1. Introduction
Significant mutualistic relationships have been established between endophytic fungi and their host plants, attracting considerable attention due to their ecological and biotechnological potential [
1]. Currently, the structural types of secondary metabolites isolated from endophytic fungi fermentation products, including alkaloids, polyketides, terpenes, etc [
2,
3]. Some compounds exhibit biological activities such as antibacterial [
4], antiviral [
5], anti-tumor [
6], and some even possess activity in promoting seed germination [
7]. Endophytic fungi can both independently produce various structural types of secondary metabolites and participate in the biosynthesis or biotransformation of secondary metabolites in host plants, making them an important source of active natural products. Active natural products derived from endophytic fungi hold vast potential applications in biopharmaceuticals, agricultural production, and industrial fermentation [
2,
8,
9,
10].
Quinazolinones have great development prospects in medicinal chemistry [
11], originating from a wide range of their antibacterial [
12], anti-inflammatory [
13,
14], antiviral [
15,
16], and antituberculosis [
17] properties. As of now, only a limited number of quinazolinones have been reported from endophytic fungi, including neosartoryadins and glyantrypines, antiviral agents from mangrove-derived fungi
Neosartorya udagawae and
Cladosporium sp., respectively [
18,
19]; chaetominine, a cytotoxic agent from an endophytic fungus
Chaetomium sp. [
20]; aniquinazolines A–D, antibacterial and cytotoxic molecules from a mangrove-derived fungus
Aspergillus nidulans [
21]; and (–)-(1
R,4
R)-1,4-(2,3)-Indolmethane-1-methyl-2,4-dihydro-1
H-pyrazino-[2,1-
b]-quinazoline-3,6-dione, an antifungal agent from an endophytic fungus
Penicillium vinaceum [
22]. In a sense, mining novel bioactive quinazoline molecules from endophytic fungi holds great promise.
As a part of our ongoing studies on the bioactive secondary metabolites from
Rhodiola tibetica endophytic fungi, we have isolated more than 30 polyketides with antiviral or anti-inflammatory activities [
23,
24,
25]. Global Natural Products Social (GNPS) molecular networking has been widely applied in the analysis of natural products to cluster compounds with similar MS/MS spectra, expediting the dereplication process of known natural products [
26,
27,
28]. We performed a GNPS molecular networking analysis of the EtOAc extract of endophytic fungus
Penicillium sp. HJT-A-6, leading to the discovery of a new quinazolinone alkaloid, peniquinazolinone A (
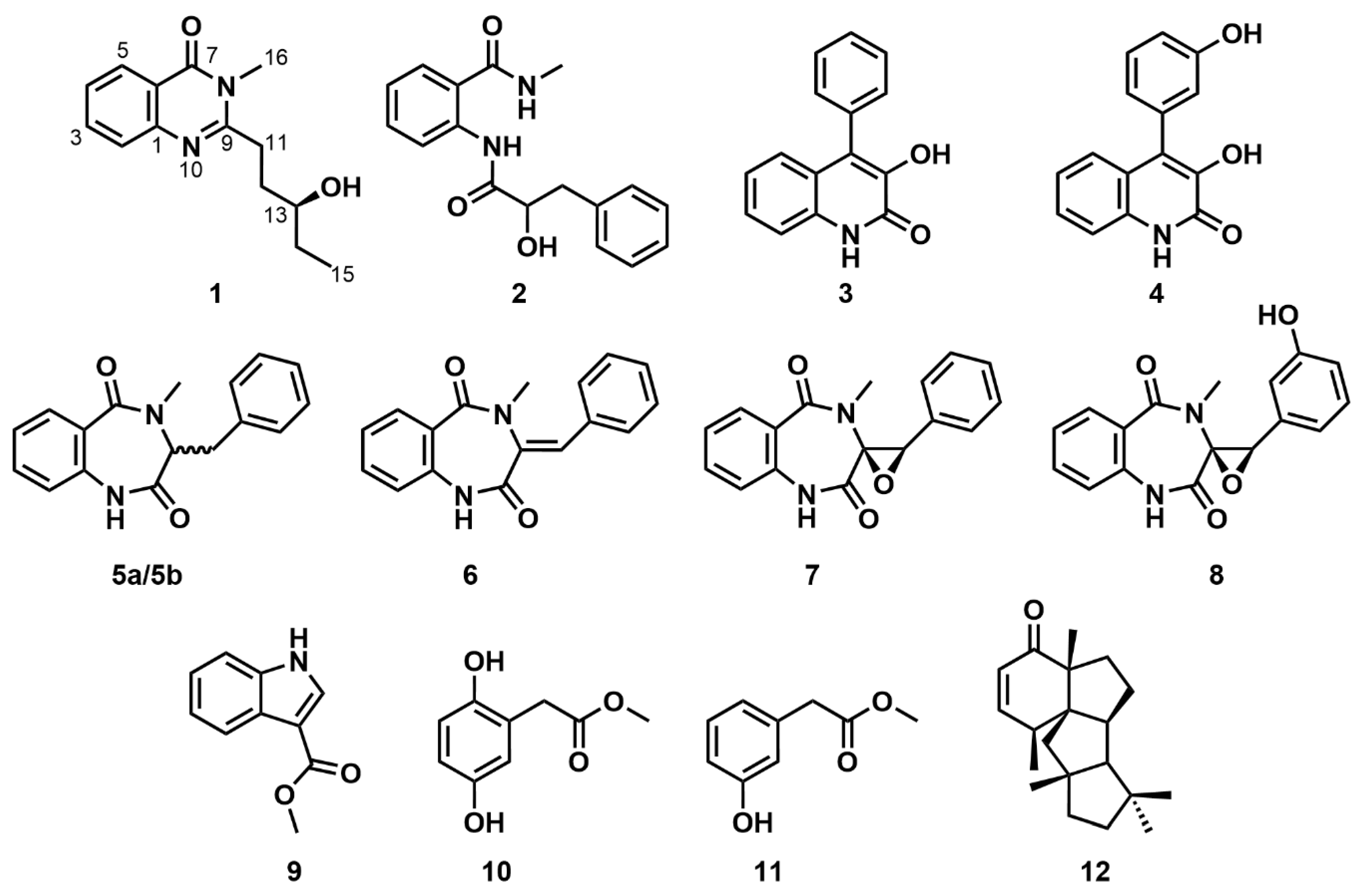
1), together with eleven known secondary metabolites including 2-(2-hydroxy-3-phenylpropionamido)-
N-methylbenzamide (
2) [
29], viridicatin (
3) [
30], viridicatol (
4) [
31], (±)-cyclopeptin (
5a/
5b) [
32,
33], dehydrocyclopeptin (
6) [
33], cyclopenin (
7) [
34], cyclopenol (
8) [
35], methyl-indole-3-carboxylate (
9) [
36], 2,5-dihydroxylbenzeneacetic acid (
10) [
37], methyl
m-hydroxyphenylacetate (
11) [
38] and conidiogenone B (
12) [
39] in
Figure 1. Herein, we describe the molecular networking analyss of the EtOAc extract, isolation and structural elucidation of the new compound, as well as the seed-germination-promoting activities of compounds
2,
3,
4,
7,
9, and
12.
