1. Introduction
Juvenile polyposis syndrome (JPS) is an autosomal dominant rare disorder characterized by multiple juvenile polyps in the gastrointestinal tract, typically beginning in childhood or adolescence[
1]. JPS is caused by mutations in certain genes that regulate the growth of cells in the colon. The most associated genes are
Smad4 and
BMPR1A[
2].
Smad4 is a tumor suppressor gene, and mutations in this gene were the most frequent among cases of JPS.
BMPR1A is another tumor suppressor gene, and mutations in this gene were less frequent among cases of JPS[
3].
PTEN is another gene that is less commonly involved in JPS[
3].
JPS is characterized by multiple prominent juvenile polyps in the gastrointestinal tract that can increase the risk of developing colon cancer if not removed [
4]. Colorectal cancer (CRC) is one of the most common cancers and the world's second leading cause of cancer-related death[
5]. CRC is caused by the accumulation of genetic and epigenetic changes that transform normal colonic mucosa into adenocarcinoma[
6]. Although modern research has shed light on the molecular mechanism of CRC and provided improved screening strategies, the prevalence of CRC is still increasing [
5].
Clinically, juvenile polyposis syndrome is diagnosed by having five or more juvenile polyps throughout the gastrointestinal tract or any number of juvenile polyps and a positive family history of juvenile polyposis[
4,
7].
Most juvenile polyps are hamartomatous, but they have the potential to turn cancerous if not removed[
8]. The risk of developing colon cancer in individuals with JPS is estimated to be between 11% to 86%, The risk is higher in individuals who have a large number of polyps, polyps that are located in the proximal (upper) part of the colon, or polyps that are larger in size. Most of this increased risk is attributed to colon cancer, but stomach, upper gastrointestinal tract, and pancreas cancers have also been reported[
3,
9,
10,
11,
12]. Even among patients of the same age, the polyp shapes and sizes vary, and it is possible to notice this even among members of the same family who have JPS[
13].
Some individuals with JPS may have a small number of polyps, while others may have hundreds or even thousands of polyps[
14]. Some polyps may be small and easily removed during a colonoscopy, while others may be large and require surgery. The size of the polyps is also an important factor in determining the risk of colon cancer[
15].The specific genetic mutation that results in the disorder can impact the variations seen in JPS. Variations in the number, size, and location of the polyps in the colon can be caused by different genetic mutations. For instance, people with JPS accompanied by
Smad4 gene mutations typically have more polyps than people with JPS accompanied by BMPR1A gene mutations[
16,
17].
JPS mouse models (transgenic and knockout models) have been developed for several genes associated with JPS, including
Smad4, BMPR1A, and PTEN[
18,
19,
20]. These models have been used to study the development and progression of polyps in JPS and the potential therapeutic effects of various drugs and treatments[
21]. This approach has been used to identify several genetic modifiers for JPS, including genes involved in the Wnt signaling pathway, which regulates cell growth and division[
22]. Another pathway identified as a modifier in JPS is the TGF-beta signaling pathway[
23]. The TGF-beta signaling pathway is involved in the regulation of cell growth, differentiation, and apoptosis[
23]. Researchers have also identified genetic variants in other pathways, such as the Hedgehog signaling pathway and the Notch signaling pathway, as modifiers of JPS, and mutations in genes that regulate these pathways are associated with the development of colon cancer[
24,
25].
The genetic diversity of Collaborative Cross (CC) mice, like that of human populations, makes them a powerful tool for biomedical research[
26]. CC mice are bred to have a wide range of genetic variation, making them an ideal model for studying complex diseases and traits that are difficult to study using traditional inbred mouse strains[
27]. CC mice are produced from intercrossing and outcrossing eight strains, including laboratory and wild-derived strains, resulting in a highly diverse population[
28]. CC mice can be used to identify genetic modifiers that modify the effects of disease-causing mutations, making them an effective tool for identifying genetic modifiers that could be related to human disease, providing new therapeutic targets for researchers[
28]. CC mice can also investigate complex traits such as behavior and immune system function[
29]. Crossing CC mice with KO mice has been a practical approach to identifying genetic modifiers[
30]. Here, we present our attempt to identify genetic modifiers for JPS that influence the number and size of polyps by crossing CC mice with
Smad4 KO mice.
2. Results
2.1. The Effect of Smad4 Heterozygous Kock Out in the General Population.
Our experiment's overall population of F1 mice shows a significant increase in intestinal polyps, as shown in
Figure 1A (p=0.000). This was seen in both the small intestine and the colon, as shown in
Figure 1B (p=0.000) and 1C (p=0.000). This matches the previous reports on
Smad4 as a model for intestinal polyposis[
31].
2.2. Sex effect
The effect of
Smad4 Knockout on polyp counts when tested in males and females separately. the polyp counts significantly increased in KO males and females compared to their WT counterparts in the small intestine (male p=0.001, female p=0.000), the colon (male p=0.001, female p=0.000), and the whole intestinal tract (male p=0.000, female p=0.000), presented in
Figure 2. Male mice tend to have more polyps than females, which is not statistically significant.
2.3. Line Genetic Effect
The results of our study reveal a nuanced impact of
Smad4 knockout across various F1 mouse lines, as depicted in
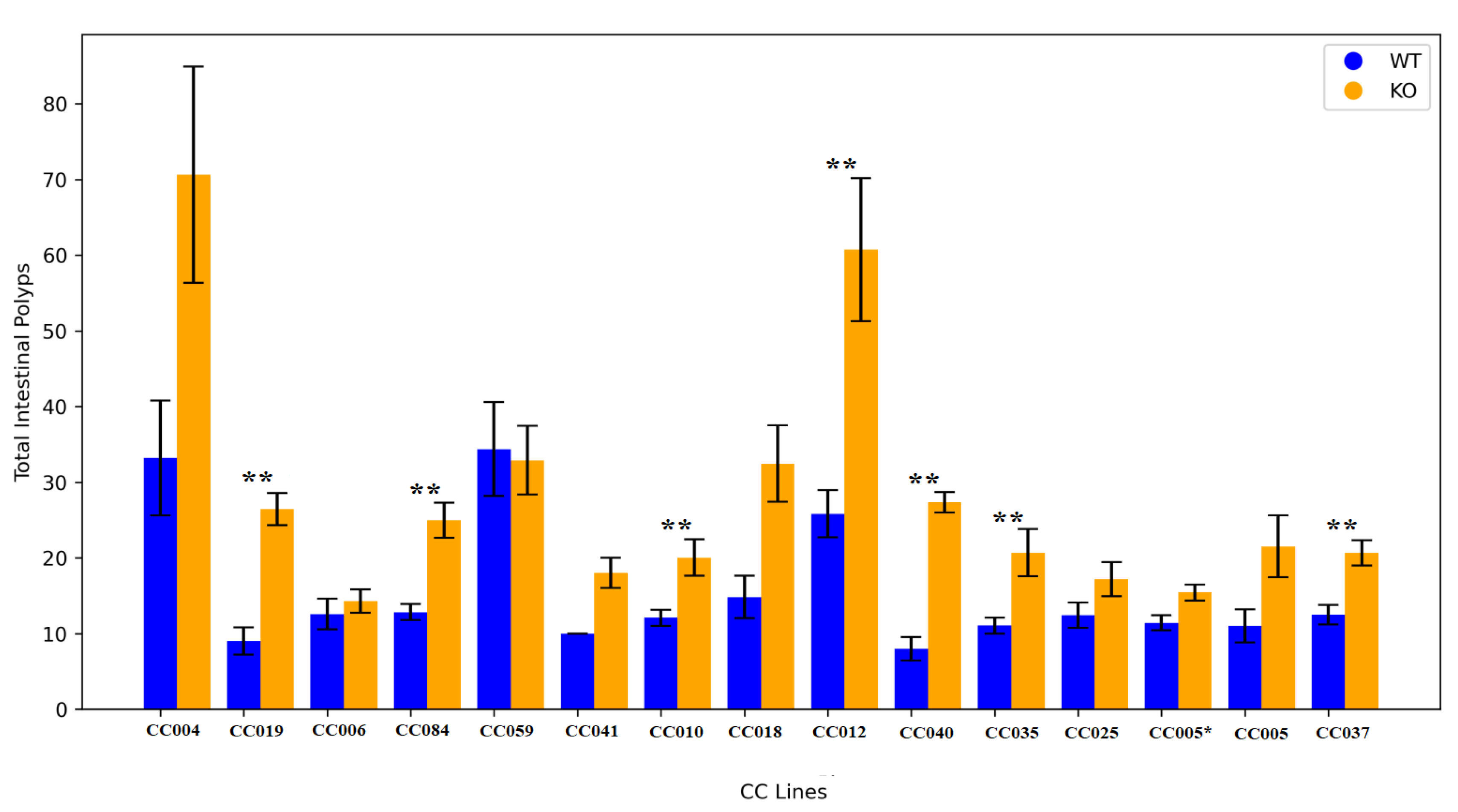
Figure 3. Notably, three lines CC006, CC059, and CC041 exhibited no statistically significant difference in polyp counts between knockout (KO) and wild-type (WT) mice in the small intestine. In contrast, lines CC004 (p=0.035), CC005 (p=0.017), and CC018 (p=0.009) demonstrated a significant increase in small intestine polyp counts in the KO group compared to the WT group.
Moreover, lines CC025 (p=0.044) and CC005* (p=0.001) displayed a statistically significant elevation in colon polyp counts. Further analysis revealed that in lines CC037 (p=0.001), CC040 (p=0.007), CC019 (p=0.000), CC084 (p=0.000), CC010 (p=0.004), CC012 (p=0.002), and CC035 (p=0.005), there was a significant increase in mean polyp counts in both the small intestine and colon of the KO mice compared to their WT counterparts, CC005 and CC005* are cousin strains formerly stated in our previous works as IL711 and IL6018 respectively.
