1. Introduction
Ningxiang pig is one of the excellent local pig breeds in China, which has the characteristics of early maturity and easy fattening, wide adaptability, and tasty meat, and thus is popular among consumers[
1]. However, it has obvious characteristics such as high fat percentage and low lean meat percentage[
2], low feed utilization efficiency, slow growth, and low meat yield, which seriously affects the efficiency and benefits of farming and becomes a key bottleneck for the development of Ningxiang pig industry.
In-depth analysis of the molecular mechanism of fat deposition is a prerequisite for regulating the rational deposition of fat in a targeted manner. In 1986, Benne et al [
3] reported for the first time RNA editing, an important and specific way of post-transcriptional modification of eukaryotic genes, which alters the genetic information conveyed by RNAs. RNA editing that occurs in miRNAs is usually referred to as miRNA editing. miRNAs [
4] are endogenous non-coding RNAs of about 22 nucleotides. Researches on miRNA editing has never stopped since it was first reported in 2004 [
5]. Increased A-to-G RNA editing during mammalian brain development has been reported to result in alterations of miR-381 and miR-376b bases, which affect their target specificity and lead to their developmental disruption [
6]. Giovanni Nigita et al. investigated the potential new roles which environmental factors play on the editing of the seed region in miRNAs, thus resulting in the alteration of their respective targetomes [
7]. In cancer research, miRNA editing also plays a crucial role, such as the downregulation of miR-17 [
8] in melanoma stem cells, which inhibits the activity of melanoma stem cells, promotes cell differentiation. Wang et al. found that edited miR-200b promotes cell invasion and migration by impairing its ability to inhibit
ZEB1/ZEB2 and gaining the ability to inhibit new targets [
9].There is limited research on miRNA editing in fat deposition. Meadows et al. found that highly edited miRNAs are closely associated with adipogenic differentiation through analyzing different stages of mouse adipose tissue [
10]. However, whether miRNA editing affects fat deposition in pigs remains unclear.
MiRNAs are widely present in pig adipose tissue and participate in regulating multiple biological processes related to pig fat deposition. Researches showed that miRNAs are extensively involved in the proliferation, differentiation, and fat metabolism of pig adipocytes through targeting specific genes related to fat deposition [
11,
12,
13,
14]. Mature miRNA combine with the 3'- untranslated region (3'-UTR) of the protein-encoded transcript through complementary seed sequences [
15](base 2-8 at that 5' end) to exert its function, thereby reducing the stability of the target transcript or inhibit its protein synthesis [
16]. Given the influence of miRNA editing on the targeting activity of host miRNAs and the significant role of miRNAs in pig fat deposition, this study utilized small RNA-seq technology to identify and analyze miRNA editing sites in subcutaneous fat tissue of Ningxiang pigs at four different stages (N30D, N90D, N150D, and N210D). We aimed to construct a miRNA editing developmental atlas in adipose tissue of Ningxiang pigs and screen functional miRNA editing sites that impact pig fat deposition, which provide novel insights into unraveling the regulatory mechanisms of pig fat deposition.
2. Materials and Methods
2.1. Experimental Animals
Our team previously conducted small RNA-seq on subcutaneous adipose tissues from 12 male Ningxiang pigs, covering four different stages (N30D, N90D, N150D, and N210D) [
17]. Each age group included three replicates, labeled as F1, F2, and F3 (all pigs were half-sibs). The experimental pigs were all provided by the Ningxiang pig breeding farm of Dalong Animal Husbandry Technology Co. Ltd. in Hunan Province, China. Whole transcriptome high-throughput sequencing data accesscode: PRJNA721288 (
https://www.ncbi.nlm.nih.gov/bioproject/PRJNA721288/).
2.2. Filtration and comparison of sequencing data
Sequencing data were filtered under the following conditions: (1) Removal of low-quality reads (where bases with a quality score Q≥30 accounted for less than 95% of the entire read); (2) Trimming of adapter sequences; (3) Exclusion of reads longer than 28 bp or shorter than 15 bp. The quality-controlled reads were aligned to the porcine reference genome (Sus scrofa 11.1) using Bowtie [
18], with the requirement for reads to align uniquely and to have no more than one mismatch (parameters as follows: -n 1 -e 50 -a -m 1 --best --strata --trim3 2). Given the common adenylation and uridylation modifications at the 3′ end of mature miRNAs [
19,
20], the 2 nt at the 3′ end were trimmed during alignment.
2.3. Identification and characterization of miRNA editing sites
The method for identifying miRNA editing sites was based on the approach reported by Alon et al. [
21], involving: (1) aligning filtered data to the pig reference genome and then to known precursor miRNA (Pre-miRNA) sequences in the miRBase database (version 21) [
22], to obtain the quantity of each of the four nucleotides at every position within all Pre-miRNA sequences; (2) performing mismatch detection based on the binomial test with Bonferroni-corrected P values ≤0.05 and mismatch base quality phred ≥30 to identify miRNA editing candidates; (3) Excluding known single nucleotide polymorphisms (SNPs) from the dbSNP database (release 150). Further selection of candidate miRNA editing sites included: (1) selecting sites present in at least three individuals; (2) excluding sites with multiple editing types; (3) considering only miRNA editing sites with a coverage ≥10 reads.
The most abundant four types of miRNA editing sites (A-to-G, C-to-U, G-to-U, and U-to-C) and their flanking sequences (5 bp upstream and downstream) were analyzed for nucleotide preferences using the Two Sample Logos online tool (
http://www.twosamplelogo.org/). RNA editing level was defined as the ratio of editing-type reads to the total reads detected at that site. Time series analysis of average editing levels across stages was conducted using the Mfuzz package [
23]. Due to the comprehensive nature of current research on human miRNA editing, with A-to-G editing types accounting for the majority of identified editing sites in humans, we download human A-to-G miRNA editing sites from the MiREDiBase database (
https://ncrnaome.osumc.edu/miredibase) [
24]. Subsequently, we used the bedtools [
25] software to extract 25 bp flanking sequences upstream and downstream of the editing sites. Following this, we utilized the makeblastdb command to construct a BLAST database for human miRNA editing sites. The flanking sequences of A-to-G miRNA editing sites extracted by bedtools were used as query sequences. We then employed BLASTN to search the human RNA editing site BLAST database, and miRNA editing sites on query sequences with an e-value < 0.001 and >85% consistency were regard as conserved sites between humans and pigs.
2.4. Screening of differential miRNA editing sites
REDITs [
26], a tool that uses a β-binomial model to identify differential editing sites. The tool takes into account both the variance in editing resulting from biological variability and the intrinsic inaccuracy arising from calculating editing from count data such as RNA-seq. As a result, it exhibits greater power and lower false positives at and below the 5% false positive threshold compared to commonly used alternatives such as the t-test, Wilcoxon's rank-sum test, or pooled Fisher's Exact test. In this study, six pairwise comparisons (N30D vs N90D, N30D vs N150D, N30D vs N210D, N90D vs N150D, N90D vs N210D, and N150D vs N210D) were conducted to investigate differences in editing levels between developmental stages.
