Submitted:
30 January 2024
Posted:
01 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction:
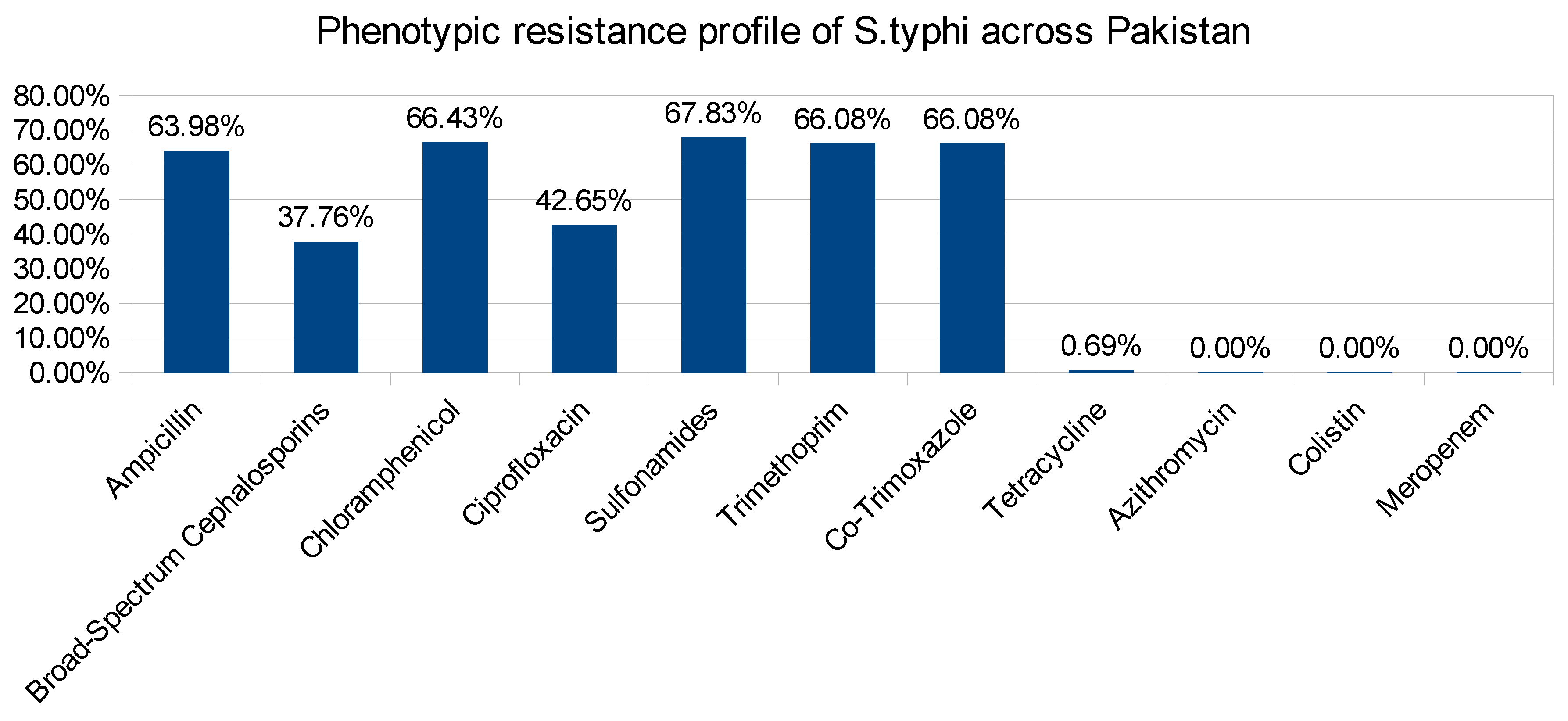
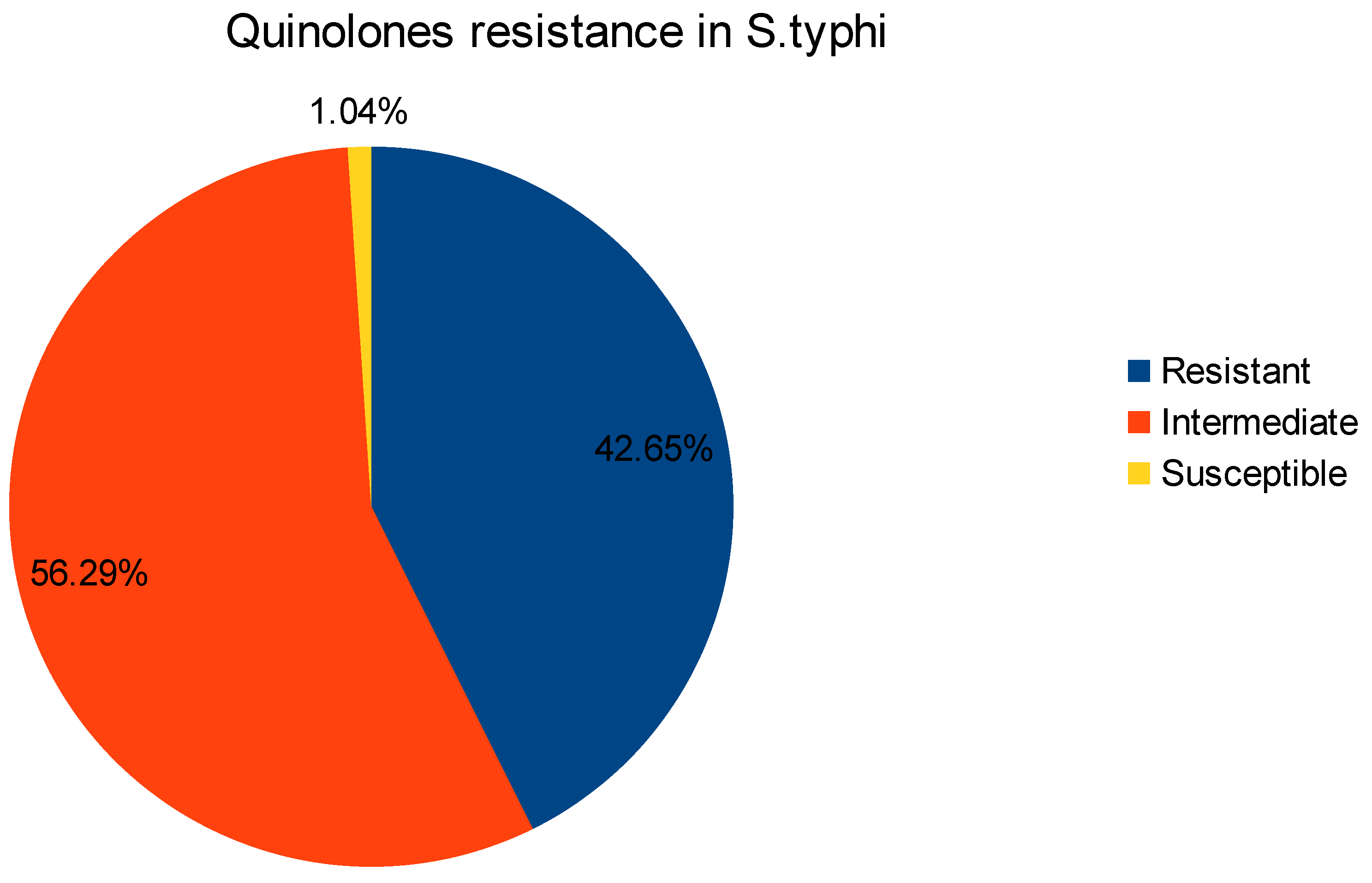
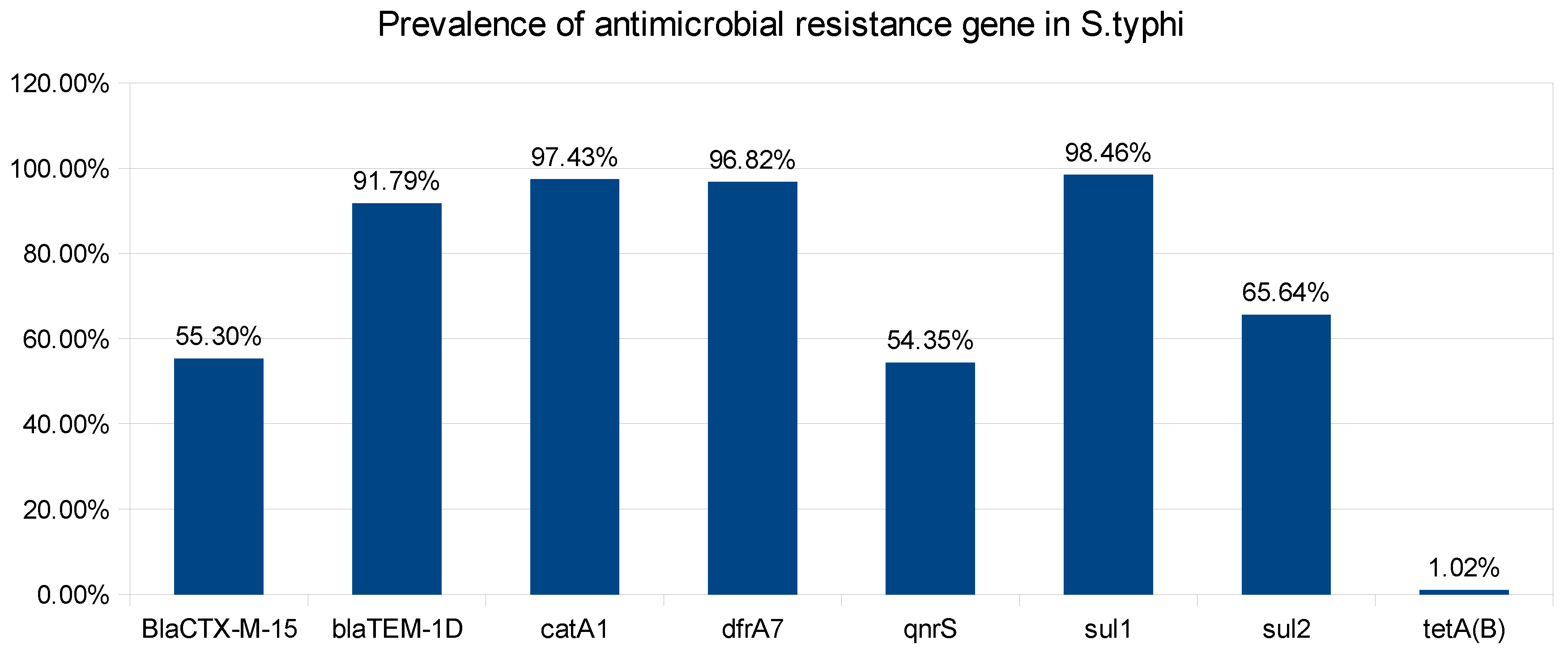
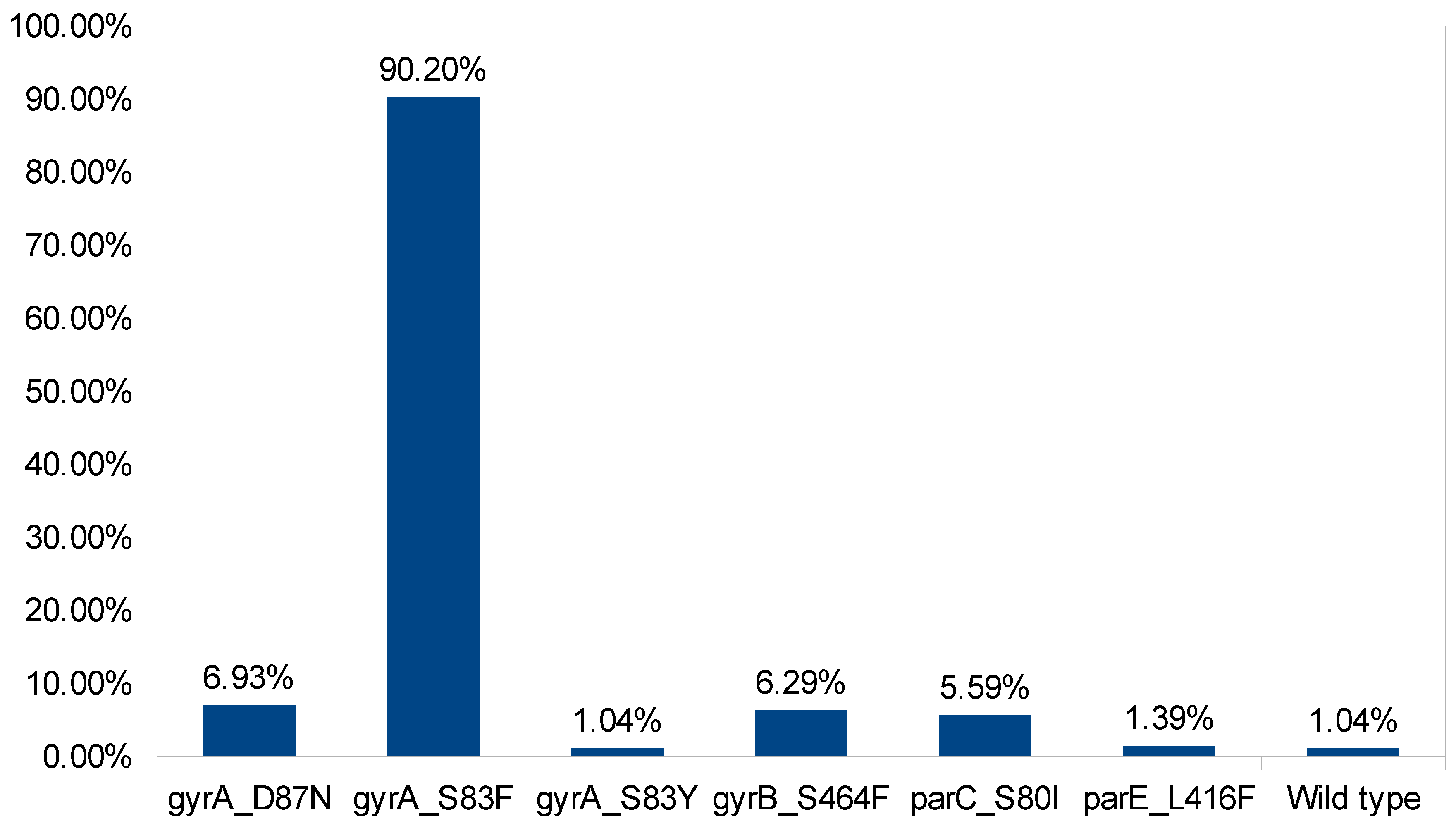
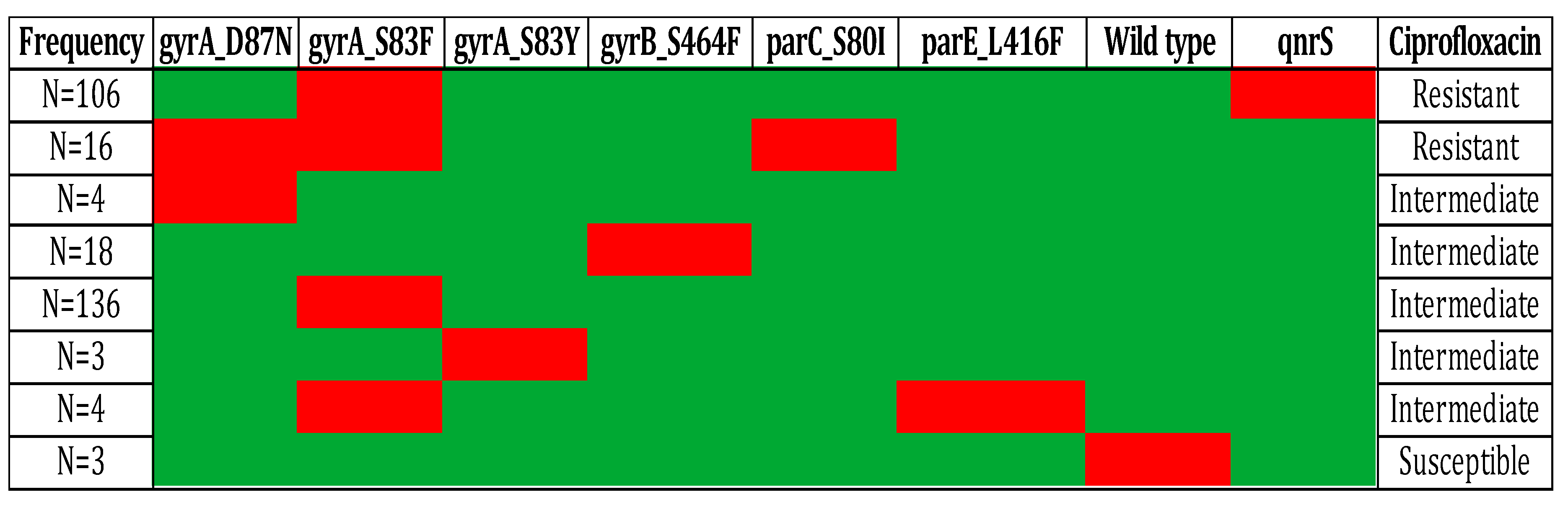
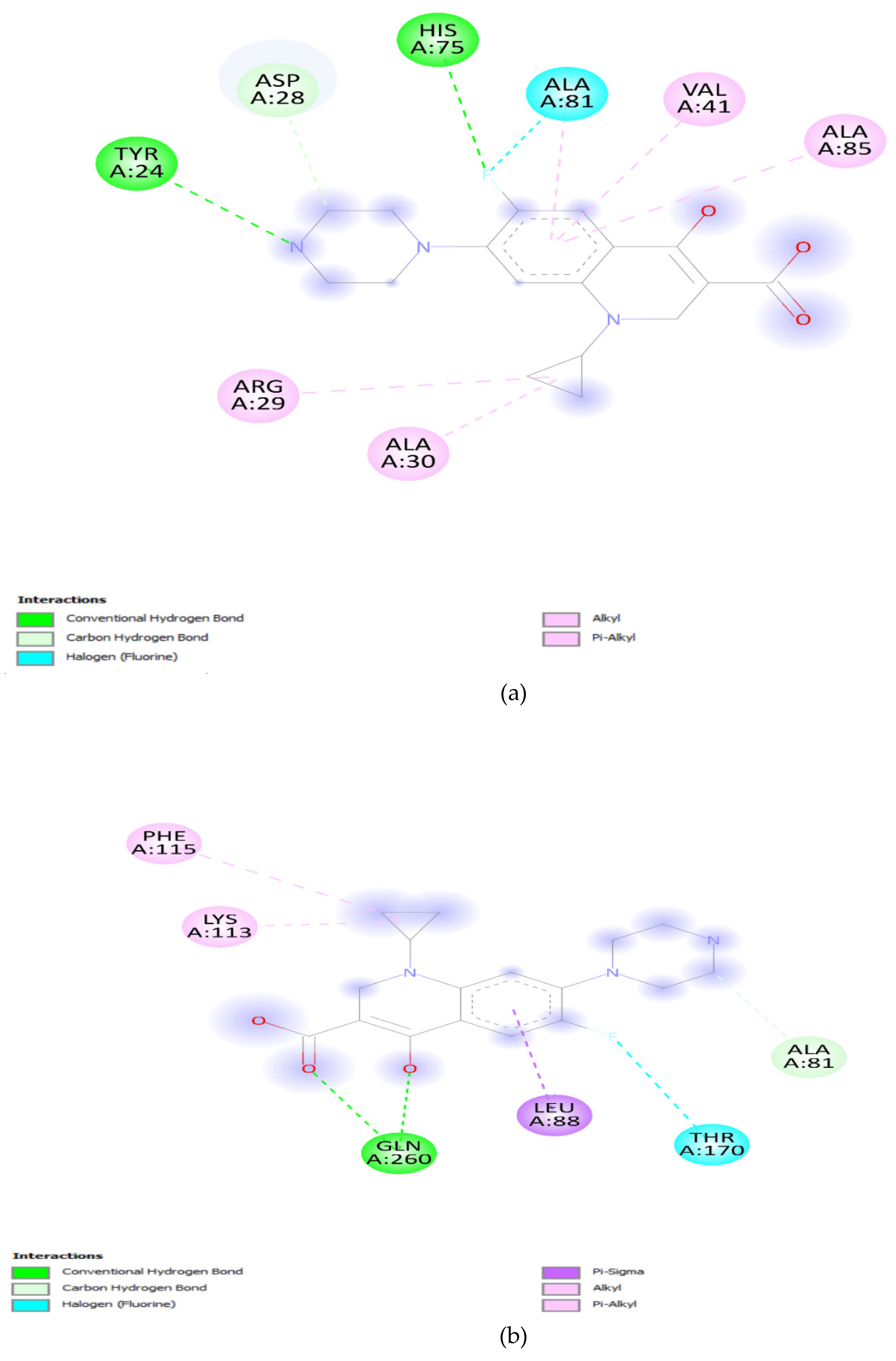
2. Results and Discussions:
3. Material and Methodology:
3.1. Data Extraction:
3.2. Genomic Analysis:
3.3. Mutation analysis in DNA gyrA, and parC genes:
3.4. Protein Structure Modeling:
3.5. Docking Analysis:
4. Conclusion:
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Siourimè, S. N., Isidore, B. O. J., Traoré, Y., & Savadogo, A. (2017). Salmonella enterica Serovar Typhi and Paratyphi Responsible of Typhoid and Paratyphoid Fevers Transmitted by Environment and Food. International Journal of Sciences, 3(05), 87–96. [CrossRef]
- Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlöf, D. L., Canals, R., Grissom, J. E., Conway, T., Hokamp, K., & Hinton, J. C. D. (2013). An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host & Microbe, 14(6), 683–695. [CrossRef]
- Gordon, M. A., Graham, S. M., Walsh, A. L., Wilson, L., Phiri, A., Molyneux, E., Zijlstra, E. E., Heyderman, R. S., Hart, C. A., & Molyneux, M. E. (2008). Epidemics of InvasiveSalmonella entericaSerovar Enteritidis andS. entericaSerovar Typhimurium Infection Associated with Multidrug Resistance among Adults and Children in Malawi. Clinical Infectious Diseases, 46(7), 963–969. [CrossRef]