These findings underscore the diverse responses to Smad4 knockout among distinct mouse lines, emphasizing the importance of genetic variability in influencing phenotypic outcomes. The observed variations in polyp counts may be attributed to genetic and environmental factors that warrant further investigation. The provided p-values indicate the degree of statistical significance, offering valuable insights into the strength of the observed effects. These results contribute to our understanding of the intricate role of Smad4 in polyp formation, shedding light on potential avenues for future research and therapeutic exploration.
2.4. Multimorbidity Heatmaps of Polyp Counts, Body Weight
Understanding the relationship between different physiological variables is crucial for determining the overall health of an organism. Studying the correlation between organ weights and disease pathology is significant in biomedical research. An essential aim of our proposed research was to investigate the effect of host genetic background interaction with Smad4 knockout on disease multimorbidity to understand better the coexistence of multiple disease states in different genetic backgrounds, so in this study, we examine the correlation between the number and size of polyps in the intestines of WT mice and KO mice from various CC lines. We aim to identify any significant differences in the correlation patterns between the two groups, which may indicate the role of specific genes in developing polyps and related diseases. These traits were converted into heat maps and then used to investigate relationships between trait intensity and transformation. An ideal positive correlation is represented by red (1), while an ideal negative correlation is represented by blue (-1). Our results provide valuable insights into the genetic basis of intestinal polyps and may have implications for developing novel therapies for related diseases.
2.5. General Population Correlations
Our investigation into the KO population compared to WT mice revealed that correlation patterns remained consistent between the two groups. A noteworthy positive correlation was observed between the development of colon C polyps and small intestinal polyps in the KO mice population. Additionally, a positive correlation was identified between total small intestinal B polyps and the overall count of intestinal polyps. This positive association extended to specifically include Small Intestinal B polyps, highlighting an interconnected relationship between these variables in
Smad4 knockout. These findings contribute to our understanding of the complex interactions between different polyp types in knockout mice, offering insights into potential factors influencing the development of polyps in distinct regions of the intestines. The data illustrating these correlation patterns is presented in
Figure 4.
2.6. Sex Variation
Distinctive correlation patterns emerged in the female subset of knockout (KO) mice. Notably, a positive correlation was established between small intestinal polyps and Small Intestinal B polyps, underscoring an interconnected relationship between these variables in
Smad4 knockout. A positive correlation was also identified between total colon polyps and Small Intestinal C polyps in female KO mice. However, it is noteworthy that in the KO group, a previously observed positive correlation between total intestinal polyps and Small Intestinal A polyps was no longer evident. These gender-specific correlations in females provide valuable insights into the nuanced effects of
Smad4 knockout on the development of polyps, shedding light on potential variations in the relationships between different polyp sizes. The specific details of these correlations are visually represented in
Figure 5A and 5B.
Our investigation revealed specific correlation patterns of particular significance in the male subset of knockout (KO) mice. A positive correlation was identified between colon polyps and the C portion of small intestinal polyps. Furthermore, a robust positive association persisted in males, linking the counts of total small intestinal B polyps with the overall number of intestinal polyps. The visual representation of these correlations is presented in
Figure 5C and 5D.
2.7. Polyp Counts Correlations in Different Lines
In our expansive exploration across diverse lines, the impact of Smad4 knockout on correlation patterns emerged as a complex and varied interplay influenced by distinct genetic backgrounds. The KO mice from different lines exhibited a range of correlations among various polyp types, revealing a nuanced landscape shaped by underlying genetic factors. The analysis illuminated unique trends in the relationships between polyp counts, providing insight into the intricate genetic interactions influencing the development of intestinal polyps. This line-specific perspective underscores the importance of comprehensively understanding the diverse effects of Smad4 knockout, offering valuable insights into the underlying genetic complexities that contribute to the manifestation of polyp-related phenotypes. The comprehensive depiction of these correlations for each line can be found in Supplementary Figures, providing a visual representation of the intricate genetic interplay in the context of Smad4 knockout across various genetic backgrounds.
2.8. Heritability
This study aimed to discover whether polyp counts and sizes phenotypic variance has a genetic basis in
Smad4 knockout F1 populations.
Table 1 summarizes the heritability (H2) values calculated to answer this question. One-way ANOVA was used to calculate the heritability of sex and genotype-specific characteristics. The different traits calculated are as follows: total polyp counts in the small intestinal and its three segments SB1, SB2 and SB3, colon polyps, the heritability was calculated for different polyp categories (A, B and C) based on size, for both sexes and genotypes.
2.9. Machine Learning
In the realm of two-class classification, the Linear Discriminant Analysis (LDA) algorithm exhibits a moderate level of performance in discerning between two distinct classes. As a linear classifier, LDA thrives when confronted with classes characterized by disparate means and akin covariances. Contrarily, the k-Nearest Neighbors (KNN) algorithm, a non-parametric approach, yields comparatively inferior results, hinting at a dataset potentially needing more robust local patterns conducive to its methodology.
On the other hand, Support Vector Machines (SVM) with a Radial Basis Function (RBF) kernel emerges as the top-performing model among its counterparts, boasting the highest accuracy. The inherent capability of SVM with RBF kernel to delineate intricate decision boundaries renders it well-suited for scenarios necessitating nuanced classification. Meanwhile, the Random Forest (RF) ensemble technique demonstrates a commendable yet slightly inferior performance when juxtaposed with SVM. RF harnesses the power of multiple decision trees to amalgamate their predictions, contributing to its competitive performance.
Transitioning to three-class classification, LDA once again presents a modest performance level akin to its two-class counterpart, relying on linear decision boundaries to discern between the three classes. Conversely, KNN's performance suffers a setback, indicative of the challenges encountered in capturing distinctions among the three classes. The parameter selection of k in KNN aids in striking a balance between bias and variance, albeit without achieving commensurate performance levels with other models.
In contrast, SVM with RBF kernel sustains its supremacy in the three-class scenario, underscoring its adeptness in handling intricate relationships within the data. While Random Forest continues to deliver respectable results in this expanded classification context, SVM remains the preeminent performer, capitalizing on the ensemble nature of RF to encapsulate the manifold patterns present within the dataset. Detailed results for the regression models are presented in
Table 2.
3. Discussion
Smad4 is a protein that is critical in the TGF-beta signaling pathway, which regulates cell growth, differentiation, and apoptosis[
32].
Smad4 knock-out (KO) mice are used as a model to study the effects of the loss of function of this protein[
33]. Our study shows that the effect of
Smad4 KO varies between different collaborative cross mice, which is essential for understanding the genetic complexity of the TGF-beta signaling pathway.
Overall, this study highlights the importance of considering genetic background when studying the effects of gene knock-out and the need for further research to fully understand the genetic basis of intestinal polyps and related diseases. Using the Genome-Wide Association Studies (GWAS), approach for modifier screening can provide valuable insights into the genes and pathways involved in these diseases. It can help identify potential therapeutic targets for preventing and treating these diseases.
This study's findings also have significant implications for understanding the genetic basis of colon polyps and related diseases. The different correlation patterns observed between different lineages and between sexes per lineage also have highlighted the importance of considering genetic background when examining the impact of gene knockout and the development of these diseases in the general population, implying that spleen weight may have a potentially protective effect on the development of colonic polyps. The positive correlation between adjusted liver weight and small bowel polyps in WT mice, on the other hand, suggests that liver weight may contribute to the development of these polyps. The absence of a positive correlation between adjusted spleen weight and adjusted organ weights in the KO population suggests that This correlation pattern is disrupted by Smad4 knockout. Both WT and KO mice indicate that the liver is important in developing this type of polyp. The correlation between adjusted liver weight and colon polyp count, on the other hand, varied between different lineages and between sexes within each lineage, implying that the liver's role in the development of colon polyps may be more complex and dependent on genetic background.
The positive correlation between adjusted heart weights and brain and kidney weights in WT and KO populations also highlights the potential relationship between heart health and brain and kidney function. However, the KO population's negative correlation between body weight and adjusted weight of all organs except adjusted spleen weight suggests that obesity may contribute to the development of polyps and related diseases. In general, our findings shed light on the genetic basis of colon polyps and related diseases, emphasizing the importance of additional research to identify specific genes and signaling pathways involved in developing these diseases from various genetic backgrounds. This knowledge could aid in the development of new therapies.
Linear Discriminant Analysis (LDA) yielded an accuracy of 0.67 and a Kappa statistic of 0.33, suggesting its effectiveness in distinguishing between classes 'I' and 'II.' The k-Nearest Neighbors (KNN) model, with an optimal k value of 9, exhibited a lower accuracy of 0.55, possibly indicating challenges in capturing complex relationships. The Support Vector Machines with Radial Basis Function Kernel (SVM-RBF) performed exceptionally well, achieving an accuracy of 0.69 and a Kappa statistic of 0.38, showcasing its robustness in handling non-linear decision boundaries in binary classification tasks in multiclass classification tasks. Linear Discriminant Analysis (LDA) demonstrated an accuracy of 0.62 and a Kappa statistic of 0.098. The k-Nearest Neighbors (KNN) model, with an optimal k value of 9, achieved an accuracy of 0.62. Support Vector Machines (SVM) with optimal parameters C = 0.25 and sigma = 0.0774 provided an accuracy of 0.63, while Random Forest (RF) achieved an accuracy of 0.62.
4. Materials and Methods
4.1. Ethical Aspects of the Project
All animal experiments in this study were compliant with national standards for the care and use of laboratory animals, and experiment was reviewed and approved by Tel Aviv University's Institutional Animal Care and Use Committee (IACUC), with an approved number (01-19-044). Mice were monitored daily for their overall health status. Mice that showed loss of around 10% of their BW between two measure points, or 20% overall of their initial body weight, or which were observed to be suffering (less movement and activity) and based on the consultation with the Veterinarian at the small animal unit, were terminated.
