1. Introduction
The most numerous microflora in soil are bacteria. Most bacteria decay organic matter, while a few cause plant diseases. Bacteria affect the chemical properties of the soil and can contribute to the production of healthy crops by making nutrients more readily available (Plaster 2013). Understanding the roles and capabilities of soil bacteria in rootzones of crops, fallow soil, and noxious weeds in a field is important for disease prevention and increasing yield.
Pseudomonas sp. are somewhat controversial, since they have the potential to be antibiotic resistant pathogens to plants or animals (Gislason 2020). However, some species also have the potential to be plant growth promoting. Other beneficial functions of Pseudomonas sp. may include antibiotic production, increase in the availability of iron to plants, degradation of contaminants, and reduction of nitrate.
Bacteria decompose organic matter and deceased organisms, breaking them down to make nutrients available to plants as well as many other important functions (Zhang 2022). Some of these other functions are Nitrogen cycling, protecting plants against harmful pathogens, and improving the soil aeration. By examining the soil for these bacteria, farmers can gain insight on ways to improve the quality of their soil, hence, improving crop yields and their profits, and reducing the use of commercial fertilizer which can have impacts on the soil itself and the surrounding environment.
For example, Pseudomonas sp. may have phosphatase enzymes that can make organic phosphates more available to plants, thus acting as a biofertilizer (Rath et al 2022, page 4, Mishra et al Eds.) According to the environmental secretary of the United Kingdom, the short-term gain from using synthetic fertilizers and pesticides may not be worth the long-term loss of soil fertility over time (Lupica 2020). Hence, it is necessary to think critically about farming practices, where the best practices maintain natural symbiotic relationships of these bacteria in the soil. According to Harrison, a professor at Cornell’s College of Agriculture and Life Sciences, in regards to soil bacteria and fungi, “If we’re right, then enriching the soil for some of these bacteria could increase crop yields and, ultimately, reduce the need for conventional fertilizers along with their associated costs and environmental impacts” (Haas 2021). Another potential benefit of natural biofertilizers is that Pseuomonas sp. activate plant defenses and prime them for pathogen attacks, as well as produce potential biocontrol agents (Tailapragada et al 2022, page 91, Mishra et al Eds.).
The identification techniques of DNA barcoding and colony PCR (Polymerase Chain Reaction) of bacteria that interact with plants will help give better understanding to plant physiology, microbial characteristics, and to anticipate potential threats from pathogens (Lebonah 2014). There is potential to identify new species from soil that may produce valuable compounds with agricultural or medical applications. The purpose of this study was to identify novel microbes performing beneficial ecological functions from soil. The hypothesis was that novel culturable species with antibiotic production potential could be readily found from soil in the Nature Canyon, Field 21, at Los Angeles Pierce College.
2. Materials and Methods
Between Fall 2021- Spring 2023 soil bacterial isolates were collected from Los Angeles Pierce College farm and local farms. One of the potentially novel isolates collected in Fall of 2022, 21-C3-ER, was isolated on nutrient agar from fallow soil in an agricultural nature preserve at Pierce College in the San Fernando Valley of Los Angeles. Barcoding isolate DNA was extracted using 10% Chelex solution heated for 10 minutes at 100 C.