2. Results and Discussion
2.1. Molecular Networking-Guided Isolation Workflow
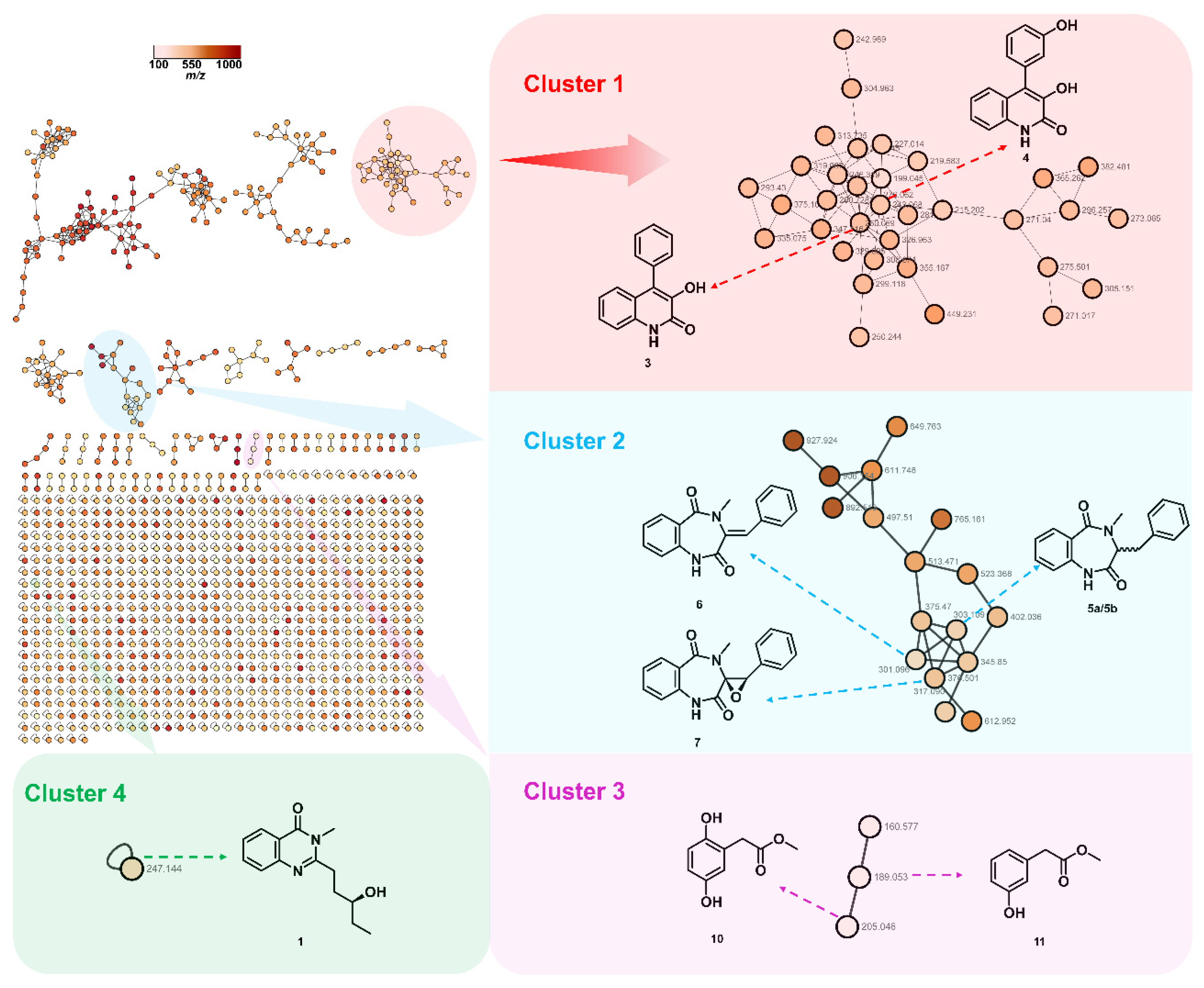
To target the isolation of quinazolinone alkaloid, the crude EtOAc extract of Penicillium sp. HJT-A-6 was subjected to full scan HPLC-Q-TOF-MS/MS analysis. The obtained MS/MS data was used to generate the initial molecular network using the Global Natural Products Social Molecular Networking (GNPS) platform following the established protocol. This initial network was visualized and analyzed via Cytoscape 3.7.1 software.
As shown in
Figure 2, the annotated nodes of total secondary metabolites were displayed, the color of the node indicates the
m/z of the parent ion. This process generated a network featuring 2373 nodes and 755 clusters, those representative highlighted clusters indicated several known natural products and a promising new compound. In cluster 1 highlighted in light red, the node (
m/z 276.062) was connected to the node (
m/z 260.069), which were supposed to be two known compounds, viridicatin (3) and viridicatol (4). Clusters 2 and 3 highlighted in light blue and light purple, respectively, identifying two classes of natural products, cyclopeptin analogues (5a/5b–7) and phenyl acetate derivates (10, 11). The orphan cluster 4 (
m/z 247.144) in light green, which was not associated with any structurally known compound, guiding the targeted isolation of a new quinazolinone alkaloid, peniquinazolinone A (1).
2.2. Structural Elucidation of Compound 1
Compound
1 was obtained as a yellow oil. Its molecular formula was determined as C
14H
18N
2O
2 based on HRESIMS at
m/z 269.1271 [M + Na]
+ (calcd for 269.1266) (
Figure S1), indicating seven degrees of unsaturation. The
1H NMR spectrum (
Table 1 and
Figure S3) revealed the resonances of four aromatic protons at
δH 8.10 (d,
J = 7.5 Hz, H-5), 7.77 (t,
J = 7.5 Hz, H-3), 7.59 (d,
J = 7.5 Hz, H-2) and 7.47 (d,
J = 7.5 Hz, H-4), which indicated the presence of one 1,2-disubstituted phenyl group in
1; three methylene protons at
δH 1.43–2.99; one oxygenated methine proton at
δH 3.48 (m, H-13); two methyl protons at
δH 3.56 (s), 0.90 (t
, J = 7.5 Hz, H-15). The
13C NMR and HSQC spectra (
Table 1 and
Figures S6–S7) of
1 displayed the presence of fourteen carbons, including one carbonyl carbon, seven aromatic/olefinic carbons, three methylenes, one oxygenated methine, and two methyls.
