Submitted:
28 January 2024
Posted:
29 January 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
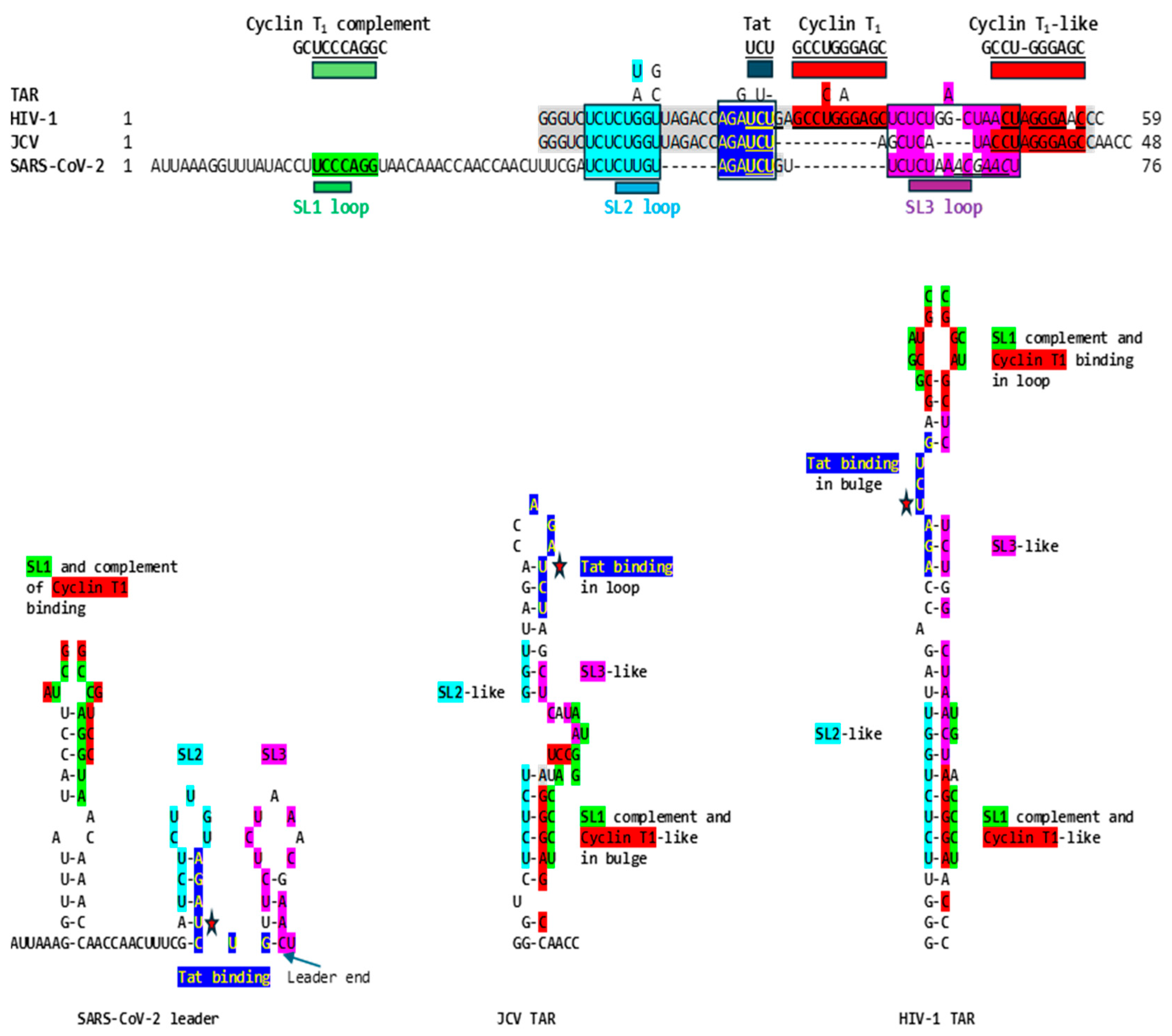
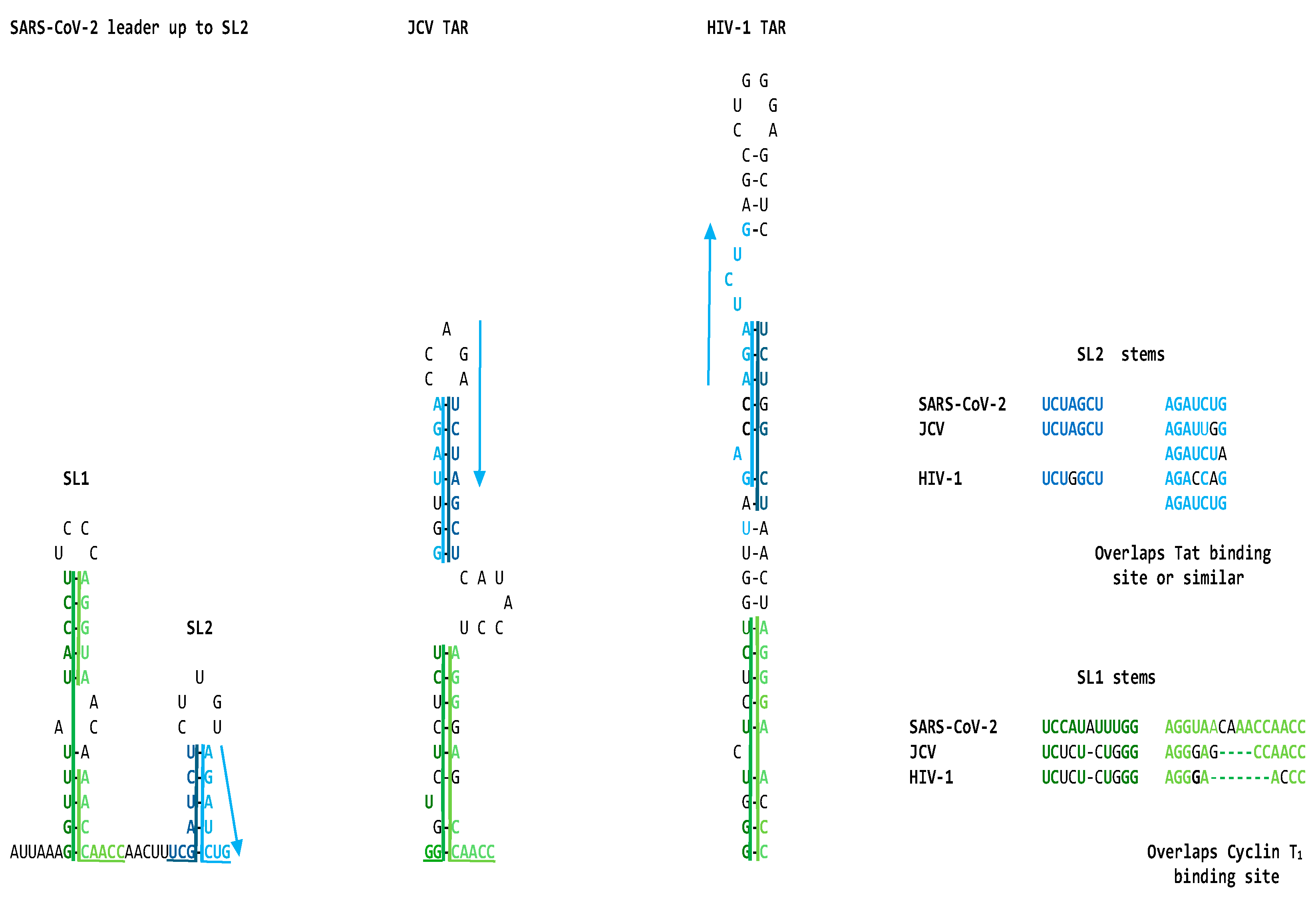
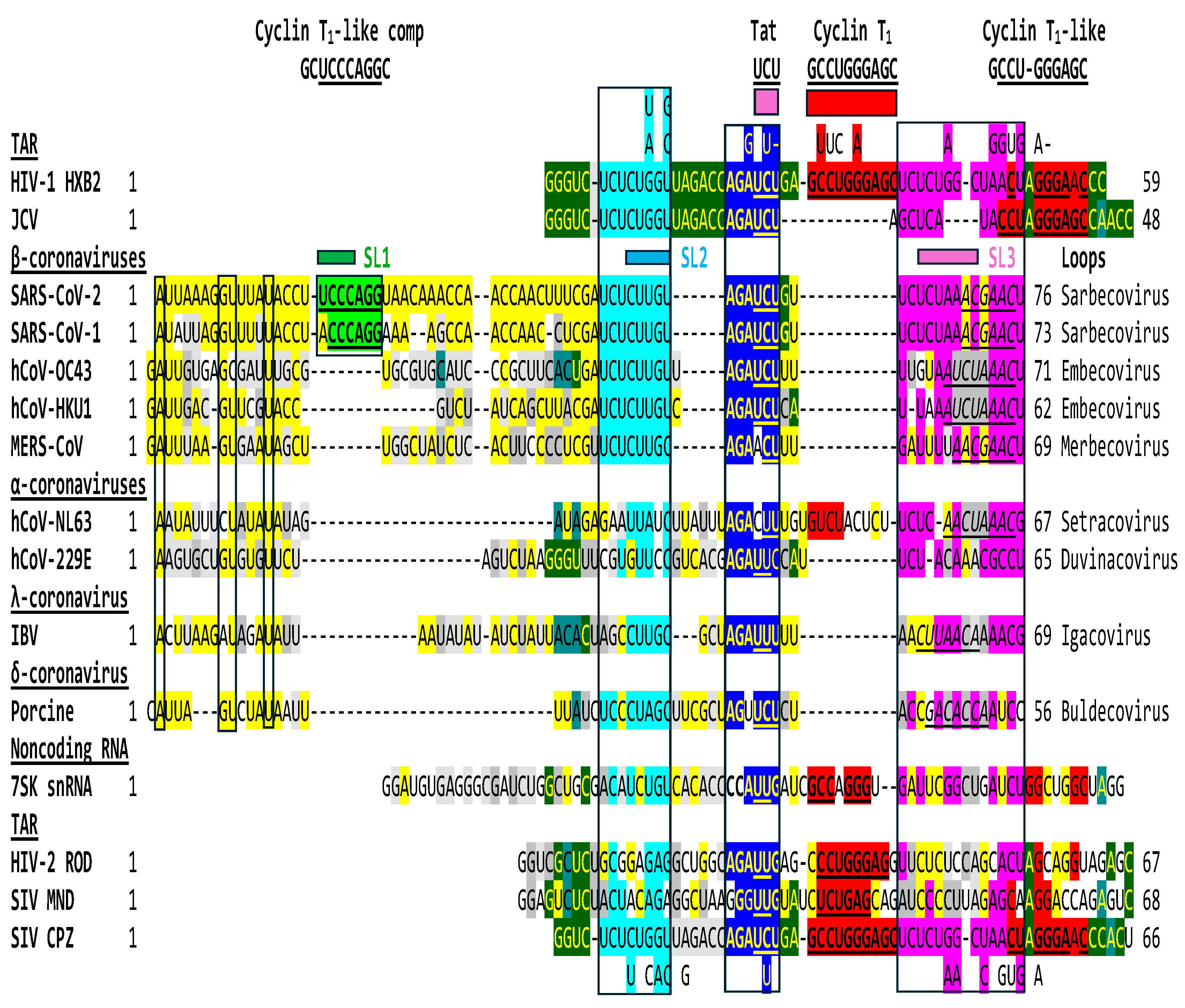
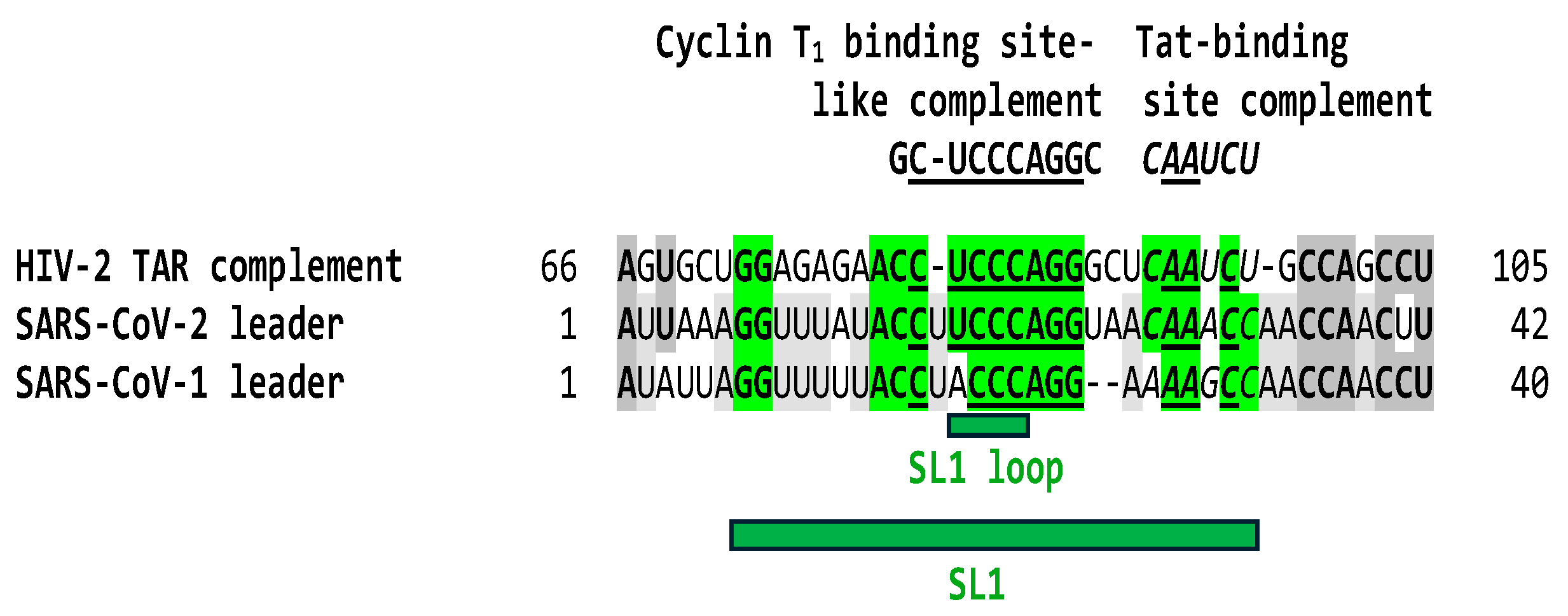
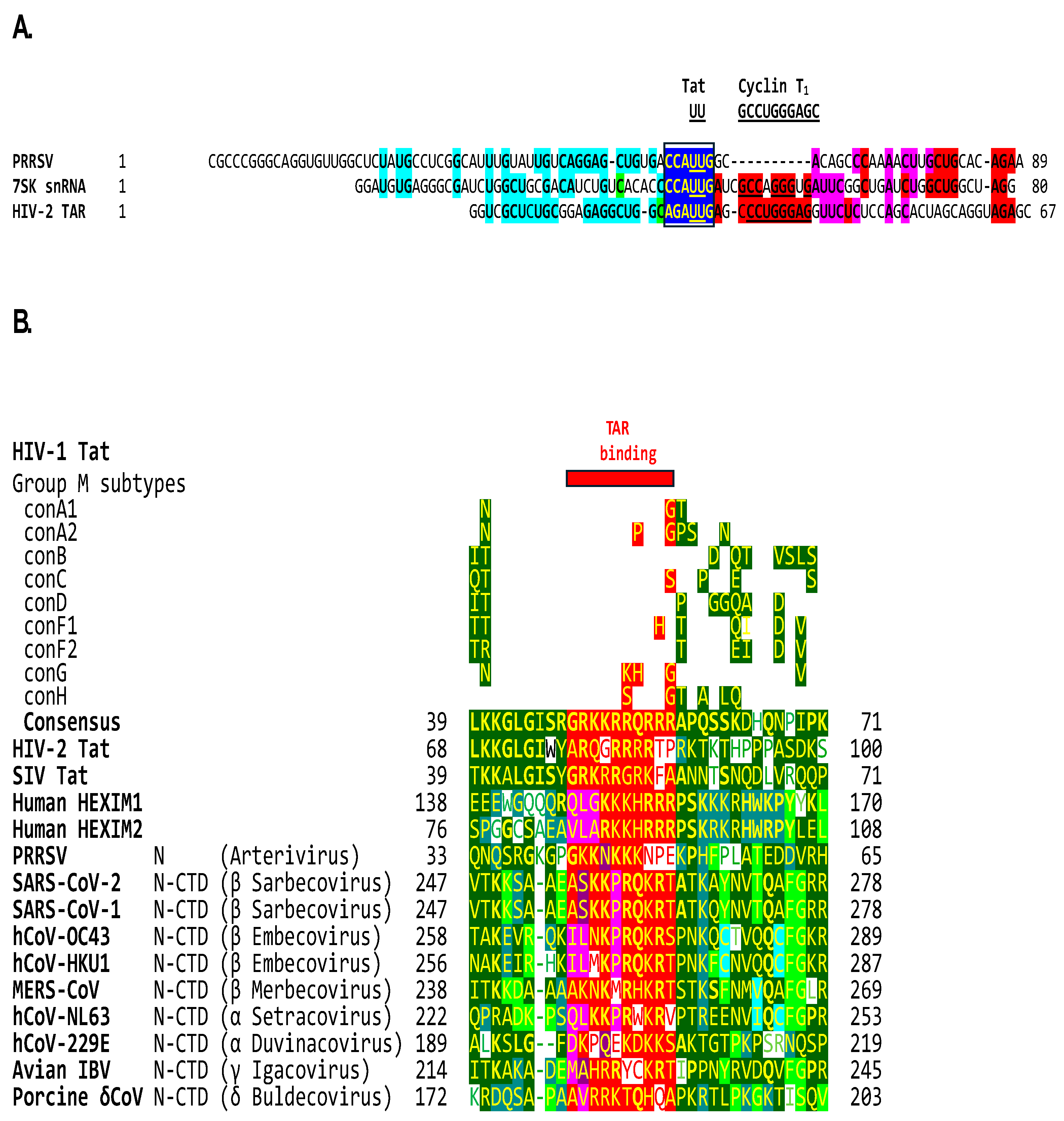
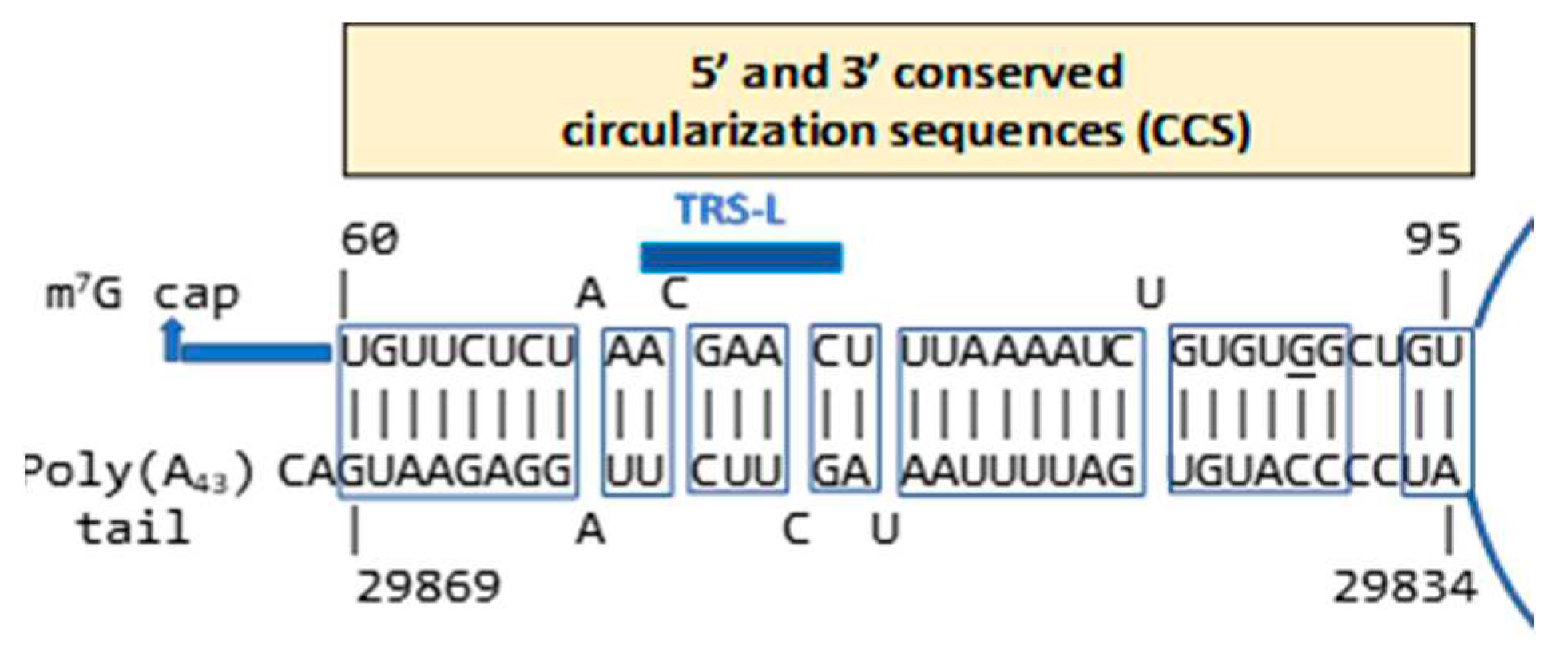
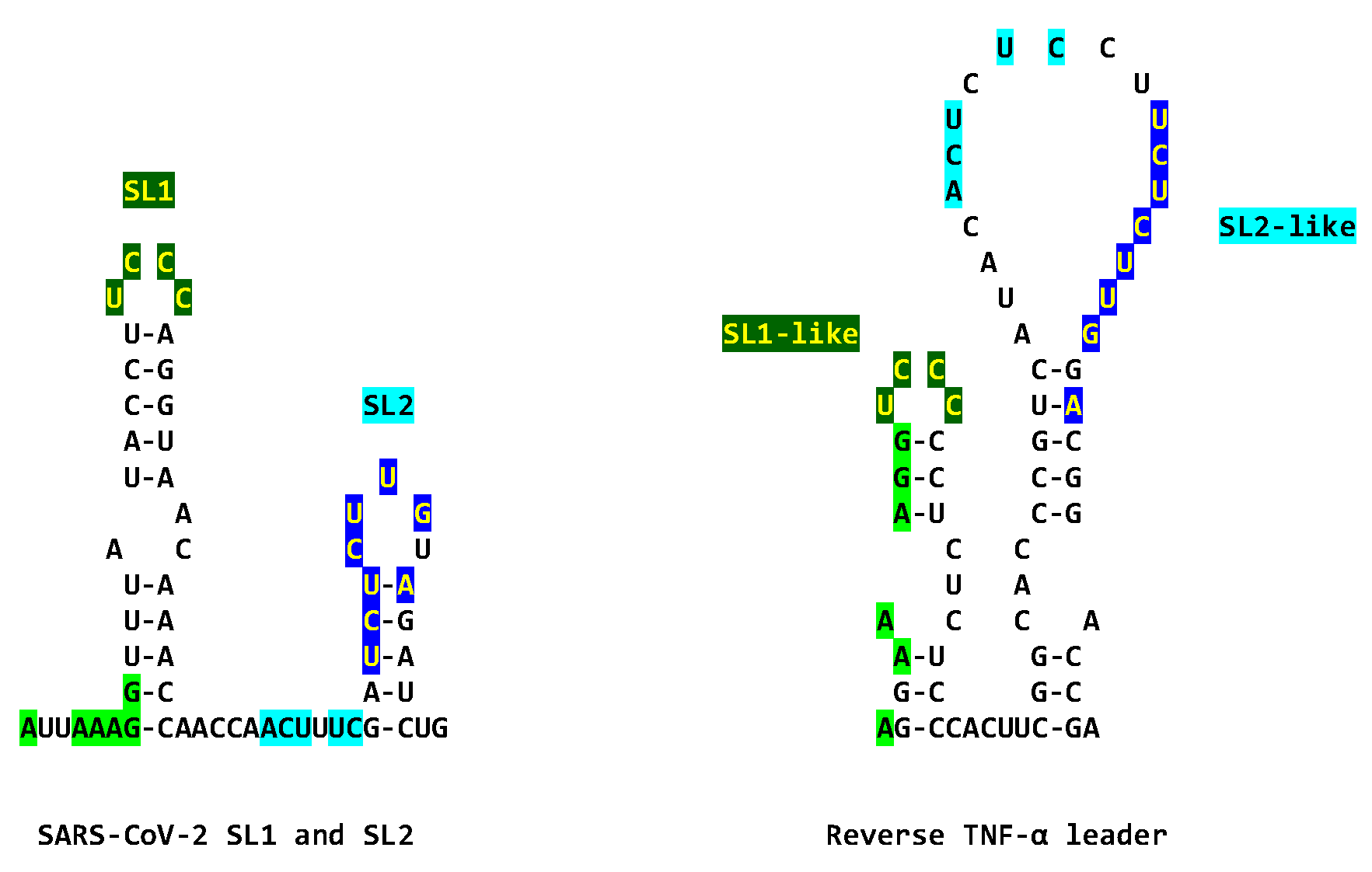
2.1. TAR-like sequences are present in the SARS-CoV-2 RNA leader
2.2. The leaders of all genera of coronaviruses share the same regions of similarities as SARS-CoV-2 with the TARs of HIV-1, HIV-2, SIV, and the human 7SK small nuclear RNA
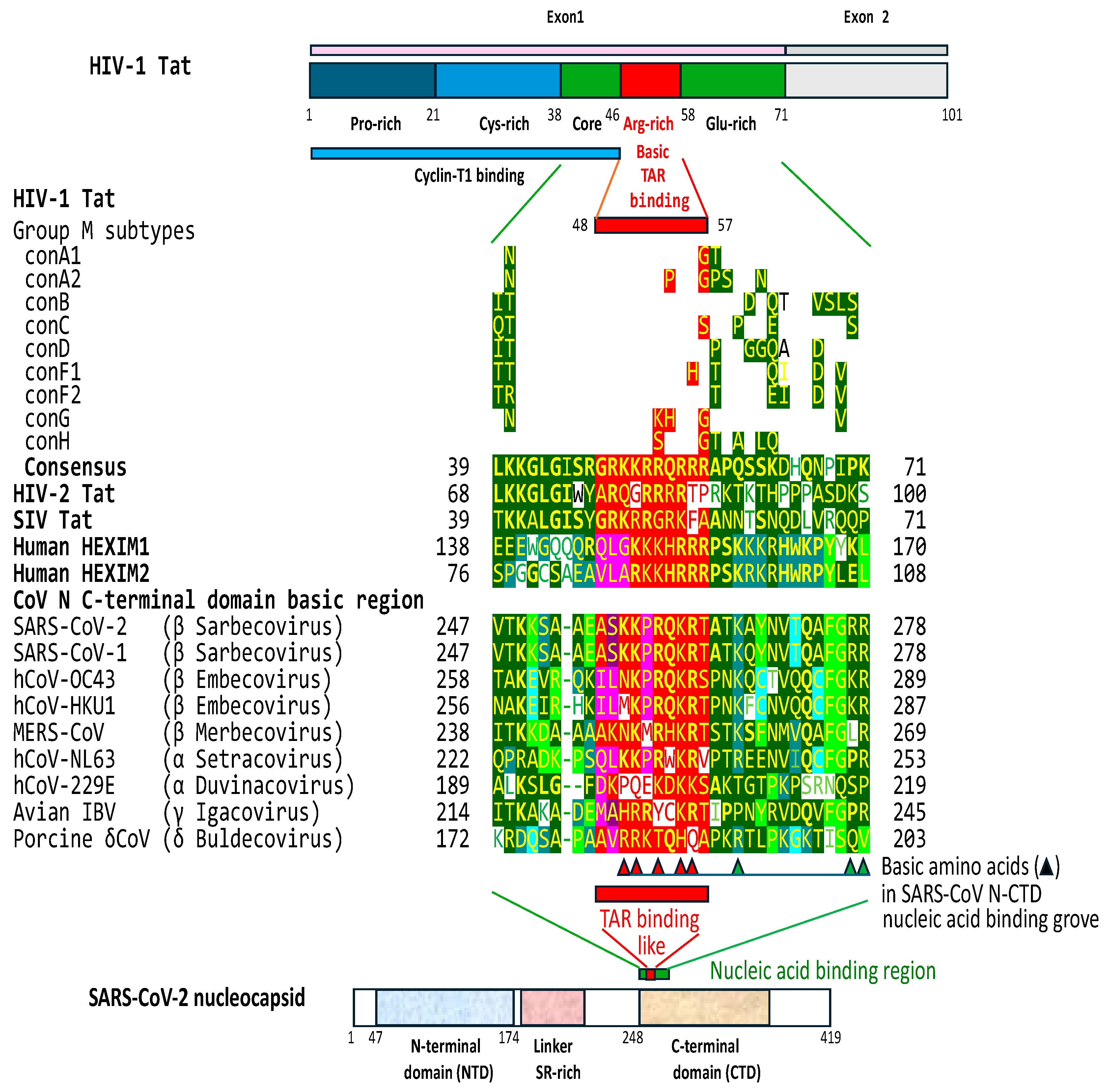
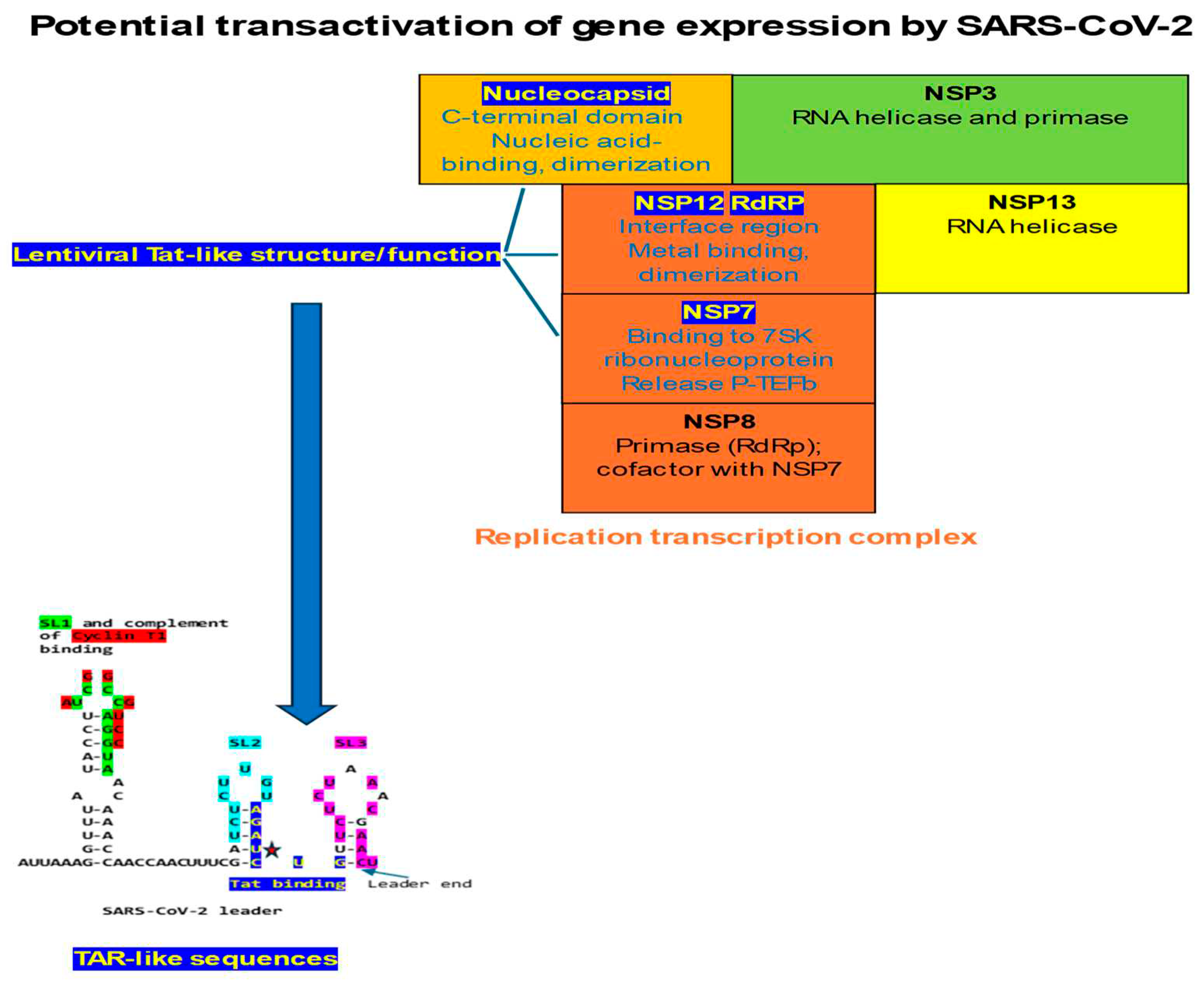
2.3. The C-terminal domain of the SARS-CoV-2 nucleocapsid protein contains a nucleotide-binding arginine-rich and adjacent regions similar to the TAR binding site and adjacent regions in HIV-1, HIV-2, and SIV Tat and the human HEXIM proteins that bind to the 7SK snRNA TAR
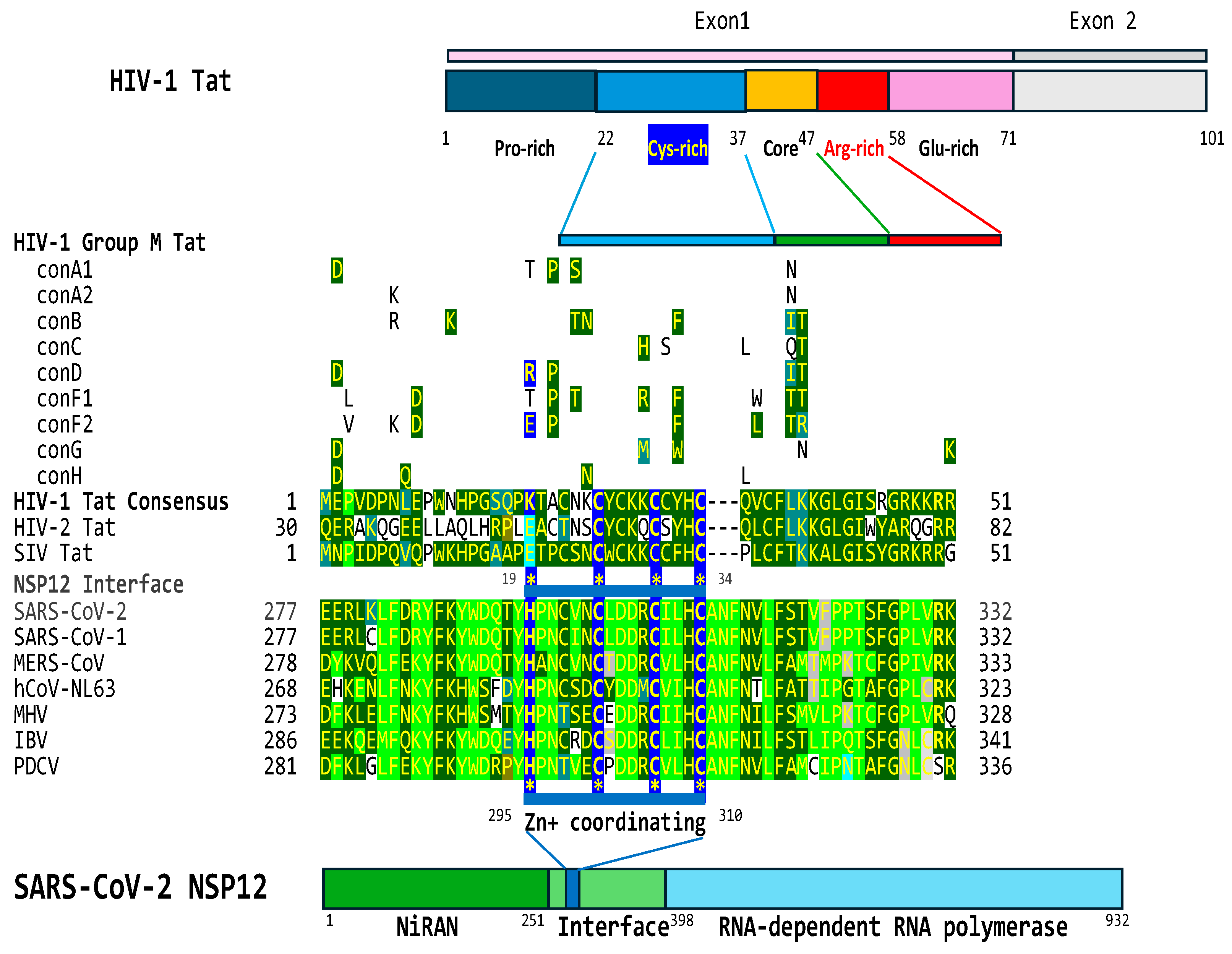
2.4. The interface region of the NSP12 protein of SARS-CoV-2 contains a cysteine-rich region that is similar to that present in HIV-1, HIV-2, and SIV Tat proteins
2.5. The β-arterivirus porcine reproductive and respiratory syndrome virus also contains a Tat-binding site identical to that in human 7SK snRNA and adjacent similarities, and its nucleocapsid has a basic amino acid-rich region similar to that in Tat
3. Discussion
3.1. Implications of potential transactivation to transcription of SARS-CoV-2 and other coronaviruses
3.2. Implications of potential transactivation to variant evolution of SARS-CoV-2 and other coronaviruses
3.3. Implications of potential intra- and trans-cellular transactivation to acute SARS-CoV-2 pathology and long COVID and their treatment
4. Materials and Methods
4.1. Detection of HIV-1 TAR-like sequences in the SARS-CoV-2 leader, and expansion to other lentiviral, JCV, and human 7SK snRNA TARs
4.2. Secondary structural mapping of the regions of similarity between HIV TAR-like and coronaviral leader sequences
4.3. Detection of HIV-1 Tat-like sequences among SARS-CoV-2 proteins
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sodroski, J.; Patarca, R.; Rosen, C.; Wong-Staal, F.; Haseltine, W. (1985). Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science (New York, N.Y.) 1985, 229, 74–77. [Google Scholar] [CrossRef]
- Sodroski, J.; Rosen, C.; Wong-Staal, F.; Salahuddin, S.Z.; Popovic, M.; Arya, S.; Gallo, R.C.; Haseltine, W. A. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science (New York, N.Y.) 1985, 227, 171–173. [Google Scholar] [CrossRef]
- Dayton, A.I.; Sodroski, J.G.; Rosen, C.A.; Goh, W.C.; Haseltine, W.A. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell 1986, 44, 941–947. [Google Scholar] [CrossRef]
- Okamoto, T.; Wong-Staal, F. Demonstration of virus-specific transcriptional activator(s) in cells infected with HTLV-III by an in vitro cell-free system. Cell 1986, 47, 29–35. [Google Scholar] [CrossRef]
- Okamoto, T.; Benter, T.; Josephs, S.F.; Sadaie, M.R.; Wong-Staal, F. Transcriptional activation from the long-terminal repeat of human immunodeficiency virus in vitro. Virology 1990, 177, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.; Kwak, Y.T.; Nee, E.; Guo, J.; García-Martínez, L.F.; Gaynor, R.B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J Mol Biol 1999, 288, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Parada, C.A.; Roeder, R.G. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 1996, 384, 375–378. [Google Scholar] [CrossRef] [PubMed]
- He, N., Zhou, Q. New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology 2011, 6, 260-268. [CrossRef]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B,R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci U S A 1999, 96, 7791-7796. [CrossRef]
- Garber, M.E.; Mayall, T.P.; Suess, E.M.; Meisenhelder, J.; Thompson, N.E.; Jones, K.A. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol 2000, 20, 6958–69. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.; Cao, H.; Rana, T.M. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1-Tat-TAR ternary complex formation. Biochemistry 2002, 41, 6391–7. [Google Scholar] [CrossRef]
- Karn, J.; Stoltzfus, C. M. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2012, 2, a006916. [Google Scholar] [CrossRef]
- Asamitsu, K.; Okamoto, T. The Tat/P-TEFb protein-protein interaction determining transcriptional activation of HIV. Curr Pharm Des 2017, 23, 4091–4097. [Google Scholar] [CrossRef]
- Asamitsu, K.; Fujinaga, K.; Okamoto, T. HIV tat/P-TEFb interaction: a potential target for novel anti-HIV therapies. Molecules 2018, 23, 933. [Google Scholar] [CrossRef]
- Chameettachal, A.; Mustafa, F.; Rizvi, T.A. Understanding Retroviral Life Cycle and its Genomic RNA Packaging. Journal of molecular biology 2023, 435, 167924. [Google Scholar] [CrossRef] [PubMed]
- Kuzmina, A.; Krasnopolsky, S.; Taube, R. Super elongation complex promotes early HIV transcription and its function is modulated by P-TEFb. Transcription 2017, 8, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Gallo, R.C.; Hahn, B.H.; Shaw, G.M.; Popovic, M.; Salahuddin, S.Z.; Wong-Staal, F. Homology of genome of AIDS-associated virus with genomes of human T-cell leukemia viruses. Science 1984, 225, 927–930. [Google Scholar] [CrossRef]
- Rosen, C.A.; Sodroski, J.G.; Haseltine, WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell 1985, 41, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Jeang, K.T. Trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol 1989, 63, 5501–5504. [Google Scholar] [CrossRef]
- Dingwall, C.; Ernberg, I.; Gait, M.J.; Green, S.M.; Heaphy, S.; Karn, J.; Lowe, A.D.; Singh, M.; Skinner, M.A.; Valerio, R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A 1989, 86, 6925–6929. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M.; Ampe, C.; Schultz, S.C.; Steitz, T.A.; Crothers, D.M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science (New York, N.Y.) 1990, 249, 1281–1285. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M.; Crothers, D.M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell 1991, 66, 577–588. [Google Scholar] [CrossRef]
- Sumner-Smith, M.; Roy, S.; Barnett, R.; Reid, L.S.; Kuperman, R.; Delling, U.; Sonenberg, N. Critical chemical features in trans-acting-responsive RNA are required for interaction with human immunodeficiency virus type 1 Tat protein. Journal of virology 1991, 65, 5196–5202. [Google Scholar] [CrossRef]
- Karn, J. Tackling Tat. J Mol Biol 1999, 293, 235-254. [CrossRef]
