1. Introduction
In the realm of diagnostic procedures, the preanalytical steps play a pivotal role in ensuring the accuracy of results. The stability of analytes over various durations and under diverse temperature conditions is a critical consideration, as it guards against the risk of false negatives. Equally important is the inactivation of pathogens during sample transport, which is essential for ensuring safety upon arrival at the test laboratory.
NAAT often involves the purification of nucleic acids from the sample matrix to eliminate potential inhibitors and potentially concentrate the analyte. While this enhances analytical sensitivity, it comes at the cost of increased expenses and turnaround time, potentially creating bottlenecks when test capacity is limited. Consequently, alternative protocols, such as extraction-free or direct PCR, have been developed to alleviate these concerns [
1].
In instances where reference NAAT testing capacity is constrained or rapid results are imperative, a triage testing strategy may be employed [
2]. This involves first conducting a rapid antigen test, followed by a reference NAAT (either or not of only the negatives). This approach aims to enhance the diagnostic yield (by increasing the test population) and/or to lower the number (and costs) of reference NAAT needed. Ideally, the same collection buffer is suitable for both rapid antigen and NAAT testing, but compatibility issues may arise.
Numerous advanced collection and transport buffers have been created to address these challenges. However, none currently combines all the desirable features, including pathogen inactivation, compatibility with downstream NAAT and rapid antigen testing, absence of inhibitors for extraction-free or direct PCR, and stability at elevated temperatures for several days. Filling this gap, InActiv Blue has developed a novel sample collection and transport buffer known as DNA/RNA Defend Pro (DRDP) [
3], specifically designed as a stabilizing medium that inactivates viruses. DRDP is compatible with both direct PCR and rapid antigen testing.
