1. Introduction
Colorectal cancer (CRC) is the third leading cause of cancer-related death and accounts for 10% of cancer diagnoses worldwide [
1]. Over the last decades, personalized decision-making has been empowered by the increasing use of comprehensive genomic profiling which has allowed the detection of novel actionable targets. As known, EGFR inhibition with specific monoclonal antibodies has become the standard of care in
RAS wild type patients, providing significant benefits in terms of survival when administered in combination with chemotherapy[
2,
3]. More recently, specific
RAS mutations such as
KRAS G12C have risen interest as possible actionable targets, following the evaluation of sotorasib or adagrasib in combination with anti-EGFR.[
4,
5]. The actionability of other mutations such as
BRAF V600E has also provided encouraging results, leading to the approval of the BRAF inhibitor encorafenib in combination with anti-EGFR cetuximab[
6]. Larger aberrations such as
HER-2 amplification have also been investigated in several studies, evaluating the role of different combination strategies and providing interesting results[
7,
8,
9]. Genomic profiling has thus widened the application of targeted therapy in digestive oncology[
10,
11] and also empowered the use of immune checkpoint inhibitors (ICI) whose efficacy is restricted to patients harboring microsatellite instability (MSI-H) and
POLE-D1 mutations[
12,
13]. Despite the encouraging results obtained in terms of survival and disease control, targeted agents employment is burdened by primary and secondary resistance [
11,
12]. For this reason, their use remains restricted to a limited proportion of patients.
The advent of high throughput techniques such as DNA and RNA sequencing has allowed the global and unbiased evaluation of the genomic landscape of tumors, enabling the identification of hotspot mutations, copy number variants and fusion transcripts.
In this study, we analyzed a real-world cohort of mCRC patients, treated in our institution. All patients underwent NGS-multigene profiling for diagnostic and therapeutic purposes. Genomic and clinical data were collected in order to provide novel insights about epidemiological prognostic features.
2. Results
2.1. Patient Characteristics
Patients with a histologically confirmed diagnosis of colorectal adenocarcinoma and an NGS assay performed in our institution from June 2019 until December 2020, were included. A total of 179 patients were considered for the analysis.
Patient characteristics are summarized in
Table 1. Male gender was the most represented (n= 105, 59%) and the median age at diagnosis was 68 years (IQ-IIIQ: 58.3-77.4, min-max: 30-89 years). Eastern Cooperative Oncology Group Performance status (ECOG PS) at metastatic disease diagnosis was recorded in 160 patients (89.3%), with 140 patients (78.2%) showing ECOG PS 0 or 1, and 20 patients (11.1%) with ECOG PS ≥ 2. Synchronous metastases were observed in 63.1% of cases (n=113), whereas metachronous lesions were identified in 36.9% of cases (n=66). A single metastatic site was identified in 115 patients (64.3%), whereas two or more metastatic sites were observed in 64 patients (35.7%). Surgical resection of metastases was performed in 28% of cases, with hepatic resections being the most common (30%), followed by peritoneal (20%) and pulmonary metastasectomies (18%). In total, 149 patients (83.2%) underwent first-line chemotherapy, with 51.9% of patients (n=93) receiving at least two lines of treatment afterwards. 44.1% of patients (n = 66) received doublet or triplet chemotherapy combined with bevacizumab, and 24.9% (37 patients) received doublet or triplet chemotherapy with an EGFR inhibitor (cetuximab or panitumumab). Chemotherapy alone was administered to 20.8% (31 patients) of cases, and 10% (15 patients) received the combination of 5-FU and bevacizumab or EGFR inhibitor. Eight patients with NGS-detected actionable alterations were administered targeted therapies. Among these patients, six harbored
BRAF V600E mutation and received a combination of encorafenib and cetuximab, while two patients with
PI3K alterations were treated with alpelisib, a PI3K inhibitor.
2.2. Molecular Alterations
No mutations were detected in 39 patients (21.8%) whereas 140 patients reported at least one mutation (78.2%),
Figure 1.