- Levintov, L.; Vashisth, H. Structural and computational studies of HIV-1 RNA. RNA biology 2024, 21, 1–32. [Google Scholar] [CrossRef]
- Kao, S.Y.; Calman, A.F.; Luciw, P.A.; Peterlin, B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987, 330, 489–493. [Google Scholar] [CrossRef]
- Rhim, H.; Rice, A.P. TAR RNA binding properties and relative transactivation activities of human immunodeficiency virus type 1 and 2 Tat proteins. J Virol 1993, 67, 1110–1121. [Google Scholar] [CrossRef]
- Cullen, B.R. RNA-sequence-mediated gene regulation in HIV-1. Infect Agents Dis 1994, 3, 68–76. [Google Scholar]
- Roebuck, K.A.; Saifuddin, M. Regulation of HIV-1 transcription. Gene Expr 1999, 8, 67–84. [Google Scholar]
- Kessler, M.; Mathews, M.B. Premature termination and processing of human immunodeficiency virus type 1-promoted transcripts. J Virol 1992, 66, 4488–4496. [Google Scholar] [CrossRef]
- Ratnasabapathy, R.; Sheldon, M.; Johal, L.; Hernandez, N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev 1990, 4, 2061–2074. [Google Scholar] [CrossRef]
- Toohey, M.G.; Jones, KA. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2 promoter-proximal downstream regions. Genes Dev 1989, 3, 265–282. [Google Scholar] [CrossRef]
- Clark, E.; Nava, B.; Caputi, M. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 2017, 8, 27569–27581. [Google Scholar] [CrossRef]
- Wada, T.; Takagi, T.; Yamaguchi, Y.; Ferdous, A.; Imai, T.; Hirose, S.; Sugimoto, S.; Yano, K.; Hartzog, G.A.; Winston, F.; Buratowski, S.; Handa, H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & development, 1998, 12, 343–356. [Google Scholar] [CrossRef]
- Wada, T.; Orphanides, G.; Hasegawa, J.; Kim, D.K.; Shima, D.; Yamaguchi, Y.; Fukuda, A.; Hisatake, K.; Oh, S.; Reinberg, D.; Handa, H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Molecular cell 2000, 5, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Kasten, M.; De Falco, G.; Micheli, P.; Khalili, K.; Giordano, A. Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression. J Cell Biochem 1999, 75, 357–368. [Google Scholar] [CrossRef]
- Marcello, A.; Zoppé, M.; Giacca, M. Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator. IUBMB Life 2001, 51, 175–181. [Google Scholar] [CrossRef]
- Isel, C.; Karn, J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol 1999, 290, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Jeang, K.T.; Xiao, H.; Rich, E.A. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 1999, 274, 28837–28840. [Google Scholar] [CrossRef]
- Zhou, M.; Halanski, M.A.; Radonovich, M.F.; Kashanchi, F.; Peng, J.; Price, D.H.; Brady, J.N. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol 2000, 20, 5077–5086. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Bourgeois, C.F.; Isel, C.; Churcher, M.J.; Karn, J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 2002, 22, 4622–4637. [Google Scholar] [CrossRef]
- Hamasaki, T.; Okamoto, M.; Baba, M. Identification of novel inhibitors of human immunodeficiency virus type 1 replication by in silico screening targeting cyclin T1/Tat interaction. Antimicrob Agents Chemother 2013, 57, 1323–1331. [Google Scholar] [CrossRef]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef]
- Yan, Y.; Tang, Y.D.; Zheng, C. When cyclin-dependent kinases meet viral infections, including SARS-CoV-2. J Med Virol 2022, 94, 2962–2968. [Google Scholar] [CrossRef]
- Fujinaga, K., Huang, F., Peterlin, B.M. P-TEFb: The master regulator of transcription elongation. Mol Cell 2023, 83, 393-403. [CrossRef]
- Fujinaga, K.; Irwin, D.; Huang, Y.; Taube, R.; Kurosu, T.; Peterlin, B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Molecular and cellular biology 2004, 24, 787–795. [Google Scholar] [CrossRef]
- Cho, S.; Schroeder, S.; Ott, M. CYCLINg through transcription: Post-translational modifications of P-TEFb regulate transcription elongation. Cell Cycle 2010, 9, 1697–1705. [Google Scholar] [CrossRef]
- Egloff, S. CDK9 keeps RNA polymerase II on track. Cellular and molecular life sciences : CMLS, 2021, 78, 5543–5567. [CrossRef]
- Yik, J.H.; Chen, R.; Pezda, A.C.; Samford, C.S.; Zhou, Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Molecular and cellular biology 2004, 24, 5094–5105. [Google Scholar] [CrossRef]
- Muniz, L.; Egloff, S.; Ughy, B.; Jády, B.E.; Kiss, T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 2010, 6, e1001152. [Google Scholar] [CrossRef]
- Sobhian, B.; Laguette, N.; Yatim, A.; Nakamura, M.; Levy, Y.; Kiernan, R.; Benkirane, M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Molecular cell, 2010, 38, 439–451. [Google Scholar] [CrossRef]
- Yang, Z.; Yik J.H.; Chen, R.; He, N; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005, 19, 535-545. [CrossRef]
- Nekhai, S.; Jeang, K.T. Transcriptional and post-transcriptional regulation of HIV-1 gene expression: role of cellular factors for Tat and Rev. Fut. Microbiol 2006, 1, 417–426. [Google Scholar] [CrossRef]
- Vardabasso, C.; Manganaro, L.; Lusic, M.; Marcello, A.; Giacca, M. The histone chaperone protein nucleosome assembly protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 2008, 5, 8. [Google Scholar] [CrossRef]
- Easley, R.; Van Duyne, R.; Coley, W.; Guendel, I.; Dadgar, S.; Kehn-Hall, K.; Kashanchi, F. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta 2010, 1799, 275–85. [Google Scholar] [CrossRef]
- Sastry, K.J.; Reddy, H.R.; Pandita, R.; Totpal, K.; Aggarwal, B.B. HIV-1 tat gene induces tumor necrosis factor-beta (lymphotoxin) in a human B-lymphoblastoid cell line. The Journal of biological chemistry 1990, 265, 20091–20093. [Google Scholar] [CrossRef]
- Buonaguro, L.; Buonaguro, F.M.; Giraldo, G.; Ensoli, B. The human immunodeficiency virus type 1 Tat protein transactivates tumor necrosis factor beta gene expression through a TAR-like structure. Journal of virology, 1994, 68, 2677–2682. [Google Scholar] [CrossRef] [PubMed]
- Brother, M.B.; Chang, H.K.; Lisziewicz, J.; Su, D.; Murty, L.C.; Ensoli, B. Block of Tat-mediated transactivation of tumor necrosis factor beta gene expression by polymeric-TAR decoys. Virology 1996, 222, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Scala, G.; Ruocco, M.R.; Ambrosino, C.; Mallardo, M; Giordano, V.; Baldassarre, F.; Dragonetti, E.; Quinto, I.; Venuta, S. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 TAT protein. J Exp Med 1994, 179, 961-971. [CrossRef]