4.2. Generation of F1 Crosses
The CC mouse lines were developed and maintained at conventional environmental conditions at the animal facility of Tel-Aviv University (TAU) by inbreeding for around 20 generations as described earlier[
34,
35]. The C57BL/6 J-
Smad4tm1Mak mouse line was purchased from the Jackson Laboratory (Bar Harbor, Maine, USA).
F1 mice were produced by a cross of females from 20 CC lines available at the Tel-Aviv animal facility with C57BL/6 J-
Smad4tm1Mak males. After PCR analysis for the
Smad4 gene genotype, 499 F1 mice from 14 lines were identified and included in the study for further assessment and analysis. The mice cohort we used is presented in
Table 3.
4.3. Mouse Housing and Diet
Mice were housed in the animal facility at the Sackler Faculty of Medicine, Tel-Aviv University (TAU), according to the standard protocol approved by the TAU Animal Use and Care Committee (01-19-044). Mice were housed on hardwood chip bedding in open-topped cages, segregated by sex and CC lineage, maintained under a 12-hour light/dark cycle (6:00 a.m. 6:00 p.m.) at 221°C and fed tap water and standard rodent chow-fed ad libitum (TD.2018SC, Teklad Global, Harlan Inc., Madison, WI, USA; contains %Kcal from fat 18%, protein 24%, carbohydrate 58%) since weaning at age three weeks until experiment termination at 80 weeks of age.F1mice were monitored for their overall health status.
4.4. Genomic DNA Extraction and Genotyping
The NaOH Extraction method was used to extract genomic DNA. As referenced in [
36]. In the DNA preparation process, 3-4 mm pieces of the tail were trimmed and then placed into an Eppendorf tube. Subsequently, a solution comprising 75ul of 25 NaOH and 0.2 mM EDTA was added to each sample. The samples were then meticulously placed within a thermocycler and subjected to a temperature of 98ºC for a duration of 1 hour, after which the temperature was lowered to 15°C and maintained at this level until the subsequent steps. Following the thermal treatment, 75ul of a 40 mM Tris HCl solution with a pH of 5.5 was precisely added to the samples. To separate the components, the samples were centrifuged at 4000rpm for a duration of 3 minutes. Finally, aliquots were extracted from the samples for PCR analysis.
4.5. Genotyping of F1 Mice
Mice were genotyped using a PCR protocol employing specific sets of primers. The primer sets utilized were as follows:
Primer 30403 (5’ - TGT AGT TCT GTC TTT CCT TCC TG – 3’)
Primer 30404 (5’ - ACT GAC CTT TAT ATA CGC GCT TG – 3’)
Primer oIMR2088 (5’ - AGA CTG CCT TGG GAA AAG CG – 3’)
PCR genotyping involved two distinct reactions denoted as Reaction A and Reaction B:
Reaction A: Primers 30403 and 30404 were employed to amplify a specific 200 bp segment from the wild-type (WT) copy of the Smad4 gene.
Reaction B: Primers 30404 and oIMR2088 were used to generate a PCR 300bp product indicative of the knockout (KO) Smad4 genotype.Both reactions constitute a touchdown phase. Afterward, the PCR resumed with denaturation at 94.0°C, annealing at 60.0°C, and extension at 72.0°C for 30 cycles. Finally, an extension step was conducted at 72.0°C, followed by a hold step at 10.0°C.
4.6. Tissue Collection
At the time of termination (80 weeks of age), the F1 mice were terminated and culled by CO2 protocol. The body weight of the mouse was recorded as the final body weight, this was used to calculate body weight change during the experiment using the following formula body weight change = (final body weight – initial body weight) x100% / final body weight. The small intestine and colon were extracted and washed with PBS. The small intestine was divided into segments (SB1-proximal, SB2-middle and SB3-distal), and the colon was kept as a whole, all segments were spread on Whatman cellulose filter papers 3 mm paper.
4.7. Intestine Whole Mounts Preparation and Assessing the Intestinal Polyp Counts
Intestines were fixed in 10 % Neutral Buffered Formalin (NBF) overnight and stained by 0.02% methylene blues described earlier [
37,
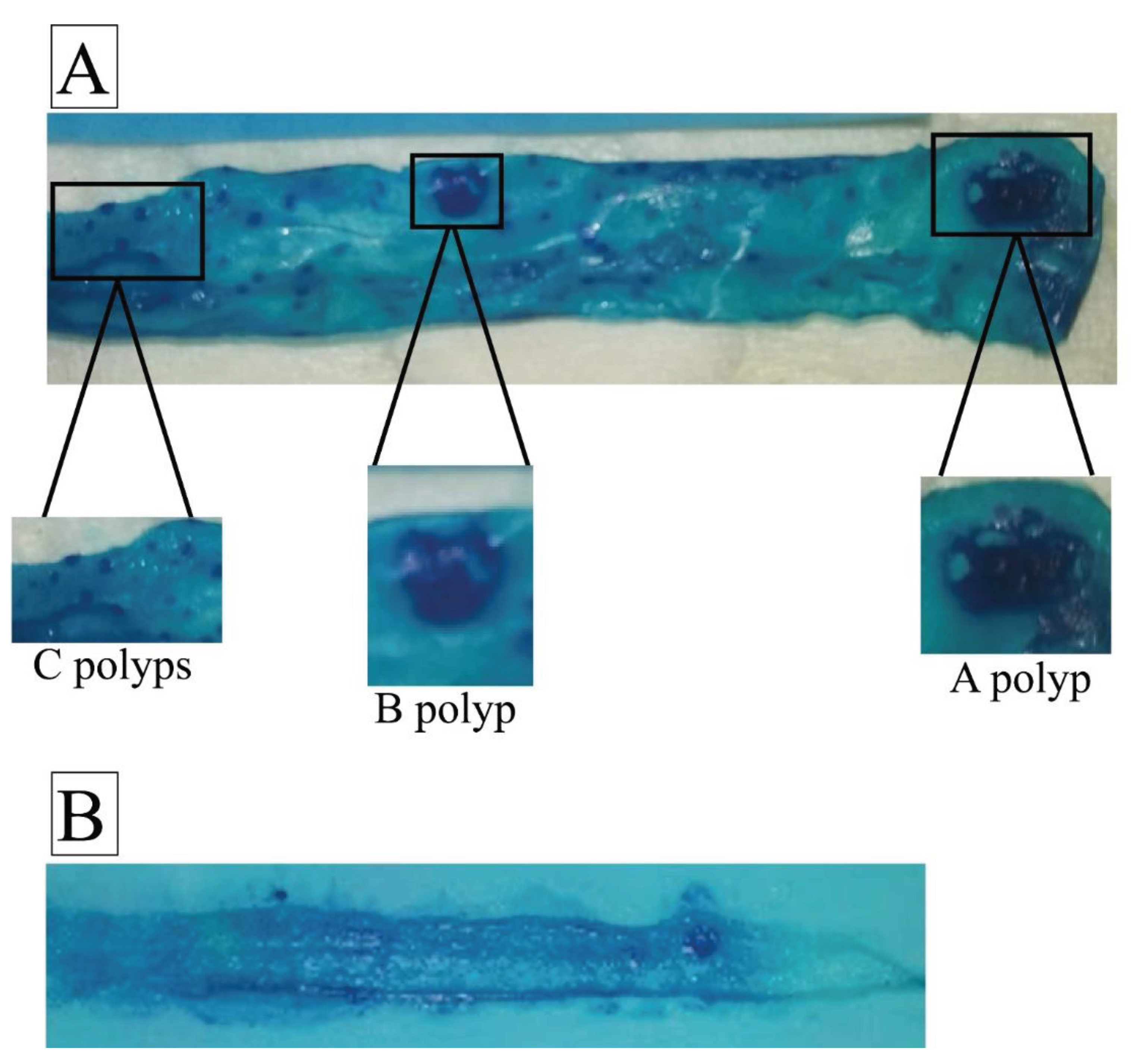
38]. A magnifying glass lens examined the stained intestinal and colon samples. Polyps were counted and categorized based on size, including those measuring greater than 3 mm (A polyps), 1-3 mm (B polyps), and less than 1 mm (C polyps).
Figure 6 visually represents the polyps observed during this experimental assessment.
4.8. Estimation of the Heritability of The Assessed Phenotypes
Heritability measures the fraction of phenotype variability attributed to genetic variation[
39]. Here, we used the ANOVA results to calculate the broad-sense heritability using the formula below:
where H2 is the heritability, Vg is the genetic variance between the CC lines, and Ve is the environment variance. Considering the heritability results, we calculated the genetic coefficient of variation (CVg), which indicates the absolute amount of genetic variation. The CVg was calculated using the standard deviation (SD) results among the CC lines and trait mean overall CC.
4.9. Statistical Analysis
Data analysis was performed using a statistical software package IBM SPSS statistic 23. An Independent sample t-test was carried out to determine if there was a significant difference in polyp counts between the whole population of KO compared with WT mi. The difference in polyp counts between different genotypes in the male and female cohorts was tested for the sex effect. Finally, the difference in mean polyp counts was tested among WT and KO in different lines to measure the line effect. Pearson product-moment correlation coefficient measured the correlation between traits (polyp counts, organ %weights, and body weight changes).
4.10. Machine Learning
Incorporating machine learning (ML) into the analysis of juvenile polyposis syndrome (JPS) provides a robust method for understanding the intricate relationships between genetic factors and polyp counts in the Collaborative Cross (CC) mouse model. The ML pipeline involves data preprocessing, model application, and performance evaluation. The analysis was conducted on a dataset comprising 304 samples, each characterized by 12 predictor variables. The response variable had three classes denoted as 'I', 'II', and 'III' in one analysis while denoted as 'I' and 'II' in the second. The dataset included information related to mouse characteristics, polyp counts, sizes, body weights at different time points, and various other features.