Quality control was achieved through electrophoresis using the E-gel system. Partial 16S amplicons were sequenced on the Sanger platform using primers for the V1-V4 regions. Barcoding data analysis was performed on the DNA Subway Blue Line (Hilgert 2014) and EZBioCloud (Yoon et al 2017). Isolates with low similarity values and high completion values were selected as putative novel isolates for WGS. Whole Genome Sequencing of the novel isolate was carried out at BGI using DNBseq platform.
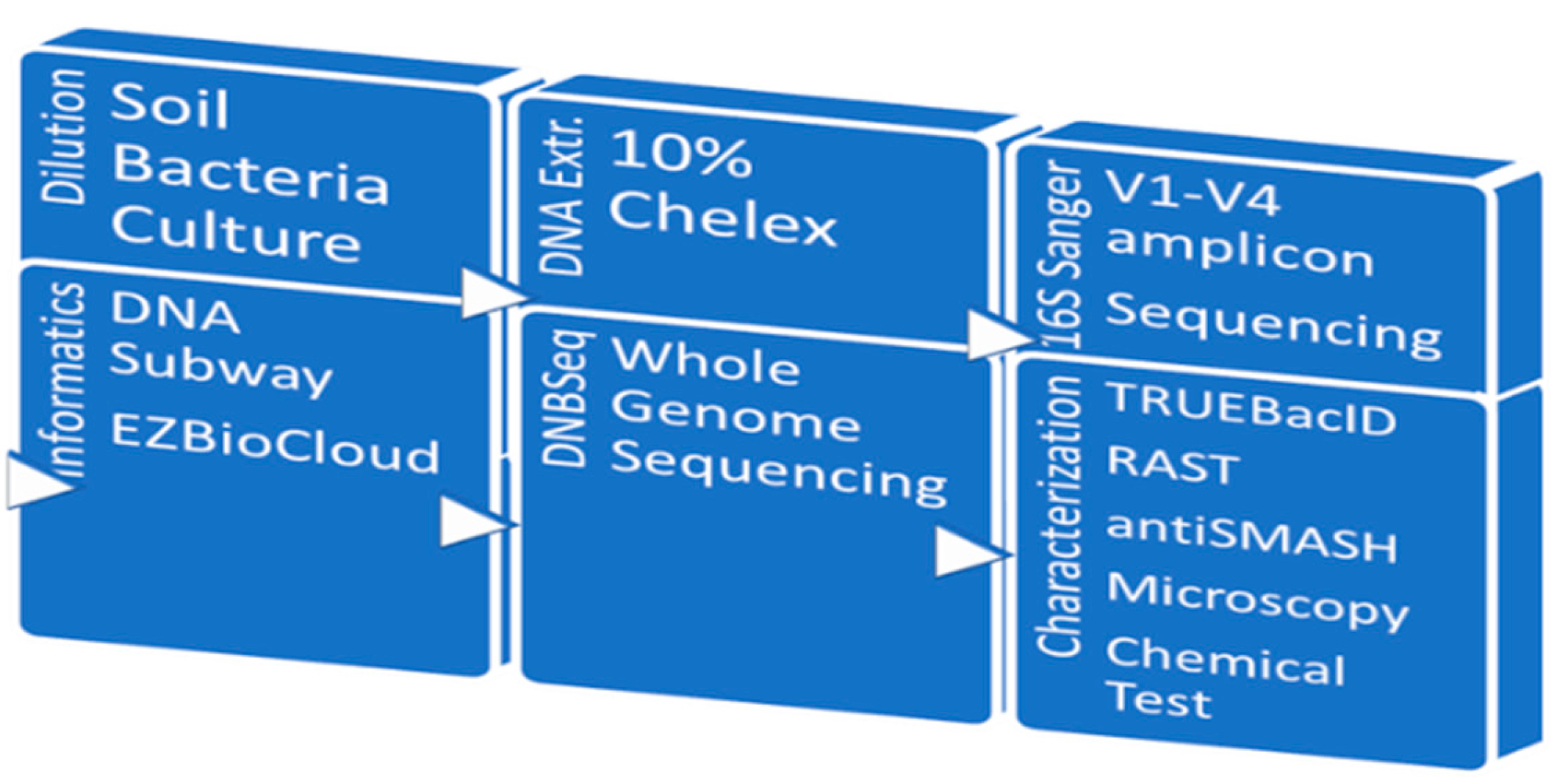
RAST was used for metabolic subsystems analysis (Aziz et al 2008). Strain identification was carried out using TRUEBacID (Ha 2019); genome assembly on this platform uses SPAdes (Bankevich 2012). Functional analysis was carried out in RAST and antiSMASH 7.0 (Blin et al 2023). A schematic representation of the experimental strategy is shown in
Figure 1.
Protein structure was simulated in SwissPROT and Protein Data Bank (PDB). Qualitative test for siderophore production used UV light to detect fluorescent pigments qualitatively (Dyer and Foy 2022, Tamang et al 2021, Banerjee et al 2017). A BLASTP search (Altschul 1990, Wang et al 2023) was conducted on domains of putative Pyoverdine synthase genes that were not well-characterized in the reference secondary metabolite gene clusters (Blin et al 2023). A template-based SwissPROT model was constructed (Waterhouse 2018) for a promysalin synthetase-like domain detected in the genome of the candidate novel species.
Using the Gram stain technique (Tripathi 2023), the bacteria was visualized on a Boreal compound light microscope at 400x. Bacteria from an agar plate was mixed with 62 µL of nuclease free water and 62 µL of 2.5% glutaraldehyde. The solution was mixed by pipetting and applied to an aluminum stub. 100 µL of ethanol was applied. The sample fan dried for 30 minutes. The Scanning electron micrograph of the bacteria was visualized on an SNE-Alpha desktop SEM (Scanning electron microscope) at 5KV, 50µM aperture size, high vacuum, and 25,000x magnification. The nitrate reduction test was carried out using the sulfanilic acid method (Buxton 2011). Pseudomonas sp. 21-C3-ER cultures were grown in nutrient broth with 64.29 ppm nitrate added for 48 hours.
Moisture, Organic matter, EC, TDS, pH, N, P, and K were determined using the methods described in the St. Clair Soil Science lab manual. Moisture % was determined by the oven dry method at 100 deg C for 24 hours. Organic matter percentage was determined by dry combustion. 1:5 soil to water ratio was used for soil pH measurements. N, P, and K were determined using Hach color changing reagents and %T was measured using a Genesys 20 spectrophotometer. Other soil physical and chemical properties from the site were measured including and TDS using a Hanna instruments probe, and texture by touch. GIS data from the USDA Web Soil Survey was reviewed (Soil Survey Staff 2024).
3. Results and Discussion
The results of initial screening using the partial 16S sequence suggested up to 30 potentially novel isolates. One of the potentially novel isolates collected in Fall of 2022, 21-C3-ER, was isolated from soil solution on nutrient agar from the 10^3 dilution. The isolate was translucent with a slight yellow color.
The soil conditions where the soil was collected were determined experimentally at Pierce College during Fall of 2022 to be 3.7% moisture, clay texture by touch, 13.37% organic matter, pH=6.63, TDS=638, 150 ppm nitrogen, 114 ppm phosphate, and 42 ppm potash. According to our experimental measurements, the organic matter was high, possibly due to the grazing of the cows and leaf litter. The soil color before combustion was dark gray. The soil color after combustion was dark reddish gray. According to the USDA NRCS Web Soil Survey (WSS), the soil map unit name was Balcom Silty Clay Loam, 15 to 30 percent slopes, Map unit symbol 104 (Soil Survey Staff 2024).