- Ambrosino, C.; Ruocco, M.R.; Chen, X.; Mallardo, M.; Baudi, F.; Trematerra, S.; Quinto, I.; Venuta, S.; Scala, G. HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein beta (NF-IL6) transcription factors. The Journal of biological chemistry, 1997, 272, 14883–14892. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Lovett, J.L.; Mueller, L.; Verdin, E. Superinduction of IL-8 in T cells by HIV-1 Tat protein is mediated through NF-kappaB factors. J Immunol 1998, 160, 2872–2880. [Google Scholar] [CrossRef]
- Bohn-Wippert, K., Tevonian, E.N., Megaridis, M.R., Dar, R.D. Similarity in viral and host promoters couples viral reactivation with host cell migration. Nature communications 2017, 8, 15006. [CrossRef]
- Lim, S.P.; Garzino-Demo, A. The human immunodeficiency virus type 1 Tat protein up-regulates the promoter activity of the beta-chemokine monocyte chemoattractant protein 1 in the human astrocytoma cell line U-87 MG: role of SP-1, AP-1, and NF-kappaB consensus sites. Journal of virology 2000, 74, 1632–1640. [Google Scholar] [CrossRef] [PubMed]
- Vacca, A.; Farina, M.; Maroder, M.; Alesse, E.; Screpanti, I.; Frati, L.; Gulino, A. Human immunodeficiency virus type-1 tat enhances interleukin-2 promoter activity through synergism with phorbol ester and calcium-mediated activation of the NF-AT cis-regulatory motif. Biochem Biophys Res Commun 1994, 205, 467-474.
- González, E.; Punzón, C.; González, M.; Fresno, M. HIV-1 Tat inhibits IL-2 gene transcription through qualitative and quantitative alterations of the cooperative Rel/AP1 complex bound to the CD28RE/AP1 composite element of the IL-2 promoter. Journal of immunology (Baltimore, Md. : 1950), 2001, 166, 4560–4569. [Google Scholar] [CrossRef] [PubMed]
- Anastasopoulou, S.; Georgakopoulos, T.; Mouzaki, A. HIV-1 Transcriptional Activator Tat Inhibits IL2 Expression by Preventing the Presence of Pol II on the IL2 Promoter. Biomolecules 2023, 13, 881. [Google Scholar] [CrossRef] [PubMed]
- Patarca, R.; Heath, C.; Goldenberg, G.J.; Rosen, C.A., Sodroski, J.G., Haseltine, W.A., Hansen, U.M. Transcription directed by the HIV long terminal repeat in vitro. AIDS research and human retroviruses 1987, 3, 41–55. [CrossRef]
- Taylor, J.P.; Pomerantz, R.J.; Raj, G.V.; Kashanchi, F.; Brady, J.N.; Amini, S.; Khalili, K. Central nervous system-derived cells express a kappa B-binding activity that enhances human immunodeficiency virus type 1 transcription in vitro and facilitates TAR-independent transactivation by Tat. Journal of virology 1994, 68, 3971–3981. [Google Scholar] [CrossRef]
- Yang, L.; Morris, G.F.; Lockyer, J.M.; Lu, M.; Wang, Z.; Morris, C.B. Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat. Virology 1997, 235, 48–64. [Google Scholar] [CrossRef]
- Iyer, K.; Mitra, A.; Mitra, D. Identification of 5' upstream sequence involved in HSPBP1 gene transcription and its downregulation during HIV-1 infection. Virus research 2023, 324, 199034. [Google Scholar] [CrossRef]
- Chaudhary, P.; Khan, S.Z.; Rawat, P.; Augustine, T.; Raynes, D.A.; Guerriero, V.; Mitra, D. HSP70 binding protein 1 (HspBP1) suppresses HIV-1 replication by inhibiting NF-κB mediated activation of viral gene expression. Nucleic Acids Res 2016, 44(4):1613-1629. [CrossRef]
- de la Vega, L.; Sánchez-Duffhues, G.; Fresno, M.; Schmitz, M.L.; Muñoz, E.; Calzado, M.A. The 73 kDa subunit of the CPSF complex binds to the HIV-1 LTR promoter and functions as a negative regulatory factor that is inhibited by the HIV-1 Tat protein. Journal of molecular biology 2007, 372, 317–330. [Google Scholar] [CrossRef]
- Verhoef, K.; Bauer, M.; Meyerhans, A.; Berkhout, B. On the role of the second coding exon of the HIV-1 Tat protein in virus replication and MHC class I downregulation. AIDS research and human retroviruses 1998, 14, 1553–1559. [Google Scholar] [CrossRef]
- Cafaro, A.; Barillari, G.; Moretti, S.; Palladino, C.; Tripiciano, A.; Falchi, M.; Picconi, O.; Pavone Cossut, M.R.; Campagna, M.; Arancio, A.; Sgadari, C.; Andreini, C.; Banci, L.; Monini, P.; Ensoli, B. HIV-1 Tat Protein Enters Dysfunctional Endothelial Cells via Integrins and Renders Them Permissive to Virus Replication. International journal of molecular sciences, 2020, 22, 317. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.; Niu, F.; Hu, G.; Guo, M.L.; Sil, S.; Buch, S. HIV Tat-mediated induction of autophagy regulates the disruption of ZO-1 in brain endothelial cells. Tissue barriers 2020, 8, 1748983. [Google Scholar] [CrossRef] [PubMed]
- Perkins, M.V.; Joseph, S.B.; Dittmer, D.P.; Mackman, N. Cardiovascular Disease and Thrombosis in HIV Infection. Arteriosclerosis, thrombosis, and vascular biology 2023, 43, 175–191. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, M.J.; Swanson, M.D.; Contreras-Galindo, R.; Cookinham, S.; King, S.R.; Noel, R.J.Jr; Kaplan, M.H.; Markovitz, D.M. Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J Virol 2012, 86, 7790–7805. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, M.J.; Cavalcoli, J.D.; Sartor, M.A.; Contreras-Galindo, R.; Meng, F.; Dai, M.; Dube, D.; Saha, A.K.; Gitlin, S.D.; Omenn, G.S.; Kaplan, M.H.; Markovitz, D.M. Regulation of the human endogenous retrovirus K (HML-2) transcriptome by the HIV-1 Tat protein. J Virol 2014, 88, 8924–8935. [Google Scholar] [CrossRef]
- Römer, C. Viruses and Endogenous Retroviruses as Roots for Neuroinflammation and Neurodegenerative Diseases. Front Neurosci 2021, 15, 648629. [Google Scholar] [CrossRef]
- Dopkins, N.; Nixon, D.F. Activation of human endogenous retroviruses and its physiological consequences. Nat Rev Mol Cell Biol 2023, Oct 23. [CrossRef]
- Chowdhury, M.; Taylor, J.P.; Chang, C.F.; Rappaport, J.; Khalili, K. Evidence that a sequence similar to TAR is important for induction of the JC virus late promoter by human immunodeficiency virus type 1 Tat. J Virol 1992, 66, 7355–7361. [Google Scholar] [CrossRef]
- Arya, S.K.; Gallo, R.C. Human immunodeficiency virus type 2 long terminal repeat: analysis of regulatory elements. Proc Natl Acad Sci U S A 1988, 85, 9753–9757. [Google Scholar] [CrossRef]
- Viglianti, G.A.; Mullins, J.I. Functional comparison of transactivation by simian immunodeficiency virus from rhesus macaques and human immunodeficiency virus type 1. J Virol 1988, 62, 4523–4532. [Google Scholar] [CrossRef] [PubMed]
- Haseltine, W.A.; Sodroski, J.; Patarca, R.; Briggs, D.; Perkins, D.; Wong-Staal, F. Structure of 3' terminal region of type II human T lymphotropic virus: evidence for new coding region. Science (New York, N.Y.) 1984, 225, 419–421. [Google Scholar] [CrossRef]
