Submitted:
18 February 2024
Posted:
19 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- When data changes, the favorite sequence requires recalculation.
- Re-comparing each database sequence with the updated favorite sequence consumes computational time and resources.
- Each database has a distinct favorite sequence, complicating the merging of databases, particularly in extensive datasets with diverse structures and access methods.
1.1. Proposed Research Objectives
1.1.1. Constant Favorite Sequence:
1.1.2. Minimizing Comparisons with Favorite Sequence:
1.1.3. Unification of Sequence Favorites across Databases:
1.2. Research Purpose and Methodology
2. Materials and Methods
2.1. Methods and Alorithms for Sequence Alignment
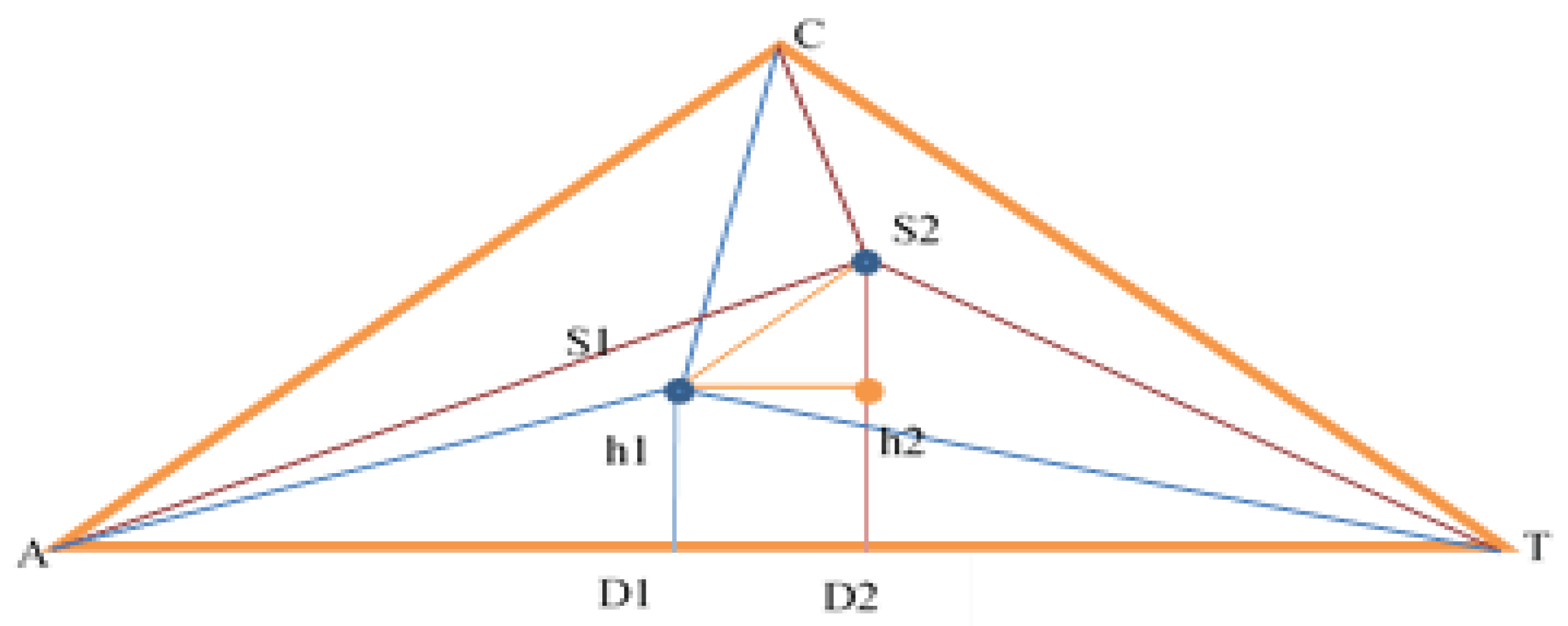
2.2. Method for DNA Sequence Alignmment Based on Trilateration
- -
- For A-benchmark and T- benchmark, we should never have matching position and base. On this way base from processed DNA could match on index and base on A or T, but not both.
- -
- For C-benchmark, we want index and base of 25% of A-benchmark and 25% of T-benchmark to match on C-benchmark.
- ACGTACGTACGTACGTACGTACGTACGTACGTACGTAC....... – A-Benchmark
- GTACGTACGTACGTACGTACGTACGTACGTACGTACGT…… – T- Benchmark
- AGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG....... – C- Benchmark
-
More than one sequence of the same length to get the same values for AD and h:
- Due to the nature of benchmark sequences and the fact that the real sequence projects at most a quarter of the bases onto the benchmark, i.e. with benchmark ACGT and projection of G at the second position, there are no matches, and no value is accumulated for match rate.