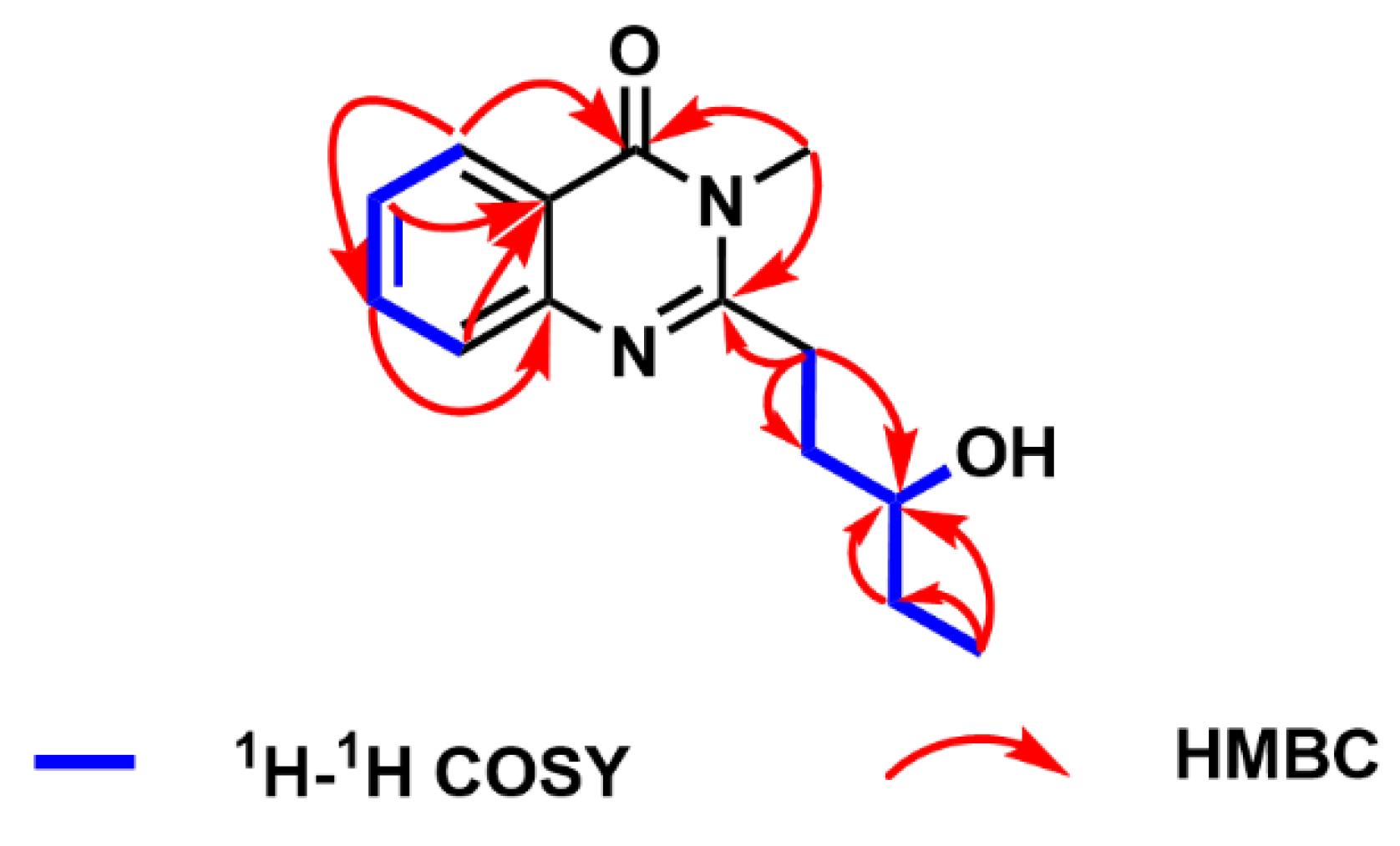
The planar structure of compound
1 was deduced by HMBC and
1H–
1H COSY spectra (
Figure 3 and
Figures S8–S9). The HMBC correlations from H-2 to C-6, from H-3 to C-1, from H-4 to C-6, from H-5 to C-3 and C-7, as well as from H-16 to C-7 and C-9, established the structural skeleton of quinazolinone. The proton spin systems of H-11/H-12/H-13/H-14/H-15 and H-13/13-OH, coupled with the HMBC correlations from H-11 to C-9, C-12, and C-13, from H-14 to C-13, and from H-15 to C-13 and C-14, confirmed the linkage of the quinazolinone group and the pentan-3-ol moiety at C-9. Consequently, the planar structure of compound
1 was constructed, named peniquinazolinone A.