This retrospective paired study design aims to comprehensively evaluate DRDP as a stabilizing medium for upper respiratory swab samples. In addition, this investigation will assess its compatibility with rapid antigen tests and its suitability for use in direct PCR applications. Through this inquiry, we aim to contribute valuable insights into the efficacy and versatility of DRDP in enhancing the reliability and efficiency of diagnostic processes
2. Results
Of the 95 positive samples according to the diagnostic test in Medisch Labo Bruyland (MBL), 88 (93%) also tested positive upon retesting a 1/10 dilution using cobas Liat in the InActiv Blue laboratory. As the samples were diluted and stored at varying temperatures and for different durations, a retest is expected to not detect the samples with the lowest signals.
2.1. Rapid Antigen Testing
Of the 88 samples that tested positive on cobas Liat, 40 (45%) were positive for rapid antigen testing. The 7 cobas Liat negative samples were also negative for rapid antigen testing. No false positive results were obtained by rapid antigen testing. Represenative examples of rapid antigen tests can be found in
Supplementary Figure S2.
Table 1.
Cross table of 1/10 diluted samples tested by cobas Liat and rapid antigen test.
Table 1.
Cross table of 1/10 diluted samples tested by cobas Liat and rapid antigen test.
| |
cobas Liat positive |
cobas Liat negative |
total |
| rapid antigen test positive |
40 |
0 |
40 |
| rapid antigen test negative |
48 |
7 |
55 |
| total |
88 |
7 |
95 |
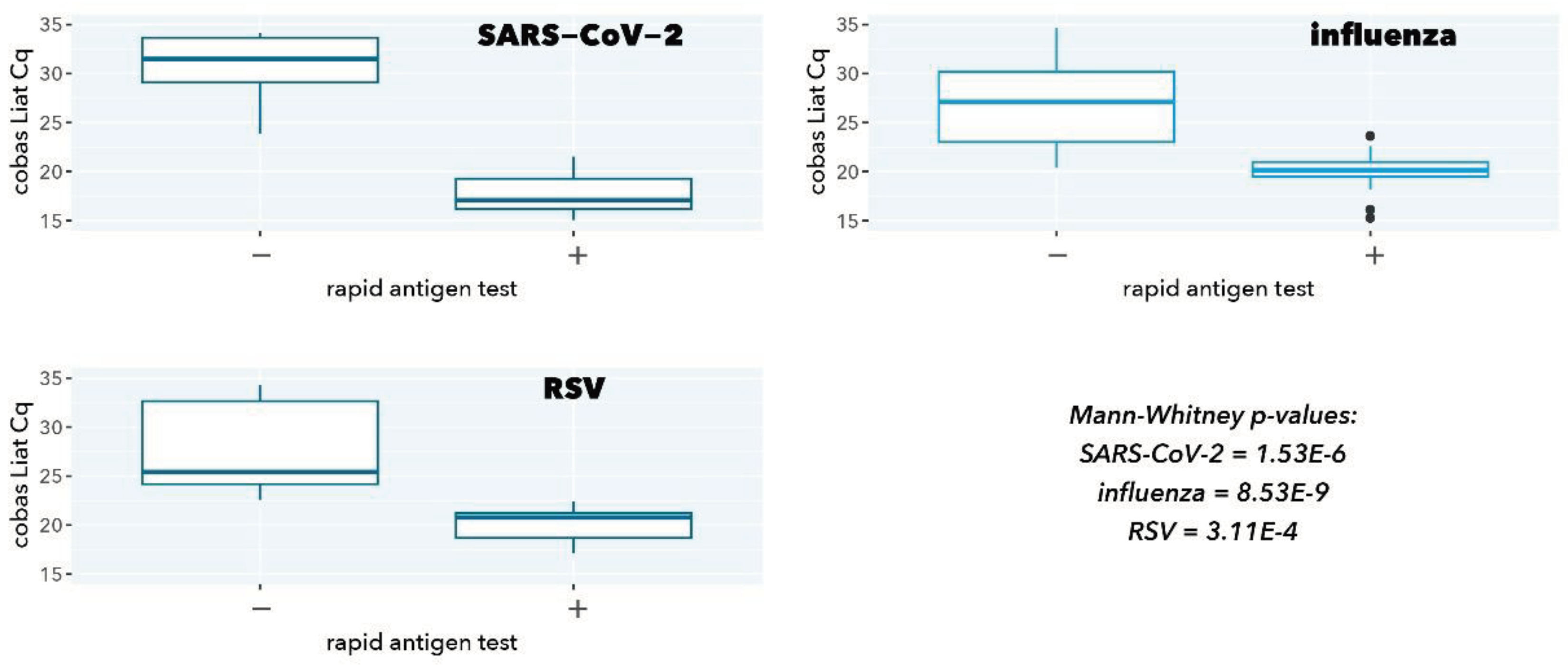
Figure 1.
cobas Liat Cq values stratified according to rapid antigen test result.
Figure 1.
cobas Liat Cq values stratified according to rapid antigen test result.
As expected, samples with a negative rapid antigen test had a significantly higher cobas Liat Cq value (low abundance of viral target), confirming the superior analytical sensitivity of a molecular test (p=1.53E-6, 8.53E-9, and 3.11E-4 for SARS-CoV-2, influenza, and RSV, respectively).
2.2. Direct PCR
Of the 88 samples positive for cobas Liat, 72 (82%) were positive for direct PCR. The 7 samples that tested negative for cobas Liat also tested negative for direct PCR. No false positive results were obtained by direct PCR.
Table 2.
Cross table of samples tested by cobas Liat and direct PCR.
Table 2.
Cross table of samples tested by cobas Liat and direct PCR.