-
During S1S2 calculation, the same values are obtained:
- Because of statistical errors accumulated when calculating AD and h.
- Because of rounding in calculations due to the range of data types, this cannot be avoided even with the use of more precise types.
- ACGT
- XGXX
- ACG_T
- X_GXX
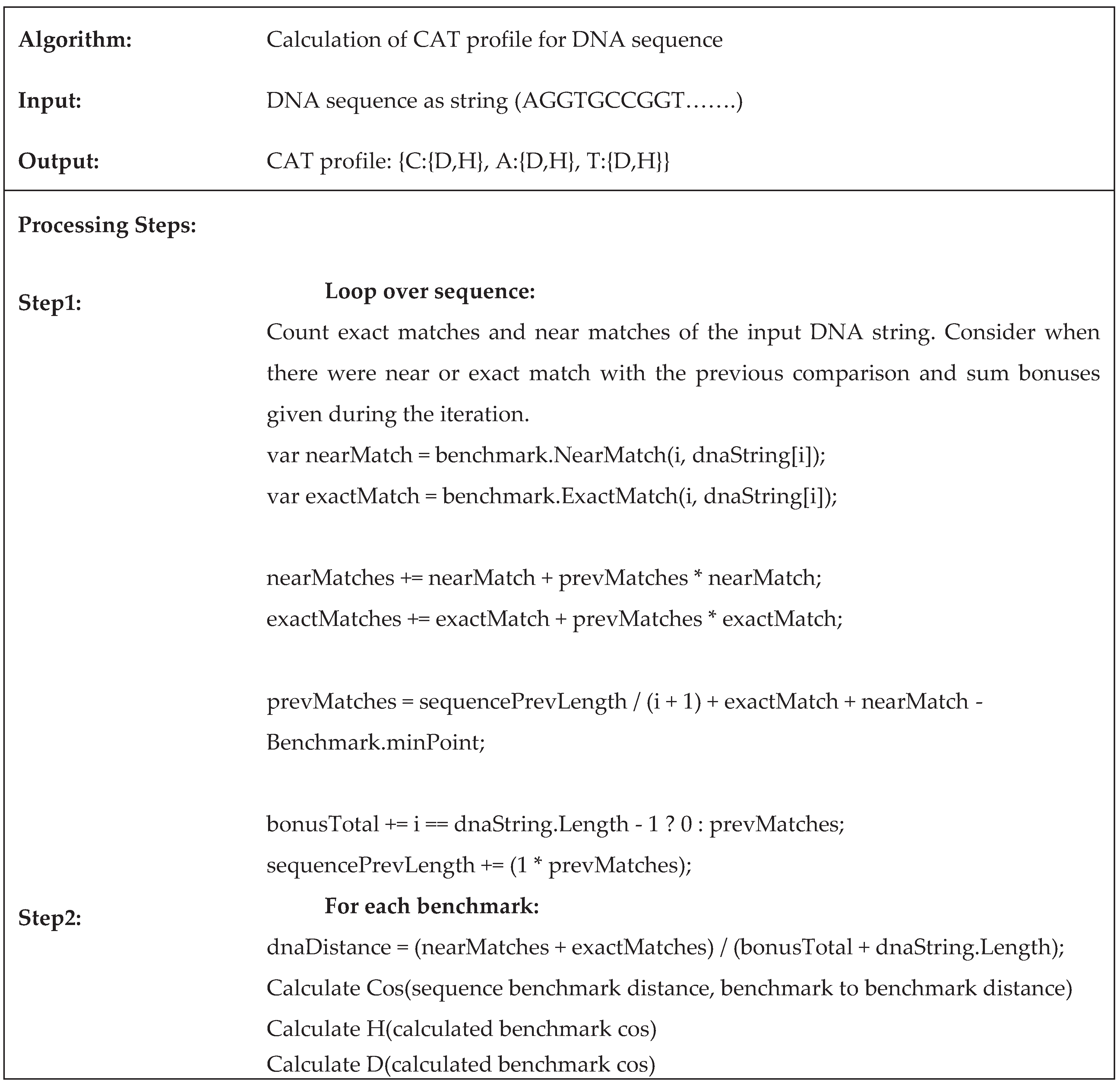