2.5. Prediction of target genes and KEGG functional enrichment analysis
To evaluate the impact of differentially edited sites within miRNA seed sequences on miRNA function, target genes were predicted for wild-type (WT) and edited-type (ET) miRNAs using the miRanda software [
27] with the following parameters: -sc 140 -en -10 -scale 4. Subsequently, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these target genes was conducted using the OmicShare online platform (
https://www.omicshare.com/tools).
3. Results
3.1. Identification of miRNA editing sites
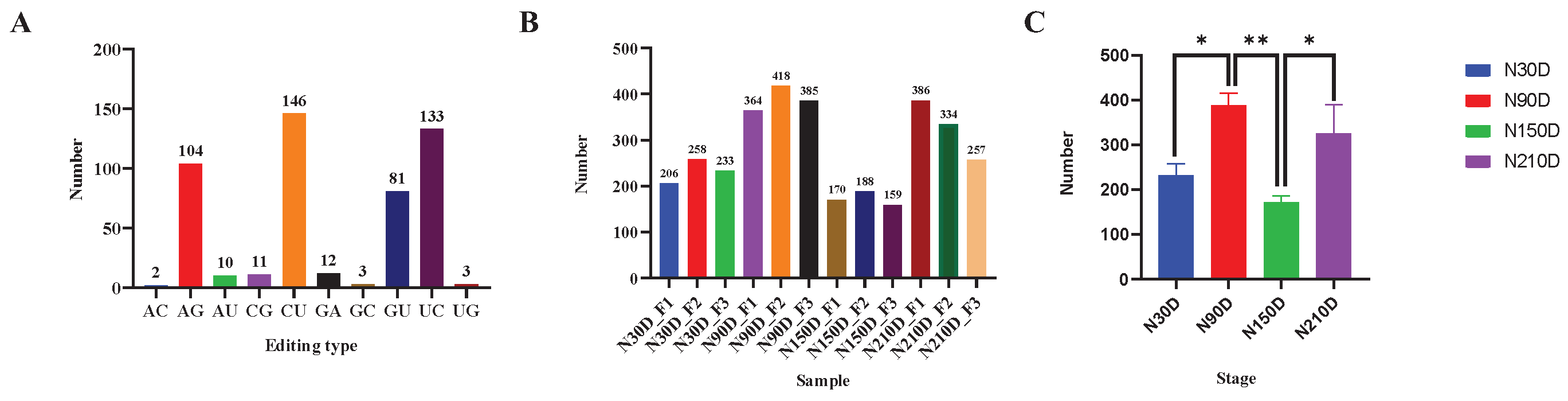
A total of 505 miRNA editing sites were identified, including 10 editing types: A-to-G, G-to-A, C-to-U, U-to-C, G-to-U, C-to-G, G-to-C, A-to-C, U-to-G, and A-to-U. C-to-U editing sites accounted for the most, with 146 (
Figure 1A). The distribution of miRNA editing sites in subcutaneous adipose tissues of Ningxiang pigs across different stages is shown in
Figure 1C. Among them, the 90-day-old had the most miRNA editing events, while the 150-day-old had the least. There was a significant difference between N30D and N90D (
P < 0.05), between N90D and N150D (
P < 0.01), between N150D and N210D (
P < 0.05), and between N30D and N150. The number of miRNA editing sites was also varied among different individuals at the same stage (
Figure 1B). The above results indicated that the distribution of miRNA editing sites is space-time specific.
3.2. miRNA editing characteristic analysis
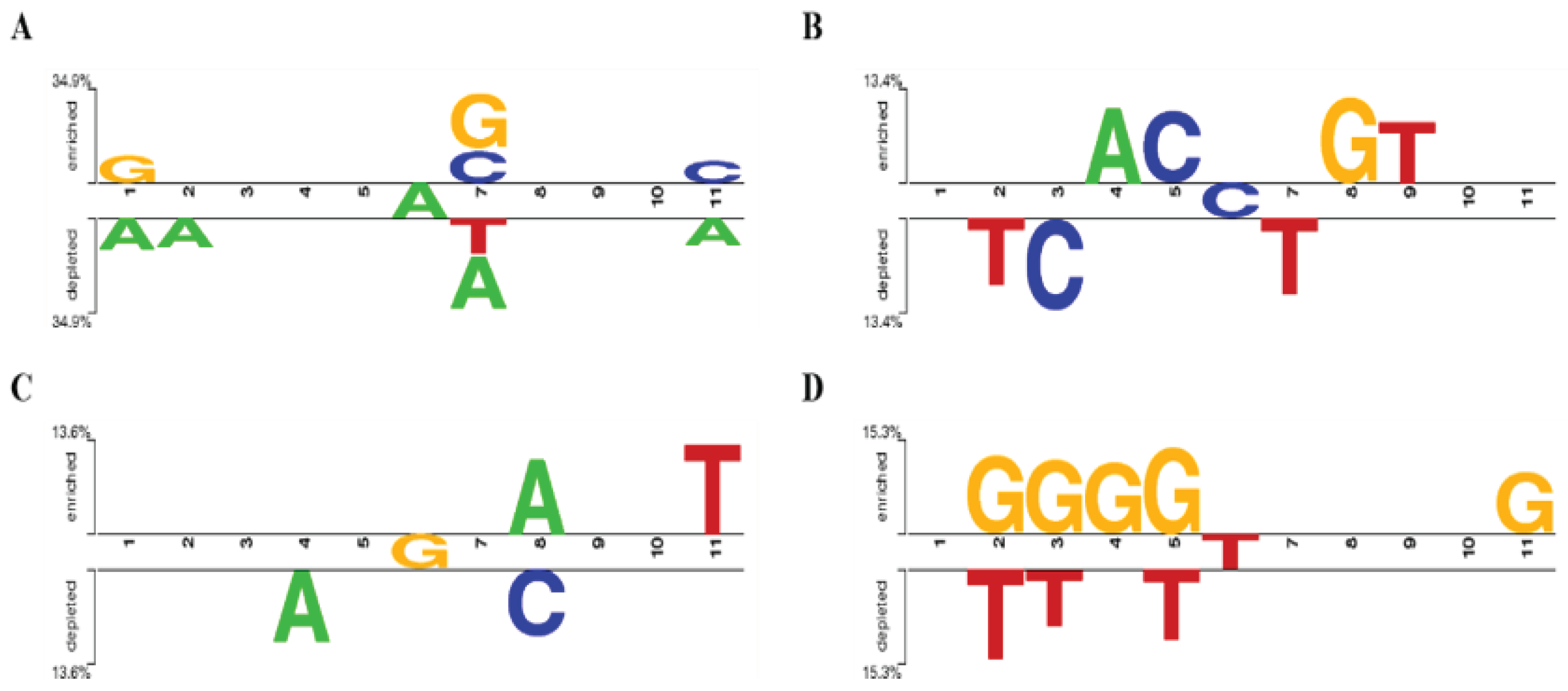
In order to explore whether there was base preference in the flank sequence of miRNA editing sites, the sequence patterns of the four most abundant miRNA editing sites (A-to-G, C-to-U, G-to-U, and U-to-C) were analyzed. As shown in
Figure 2A, there was no preference at 1bp upstream of the A-to-G editing site, while G and C were preferred but A and T were excluded at 1bp downstream. As shown in
Figure 2B, 1bp upstream of the C-to-U editing site favored C, and T was rejected at 1bp downstream. As shown in
Figure 2C, there is no preference within 1bp upstream and downstream of the G-to-U editing site, with the upstream 2bp rejecting A, and the downstream 2bp favoring A and rejecting C. As shown in
Figure 2D, 1bp upstream of the U-to-C editing site preferred G and rejected T, while there was no preference within 4bp downstream, and G was preferred at 5bp downstream.