- Sodroski, J.; Trus, M.; Perkins, D.; Patarca, R.; Wong-Staal, F.; Gelmann, E.; Gallo, R.; Haseltine, W.A. Repetitive structure in the long-terminal-repeat element of a type II human T-cell leukemia virus. Proceedings of the National Academy of Sciences of the United States of America 1984, 81, 4617–4621. [Google Scholar] [CrossRef]
- Nevins, J.R.; Raychaudhuri, P.; Yee, A.S.; Rooney, R.J.; Kovesdi, I.; Reichel, R. Transactivation by the adenovirus E1A gene. Biochemistry and cell biology = Biochimie et biologie cellulaire 1988, 66, 578–583. [Google Scholar] [CrossRef]
- Brady, J.; Bolen, J.B.; Radonovich, M.; Salzman, N.; Khoury, G. Stimulation of simian virus 40 late gene expression by simian virus 40 tumor antigen. Proceedings of the National Academy of Sciences of the United States of America 1984, 81, 2040–2044. [Google Scholar] [CrossRef]
- Diaz, J.J.; Dodon, M.D.; Schaerer-Uthurralt, N.; Simonin, D.; Kindbeiter, K.; Gazzolo, L.; Madjar, J.J. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 1996, 379, 273–277. [Google Scholar] [CrossRef]
- Scala, G.; Quinto, I.; Ruocco, M.R.; Mallardo, M.; Ambrosino, C.; Squitieri, B.; Tassone, P.; Venuta, S. Epstein-Barr virus nuclear antigen 2 transactivates the long terminal repeat of human immunodeficiency virus type 1. Journal of virology 1993, 67, 2853–2861. [Google Scholar] [CrossRef]
- Hung, C.C.; Kuo, CW; Wang, WH; Chang, T.H.; Chang, P.J.; Chang L.K.; Liu, S.T. Transcriptional activation of Epstein-Barr virus BRLF1 by USF1 and Rta. The Journal of general virology 2015, 96, 2855–2866. [CrossRef]
- Roy, S.; Delling, U.; Chen, C.H.; Rosen, C.A.; Sonenberg, N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes & development 1990, 4, 1365–1373. [Google Scholar] [CrossRef]
- Delling, U.; Reid, L.S.; Barnett, R.W.; Ma, M.Y.; Climie, S.; Sumner-Smith, M.; Sonenberg, N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: model for TAR RNA tertiary structure. Journal of virology 1992, 66, 3018–3025. [Google Scholar] [CrossRef]
- Churcher, M.J.; Lamont, C.; Hamy, F.; Dingwall, C.; Green, S.M.; Lowe, A.D.; Butler, J.G.; Gait, M.J.; Karn, J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol 1993, 230, 90–110. [Google Scholar] [CrossRef]
- Baker, B.; Muckenthaler, M.; Vives, E.; Blanchard, A.; Braddock, M.; Nacken, W.; Kingsman, A.J.; Kingsman, S.M. Identification of a novel HIV-1 TAR RNA bulge binding protein. Nucleic acids research 1994, 22, 3365–3372. [Google Scholar] [CrossRef]
- Aboul-ela, F.; Karn, J.; Varani, G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. Journal of molecular biology 1995, 253, 313–332. [Google Scholar] [CrossRef]
- Naryshkin, N.A.; Gait, M.J.; Ivanovskaya, M.G. RNA recognition and regulation of HIV-1 gene expression by viral factor Tat. Biochemistry. Biokhimiia 1998, 63, 489–503. [Google Scholar] [PubMed]
- Lalonde, M.S.; Lobritz, M.A.; Ratcliff, A.; Chamanian, M.; Athanassiou, Z.; Tyagi, M.; Wong, J.; Robinson, J.A.; Karn, J.; Varani, G.; Arts, E.J. Inhibition of both HIV-1 reverse transcription and gene expression by a cyclic peptide that binds the Tat-transactivating response element (TAR) RNA. PLoS Pathog 2011, 7, e1002038. [Google Scholar] [CrossRef] [PubMed]
- Chavali, S.S.; Bonn-Breach, R.; Wedekind, J.E. Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery. J Biol Chem 2019, 294, 9326–9341. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y., Yan, W., Hall, A.B., Jiang, X. Characterizing Transcriptional Regulatory Sequences in Coronaviruses and Their Role in Recombination. Mol Biol Evol 2021, 38, 1241-1248. [CrossRef]
- Berkhout, B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic acids research 1992, 20, 27–31. [Google Scholar] [CrossRef]
- Berkhout, B.; Gatignol, A.; Silver, J.; Jeang, K.T. Efficient trans-activation by the HIV-2 Tat protein requires a duplicated TAR RNA structure. Nucleic Acids Res 1990, 18, 1839–1846. [Google Scholar] [CrossRef]
- Pachulska-Wieczorek, K.; Purzycka, K.J.; Adamiak, R.W. New, extended hairpin form of the TAR-2 RNA domain points to the structural polymorphism at the 5' end of the HIV-2 leader RNA. Nucleic Acids Res 2006, 34, 2984–2997. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Guo, C.; Josephs, S.F.; Wong-Staal, F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science 1985, 229, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Laspia, M.F.; Rice, A.P.; Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 1989, 59, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Shida, H.I.; Maki, M.A.; Hatanaka, M.A. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol 1990, 64, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Calnan, B.J.; Biancalana, S.; Hudson, D.; Frankel, A.D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes & development 1991, 5, 201-210. [CrossRef]
- Calnan, B.J.; Tidor, B.; Biancalana, S.; Hudson, D.; Frankel, A.D. Arginine-mediated RNA recognition: the arginine fork. Science 1991, 252, 1167–1171. [Google Scholar] [CrossRef]
- Chiozzini, C.; Toschi, E. HIV-1 TAT and immune dysregulation in aids pathogenesis: a therapeutic target. Curr. Drug Targets 2016, 17, 33–45. [Google Scholar] [CrossRef]
- Tan, R.; Chen, L.; Buettner, J.A.; Hudson, D.; Frankel, A.D. RNA recognition by an isolated alpha helix. Cell 1993, 73, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.N.; Jin, H. The RGG motif proteins: Interactions, functions, and regulations. Wiley Interdiscip Rev RNA 2023, 14, e1748. [Google Scholar] [CrossRef]
- Gotora, P.T.; van der Sluis, R.; Williams, M.E. HIV-1 Tat amino acid residues that influence Tat-TAR binding affinity: a scoping review. BMC Infect Dis 2023, 23, 164. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zeng, R.; von Brunn, A.; Lei, J. Structural characterization of the C-terminal domain of SARS-CoV-2 nucleocapsid protein. Mol Biomed 2020, 1, 2. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chang, C.K.; Chang, Y.W.; Sue, S.C.; Bai, H.I.; Riang, L.; Hsiao, C.D.; Huang, T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. Journal of molecular biology 2007, 368, 1075–1086. [Google Scholar] [CrossRef]
- Huang, Q.; Yu, L.; Petros, A.M.; Gunasekera, A.; Liu, Z.; Xu, N.; Hajduk, P.; Mack, J.; Fesik, S.W.; Olejniczak, E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 2004, 43, 6059–6063. [Google Scholar] [CrossRef]
- Zinzula, L.; Basquin, J.; Bohn, S.; Beck, F.; Klumpe, S.; Pfeifer, G.; Nagy, I.; Bracher, A.; Hartl, F.U.; Baumeister, W. High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2. Biochem Biophys Res Commun 2021, 538, 54–62. [Google Scholar] [CrossRef]