- Britto, C., Wong, V., Dougan, G., & Pollard, A. J. (2018). A systematic review of antimicrobial resistance in Salmonella enterica serovar Typhi, the etiological agent of typhoid. PLOS Neglected Tropical Diseases, 12(10), e0006779. [CrossRef]
- Crump, J. A., Barrett, T. J., Nelson, J., & Angulo, F. J. (2003). Reevaluating Fluoroquinolone Breakpoints for Salmonella enterica Serotype Typhi and for Non-Typhi Salmonellae. Clinical Infectious Diseases, 37(1), 75–81. [CrossRef]
- Akhtar, S., Sarker, M. R., Jabeen, K., Sattar, A., Qamar, A., & Fasih, N. (2014). Antimicrobial resistance in Salmonella entericaserovar typhi and paratyphi in South Asia-current status, issues and prospects. Critical Reviews in Microbiology, 41(4), 536–545. [CrossRef]
- Krishnan, P., Stalin, M., & Balasubramanian, S. (2009). Changing trends in antimicrobial resistance ofSalmonellaentericaserovar typhi andsalmonella entericaserovar paratyphi A in Chennai. Indian Journal of Pathology & Microbiology, 52(4), 505. [CrossRef]
- Karkey, A., Thwaites, G., & Baker, S. (2018). The evolution of antimicrobial resistance in Salmonella Typhi. Current Opinion in Gastroenterology, 34(1), 25–30. [CrossRef]
- Akinyemi, K. O., & Ajoseh, S. O. (2017). Factors contributing to the emergence and spread of antibiotics resistance in salmonella species. In InTech eBooks. [CrossRef]
- Bengtsson-Palme, J., Kristiansson, E., & Larsson, D. G. J. (2017). Environmental factors influencing the development and spread of antibiotic resistance. Fems Microbiology Reviews, 42(1). [CrossRef]
- Beceiro, A., Tomás, M., & Bou, G. (2013). Antimicrobial Resistance and Virulence: a Successful or Deleterious Association in the Bacterial World? Clinical Microbiology Reviews, 26(2), 185–230. [CrossRef]