The 16S partial sequence of the isolate was 94.21% similar to
P. bijeensis and 36.3% complete. According to Whole Genome Sequencing results process with TRUEBacID, the closest relative was
Pseudomonas brassicacearum subsp.
neoaurantiaca which had 92.05% ANI (Average Nucleotide Identity) by genomic evidence. The summary is shown in
Table 1.
P. brassicacearum is also in the
P. fluorscens group and at least one strain is known for anti-nematode properties (Nandi 2016). Typically, individuals in the same species will have greater than 95% ANI shared between them (Jain 2018). The recA gene was 96.3% similar. The recA gene is useful for differentiating between species with high similarity, since some species cannot be differentiated by the full 16S sequence alone (Blackwood 2000).
197 virulence factors were identified by TRUEBacID including flagellar biosynthesis, motor proteins, Beta-lactamase, Fosfomycin, copper, and sulfur drug resistance. For example, alginate biosynthetic capability was identified, which can used by pathogenic Pseudomonas to provide a protective covering when the environment is hostile (Muhammadi and Nuzhat 2007).
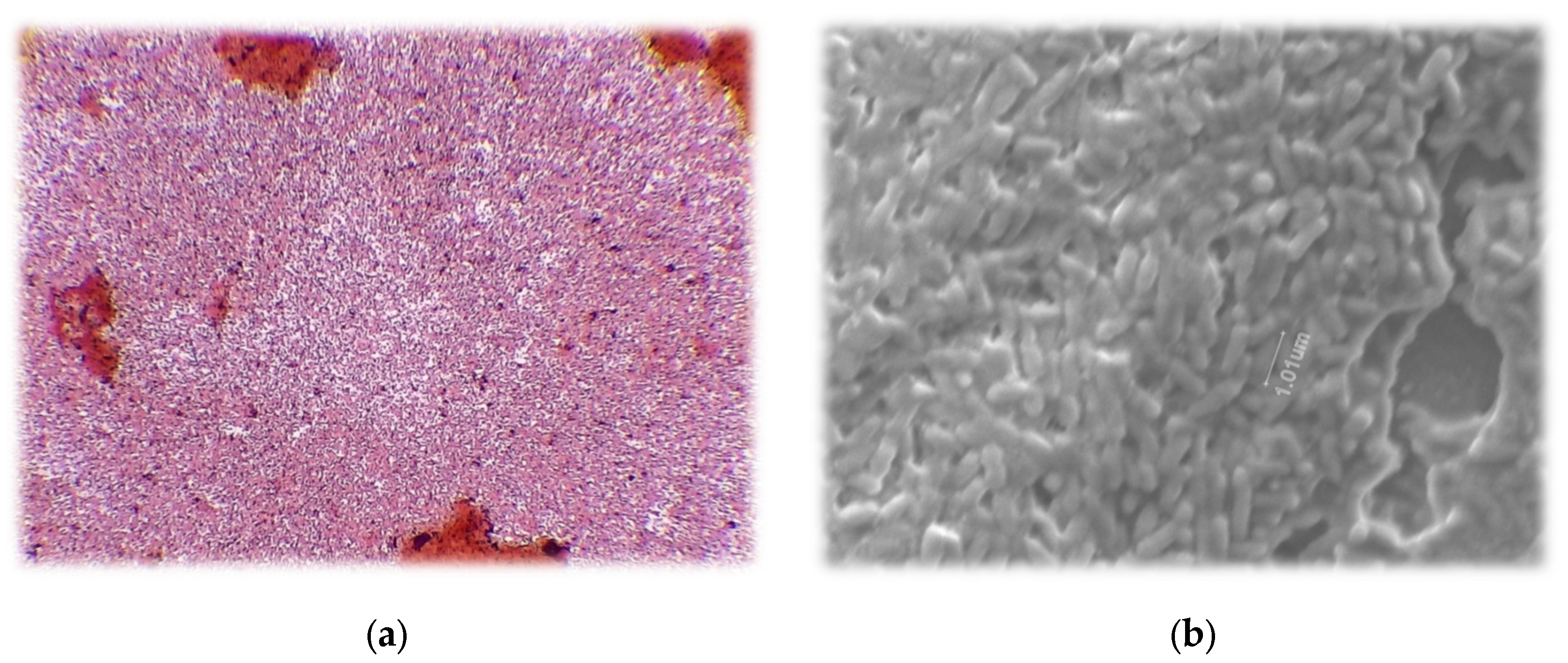
Gram stain of strain 21-C3-ER revealed gram negative rods. The individuals were pill-shaped. The red color indicated gram negative classification (
Figure 3).
Rods were visualized with scanning electron microscopy, revealing individuals approximately 1 µm in length. The small size makes sense when considering that these bacteria were found in fallow soil with a clay texture and high organic matter. They were likely able to hide in the soil capillaries and exude alginate in order to prevent desiccation.
Secondary metabolite analysis using antiSMASH showed a variety of RIPP-like proteins and a promysalin analog. There were 19 putative secondary metabolite gene clusters identified in the proposed novel bacterium. One example is an NRP-metallophore (Non-Ribosomal Protein) that was 84% similar to histocorrugatin. There was a hydrogen cyanide generating region and a brabantamide NRP gene cluster that were each 100% similar to the reference genes. There was also a gene cluster that was 88% similar to the reference pathway of syringomycin.