4.11. Data Preprocessing
Before model training, the dataset was examined for missing values and outliers. Any necessary data cleaning or imputation was performed to ensure the integrity of the dataset. The dataset underwent pre-processing steps, including centering and scaling the ten predictor variables. This was done to standardize the features and enhance the performance of specific algorithms.Descriptive statistics were computed to gain insights into the distribution and characteristics of the dataset. This involved calculating summary statistics such as mean, median, minimum, maximum, and quartiles for continuous variables and frequency distributions for categorical variables.
Classification Algorithms:
Several machine-learning classification algorithms were employed to predict the class labels of the samples [
40]. The primary algorithms used were:
a. Linear Discriminant Analysis (LDA):
LDA is a linear classification technique that aims to find a linear combination of predictors that best separates the classes [
40,
41].
b. k-Nearest Neighbors (KNN):
KNN is a non-parametric algorithm that classifies a data point based on the majority class of its k-nearest neighbors in the feature space.
c. Support Vector Machines with Radial Basis Function Kernel (SVM-RBF):
SVM with an RBF kernel is a powerful algorithm for non-linear classification. The hyperparameters C and sigma were tuned to optimize model performance.
d. Random Forest (RF):
RF is an ensemble learning method that constructs a multitude of decision trees during training and outputs the class that is the mode of the classes (classification) of the individual trees [
42]. Additionally, RF is based on bagging and plays an important role in ensemble ML [
42]. RF thus has been implemented in biomedicine research vastly [
43,
44]. In this study, we used “rf” default implementation for RF with 100 trees. Also, in this model, RMSE was used to select the optimal model using the smallest value.
4.12. Model Evaluation
A robust evaluation process was implemented to gauge the models' predictive capabilities. A 70%-30% train-test split ensured unbiased evaluations of unseen data. Key metrics, including RMSE, R-squared, and Mean Absolute Error (MAE), were employed for comprehensive performance analysis [
45].
5. Conclusions
In conclusion, our study delves into the intricate genetic interplay associated with Smad4 knockout in Juvenile Polyposis Syndrome (JPS) using Collaborative Cross (CC) mice, a genetically diverse model. Our findings reveal a significant increase in intestinal polyps in Smad4 knockout mice across the entire population, emphasizing the broad influence of Smad4 on polyposis. Sex-specific analyses demonstrate higher polyp counts in knockout males and females, with distinct correlation patterns. Line-specific effects highlight the nuanced response to Smad4 knockout, underscoring the importance of genetic variability. The heritability analysis underscores a significant genetic basis for polyp counts and sizes, reaffirming the importance of considering the genetic background when studying the effects of gene knockout. Machine learning models, including k-Nearest Neighbors and Linear Regression, identify key predictors, enhancing our understanding of juvenile polyposis genetics. Our comprehensive investigation extends to multimorbidity heat maps, revealing complex relationships between polyp counts, locations, and sizes. The correlation patterns provide valuable insights into the interconnected nature of different polyp types, shedding light on potential factors influencing their development in distinct regions of the intestines. Moreover, our study explores the impact of Smad4 knockout across various CC mouse lines, highlighting diverse responses and emphasizing the need to consider genetic variability in influencing phenotypic outcomes. The observed variations in polyp counts may result from a combination of genetic and environmental factors that warrant further investigation.
The machine learning analysis, employing Linear Discriminant Analysis, k-Nearest Neighbors, Support Vector Machines, and Random Forest, adds a predictive dimension to our understanding. These models showcase varying accuracies in classifying different polyp categories, reinforcing the complexity of the genetic landscape in the context of Smad4 knockout. In essence, our study provides a comprehensive understanding of the intricate genetic factors at play in Smad4 knockout, offering valuable insights into potential therapeutic targets for juvenile polyposis and related diseases. The consideration of genetic variability, as highlighted throughout our research, underscores the importance of personalized and precise approaches in addressing the complexities of polyposis syndromes. Further research into specific genes and signaling pathways involved in these diseases from various genetic backgrounds could pave the way for innovative therapies and preventive strategies.
Author Contributions
Conceptualization, F.A.I., A.N. and I.A.E.-N.; Methodology, O.Z., N.Q., I.M.L.; Validation, F.A.I.; Investigation, O.Z. and I.M.L.; Resources, F.A.I.; Data Curation, O.Z., I.M.L., K.M. and O.Z.; Writing—Original Draft Preparation, O.Z., K.M. and I.M.L.; Writing—Review and Editing, O.Z. and F.A.I.; Supervision, F.A.I. and A.N.; Project Administration, F.A.I.; Funding Acquisition, F.A.I. and I.A.E.-N. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by a core fund from Tel Aviv University and the Department of Oral and Maxillofacial Surgery, Baruch Padeh Medical Center, Poriya, Israel and Ministry of the Negev, Galilee, and National Resilience.
Institutional Review Board Statement
All animal experiments in this study were compliant with national standards for the care and use of laboratory animals, and the experiment was reviewed and approved by Tel Aviv University’s Institutional Animal Care and Use Committee (IACUC), with an approved number (01–19–044).
Conflict of Interest
The authors declare no conflict of interest.
Ethics Statements
All animal experiments conducted in this study adhered to national standards governing the care and utilization of laboratory animals. The experiment was thoroughly reviewed and received approval from Tel Aviv University's Institutional Animal Care and Use Committee (IACUC) under the designated reference number 01–19–044.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
References
- Van Hattem, W.A.; Brosens, L.A.A.; De Leng, W.W.J.; Morsink, F.H.; Lens, S.; Carvalho, R.; Giardiello, F.M.; Offerhaus, G.J.A. Large Genomic Deletions of SMAD4, BMPR1A and PTEN in Juvenile Polyposis. Gut 2008, 57, 623–627. [Google Scholar] [CrossRef]
- van Hattem, W.A.; Langeveld, D.; de Leng, W.W.J.; Morsink, F.H.; van Diest, P.J.; Iacobuzio-Donahue, C.A.; Giardiello, F.M.; Offerhaus, G.J.A.; Brosens, L.A.A. Histological Variations in Juvenile Polyp Phenotype Correlate with Genetic Defect Underlying Juvenile Polyposis. Am J Surg Pathol 2011, 35, 530. [Google Scholar] [CrossRef] [PubMed]
- Blatter, R.; Tschupp, B.; Aretz, S.; Bernstein, I.; Colas, C.; Evans, D.G.; Genuardi, M.; Hes, F.J.; Hüneburg, R.; Järvinen, H.; et al. Disease Expression in Juvenile Polyposis Syndrome: A Retrospective Survey on a Cohort of 221 European Patients and Comparison with a Literature-Derived Cohort of 473 SMAD4/BMPR1A Pathogenic Variant Carriers. Genetics in Medicine 2020 22:9 2020, 22, 1524–1532. [Google Scholar] [CrossRef] [PubMed]
- Brosens, L.A.A.; Langeveld, D.; van Hattem, W.A.; Giardiello, F.M.; Offerhaus, G.J.A. Juvenile Polyposis Syndrome. World Journal of Gastroenterology : WJG 2011, 17, 4839. [Google Scholar] [CrossRef] [PubMed]
- Malki, A.; Abu Elruz, R.; Gupta, I.; Allouch, A.; Vranic, S.; al Moustafa, A.-E. Molecular Sciences Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. 2020. [CrossRef]
- Yamagishi, H.; Kuroda, H.; Imai, Y.; Hiraishi, H. Molecular Pathogenesis of Sporadic Colorectal Cancers. Chin J Cancer 2016, 35, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jelsig, A.M.; Ousager, L.B.; Brusgaard, K.; Qvist, N. Juvenile Polyps in Denmark from 1995 to 2014. Dis Colon Rectum 2016, 59, 751–757. [Google Scholar] [CrossRef]