- Silva, C., Wiesner, M., & Calva, E. (2012). The importance of mobile genetic elements in the evolution of salmonella: pathogenesis, antibiotic resistance and host adaptation. In InTech eBooks. [CrossRef]
- Dasgupta, N., Paul, D., Chanda, D. D., Chetri, S., Chakravarty, A., & Bhattacharjee, A. (2018). Observation of a New Pattern of Mutations in gyrA and parC within Escherichia coli Exhibiting Fluroquinolone Resistance. Indian Journal of Medical Microbiology, 36(1), 131–135. [CrossRef]
- Bagel, S., Hüllen, V., Wiedemann, B., & Heisig, P. (1999). Impact of gyrA and parC Mutations on Quinolone Resistance, Doubling Time, and Supercoiling Degree of Escherichia coli. Antimicrobial Agents and Chemotherapy, 43(4), 868–875. [CrossRef]
- Gopal, M., Elumalai, S., Suresh, A., Durairajpandian, V., Kannan, M. A., Selvam, E., & Srivani, S. (2016). GyrA ser83 and ParC trp106 Mutations in Salmonella enterica Serovar Typhi Isolated from Typhoid Fever Patients in Tertiary Care Hospital. Journal of Clinical and Diagnostic Research. [CrossRef]
- Tadesse, D. A., Singh, A., Zhao, S., Bartholomew, M. J., Womack, N., Ayers, S., Fields, P. I., & McDermott, P. F. (2016). Antimicrobial Resistance in Salmonella in the United States from 1948 to 1995. Antimicrobial Agents and Chemotherapy, 60(4), 2567–2571. [CrossRef]
- Kariuki, S., Gordon, M. A., Feasey, N. A., & Parry, C. M. (2015). Antimicrobial resistance and management of invasive Salmonella disease. Vaccine, 33, C21–C29. [CrossRef]
- Cuypers, W. L., Jacobs, J., Wong, V., Klemm, E. J., Deborggraeve, S., & Van Puyvelde, S. (2018). Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microbial Genomics, 4(7). [CrossRef]