- Khare, S., Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N; Ho, J.; Lee, R.T.; Yeo, W.; Curation Team GC; Maurer-Stroh, S. GISAID’s Role in Pandemic Response. China CDC Weekly 2021, 3, 1049-1051. [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: from vision to reality. EuroSurveillance 2017, 22. [Google Scholar] [CrossRef]
- Surjit, M.; Kumar, R.; Mishra, R.N.; Reddy, M.K.; Chow, V.T.; Lal, S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. Journal of virology 2005, 79, 11476–11486. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O'Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol Cell 2013, 50, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Ozdilek, B.A.; Thompson, V.F.; Ahmed, N.S.; White, C.I.; Batey, R.T.; Schwartz, J.C. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res 2017, 45, 7984–7996. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zandi, R. Biophysical Modeling of SARS-CoV-2 Assembly: Genome Condensation and Budding. Viruses 2022, 14, 2089. [Google Scholar] [CrossRef]
- Yu, I.M.; Oldham, M.L.; Zhang, J.; Chen, J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. The Journal of biological chemistry 2006, 281, 17134–17139. [Google Scholar] [CrossRef]
- Hauber, J.; Malim, M.H.; Cullen, B.R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol 1989, 63, 1181–1187. [Google Scholar] [CrossRef]
- Peng, T.Y.; Lee, K.R.; Tarn, W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. The FEBS journal 2008, 275, 4152–4163. [Google Scholar] [CrossRef]
- Adly, A.N.; Bi, M.; Carlson, C.R.; Syed, A.M.; Ciling, A.; Doudna, J.A.; Cheng, Y.; Morgan, D.O. Assembly of SARS-CoV-2 ribonucleosomes by truncated N∗ variant of the nucleocapsid protein. J Biol Chem 2023, 299, 105362. [Google Scholar] [CrossRef]
- Stertz, S.; Reichelt, M.; Spiegel, M.; Kuri, T.; Martínez-Sobrido, L.; García-Sastre, A.; Weber, F.; Kochs, G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 2007, 361, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S.; Koetzner, C.A.; Kerr, C.A.; Heo, Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. Journal of virology 1994, 68, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, S.; Cruz, J.L.; Sola, I.; Mateos-Gómez, P.A.; Palacio, L.; Enjuanes, L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. Journal of virology 2010, 84, 2169–2175. [Google Scholar] [CrossRef] [PubMed]
- Almazán, F.; Galán, C.; Enjuanes, L. The nucleoprotein is required for efficient coronavirus genome replication. Journal of virology 2004, 78, 12683–12688. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.A.; Harrich, D.; Pearson, L.; Mitsuyasu, R.; Gaynor, R.B. Functional domains required for tat-induced transcriptional activation of the HIV-1 long terminal repeat. EMBO J 1988, 7, 3143–3147. [Google Scholar] [CrossRef] [PubMed]
- Sadaie, M.R.; Mukhopadhyaya, R.; Benaissa, Z.N.; Pavlakis, G.N.; Wong-Staal, F. Conservative mutations in the putative metal-binding region of human immunodeficiency virus tat disrupt virus replication. AIDS Res Hum Retroviruses 1990, 6, 1257–1263. [Google Scholar] [CrossRef] [PubMed]
- Kuppuswamy, M.; Subramanian, T.; Srinivasan, A.; Chinnadurai, G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res 1989, 17, 3551–3561. [Google Scholar] [CrossRef]
- Ruben, S.; Perkins, A.; Purcell, R.; Joung, K.; Sia, R.; Burghoff, R.; Haseltine, W.A.; Rosen, C.A. Structural and functional characterization of human immunodeficiency virus tat protein. J Virol 1989, 63, 1–8. [Google Scholar] [CrossRef]
- Rice, A.P.; Carlotti, F. Structural analysis of wild-type and mutant human immunodeficiency virus type 1 Tat proteins. J Virol 1990, 64, 6018–6026. [Google Scholar] [CrossRef]
- Frankel, A.D.; Bredt, D.S.; Pabo, C.O. Tat protein from human immunodeficiency virus forms a metal-linked dimer. Science 1988, 240, 70–73. [Google Scholar] [CrossRef]
- Frankel, A.D.; Chen, L.; Cotter, R.J.; Pabo, C.O. Dimerization of the tat protein from human immunodeficiency virus: a cysteine-rich peptide mimics the normal metal-linked dimer interface. Proc Natl Acad Sci U S A 1988, 85, 6297–6300. [Google Scholar] [CrossRef]
- Cong, Y.; Ulasli, M.; Schepers, H.; Mauthe, M.; V'kovski, P.; Kriegenburg, F.; Thiel, V.; de Haan, C.A.M.; Reggiori, F. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. Journal of virology 2020, 94, e01925–19. [Google Scholar] [CrossRef]
- Mulabbi, E.N.; Tweyongyere, R.; Byarugaba, DK. The history of the emergence and transmission of human coronaviruses. Onderstepoort J Vet Res 2021, 88, e1–e8. [Google Scholar] [CrossRef]
- Lai, M.M.; Cavanagh, D. The molecular biology of coronaviruses. Advances in virus research 1997, 48, 1–100. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Chu, D.K.; Peiris, J.S.; Kosakovsky Pond, S.L.; Poon, L.L. A case for the ancient origin of coronaviruses. J Virol 2013, 87, 7039–7045. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Forero-Muñoz, N.R.; Muylaert, R.L.; Seifert, S.N.; Albery, G.F.; Becker, D.J.; Carlson, C.J.; Poisot T. The coevolutionary mosaic of bat betacoronavirus emergence risk, Virus Evolution, 2023, vead079. [CrossRef]
- Woo, P.C.; Lau, S.K.; Huang, Y.; Yuen, K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Experimental Biology and medicine 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Wille, M.; Holmes, E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS microbiology reviews 2020, 44, 631–644. [Google Scholar] [CrossRef]
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annual review of virology 2015, 2, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Olsthoorn, R.C. Group-specific structural features of the 5'-proximal sequences of coronavirus genomic RNAs. Virology 2010, 401, 29–41. [Google Scholar] [CrossRef]
- Madhugiri, R.; Karl, N.; Petersen, D.; Lamkiewicz, K.; Fricke, M.; Wend, U.; Scheuer, R.; Marz, M.; Ziebuhr, J. Structural and functional conservation of cis-acting RNA elements in coronavirus 5'-terminal genome regions. Virology 2018, 517, 44–55. [Google Scholar] [CrossRef]
- Ma, Y.; Tong, X.; Xu, X.; Li, X.; Lou, Z.; Rao, Z. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell 2010, 1, 688–697. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Takeda, M.; Chang, C.K.; Ikeya, T.; Güntert, P.; Chang, Y.H.; Hsu, Y.L.; Huang, T.H.; Kainosho, M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. Journal of molecular biology, 2008, 380, 608–622. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.K.; Sue, S.C.; Yu, T.H.; Hsieh, C.M.; Tsai, C.K.; Chiang, Y.C.; Lee, S.J.; Hsiao, H.H.; Wu, W.J.; Chang, W.L.; Lin, C.H.; Huang, T.H. Modular organization of SARS coronavirus nucleocapsid protein. Journal of biomedical science 2006, 13, 59–72. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. Journal of virology 2009, 83, 7221–7234. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Syed, A.M.; Khalid, M.M.; Nguyen, A.; Ciling, A.; Wu, D.; Yau, W.M.; Srinivasan, S.; Esposito, D.; Doudna, J.A.; Piszczek, G.; Ott, M.; Schuck, P. Assembly reactions of SARS-CoV-2 nucleocapsid protein with nucleic acid. bioRxiv [Preprint]. 2023 Nov 23:2023.11.22.568361. [CrossRef]