- Phen, C.; Rojas, I. Paediatric Polyposis Syndromes: Burden of Disease and Current Concepts. Curr Opin Pediatr 2021, 33, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Latchford, A.R.; Neale, K.; Phillips, R.K.S.; Clark, S.K. Juvenile Polyposis Syndrome: A Study of Genotype, Phenotype, and Long-Term Outcome. Dis Colon Rectum 2012, 55, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Aytac, E.; Sulu, B.; Heald, B.; O’Malley, M.; Laguardia, L.; Remzi, F.H.; Kalady, M.F.; Burke, C.A.; Church, J.M. Genotype-Defined Cancer Risk in Juvenile Polyposis Syndrome. British Journal of Surgery 2014, 102, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Ishibashi, K.; Iwama, T. Malignant Tumors Associated with Juvenile Polyposis Syndrome in Japan. Surg Today 2018, 48, 253–263. [Google Scholar] [CrossRef] [PubMed]
- MacFarland, S.P.; Ebrahimzadeh, J.E.; Zelley, K.; Begum, L.; Bass, L.M.; Brand, R.E.; Dudley, B.; Fishman, D.S.; Ganzak, A.; Karloski, E.; et al. Phenotypic Differences in Juvenile Polyposis Syndrome with or without a Disease-Causing SMAD4/BMPR1A Variant. Cancer Prev Res (Phila) 2021, 14, 215. [Google Scholar] [CrossRef] [PubMed]
- Micolonghi, C.; Piane, M.; Germani, A.; Sadeghi, S.; Libi, F.; Savio, C.; Fabiani, M.; Mancini, R.; Ranieri, D.; Pizzuti, A.; et al. A New SMAD4 Splice Site Variant in a Three-Generation Italian Family with Juvenile Polyposis Syndrome. Diagnostics 2022, 12, 2684. [Google Scholar] [CrossRef] [PubMed]
- Samadder, N.J.; Baffy, N.; Giridhar, K. V.; Couch, F.J.; Riegert-Johnson, D. Hereditary Cancer Syndromes-A Primer on Diagnosis and Management, Part 2: Gastrointestinal Cancer Syndromes. Mayo Clin Proc 2019, 94, 1099–1116. [Google Scholar] [CrossRef] [PubMed]
- Vasen, H.F.A.; Tomlinson, I.; Castells, A. Clinical Management of Hereditary Colorectal Cancer Syndromes. Nature Reviews Gastroenterology & Hepatology 2015 12:2 2015, 12, 88–97. [Google Scholar] [CrossRef]
- Latchford, A.R.; Neale, K.; Phillips, R.K.S.; Clark, S.K. Juvenile Polyposis Syndrome: A Study of Genotype, Phenotype, and Long-Term Outcome. Dis Colon Rectum 2012, 55, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.H.; Gingold-Belfer, R.; Vainer, E.; Hegger, S.; Laish, I.; Derazne, E.; Weintraub, I.; Reznick-Levi, G.; Goldberg, Y.; Levi, Z.; et al. Phenotypic Diversity among Juvenile Polyposis Syndrome Patients from Different Ethnic Background. Hered Cancer Clin Pract 2022, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Brodie, S.G.; Yang, X.; Im, Y.-H.; Parks, W.T.; Chen, L.; Zhou, Y.-X.; Weinstein, M.; Kim, S.-J.; Deng, C.-X. Haploid Loss of the Tumor Suppressor Smad4/Dpc4 Initiates Gastric Polyposis and Cancer in Mice. Oncogene 2000, 19, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- He, X.C.; Zhang, J.; Tong, W.G.; Tawfik, O.; Ross, J.; Scoville, D.H.; Tian, Q.; Zeng, X.; He, X.; Wiedemann, L.M.; et al. BMP Signaling Inhibits Intestinal Stem Cell Self-Renewal through Suppression of Wnt–β-Catenin Signaling. Nature Genetics 2004 36:10 2004, 36, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Marsh Durban, V.; Jansen, M.; Davies, E.J.; Morsink, F.H.; Offerhaus, G.J.A.; Clarke, A.R. Epithelial-Specific Loss of PTEN Results in Colorectal Juvenile Polyp Formation and Invasive Cancer. Am J Pathol 2014, 184, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, A.; Feinberg, A.P. Loss of Imprinting of IGF2: A Common Epigenetic Modifier of Intestinal Tumor Risk. Cancer Res 2005, 65, 11236–11240. [Google Scholar] [CrossRef]
- Oshima, H.; Matsunaga, A.; Fujimura, T.; Tsukamoto, T.; Taketo, M.M.; Oshima, M. Carcinogenesis in Mouse Stomach by Simultaneous Activation of the Wnt Signaling and Prostaglandin E2 Pathway. Gastroenterology 2006, 131, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Haramis, A.P.G.; Begthel, H.; Van Den Born, M.; Van Es, J.; Jonkheer, S.; Offerhaus, G.J.A.; Clevers, H. De Novo Crypt Formation and Juvenile Polyposis on BMP Inhibition in Mouse Intestine. Science (1979) 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Büller, N.V.J.A.; Rosekrans, S.L.; Metcalfe, C.; Heijmans, J.; Van Dop, W.A.; Fessler, E.; Jansen, M.; Ahn, C.; Vermeulen, J.L.M.; Westendorp, B.F.; et al. Stromal Indian Hedgehog Signaling Is Required for Intestinal Adenoma Formation in Mice. Gastroenterology 2015, 148. [Google Scholar] [CrossRef] [PubMed]
- Van Es, J.H.; Clevers, H. Notch and Wnt Inhibitors as Potential New Drugs for Intestinal Neoplastic Disease. Trends Mol Med 2005, 11, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Threadgill, D.W.; Churchill, G.A. Ten Years of the Collaborative Cross. Genetics 2012, 190, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Nashef, A.; Qahaz, N.; El-Naaj, I.A.; Iraqi, F.A. Systems Genetics Analysis of Oral Squamous Cell Carcinoma Susceptibility Using the Mouse Model: Current Position and New Perspective. Mammalian Genome 2021, 32, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Churchill, G.A.; Airey, D.C.; Allayee, H.; Angel, J.M.; Attie, A.D.; Beatty, J.; Beavis, W.D.; Belknap, J.K.; Bennett, B.; Berrettini, W.; et al. The Collaborative Cross, a Community Resource for the Genetic Analysis of Complex Traits. Nat Genet 2004, 36, 1133–1137. [Google Scholar] [CrossRef]
- Atamni, H.J.A.T.; Mott, R.; Soller, M.; Iraqi, F.A. High-Fat-Diet Induced Development of Increased Fasting Glucose Levels and Impaired Response to Intraperitoneal Glucose Challenge in the Collaborative Cross Mouse Genetic Reference Population. BMC Genet 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Dorman, A.; Binenbaum, I.; Abu-Toamih Atamni, H.J.; Chatziioannou, A.; Tomlinson, I.; Mott, R.; Iraqi, F.A. Genetic Mapping of Novel Modifiers for Apc Min Induced Intestinal Polyps’ Development Using the Genetic Architecture Power of the Collaborative Cross Mice. BMC Genomics 2021, 22. [Google Scholar] [CrossRef]
- Takaku, K.; Miyoshi, H.; Matsunaga, A.; Oshima, M.; Sasaki, N.; Taketo, M.M. Gastric and Duodenal Polyps in Smad4 (Dpc4) Knockout Mice. Cancer Res 1999, 59. [Google Scholar]
- Langeveld, D.; Van Hattem, W.A.; De Leng, W.W.J.; Morsink, F.H.; Ten Kate, F.J.W.; Giardiello, F.M.; Offerhaus, G.J.A.; Brosens, L.A.A. SMAD4 Immunohistochemistry Reflects Genetic Status in Juvenile Polyposis Syndrome. Clinical Cancer Research 2010, 16, 4126–4134. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yang, X. Smad4-Mediated TGF-β Signaling in Tumorigenesis. Int J Biol Sci 2010, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Iraqi, F.A.; Churchill, G.; Mott, R. The Collaborative Cross, Developing a Resource for Mammalian Systems Genetics: A Status Report of the Wellcome Trust Cohort. Mammalian Genome 2008, 19. [Google Scholar] [CrossRef]
- Iraqi, F.A.; Mahajne, M.; Salaymah, Y.; Sandovski, H.; Tayem, H.; Vered, K.; Balmer, L.; Hall, M.; Manship, G.; Morahan, G.; et al. The Genome Architecture of the Collaborative Cross Mouse Genetic Reference Population. Genetics 2012, 190. [Google Scholar] [CrossRef]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-Quality Mouse Genomic Dna with Hot Sodium Hydroxide and Tris (HotSHOT). Biotechniques 2000, 29. [Google Scholar] [CrossRef]
- Dorman, A.; Binenbaum, I.; Abu-Toamih Atamni, H.J.; Chatziioannou, A.; Tomlinson, I.; Mott, R.; Iraqi, F.A. Genetic Mapping of Novel Modifiers for Apc Min Induced Intestinal Polyps’ Development Using the Genetic Architecture Power of the Collaborative Cross Mice. BMC Genomics 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Rudling, R.; Hassan, A.B.; Kitau, J.; Mandir, N.; Goodlad, R.A. A Simple Device to Rapidly Prepare Whole Mounts of Murine Intestine. Cell Prolif 2006, 39. [Google Scholar] [CrossRef] [PubMed]
- Iraqi, F.A.; Athamni, H.; Dorman, A.; Salymah, Y.; Tomlinson, I.; Nashif, A.; Shusterman, A.; Weiss, E.; Houri-Haddad, Y.; Mott, R.; et al. Heritability and Coefficient of Genetic Variation Analyses of Phenotypic Traits Provide Strong Basis for High-Resolution QTL Mapping in the Collaborative Cross Mouse Genetic Reference Population. Mammalian Genome 2014, 25, 109–119. [Google Scholar] [CrossRef]
- Quinlan, J.R. Induction of Decision Trees. Mach Learn 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Krzywinksi, M.; Altman, N. Points of Significance Classification and Regression Trees. Nat Methods 2017, 14. [Google Scholar] [CrossRef]
- Breiman, L. (Impo)Random Forests(Book). Mach Learn 2001. [Google Scholar]
- Zhao, X.; Zou, Q.; Liu, B.; Liu, X. Exploratory Predicting Protein Folding Model with Random Forest and Hybrid Features. Curr Proteomics 2015, 11. [Google Scholar] [CrossRef]
- Liao, Z.; Ju, Y.; Zou, Q. Prediction of G Protein-Coupled Receptors with SVM-Prot Features and Random Forest. Scientifica (Cairo) 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Cawley, G.C.; Talbot, N.L.C. On Over-Fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. Journal of Machine Learning Research 2010, 11. [Google Scholar]
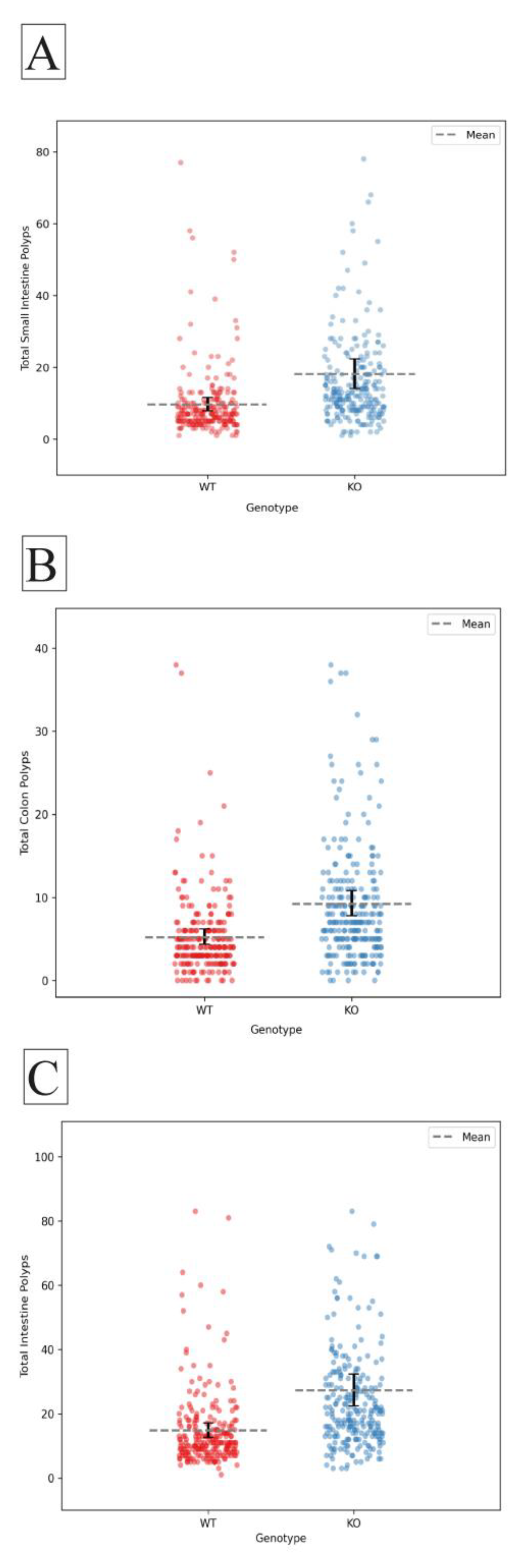
Figure 1.