- Gangathraprabhu, B., Kannan, S. S., Santhanam, G., Suryadevara, N., & Murugan, M. (2020). A review on the origin of multidrug-resistant Salmonella and perspective of tailored phoP gene towards avirulence. Microbial Pathogenesis, 147, 104352. [CrossRef]
- Oo, K. M., Myat, T. O., Htike, W. W., Biswas, A., Hannaway, R. F., Murdoch, D. R., Crump, J. A., & Ussher, J. E. (2019). Molecular mechanisms of antimicrobial resistance and phylogenetic relationships of Salmonella enterica isolates from febrile patients in Yangon, Myanmar. Transactions of the Royal Society of Tropical Medicine and Hygiene, 113(10), 641–648. [CrossRef]
- Van Hoek, A. H., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., & Aarts, H. (2011). Acquired Antibiotic Resistance Genes: An Overview. Frontiers in Microbiology, 2. [CrossRef]
- Gessese, Y. A., Damessa, D. L., Amare, M. M., Bahta, Y. H., Shifera, A. D., Tasew, F. S., & Gebremedhin, E. Z. (2017). Urinary pathogenic bacterial profile, antibiogram of isolates and associated risk factors among pregnant women in Ambo town, Central Ethiopia: a cross-sectional study. Antimicrobial Resistance and Infection Control, 6(1). [CrossRef]
- Kibret, M., & Abera, B. (2014). Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pacific Journal of Tropical Biomedicine, 4(2), 164–168. [CrossRef]
- Bush, N. G., Diez-Santos, I., Abbott, L., & Maxwell, A. (2020). Quinolones: mechanism, lethality and their contributions to antibiotic resistance. Molecules, 25(23), 5662. [CrossRef]