- Korn, M.; Dhamotharan, K.; Jeffries, C.M.; Schlundt, A. The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5’-genomic RNA elements. Nat Commun 2023, 14, 3331. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.R.; Asfaha, J.B.; Ghent, C.M.; Howard, C.J.; Hartooni, N.; Safari, M.; Frankel, A.D.; Morgan, D.O. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions. Molecular cell 2020, 80, 1092–1103.e4. [Google Scholar] [CrossRef]
- Estelle, A.B.; Forsythe, H.M.; Yu, Z.; Hughes, K.; Lasher, B.; Allen, P.; Reardon, P.N.; Hendrix, D.A.; Barbar, E.J. RNA structure and multiple weak interactions balance the interplay between RNA binding and phase separation of SARS-CoV-2 nucleocapsid. PNAS nexus 2023, 2, pgad333. [Google Scholar] [CrossRef]
- Grossoehme, N.E.; Li, L.; Keane, S.C.; Liu, P.; Dann, C.E. 3rd.; Leibowitz, J.L.; Giedroc, D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J Mol Biol 2009, 394, 544–557. [Google Scholar] [CrossRef]
- Carlson, C.R.; Adly, A.N.; Bi, M.; Howard, C.J.; Frost, A.; Cheng, Y.; Morgan, D.O. Reconstitution of the SARS-CoV-2 ribonucleosome provides insights into genomic RNA packaging and regulation by phosphorylation. The Journal of biological chemistry 2022, 298, 102560. [Google Scholar] [CrossRef]
- Sánchez-Jiménez, C.; Izquierdo, J.M. T-cell intracellular antigens in health and disease. Cell cycle (Georgetown, Tex.) 2015, 14, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Leibowitz, J.L. The structure and functions of coronavirus genomic 3' and 5' ends. Virus Res 2015, 206, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.C.; Sexton, N.R.; Denison, M.R. Thinking Outside the Triangle: Replication Fidelity of the Largest RNA Viruses. Annu Rev Virol 2014, 1, 111–132. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Lu, S.; Wang, Y.; He, L.; Wang, M.; Jia, R.; Chen, S.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; Wu, Y.; Zhang, S.; Huang, J.; Mao, S.; Ou, X.; Sun, D.; Tian, B.; Cheng, A. Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria. Frontiers in microbiology 2023, 14, 1129437. [Google Scholar] [CrossRef] [PubMed]
- Spencer, K.A.; Hiscox, J.A. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS letters 2006, 580, 5993–5998. [Google Scholar] [CrossRef] [PubMed]
- Caruso, Í.P.; Sanches, K.; Da Poian, A.T.; Pinheiro, A.S.; Almeida, F.C.L. Dynamics of the SARS-CoV-2 nucleoprotein N-terminal domain triggers RNA duplex destabilization. Biophys J 2021, 120, 2814–2827. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, A.; Feng, J.; Li, G.; Guo, D.; Sajid, M.; Wu, K.; Zhang, Q.; Ponty, Y.; Will, S.; Liu, F.; Yu, X.; Li, S.; Liu, Q.; Yang, X.L.; Guo, M.; Li, X.; Chen, M.; Shi, Z.L.; Lan, K.; Chen, Y.; Zhou, Y. The SARS-CoV-2 subgenome landscape and its novel regulatory features. Molecular cell 2021, 81, 2135–2147.e5. [Google Scholar] [CrossRef]
- Ziv, O.; Gabryelska, M.M.; Lun, A.T.L.; Gebert, L.F.R.; Sheu-Gruttadauria, J.; Meredith, L.W.; Liu, Z.Y.; Kwok, C.K.; Qin, C.F.; MacRae, I.J.; Goodfellow, I.; Marioni, J.C.; Kudla, G.; Miska, E.A. COMRADES determines in vivo RNA structures and interactions. Nat Methods 2018, 15, 785–788. [Google Scholar] [CrossRef]
- Li, L.; Kang, H.; Liu, P.; Makkinje, N.; Williamson, S.T.; Leibowitz, J.L.; Giedroc, D.P. Structural lability in stem-loop 1 drives a 5' UTR-3' UTR interaction in coronavirus replication. J Mol Biol 2008, 377, 790–803. [Google Scholar] [CrossRef]
- Miao, Z.; Tidu, A.; Eriani, G.; Martin, F. Secondary structure of the SARS-CoV-2 5'-UTR. RNA biology 2021, 18, 447–456. [Google Scholar] [CrossRef]
- Hahn, C.S.; Hahn, Y.S.; Rice, C.M.; Lee, E.; Dalgarno, L.; Strauss, E.G.; Strauss, J.H. Conserved elements in the 3' untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 1987, 198, 33–41. [Google Scholar] [CrossRef]
- Lo, C.Y.; Tsai, T.L.; Lin, C.N.; Lin, C.H.; Wu, H.Y. Interaction of coronavirus nucleocapsid protein with the 5'- and 3'-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. The FEBS journal 2019, 286, 3222–3239. [Google Scholar] [CrossRef]
- Goebel, S.J.; Hsue, B.; Dombrowski, T.F.; Masters, P.S. Characterization of the RNA components of a putative molecular switch in the 3' untranslated region of the murine coronavirus genome. J Virol 2004, 78, 669–682. [Google Scholar] [CrossRef]
- Xue, X.; Yang, H.; Shen, W.; Zhao, Q.; Li, J.; Yang, K.; Chen, C.; Jin, Y.; Bartlam, M.; Rao, Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of molecular biology 2007, 366, 965–975. [Google Scholar] [CrossRef]
- Jonassen, C.M.; Jonassen, T.O.; Grinde, B. A common RNA motif in the 3' end of the genomes of astroviruses, avian infectious bronchitis virus and an equine rhinovirus. The Journal of general virology 1998, 79, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.P.; Igel, H.; Baertsch, R.; Haussler, D.; Ares, M.Jr.; Scott, W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS biology 2005, 3, e5. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, J.F.; Hogue, B.G. Host protein interactions with the 3' end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. Journal of virology 2000, 74, 5053–5065. [Google Scholar] [CrossRef] [PubMed]
- Tarun, S.Z.Jr.; Wells, S.E.; Deardorff, J.A.; Sachs, A.B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proceedings of the National Academy of Sciences of the United States of America 1997, 94, 9046–9051. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, T.; Arya, S.; Chan, S.H.; Qi, S.; Dai, N.; Misra, A.; Park, J.G.; Oladunni, F.; Kovalskyy, D.; Hromas, R.A.; Martinez-Sobrido, L.; Gupta, Y.K. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun 2020, 11, 3718. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Ivanov, K.A.; Putics, Á.; Hertzig, T.; Schelle, B.; Bayer, S.; Weißbrich, B.; Snijder, E.J.; Rabenau, H.; Doerr, H.W.; Gorbalenya, A.E.; Ziebuhr, J. Mechanisms and enzymes involved in SARS coronavirus genome expression. The Journal of general virology 2003, 84, 2305–2315. [Google Scholar] [CrossRef]
