early Breast Cancer. Int. J. cancer 2020, 147, 266–276. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and Interpreting Cancer Genomics Data via the Xena Platform. Nat. Biotechnol. 2020 386 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Hadley Wickham Ggplot2: Elegant Graphics for Data Analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 216AD, 174, 245–246.
- Imbroisi Filho, R.; Ochioni, A.C.; Esteves, A.M.; Leandro, J.G.B.; Demaria, T.M.; Sola-Penna, M.; Zancan, P. Western Diet Leads to Aging-Related Tumorigenesis via Activation of the Inflammatory, UPR, and EMT Pathways. Cell Death Dis. 2021 127 2021, 12, 1–9. [Google Scholar] [CrossRef]
- Gray, A.; Dang, B.N.; Moore, T.B.; Clemens, R.; Pressman, P. A Review of Nutrition and Dietary Interventions in Oncology. SAGE Open Med. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Mercier, B.D.; Tizpa, E.; Philip, E.J.; Feng, Q.; Huang, Z.; Thomas, R.M.; Pal, S.K.; Dorff, T.B.; Li, Y.R. Dietary Interventions in Cancer Treatment and Response: A Comprehensive Review. Cancers (Basel). 2022, 14, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Safdie, F.; Brandhorst, S.; Wei, M.; Wang, W.; Lee, C.; Hwang, S.; Conti, P.S.; Chen, T.C.; Longo, V.D. Fasting Enhances the Response of Glioma to Chemo- and Radiotherapy. PLoS One 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Naji, H.S. The Role of Calorie Restriction in Cancer Prevention, Cancer Treatment, Longevity, and in Reducing Cellular Stress. Eur. J. Med. Heal. Sci. 2023, 5, 1–5. [Google Scholar] [CrossRef]
- Pomatto-Watson, L.C.D.; Bodogai, M.; Bosompra, O.; Kato, J.; Wong, S.; Carpenter, M.; Duregon, E.; Chowdhury, D.; Krishna, P.; Ng, S.; et al. Daily Caloric Restriction Limits Tumor Growth More Effectively than Caloric Cycling Regardless of Dietary Composition. Nat. Commun. 2021 121 2021, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Meynet, O.; Ricci, J.E. Caloric Restriction and Cancer: Molecular Mechanisms and Clinical Implications. Trends Mol. Med. 2014, 20, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Vernieri, C.; Fucà, G.; Ligorio, F.; Huber, V.; Vingiani, A.; Iannelli, F.; Raimondi, A.; Rinchai, D.; Frigè, G.; Belfiore, A.; et al. Fasting-Mimicking Diet Is Safe and Reshapes Metabolism and Antitumor Immunity in Patients with Cancer. Cancer Discov. 2022, 12, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Wada-Hiraike, O.; Yano, T.; Shirane, A.; Hirano, M.; Hiraike, H.; Koyama, S.; Oishi, H.; Yoshino, O.; Miyamoto, Y.; et al. Resveratrol Promotes Expression of SIRT1 and StAR in Rat Ovarian Granulosa Cells: An Implicative Role of SIRT1 in the Ovary. Reprod. Biol. Endocrinol. 2012, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Iside, C.; Scafuro, M.; Nebbioso, A.; Altucci, L. SIRT1 Activation by Natural Phytochemicals: An Overview. Front. Pharmacol. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Clifton, K.K.; Ma, C.X.; Fontana, L.; Peterson, L.L. Intermittent Fasting in the Prevention and Treatment of Cancer. CA. Cancer J. Clin. 2021, 71, 527–546. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.M.; Al-Foheidi, M.H.; Al-Mansour, M.M. Energy and Caloric Restriction, and Fasting and Cancer: A Narrative Review. Support. Care Cancer 2021, 29, 2299–2304. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, X.; Wang, H.; Wang, F.; Guan, W. Roles of Caloric Restriction, Ketogenic Diet and Intermittent Fasting during Initiation, Progression and Metastasis of Cancer in Animal Models: A Systematic Review and Meta-Analysis. PLoS One 2014, 9, 1–17. [Google Scholar] [CrossRef]
