Submitted:
19 April 2024
Posted:
22 April 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
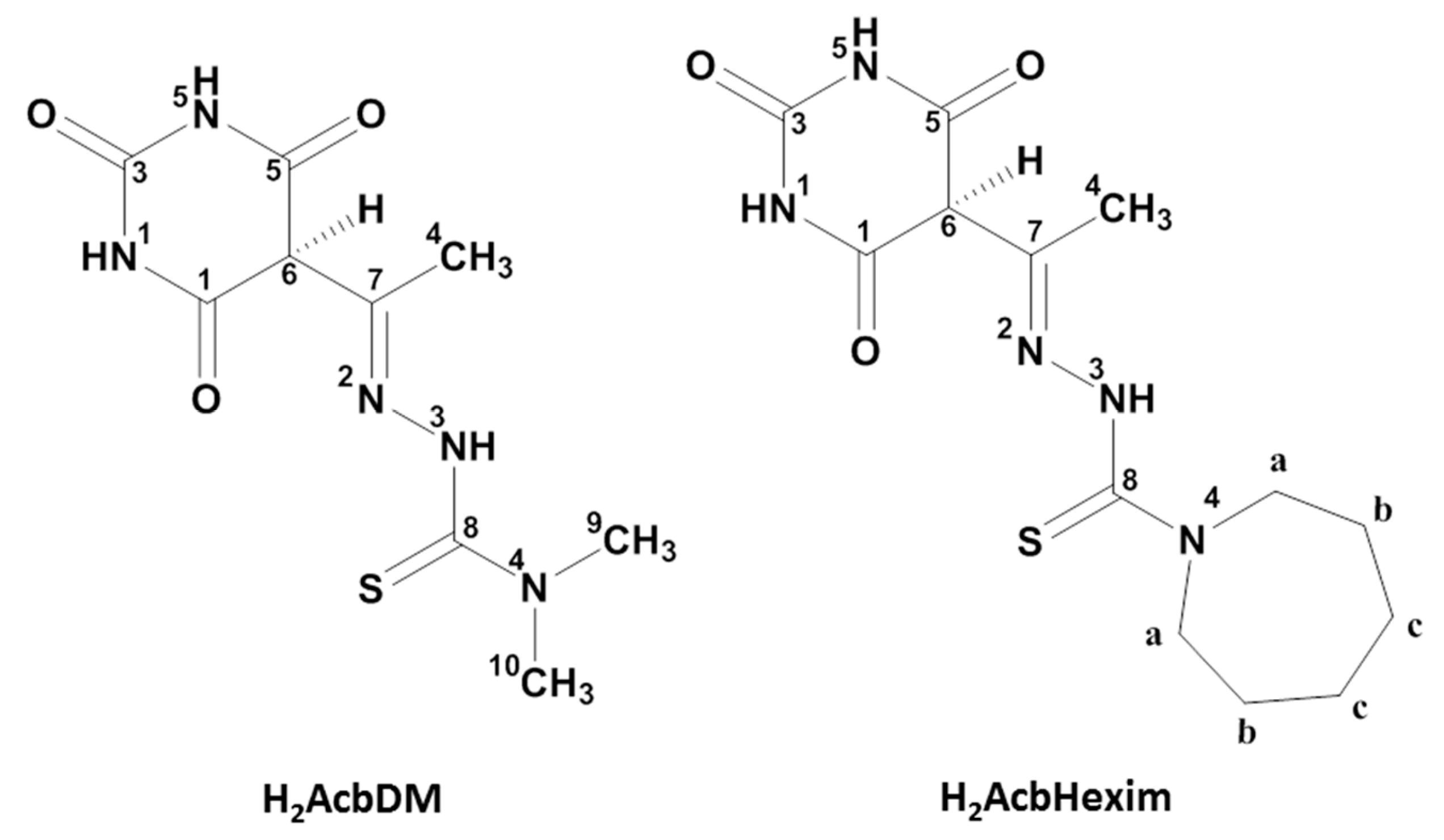
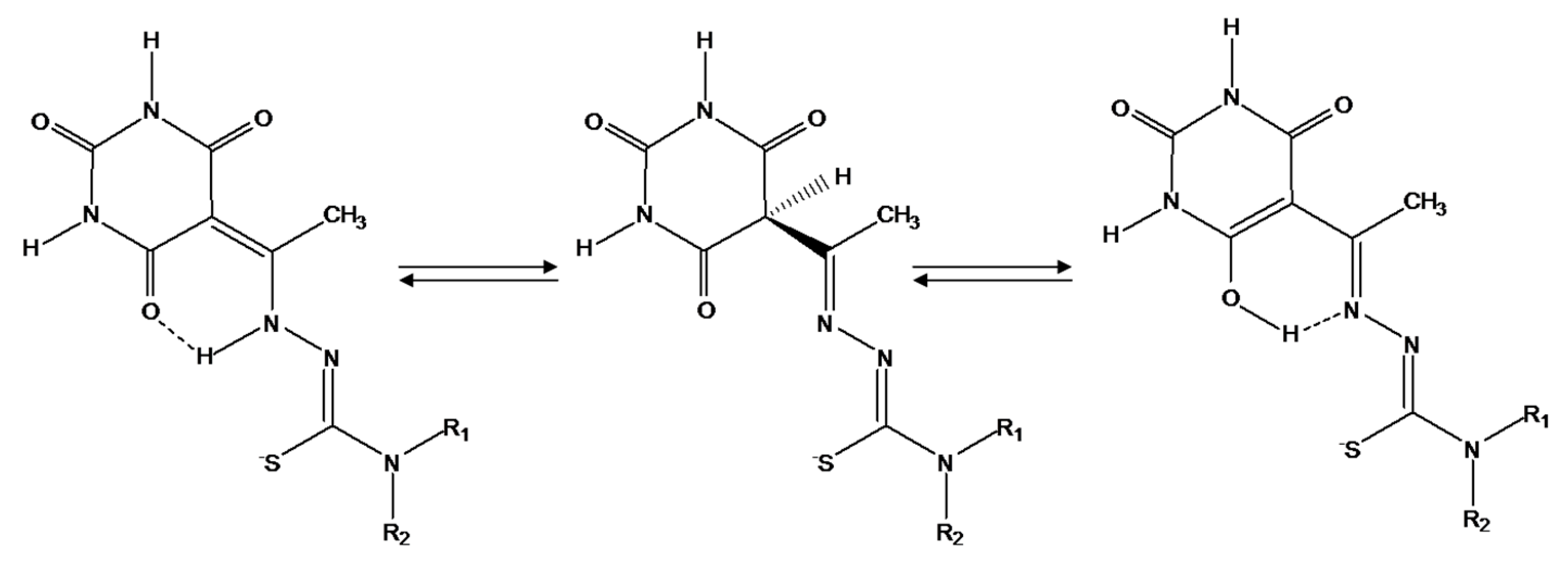
2.1. Synthesis and Characterization
2.2. FT-IR Spectra
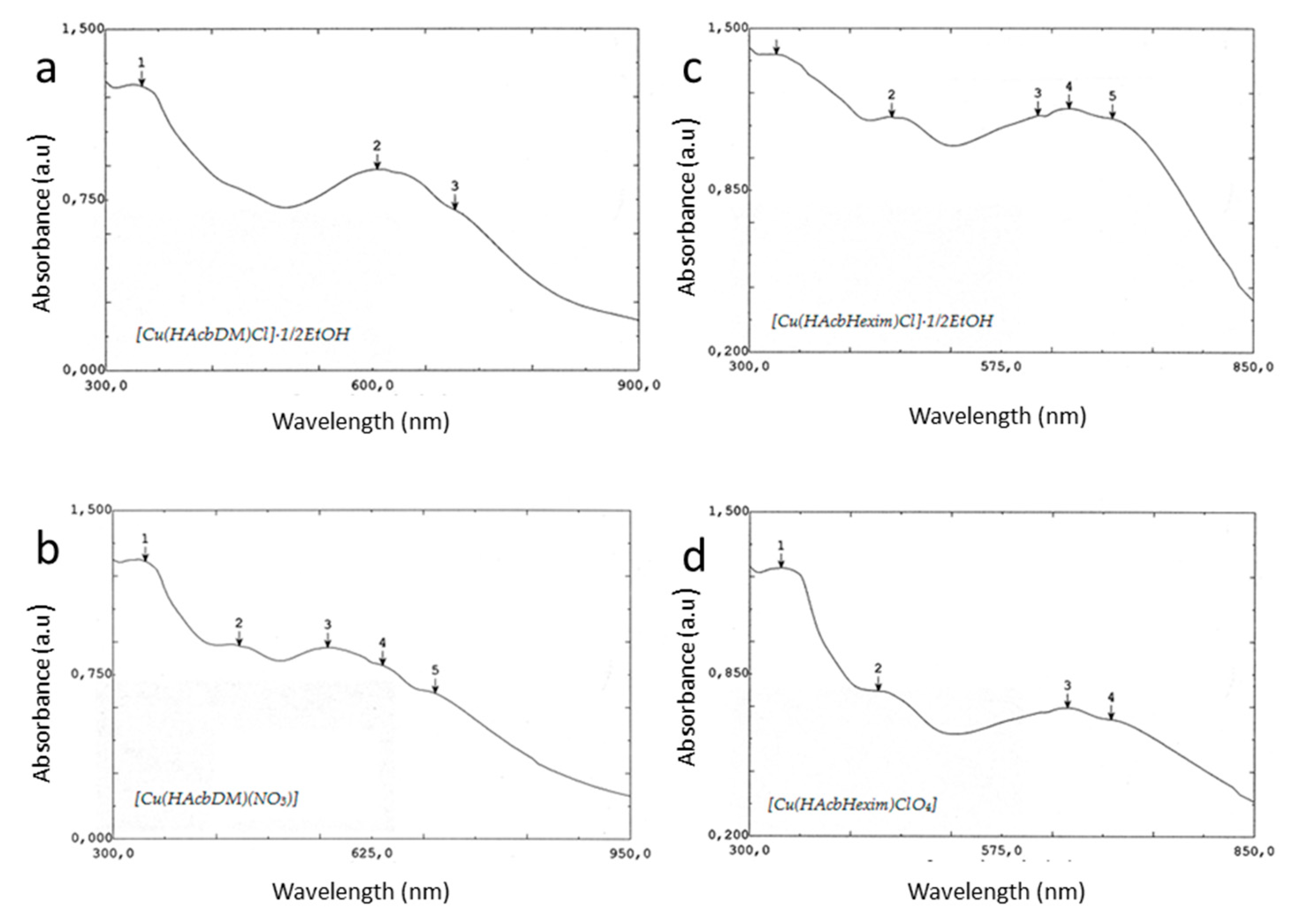
2.3. UV-Visible Spectra
2.4. Magnetic Susceptibilities
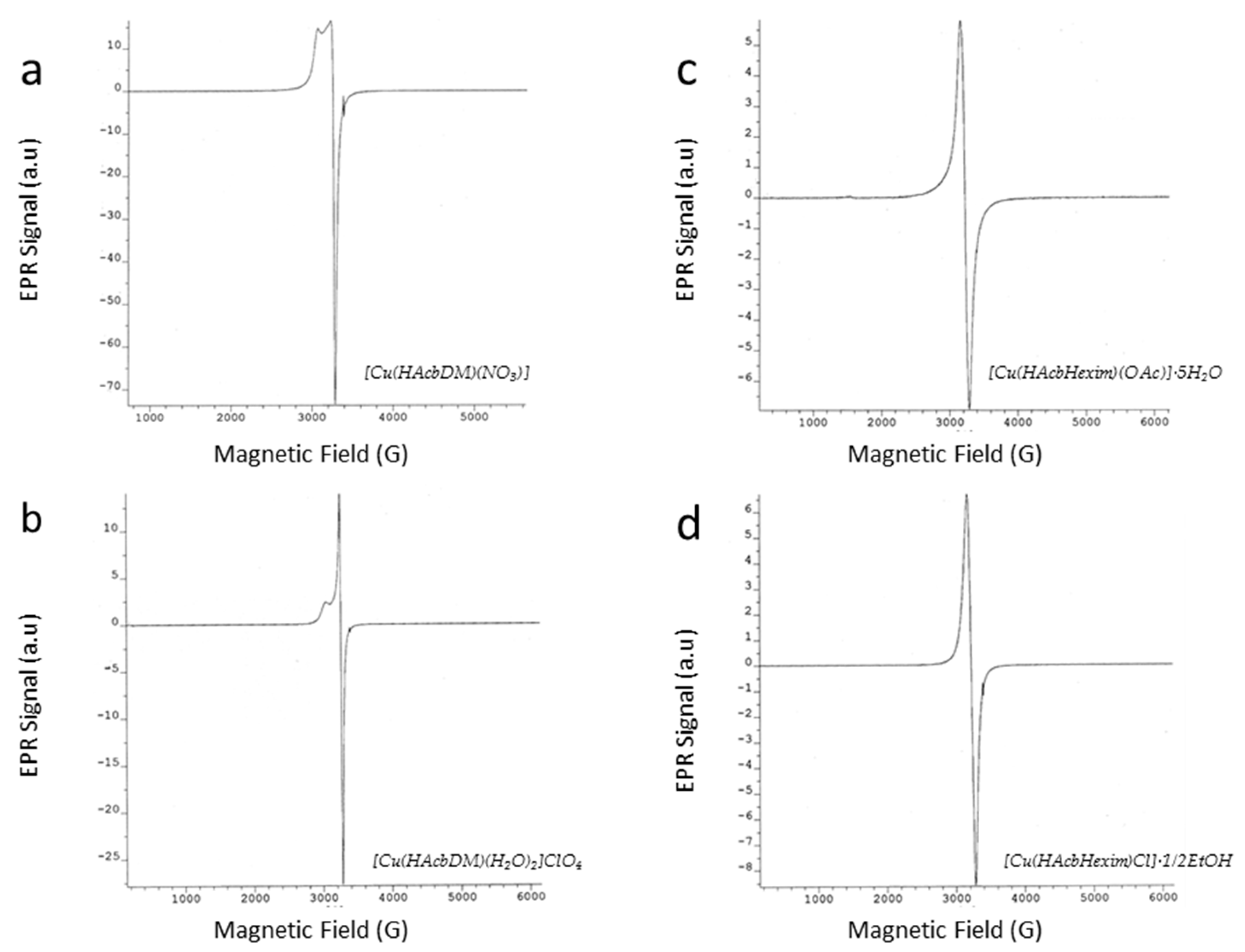
2.5. EPR analysis
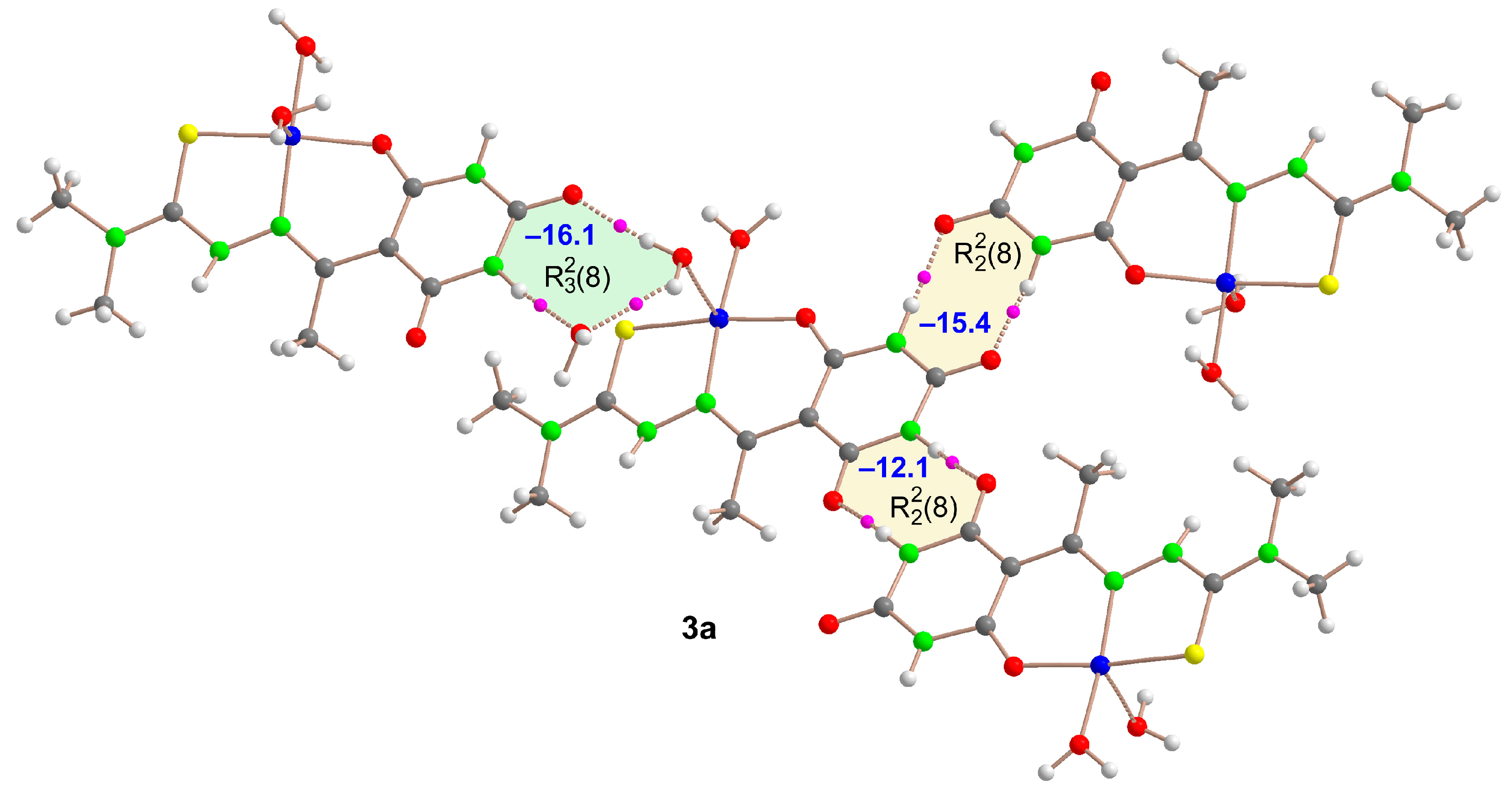
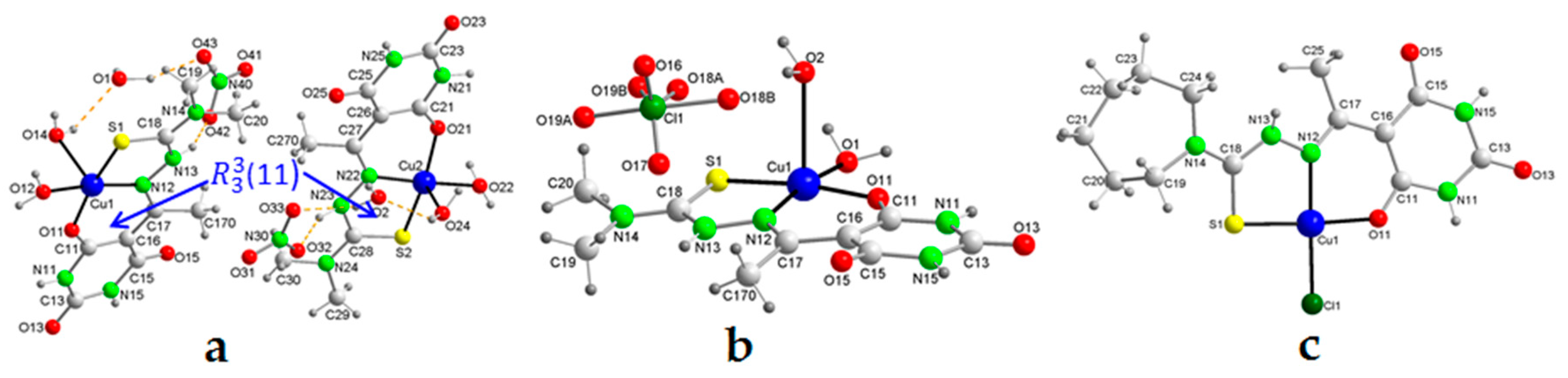
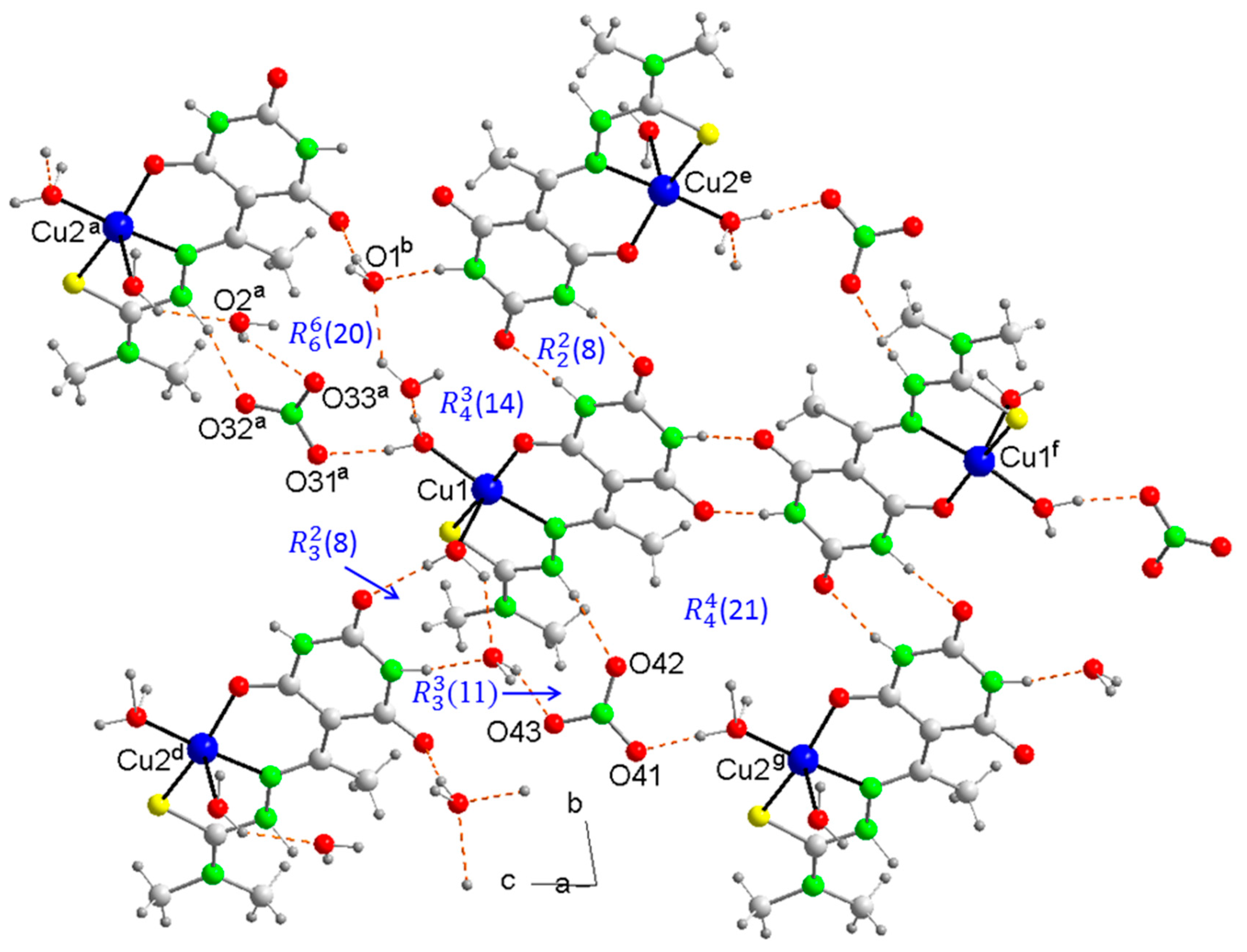
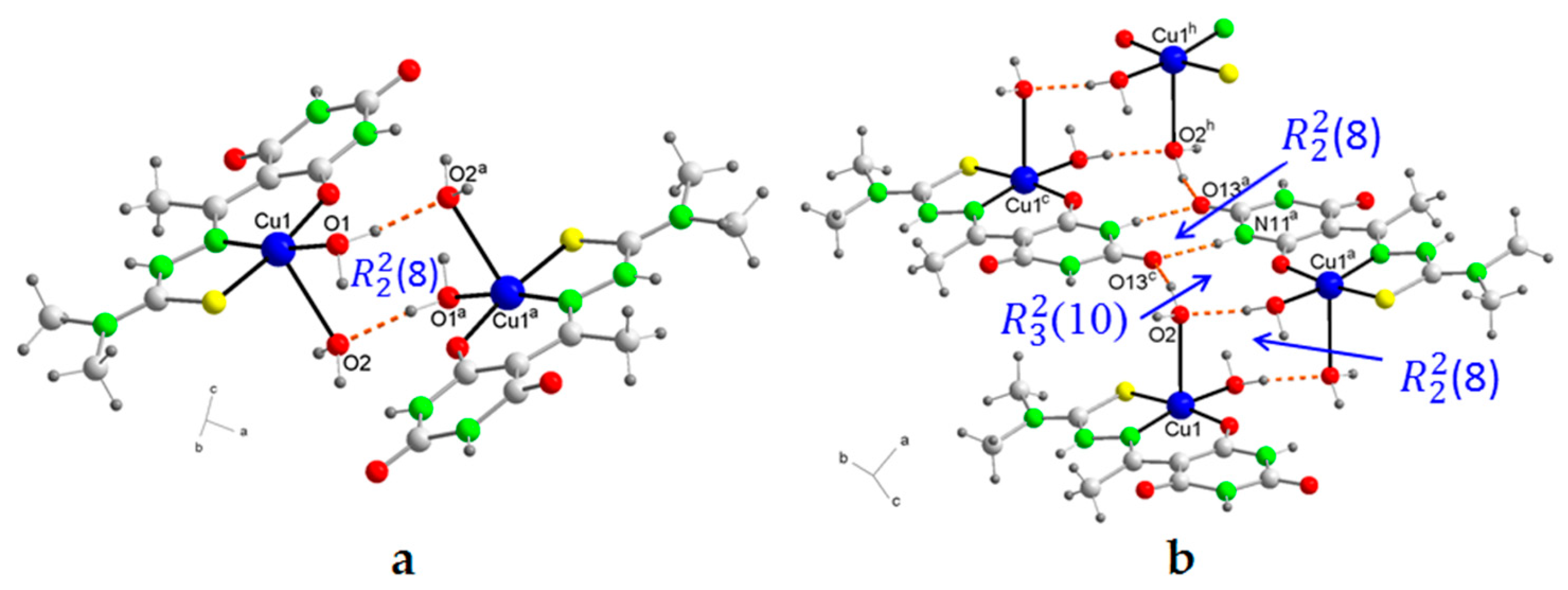
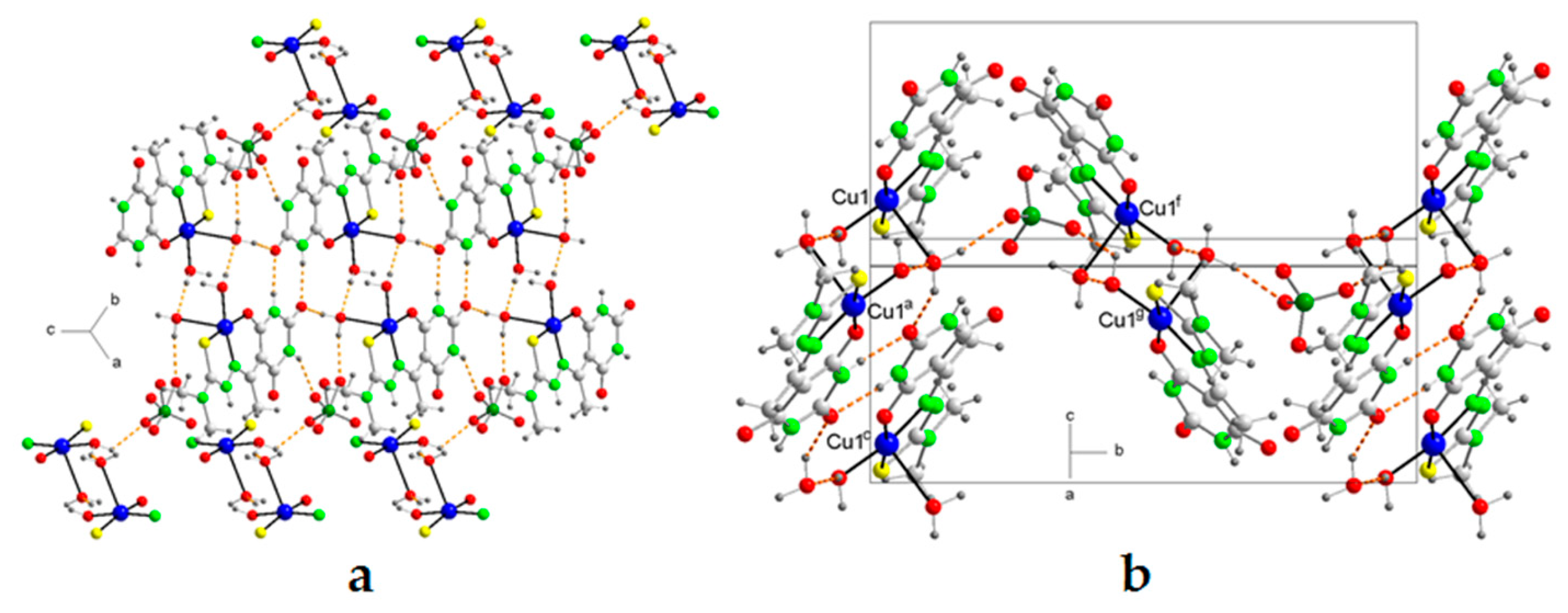
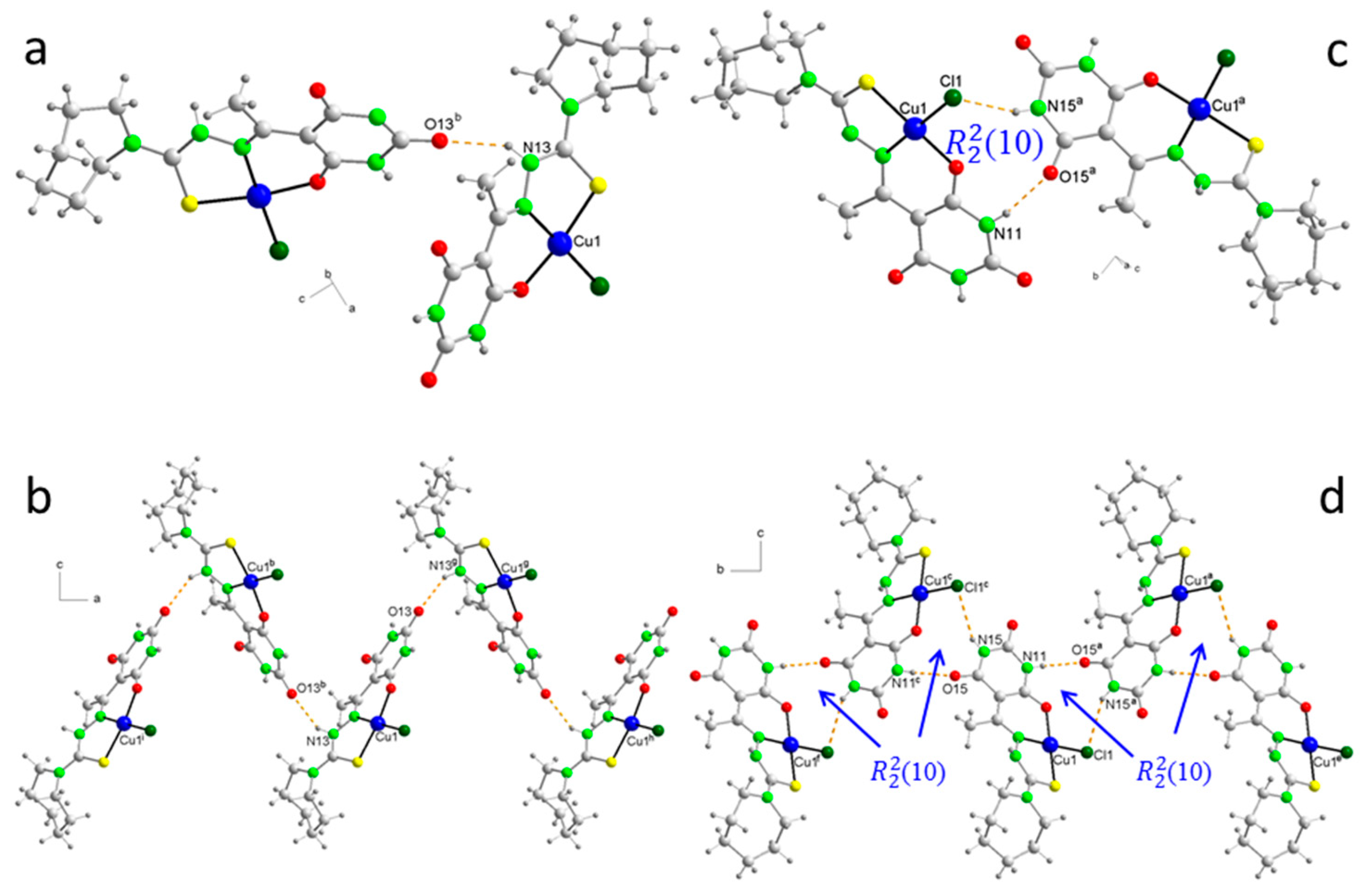
2.5. Molecular Structures and Supramolecular Analysis
2.6. Cyclic Voltammetry Studies
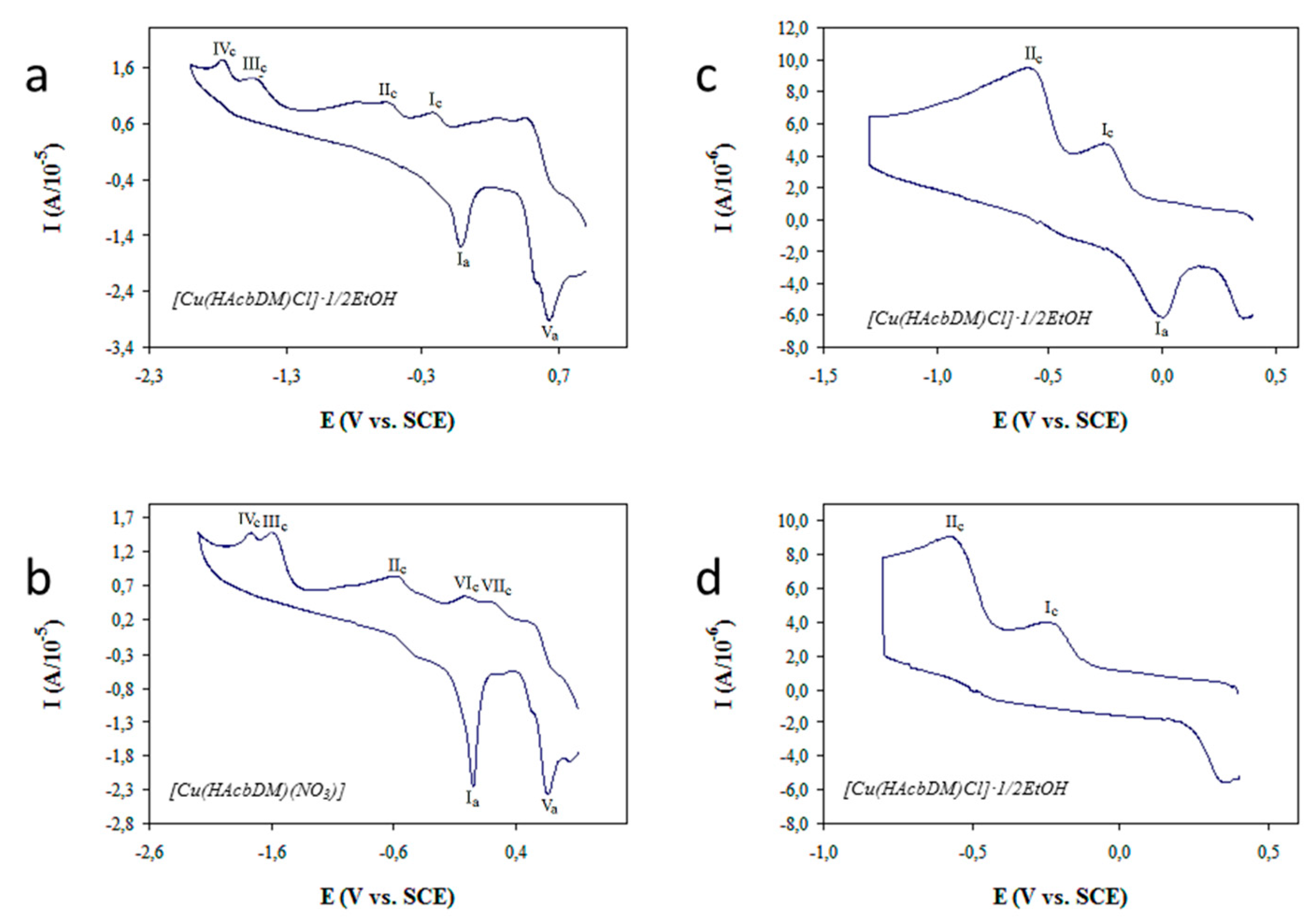
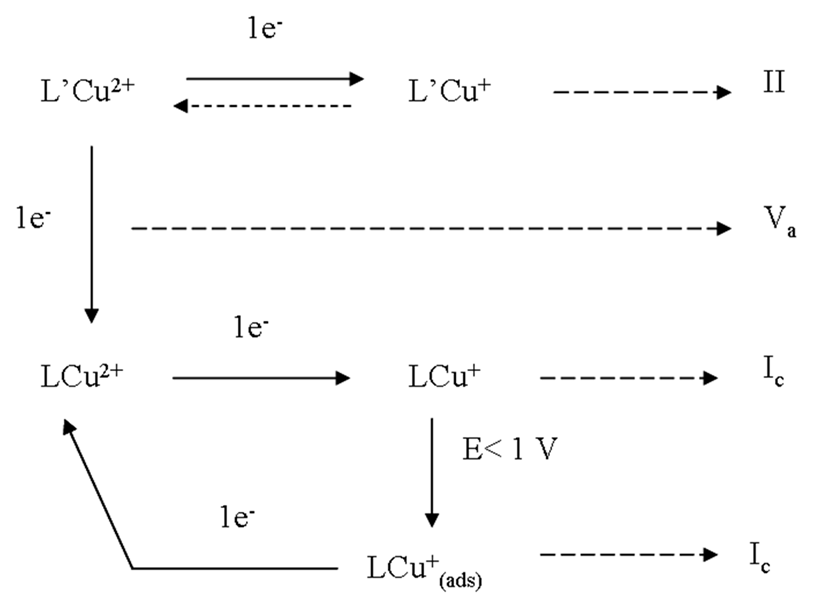
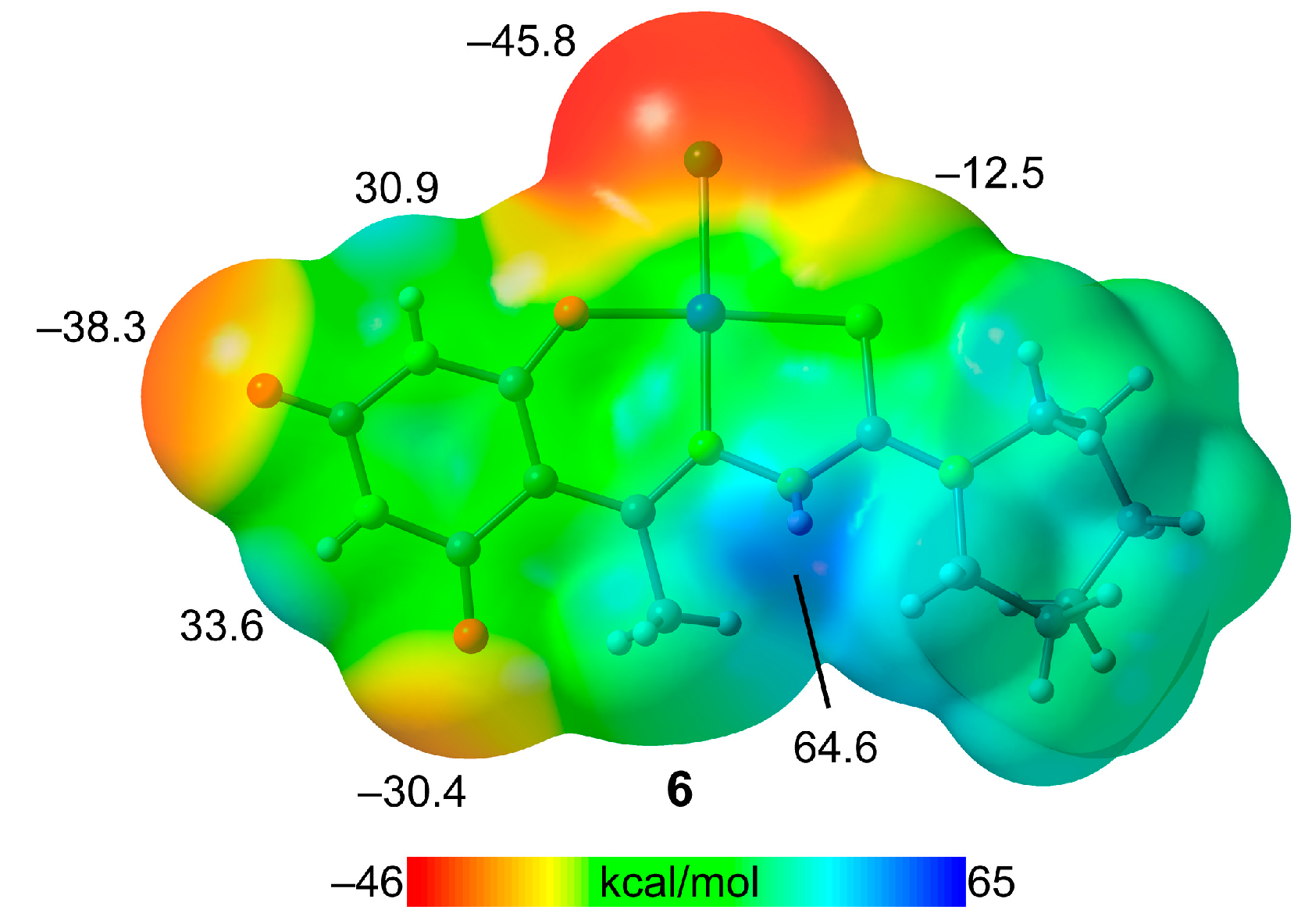
2.7. DFT Study

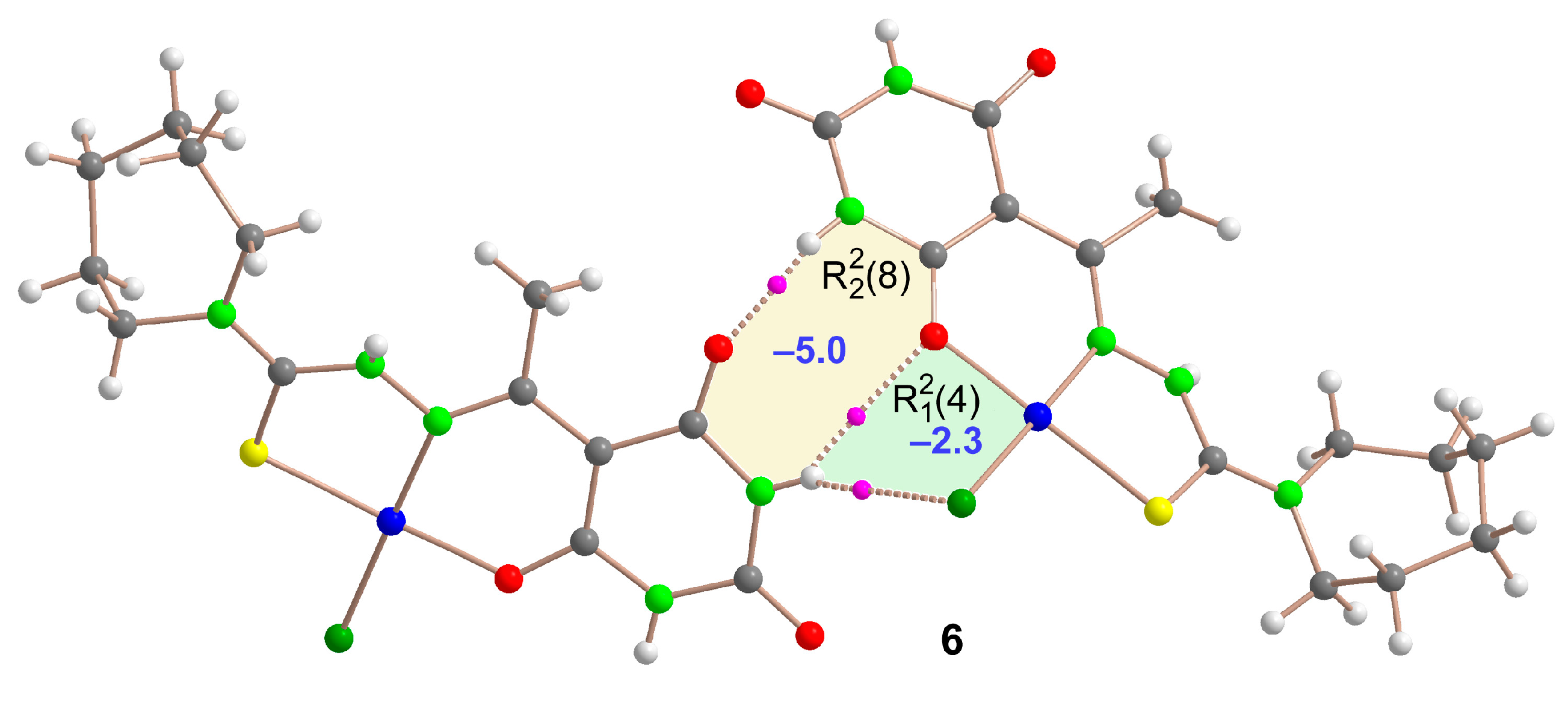
3. Materials and Methods
3.1. Physical Measurements
3.2. Synthesis of Thiosemicarbazone Ligands
3.3. Synthesis and Crystallization of Complexes
4.4. Single-Crystal X-ray Diffraction
4.5. Theoretical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Benesperi, I.; Singh, R.; Freitag, M. Copper Coordination Complexes for Enżrgy-Relevant Applications. Energies 2020, 13, 2198. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, V.K.; Mishra, A.; Varsha, *!!! REPLACE !!!