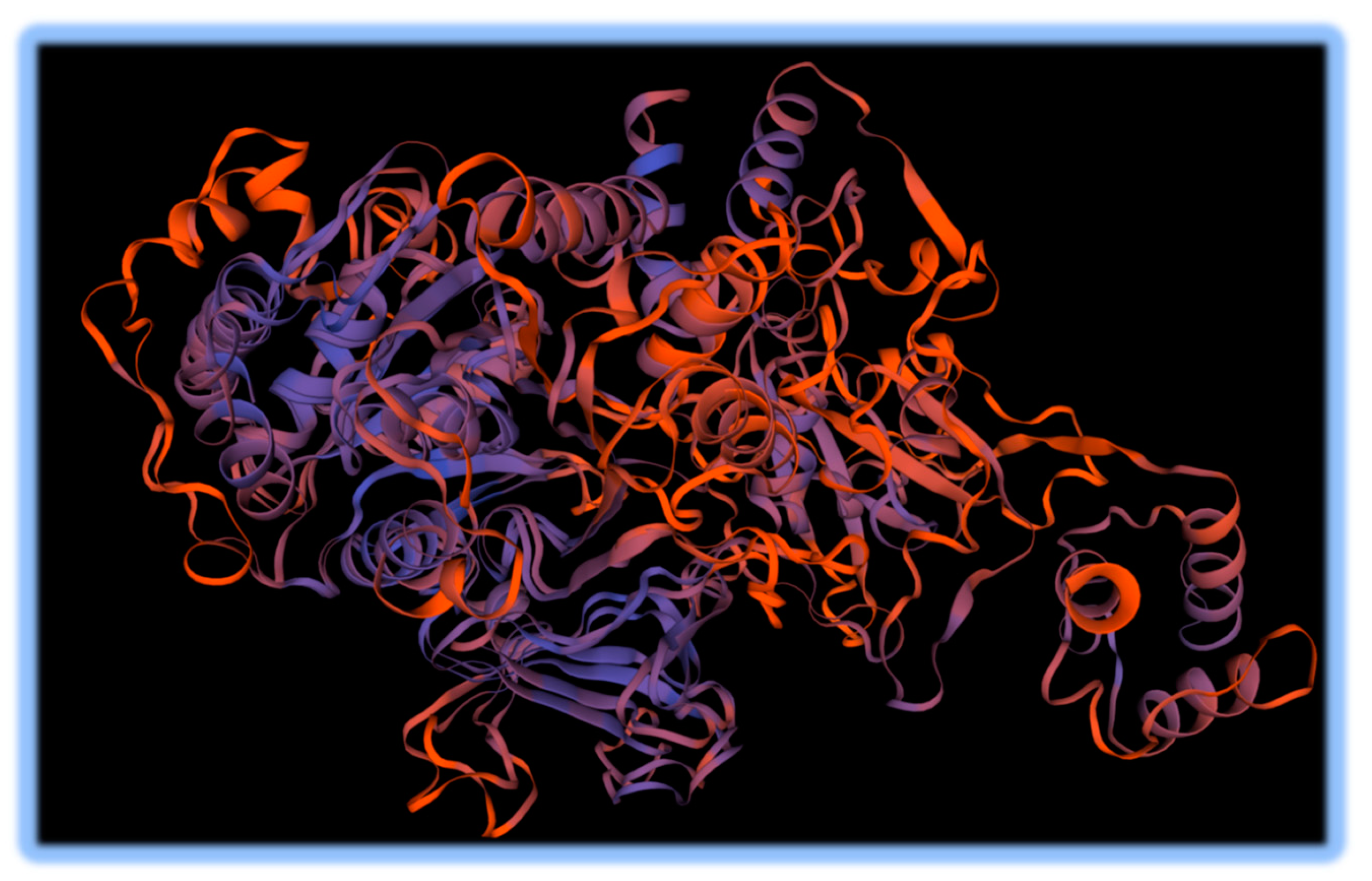
A promysalin synthase- like gene cluster was detected using antiSMASH, which is a Shikimate-derived NRP polyketide. The region was 66% similar to the reference. The gene cluster consisted of 1 core biosynthetic gene and one regulatory gene. Promysalin is a salicylic acid antibiotic compound. To further investigate this promysalin synthase-like gene that differed from the reference and is relevant to plants, a template-based SwissPROT model was constructed (Waterhouse 2018). The result is shown in
Figure 4. Although the sequence identity of the thioester forming conformation of the Enterobactin synthase translated protein template was only 26.29%, the QMEANDisCo (Quality score using the Distance constraint) value for the Enterobactin model was the highest at 0.66. There were higher %ID values for other
Pseudomonas-related templates, however given the distance constraints the Promysalin synthetase from the novel species was less likely to fold in the same way, based on the QMEANDisCo. Enterobactin is a siderophore that is selectively antimicrobial and facilitates infections by other pathogens (Keough 2016).
The gene for promysalin synthase in the candidate novel bacterium was found to be homologous to the gene in Pseudomonas putida and Pseudomonas fluorescens when high sequence similarity was prioritized. The product is known to inhibit growth of Pseudomonas aeruginosa (Keohane 2018, Li 2011). Promysalin disrupts cell membrane of gram-positive bacteria (Kaduskar 2017). It has a synergistic effect with chlorhexidine against gram negative bacteria. Plants such as Arabidopsis thaliana, Vitis vinifera, Nicotiana tabacum, and Populus tomentosa were also found to produce similar proteins according to the RCSB PDB results (Berman et al 2000).