- Murakami, R.; Matsumura, N.; Mandai, M.; Yoshihara, K.; Tanabe, H.; Nakai, H.; Yamanoi, K.; Abiko, K.; Yoshioka, Y.; Hamanishi, J.; et al. Establishment of a Novel Histopathological Classification of High-Grade Serous Ovarian Carcinoma Correlated with Prognostically Distinct Gene Expression Subtypes. Am. J. Pathol. 2016, 186, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.; Price, Z.K.; Lokman, N.A.; Wang, W.; Goonetilleke, L.; Kadife, E.; Oehler, M.K.; Ricciardelli, C.; Kannourakis, G.; Ahmed, N. Platinum-Resistance in Epithelial Ovarian Cancer: An Interplay of Epithelial–Mesenchymal Transition Interlinked with Reprogrammed Metabolism. J. Transl. Med. 2022, 20, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Fagotti, A.; Cintoni, M.; Raoul, P.; Scaletta, G.; Quagliozzi, L.; Miggiano, G.A.D.; Scambia, G.; Gasbarrini, A.; Mele, M.C. Nutritional Interventions to Improve Clinical Outcomes in Ovarian Cancer: A Systematic Review of Randomized Controlled Trials. Nutrients 2019, 11. [Google Scholar] [CrossRef] [PubMed]
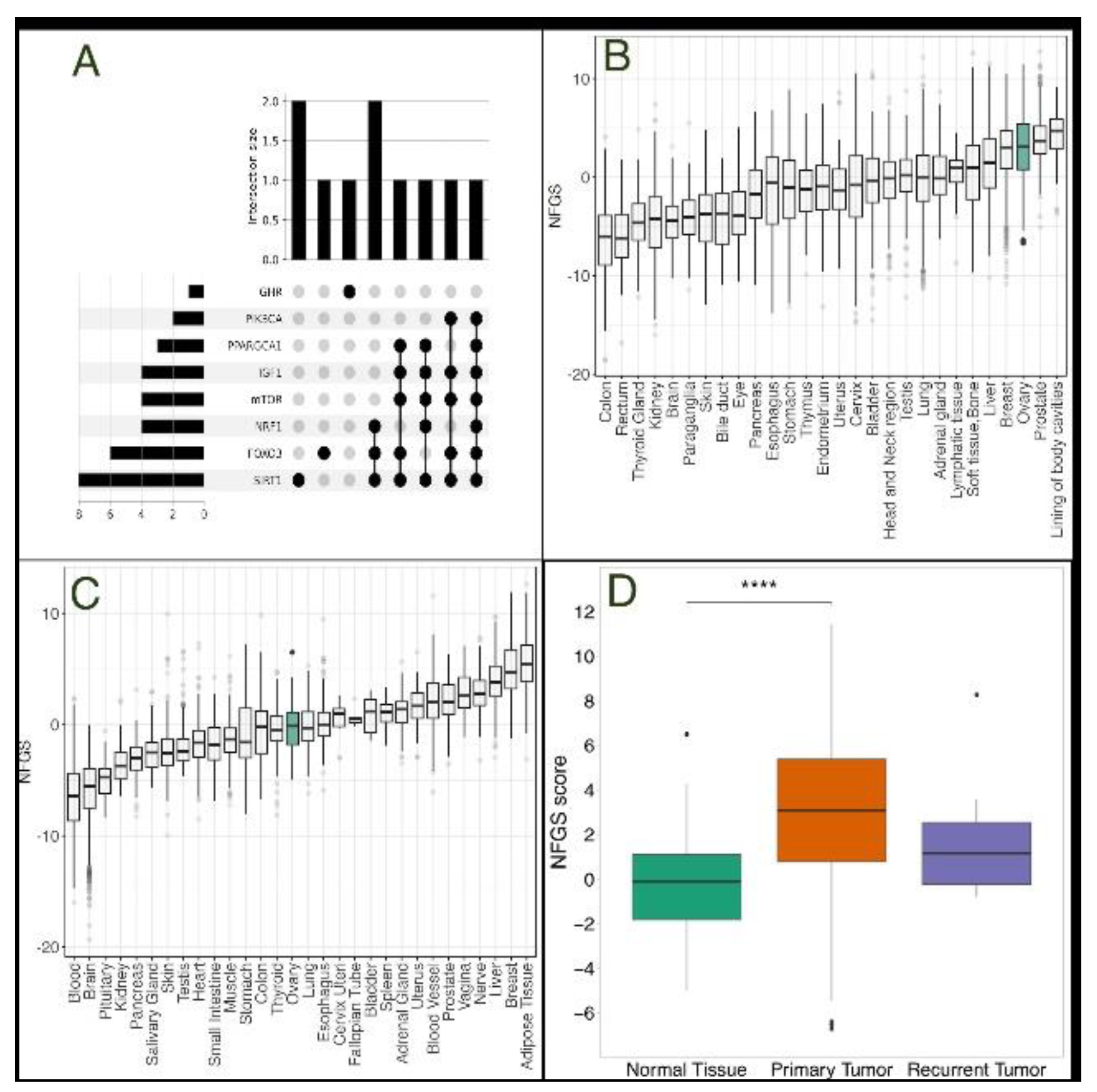
| Protein | Description | Up/Down-Regulated under a “Non-Fasting” State |
|---|---|---|
| SIRT1 | NAD-dependent deacetylase that plays a key role in multiple biological processes, including cellular senescence, apoptosis, sugar metabolism, inflammation, and fatty liver diseases | downregulated |
| FOXO3 | Transcription factor that regulates genes involved in stress resistance, metabolism, cell cycle arrest, and autophagy | downregulated |
| NRF1 | Transcription factor that maintainins cellular homeostasis, embryonic development, and mitochondrial homeostasis; directly regulates PGC1α | downregulated |
| PPARGC1A | Transcriptional coactivator which has been linked to the pathogenesis of type 2 diabetes and is involved in the antioxidant response upon mild redox and metabolic imbalance | downregulated |
| IGF-1 | A growth hormone that mediates the anabolic and linear growth promoting effects of growth hormone, while independently affecting tissue growth and development, proliferation and lipid metabolism | upregulated |
| GHR | Mediates the effects of growth hormone (GH) by activating the JAK-STAT signaling pathway, leading to the regulation of various physiological processes such as growth, metabolism, and immune function | upregulated |
| mTOR | Protein kinase that plays a crucial role in regulating cell growth, survival, metabolism, and immunity, acting as a master regulator of a cell’s growth and metabolic state in response to nutrients and growth factors | upregulated |
| PIK3CA | Encodes for a subunit of an enzyme called phosphatidylinositol 3-kinase (PI3K), which plays a role in mediating cell survival, differentiation, and proliferation, and has been linked to the development of cancer | upregulated |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