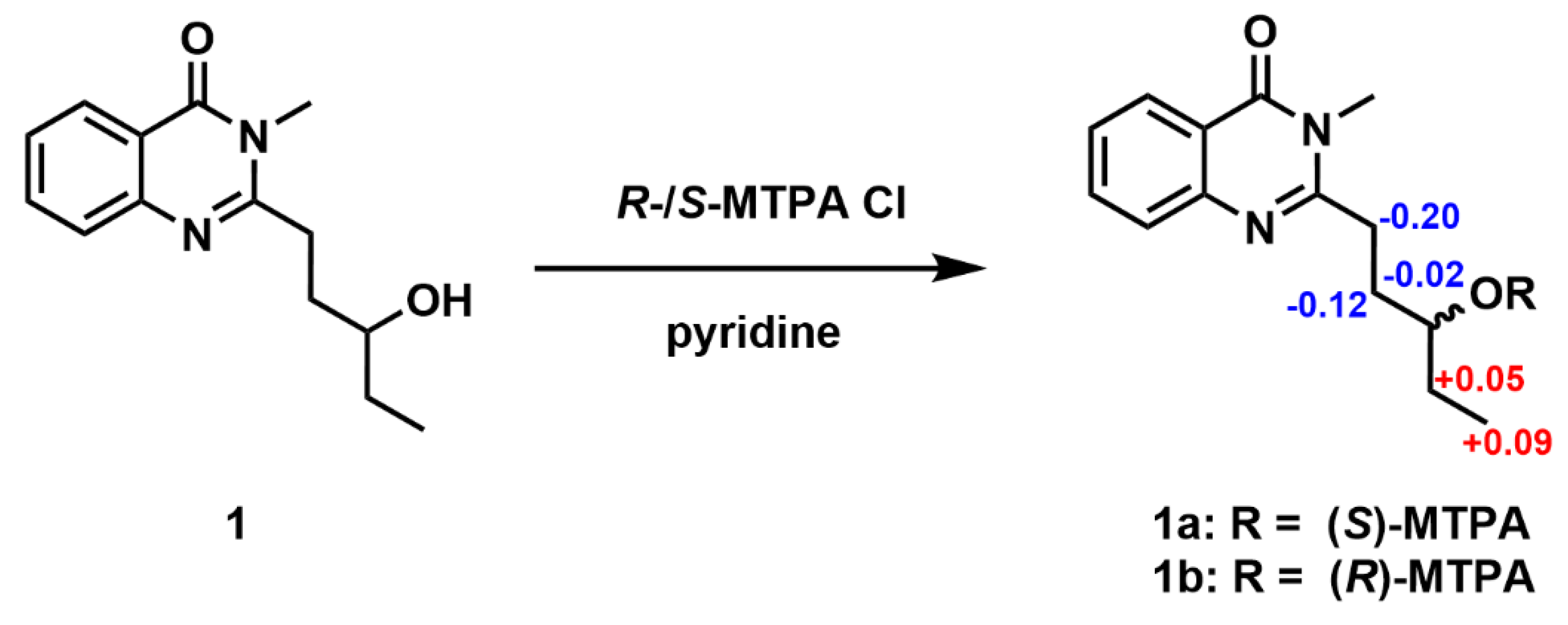
The absolute configuration of C-13 was defined by the application of the Mosher method. Compound
1 was reacted with both (
R)-(–) and
S-(+)-α-methoxy-α-(trifluoromethyl) phenylacetyl chlorides (MTPA-Cl) to afford the corresponding (
S)- and (
R)-Mosher esters, respectively (
1a and
1b), respectively. The absolute configuration at C-13 in
1 was determined to be
R by the observed chemical shift differences Δδ(δ
S-δ
R) (
Figure 4 and
Figures S4–S5)
.
2.3. Seed-Germination-Promoting Activity of the Isolated Compounds
Compounds
2,
3,
4,
7,
9, and
12 were tested for their seed-germination-promoting activity. As shown in
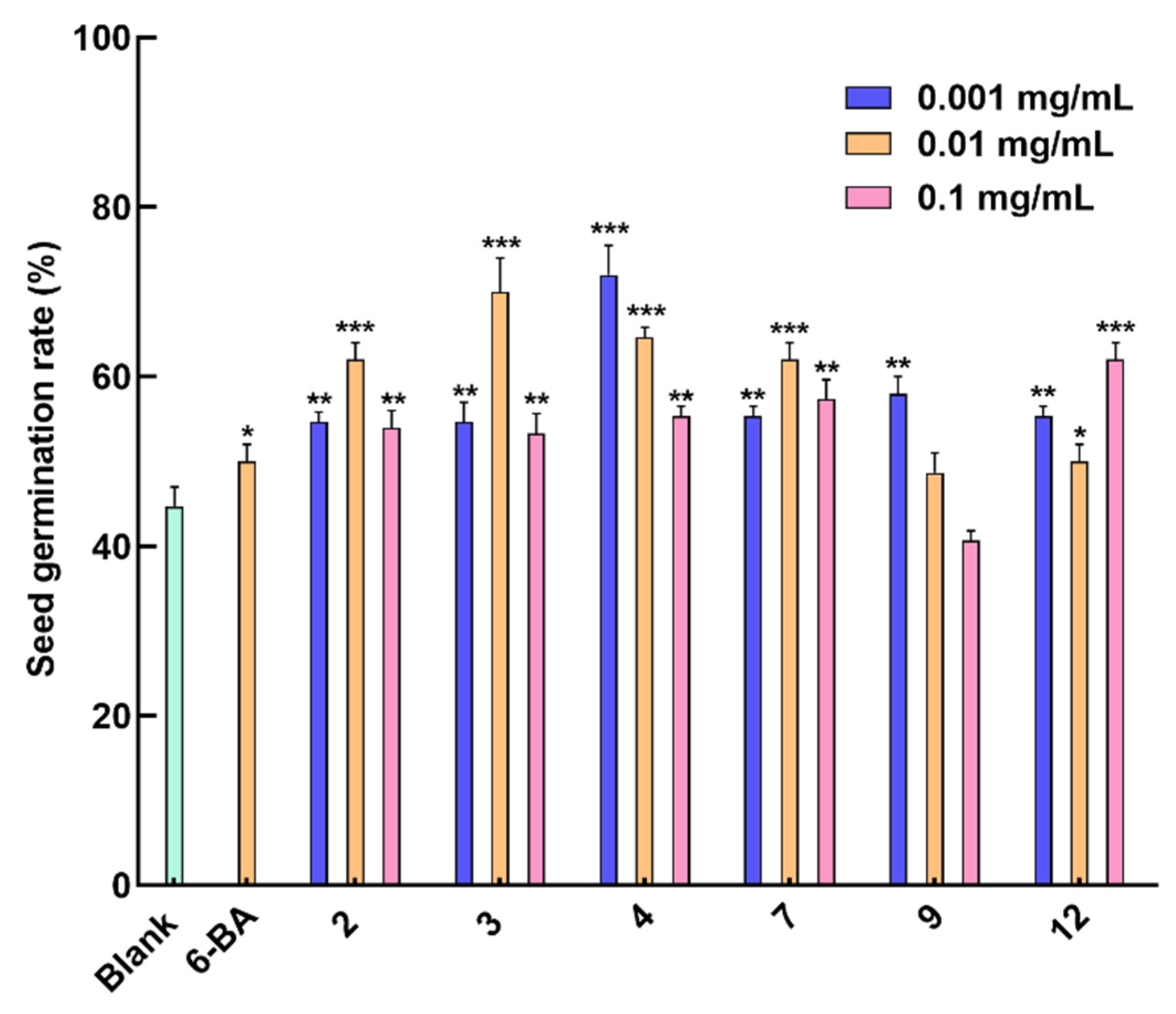
Figure 5, Compounds
2,
3, and
7 showed an optimal promoting effect on the seed germination of
Rhodiola tibetica at a concentration of 0.01 mg/mL, with a germination rate of about 62%, 70%, and 62%, respectively, compared to the germination rate of about 52% of the positive drug 6-BA. Compounds
4 and
9 exhibited a germination rate of about 72% and 58%, respectively, at the optimal concentration of 0.001 mg/mL, indicating a negative correlation between concentration and germination rate. Compared with other tested compounds, compound
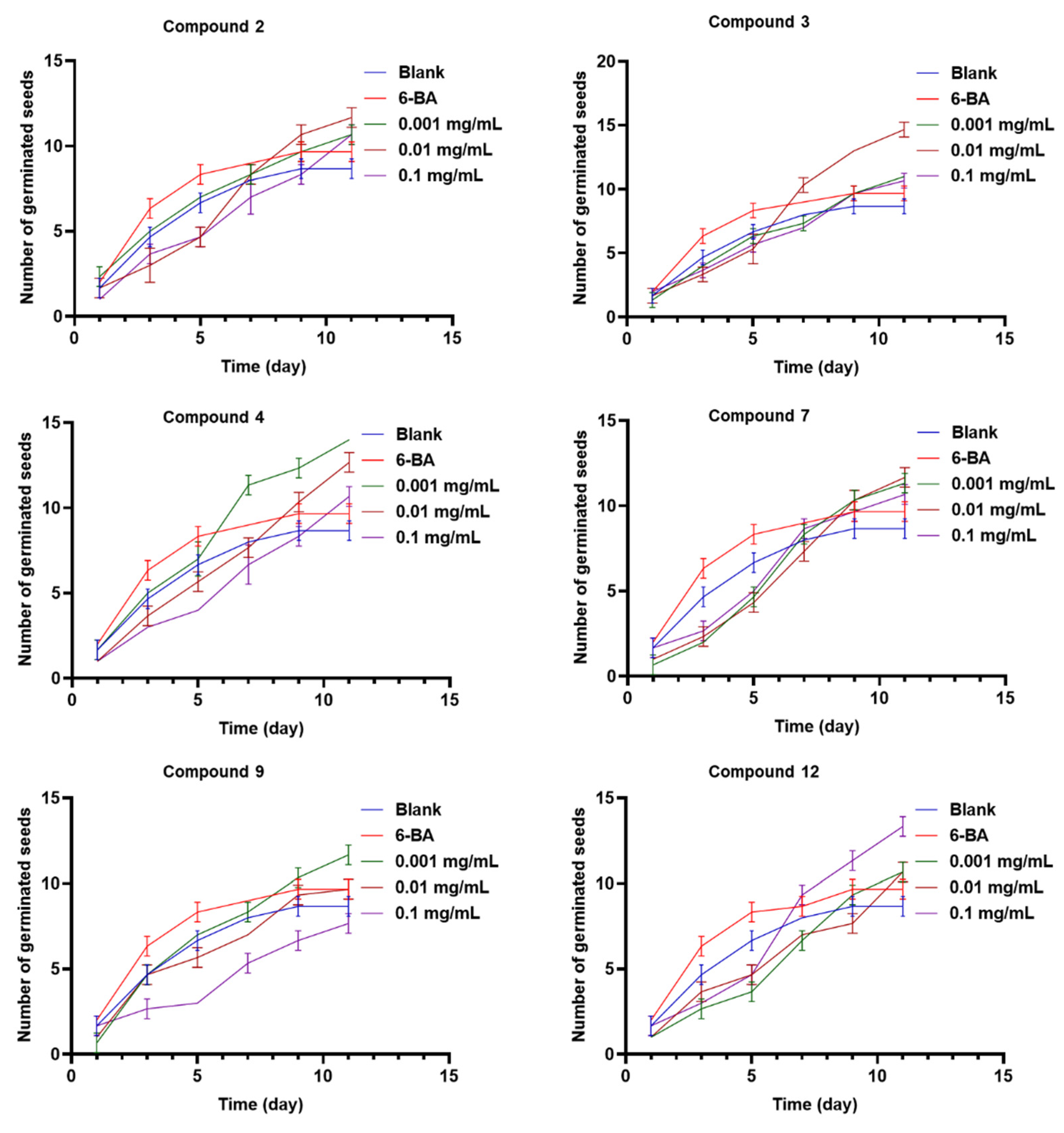
12 showed the best germination rate of about 62% at the same concentration level (0.1 mg/mL). In addition, compounds
2,
3,
4,
7,
9, and
12 could prolong the seed-germination period of
Rhodiola tibetica up to the 11
th day, which was relatively better than the positive drug 6-BA (
Figure 6). Compound