| |
cobas Liat positive |
cobas Liat negative |
total |
| direct PCR positive |
72 |
0 |
72 |
| direct PCR negative |
16 |
7 |
23 |
| total |
88 |
7 |
95 |
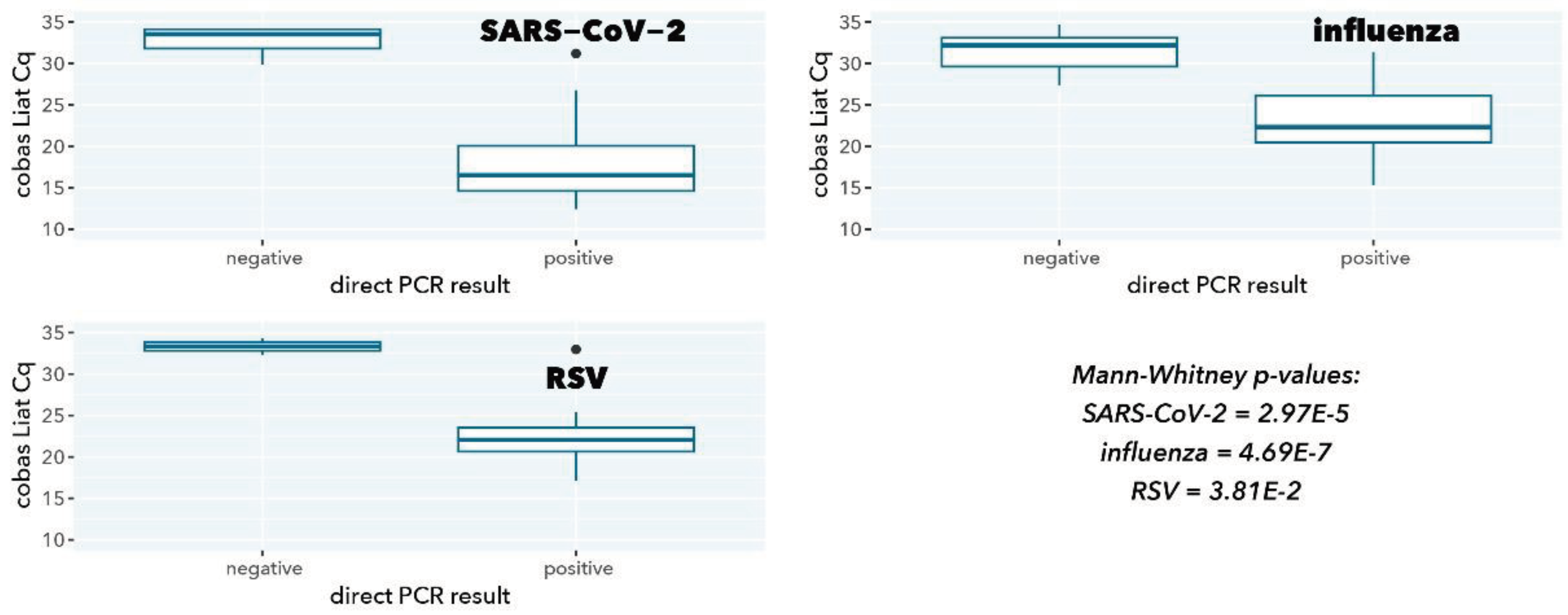
Figure 2.
cobas Liat Cq values stratified according to direct PCR result.
Figure 2.
cobas Liat Cq values stratified according to direct PCR result.
As expected, samples with a negative direct PCR result had a significantly higher cobas Liat Cq value (low abundance of viral target, p=2.97E-5, 4.69E-7, and 0.038 for SARS-CoV-2, influenza A, and RSV A, respectively), in line with the higher sensitivity because of a 200-fold higher input in cobas Liat testing compared to direct PCR (200 µL vs. 1 µL).
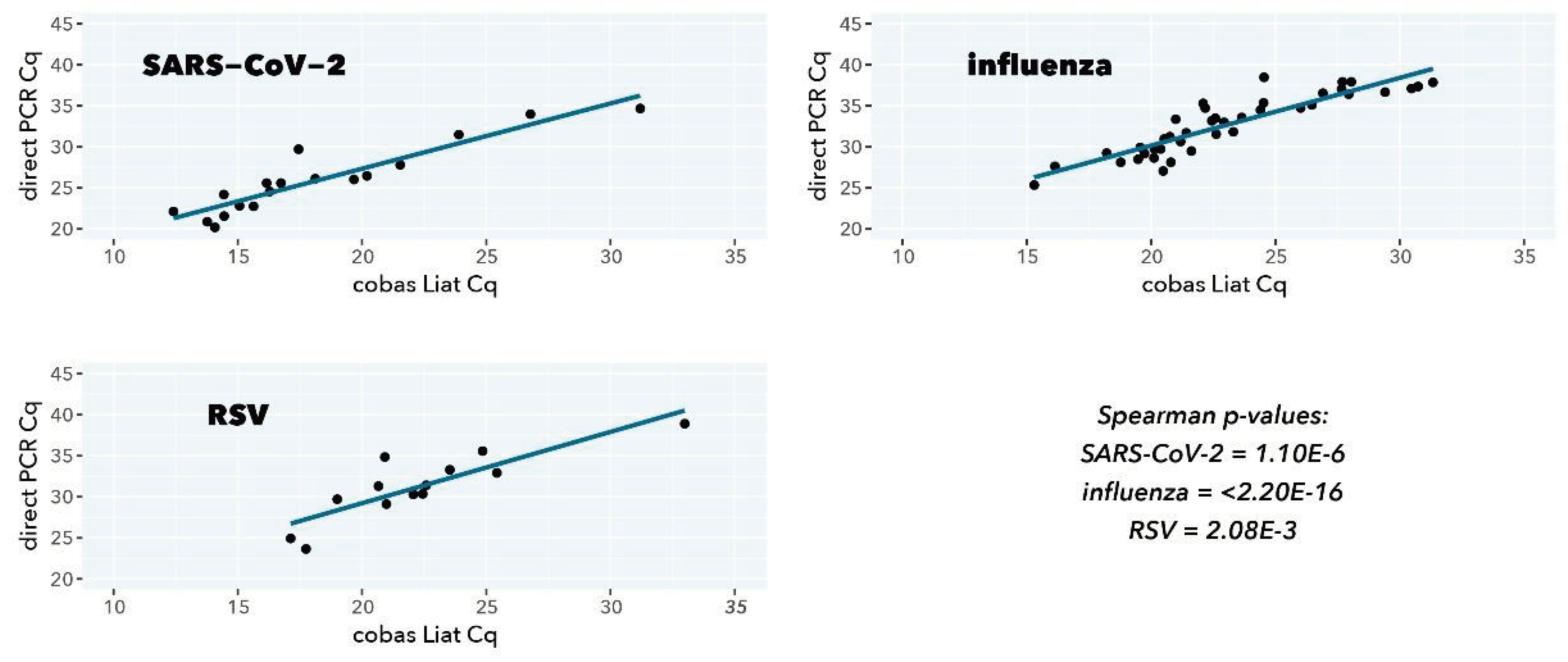
Table 3.
Mean delta-Cq, standard error of the mean (SEM), Spearman rank correlation, and p-value of Cq-value correlation in
Figure 3.
Table 3.
Mean delta-Cq, standard error of the mean (SEM), Spearman rank correlation, and p-value of Cq-value correlation in
Figure 3.
| |
SARS-CoV-2 |
influenza A |
RSV A |
| number of samples |
19 |
40 |
13 |
| mean delta Cq (SEM) |
-7.7 (0.45) |
-9.61 (0.28) |
-8.92 (0.61) |
| r (Spearman) |
0.940 |
0.916 |
0.791 |
| p (Spearman) |
1.10E-6 |
2.20E-16 |
2.08E-3 |
Figure 3.
Scatterplot correlation and ordinary least-squares regression of Cq values from direct PCR (y-axis) and cobas Liat (x-axis).
Figure 3.