Polyp counts in Smad4 heterozygous knockout (KO) and wild-type (WT) mice. (A) Mean polyp count in the entire intestine is significantly higher in KO mice compared to WT mice (p < 0.001). (B) Variation in mean polyp count in the small intestine is significantly elevated in KO mice compared to WT mice (p < 0.001). (C) Mean polyp count in the colon is markedly higher in KO mice compared to WT mice (p < 0.001) within the general mouse population. The X-axis represents the genotype, while the Y-axis represents the number of polyps.
Figure 1.
Polyp counts in Smad4 heterozygous knockout (KO) and wild-type (WT) mice. (A) Mean polyp count in the entire intestine is significantly higher in KO mice compared to WT mice (p < 0.001). (B) Variation in mean polyp count in the small intestine is significantly elevated in KO mice compared to WT mice (p < 0.001). (C) Mean polyp count in the colon is markedly higher in KO mice compared to WT mice (p < 0.001) within the general mouse population. The X-axis represents the genotype, while the Y-axis represents the number of polyps.
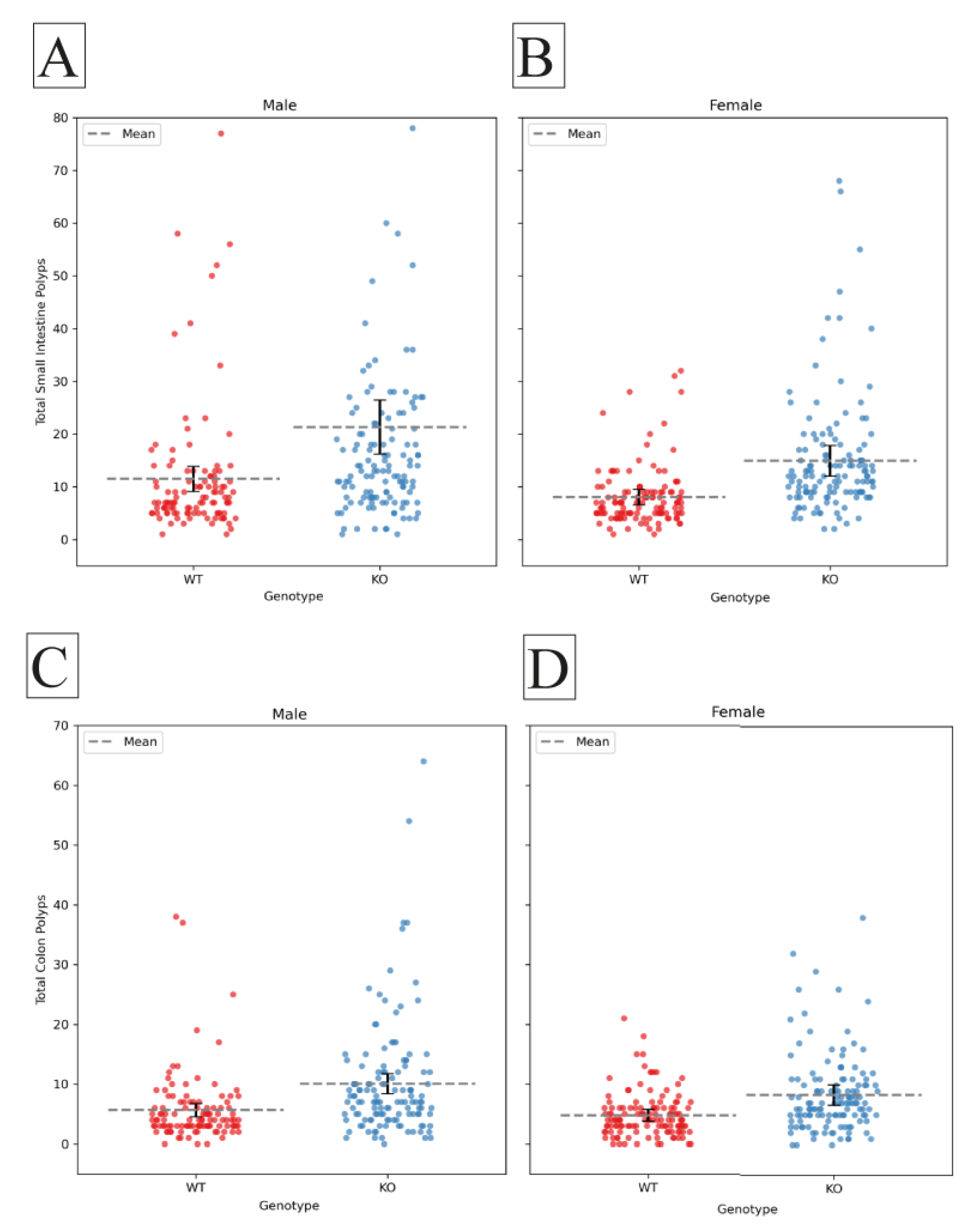
Figure 2.
Differential impact of Smad4 heterozygous knockout on polyp counts in male and female mice. (A) Male mice with Smad4 heterozygous knockout exhibit a significant increase in polyp count in the small intestine compared to wild-type (p = 0.001). (B) Female mice with Smad4 heterozygous knockout demonstrate a significant increase in polyp count in the small intestine compared to wild-type (p < 0.001). (C) Smad4 heterozygous knockout in male mice leads to a significant increase in polyp count in the colon (p = 0.001). (D) Female mice with Smad4 heterozygous knockout show a significant increase in polyp count in the colon compared to wild-type (p < 0.001). The X-axis represents the genotype, while the Y-axis represents the number of polyps.
Figure 2.
Differential impact of Smad4 heterozygous knockout on polyp counts in male and female mice. (A) Male mice with Smad4 heterozygous knockout exhibit a significant increase in polyp count in the small intestine compared to wild-type (p = 0.001). (B) Female mice with Smad4 heterozygous knockout demonstrate a significant increase in polyp count in the small intestine compared to wild-type (p < 0.001). (C) Smad4 heterozygous knockout in male mice leads to a significant increase in polyp count in the colon (p = 0.001). (D) Female mice with Smad4 heterozygous knockout show a significant increase in polyp count in the colon compared to wild-type (p < 0.001). The X-axis represents the genotype, while the Y-axis represents the number of polyps.
Figure 3.
Comparison of polyp counts in F1 CC-C57BL/6 and F1 CC-C57BL/6 J-Smad4^tm1Mak lines. The average number of polyps (±SE) is shown for 14 F1 CC-C57BL/6 wild-type mice (blue colored bars) and 14 F1 CC-C57BL/6 J-Smad4^tm1Mak heterozygous knockout lines (orange-colored bars). The X-axis represents different Collaborative Cross (CC) lines, while the Y-axis shows the number of polyps. Statistical significance of differences in the average number of polyps between the two groups is denoted as follows: (*) indicates a significant difference at p < 0.05, and (**) indicates a highly significant difference at p < 0.01.
Figure 3.
Comparison of polyp counts in F1 CC-C57BL/6 and F1 CC-C57BL/6 J-Smad4^tm1Mak lines. The average number of polyps (±SE) is shown for 14 F1 CC-C57BL/6 wild-type mice (blue colored bars) and 14 F1 CC-C57BL/6 J-Smad4^tm1Mak heterozygous knockout lines (orange-colored bars). The X-axis represents different Collaborative Cross (CC) lines, while the Y-axis shows the number of polyps. Statistical significance of differences in the average number of polyps between the two groups is denoted as follows: (*) indicates a significant difference at p < 0.05, and (**) indicates a highly significant difference at p < 0.01.
Figure 4.
Correlation Analysis of Polyp Development Patterns in Smad4 Heterozygous Knockout and Wild-Type Mice Populations. This figure presents the correlation analysis results for polyp development within the gastrointestinal tract of wild-type (WT) controls (section A) compared to Smad4 heterozygous knockout (KO) mice (section B). The analysis reveals a significant positive correlation between the occurrence of Type C polyps in the colon and the presence of polyps in the small intestine within the KO population, suggesting a potential systemic effect or shared susceptibility factors. Furthermore, a marked positive correlation is observed between the total number of Type B polyps in the small intestine and the aggregate count of intestinal polyps in KO mice. This indicates that Type B polyps may be a predominant factor in the overall polyp burden. Additionally, a strong positive association is highlighted between the occurrence of Type B polyps in the small intestine and the total intestinal polyp count in KO mice, underscoring the significance of this polyp subtype in the observed pathology. These findings underscore the complex interplay between different polyp types in the intestines of Smad4 heterozygous knockout mice and contribute to our broader understanding of polyp development dynamics in genetic models of intestinal tumorigenesis. The data include correlation coefficients and p-values, delineating statistical significance and facilitating a nuanced interpretation of polyp distribution and frequency patterns in relation to genetic modifications.