- Viehweger, A.; Krautwurst, S.; Lamkiewicz, K.; Madhugiri, R.; Ziebuhr, J.; Hölzer, M.; Marz, M. Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome research 2019, 29, 1545–1554. [Google Scholar] [CrossRef]
- Pasternak, A.O.; Spaan, W.J.; Snijder, E.J. Regulation of relative abundance of arterivirus subgenomic mRNAs. Journal of virology 2004, 78, 8102–8113. [Google Scholar] [CrossRef]
- Sola, I.; Mateos-Gomez, P.A.; Almazan, F.; Zuñiga, S.; Enjuanes, L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA biology 2011, 8, 237–248. [Google Scholar] [CrossRef]
- Wu, H.Y.; Guan, B.J.; Su, Y.P.; Fan, Y.H.; Brian, D.A. Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5'-untranslated-region mutants. Journal of virology 2014, 88, 846–858. [Google Scholar] [CrossRef]
- Byrd, A.K.; Raney, K.D. Superfamily 2 helicases. Frontiers in bioscience (Landmark edition) 2012, 17, 2070–2088. [Google Scholar] [CrossRef] [PubMed]
- Emmott, E.; Munday, D.; Bickerton, E.; Britton, P.; Rodgers, M.A.; Whitehouse, A.; Zhou, E.M.; Hiscox, J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J Virol 2013, 87, 9486–9500. [Google Scholar] [CrossRef] [PubMed]
- Cartier, C.; Sivard, P.; Tranchat, C.; Decimo, D.; Desgranges, C.; Boyer, V. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. The Journal of biological chemistry 1999, 274, 19434–19440. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Rönnstrand, L.; Valmu, L.; Kalkkinen, N.; Mäkinen, K. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. The Plant cell 2003, 15, 2124–2139. [Google Scholar] [CrossRef] [PubMed]
- Law, L.M.; Everitt, J.C.; Beatch, M.D.; Holmes, C.F.; Hobman, T.C. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J Virol 2003, 77, 1764–1771. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lei, X.; Jiang, Z.; Humphries, F.; Parsi, K.M.; Mustone, N.J.; Ramos, I.; Mutetwa, T.; Fernandez-Sesma, A.; Maehr, R.; Caffrey, D.R.; Fitzgerald, K.A. Cellular nucleic acid-binding protein restricts SARS-CoV-2 by regulating interferon and disrupting RNA-protein condensates. Proc Natl Acad Sci U S A 2023, 120, e2308355120. [Google Scholar] [CrossRef]
- Lu, X.; Pan, J.; Tao, J.; Guo, D. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus genes 2011, 42, 37–45. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, L.; Cai, S.; Zhuang, Z.; Zhao, Z.; Jin, S.; Xie, W.; Zhou, L.; Zhang, L.; Zhao, J.; Cui, J. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-κB hyper-activation and inflammation. Signal transduction and targeted therapy 2021, 6, 167. [Google Scholar] [CrossRef]
- Lai, F.W.; Stephenson, K.B.; Mahony, J.; Lichty, B.D. Human coronavirus OC43 nucleocapsid protein binds microRNA 9 and potentiates NF-κB activation. Journal of virology, 2014, 88, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.X.; Liang, J.Q.; Zhu, Q.C.; Dai, G.; Li, S.; Fung, T.S. Liu, D.X. Gammacoronavirus Avian Infectious Bronchitis Virus and Alphacoronavirus Porcine Epidemic Diarrhea Virus Exploit a Cell-Survival Strategy via Upregulation of cFOS to Promote Viral Replication. Journal of virology 2021, 95, e02107-20. [CrossRef]
- Pan, J.; Peng, X.; Gao, Y.; Li, Z.; Lu, X.; Chen, Y.; Ishaq, M.; Liu, D.; Dediego, M.L.; Enjuanes, L.; Guo, D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PloS one 2008, 3, e3299. [Google Scholar] [CrossRef] [PubMed]
- Patarca, R.; Haseltine, W.A. Intragenomic rearrangements involving 5'-untranslated region segments in SARS-CoV-2, other betacoronaviruses, and alphacoronaviruses. Virology journal 2023, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; Ragazzini, R.; Jungreis, I.; Ummadi, M.; Rojc, A.; Turner, J.; Bischof, M.L.; Obernier, K.; Braberg, H.; Soucheray, M.; Richards, A.; … Krogan, N.J. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Reuschl, A.K.; Thorne, L.G.; Whelan, M.V.X.; Ragazzini, R.; Furnon, W.; Cowton, V.M.; De Lorenzo, G.; Mesner, D.; Turner, J.L.E.; Dowgier, G.; Bogoda, N.; Bonfanti, P.; Palmarini, M.; Patel, A.H.; Jolly, C.; Towers, G.J. Evolution of enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants. Nature microbiology 2024, Advance online publication. [CrossRef]
- Emerman, M.; Guyader, M.; Montagnier, L.; Baltimore, D.; Muesing, MA. The specificity of the human immunodeficiency virus type 2 transactivator is different from that of human immunodeficiency virus type 1. EMBO J 1987, 6, 3755–60. [Google Scholar] [CrossRef] [PubMed]
- Selby, M.J.; Bain, E.S.; Luciw, P.A.; Peterlin, B.M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev 1989, 3, 547–558. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, L.F.; Mavankal, G.; Peters, P.; Wu-Baer, F.; Gaynor, R.B. Tat functions to stimulate the elongation properties of transcription complexes paused by the duplicated TAR RNA element of human immunodeficiency virus 2. J Mol Biol 1995, 254, 350–63. [Google Scholar] [CrossRef] [PubMed]
- Jeang, K.T.; Chun, R.; Lin, N.H.; Gatignol, A.; Glabe, C.G.; Fan, H. In vitro and in vivo binding of human immunodeficiency virus type 1 Tat protein and Sp1 transcription factor. J Virol 1993, 67, 6224–6233. [Google Scholar] [CrossRef]
- Chun, R.F.; Semmes, O.J.; Neuveut, C.; Jeang, K.T. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J Virol 1998, 72, 2615–2629. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Liu, J.; Wang, Q.; Liu, X.; Li, X.; Li, P.; Ma, Q.; Cao, C. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1alpha. J Virol 2008, 82, 6962–6971. [Google Scholar] [CrossRef] [PubMed]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Herbein, G. Management of epigenomic networks entailed in coronavirus infections and COVID-19. Clin Epigenetics 2020, 12, 118. [Google Scholar] [CrossRef]
- Zaborowska, J.; Isa, N.F.; Murphy, S. P-TEFb goes viral. BioEssays : news and reviews in molecular, cellular and developmental biology, 2016, 38 Suppl 1, S75–S85. [CrossRef]
- López-Muñoz, A.D.; Santos, J.J.S.; Yewdell, J.W. Cell surface nucleocapsid protein expression: A betacoronavirus immunomodulatory strategy. Proc Natl Acad Sci U S A 2023, 120, e2304087120. [Google Scholar] [CrossRef]