*; Singh, A.A.; Prasad, G.; Singh, A.K. Recent advancements in coordination compounds and their potential clinical application in the management of diseases: An up-to-date review. Polyhedron 2023, 241, 116485. [Google Scholar] [CrossRef]
- Malinowski, J.; Zych, D.; Jacewicz, D.; Gawdzik, B.; Drzeżdżon, J. Application of Coordination Compounds with Transition Metal Ions in the Chemical Industry—A Review. Int. J. Mol. Sci. 2020, 21, 5443. [Google Scholar] [CrossRef]
- Chebout, O.; Bouchene, R.; Bouacida, S.; Boudraa, M.; Mazouz, W.; Merzougui, M.; Ouari, K.; Boudaren, C.; Merazig, H. Two copper(II) coordination complexes based on sulfanilamide ligand: Synthesis, structure, thermal analyzes, electrochemical properties and biological activities. J. Mol. Struct. 2022, 1247, 131346. [Google Scholar] [CrossRef]
- Borthakur, R.; Kumar, A.; De, A.K.; Lal, R.A. Synthesis, characterization and electrochemical properties of copper(II) complexes derived from succinoyldihydrazine Schiff base ligands. Arab. J. Chem. 2019, 12, 2192–2205. [Google Scholar] [CrossRef]
- Peng, Y.-D.; Huang, F.; Du, M.-X.; La, Y.-T.; Yu, J.; Hu, X.; Dong, W.-K. Rare formyl-coordinated homotrinuclear copper(II) salamo-based N2O3-donor complex: Experimental and theoretical studies. Inorg. Chim. Acta 2024, 560, 121815. [Google Scholar] [CrossRef]