- Drlica, K., & Zhao, X. (2020). Bacterial death from treatment with fluoroquinolones and other lethal stressors. Expert Review of Anti-infective Therapy, 19(5), 601–618. [CrossRef]
- Kassab, A. E., & Gedawy, E. M. (2018). Novel ciprofloxacin hybrids using biology oriented drug synthesis (BIODS) approach: Anticancer activity, effects on cell cycle profile, caspase-3 mediated apoptosis, topoisomerase II inhibition, and antibacterial activity. European Journal of Medicinal Chemistry, 150, 403–418. [CrossRef]
- Ashley, R. E., Dittmore, A., McPherson, S. A., Turnbough, C. L., Neuman, K. C., & Osheroff, N. (2017). Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Research, 45(16), 9611–9624. [CrossRef]
- Urban-Chmiel, R., Marek, A., Stępień-Pyśniak, D., Wieczorek, K., Dec, M., Nowaczek, A., & Osek, J. (2022). Antibiotic Resistance in Bacteria—A Review. Antibiotics, 11(8), 1079. [CrossRef]
- De Alcântara Rodrigues, I., Ferrari, R. G., Panzenhagen, P., Mano, S. B., & Conté-Júnior, C. A. (2020). Antimicrobial resistance genes in bacteria from animal-based foods. In Advances in Applied Microbiology (pp. 143–183). [CrossRef]
- Debabza, M., Dziri, R., Mechai, A., Bouguessa, A., Klibi, N., & Ouzari, H. (2023). Characterization of carbapenemase-producing Gram-negative bacilli: first report of blaNDM-1 in Enterobacter cloacae. Journal of Infection in Developing Countries, 17(09), 1300–1309. [CrossRef]
- Sumbana, J. J., Santona, A., Fiamma, M., Taviani, E., Deligios, M., Zimba, T. F., Lucas, G., Sacarlal, J., Rubino, S., & Paglietti, B. (2021). Extraintestinal Pathogenic Escherichia coli ST405 Isolate Coharboring blaNDM-5 and blaCTXM-15: A New Threat in Mozambique. Microbial Drug Resistance, 27(12), 1633–1640. [CrossRef]