Scatterplot correlation and ordinary least-squares regression of Cq values from direct PCR (y-axis) and cobas Liat (x-axis).
For all viral targets, a good to excellent correlation is observed between the direct PCR test and the CE-IVDR cobas Liat test (Spearman r=0.940 (p=1.10E-6), Spearman r=0.916 (p=2.20E-16), Spearman r=0.791 (p=2.08E-3) for SARS-CoV-2, influenza A, and RSV A, respectively). The mean differences in Cq value between the direct PCR and cobas Liat are closely in line with the approximately 200-fold difference in input (log2(200)=7.64), also considering that different primers and probes sequences and PCR reaction conditions are used.
2.3. RNA Stability
Of the 72 samples that tested positive with direct PCR and cobas Liat, a semi-random subset of 31 (43%) was selected for stability testing. As the DRDP instructions for use define stability as the maximum time that the delta-Cq remains ≤ 2 cycles compared to day 0, the inclusion cut-off for a sample in our stability study was a median Cq value 2 cycles lower than an ad hoc detection limit. This detection limit was defined as the highest median Cq of a sample with all 3 replicates positive and subtracted by 2 (i.e., 33.34, 33.57, and 31.99, for influenza A, RSV A, and SARS-CoV-2, respectively). From all 41 (57%) samples with the median Cq lower than the cut-off value at timepoint 0, a random subset of 31 (76%) was selected for stability testing (i.e., 9 SARS-CoV-2, 13 influenza A, 9 RSV A).