1 was not tested for its seed-germination-promoting activity due to the trace amount. Compound
2 is a structural analogue of compound
1, generated by ring-opening of the quinazoline moiety of compound
1, further seed germination assays of compound
1 may verify whether the existence of the quinazoline moiety affects its seed-germination-promoting activity.
3. Materials and Methods
3.1. General Experimental Procedures
UV spectrum was recorded on a Jasco V-560 spectrophotometer (JASCO Corporation, Japan). Optical rotation was obtained on an Autopol IV Polarimeter (Rudolph Research Analytical, Flanders, NJ, USA). CD spectrum was acquired on a Jasco J-810-150S spectropolarimeter (JASCO Corporation, Japan). High-resolution electrospray ionization mass spectrometry (HRESIMS) data was collected on an AB Sciex Triple TOF 4600 mass spectrometer (AB SCIEX, Framingham, MA, USA). NMR spectra were carried out on a Bruker Avance II 500 MHz NMR spectrometer (Bruker, Karlsruhe, Germany) with tetramethylsilane (TMS) as an internal standard. Agilent 1260 Infinity (Agilent Technologies Inc., Santa Clara, CA, USA), Waters 2535 (Waters Corporation, Milford, MA, USA) and Shimadzu LC-20AR (Shimadzu Corporation, Kyoto, Japan) semi-preparative HPLC systems were performed using a Welch Ultimate XB-C18 column, (250 mm × 10.0 mm, 5 μm). Silica gel (100−200 mesh & 200−300 mesh, Qingdao Marine Chemical Ltd., Qingdao, China) and Sephadex LH-20 (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) were used for column chromatography. The silica gel GF254 (Qingdao Marine Chemical Co., Ltd., Qingdao, China) was used for analytical and preparative thin-layer chromatography (TLC).