Figure 4.
Correlation Analysis of Polyp Development Patterns in Smad4 Heterozygous Knockout and Wild-Type Mice Populations. This figure presents the correlation analysis results for polyp development within the gastrointestinal tract of wild-type (WT) controls (section A) compared to Smad4 heterozygous knockout (KO) mice (section B). The analysis reveals a significant positive correlation between the occurrence of Type C polyps in the colon and the presence of polyps in the small intestine within the KO population, suggesting a potential systemic effect or shared susceptibility factors. Furthermore, a marked positive correlation is observed between the total number of Type B polyps in the small intestine and the aggregate count of intestinal polyps in KO mice. This indicates that Type B polyps may be a predominant factor in the overall polyp burden. Additionally, a strong positive association is highlighted between the occurrence of Type B polyps in the small intestine and the total intestinal polyp count in KO mice, underscoring the significance of this polyp subtype in the observed pathology. These findings underscore the complex interplay between different polyp types in the intestines of Smad4 heterozygous knockout mice and contribute to our broader understanding of polyp development dynamics in genetic models of intestinal tumorigenesis. The data include correlation coefficients and p-values, delineating statistical significance and facilitating a nuanced interpretation of polyp distribution and frequency patterns in relation to genetic modifications.
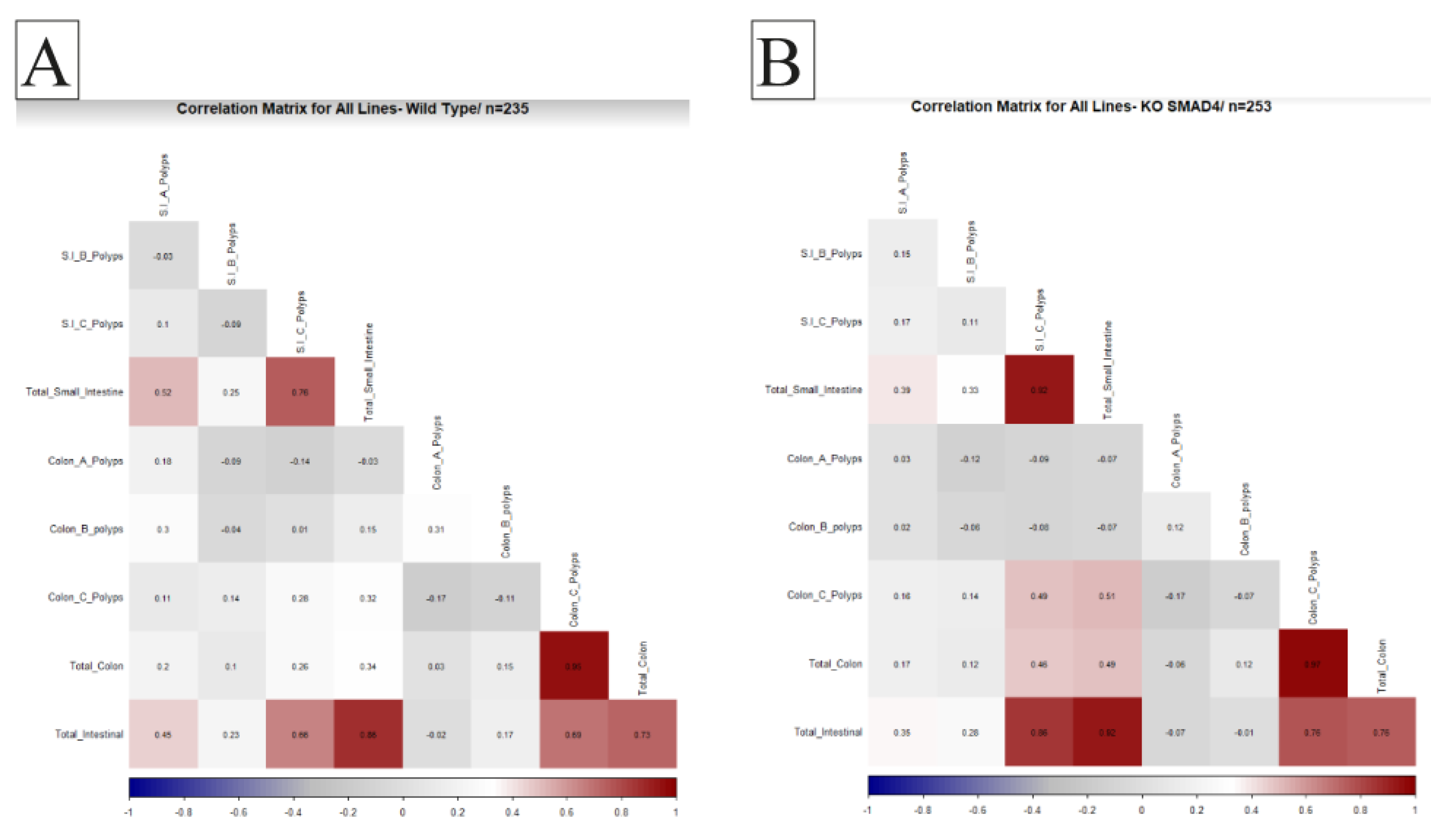
Figure 5.
Gender-Specific Correlation Patterns of Polyp Development in Smad4 Heterozygous Knockout Mice compared to wild type Mice. Correlation matrixes for (A) KO male mice lines (B) WT male mice lines (C) KO female mice lines (D) WT female mice lines. In male KO mice, a positive correlation is evident between colon polyps and the C portion of small intestinal polyps. Additionally, robust positive association exists between counts of total small intestinal B polyps and the overall number of intestinal polyps. In female KO mice, a positive correlation is observed between small intestinal polyps and Small Intestinal B polyps. Moreover, a positive correlation exists between total colon polyps and Small Intestinal C polyps. Notably, the previously observed positive correlation between total intestinal polyps and Small Intestinal A polyps in the KO group is absent in females. These gender-specific correlations provide valuable insights into the effects of Smad4 knockout on polyp development, highlighting variations in the relationships between different polyp sizes in male and female mice.
Figure 5.
Gender-Specific Correlation Patterns of Polyp Development in Smad4 Heterozygous Knockout Mice compared to wild type Mice. Correlation matrixes for (A) KO male mice lines (B) WT male mice lines (C) KO female mice lines (D) WT female mice lines. In male KO mice, a positive correlation is evident between colon polyps and the C portion of small intestinal polyps. Additionally, robust positive association exists between counts of total small intestinal B polyps and the overall number of intestinal polyps. In female KO mice, a positive correlation is observed between small intestinal polyps and Small Intestinal B polyps. Moreover, a positive correlation exists between total colon polyps and Small Intestinal C polyps. Notably, the previously observed positive correlation between total intestinal polyps and Small Intestinal A polyps in the KO group is absent in females. These gender-specific correlations provide valuable insights into the effects of Smad4 knockout on polyp development, highlighting variations in the relationships between different polyp sizes in male and female mice.

Figure 6.
Representative whole mounts of mouse intestine highlighting polyp classification. (A) Whole mounts stained to visualize polyps in a mouse with heterozygous Smad4 knockout genotype. Multiple polyps are visible, categorized into three classes (A, B, and C) based on size criteria. (B) whole mount from a mouse with the wild-type genotype, showing fewer polyps in the same intestinal segment compared to the Smad4 knockout mouse.
Figure 6.
Representative whole mounts of mouse intestine highlighting polyp classification. (A) Whole mounts stained to visualize polyps in a mouse with heterozygous Smad4 knockout genotype. Multiple polyps are visible, categorized into three classes (A, B, and C) based on size criteria. (B) whole mount from a mouse with the wild-type genotype, showing fewer polyps in the same intestinal segment compared to the Smad4 knockout mouse.
Table 1.
Results of calculating heritability (H2) values. The heritability was calculated using one-way ANOVA for the traits in our study that were calculated separately by sex and genotype.
Table 1.
Results of calculating heritability (H2) values. The heritability was calculated using one-way ANOVA for the traits in our study that were calculated separately by sex and genotype.