Table 2 shows the distribution of molecular alterations observed in the study population. The most frequent mutations were those in the
KRAS, PIK3CA, and
BRAF V600E genes. Some mutations were observed only in 3 patients such as those in the
CTNNB1,
MYC, and
MAP2K1 genes. Other mutations where even more rare such as those in
ERBB3,
RAF1,
MTOR,
JAK1, and
FGFR2 genes detected each in only two patients or those in the in
CDK4,
MET,
FGFR3,
GNA11,
EGFR,
ALK,
ROS1,
DDR2, and
KIT genes found in only one patient each. It is worth noting that no amplifications or fusions were identified and hotspot mutations were the only type of detected aberration (Single-Nucleotide Variants SNVs, and insertions-deletions INDELs). No mutations were observed for 28 genes of the Oncomine Focus panel, not shown.
2.3. Association between Mutated Genes and Survival (PFS, OS)
Overall, the median follow-up time was 33 months (95% CI 28.45 – not reached - NR), with a median PFS of 10.3 months (95% CI 8.8 - 12.3) and a median OS of 32.7 months (95% CI 24.8 - 39.2). The study investigated the association between the most frequent mutations, that is those occurring in at least 5% of patients, and PFS during first-line chemotherapy. Patients with a
KRAS mutation exhibited a median PFS of 10.4 months (95% CI 8.84 – 12.25), compared to 10.1 months (95% CI 7.29 – 13.70) in
KRAS wild type patients (p=0.737; HR 0.95, 95% CI 0.69 – 1.29). Patients harboring