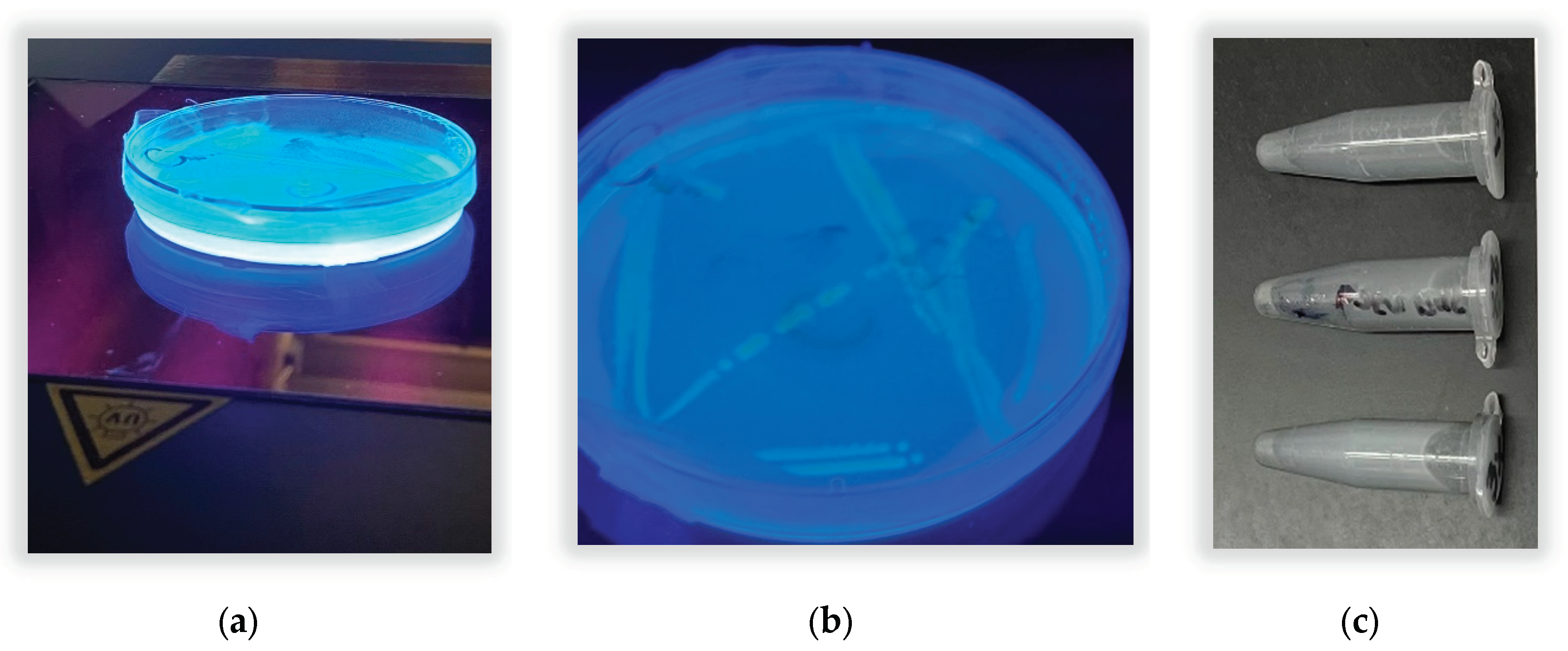
There were also 3 putative pyoverdine domains detected using antiSMASH. The qualitative test for pyoverdine siderophore production using UV light was positive. The result is shown in
Figure 5. Fluorescence under UV light indicated that the novel bacterium produced a blue- green fluorescent compound, giving similar glowing results as reported by Banerjee et al 2017 and Martin et al 2011 in their studies of
P. aeruginosa and
P. fluorescens. Pyoverdine is key for iron acquisition and could improve iron solubilization and availability to plants.
Siderophore production in Pseudomonas sp. 21-C3-ER may benefit plant roots because it assists with iron solubilization in soil; pyoverdine is also a strong antimicrobial compound (Bonneau et al 2020). Chelation of iron by siderophores are especially relevant at high pH (Rajkumar 2010), e.g., in calciferous soils in arid regions. In one study, Pseudomonas fluorescens was able to take up small quantities of Uranium from U ores (Rajkumar 2010). The potential for increasing heavy metal extraction by plants with bacterial inoculation is of interest.
The Pyoverdine domain is also related to virulence because it is used as a chemical signal. Interestingly, in the biosynthetic genes of the pyoverdine gene clusters that differed from P. fluorescens Pf-5 and Pseudomonas sp. SXM-1 in the RAST analysis, one of the top BLASTP hits was L-ornithine 5-monooxygenase from P. fluorescens. Another interesting BLASTP hit from the pyoverdine gene cluster that differed from the reference was the formylglycene-generating enzyme from P. brassicacearum and a Sulfatase-modifying factor enzyme (Wang et al Conserved Domain Database). There were two medium confidence hits for transcription factor binding sites for the Pyoverdine synthase gene clusters: Ctg2_25 ZuR Zinc-responsive repressor and Ctg2_36 Pleiotropic regulatory for antibiotic production.