- Obayiuwana, A., & Ibekwe, A. M. (2020). Antibiotic Resistance Genes Occurrence in Wastewaters from Selected Pharmaceutical Facilities in Nigeria. Water, 12(7), 1897. [CrossRef]
- Paulshus, E., Colque, P., Kühn, I., Tauhid, T., Hu, Y., Zhou, Y., Thorell, K., Möllby, R., Sørum, H., Sjöling, Å., & Joffré, E. (2023). Escherichia coli ST2797 Is Abundant in Wastewater and Might Be a Novel Emerging Extended-Spectrum Beta-Lactamase E. coli. Microbiology Spectrum, 11(4). [CrossRef]
- Vázquez, X., Fernández, J., Hernáez, S., Rodicio, R., & Rodicio, M. R. (2022). Plasmid-Mediated Quinolone Resistance (PMQR) in Two Clinical Strains of Salmonella enterica Serovar Corvallis. Microorganisms, 10(3), 579. [CrossRef]
- Venkatesan, M., Fruci, M., Verellen, L. A., Skarina, T., Mesa, N., Flick, R., Pham, C., Mahadevan, R., Stogios, P., & Savchenko, A. (2023). Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nature Communications, 14(1). [CrossRef]
- Foong, W., Wilhelm, J., Tam, H., & Pos, K. M. (2020b). Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. Journal of Antimicrobial Chemotherapy, 75(5), 1135–1139. [CrossRef]
- Gajić, I., Kabić, J., Kekić, D., Jovićević, M., Milenković, M., Mitić-Ćulafić, D., Trudić, A., Ranin, L., & Opavski, N. (2022). Antimicrobial susceptibility testing: A comprehensive review of currently used methods. Antibiotics, 11(4), 427. [CrossRef]
- Van Belkum, A., Burnham, C. D., Rossen, J. W. A., Mallard, F., Rochas, O., & Dunne, W. M. (2020). Innovative and rapid antimicrobial susceptibility testing systems. Nature Reviews Microbiology, 18(5), 299–311. [CrossRef]