Figure 2.
Characteristics of upstream and downstream bases of A-to-G (A)、C-to-U (B)、G-to-U (C), and U-to-C (D) miRNA editing sites.
Figure 2.
Characteristics of upstream and downstream bases of A-to-G (A)、C-to-U (B)、G-to-U (C), and U-to-C (D) miRNA editing sites.
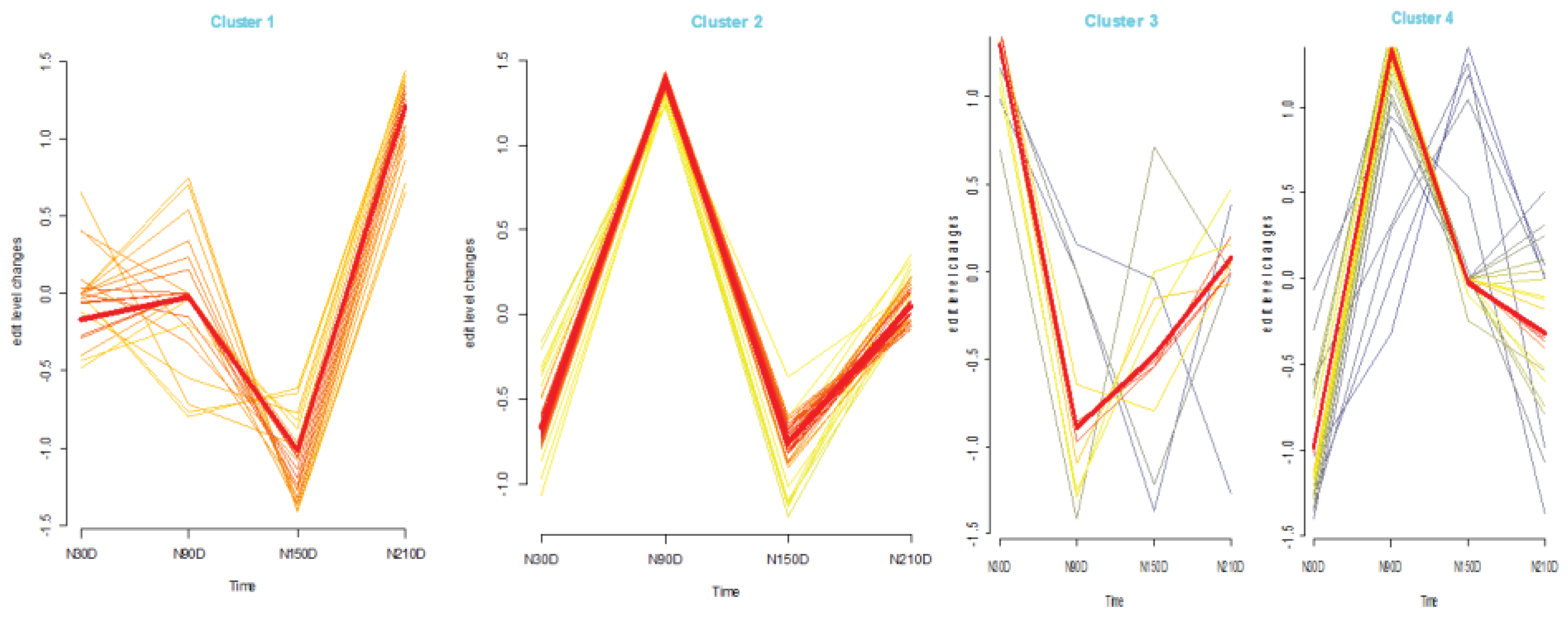
To investigate the dynamic changes in average editing levels across different stages during fat development (Figure 3), Mfuzz was used to categorize the average editing levels into four patterns (Cluster 1, Cluster 2, Cluster 3, and Cluster 4). For Cluster 1, the average editing levels exhibited a gradual increase from N30D to N90D and a rapid decrease followed by an increase from N90D to N210D. In Cluster 2, the average editing levels showed a rapid increase, decrease, and then increase again. Cluster 3 displayed a rapid decrease and increase in average editing levels from N30D to N90D and from N90D to N210D. Meanwhile, for Cluster 4, the average editing levels rapidly increased from N30D to N90D, followed by a progressively slower decrease from N90D to N150D and N150D to N210D. Based on these findings, it is evident that the miRNA editing levels during fat development do not change smoothly but rather undergo distinct transitions. This suggests a close association between fat development and miRNA editing.
Figure 3.
Time series analysis of average editing level among stages.
Figure 3.
Time series analysis of average editing level among stages.
To investigate the conservation of the identified miRNA editing sites, a cross-species analysis was performed between the A-to-G miRNA editing sites identified in Ningxiang pigs and those reported in humans. A total of 67 conserved sites were identified between the two species (Table S1).