Metabolic analysis using RAST indicated genes were present for ammonification, pyoverdine production, and antimicrobial resistance. The detected subsystems covered 27% of the genome. There were 33 features identified related to phosphate metabolism. There were features detected related to sulfate assimilation, Thioredoxin-disulfide reductase, and inorganic sulfur assimilation. Tolerance to colicin E2 (a bacteriocin), resistance to fluoroquinolones, cobalt-zinc-cadmium resistance, chromium, copper, multidrug resistance efflux pumps, and Streptothricin resistance features were also detected. Within the detected subsystems, gentisate degradation pathways were detected as well as degradation of Benzoate, quinate, n-Phenylalkanoic acid, and Hydroxybenzoate. There were also 49 hits for metabolism of central aromatic intermediates. The proposed novel genome contains 151 functions not found the reference genome Pseudomonas fluorescens Pf-5.
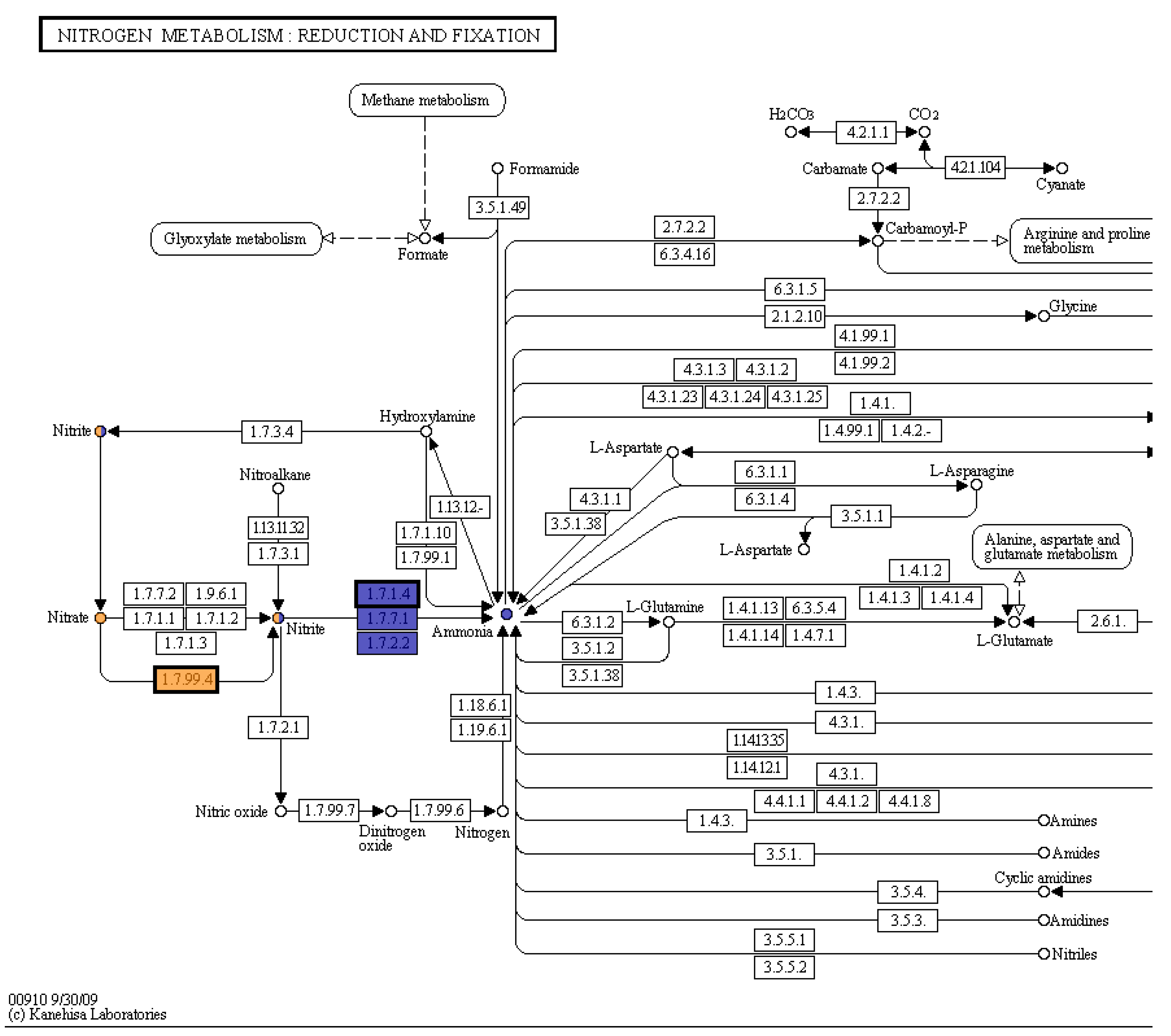
The nitrate reduction test using the sulfanilic acid method was positive (
Figure 5). There was no color change at time point 2, after adding zinc powder. This indicated that the bacteria had an active nitrate reductase. The KEGG pathway map (Kanehisa and Goto 2000) from the whole genome sequence analysis in RAST shows the genes for reduction of nitrate and nitrite to ammonia are present in this organism (
Figure 6).
The KEGG database gives organized functional information about genes. The process in Pseudomonas sp. 21-C3-ER is considered incomplete denitrification since nitrate is not reduced all the way to nitrogen gas. This bacterium is involved with ammonification, which could be helpful to plants. Denitrifying bacteria may prevent leaching of nitrates from soil, since ammonia has a higher affinity to soil and organic matter than nitrate (University of Minnesota Extension 2021).