PIK3CA mutation showed a median PFS of 10.7 months (95% CI 7.75 – 13.70), versus 10.0 months (95% CI 8.57 – 12.32) in
PIK3CA wild type cases (p=0.942; HR 1.01, 95% CI 0.70 – 1.47).
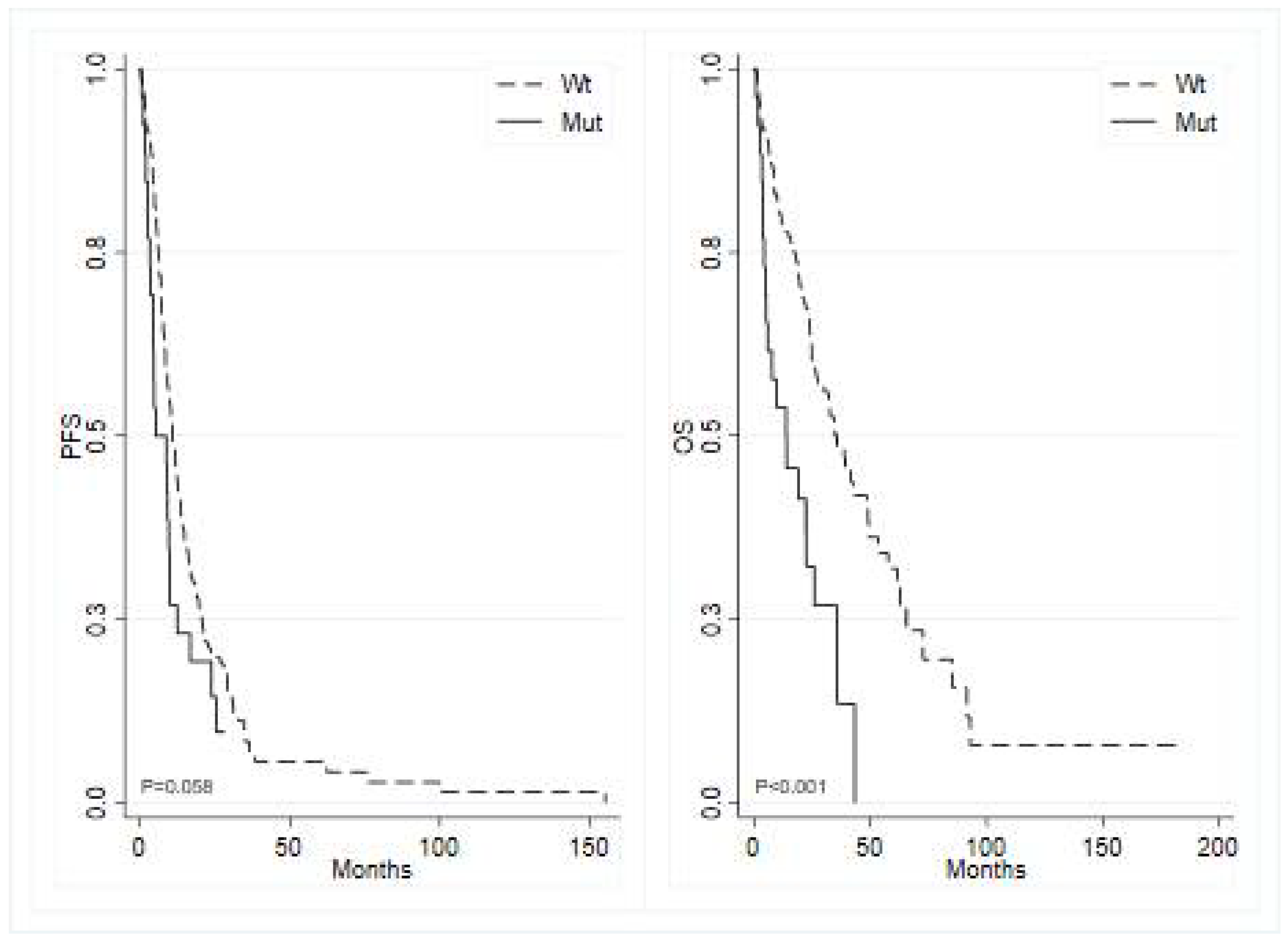
BRAF mutated tumors showed a median PFS of 5.2 months (95% CI 3.52 – 9.86) compared to 10.7 months (95% CI 9.00 – 12.94) in
BRAF wild type (p=0.060; HR 1.54, 95% CI 0.98 – 2.40) (
Figure 2, left panel), and
APC mutated tumors demonstrated a median PFS of 9.0 months (95% CI 1.81 – 12.32) versus 10.3 months (95% CI 8.84 – 12.52) in
APC wild type (p=0.330; HR 1.33, 95% CI 0.75 – 2.35).
The study also investigated the potential association between OS and the most frequently mutated genes. Analogous analyses for OS showed that patients with mutated
KRAS exhibited a median survival of 34.9 months (95% CI 24.77 – 65.60) compared to 26.9 months (95% CI 22.37 – 39.19) in wild-type
KRAS (p=0.084; HR 0.71, 95% CI 0.48 – 1.05). Similarly, mutated
PIK3CA displayed a median OS of 35.8 months (95% CI 23.85 - NR) versus 30.2 months (95% CI 24.01 – 39.19) in wild-type
PIK3CA (p=0.270; HR 0.76, 95% CI 0.47 – 1.24). In contrast, mutated
BRAF showed a median OS of 13.9 months (95% CI 4.89 – 26.05) versus 35.8 months (95% CI 26.91 – 48.59) in wild-type
BRAF (p<0.001; HR 2.62, 95% CI 1.59 – 4.32) (
Figure 2, right panel). Additionally, mutated
APC demonstrated a median OS of 24.8 months ( 95% CI 2.3 - MR) versus 32.8 months (95% CI 24.8 – 41.8) in wild-type
APC (p=0.354; HR 1.36, 95% CI 0.71 – 2.62).