3.2. Fungal Material
The fungus strain Penicillium sp. HJT-A-6 was obtained from the stem of Rhodiola tibetica collected in Langkazi County, Shannan City, Tibet, China, in July 2021. It was identified based on its morphological characteristics and its sequence of the internal transcribed spacer (ITS) analysis of rDNA, and the BLAST search result showed that the sequence was the most similar (99%) to the sequence of Penicillium sp. (compared to MN634462.1), The sequence data of the fungus was submitted to GenBank database, accession number: OR346333.1. The fungus was deposited in the College of Life and Health, Dalian University, Dalian, China.
3.3. Fermentation and Isolation
The fungal strain was cultured on autoclaved rice medium (one hundred 500 mL Erlenmeyer flasks, each containing 80 g rice, 110 mL water) in the stationary phase at 28 oC for 40 days. After 40 days, the fermentation was cut into small pieces, and extracted with 95% EtOH three times. The extract was concentrated under reduced pressure to afford an aqueous solution, and then partitioned with petroleum ether, EtOAc, and n-BuOH to obtain the EtOAc-soluble extract (64 g). The extract was subjected to silica gel column chromatography with CH2Cl2/MeOH (100:0–0:100) to afford fourteen fractions (Fr. A–N).
Fr. B (10.5 g) was chromatographed on a silica gel column with gradient elution (PE/EtOAc, 5:1–1:2) to yield 8 subfractions (Fr. B1–Fr. B8). Fr. B5 was further purified by semi-preparative HPLC with MeOH/H2O (40:60, 0–35 min, 3 mL/min) to obtain compound 11 (1.6 mg, tR = 29 min).
Fr. C (6.3 g) was eluting with gradient petroleum (PE/EtOAc, 5:1–1:1) to afford 9 subfractions (Fr. C1–Fr. C9). Fr. C1 and Fr. C4 were subjected to Sephadex LH-20 gel column and preparative TLC, respectively, to obtain compounds 3 (250 mg) and 12 (4.6 mg). Fr. C5 was chromatographed by Sephadex LH-20 gel column using isocratic elution with CH2Cl2/MeOH (1:1), yielding Fr. C5b, which was purified by semi-preparative HPLC with CH3CN /H2O (30:70, 0–30 min, 3 mL/min) to give compound 6 (13.6 mg, tR = 22 min). Fr. C7 was purified by semi-preparative HPLC with MeOH/H2O (60:40, 0–40 min, 3 mL/min) to obtain compound 7 (193 mg, tR = 14 min) and the mixture of 5a and 5b in a 1:1.7 molar ratio (13.8 mg, tR = 17 min). Fr. C8 was applied to Sephadex LH-20 gel column with CH2Cl2/MeOH (1:1) to give Fr. C8e, which was purified by semi-preparative HPLC with gradient MeOH/H2O (20:80–60:40, 0–30 min, 3 mL/min) to obtain compound 9 (20 mg, tR = 25 min).
Fr. E (6.3 g) was chromatographed on a silica gel column with gradient elution (PE/EtOAc, 7:1–1:2), affording 20 fractions (Fr. E1–Fr. E20). Fr. E13 was purified by semi-preparative HPLC with gradient MeOH/H2O (20:80–95:5, 0–40 min, 3 mL/min) to yield compound 1 (1.6 mg, tR = 26 min). Fr. E14 was purified by semi-preparative HPLC with MeOH/H2O (40:60, 0–40 min, 3 mL/min) to obtain compound 2 (3.4 mg, tR = 16 min) and compound 4 (120 mg, tR = 32 min). Fr. E16 was purified by semi-preparative HPLC with CH3CN/H2O (45:55, 0–25 min, 3 mL/min) to afford compound 8 (6 mg, tR = 12 min).
Fr. G (6.7g) was chromatographed on a silica gel column with gradient elution (PE/EtOAc, 10:1–1:1) to afford 5 subfractions (Fr. G1–Fr. G5). Fr. G4 was applied to Sephadex LH-20 gel column with CH2Cl2/MeOH (1:1) to yield Fr. G4d, which was purified by semi-preparative HPLC with gradient MeOH/H2O (20:80–95:5, 0–30 min, 3 mL/min) to obtain compound 10 (5.8 mg, tR = 10 min).
Peniquinazolinone A (
1): yellow oil;
+20 (
c 0.2, MeOH); UV (CH
3OH)
λmax (log
ε) 223 (4.24), 267 (3.74) nm ; ECD (CH
3OH)
λmax (Δ
ε) 220 (–7.0) nm;
1H NMR (DMSO-
d6, 500 MHz) and
13C NMR (DMSO-
d6, 125 MHz) data (see
Table 1); HRESIMS
m/z 269.1271 [M + Na]
+ (calculated for C
14H
18N
2O
2Na, 269.1266).
3.4. Mosher Esterification of Compound 1
Compound 1 (0.5 mg) was dissolved in 100 μL CDCl3 in an NMR tube, sequentially added 9 μL pyridine and 15 μL (R)-(–)-α-methoxy-α-(trifluoromethyl)phenylacetyl chloride ((R)-(–)-MTPA-Cl). The mixture was stirred at room temperature for 1 h to afford the corresponding (S)-Mosher ester (1a), and subsequently diluted with 300 μL CDCl3 to acquire 1H NMR spectrum. The (R)-Mosher ester (1b) of 1 was prepared from (S)-(+)-MTPA-Cl using the same method.
3.5. Seed-Germination-Promoting Assay
Compounds 2, 3, 4, 7, 9 and 12 was dissolved in 0.2% DMSO aqueous solution to yield the stock solution with a concentration of 0.06 mg/mL. 5 µL, 50 µL and 500 µL of compounds 2, 3, 4, 7, 9 and 12 was added to the 30 mm filter paper placed in the 6-well plate. After evaporation of the solvent, the filter paper was immersed in 300 µL of distilled water, and then 20 seeds of Rhodiola tibetica were displayed in each 30 mm filter paper and incubated at a light-dark regime of 16:8 h condition, 20 °C for 7 days. The germination rate of the seeds was calculated after incubation. The experimental data were collected from three independent experiments. Further experiments of the relationship between the number of germinated seeds and germination time for compounds 2, 3, 4, 7, 9 and 12 were also conducted, the germination period was set up to 11 days.