Table 4.
Cross table of 31 samples tested with direct RT-qPCR after incubation for 0, 1, 3, and 7 days at 4 °C, 20 °C, and 37 °C.
Table 4.
Cross table of 31 samples tested with direct RT-qPCR after incubation for 0, 1, 3, and 7 days at 4 °C, 20 °C, and 37 °C.
| samples positive |
|---|
| |
day |
| target |
temperature |
0 |
1 |
3 |
7 |
| SARS-CoV-2 |
4 °C |
1 |
9 |
9 |
9 |
| 20 °C |
1 |
9 |
9 |
9 |
| 37 °C |
1 |
9 |
9 |
9 |
| influenza A |
4 °C |
13 |
13 |
13 |
13 |
| 20 °C |
13 |
13 |
13 |
13 |
| 37 °C |
13 |
13 |
13 |
13 |
| RSV A |
4 °C |
9 |
9 |
9 |
9 |
| 20 °C |
9 |
9 |
9 |
9 |
| 37 °C |
9 |
9 |
8 |
9 |
Target was defined as detected if the Cq < 40. All targets in all samples were detected on day 7 at all temperatures. For target RSV A, 1 sample was not detected on day 3 but detected again on day 7. This sample (ID 24-0013) had a high Cq on day 7 (37.07).
Table 5.
Mean delta-Cq (and standard error of the mean, SEM) of samples after incubation for 1, 3, and 7 days at 4 °C, 20 °C, and 37 °C compared to day 0 (bold indicates conditions that have a delta-Cq > 2).
Table 5.
Mean delta-Cq (and standard error of the mean, SEM) of samples after incubation for 1, 3, and 7 days at 4 °C, 20 °C, and 37 °C compared to day 0 (bold indicates conditions that have a delta-Cq > 2).
| delta-Cq (SEM) |
|---|
| |
day |
| target |
temperature |
0 |
1 |
3 |
7 |
| SARS-CoV-2 |
4 °C |
0.00 (0.00) |
0.29 (0.59) |
-0.10 (0.53) |
1.84 (0.35) |
| 20 °C |
0.00 (0.00) |
1.51 (0.62) |
1.51 (0.43) |
2.73 (0.58) |
| 37 °C |
0.00 (0.00) |
1.66 (0.71) |
1.22 (0.63) |
2.01 (0.62) |
| influenza A |
4 °C |
0.00 (0.00) |
0.04 (0.51) |
-0.61 (0.49) |
0.92 (1.00) |
| 20 °C |
0.00 (0.00) |
-1.36 (0.78) |
2.35 (0.43) |
1.09 (0.88) |
| 37 °C |
0.00 (0.00) |
0.03 (0.77) |
1.19 (0.82) |
-0.54 (1.02) |
| RSV A |
4 °C |
0.00 (0.00) |
0.08 (0.70) |
1.00 (0.45) |
0.85 (0.69) |
| 20 °C |
0.00 (0.00) |
0.88 (0.70) |
0.28 (0.90) |
2.51 (0.61) |
| 37 °C |
0.00 (0.00) |
-0.67 (0.95) |
0.76 (0.90) |
6.59 (1.96) |
For stability based on the mean Cq of all samples within a target, a cut-off was set at 2 Cq per the instructions of use of DRDP. If the delta Cq of the time points exceeded 2 compared to timepoint 0, it was determined no longer stable.
Influenza samples remained stable for up to 7 days at all tested temperatures. SARS-CoV-2 and RSV A samples remained stable up to 3 days at all tested temperatures, and up to 7 days at 4 °C.
3. Discussion and Conclusions
DNA/RNA Defend Pro (DRDP) is a unique sample collection and transport buffer that inactivates pathogens [
3], enables triage and reflex testing using rapid antigen tests, stabilizes viral RNA for several days at elevated temperatures, and can reduce costs by not necessarily requiring nucleic acid purification prior to a molecular NAAT test.
Collecting patients’ swab samples directly in DRDP would have been the preferred method for the evaluation of its performance. However, to minimize patient discomfort, anonymized nasopharyngeal swab samples provided by Medisch Labo Bruyland were diluted 1/10 in DRDP, as done in a previous study [
4].