Among the less mutated genes, no differences were observed except for
CTNNB1 gene, however mutated in only three patients. Patients harboring a mutation in such gene showed a statistically significant shorter PFS and OS compared to wild type patients. In detail, patients with mutated
CTNNB1, had a median PFS of 1.8 months (95% CI 1.15 - NR) compared to 10.4 months (95% CI 9.0 – 12.32) in
CTNBB1 wild type patients (p=0.001; HR 6.85, 95% CI 2.13 – 22.03). Similarly, the median OS in patients with mutated
CTNNB1 was 2.1 months (95% CI 1.87 - NR) versus 32.8 months (95% CI 24.8 – 41.8) in
CTNBB1 wild type (p<0.001; HR 9.82, 95% CI 3.00 – 32.12),
Figure S1.
No significant differences in terms of OS and PFS were found when comparing patients with high KRAS or BRAF allele frequency, defined as Variant Allele Frequency (VAF) more than median value, versus low allele frequency (
Figure S2,
Figure S3).
No significant relationship was found between the number of mutated genes and survival, results not shown.
2.4. Association between RAS Mutation Allele Frequency and Survival
We also investigated the association between
RAS mutation (
KRAS and
NRAS) allele frequency and survival outcomes in patients treated with first-line chemotherapy (n= 148). Patients with a high
RAS allele frequency, defined as Variant Allele Frequency (VAF) > 20%, showed a trend for lower survival rates with a median OS of 30.95 months (95% CI 18.2-47.4) compared to 37.42 months (95% CI 26.97-NR) for patients with low VAF (p-=0.091; HR=2.13, 95% CI 0.89-5.09), as shown in
Figure S4.
3. Discussion
To the best of our knowledge, there are currently few real world evidence studies based on routinely collected data from standard clinical practice that investigate the impact of NGS on a large cohort of patients diagnosed with mCRC. One of the main objectives of this study was to assess the clinical value of genomic profiling using NGS techniques in a sample of treated patients referred to our oncology units.
No new genomic prognostic factors were identified with this NGS panel, and
BRAF V600E mutation is confirmed as a negative prognostic factor as previously reported in the literature [
14,
15,
16]. While no statistically significant correlation was identified between the number of mutated genes and survival, there appears to be a negative survival trend in patients with at least one mutation. A statistically significant decrease in both PFS and OS was noted among patients harboring a mutation in the CTNNB1 gene. Nonetheless, due to the limited number of patients exhibiting this mutation, the reliability of the data, despite its statistical significance, has to be carefully interpreted, and no conclusive inferences can be drawn concerning its prognostic role.
The choice of the first-line therapeutic strategy was not influenced by NGS results. Indeed, in daily practice, the choice of the first line treatment is based only on the mutational status of KRAS and BRAF as well as microsatellite instability. Therefore, NGS data were taken in consideration only at progression following the discontinuation of standard treatments, in order to allow enrollment in a clinical trial or the use of off-label or expanded-access therapies. Unfortunately, many of these heavily pretreated patients were unfit for subsequent lines of treatment. This may explain the low number of patients treated with molecular-targeted therapies.
Molecular-targeted therapy in mCRC is increasingly becoming a therapeutic option due to the availability of selective inhibitors. Therefore, centralizing NGS genomic profiling in high-volume reference laboratories and establishing teams of professionals capable of evaluating and interpreting complex molecular test results (Molecular Tumor Board) is increasingly necessary to identify molecular alterations that provide sensitivity to specific molecular-targeted therapies in subpopulations of otherwise difficult-to-treat patients.
This study has some limitations that are worth noting. First, molecular analysis was performed on the primary tumor or metastasis without taking into account, as per clinical practice, the spatial and temporal heterogeneity of colorectal cancer. One limitation of this study is also related to the employed NGS panel which was restricted to 52 genes. Broader panels with the ability to analyze hundreds of genes can indeed identify a greater number of druggable molecular alterations, define the tumor mutational burden, and detect increasingly informative mutations such as POLE/D1 mutations which are predictive of sensitivity to immunotherapies.