- Gerhards, L.; Werr, M.; Hübner, O.; Solov’yov, I.A.; Himmel, H.-J. Peculiar Differences between Two Copper Complexes Containing Similar Redox-Active Ligands: Density Functional and Multiconfigurational Calculations. Inorg. Chem. 2024, 63, 961–975. [Google Scholar] [CrossRef]
- Yusuf, T.L.; Oladipo, S.D.; Zamisa, S.; Kumalo, H.M.; Lawal, I.A.; Lawal, M.M.; Mabuba, N. Design of New Schiff-Base Copper(II) Complexes: Synthesis, Crystal Structures, DFT Study, and Binding Potency toward Cytochrome P450 3A4. ACS Omega 2021, 6, 13704–13718. [Google Scholar] [CrossRef] [PubMed]
- Misigo, W.O.; Njenga, L.W.; Odhiambo, R.A.; Meyer, M.; Julius, L.; Sibuyi, N.; Lalancette, R.A.; Onani, M.O. New thiosemicarbazones and their palladium(II) complexes: Synthesis, spectroscopic characterization, X-ray structure and anticancer evaluation. Inorg. Chim. Acta 2023, 558, 121746. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, M.; Li, S.; Xu, G.; Zhang, Z.; Yang, F. Novel mono-, bi-, tri- and tetra-nuclear copper complexes that inhibit tumor growth through apoptosis and anti-angiogenesis. J. Inorg. Biochem. 2024, 250, 112403. [Google Scholar] [CrossRef]
- Normaya, E.; Ahmad, M.N.; Farinab, Y.; Bulat, K.H.K. Synthesis, Characterization and Preliminary Study on Acetylpyrazine N(4)Butylthiosemicarbazone as a Potential CDK2 Inhibitor Combined with DFT Calculations. J. Braz. Chem. Soc. 2018, 29, 2197–2206. [Google Scholar] [CrossRef]