- Onishi, R., Shigemura, K., Osawa, K., Yang, Y., Maeda, K., Tanimoto, H., Kado, M., Fang, S., Sung, S., Miyara, T., & Fujisawa, M. (2022). Impact on quinolone resistance of plasmid-mediated quinolone resistance gene and mutations in quinolone resistance-determining regions in extended spectrum beta lactamase-producing Klebsiella pneumoniae isolated from urinary tract infection patients. Pathogens and Disease, 80(1). [CrossRef]
- Shaheen, A., Tariq, A., Iqbal, M., Mirza, O., Haque, A., Walz, T., & Rahman, M. (2021). Mutational diversity in the quinolone Resistance-Determining regions of Type-II topoisomerases of salmonella serovars. Antibiotics, 10(12), 1455. [CrossRef]
- Azargun, R., Gholizadeh, P., Sadeghi, V., Hosainzadegan, H., Tarhriz, V., Memar, M. Y., Pormohammad, A., & Eyvazi, S. (2020). Molecular mechanisms associated with quinolone resistance in Enterobacteriaceae: review and update. Transactions of the Royal Society of Tropical Medicine and Hygiene, 114(10), 770–781. [CrossRef]
- Wang, M., Cang, Z., & Wei, G. (2020). A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nature Machine Intelligence, 2(2), 116–123. [CrossRef]
- Teng, S., Sobitan, A., Rhoades, R., Liu, D., & Tang, Q. (2020). Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Briefings in Bioinformatics, 22(2), 1239–1253. [CrossRef]
- Moses, D., Ginell, G. M., Holehouse, A. S., & Sukenik, S. (2023). Intrinsically disordered regions are poised to act as sensors of cellular chemistry. Trends in Biochemical Sciences, 48(12), 1019–1034. [CrossRef]