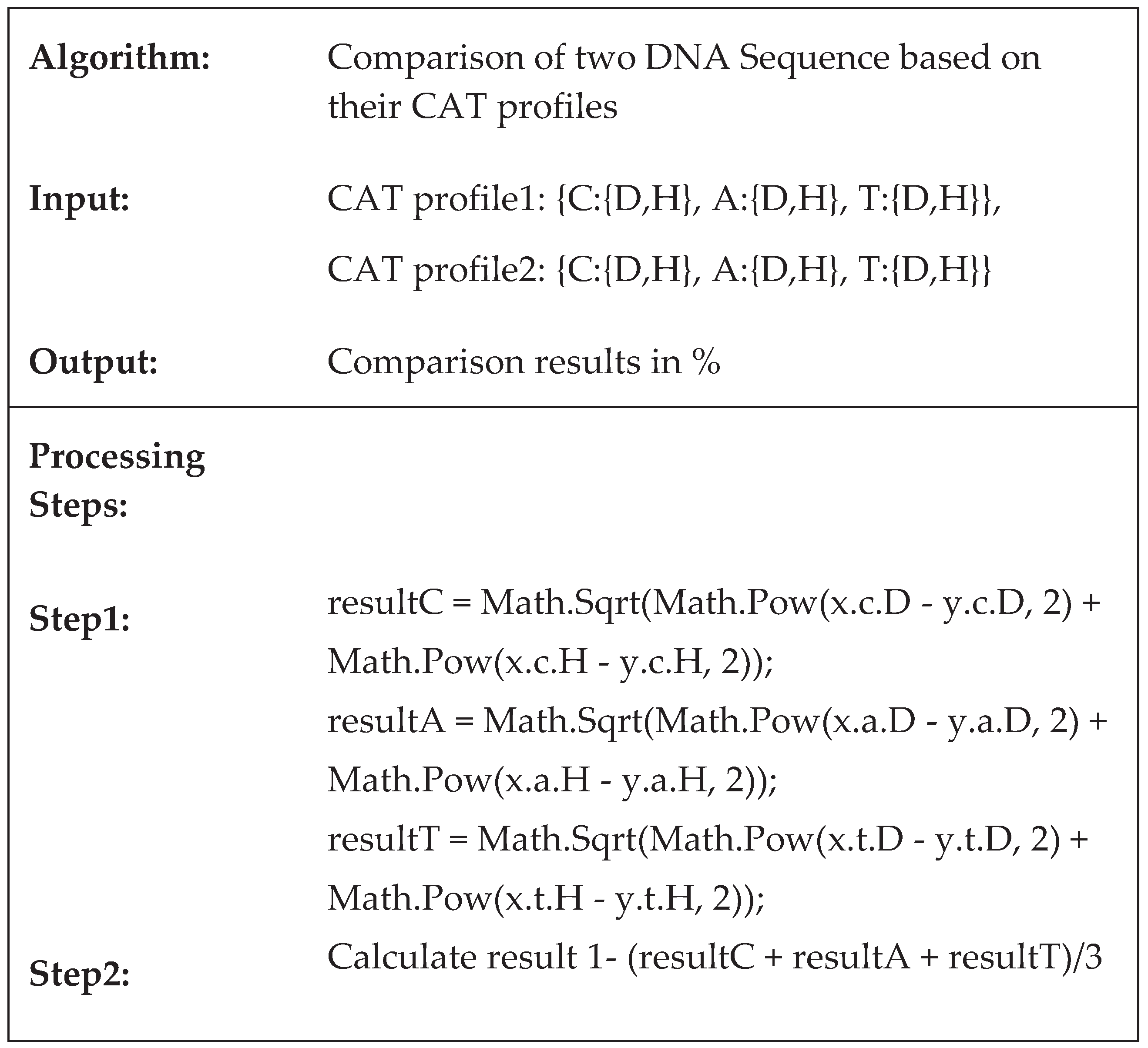
2.3. An Algorithm for Pairwise DNA Sequences Alignment Based on the Proposed CAT Method
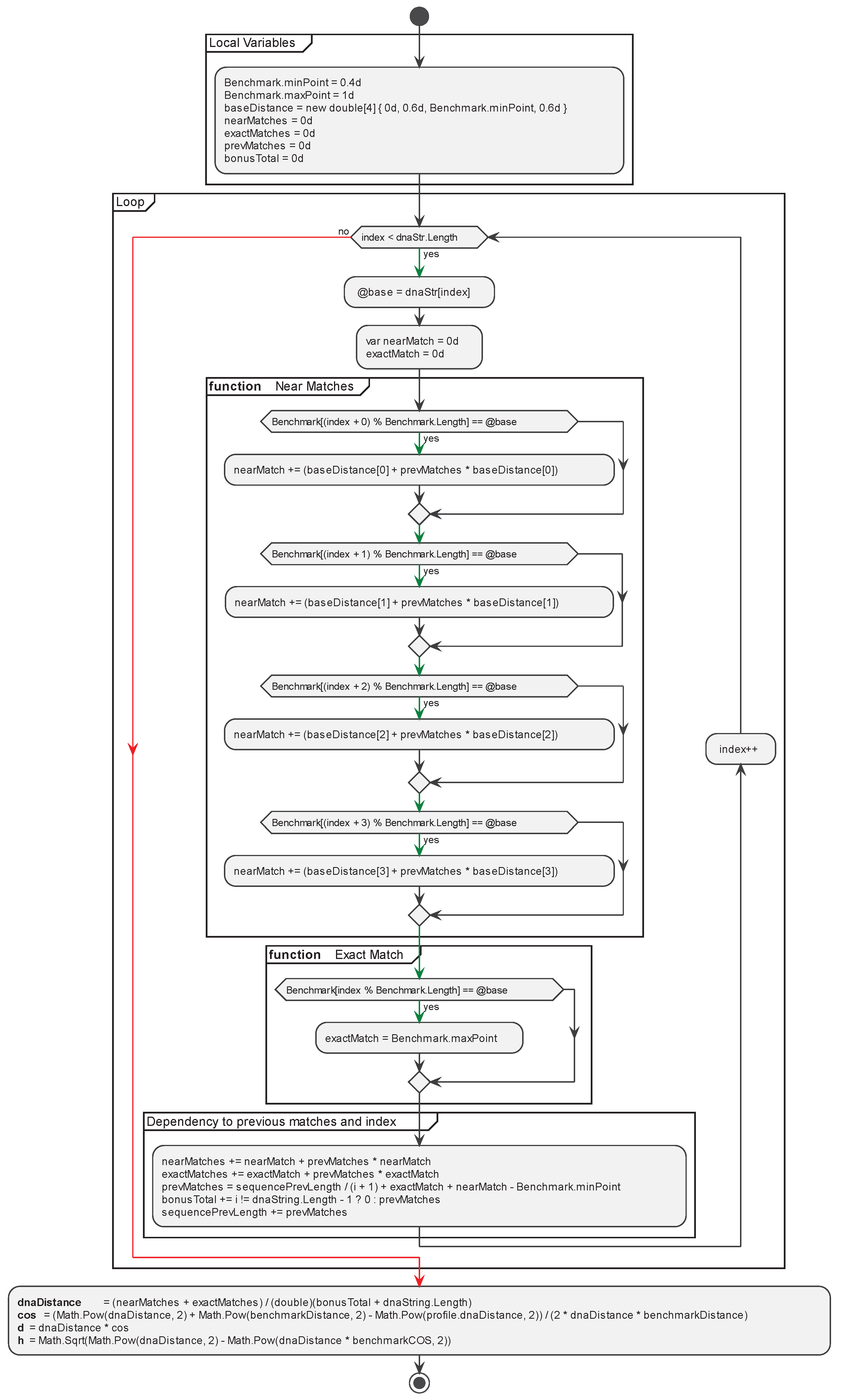
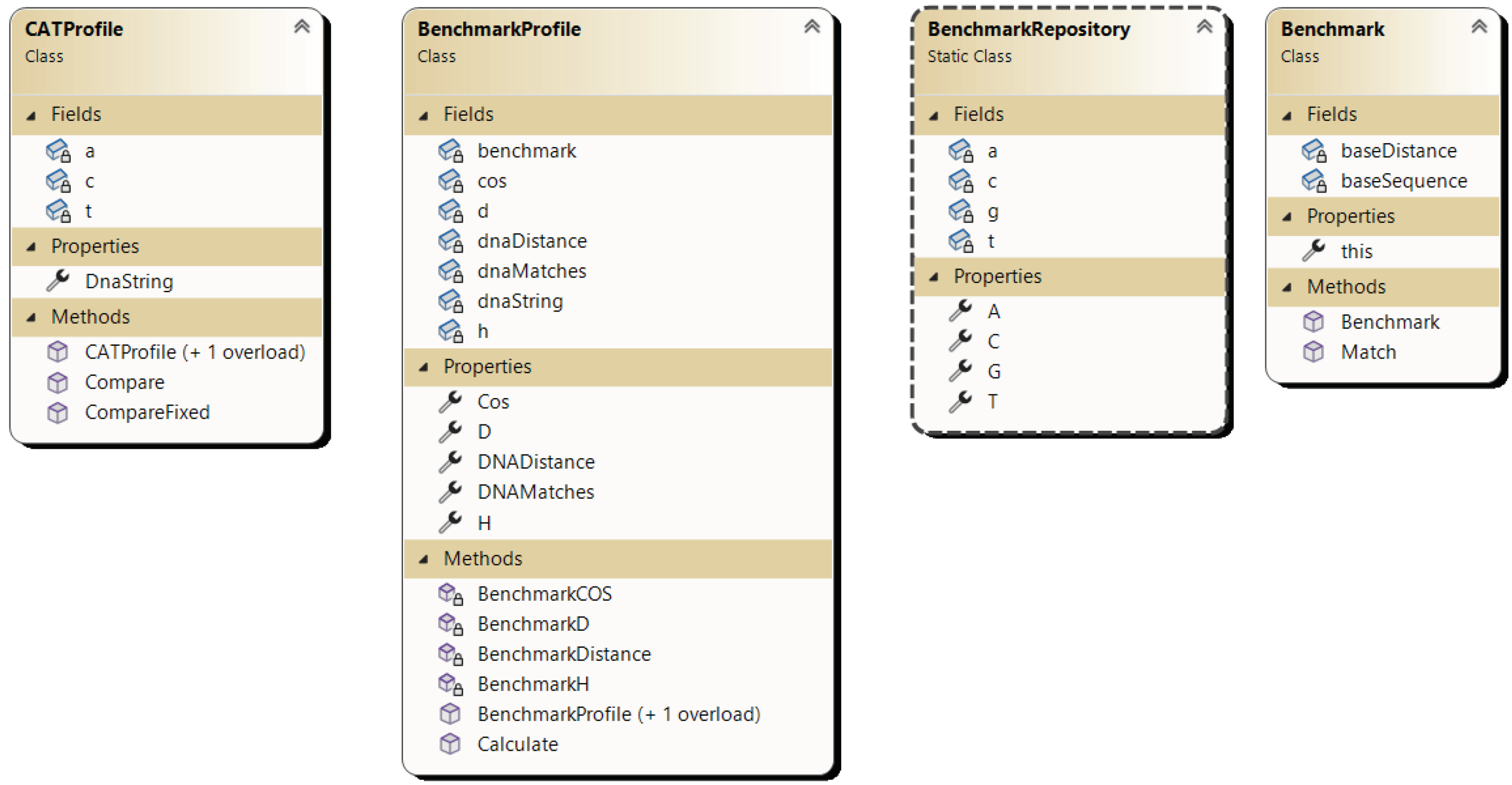
2.4. Implementation of the Proposed Method CAT for Sequences Alignment
- Benchmark: Base class serving as an abstraction for representing benchmark sequences.
- BenchmarkRepo: Repository containing predefined benchmark sequences.
- BenchmarkProfile: Abstraction for plotting a DNA sequence against a benchmark sequence, calculating base parameters for the CAT comparison method such as Cos, D, H.
- CatProfile: Abstraction representing a DNA sequence with pre-calculated parameters for each benchmark sequence from the CAT method.
3. Results
3.1. Collision Analysis
3.2. Performance Analysis
4. Discussion
5. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- EMBL’s European Bioinformatics Institute (EMBL-EBI), Available online:. Available online: https://www.ebi.ac.uk/about/our-impact (accessed on 31 November 2023).