| Sex |
Genotype |
Trait |
df between |
df within |
n |
MS between |
MS within |
VG |
H2 |
Trait mean |
CVg |
Anova Sig |
| Female |
WT |
SB1_C |
12 |
96 |
7.46 |
14.04 |
3.11 |
1.47 |
0.32 |
1.71 |
0.71 |
0.000 |
| Female |
WT |
SB2_A |
12 |
96 |
7.46 |
1.31 |
0.70 |
0.08 |
0.11 |
0.66 |
0.43 |
0.046 |
| Female |
WT |
SB2_B |
12 |
96 |
7.46 |
0.49 |
0.41 |
0.01 |
0.03 |
0.52 |
0.20 |
0.298 |
| Female |
WT |
SB3_A |
12 |
96 |
7.46 |
1.92 |
0.86 |
0.14 |
0.14 |
0.80 |
0.47 |
0.016 |
| Female |
WT |
SB3_C |
12 |
96 |
7.46 |
26.55 |
7.09 |
2.61 |
0.27 |
1.70 |
0.95 |
0.000 |
| Female |
WT |
S.I_A_Polyps |
12 |
96 |
7.46 |
7.21 |
3.55 |
0.49 |
0.12 |
2.04 |
0.34 |
0.029 |
| Female |
WT |
S.I_C_Polyps |
12 |
96 |
7.46 |
68.87 |
18.15 |
6.80 |
0.27 |
4.59 |
0.57 |
0.000 |
| Female |
WT |
Total_Small_Intestine |
12 |
96 |
7.46 |
96.44 |
26.12 |
9.42 |
0.27 |
8.48 |
0.36 |
0.000 |
| Female |
WT |
Colon_A_Polyps |
12 |
96 |
7.46 |
0.15 |
0.08 |
0.01 |
0.10 |
0.10 |
0.96 |
0.054 |
| Female |
WT |
Colon_B_polyps |
12 |
96 |
7.46 |
0.70 |
0.52 |
0.02 |
0.04 |
0.40 |
0.39 |
0.204 |
| Female |
WT |
Colon_C_Polyps |
12 |
96 |
7.46 |
50.97 |
9.60 |
5.54 |
0.37 |
4.64 |
0.51 |
0.000 |
| Female |
WT |
Total_Colon |
12 |
96 |
7.46 |
46.08 |
9.92 |
4.85 |
0.33 |
5.15 |
0.43 |
0.000 |
| Female |
WT |
Total_Intestinal |
12 |
96 |
7.46 |
191.35 |
43.68 |
19.79 |
0.31 |
13.62 |
0.33 |
0.000 |
| Female |
KO |
SB1_A |
13 |
100 |
7.21 |
0.96 |
0.85 |
0.02 |
0.02 |
0.87 |
0.14 |
0.340 |
| Female |
KO |
SB1_B |
13 |
100 |
7.21 |
0.77 |
0.74 |
0.00 |
0.01 |
0.92 |
0.07 |
0.417 |
| Female |
KO |
SB1_C |
13 |
100 |
7.21 |
43.26 |
29.78 |
1.87 |
0.06 |
3.37 |
0.41 |
0.149 |
| Female |
KO |
SB2_A |
13 |
100 |
7.21 |
0.93 |
0.84 |
0.01 |
0.02 |
1.00 |
0.11 |
0.358 |
| Female |
KO |
SB2_C |
13 |
100 |
7.21 |
16.22 |
7.22 |
1.25 |
0.15 |
2.39 |
0.47 |
0.013 |
| Female |
KO |
SB3_A |
13 |
100 |
7.21 |
2.14 |
1.00 |
0.16 |
0.14 |
1.20 |
0.33 |
0.018 |
| Female |
KO |
SB3_C |
13 |
100 |
7.21 |
118.77 |
36.60 |
11.39 |
0.24 |
3.96 |
0.85 |
0.000 |
| Female |
KO |
S.I_A_Polyps |
13 |
100 |
7.21 |
5.74 |
3.05 |
0.37 |
0.11 |
3.07 |
0.20 |
0.041 |
| Female |
KO |
S.I_B_Polyps |
13 |
100 |
7.21 |
3.35 |
3.00 |
0.05 |
0.02 |
2.57 |
0.09 |
0.355 |
| Female |
KO |
S.I_C_Polyps |
13 |
100 |
7.21 |
348.49 |
88.36 |
36.06 |
0.29 |
9.73 |
0.62 |
0.000 |
| Female |
KO |
Total_Small_Intestine |
13 |
100 |
7.21 |
394.98 |
92.78 |
41.89 |
0.31 |
15.37 |
0.42 |
0.000 |
| Female |
KO |
Colon_B_polyps |
13 |
100 |
7.21 |
1.90 |
0.88 |
0.14 |
0.14 |
0.62 |
0.60 |
0.017 |
| Female |
KO |
Colon_C_Polyps |
13 |
100 |
7.21 |
87.11 |
39.20 |
6.64 |
0.14 |
8.23 |
0.31 |
0.014 |
| Female |
KO |
Total_Colon |
13 |
100 |
7.21 |
81.18 |
38.90 |
5.86 |
0.13 |
8.96 |
0.27 |
0.021 |
| Female |
KO |
Total_Intestinal |
13 |
100 |
7.21 |
650.63 |
153.55 |
68.90 |
0.31 |
24.33 |
0.34 |
0.000 |
| Male |
WT |
SB1_A |
13 |
87 |
6.29 |
0.83 |
0.60 |
0.04 |
0.06 |
0.50 |
0.38 |
0.187 |
| Male |
WT |
SB1_C |
13 |
87 |
6.29 |
10.50 |
4.77 |
0.91 |
0.16 |
1.64 |
0.58 |
0.016 |
| Male |
WT |
SB2_A |
13 |
87 |
6.29 |
1.77 |
0.76 |
0.16 |
0.17 |
0.74 |
0.54 |
0.011 |
| Male |
WT |
SB2_C |
13 |
87 |
6.29 |
35.20 |
15.05 |
3.21 |
0.18 |
2.21 |
0.81 |
0.010 |
| Male |
WT |
SB3_B |
13 |
87 |
6.29 |
0.90 |
0.81 |
0.01 |
0.02 |
0.76 |
0.16 |
0.359 |
| Male |
WT |
SB3_C |
13 |
87 |
6.29 |
220.63 |
92.31 |
20.41 |
0.18 |
3.84 |
1.18 |
0.009 |
| Male |
WT |
S.I_A_Polyps |
13 |
87 |
6.29 |
5.49 |
3.48 |
0.32 |
0.08 |
1.98 |
0.29 |
0.107 |
| Male |
WT |
S.I_C_Polyps |
13 |
87 |
6.29 |
380.31 |
123.12 |
40.92 |
0.25 |
7.69 |
0.83 |
0.001 |
| Male |
WT |
Total_Small_Intestine |
13 |
87 |
6.29 |
445.20 |
122.28 |
51.37 |
0.30 |
11.67 |
0.61 |
0.000 |
| Male |
WT |
Colon_A_Polyps |
13 |
87 |
6.29 |
0.08 |
0.07 |
0.00 |
0.01 |
0.08 |
0.35 |
0.400 |
| Male |
WT |
Colon_B_polyps |
13 |
87 |
6.29 |
0.94 |
0.46 |
0.08 |
0.14 |
0.39 |
0.72 |
0.025 |
| Male |
WT |
Colon_C_Polyps |
13 |
87 |
6.29 |
72.84 |
28.17 |
7.11 |
0.20 |
5.38 |
0.50 |
0.004 |
| Male |
WT |
Total_Colon |
13 |
87 |
6.29 |
68.33 |
30.91 |
5.95 |
0.16 |
5.84 |
0.42 |
0.015 |
| Male |
WT |
Total_Intestinal |
13 |
87 |
6.29 |
699.96 |
172.11 |
83.98 |
0.33 |
17.51 |
0.52 |
0.000 |
| Male |
KO |
SB1_A |
13 |
95 |
6.86 |
2.07 |
1.12 |
0.14 |
0.11 |
1.19 |
0.31 |
0.046 |
Table 2.
Summary of Machine-Learning Classification Models Tested, Featuring Linear Discriminant Analysis (LDA), Support Vector Machine (SVM), K-Nearest Neighbors, and Random Forest (RF) in Two-Class and Three-Class Classifications.
Table 2.
Summary of Machine-Learning Classification Models Tested, Featuring Linear Discriminant Analysis (LDA), Support Vector Machine (SVM), K-Nearest Neighbors, and Random Forest (RF) in Two-Class and Three-Class Classifications.
| Two classes |
| |
LDA |
KNN |
SVM |
RF |
| Accuracy |
0.67 |
0.55 |
0.69 |
0.64 |
| Kappa |
0.33 |
0.1 |
0.38 |
0.27 |
| Three classes |
| Accuracy |
0.62 |
0.62 |
0.63 |
0.62 |
| Kappa |
0.1 |
0.07 |
0.0 |
0.1 |
Table 3.
Summary of the sample size of male and female mice used from the 14 different lines backgrounds of the Collaborative Cross mouse population.
Table 3.
Summary of the sample size of male and female mice used from the 14 different lines backgrounds of the Collaborative Cross mouse population.
| F1 (Samd4KO X CCxxx) |
Sex |
Total |
| F |
M |
| CC037 |
Genotype |
WT |
17 |
15 |
32 |
| KO |
14 |
19 |
33 |
| Total |
31 |
34 |
65 |
| CC004 |
Genotype |
WT |
4 |
5 |
9 |
| KO |
11 |
8 |
19 |
| Total |
15 |
13 |
28 |
| CC040 |
Genotype |
WT |
1 |
2 |
3 |
| KO |
1 |
2 |
3 |
| Total |
2 |
4 |
6 |
| CC005 |
Genotype |
WT |
4 |
4 |
8 |
| KO |
3 |
3 |
6 |
| Total |
7 |
7 |
14 |
| CC019 |
Genotype |
WT |
4 |
3 |
7 |
| KO |
12 |
4 |
16 |
| Total |
16 |
7 |
23 |
| CC006 |
Genotype |
WT |
7 |
5 |
12 |
| KO |
10 |
7 |
17 |
| Total |
17 |
12 |
29 |
| CC084 |
Genotype |
WT |
12 |
12 |
24 |
| KO |
13 |
11 |
24 |
| Total |
25 |
23 |
48 |
| CC059 |
Genotype |
WT |
4 |
9 |
13 |
| KO |
4 |
6 |
10 |
| Total |
8 |
15 |
23 |
| CC041 |
Genotype |
WT |
2 |
0 |
2 |
| KO |
2 |
0 |
2 |
| Total |
4 |
|
4 |
| CC010 |
Genotype |
WT |
11 |
12 |
23 |
| KO |
14 |
10 |
24 |
| Total |
25 |
22 |
47 |
| CC018 |
Genotype |
WT |
16 |
8 |
24 |
| KO |
14 |
15 |
29 |
| Total |
30 |
23 |
53 |
| CC012 |
Genotype |
WT |
7 |
4 |
11 |
| KO |
5 |
6 |
11 |
| Total |
12 |
10 |
22 |
| CC035 |
Genotype |
WT |
11 |
8 |
19 |
| KO |
5 |
13 |
18 |
| Total |
16 |
21 |
37 |
| CC025 |
Genotype |
WT |
9 |
11 |
20 |
| KO |
8 |
11 |
19 |
| Total |
17 |
22 |
39 |
| CC005* |
Genotype |
WT |
20 |
12 |
32 |
| KO |
15 |
14 |
29 |
| Total |
35 |
26 |
61 |
| Total |
Genotype |
WT |
129 |
110 |
239 |
| KO |
131 |
129 |
260 |
| Total |
260 |
239 |
499 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).