- Potts, D. S., Bregante, D. T., Adams, J. S., Torres, C., & Flaherty, D. W. (2021). Influence of solvent structure and hydrogen bonding on catalysis at solid–liquid interfaces. Chemical Society Reviews, 50(22), 12308–12337. [CrossRef]
- Bhanja, K., Sharma, M., & Patra, N. (2023). Uncovering the structural and binding insights of dual inhibitors simultaneously targeting two distinct sites on EGFR kinase. The Journal of Physical Chemistry B, 127(50), 10749–10765. [CrossRef]
- Wang, R., & Zheng, Q. (2020). Multiple Molecular Dynamics Simulations of the Inhibitor GRL-02031 Complex with Wild Type and Mutant HIV-1 Protease Reveal the Binding and Drug-Resistance Mechanism. Langmuir, 36(46), 13817–13832. [CrossRef]
- Florensa, A. F., Kaas, R. S., Clausen, P. T. L. C., Aytan-Aktug, D., & Aarestrup, F. M. (2022). ResFinder – an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microbial Genomics, 8(1). [CrossRef]
- Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology and Evolution, 38(7), 3022–3027. [CrossRef]
- Trott, O., & Olson, A. J. (2009). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. [CrossRef]
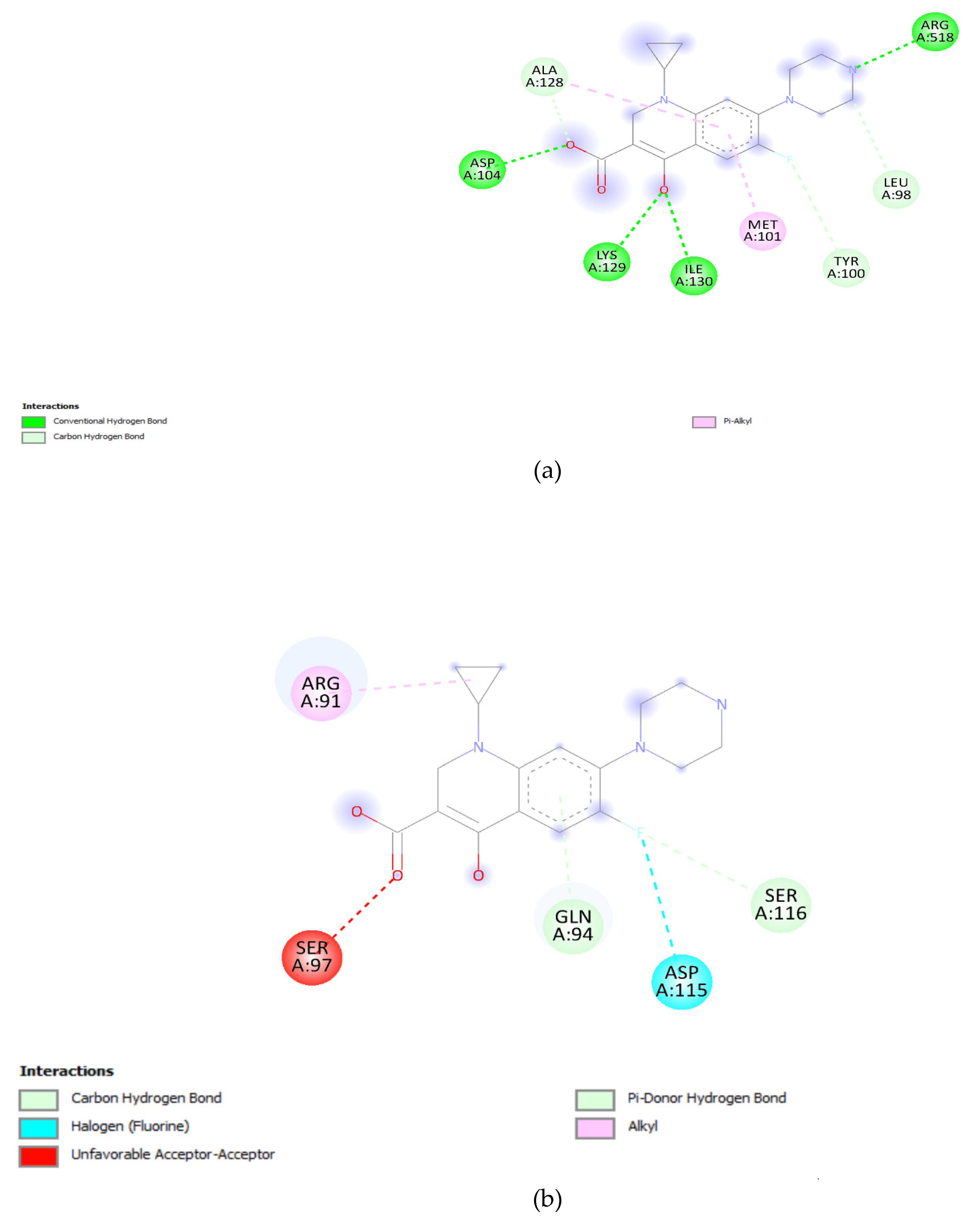
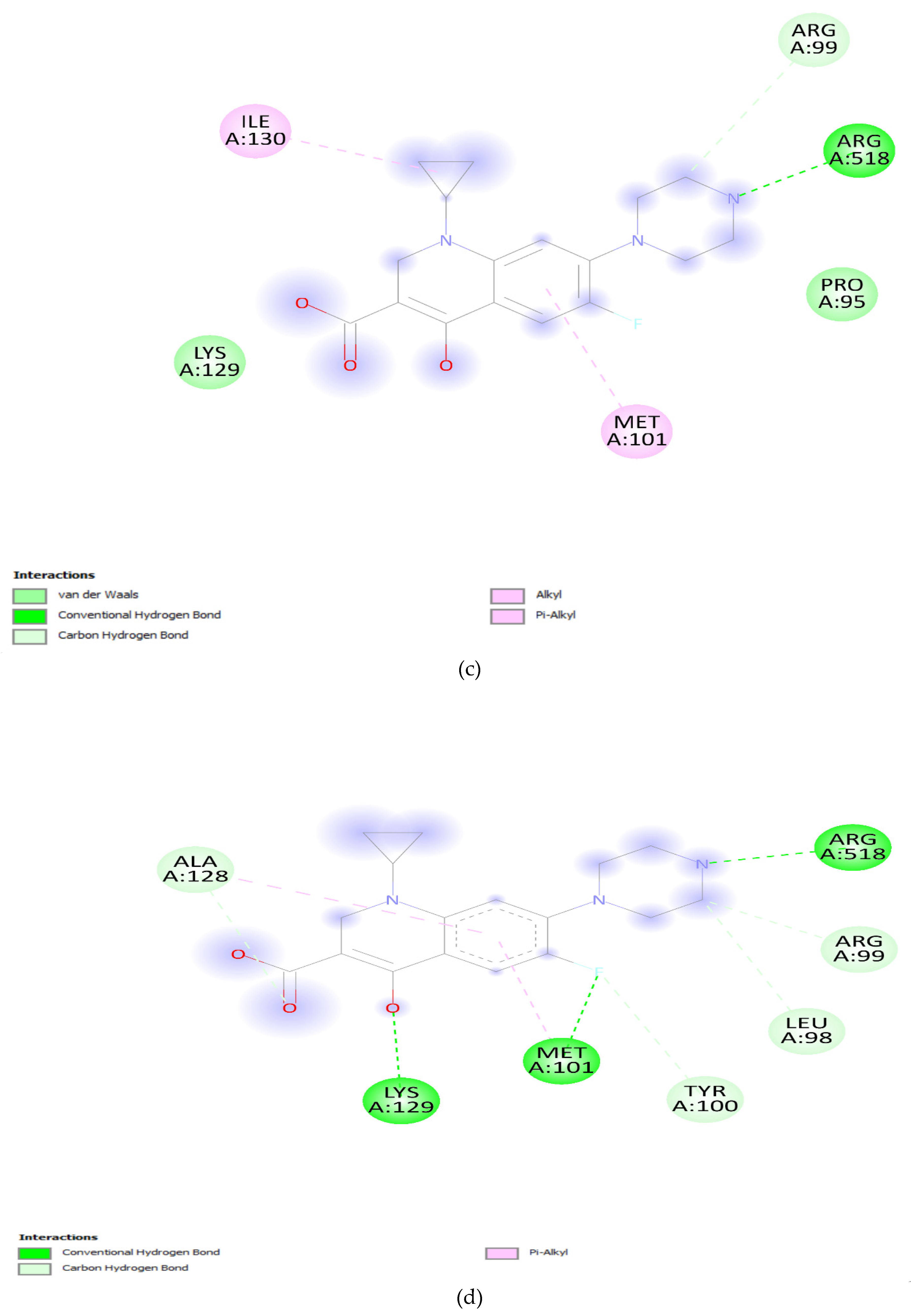
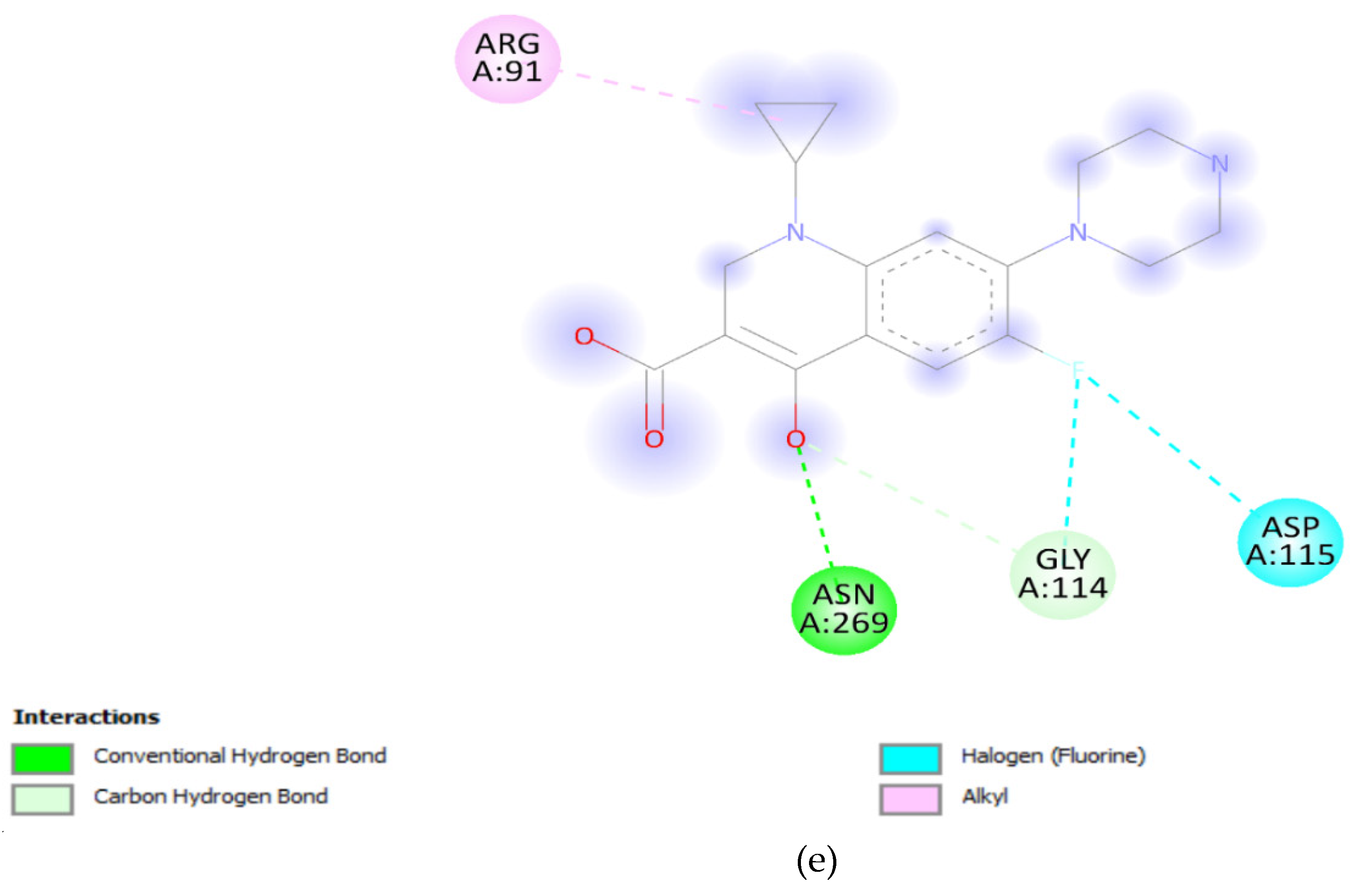
| Gene Variants | Binding Affinity in QRDR (Kcal/mol) |
Solvent Accessbility in QRDR (Ao) |
|---|---|---|
| GyrA Wild type | -7.1 | 2830.99 |
| gyrA_S83F | -6.2 | 2731.51 |
| gyrA_S83Y | -6.4 | 2741.95 |
| gyrA_D87N | -6.4 | 2756.78 |
| gyrA_S83F, D87N | -6.3 | 2758.2 |
| ParC Wild type | -6.7 | 3326.01 |
| parC_S80I | -6.0 | 3014.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