- Luo, R., Liu, B., Xie, Y. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci 2012, 1, 18. [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology 1970;48:443–453. [CrossRef]
- Smith, T.F.; Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. [CrossRef]
- Regnier, M. Knuth-Morris-Pratt algorithm: An analysis. In: Kreczmar, A., Mirkowska, G. (eds) Mathematical Foundations of Computer Science, Lecture Notes in Computer Science 1989, vol 379. Springer. [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. [CrossRef]
- Altschul, S.; et al. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Research 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Borovska, P.; Gancheva, V.; Landzhev, N. Massively parallel algorithm for multiple biological sequences alignment. Proceeding of 36th IEEE International Conference on Telecommunications and Signal Processing 2013, pp. 638-642. [CrossRef]
- Gancheva, V.; Stoev, H. An algorithm for pairwise DNA sequences alignment. Bioinformatics and Biomedical Engineering. IWBBIO 2023. Lecture Notes in Computer Science 2023, vol 13919. Springer, Cham. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, Y.; Ren, J.; Marshall, S. Sequence similarity alignment algorithm in bioinformatics: techniques and challenges. In: Ren, J. et al. Advances in Brain Inspired Cognitive Systems. Lecture Notes in Computer Science 2020, vol 11691. Springer, Cham. [CrossRef]
- Karp, R.M.; Rabin, M.O. Efficient randomized pattern-matching algorithms. IBM J. Res. Dev. 1987;31:249–260. [CrossRef]
- Harde, P. Comparative study of string matching algorithms for DNA dataset. International Journal of Computer Sciences and Engineering 2018, 6. [CrossRef]
- Tun, N.; Thin, M. Comparison of three pattern matching algorithms using DNA Sequences. International Journal of Scientific Engineering and technology Research 2014, 3, 6916–6920. [Google Scholar]
- Chao, J.; Tang, F.; Xu L. Developments in algorithms for sequence alignment: a review. Biomolecules. 2022 Apr 6;12(4):546. [CrossRef]
- Spouge, J.L. Speeding up dynamic programming algorithms for finding optimal lattice paths. SIAM J. Appl. Math. 1989;49:1552–1566. [CrossRef]