4. Conclusions
Partial 16S sequences are an effective primary screening method for novel soil bacteria, followed by whole genome sequencing. Barcoding methods are helpful for monitoring for potential pathogens and plant growth promoting bacteria, antibiotic resistant bacteria and antibiotic producers.
The purpose of this study was to identify novel microbes performing beneficial ecological functions from agricultural soil. The hypothesis was that novel culturable species with antibiotic production potential could be readily identified from soil in the Nature Canyon at Los Angeles Pierce College. Our proposed novel Pseudomonas sp. strain 21-C3-ER has 92.01% ANI to the nearest species match. Therefore, our hypothesis that the isolate was a novel species was supported by genomic evidence. We propose the name Pseudomonas fernandeno, a candidate novel nitrogen reducing species with genomic features related to polyketide synthesis and post translationally modified proteins. The rods are approximately 1 µm in length. The bacterium was isolated from fallow agricultural soil in a naturalized preserve with clay texture and high organic matter at Pierce College. The bacterium was named in honor of native people of the San Fernando Valley. The bacterium was approved for deposit at USDA NRRL. The type stain is 21-C3-ER.
Author Contributions
Conceptualization, S.S.; methodology, B.N., S.S..; formal analysis, S.S., E.R., G.W., J.M.F.; investigation, S.S.,T.S., M.B., E.C., M.M., L.V., E.R., M.K., D.M., K.S., R.P., A.L.B., G.W., J.M.F.; resources, B.N.; data curation, S.S.; writing—original draft preparation, S.S., M.B., M.K., L.V., A.L.B; writing—review and editing, S.S., T.S., J.M.F.; visualization, S.S., E.C., M.K., B.P.; supervision, S.S.; funding acquisition, B.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by training and resources from the National Science Foundation IUSE grant, NSF DUE-1821657 awarded to Bruce Nash and Ray Enke in part to support Community College faculty. This material is based upon work supported by the National Science Foundation under Grant Nos. DBI-0735191, DBI-1265383, and DBI-1743442 which supported iPlant Collaborative and DNA Subway development. .
Data Availability Statement
The full 16S sequence is available on NCBI GenBank: PP087912.1. The Sequence Read Archive for the Whole Genome Sequence is available on NCBI GenBank: BioProject PRJNA105.
Acknowledgments
Thank you to Pierce College Chemistry Department, Pierce College Life Sciences Department, Amgen Biotech Experience, and Amgen Foundation.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Haas, Michael J., et al. “Soil Bacteria Could Improve Crop Yields, via Fungi.” Cornell Chronicle, 6 Apr. 2021. https://news.cornell.edu/stories/2021/04/soil-bacteria-couldimprove-crop-yields-fungi.
- Lupica, Diana. “UK to Lose Soil Fertility in 30 Years, Says Environment Secretary.” Plant Based News, 1 Oct. 2020. https://plantbasednews.
- Savanah St. Clair, S., Saraylou, M. and Maine, E. 2021. Introduction to Soil Science Lab Manual. 3rd ed.
- Blackwood, K S et al. “Evaluation of recA sequences for identification of Mycobacterium species.” Journal of clinical microbiology vol. 38,8 (2000): 2846-52. [CrossRef]
- Fernandez, F. G. 2021. Understanding nitrogen in soils. Extension at the University of Minnesota. https://extension.umn.edu/nitrogen/understanding-nitrogen-soils.
- Soil Survey Staff, Natural Resources Conservation Service, United States Department of Agriculture, Web Soil Survey. Available online: https://websoilsurvey.sc.egov.usda.gov/ (accessed on 3 April 2024).
- Aziz, R.K., Bartels, D., Best, A.A. et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9, 75 (2008). [CrossRef]
- Bankevich, Anton et al. “SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.” Journal of computational biology: a journal of computational molecular cell biology vol. 19,5 (2012): 455-77. [CrossRef]
- Bonneau, A., Roche, B. & Schalk, I.J. Iron acquisition in Pseudomonas aeruginosa by the siderophore pyoverdine: an intricate interacting network including periplasmic and membrane proteins. Sci Rep 10, 120 (2020). [CrossRef]
- Boyd A, Chakrabarty AM. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. JInd Microbiol. 1995 Sep;15(3):162-8. [CrossRef] [PubMed]
- Dyer, Joseph M, and Valerie M Foy. “Revealing The Unseen: A Review of Wood's Lamp in Dermatology. “The Journal of clinical and aesthetic dermatology vol. 15,6 (2022): 25-30.
- Genome Analysis Egalitarian. In Proceedings of the 2014 Annual Conference on Extreme Science and Engineering Discovery Environment (XSEDE '14). Association for Computing Machinery, New York, NY, USA, Article 70, 1–3. [CrossRef]
- H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne, The Protein Data Bank (2000)Nucleic Acids Research 28: 235-242. [CrossRef]
- Haas, Michael J., et al. “Soil Bacteria Could Improve Crop Yields, via Fungi.” Cornell Chronicle, 6 Apr. 2021. https://news.cornell.edu/stories/2021/04/soil-bacteria-couldimprove-crop-yields-fungi.