3.3. Identification of differential editing sites
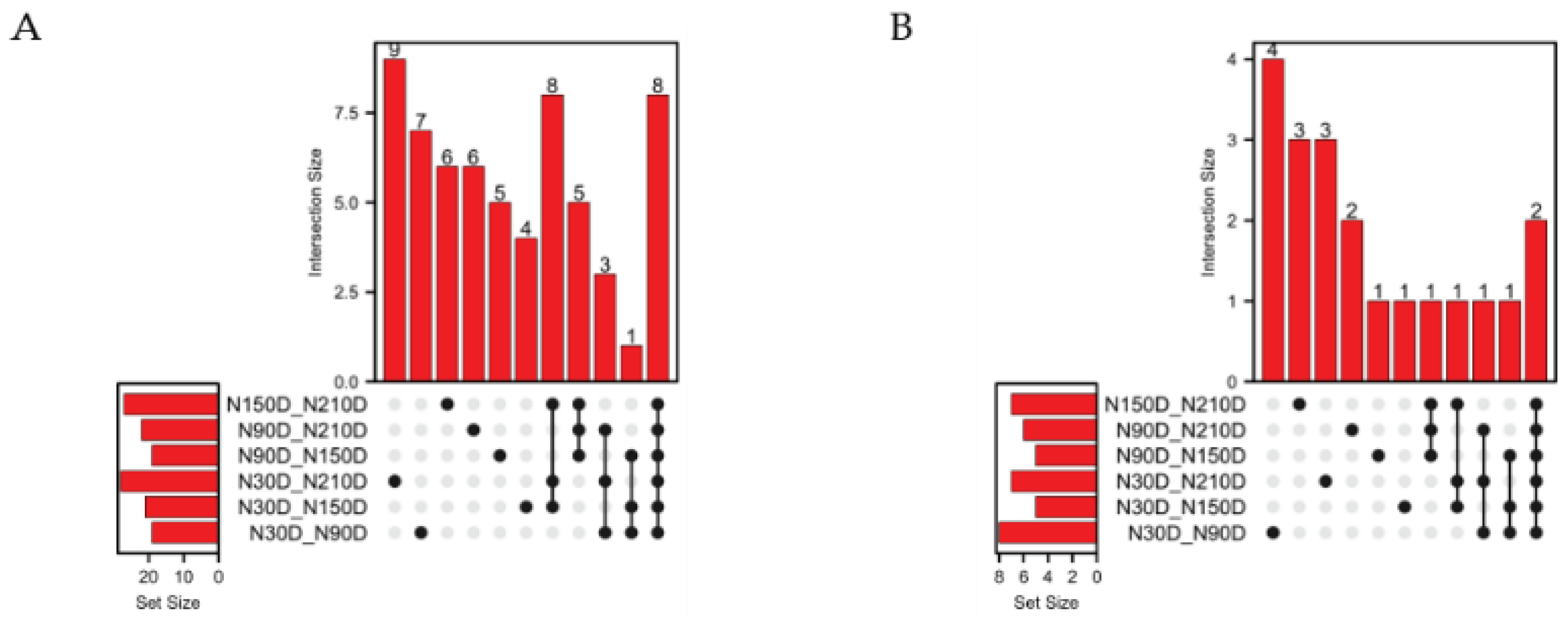
To select miRNA editing sites with significantly different editing levels in the seed region, pairwise comparisons of the editing levels at each site were conducted across the 4 stages. Results revealed 37 sites with differential editing level in only one comparison group, none in two comparison groups, 17 in three comparison groups, 39 in four comparison groups, and 8 sites exhibiting differential editing across all comparison groups (Figure 4A). Among the different comparison groups, there are 62 miRNA editing sites with significant differences in editing levels. After excluding the sites located on sex chromosomes, 20 of these are located in the seed region (Table 1). In the analysis of editing level of the 20 editing sites, there are 14 sites with differences in only one comparison group, 4 sites with differences in three comparison groups, and 2 sites with differences across all comparison groups (Figure 4B).
Figure 4.
Distribution of miRNA editing sites with significant differences in editing levels. (A) Distribution of differential editing sites among compared groups; (B) Distribution of differential editing sites in seed region among compared groups.
Figure 4.
Distribution of miRNA editing sites with significant differences in editing levels. (A) Distribution of differential editing sites among compared groups; (B) Distribution of differential editing sites in seed region among compared groups.
Table 1.
Differential editing sites in miRNA seed region.
Table 1.
Differential editing sites in miRNA seed region.
| Chromosome |
Position |
Mature miRNA name |
Position in miRNA |
Editing type |
| 2 |
65308161 |
ssc-miR-23a |
4 |
A-to-G |
| 2 |
65308350 |
ssc-miR-27a |
4 |
A-to-G |
| 4 |
6952809 |
ssc-miR-30b-5p |
5 |
A-to-G |
| 4 |
6952808 |
ssc-miR-30b-5p |
5 |
A-to-G |
| 11 |
17757478 |
ssc-miR-15a |
5 |
A-to-G |
| 12 |
52422400 |
ssc-miR-497 |
2 |
A-to-G |
| 12 |
52422397 |
ssc-miR-497 |
5 |
A-to-G |
| 13 |
100083195 |
ssc-miR-15b |
5 |
A-to-G |
| 13 |
31655056 |
ssc-miR-425-5p |
7 |
A-to-G |
| 2 |
65308351 |
ssc-miR-27a |
5 |
C-to-U |
| 12 |
46211541 |
ssc-miR-423-5p |
8 |
C-to-U |
| 14 |
6520933 |
ssc-miR-320 |
6 |
C-to-U |
| 1 |
224065570 |
ssc-miR-204 |
2 |
U-to-C |
| 2 |
150580147 |
ssc-miR-145-5p |
7 |
U-to-C |
| 2 |
65308354 |
ssc-miR-27a |
8 |
U-to-C |
| 6 |
58332107 |
ssc-let-7e |
6 |
U-to-C |
| 12 |
45088852 |
ssc-miR-451 |
8 |
U-to-C |
| 12 |
43337029 |
ssc-miR-193a-5p |
5 |
U-to-C |
| 13 |
189138833 |
ssc-miR-155-5p |
2 |
U-to-C |
| 15 |
120453426 |
ssc-miR-26b-5p |
7 |
U-to-C |
3.4. Target gene Prediction and functional enrichment analysis for A-to-G editing site host
Among the 20 differential editing sites located in the seed region, there are 9 editing sites with A-to-G type, 2_65308161_A-to-G, 2_65308350_A-to-G, 4_6952809_A-to-G, 4_6952808_A-to-G, 11_17757478_A-to-G, 12_52422400_A-to-G, 12_52422397_A-to-G, 13_100083195_A-to-G, and 13_31655056_A-to-G, which are respectively located in ssc-miR-23a, ssc-miR-27a, ssc-miR-30b-5p, ssc-miR-15a, ssc-miR-497, ssc-miR-15b, and ssc-miR-425-5p. Notably, ssc-miR-497_2 and ssc-miR-497_5 indicate that editing occurs in bases 2 and 5 of ssc-miR-497, and ssc-miR-30b-5p_5 and ssc-miR-30b-5p_6 indicate that editing occurs in bases 5 and 6 of ssc-miR-30b-5p. These miRNA editing sites led to significant changes in the target gene profiles of their host miRNAs. Specifically, the editing at these sites resulted in a loss of 2,310 to 3,591 target genes, and concurrently a gain of 187 to 2,078 new target genes. (
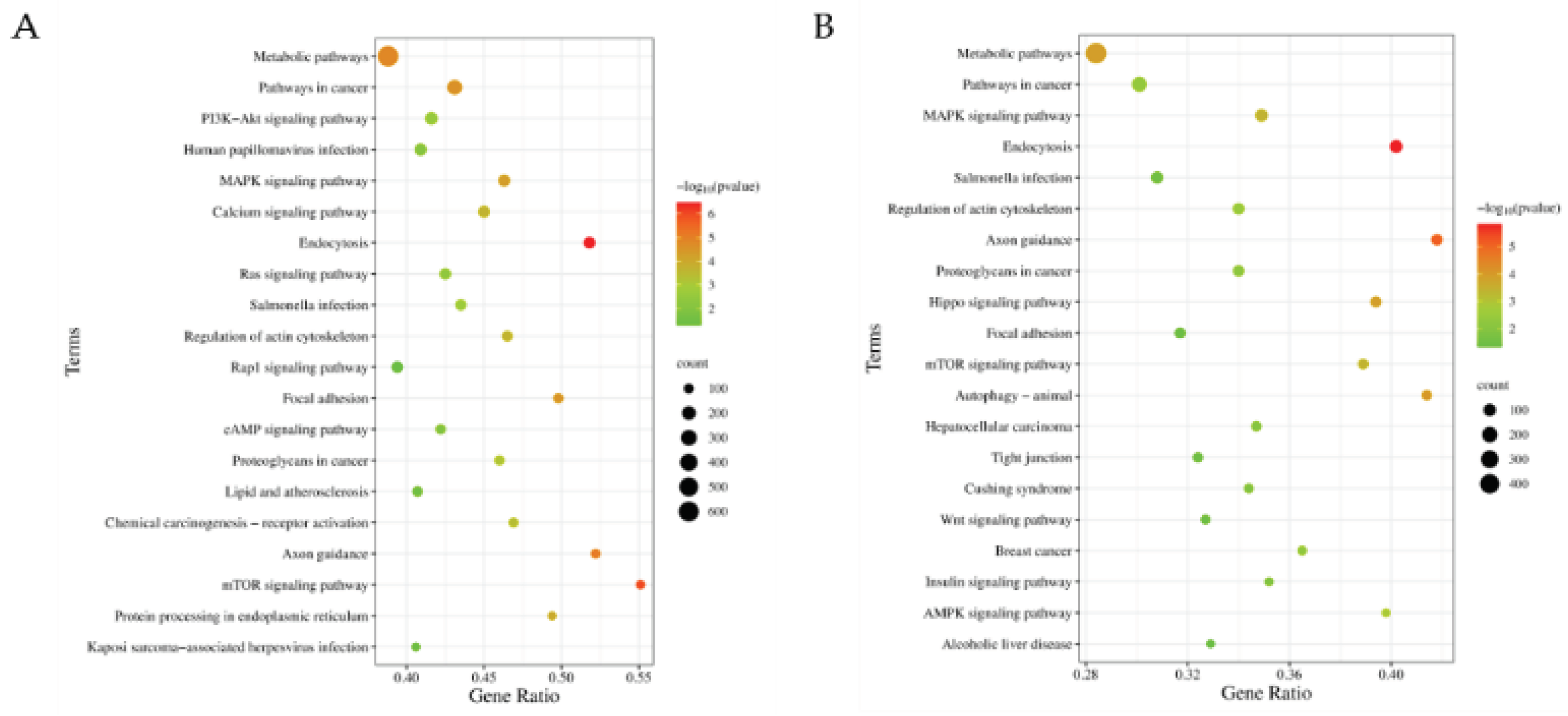
Figure 5). Comparative KEGG enrichment analysis of all wild-type (WT) and edited-type (ET) miRNA target genes revealed the changes of the enriched pathways related to lipid metabolism including a loss of PI3K-Akt signaling pathway, and two new pathways, AMPK signaling pathway and insulin signaling pathway (