- Zhang, F.; Qiao, X. Z.; Liu, Z. Y. A parallel Smith-Waterman algorithm based on divide and conquer, Proceedings of the Fifth International Conference on Algorithms and Architectures for Parallel Processing ICA3PP 2002. [CrossRef]
- Lipman, D.J.; Pearson, W.R. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. [CrossRef]
- Pearson, W.R.; Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA. 1988 ;85:2444–2448. [CrossRef]
- Braun, R.; et al. Three complementary approaches to parallelization of local BLAST service on workstation clusters. Fifth International Conference on Parallel Computing Technologies (PaCT). Lecture Notes in Computer Science (LNCS) 1999, 1662. [Google Scholar]
- Costa, R.; Lifschitz, S. Database allocation strategies for parallel BLAST evaluation on clusters Distributed Parallel Databases 2003, 13, 99–127.
- Oehmen, C.; Nieplocha, J. ScalaBLAST: A scalable implementation of BLAST for high-performance data-intensive bioinformatics analysis. IEEE Transactions on Parallel and Distributed Systems 2006, 17, 740–749. [Google Scholar] [CrossRef]
- Thorsen, O.; Jiang, K.; Peters, A.; Smith, B.; Lin, H.; Feng, W.; Sosa, C. Parallel genomic sequence-search on a massively parallel system. ACM International Conference on Computing Frontiers 2007, New York, USA. [Google Scholar]
- Lin, H.; et al. Massively parallel genomic sequence search on the Blue Gene/P architecture. In Proceedings of the ACM/IEEE conference on Supercomputing, 2008.
- Farrar, M. Striped Smith-Waterman speeds database searches six times over other SIMD implementations. Bioinformatics 2007, 23, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Sathe S.R.; Shrimankar, D.D. Parallelizing and analyzing the behavior of sequence alignment algorithm on a cluster of workstations for large datasets, Int. J. Comput. Appl. 2013, vol. 74, no. 21, pp. 1-13.
- Kaur, K.; Chakraborty, S.; Gupta, M.K. Accelerating Smith-Waterman algorithm for faster sequence alignment using graphical processing unit, Phys.: Conf. Ser. 2022, 2161 012028. [CrossRef]
- Lipták, P.; Kiss, A.; Szalai-Gindl, J.M. Heuristic pairwise alignment in database environments. Genes 2022, 13, 2005. [Google Scholar] [CrossRef] [PubMed]