4. Materials and Methods
4.1. Patient Selection and Data Collection
Patients aged 18 years and older diagnosed with mCRC who were NGS-tested with the Oncomine Focus Assay between June 2019 and December 2020 at the Biosciences Laboratory of IRCCS Istituto Romagnolo per lo studio dei Tumori (IRST)were included. Patients with other concurrent neoplasms or localized colorectal carcinoma were excluded.
For each included patient, demographic and clinical data were collected. Additionally, molecular information such as the quantity and type of molecular alterations detected as well as the variations of their allelic frequency (VAF) were recorded.
All participants provided written informed consent, and the study was conducted in compliance with the Declaration of Helsinki under good clinical practice conditions and with approval from the Local Ethics Committee (Comitato Etico Area Vasta Romagna e IRST). Information was extracted from patients' electronic medical records in accordance with stringent privacy standards, and the data were anonymized and recorded in an Excel database.
4.2. Molecular Analyses
Formalin-fixed-paraffin-embedded (FFPE) tumor samples were used for molecular analysis. For each sample, tumoral areas were defined by the pathologists, macrodissected and collected in specific tubes. Nucleic acids were extracted using MagMAX FFPE DNA/RNA Ultra Kit (Applied Biosystems, Waltham, MA, USA) following the manufacturer’s protocol. DNA and RNA concentrations were determined by fluorometric quantitation using a Qubit 4.0 Fluorometer with Qubit DNA dsDNA HS Assay Kit and Qubit RNA HS Assay Kit (Thermo Fisher), as appropriate.
NGS analyses were performed by Oncomine™ Focus Assay panel (Thermo Fisher Scientific Waltham, MA, USA), an amplicon-based DNA/RNA NGS assay that covers 52 cancer-associated genes. The DNA panel could identify hotspot mutations in 35 genes and copy number variants in 19 genes. The RNA panel was able to detect fusion drivers in 23 genes.
DNA and RNA libraries preparation were performed automatically using the library preparer “Ion ChefTM System” (Thermo Fisher Scientific) following the manufacturer’s instructions, with 10 ng of input DNA and RNA per sample.
Prior to RNA library preparation, complementary DNA (cDNA) synthesis was carried out using SuperScript™ VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific). The libraries were loaded onto Ion Chef System (Thermo Fisher Scientific) for template preparation and finally sequenced on the Ion S5 Plus platform (Thermo Fisher Scientific) using the Ion 520 Chips (Thermo Fisher Scientific). Primary analysis was carried out using a Torrent Suite Server™ to perform initial quality control, including chip loading density, median read length and number of mapped reads. Afterwards, a second analysis was performed by Ion Reporter™ Software, hosting informatics tools for variants, filtering, and annotations. Only variants with Variant Allele Frequency (VAF) greater than or equal to 5% and coverage greater than 500X have been reported. In addition, reliable variants are also reported if they have a relevant therapeutic significance, based on the sample pathologies in which they have been detected.
4.3. Statistical Analysis
A dedicated database was created to retrospectively collect demographic, clinical, histological, and treatment data. Patients characteristics were summarized using median and first- and third quartiles for continuous variables, and frequencies and percentages for categorical variables. PFS was defined as the time in months from the date of metastatic tumor diagnosis to progression or death from any cause, whichever occurred first, during the course of first-line chemotherapy. For patients alive without progression, the date of the last follow-up was recorded. OS was determined as the time in months from the date of metastatic tumor diagnosis to death from any cause or the date of the last follow-up. The median follow-up time was computed using the reverse Kaplan-Meier method whereas the survival curves were estimated using the Kaplan-Meier method, and comparisons were made using the log-rank test.. Univariate Cox regression models were employed to estimate hazard ratios (HR) and their corresponding 95% confidence intervals (CIs). With regard to the analysis of the association between the mutational status together with the variant allelic frequency (VAF) and survival, the cut-off were based on the median value of the VAF distribtuion of a specific gene excluding the wild type patients. Results were considered statistically significant if the two-sided p-values were <0.05. Statistical analyses were conducted using STATA 15.0 (College Station, TX, USA).