- Kaduskar, R.D., Scala, G.D., Al Jabri, Z.J.H. et al. Promysalin is a salicylate-containing antimicrobial with a cell-membrane-disrupting mechanism of action on Gram-positive bacteria. Sci Rep 7, 8861 (2017). [CrossRef]
- Keogh, Damien et al. “Enterococcal Metabolite Cues Facilitate Interspecies Niche Modulation and Polymicrobial Infection. “Cell host & microbe vol. 20,4 (2016): 493-503. [CrossRef]
- Li, Wen et al. “Promysalin, a salicylate-containing Pseudomonas putida antibiotic, promotes surface colonization and selectively targets other Pseudomonas.” Chemistry & biology vol. 18,10 (2011): 1320-30. [CrossRef]
- Lupica, Diana. “UK to Lose Soil Fertility in 30 Years, Says Environment Secretary.” Plant Based News, 1 Oct. 2020. https://plantbasednews.org/news/uk-lose-soil-fertility-saysenvironment-secretary/.
- Martin, N. H., et al. "When cheese gets the blues: Pseudomonas fluorescens as the causative agent of cheese spoilage. “Journal of Dairy Science 94.6 (2011): 3176-3183.
- Muhammadi, and Nuzhat Ahmed. “Genetics of bacterial alginate: alginate genes distribution, organization and biosynthesis in bacteria “Current genomics vol 83 (2007): 191-202.
- Promysalin Elicits Species-Selective Inhibition of Pseudomonas aeruginosa by Targeting Succinate Dehydrogenase. Colleen E. Keohane, Andrew D. Steele, Christian Fetzer, JittasakKhowsathit, Daria Van Tyne, Lucile Moynié, Michael S. Gilmore, John Karanicolas, Stephan A. Sieber, and William M. Wuest. Journal of the American Chemical Society 2018 140 (5), 1774-1782. [CrossRef]
- Tamang, M., et al. 2021. Unravelling the Role of PGPR Pseudomonas fluorescens in Semi-Arid Soils of the Rio Grande Valley. UTRGV ScholarWorks. https://scholarworks.utrgv.edu/cgi/viewcontent.cgi?article=1980&context=etd.
- Uwe Hilgert, Sheldon McKay, Mohammed Khalfan, Jason Williams, Cornel Ghiban, and David Micklos. 2014. M: DNA Subway.
- et al. (2023),"The conserved domain database in 2023”, Nucleic Acids Res.51(D)384-8.
- Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F.T., de Beer, T.A.P., Rempfer, C., Bordoli, L., Lepore, R., Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296-W303 (2018).
- Yoon, Seok-Hwan, et al. "Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. “International journal of systematic and evolutionary microbiology 67.5 (2017): 1613.
- Kanehisa, M, and S Goto. “KEGG: kyoto encyclopedia of genes and genomes.” Nucleic acids research vol. 28,1 (2000): 27-30. [CrossRef]
- Ha, Sung Min et al. “Application of the Whole Genome-Based Bacterial Identification System, TrueBac ID, Using Clinical Isolates That Were Not Identified With Three Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Systems.” Annals of laboratory medicine vol. 39,6 (2019): 530-536. [CrossRef]
- Banerjee S, Batabyal K, Joardar SN, Isore DP, Dey S, Samanta I, Samanta TK, Murmu S (2017) Detection and characterization of pathogenic Pseudomonas aeruginosa from bovine subclinical mastitis in West Bengal, India, Veterinary World, 10(7): 738-742.
- Kai Blin, Simon Shaw, Hannah E Augustijn, Zachary L Reitz, Friederike Biermann, Mohammad Alanjary, Artem Fetter, Barbara R Terlouw, William W. Metcalf, Eric JN Helfrich, Gilles P van Wezel, Marnix. H Medema & Tilmann Weber. Nucleic Acids Research (2023). [CrossRef]
- Buxton, Rebecca. "Nitrate and nitrite reduction test protocols." American Society for Microbiology (2011): 1-20.
- Rajkumar, Mani, et al. "Potential of siderophore-producing bacteria for improving heavy metal phytoextraction." Trends in biotechnology 28.3 (2010): 142-149.
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. (1990) “Basic local alignment search tool.” J. Mol. Biol. 215:403-410. PubMed.
- Gislason, April S, and Teri R de Kievit. “Friend or foe? Exploring the fine line between Pseudomonas brassicacearum and phytopathogens.” Journal of medical microbiology vol. 69,3 (2020): 347-360. [CrossRef]
- Plaster, Edward J. “Physical Properties of Soil.” Soil Science & Management, Delmar, Cengage Learning, 2013.
- Zhang, Huanhuan et al. “Simultaneous Nitrogen Removal and Plant Growth Promotion Using Salt-tolerant Denitrifying Bacteria in Agricultural Wastewater.” Microbes and environments vol. 37,3 (2022): ME22025. [CrossRef]
- Lebonah, D. E., et al. "DNA barcoding on bacteria: A review." Advances in Biology 2014 (2014).
- Jain, C., Rodriguez-R, L.M., Phillippy, A.M. et al. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9, 5114 (2018). [CrossRef]
- Nandi, Munmun et al. “Pseudomonas brassicacearum strain DF41 kills Caenorhabditis elegans through biofilm-dependent and biofilm-independent mechanisms.” Applied and environmental microbiology vol. 82,23 (2016): 6889-6898. [CrossRef]
- Tripathi N, Sapra A. Gram Staining. [Updated 2023 Aug 14]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK562156/.
- Mishra, Bibhuti Bhusan, et al., editors. Agriculturally Important Microorganisms : Mechanisms and Applications for Sustainable Agriculture. CRC Press, 2022.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).