- Bermejo, E.; Castiñeiras, A.; García, I.; West, D.X. Spectral and structural studies of mercury(II) complexes of 2-pyridineformamide N(4)-dimethylthiosemicarbazone. Polyhedron 2003, 22, 1147–1154. [Google Scholar] [CrossRef]
- Sripathi, M.P.; Berely, S.; Reddy, C.V.R. Computational Studies of 4-Formylpyridinethiosemicarbazone and Structural and Biological Studies of Its Ni(II) and Cu(II) Complexes. Heteroatom Chem. 5078. [Google Scholar] [CrossRef]
- Castiñeiras, A.; Fernández-Hermida, N.; García-Santos, I.; Gómez-Rodríguez, L.; Gil, D.M.; Frontera, A. Supramolecular, spectroscopic and computational analysis of weak interactions in some thiosemicarbazones derived from 5-acetylbarbituric acid. J. Mol. Struct. 2021, 1245, 131031. [Google Scholar] [CrossRef]
- Bermejo, E.; Castiñeiras, A.; García-Santos, I.; Gómez-Rodríguez, L.; Sevillano, P. Pt4S4 Clusters with Functionalized Thiosemicarbazones derived from 5-Acetylbarbituric Acid. Z. Anorg. Allg. Chem. 2007, 633, 2255–2261. [Google Scholar] [CrossRef]
- Castiñeiras, A.; Fernández-Hermida, N.; García-Santos, I.; Gómez-Rodríguez, L. Neutral NiII, PdII and PtII ONS-pincer complexes of 5-acetylbarbituric-4Ndimethylthiosemicarbazone: synthesis, characterization and properties. Dalton Trans. 2012, 41, 13486–13495. [Google Scholar] [CrossRef] [PubMed]