5. Conclusions
At present, molecular testing for MMR status as well as
KRAS,
NRAS, and
BRAF mutations is recommended for all mCRC patients at diagnosis, according to ESMO Guidelines[
17]. For what concerns CRC, all of the level 1 genomic alterations according to ESCAT (ESMO Scale for Clinical Actionability of molecular Targets), are represented by point mutations which can be easily identified by PCR. For this reason, ESMO recommendations for the use of NGS suggest genomic sequencing as a possible alternative to PCR only when it does not generate extra costs. Indeed, potentially actionable alterations are considered a rare event in CRC and the systematic use of NGS would not result cost-effective from a large-scale perspective. Nonetheless, unlike PCR, comprehensive genomic profiling increases multiplexing ability, analyzing multiple genes in a single test and providing information about different types of genomic aberrations. In this manner, NGS can provide vital information both for clinical and for research purposes by providing novel insights into patients’ prognosis and resistance mechanisms. Even the enrollment in clinical trials could be hopefully increased following a wider accessibility to NGS platform, providing patients an easier access to novel, promising drugs. Comprehensive genomic profiling is feasible and cost-effective, when performed in reference centers. Thus, a major effort is requested to local authorities in order to promote collaboration networks and facilitate information and tissue exchanges between peripheral and reference institutions.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: title; Table S1: title; Video S1: title.
Author Contributions
Conceptualization, L.M., A.P. and A.B.; writing—original draft preparation, L.M., F.G.S., A.P., A.B..; writing—review and editing, C.G.,L.E., M.M., I.G.R., D.C., E.P., C.R., L.C., E.C., P.U..; supervision, A.P., A.B.; All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly supported thanks to the contribution of Ricerca Corrente by the Italian Ministry of Health.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209-249. [CrossRef]
- Cervantes A, Adam R, Roselló S, Arnold D, Normanno N, Taïeb J, Seligmann J, De Baere T, Osterlund P, Yoshino T, Martinelli E; ESMO Guidelines Committee. Electronic address: clinicalguidelines@esmo.org. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023 Jan;34(1):10-32. [CrossRef]
- Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006 Apr 15;66(8):3992-5. [CrossRef]
- Fakih MG, Salvatore L, Esaki T, Modest DP, Lopez-Bravo DP, Taieb J, Karamouzis MV, Ruiz-Garcia E, Kim TW, Kuboki Y, Meriggi F, Cunningham D, Yeh KH, Chan E, Chao J, Saportas Y, Tran Q, Cremolini C, Pietrantonio F. Sotorasib plus Panitumumab in Refractory Colorectal Cancer with Mutated KRAS G12C. N Engl J Med. 2023 Dec 7;389(23):2125-2139. [CrossRef]
- Yaeger R, Weiss J, Pelster MS, Spira AI, Barve M, Ou SI, Leal TA, Bekaii-Saab TS, Paweletz CP, Heavey GA, Christensen JG, Velastegui K, Kheoh T, Der-Torossian H, Klempner SJ. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N Engl J Med. 2023 Jan 5;388(1):44-54. [CrossRef]
- Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, Wasan H, Ciardiello F, Loupakis F, Hong YS, Steeghs N, Guren TK, Arkenau HT, Garcia-Alfonso P, Pfeiffer P, Orlov S, Lonardi S, Elez E, Kim TW, Schellens JHM, Guo C, Krishnan A, Dekervel J, Morris V, Calvo Ferrandiz A, Tarpgaard LS, Braun M, Gollerkeri A, Keir C, Maharry K, Pickard M, Christy-Bittel J, Anderson L, Sandor V, Tabernero J. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med. 2019 Oct 24;381(17):1632-1643. [CrossRef]