- Grešová, K.; Vaculík, O.; Alexiou, P. Using attribution sequence alignment to interpret deep learning models for miRNA binding site prediction. Biology 2023, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Petty, T.; Hannig, J.; Huszar, T.I.; Iyer, H. A New string edit distance and applications. Algorithms 2022, 15, 242. [Google Scholar] [CrossRef]
- Gancheva, V.; Stoev, H. DNA sequence alignment method based on trilateration. Bioinformatics and Biomedical Engineering, Lecture Notes in Computer Science 2019, vol. 11466, Springer, Cham, pp. 271-283. [CrossRef]
| DNA length | Average Collisions | Total permutations | Rate of collision‰ 0 - 1000 |
|---|---|---|---|
| 10 | 1 | 1048576 | NaN |
| 11 | 1 | 4194304 | NaN |
| 12 | 1 | 16777216 | NaN |
| 13 | 1 | 67108864 | NaN |
| 15 | 1 | 1073741824 | NaN |
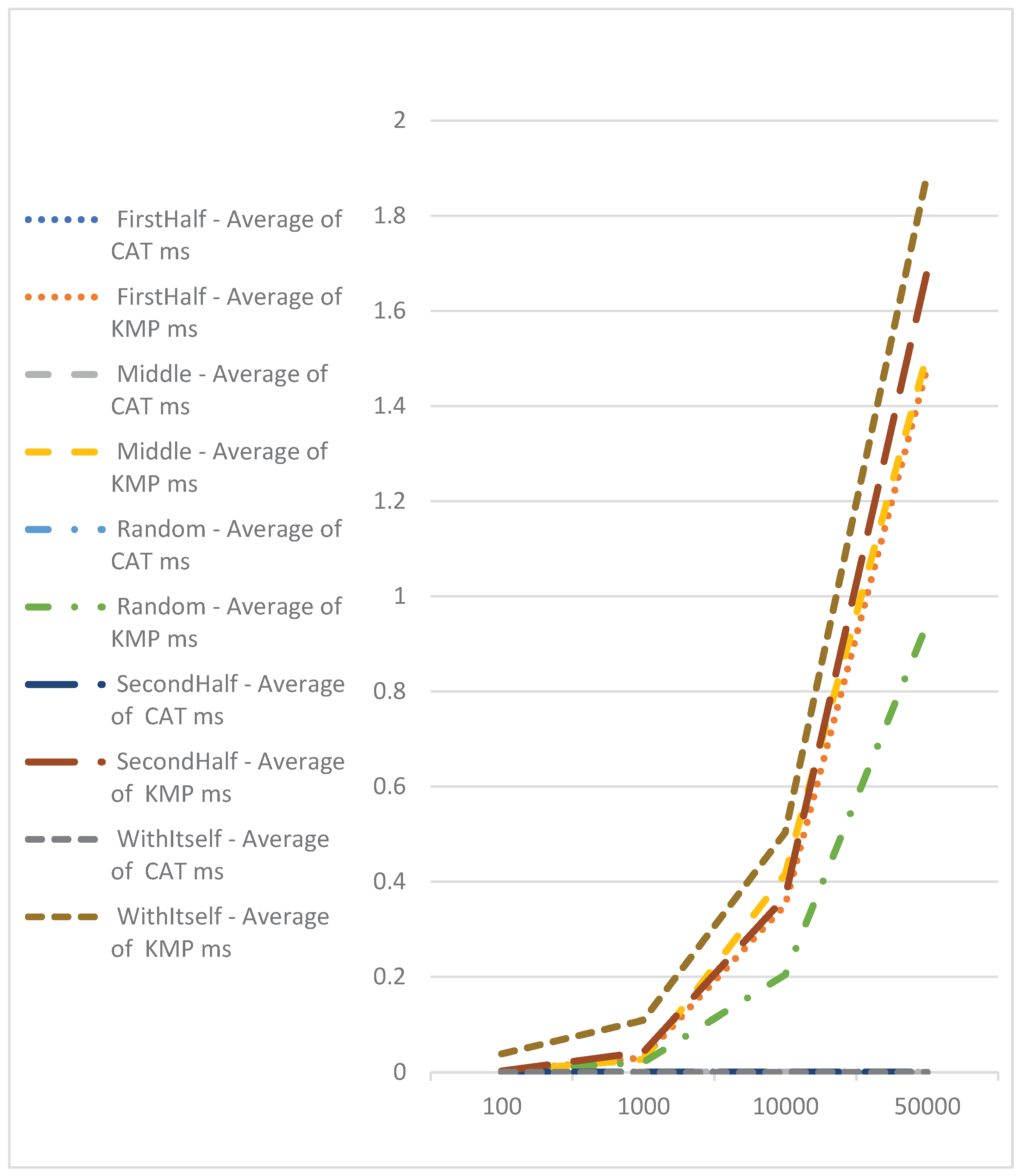
| Average of CAT Elapsed Time | |||||
| DNA Length | FirstHalf | Middle | Random | SecondHalf | WithItself |
| 100 | 0.0004025 | 0.000396 | 0.0004144 | 0.0003975 | 0.000254 |
| 1000 | 0.000466 | 0.0004495 | 0.0005036 | 0.0004555 | 0.0003 |
| 10000 | 0.0007015 | 0.000644 | 0.0007408 | 0.0006945 | 0.00036 |
| 50000 | 0.0003645 | 0.0003805 | 0.0003788 | 0.000356 | 0.000388 |
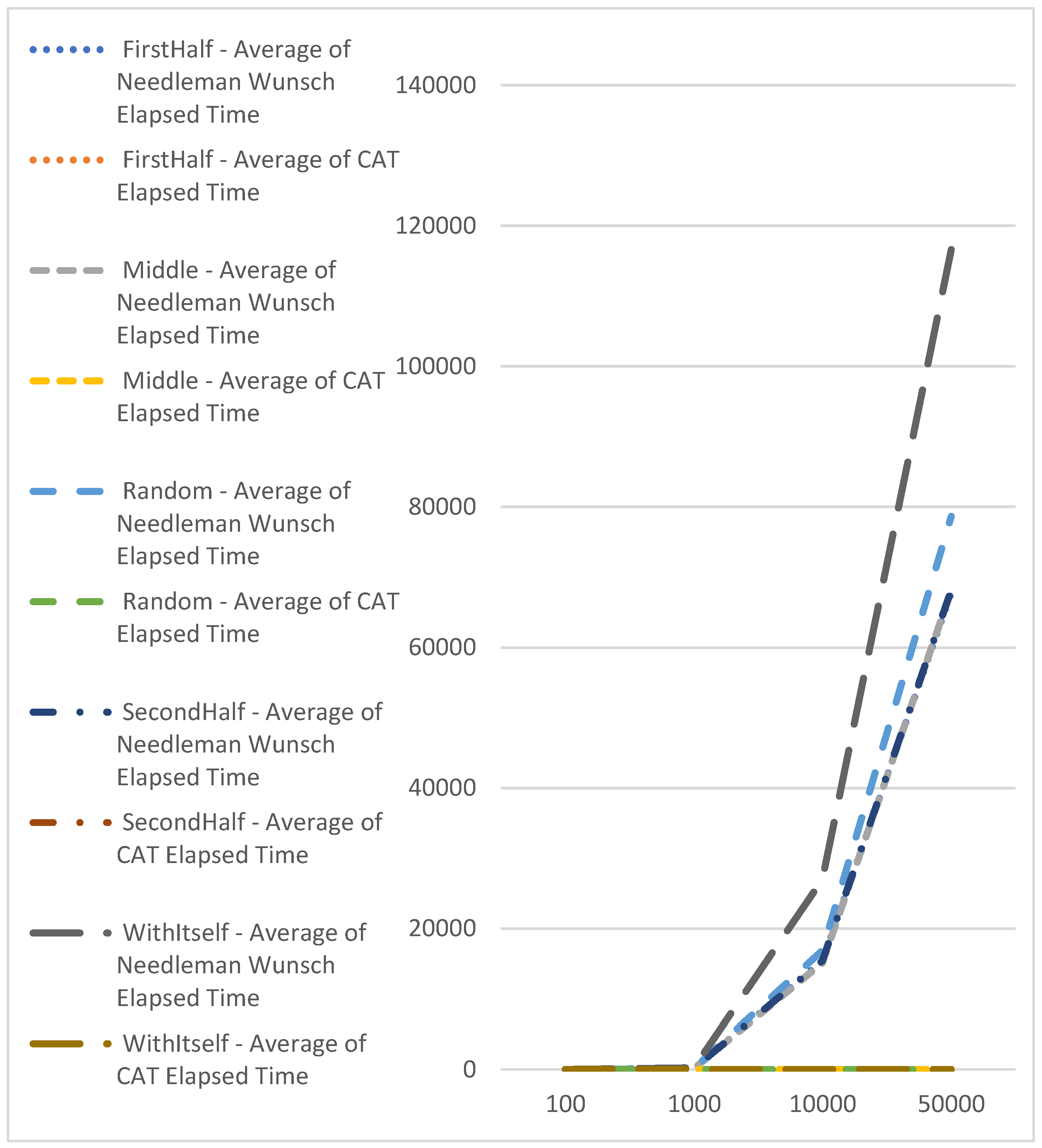
| Average of Needleman Wunsch Elapsed Time | |||||
| FirstHalf | Middle | Random | SecondHalf | WithItself | |
| 100 | 0.810224833 | 0.8224725 | 0.875811143 | 0.807499499 | 1.59419 |
| 1000 | 137.5561462 | 137.9288827 | 148.2296519 | 136.8088905 | 238.639932 |
| 10000 | 15351.73013 | 15130.27244 | 16907.69611 | 15416.20968 | 26981.14435 |
| 50000 | 67806.26151 | 68099.07336 | 78652.58283 | 68162.06354 | 116611.3085 |
| Average of Knuth–Morris–Pratt Elapsed Time | |||||
| FirstHalf | Middle | Random | SecondHalf | WithItself | |
| 100 | 0.002494 | 0.002726 | 0.0018664 | 0.002865 | 0.038884 |
| 1000 | 0.0312275 | 0.0270915 | 0.0212584 | 0.043221 | 0.109476 |
| 10000 | 0.350831 | 0.416332 | 0.2042432 | 0.368386 | 0.503456 |
| 50000 | 1.4803245 | 1.507671 | 0.9423872 | 1.683394 | 1.88701 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