Figure 6). The KEGG enrichment analysis results of WT and ET miRNAs target genes of the 9 A-to-G miRNA editing sites are shown in
Figure S1.
Figure 5.
Target genes of WT and ET miRNAs. Target genes of wild (WT) and edited type (ET) ssc-miR-23a (A), ssc-miR-27a (B), ssc-miR-30b-5p_5 (C), ssc-miR-30b-5p_6(D), ssc-miR-15a (E), ssc-miR-497_2, (F) ssc-miR-497_5(G), ssc-miR-15b (H), and ssc-miR-425-5p (I).
Figure 5.
Target genes of WT and ET miRNAs. Target genes of wild (WT) and edited type (ET) ssc-miR-23a (A), ssc-miR-27a (B), ssc-miR-30b-5p_5 (C), ssc-miR-30b-5p_6(D), ssc-miR-15a (E), ssc-miR-497_2, (F) ssc-miR-497_5(G), ssc-miR-15b (H), and ssc-miR-425-5p (I).
Figure 6.
KEGG enrichment analysis results for miRNAs with A-to-G differential editing sites in seed region. (A) KEGG enrichment analysis results for WT miRNAs; (B) KEGG enrichment analysis results for ET miRNAs.
Figure 6.
KEGG enrichment analysis results for miRNAs with A-to-G differential editing sites in seed region. (A) KEGG enrichment analysis results for WT miRNAs; (B) KEGG enrichment analysis results for ET miRNAs.
4. Discussion
Previous studies on miRNA editing have primarily focused on cancer. This study is the first to link pig fat deposition with miRNA editing, analyzing its impact on fat development. Utilizing high-throughput sequencing data, the study explores the potential roles of RNA editing in the fat deposition process of Ningxiang pigs. Identifying miRNA editing sites at various stages (30, 90, 150, and 210 days) revealed fewer miRNA editing sites in Ningxiang pig adipose tissue compared to humans [
21]. Unexpectedly, C-to-U is the most common type in pigs, while A-to-G in humans. However, the result of this study are highly consistent with the study of Wang et al. [
28] which analyzed miRNA editing sites during pig sperm development.
Furthermore, miRNA editing in the seed region can lead to changes in seed sequences, which in turn affect the targeting effect of host miRNAs. Therefore, this study screened miRNA editing sites in the seed region with significantly different editing levels. It has been found that there are numerous RNA editing events in pig fat tissue, with the majority of editing events being A-to-G edits, which might play an important role in pig fat deposition [
29]. Furthermore, miRNA editing sites that have been functionally characterized are almost exclusively of the A-to-G editing type [
30,
31]. Therefore, this study focuses on A-to-G editing sites to analyze the regulatory role of miRNA editing more accurately and specifically on fat deposition in Ningxiang pigs.
In miRNA seed region, 9 differentially edited A-to-G miRNA sites (2_65308161_A-to-G, 2_65308350_A-to-G, 4_6952809_A-to-G, 4_6952808_A-to-G, 11_17757478_A-to-G, 12_52422400_A-to-G, 12_52422397_A-to-G, 13_100083195_A-to-G, and 13_31655056_A-to-G) were ultimately selected, which were located in ssc-miR-23a, ssc-miR-27a, ssc-miR-30b-5p, ssc-miR-15a, ssc-miR-497, ssc-miR-15b, and ssc-miR-425-5p , respectively. These host miRNAs are all related to fat deposition. For instance, miR-23a promotes lipid accumulation [
32]; several studies have reported that miR-27a affects apoptosis and insulin resistance in adipocytes [
33,
34,
35]; miR-30b-5p regulates intracellular lipid metabolism by targeting PPARGC1 [
36]; miR-15a regulates the differentiation of preadipocytes in Yanbian cattle by inhibiting the expression of ABAT [
37]; miR-497 is involved in regulating fatty acid synthesis and affecting insulin resistance [
38]; miR-15b participates in lipid synthesis [
39]; and miR-425-5p inhibits the differentiation and proliferation of preadipocytes [
40].