Compared to a CE-IVDR NAAT with excellent analytical sensitivity incorporating a nucleic acid extraction and purification [
5], a somewhat lower sensitivity of direct (or extraction-free) NAAT is observed in our study. As much less sample is used in direct NAAT, this lower sensitivity is expected. The sensitivity could be increased by using an RT-qPCR kit that is even more tolerable to inhibitors allowing more than 10% buffer input and/or performing PCR in larger volumes with more sample input (e.g., 10 µL sample in a 50 µL PCR reaction, a 10-fold increase compared to the current procedure). Detection of all positive samples by direct NAAT under a certain CE-IVDR extraction-based NAAT Cq supports the notion that decreased sensitivity is attributed to the dilution factor and that DRDP is perfectly compatible with direct NAAT. While several studies indicated the suitability of extraction-free SARS-CoV-2 molecular testing, the analytical sensitivity compared to an extraction-based procedure ranged from 80% to 100% [
6,
7,
8]. Importantly, this is partly related to the magnitude of the concentration effect in the extraction method: some procedures only purify nucleic acids (i.e., 200 µL sample input in 200 µL elution buffer) while others purify and concentrate (i.e., 400 µL sample input in 50 µL elution buffer, an 8-fold enrichment).
The DRDP buffer preserves antigens in their native state, as evidenced by the perfect correlation between rapid antigen testing and the quantitative values of a NAAT. As expected, the rapid antigen test displays lower sensitivity, as this method requires a substantial number of viral antigens for a positive result. Detection of all positive samples by rapid antigen testing under a certain CE-IVDR extraction-based NAAT Cq again strongly suggests that decreased sensitivity is attributed to the technique, that DRDP is compatible with rapid antigen testing, and that antigens are preserved in the inactivating buffer.
Finally, viral RNA in swab samples stabilized in DRDP shows stability for several days. Of note, the stability of viral RNA in DRDP at higher temperatures seems to be virus dependent. Qualitative presence/absence and quantitative Cq data show stability of viral RNA from nasopharyngeal swabs in DRDP between 3 and 7 days at all temperatures, but in a target dependent manner. For longer stability, storage at 4 °C is recommended (21 days according to DRDP Instructions for Use [
9]).
4. Materials and Methods
Over a period from December 2023 to February 2024, left-over nasopharyngeal swab samples from Medisch Labo Bruyland (MLB) were set aside at 4 °C or -20 °C for one day up to 2 weeks. The study of stabilizing buffers for transport and storage of human body specimens was approved by the Ethics Committee of University Hospital Antwerp (ID 5931). A total of 95 samples were stored that had tested positive for influenza A, RSV A, and/or SARS-CoV-2.
Diagnostic swabs were transported in PBS or Amies buffer to MLB. To ensure that results obtained in this study reflect the capabilities of DRDP and not the original collection buffers, 0.5 mL of left-over buffer samples were anonymized and diluted 1/10 with DRDP at MLB immediately prior to transport at room temperature to InActiv Blue.
To compensate for variations in storage conditions, all diluted samples that originally tested positive in MLB by Flow (Roche) or cobas Liat (Roche) were re-analyzed using the cobas Liat at InActiv Blue to generate a baseline value (cobas influenza A/B & RSV # 08160104190 and/or cobas SARS-CoV-2 kit # 09408592190).
In addition, all samples that were diluted with DRDP were analyzed with a quick antigen test (fluorecare, SARS-CoV-2 & Influenza A/B & RSV Antigen Combo test kit, # 5501788) and by direct RT-qPCR (i.e., no extraction of nucleic acids was performed prior to RT-qPCR) (Takara, One Step PrimeScript III RT-qPCR, # RR60TW). For stability testing, 31 semi-random samples, positive for direct PCR, were selected.
Cobas Liat runs were performed according to the manufacturer’s instructions.
Because of low pH, DRDP is not directly compatible with quick antigen tests containing colloidal gold. Therefore, 7 µL of 1 M NaOH was added to 100 µL of DRDP-diluted samples and homogenized before pipetting onto the lateral flow cartridge.