- Geary, W.J. The use of conductivity measurements in organic solvents for the characterisation of coordination compounds. Coord. Chem. Rev. 1971, 7, 81–122. [Google Scholar] [CrossRef]
- Nakamoto, K. Infrared and Raman Spectra of Inorganic and Coordination Compounds, 6th ed.; Wiley: Hoboken, NJ, 2009. [Google Scholar]
- Addison, C.C.; Logan, N.; Wallwork, C.S.; Garner, C.D. Structural aspects of co-ordinated nitrate groups. Q. Rev. Chem. Soc. 1971, 25, 289–322. [Google Scholar] [CrossRef]
- Deacon, G.B.; Phillips, R.J. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Bresolin, L.; Burrow, R.A.; Hörner, M.; Bermejo, E.; Castiñeiras, A. Synthesis and crystal structure of di[(μ-acetato) (2-acetylpyridine 4Nethylthiosemicarbazonato)zinc(II)]. Polyhedron 1997, 16, 3947–3951. [Google Scholar] [CrossRef]
- Weidlein, J.; Müller, U.; Deknicke, K. Schwingungsspektroskopie, G. Thieme Verlag, Stuttgart, 2nd edn, 1988.
- West, D.X.; Beraldo, H.; Nassar, A.A. Copper(II) complexes of 4-acetamidobenzaldehyde N(4)-substituted thiosemicarbazones. Transition Met. Chem. 1999, 24, 421–424. [Google Scholar] [CrossRef]
- Chandra, S.; Gupta, L.; Tyagi, V.P. Co(II), Ni(II), Cu(II) and Zn(II) complexes of Thiosemicarbazones. Synth. React. Inorg. Met.-Org. Chem. 1996, 26, 125–137. [Google Scholar] [CrossRef]