Additionally, comparative KEGG enrichment analysis of all wild-type (WT) and edited-type (ET) miRNA target genes revealed the changes of the enriched pathways related to lipid metabolism that PI3K-Akt signaling pathway was lost, while AMPK signaling pathway and insulin signaling pathway were newly enriched. The AMPK signaling pathway is typically considered as a metabolic regulatory hub under conditions of energy expenditure, with its activation inhibiting fatty acid synthesis and promoting fatty acid oxidation, thereby reducing fat deposition [
41,
42,
43,
44]. Conversely, the PI3K-Akt signaling pathway promotes the growth, differentiation, and fatty acid synthesis of adipocytes, leading to increased fat deposition [
45,
46,
47]. Therefore, miRNA editing may play a significant role in regulating fat deposition.
Of note, the edited ssc-miR-497_2 newly gained the most target genes, and they significantly enriched in a new pathway, Wnt signaling pathway, which is associated with the regulation of fat deposition [
48] (
Supplementary Figure S1). These findings suggest that miR-497 editing may introduce a novel regulatory role and function in fat deposition. Furthermore, studies have shown that the A-to-G editing site located within miR-497 (12_52422400_A-to-G) is conserved among humans, mice, kangaroos, rhesus monkeys, and pigs [
49], which is of the highest editing level in our study. Therefore, this site was chosen as a key candidate.
A comparative analysis of target genes significantly enriched in the lipid metabolism pathways reveals that the edited ssc-miR-497 loses target genes such as
SCD [
50],
PLAAT3 [
51]
、PNPLA6 [
52]
、ACSL6 [
53]
、ASAH2 [
54]
、CHPT1 [
55]
、FADS2 [
56] and
ACSL4 [
57], all of which promote fat synthesis and differentiation. Conversely, the newly acquired target genes are primarily involved in fat metabolism, including
PLA2G12A [
58]
、LPGAT1 [
54]
、GGT1 [
59]
、HADH [
60]
、MGLL [
61]
、CPT1B [
62], etc. Therefore, we speculated that ssc-miR-497 editing may play an inhibition role in regulating fat deposition in Ningxiang pigs. Based on this speculation, corresponding validation experiments will be conducted in animals or cells in the future.
MiRNA editing, as a novel molecular regulatory mechanism, may not only regulate fat deposition but also impact many biological processes. It might play a crucial role in improving the production performance and genetic improvement of pigs. However, compared to the more abundant results of human miRNA editing, few studies were performed in pigs. It is worth to pay more attention to pig RNA editing and conduct further comparative studies.
5. Conclusions
This study is the first to report miRNA editing phenomenon in pig adipose tissue, which identified and analyzed miRNA editing sites during fat development. We further selected functionally relevant miRNA editing sites and their target genes. Combined with target gene annotation and functional enrichment analysis, ssc-miR-497 editing may play an important role in regulating fat deposition in Ningxiang pigs. These results provide new insights into the regulatory mechanism of pig fat deposition.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: Conservative analysis; Figure S1: KEGG enrichment analysis results of WT and ET miRNAs target genes. ssc-miR-23a (A), ssc-miR-27a (B), ssc-miR-30b-5p_5 (C), ssc-miR-30b-5p_6(D), ssc-miR-15a (E), ssc-miR-497_2 (F); ssc-miR-497_5(G), ssc-miR-15b (H), ssc-miR-425-5p (I).
Author Contributions
Conceptualization, J.H., Y.Z.; Methodology, J.L., Y.Z. and F.Y.; Software, Y.Z., J.L., F.Y. and Y.L.; Resources, Q.Z. and H.M.; Data curation, J.L., F.Y. and Y.L.; Writing—original draft, J.L.; Writing—review and editing, J.L., F.Y., Y.L., J.H. and Y.Z.; Supervision, Y.Z., N.G. and J.H.; Project administration, Y.Z., N.G. and J.H.; Funding acquisition, Y.Z., N.G. and J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Natural Science Foundation of Hunan Province (2021JJ40254), the Excellent Youth Project of Hunan Provincial Department of Education (22B0209) and the Special Fund for the Construction of Innovative Provinces in Hunan (2021NK1009).
Institutional Review Board Statement
The Institutional Animal Care and Use Committee of Hunan Agricultural University evaluated and approved the experimental protocols under approval number 2013-06.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhu, B.; Gao, H.; Yang, F.; Li, Y.; Yang, Q.; Liao, Y.; Guo, H.; Xu, K.; Tang, Z.; Gao, N.; et al. Comparative Characterization of Volatile Compounds of Ningxiang Pig, Duroc and Their Crosses (Duroc x Ningxiang) by Using SPME-GC-MS. Foods 2023, 12. [Google Scholar] [CrossRef]
- Lei, L.; Wang, Z.; Li, J.; Yang, H.; Yin, Y.; Tan, B.; Chen, J. Comparative Microbial Profiles of Colonic Digesta between Ningxiang Pig and Large White Pig. Animals (Basel) 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Sandhoff, R.; Sandhoff, K. Emerging concepts of ganglioside metabolism. Febs Lett 2018, 592, 3835–3864. [Google Scholar] [CrossRef]
- Luciano, D.J.; Mirsky, H.; Vendetti, N.J.; Maas, S. RNA editing of a miRNA precursor. Rna 2004, 10, 1174–1177. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, Y.; Farahani, H.S.; Behm, M.; Lagergren, J.; Ohman, M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res 2012, 22, 1477–1487. [Google Scholar] [CrossRef]
- Nigita, G.; Acunzo, M.; Romano, G.; Veneziano, D.; Lagana, A.; Vitiello, M.; Wernicke, D.; Ferro, A.; Croce, C.M. microRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res 2016, 44, 6298–6308. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, X.; Cui, Y.; Zhang, X. Suppression of RNA editing by miR-17 inhibits the stemness of melanoma stem cells. Mol Ther Nucleic Acids 2022, 27, 439–455. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res 2017, 27, 1112–1125. [Google Scholar] [CrossRef]