Direct PCR was performed using the Takara One step PrimeScript III RT-qPCR kit as described in the manual. According to the DRDP instructions for use, the buffer is compatible with direct PCR if the volume of the sample does not exceed 10% of the total RT-qPCR volume: 5 µL 2x Takara mix + 1 µL primer/probe mix (250 nM of each primer, 125 nM probe, oligonucleotide sequences of SARS-CoV-2, influenza A/B and RSVA/B to be found in
Supplementary Table S1) + 3 µL nuclease-free water (InActiv Blue, # NFW_0015) + 1 µL sample. RT-qPCR was performed using the Roche LightCycler II in a 96-well plate. The following cycling protocol was used: reverse transcription (52 °C for 5 min, 95 °C for 10s), followed by 40 cycles (95 °C for 5 s, 60 °C for 30 s).
Of the 31 selected samples for stability testing, 9 were positive for RSV A, 13 positive for influenza A, and 9 positive for SARS-CoV-2. Samples were divided into three 100 µL aliquots and subjected to incubation at 4 °C, 20 °C, or 37 °C. Direct RT-qPCR measurements were performed on incubation day 0, 1, 3 and 7.
Supplementary Materials
The following supporting information can be downloaded at: Preprints.org: primer and probe sequences used in direct RT-qPCR, Figure S2: representative examples of rapid antigen test results, Table S3: data table containing diagnostic, cobas Liat, direct PCR and rapid antigen results.
Funding
This study did not receive any funding.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Acknowledgments
We would like to thank the InActiv Blue team members for help during DNA/RNA Defend Pro product development and critical reading of the manuscript: Sven Comhaire, Ilse Jonckheere, Liesbeth Faes, Dennis Berteloot. We also thank Michiel Vandewalle for the rapid antigen test photo collage.
Conflicts of Interest
DNA/RNA Defend Pro medium was provided by InActiv Blue. M.C. and J.V. are affiliated to InActiv Blue.
References
- World Health Organization (WHO). Diagnostic testing for SARS-CoV-2, Interim guidance. Available online: https://www.who.int/publications/i/item/diagnostic-testing-for-sars-cov-2 (accessed on 10 March 2024).
- Bouttell, J.; Hawkins, N. Evaluation of Triage Tests When Existing Test Capacity Is Constrained: Application to Rapid Diagnostic Testing in COVID-19. Med Decis Making 2021, 41, 978–987. [Google Scholar] [CrossRef]
- InActiv Blue. DNA/RNA Defend Pro buffer. Available online: https://www.inactivblue.com/product-dna-rna-defend-pro/ (accessed on 10 March 2024).
- Deprez, S.; Steyaert, S. Evaluation of a novel respiratory virus inactivating buffer for parallel RT-qPCR and quick antigen testing. (Preprint). [CrossRef]
- cobas SARS-CoV-2 & Influenza A/B [package insert V01]. Roche Molecular Systems, Inc.: Pleasanton, CA, 2020.
- Fomsgaard, A.S.; Rosenstierne, M.W. An alternative workflow for molecular detection of SARS-CoV-2—escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill 2020, 25, 2000398. [Google Scholar] [CrossRef]
- Chu, A.W.; Chan, W.M. Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. J Clin Virol 2020, 129, 104519. [Google Scholar] [CrossRef]
- Samantha, A. Byrnes, Multiplexed and Extraction-Free Amplification for Simplified SARS-CoV-2 RT-PCR Tests. Analytical Chemistry 2021, 93, 4160–4165. [Google Scholar] [CrossRef]
- InActiv Blue. DNA/RNA Defend Pro instructions for use (IFU). Available online: https://www.inactivblue.com/files/Instructions%20for%20use%20-%20RUO%20-%20DRDP%20-%20final%20version.pdf (accessed on 10 March 2024).
- Corman, V.M.; Landt, O. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020, 25, 2000045. [Google Scholar] [CrossRef]
- LeBlanc, J.J.; Li, Y. Switching gears for an influenza pandemic: validation of a duplex reverse transcriptase PCR assay for simultaneous detection and confirmatory identification of pandemic (H1N1) 2009 influenza virus. J Clin Microbiol 2009, 47, 3805–3813. [Google Scholar] [CrossRef]
- van Elden, L.J.; Nijhuis, M. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J Clin Microbiol 2001, 39, 196–200. [Google Scholar] [CrossRef]
- Hu, A.; Colella, M. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real-time PCR. J Clin Microbiol 2003, 41, 149–154. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).