- Lobana, T.S.; Kaushal, M.; Virk, R.K.; Garcia-Santos, I.; Jasinski, J.P. Thiosemicarbazonates of copper: Crystal structures of [(furan-2-acetaldehyde-N-phenyl-thiosemicarbazonato)][bis(triphenylphosphine)]copper(I) and [bis(furan-2-formaldehyde-N-phenyl-thiosemicarbazonato)]copper(II). Polyhedron 2018, 152, 49–54. [Google Scholar] [CrossRef]
- Hathway, B.J.; Billing, D.E. The Electronic Properties and Stereochemistry of Mono-Nuclear Complexes of the Copper(II) Ion. Coord. Chem. Rev. 1970, 5, 143–207. [Google Scholar] [CrossRef]
- García-Tojal, J.; Lezama, L.; Pizarro, J.L.; Insausti, M.; Arriourtia, M.I.; Rojo, T. Spectroscopic and magnetic properties of copper(II) complexes derived from pyridine-2-carbaldehyde thiosemicarbazone. Structures of [Cu(NO3)(C7H8N4S)(H2O)](NO3) and [{Cu(NCS)(C7H7N4S)}2]. Polyhedron 1999, 18, 3703–3711. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, C.G. Synthesis, structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Yang, L.; Powell, D.R.; Houser, R.P. Structural variation in copper(I) complexes with pyridylmethylamide ligands: structural analysis with a new four-coordinate geometry index, τ4. Dalton Trans. 2007, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lewis, T.C.; Tocher, D.A.; Price, S.L. An Experimental and Theoretical Search for Polymorphs of Barbituric Acid: The Challenges of Even Limited Conformational Flexibility. Cryst. Growth. Des. 2004, 4, 979–987. [Google Scholar] [CrossRef]