- Meadows, S.; Seidler, A.; Wall, M.; Page, J.; Taylor, C.; Flinn, B.; Turner, R.; Santanam, N. Altered Regulation of adipomiR Editing with Aging. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Wang, K.; Li, W.; Bai, Y.; Yang, W.; Ling, Y.; Fang, M. ssc-miR-7134-3p regulates fat accumulation in castrated male pigs by targeting MARK4 gene. Int J Biol Sci 2017, 13, 189–197. [Google Scholar] [CrossRef]
- Feng, H.; Liu, T.; Yousuf, S.; Zhang, X.; Huang, W.; Li, A.; Xie, L.; Miao, X. Identification of potential miRNA-mRNA regulatory network and the key miRNAs in intramuscular and subcutaneous adipose. Front Vet Sci 2022, 9, 976603. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, Y.; Chen, W.; Zhang, Y.; Zeng, Y. miR-34a Regulates Lipid Droplet Deposition in 3T3-L1 and C2C12 Cells by Targeting LEF1. Cells-Basel 2022, 12. [Google Scholar] [CrossRef]
- Cai, R.; Chao, M.; Zhao, T.; Li, R.; Zhang, Z.; Yan, W.; Pang, W. miR-503 targets MafK to inhibit subcutaneous preadipocyte adipogenesis causing a decrease of backfat thickness in Guanzhong Black pigs. Meat Sci 2023, 198, 109116. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Vesely, C.; Tauber, S.; Sedlazeck, F.J.; Tajaddod, M.; von Haeseler, A.; Jantsch, M.F. ADAR2 induces reproducible changes in sequence and abundance of mature microRNAs in the mouse brain. Nucleic Acids Res 2014, 42, 12155–12168. [Google Scholar] [CrossRef]
- Li, B.; Yang, J.; He, J.; Gong, Y.; Xiao, Y.; Zeng, Q.; Xu, K.; Duan, Y.; He, J.; Ma, H. Spatiotemporal Regulation and Functional Analysis of Circular RNAs in Skeletal Muscle and Subcutaneous Fat during Pig Growth. Biology (Basel) 2021, 10. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009, 10, R25. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Nishibu, T.; Ukekawa, R.; Funakoshi, T.; Kurokawa, T.; Suzuki, H.; Hayashizaki, Y.; et al. A comprehensive survey of 3' animal miRNA modification events and a possible role for 3' adenylation in modulating miRNA targeting effectiveness. Genome Res 2010, 20, 1398–1410. [Google Scholar] [CrossRef]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- Alon, S.; Eisenberg, E. Identifying RNA editing sites in miRNAs by deep sequencing. Methods Mol Biol 2013, 1038, 159–170. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kumar, L.F.M. Mfuzz: Soft clustering of time series gene expression data. Bioinformation 2015, 5–7. [Google Scholar]
- Marceca, G.P.; Distefano, R.; Tomasello, L.; Lagana, A.; Russo, F.; Calore, F.; Romano, G.; Bagnoli, M.; Gasparini, P.; Ferro, A.; et al. MiREDiBase, a manually curated database of validated and putative editing events in microRNAs. Sci Data 2021, 8, 199. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 2014, 47, 11–12. [Google Scholar] [CrossRef]
- Picardi, E.; Pesole, G. REDItools: high-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, P.; Li, L.; Che, D.; Li, T.; Li, H.; Li, Q.; Jia, H.; Tao, S.; Hua, J.; et al. miRNA editing landscape reveals miR-34c regulated spermatogenesis through structure and target change in pig and mouse. Biochem Biophys Res Commun 2018, 502, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Zhang, L.; Wang, L.; He, J.; Ma, H.; Wang, L. Preliminary identification and analysis of differential RNA editing between higher and lower backfat thickness pigs using DNA-seq and RNA-seq data. Anim Genet 2022, 53, 327–339. [Google Scholar] [CrossRef]
- Pinto, Y.; Buchumenski, I.; Levanon, E.Y.; Eisenberg, E. Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets. Nucleic Acids Res 2018, 46, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Li, H.; Yang, J.; Shen, X.; Song, C.; Yang, Z.; Wang, X.; Huang, Y.; Lan, X.; Lei, C.; et al. circRNA Profiling Reveals an Abundant circFUT10 that Promotes Adipocyte Proliferation and Inhibits Adipocyte Differentiation via Sponging let-7. Mol Ther Nucleic Acids 2020, 20, 491–501. [Google Scholar] [CrossRef]
- Zheng, Y.; Ji, B.; Song, R.; Wang, S.; Li, T.; Zhang, X.; Chen, K.; Li, T.; Li, J. Accurate detection for a wide range of mutation and editing sites of microRNAs from small RNA high-throughput sequencing profiles. Nucleic Acids Res 2016, 44, e123. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun 2010, 392, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, X.; Li, Y.; Jia, L.; Zhai, L.; Wei, W.; Zhang, L.; Jiang, H.; Bai, Y. Exercise-Induced Browning of White Adipose Tissue and Improving Skeletal Muscle Insulin Sensitivity in Obese/Non-obese Growing Mice: Do Not Neglect Exosomal miR-27a. Front Nutr 2022, 9, 940673. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, D.; Peng, C.; Gao, R.; Li, X.; Zhang, L.; Lv, Q.; Xiao, X.; Li, Q. MicroRNA-27a, downregulated in human obesity, exerts an antiapoptotic function in adipocytes. Endocr J 2023, 70, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, X.F.; Dong, M.Z.; Tan, J.; Zhang, J.; Zhuang, L.K.; Liu, S.S.; Xin, Y.N. MiR-30b-5p regulates the lipid metabolism by targeting PPARGC1A in Huh-7 cell line. Lipids Health Dis 2020, 19, 76. [Google Scholar] [CrossRef]
- Bai, J.; Xu, H.; Fang, J.; Zhang, C.; Song, J.; Zhang, X.; Hao, B.; Yin, B.; Xia, G. miR-15a regulates the preadipocyte differentiation by targeting ABAT gene in Yanbian yellow cattle. Anim Biotechnol 2023, 34, 2343–2352. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yang, H.; Tai, R.; Li, C.; Xia, T.; Liu, Y.; Sun, C. Lnc-hipk1 inhibits mouse adipocyte apoptosis as a sponge of miR-497. Biofactors 2022, 48, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Zhao, Y.; Yu, S.; Hao, Y.; Zhang, P.; Feng, Y.; Zhang, H.; Ma, D.; Liu, J.; Cheng, M.; et al. miR-15b negatively correlates with lipid metabolism in mammary epithelial cells. Am J Physiol Cell Physiol 2018, 314, C43–C52. [Google Scholar] [CrossRef]