4. Conclusions
In this work, the chemical investigation of the Rhodiola tibetica endophytic fungus Penicillium sp. HJT-A-6 led to the isolation and identification of a new quinazolinone alkaloid, named peniquinazolinone A (1), and eleven known compounds, 2-(2-hydroxy-3-phenylpropionamido)-N-methylbenzamide (2), viridicatin (3), viridicatol (4), (±)-cyclopeptin (5a/5b), dehydrocyclopeptin (6), cyclopenin (7), cyclopenol (8), methyl-indole-3-carboxylate (9), 2,5-dihydroxyphenyl acetate (10), methyl m-hydroxyphenylacetate (11), and conidiogenone B (12). Compound 1 was identified by extensive spectroscopic analysis and the modified Mosher’s method. Compounds 2, 3, 4, 7, 9, and 12 were evaluated for their seed germination activity. Compounds 2, 3, and 7 showed an optimal promoting effect on the seed germination of Rhodiola tibetica at a concentration of 0.01 mg/mL. Compounds 4 and 9 optimal seed-germination-promoting activity at a concentration of 0.001 mg/mL, while the optimal concentration for compound 12 to promote Rhodiola tibetica seed germination is 0.1 mg/mL. Additionally, all the tested compounds assuredly prolonged the seed-germination period of the host plant. These results not only broadened the structural diversity of quinazoline metabolites derived from fungi, but also provided data support for understanding the interactive relationship between endophytic fungi and host plants.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Positive mode HRESIMS spectrum of 1; Figure S2: UV spectrum of 1; Figure S3: 1H NMR (DMSO-d6, 500 MHz) spectrum of 1; Figure S4: 1H NMR (DMSO-d6, 500 MHz) spectrum of 1a; Figure S5: 1H NMR (DMSO-d6, 500 MHz) spectrum of 1b; Figure S6: 13C NMR (DMSO-d6, 125 MHz) spectrum of 1; Figure S7: HSQC (DMSO-d6, 500 MHz) spectrum of 1; Figure S8: HMBC (DMSO-d6, 500 MHz) spectrum of 1; Figure S9: 1H-1H COSY (DMSO-d6, 500 MHz) spectrum of 1; Figure S10: NOESY (DMSO-d6, 500 MHz) spectrum of 1; Figure S11: CD spectrum of 1.
Author Contributions
Conceptualization, D.X., X.L. and B.F.; methodology, D.X., Y.W., X.L. and B.F.; validation, D.X., Y.W., X.L. and B.F.; formal analysis, D.X., X.L. and B.F.; investigation, D.X., Y.W., C.G., X.Z., W.F., X.L. and B.F.; resources, X.L. and B.F.; data curation, D.X., X.L. and B.F.; writing—original draft preparation, D.X. and Y.W.; writing—review and editing, D.X. and X.L.; visualization, X.L. and Y.W.; supervision, D.X., X.L. and B.F.; project administration, X.L. and B.F.; funding acquisition, D.X., C.G. and X.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grants from the Dalian Science and Technology Innovation Fund Project (2022JJ12WZ059), Dalian University Research Platform Project (202101YB08), and Liaoning Provincial Department of Education Basic Scientific Research Project (JYTQN2023100, LJKFZ20220287).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Not applicable.
References
- Kusari, S.; Spiteller, M. Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat. Prod. Rep. 2011, 28, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.H.; Li, S.J.; Zhang, X.; Zhao, C.Q. Biological activities of some new secondary metabolites isolated from endophytic fungi: a review study. Int. J. Mol. Sci. 2021, 22, 959. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Ai, H. L.; Liu, J.K. Identification and bioactivities of secondary metabolites derived from endophytic fungi isolated from ethnomedicinal plants of Tujia in Hubei province: a review. Nat. Prod. Bioprospect. 2021, 11, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Keswani, C.; Singh, H.B.; Hermosa, R.; García-Estrada, C.; Caradus, J.; He, Y.W.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Vinale, F.; et al. Antimicrobial secondary metabolites from agriculturally important fungi as next biocontrol agents. Appl. Microbiol. Biotechnol. 2019, 103, 9287–9303. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, Í.C.D.S.; Polonio, J.C.; Golias, H.C. Endophytic fungi as a source of antiviral compounds - a review. Chem. Biodivers. 2022, 19, e202100971. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Ma, N.; Lu, Y.F.; Dai, S.Y.; Song, X.; Li, C.; Sun, Y.; Pei, Y.H. Structure elucidation and anti-tumor activities of trichothecenes from endophytic fungus Fusarium sporotrichioides. Biomolecules 2022, 12, 778. [Google Scholar]
- Bie, Q.; Chen, C.M.; Yu, M.Y.; Guo, J.R.; Wang, J.P.; Liu, J.J.; Zhou, Y.; Zhu, H.C.; Zhang, Y.H. Dongtingnoids A-G: fusicoccane diterpenoids from a Penicillium species. J. Nat. Prod. 2019, 82, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Daley, S.K.; Cordell, G.A. Biologically significant and recently isolated alkaloids from endophytic fungi. J. Nat. Prod. 2021, 84, 871–897. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, J.; Liu, M.Y.J.; Zhao, L. Progress on terpenoids with biological activities produced by plant endophytic fungi in China between 2017 and 2019. Nat. Prod. Commun. 2020, 15, 1–18. [CrossRef]
- Jafari, E.; Khajouei, M.R.; Hassanzadeh, F.; Hakimelahi, G.H.; Khodarahmi, G.A. Quinazolinone and quinazoline derivatives: recent structures with potent antimicrobial and cytotoxic activities. Res. Pharm. Sci. 2016, 11, 1–14. [Google Scholar]
- Habib, O.M.O.; Hassan, H.M.; El-Mekabaty, A. Novel quinazolinone derivatives: synthesis and antimicrobial activity. Med. Chem. Res. 2013, 22, 507–519. [Google Scholar] [CrossRef]