- Malenov, D.P.; Zarić, S.D. Recognizing New Types of Stacking Interactions by Analyzing Data in the Cambridge Structural Database. Chemistry 2023, 5, 2513–2541. [Google Scholar] [CrossRef]
- Kumbhar, A.S.; Padhye, S.B.; West, D.X.; Liberta, A.E. Electrochemical studies of copper(II) 2-acetylpyridine4 N-dialkylthiosemicarbazones. Relation to their spectral, magnetic, and biological properties. Transition Met. Chem. 1991, 16, 276–279. [Google Scholar] [CrossRef]
- Eisner, V.; Eisner, E.K.; Bard, A.J.E.; Lund, H. Encyclopedia of Electrochemistry of Elements; Marcel Dekker: New York, 1979; Volume 13. [Google Scholar]
- Kitaev, Y.P.; Budnikov, G.K.; Skrebkova, I.M. Tantomerism and geometrical isomerism of nitrogen- containing derivatives of carbonyl compounds. VI. The polarographic investigation of semi- and thiosemicarbazones of some aliphatic and alicyclic ketones Izv. Akad. Nauk SSSR, Otd. Khim. Nauk 2 (1962) 244-252. CA 56, 15287e.
- Kitaev, Y.P.; Budnikov, G.K.; Arbuzov, A.E. Tautomerism and geometric isomerism and nitrogenous derivatives of carbonyl compounds. III. Polarographic study of some semi- and thiosemicarbazones in aqueous alcohol solutions. Izv. Akad. Nauk SSSR, Otd. Khim. Nauk 5 (1961) 824-31. CA 55, 440f. 55.