- Chen, F.F.; Xiong, Y.; Peng, Y.; Gao, Y.; Qin, J.; Chu, G.Y.; Pang, W.J.; Yang, G.S. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int J Mol Sci 2017, 18. [Google Scholar] [CrossRef]
- Diniz, T.A.; de Lima, J.E.; Teixeira, A.A.; Biondo, L.A.; Da, R.L.; Valadao, I.C.; Silveira, L.S.; Cabral-Santos, C.; de Souza, C.O.; Rosa, N.J. Aerobic training improves NAFLD markers and insulin resistance through AMPK-PPAR-alpha signaling in obese mice. Life Sci 2021, 266, 118868. [Google Scholar] [CrossRef]
- Neopane, K.; Kozlov, N.; Negoita, F.; Murray-Segal, L.; Brink, R.; Hoque, A.; Ovens, A.J.; Tjin, G.; McAloon, L.M.; Yu, D.; et al. Blocking AMPK beta1 myristoylation enhances AMPK activity and protects mice from high-fat diet-induced obesity and hepatic steatosis. Cell Rep 2022, 41, 111862. [Google Scholar] [CrossRef]
- Jung, T.W.; Park, H.S.; Choi, G.H.; Kim, D.; Lee, T. beta-aminoisobutyric acid attenuates LPS-induced inflammation and insulin resistance in adipocytes through AMPK-mediated pathway. J Biomed Sci 2018, 25, 27. [Google Scholar] [CrossRef]
- Frosig, C.; Jensen, T.E.; Jeppesen, J.; Pehmoller, C.; Treebak, J.T.; Maarbjerg, S.J.; Kristensen, J.M.; Sylow, L.; Alsted, T.J.; Schjerling, P.; et al. AMPK and insulin action--responses to ageing and high fat diet. Plos One 2013, 8, e62338. [Google Scholar] [CrossRef]
- Xiao, H.; Sun, X.; Lin, Z.; Yang, Y.; Zhang, M.; Xu, Z.; Liu, P.; Liu, Z.; Huang, H. Gentiopicroside targets PAQR3 to activate the PI3K/AKT signaling pathway and ameliorate disordered glucose and lipid metabolism. Acta Pharm Sin B 2022, 12, 2887–2904. [Google Scholar] [CrossRef]
- Xu, Z.; You, W.; Chen, W.; Zhou, Y.; Nong, Q.; Valencak, T.G.; Wang, Y.; Shan, T. Single-cell RNA sequencing and lipidomics reveal cell and lipid dynamics of fat infiltration in skeletal muscle. J Cachexia Sarcopenia Muscle 2021, 12, 109–129. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a promotes insulin resistance and mediates glucose metabolism by targeting PPAR-gamma-mediated PI3K/AKT signaling. Aging (Albany Ny) 2019, 11, 7510–7524. [Google Scholar] [CrossRef]
- Chen, N.; Wang, J. Wnt/beta-Catenin Signaling and Obesity. Front Physiol 2018, 9, 792. [Google Scholar] [CrossRef]
- Warnefors, M.; Liechti, A.; Halbert, J.; Valloton, D.; Kaessmann, H. Conserved microRNA editing in mammalian evolution, development and disease. Genome Biol 2014, 15, R83. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Liang, X.; Wu, X.; Liu, J.; Yang, S.; Tao, C.; Zhang, J.; Tian, J.; Zhao, J.; et al. Stearoyl-CoA Desaturase is Essential for Porcine Adipocyte Differentiation. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Wang, Z.; Chai, J.; Wang, Y.; Gu, Y.; Long, K.; Li, M.; Jin, L. LncPLAAT3-AS Regulates PLAAT3-Mediated Adipocyte Differentiation and Lipogenesis in Pigs through miR-503-5p. Genes (Basel) 2023, 14. [Google Scholar] [CrossRef]
- Lan, Q.; Liufu, S.; Liu, X.; Ai, N.; Xu, X.; Li, X.; Yu, Z.; Yin, Y.; Liu, M.; Ma, H. Comprehensive analysis of transcriptomic and metabolomic profiles uncovered the age-induced dynamic development pattern of subcutaneous fat in Ningxiang pig. Gene 2023, 880, 147624. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yang, Z.; Zhang, Q.; Hao, W.; Pang, Y.; Zhang, D.; Liu, D. Transcriptional Regulation Associated with Subcutaneous Adipogenesis in Porcine ACSL1 Gene. Biomolecules 2023, 13. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, S.; Zhang, K.; Zhan, H.; Peng, X.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Identifying Selection Signatures for Backfat Thickness in Yorkshire Pigs Highlights New Regions Affecting Fat Metabolism. Genes (Basel) 2019, 10. [Google Scholar] [CrossRef]
- Gong, X.; Zheng, M.; Zhang, J.; Ye, Y.; Duan, M.; Chamba, Y.; Wang, Z.; Shang, P. Transcriptomics-Based Study of Differentially Expressed Genes Related to Fat Deposition in Tibetan and Yorkshire Pigs. Front Vet Sci 2022, 9, 919904. [Google Scholar] [CrossRef]
- Renaville, B.; Prandi, A.; Fan, B.; Sepulcri, A.; Rothschild, M.F.; Piasentier, E. Candidate gene marker associations with fatty acid profiles in heavy pigs. Meat Sci 2013, 93, 495–500. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, H.; Hua, Z.; Zhu, Z.; Tao, J.; Xiao, H.; Zhang, L.; Bi, Y.; Wang, H. ACSL4 Directs Intramuscular Adipogenesis and Fatty Acid Composition in Pigs. Animals (Basel) 2022, 12. [Google Scholar] [CrossRef]
- Ballester, M.; Ramayo-Caldas, Y.; Revilla, M.; Corominas, J.; Castello, A.; Estelle, J.; Fernandez, A.I.; Folch, J.M. Integration of liver gene co-expression networks and eGWAs analyses highlighted candidate regulators implicated in lipid metabolism in pigs. Sci Rep 2017, 7, 46539. [Google Scholar] [CrossRef]
- Fan, B.; Onteru, S.K.; Rothschild, M.F. The GGT1 and IGFBP5 genes are associated with fat deposition traits in the pig (Brief Report). Arch. Anim. Breed. 2009, 52, 337–339. [Google Scholar] [CrossRef]
- Shang, P.; Li, W.; Liu, G.; Zhang, J.; Li, M.; Wu, L.; Wang, K.; Chamba, Y. Identification of lncRNAs and Genes Responsible for Fatness and Fatty Acid Composition Traits between the Tibetan and Yorkshire Pigs. Int J Genomics 2019, 2019, 5070975. [Google Scholar] [CrossRef] [PubMed]
- Dione, N.; Lacroix, S.; Taschler, U.; Deschenes, T.; Abolghasemi, A.; Leblanc, N.; Di Marzo, V.; Silvestri, C. Mgll Knockout Mouse Resistance to Diet-Induced Dysmetabolism Is Associated with Altered Gut Microbiota. Cells-Basel 2020, 9. [Google Scholar] [CrossRef]
- Watanabe, L.M.; Pereira, V.; Noronha, N.Y.; de Souza, P.M.; Wolf, L.S.; de Oliveira, C.C.; Placa, J.R.; Noma, I.; Da, S.R.G.; de Souza, V.; et al. The influence of serum selenium in differential epigenetic and transcriptional regulation of CPT1B gene in women with obesity. J Trace Elem Med Biol 2023, 83, 127376. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).