- Manivannan, E.; Chaturvedi, S.C. Analogue-based design, synthesis and molecular docking analysis of 2,3-diaryl quinazolinones as non-ulcerogenic anti-inflammatory agents. Bioorg. Med. Chem., 2011, 19, 4520–4528. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Aziz, A.A.; Abou-Zeid, L.A.; ElTahir, K.E.H.; Ayyad, R.R.; El-Sayed, M.A.; El-Azab, A.S. Synthesis, anti-inflammatory, analgesic, COX-1/2 inhibitory activities and molecular docking studies of substituted 2-mercapto-4(3H)-quinazolinones. Eur. J. Med. Chem. 2016, 121, 410–421. [Google Scholar] [CrossRef]
- Brown, C.E.; Kong, T.; McNulty, J.; D'Aiuto, L.; Williamson, K.; McClain, L.; Piazza, P.; Nimgaonkar, V.L. Discovery of potent antiviral (HSV-1) quinazolinones and initial structure-activity relationship studies. Bioorg. Med. Chem. Lett. 2017, 27, 4601–4605. [Google Scholar] [CrossRef]
- Wang, Z.W.; Wang, M.X.; Yao, X.; Li, Y.; Tan, J.; Wang, L.Z.; Qiao, W.T.; Geng, Y.Q.; Liu, Y.X.; Wang, Q.M. Design, synthesis and antiviral activity of novel quinazolinones. Eur. J. Med. Chem. 2012, 53, 275–282. [Google Scholar] [CrossRef]
- Zhu, S.R.; Wang, J.; Chandrashekar, G.; Smith, E.; Liu, X.J.; Zhang, Y.S. Synthesis and evaluation of 4-quinazolinone compounds as potential antimalarial agents. Eur. J. Med. Chem. 2010, 45, 3864–3869. [Google Scholar] [CrossRef]
- Yu, G.H.; Zhou, G.L.; Zhu, M.L.; Wang, W.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Neosartoryadins A and B, fumiquinazoline alkaloids from a mangrove-derived fungus Neosartorya udagawae HDN13-313. Org. Lett., 2016, 18, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.X.; Lin, T.; Wang, W.; Xin, Z.H.; Zhu, T.J.; Gu, Q.Q.; Li, D.H. Antiviral alkaloids produced by the mangrove-derived fungus Cladosporium sp. PJX-41. J. Nat. Prod. 2013, 76, 1133–1140. [Google Scholar] [CrossRef]
- Jiao, R.H.; Xu, S.; Liu, J.Y.; Ge, H.M.; Ding, H.; Xu, C.; Zhu, H.L.; Tan, R.X. Chaetominine, a cytotoxic alkaloid produced by endophytic Chaetomium sp. IFB-E015. Org. Lett. 2006, 8, 5709–5712. [CrossRef]
- An, C.Y.; Li, X.M.; Li, C.S.; Wang, M.H.; Xu, G.M.; Wang, B.G. Aniquinazolines A-D, four new quinazolinone alkaloids from marine-derived endophytic fungus Aspergillus nidulans. Mar. Drugs. 2013, 11, 2682–2694. [Google Scholar] [CrossRef]
- Zheng, C.J.; Li, L.; Zou, J.P.; Han, T.; Qin, L.P. Identification of a quinazoline alkaloid produced by Penicillium vinaceum, an endophytic fungus from Crocus sativus. Pharm. Biol. [CrossRef]
- Lu, X.; Tang, X.Y.; Wang, H.X.; Huang, W.J.; Feng, W.X.; Feng, B.M. Polyketone metabolites isolated from Rhodiola tibetica endophytic fungus Alternaria sp. HJT-Y7 and their SARS-CoV-2 virus inhibitory activities. Bioorg. Chem. [CrossRef]
- Tang, X.Y.; Zhao, Y.X.; Wang, X.D.; Ye, C.T.; Feng, B.M.; Tang, C.; Lu, X. Polyketides with anti-inflammatory activity from Rhodiola tibetica endophytic fungus Alternaria sp. HJT-Y7. Phytochemistry 2022, 203, 113383. [Google Scholar] [CrossRef]
- Lu, X.; Qi, J.K.; Tang, X.Y.; Wang, X.D.; Ye, C.T.; Bai, J.; Tang, C.; Xi, Y.F.; Wu, B.; Wan, X.; Feng, B.M. Polyketides with anti-inflammatory activity isolated from Rhodiola tibetica endophytic fungus Penicillium sp. HJT-A-10. Fitoterapia, 2023, 164, 105361. [Google Scholar] [CrossRef] [PubMed]
- Gaudêncio, S.P.; Pereira, F. Dereplication: racing to speed up the natural products discovery process. Nat. Prod. Rep. 2015, 32, 779–810. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.W.; Rapoport, H. Mechanism of the transformation of cyclopenin to viridicatin. J. Am. Chem. Soc. 1969, 91, 6083–6089. [Google Scholar] [CrossRef]
- Cunningham, K.G.; Freeman, G.G. The isolation and some chemical properties of viridicatin, a metabolic product of Penicillium viridicatum Westling. Biochem. J. 1953, 53, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Harayama, T. A concise and versatile synthesis of viridicatin alkaloids from cyanoacetanilides. Org. Lett. 2009, 11, 1603–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Ding, S.S.; Shi, W.S.; Cao, F.; Zhu, H.J.; Wen, M.L. A new quinolinone from freshwater lake-derived fungus Myrothecium verrucaria. Nat. Prod. Res. 2017, 31, 99–103. [Google Scholar] [CrossRef]
- Framm, J.; Nover, L.; el-Azzouny, A.; Richter, H.; Winter, K.; Werner, S.; Luckner, M. Cyclopeptin and dehydrocyclopeptin. Intermediates in the biosynthesis of cyclopeptin-viridicatin alkaloids in Penicillium cyclopium Westling. Eur. J. Biochem. 1973, 37, 78–85. [Google Scholar] [CrossRef]
- Pan, C.; Shi, Y.; Chen, X.; Chen, C.A.; Tao, X.; Wu, B. New compounds from a hydrothermal vent crab-associated fungus Aspergillus versicolor XZ-4. Org. Biomol. Chem. 2017, 15, 1155–1163. [Google Scholar] [CrossRef]
- Cai, X.Y.; Wang, J.P.; Shu, Y.; Hu, J.T.; Sun, C.T.; Cai, L.; Ding, Z.T. A new cytotoxic indole alkaloid from the fungus Penicillium polonicum TY12. Nat. Prod. Res. 2022, 36, 2270–2276. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Ahmad, V.U.; Perveen, S.; Bano, N.; Shafiuddin, S.; Shameel, M. Marine natural products; II. Chemical constituents of red alga Botryocladia leptopoda. Planta. Med. 1987, 53, 117–118. [Google Scholar] [CrossRef]
- Tan, D.P.; Chou, G.; Wang, Z.T. Phenolic compounds from Senecio scandens. Biochem. Syst. Ecol. 2010, 38, 122–124. [Google Scholar] [CrossRef]
- Xu, L.; Wang, X.H.; Luo, R.Y.; Lu, S.Q.; Guo, Z.J.; Wang, M.A.; Zhou, L.G. Secondary metabolites of rice sheath blight pathogen Rhizoctonia solani Kühn and their biological activities. J. Integr. Agr. 2015, 14, 80–87. [Google Scholar] [CrossRef]
- Gao, S.S.; Li, X.M.; Zhang, Y.; Li, C.S.; Wang, B.G. Conidiogenones H and I, two new diterpenes of Cyclopiane class from a marine-derived endophytic fungus Penicillium chrysogenum QEN-24S. Chem. Biodivers. 2011, 8, 1748–1753. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).