- Biltz, H.; Wittek, H. Über alkylierte und acylierte Barbitursäuren. Chem. Ber. 1991, 54, 1035–1058. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SADABS, Program for Empirical Absorption Correction of Area Detector Data; University of Göttingen: Germany, 2001. [Google Scholar]
- Sheldrick, G.M. SHELXT – Integrated space-group and crystal structure Determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Putz, H.; Brandenburg, K. Diamond - Crystal and Molecular Structure Visualization Version 4.6.2; Crystal Impact GbR: Bonn, Germany, 2020. [Google Scholar]
- Espinosa, E.; Molins, E.; Lecomte, C. Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem. Phys. Lett. 1998, 285, 170. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; Revision, A. Gaussian 16; Revision A.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A Consistent and Accurate ab Initio Parametrization of Density Functional Dispersion Correction (DFT-D) for the 94 Elements H-Pu. J. Chem. Phys. 2010, 132, 154104–154122. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158–6169. [Google Scholar] [CrossRef]
- Bader, R.F.W. A quantum theory of molecular structure and its applications. Chem. Rev. 1991, 91, 893928. [Google Scholar] [CrossRef]
- Keith, T.A. TK Gristmill Software, version 13.05.06; AIMAll: Overland Park, KS, USA, 2013. [Google Scholar]
- Mousavi, H.; García-Rubiño, M.E.; Choquesillo-Lazarte, D.; Castiñeiras, A.; Lezama, L.; Frontera, A.; Niclós-Gutiérrez, J. H(N3)dap (Hdap = 2,6-Diaminopurine) Recognition by Cu2(EGTA): Structure, Physical Properties, and Density Functional Theory Calculations of [Cu4(μ-EGTA)2(μ-H(N3)dap)2(H2O)2]·7H2O. Molecules 2023, 28, 6263. [Google Scholar] [CrossRef] [PubMed]
- Belmont-Sánchez, J.C.; Choquesillo-Lazarte, D.; García-Rubiño, M.E.; Matilla-Hernández, A.; Niclós-Gutiérrez, J.; Castiñeiras, A.; Frontera, A. Supramolecular Nature of Multicomponent Crystals Formed from 2,2′-Thiodiacetic Acid with 2,6-Diaminopurine or N9-(2-Hydroxyethyl)adenine. Int. J. Mol. Sci. 2023, 24, 17381. [Google Scholar] [CrossRef]
| Compound | 3a | 4 | 6 |
| Empirical formula | C9H18CuN6O9S | C9H16ClCuN5O9S | C13H18ClCuN5O3S |
| Formula weight | 449.89 | 469.32 | 423.37 |
| Temperature / K | 293(2) | 293(2) | 293(2) |
| Wavelength / Å | 1.54184 | 1.54184 | 1.54184 |
| Crystal system | Triclinic | Monoclinic | Orthorhombic |
| Space group | P | P21/n | Pbca |
| Unit cell dimensions | |||
| a / Å | 9.039(4) | 9.747 | 12.8807(7) |
| b / Å | 13.289(2) | 17.318 | 12.3524(5) |
| c / Å | 14.880(2) | 9.807 | 20.9062(8) |
| α / º | 81.526(8) | 90 | 90 |
| β / º | 78.678(17) | 96.26 | 90 |
| γ / º | 76.91(2) | 90 | 90 |
| Volume / Å–3 | 1697.2(8) | 1645.5 | 3326.3(3) |
| Z | 4 | 4 | 8 |
| Calc. density / Mg/m3 | 1.761 | 1.894 | 1.691 |
| Absorp. coefc. / mm–1 | 3.543 | 5.129 | 4.723 |
| F(000) | 924 | 956 | 1736 |
| Crystal size | 0.52 × 0.16 × 0.08 | 0.24 × 0.16 × 0.16 | 0.40 × 0.32 × 0.16 |
| θ range / º | 3.046 − 71.935 | 5.108 − 72.910 | 4.229 − 72.923 |
| Limiting indices / h,k,l | 0/11, −15/16, −17/18 | −12/12, 0/21, 0/12 | 0/15, 0/15, 0/25 |
| Refl. collect / unique | 7116 / 6674 [Rint = 0.1085] |
2893 / 2729 [Rint = 0.0377] |
3557 / 3319 [Rint = 0.0258] |
| Completeness 67.684 º | 100.0 | 81.4 | 100.0 |
| Absorp. correct. | Psi-scan | Psi-scan | Psi-scan |
| Max. /min. transm. | 1.000 / 0.340 | 1.000 / 0.753 | 1.000 / 0.488 |
| Data / parameters | 6674 / 469 | 2729 / 235 | 3319 / 227 |
| Goodness-of-fit on F2 | 1.028 | 1.190 | 1.050 |
| Final R indices |
R1 = 0.0580, wR2 = 0.1577 |
R1 = 0.1069, wR2 = 0.2123 |
R1 = 0.0392, wR2 = 0.1059 |
| R indices (all data) |
R1 = 0.1168, wR2 = 0.1871 |
R1 = 0.1370, wR2 = 0.2237 |
R1 = 0.0524, wR2 = 0.1123 |
| Largest dif. peak/hole e.Å−3 | 0.896/−0.780 | 0.635/−0.733 | 0.556/-0.699 |
| CCDC number |
| 3a | 4 | 6 | |
| Cu(1)-O(11) | 1.912(3) | 1.897(8) | 1.9114(19) |
| Cu(1)-N(12) | 1.961(4) | 1.929(9) | 1.959(2) |
| Cu(1)-O(12) /O(1) /Cl(1) | 1.964(3) | 1.963(8) | 2.2338(7) |
| Cu(1)-S(1) | 2.2487(14) | 2.247(3) | 2.2311(7) |
| Cu(1)-O(14) /O(2) | 2.441(4) | 2.609(10) | |
| Cu(2)-O(21) | 1.913(3) | ||
| Cu(2)-N(22) | 1.947(4) | ||
| Cu(2)-O(22) | 1.945(4) | ||
| Cu(2)-S(2) | 2.2470(14) | ||
| Cu(2)-O(24) | 2.477(4) | ||
| O(11)-Cu(1)-N(12) | 91.03(14) | 91.6(3) | 87.90(8) |
| O(11)-Cu(1)-O(12) /O(1) /Cl(1) | 85.98(14) | 86.1(3) | 91.19(6) |
| N(12)-Cu(1)-O(12) /O(1) /Cl(1) | 173.12(17) | 170.1(4) | 178.41(7) |
| O(11)-Cu(1)-S(1) | 168.82(12) | 171.4(3) | 172.67(7) |
| N(12)-Cu(1)-S(1) | 88.39(11) | 88.6(3) | 87.50(6) |
| O(12) /O(1) /Cl(1)-Cu(1)-S(1) | 93.34(10) | 92.3(3) | 93.28(3) |
| O(11)-Cu(1)-O(14) /O(2) | 92.73(14) | 95.0(3) | |
| N(12)-Cu(1)-O(14) /O(2) | 96.05(15) | 104.6(3) | |
| O(12) /O(1) /Cl(1)-Cu(1)-O(14) /O(2) | 90.28(14 | 85.2(4) | |
| S(1)-Cu(1)-O(14) /O(2) | 98.43(9) | 93.3(2) | |
| O(21)-Cu(2)-O(22) | 85.97(16) | ||
| O(21)-Cu(2)-N(22) | 91.38(15) | ||
| O(22)-Cu(2)-N(22) | 177.17(19) | ||
| O(21)-Cu(2)-S(2) | 162.62(13) | ||
| O(22)-Cu(2)-S(2) | 94.18(12) | ||
| N(22)-Cu(2)-S(2) | 88.04(12) | ||
| O(21)-Cu(2)-O(24) | 95.59(14) | ||
| O(22)-Cu(2)-O(24) | 93.39(19) | ||
| N(22)-Cu(2)-O(24) | 87.87(15) | ||
| S(2)-Cu(2)-O(24) | 101.74(10) |
| System | (Ep)c[a] | (Ep)a[a] | (Ep)a -(Ep)c[b] | [(Ep)a +(Ep)c]/2[b] | (Ep/2)c[a] | (Ep/2)c + (Ep)c[b] | Eº’[a] | |
|---|---|---|---|---|---|---|---|---|
| 1 | II | -0.708 | -0.371 | 0.337 | -0.539 | -0.518 | 0.190 | -0.679 |
| 2 | I | -0.234 | -0.170 | 0.064 | -0.205 | |||
| II | -0.582 | -0.494 | 0.088 | -0.553 | ||||
| 3 | I | 0.057 | ||||||
| II | -0.580 | -0.482 | 0.098 | -0.551 | ||||
| 6 | I | -0.250 | -0.184 | 0.066 | -0.221 | |||
| II | -0.604 | -0.516 | 0.088 | -0.575 | ||||
| 7 | I | 0.074 | ||||||
| II | -0.628 | -0.502 | 0.126 | -0.599 | ||||
| 8 | I | 0.085 | ||||||
| II | -0.604 | -0.506 | 0.098 | -0.576 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