- Strickler JH, Cercek A, Siena S, André T, Ng K, Van Cutsem E, Wu C, Paulson AS, Hubbard JM, Coveler AL, Fountzilas C, Kardosh A, Kasi PM, Lenz HJ, Ciombor KK, Elez E, Bajor DL, Cremolini C, Sanchez F, Stecher M, Feng W, Bekaii-Saab TS; MOUNTAINEER investigators. Tucatinib plus trastuzumab for chemotherapy-refractory, HER2-positive, RAS wild-type unresectable or metastatic colorectal cancer (MOUNTAINEER): a multicentre, open-label, phase 2 study. Lancet Oncol. 2023 May;24(5):496-508. [CrossRef]
- Siena S, Di Bartolomeo M, Raghav K, Masuishi T, Loupakis F, Kawakami H, Yamaguchi K, Nishina T, Fakih M, Elez E, Rodriguez J, Ciardiello F, Komatsu Y, Esaki T, Chung K, Wainberg Z, Sartore-Bianchi A, Saxena K, Yamamoto E, Bako E, Okuda Y, Shahidi J, Grothey A, Yoshino T; DESTINY-CRC01 investigators. Trastuzumab deruxtecan (DS-8201) in patients with HER2-expressing metastatic colorectal cancer (DESTINY-CRC01): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2021 Jun;22(6):779-789. [CrossRef]
- Yoshino T, Di Bartolomeo M, Raghav K, Masuishi T, Loupakis F, Kawakami H, Yamaguchi K, Nishina T, Wainberg Z, Elez E, Rodriguez J, Fakih M, Ciardiello F, Saxena K, Kobayashi K, Bako E, Okuda Y, Meinhardt G, Grothey A, Siena S; DESTINY-CRC01 investigators. Final results of DESTINY-CRC01 investigating trastuzumab deruxtecan in patients with HER2-expressing metastatic colorectal cancer. Nat Commun. 2023 Jun 7;14(1):3332. [CrossRef]
- Li F, Lin Y, Li R, Shen X, Xiang M, Xiong G, Zhang K, Xia T, Guo J, Miao Z, Liao Y, Zhang X, Xie L. Molecular targeted therapy for metastatic colorectal cancer: current and evolving approaches. Front Pharmacol. 2023 Oct 20;14:1165666. [CrossRef]
- Passardi A, Gibbons D. Editorial: Molecular targets for the treatment of metastatic colorectal cancer. Front Oncol. 2023 Dec 12;13:1341594. [CrossRef]
- Diaz LA Jr, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fourchardiere C, Rivera F, Elez E, Le DT, Yoshino T, Zhong WY, Fogelman D, Marinello P, Andre T; KEYNOTE-177 Investigators. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022 May;23(5):659-670. [CrossRef]
- Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ, van Krieken JH. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009 Jan 27;100(2):266-73. [CrossRef]
- Nakanishi R, Harada J, Tuul M, Zhao Y, Ando K, Saeki H, Oki E, Ohga T, Kitao H, Kakeji Y, Maehara Y. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013 Dec;18(6):1042-8. [CrossRef]
- Popovici V, Budinska E, Bosman FT, Tejpar S, Roth AD, Delorenzi M. Context-dependent interpretation of the prognostic value of BRAF and KRAS mutations in colorectal cancer. BMC Cancer. 2013 Sep 27;13:439. [CrossRef]
- Morris V, Overman MJ, Jiang ZQ, Garrett C, Agarwal S, Eng C, Kee B, Fogelman D, Dasari A, Wolff R, Maru D, Kopetz S. Progression-free survival remains poor over sequential lines of systemic therapy in patients with BRAF-mutated colorectal cancer. Clin Colorectal Cancer. 2014 Sep;13(3):164-71. [CrossRef]
- Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, Normanno N, Scarpa A, Robson M, Meric-Bernstam F, Wagle N, Stenzinger A, Bonastre J, Bayle A, Michiels S, Bièche I, Rouleau E, Jezdic S, Douillard JY, Reis-Filho JS, Dienstmann R, André F. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020 Nov;31(11):1491-1505. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).