- Nakanaga, K.; Yamanouchi, K.; Fujiwara, K. Protective effect of monoclonal antibodies on lethal mouse hepatitis virus infection in mice. Journal of virology, 1986, 59, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, J.; Cainelli-Gebara, V.; Mercier, G.; Mansour, S.; Talbot, P.J.; Lussier, G.; Oth, D. Protection from mouse hepatitis virus type 3-induced acute disease by an anti-nucleoprotein monoclonal antibody. Brief report. Archives of virology 1987, 97, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Sparn, C.; Meyer, A.; Saleppico, R.; Nickel, W. Unconventional secretion mediated by direct protein self-translocation across the plasma membranes of mammalian cells. Trends in biochemical sciences 2022, 47, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, A.; Giacca, M. Transcellular protein transduction using the Tat protein of HIV-1. Advanced drug delivery reviews, 2005, 57, 597–608. [Google Scholar] [CrossRef]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J Biol Chem 2001, 276, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.P.; Ajasin, D.O.; Ramasamy, S.; DesMarais, V.; Eugenin, E.A.; Prasad, V.R. A naturally occurring polymorphism in the HIV-1 tat basic domain inhibits uptake by bystander cells and leads to reduced neuroinflammation. Sci. Rep 2019, 9, 3308. [Google Scholar] [CrossRef] [PubMed]
- Ajasin, D.; Eugenin, E.A. HIV-1 Tat: role in bystander toxicity. Frontiers in Cellular and Infection Microbiology 2020, 10, 61. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Nema, V. HIV-Associated Neurotoxicity: The Interplay of Host and Viral Proteins. Mediators Inflamm 2021, 2021, 1267041. [Google Scholar] [CrossRef] [PubMed]
- Buscemi, L.; Ramonet, D.; Geiger, J.D. Human immunodeficiency virus type-1 protein Tat induces tumor necrosis factor- α-mediated neurotoxicity. Neurobiology of Disease 2007, 26, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Zheng, J.; Okamoto, S.; Gendelman, HE; Lipton, S.A. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death and Differentiation 2005, 12, 878–892. [CrossRef]
- Nedwin, G.E.; Naylor, S.L.; Sakaguchi, A.Y.; Smith, D.; Jarrett-Nedwin, J.; Pennica, D.; Goeddel, D.V.; Gray, P.W. Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic acids research 1985, 13, 6361–6373. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.R.; Ramelli, S.C.; Grazioli. A; Chung, J.Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; Ko, S.H.; Platt, A.P.; Burbelo, P.D.; Quezado, M.; Pittaluga, S.; Purcell, M.; Munster, V.J.; Belinky, F.; Ramos-Benitez, M.J.; Boritz, E.A.; Lach, I.A.; Herr, D.L.; Rabin, J.; Saharia, K.K.; Madathil, R.J.; Tabatabai, A.; Soherwardi, S.; McCurdy, M.T.; NIH COVID-19 Autopsy Consortium; Peterson, K.E.; Cohen, J.I.; de Wit, E.; Vannella, K.M.; Hewitt, S.M.; Kleiner, D.E.; Chertow, D.S. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 2022, 612, 758-763. [CrossRef]
- Patel, K.P.; Vunnam, S.R.; Patel, P.A.; Krill, K.L.; Korbitz, P.M.; Gallagher, J.P.; Suh, J.E.; Vunnam, R.R. Transmission of SARS-CoV-2: An update of current literature. Eur. J. Clin. Microbiol. Infect. Dis 2020, 39, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol 2020, 5, 335–337. [Google Scholar] [CrossRef]
- Su, Y.; Yuan, D.; Chen, D.G.; Ng, R.H.; Wang, K.; Choi, J.; Li, S.; Hong, S.; Zhang, R.; Xie, J.; Kornilov, S.A.; Scherler, K.; Pavlovitch-Bedzyk, A.J.; Dong, S.; Lausted, C.; Lee, I.; Fallen, S.; Dai, C.L.; Baloni, P.; Smith, B.; Duvvuri, V.R.; Anderson, K.G.; Li, J.; Yang, F.; Duncombe, C.J.; McCulloch, D.J.; Rostomily, C.; Troisch, P.; Zhou, J.; Mackay, S.; DeGottardi, Q.; May, D.H.; Taniguchi, R.; Gittelman, R.M.; Klinger, M.; Snyder, T.M.; Roper, R.; Wojciechowska, G.; Murray, K.; Edmark, R.; Evans, S.; Jones, L.; Zhou, Y.; Rowen, L.; Liu, R.; Chour, W.; Algren, H.A.; Berrington, W.R.; Wallick, J.A.; Cochran, R.A.; Micikas, M.E.; ISB-Swedish COVID-19 Biobanking Unit; Wrin, T.; Petropoulos, C.J.; Cole, H.R.; Fischer, T.D.; Wei, W.; Hoon, D.S.B.; Price, N.D.; Subramanian, N.; Hill, J.A.; Hadlock, J.; Magis, A.T.; Ribas, A.; Lanier, L.L.; Boyd, S.D.; Bluestone, J.A.; Chu, H.; Hood, L.; Gottardo, R,; Greenberg, P.D.; Davis, M.M.; Goldman, J.D.; Heath, J.R. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 2022, 185, 881-895.e20. [CrossRef]
- Castanares-Zapatero, D.; Chalonm, P.; Kohn, L.; Kohn, L.; Dauvrin, M.; Detollenaere, J.; Maertens de Noordhout, C.; Primus-de Jong, C.; Cleemput, I.; Van den Heede, K. Pathophysiology and mechanism of long COVID: a comprehensive review. Ann Med 2022, 54, 1473–1487. [Google Scholar] [CrossRef]
- Perumal, R.; Shunmugam, L.; Naidoo, K.; Abdool Karim, S.S.; Wilkins, D.; Garzino-Demo, A.; Brechot, C.; Parthasarathy, S.; Vahlne, A.; Nikolich, J.Ž. Long COVID: a review and proposed visualization of the complexity of long COVID. Frontiers in immunology 2023, 14, 1117464. [Google Scholar] [CrossRef]
- Buonsenso, D.; Martino, L.; Morello, R.; Mariani, F.; Fearnley, K.; Valentini, P. Viral persistence in children infected with SARS-CoV-2: current evidence and future research strategies. The Lancet. Microbe 2023, S2666-5247(23)00115-5. [CrossRef]
- Cupelli, L.A.; Hsu, M.C. The human immunodeficiency virus type 1 Tat antagonist, Ro 5-3335, predominantly inhibits transcription initiation from the viral promoter. J Virol 1995, 69, 2640–2643. [Google Scholar] [CrossRef]
- Hwang, S.; Tamilarasu, N.; Kibler, K.; Cao, H.; Ali, A.; Ping, Y.H.; Jeang, K.T.; Rana, T.M. Discovery of a small molecule Tat-trans-activation-responsive RNA antagonist that potently inhibits human immunodeficiency virus-1 replication. J Biol Chem 2003, 278, 39092–39103. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.; Afshar, M.; Varani, G.; Murchie, A.I.; Karn, J.; Lentzen, G.; Drysdale, M.; Bower, J.; Potter, A.J.; Starkey, I.D.; Swarbrick, T.; Aboul-ela, F. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic "hot spots". J Mol Biol 2004, 336, 343–56. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.C.: Schutt, A.D.; Holly, M.; Slice, L.W.; Sherman, M.I.; Richman, D.D.; Potash, M.J.; Volsky, D.J. Inhibition of HIV replication in acute and chronic infections in vitro by a Tat antagonist. Science 1991, 254, 1799-802. [CrossRef]
- Kim, H.I.; Kim, G.N.; Yu, K.L.; Park, S.H.; You, J.C. Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication. Int J Mol Sci 2022, 23, 12340. [Google Scholar] [CrossRef] [PubMed]
- Ait-Ammar, A.; Kula, A.; Darcis, G.; Verdikt, R.; De Wit, S.; Gautier, V.; Mallon, P.W.G.; Marcello, A.; Rohr, O.; Van Lint, C. Current Status of Latency Reversing Agents Facing the Heterogeneity of HIV-1 Cellular and Tissue Reservoirs. Frontiers in microbiology 2020, 10, 3060. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: a better web interface. Nucleic Acids Res 2008, 36(Web Server issue): W5-W9. [CrossRef]
- ICTV Coronaviridae Study Group. International Committee on Taxonomy of Viruses (ICTV). 2021. Available from. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/223/coronaviridae-figures.
- Nikolaidis, M.; Markoulatos, P.; van de Peer, Y.; Oliver, S.G.; Amoutzias, G.D. The neighborhood of the spike gene is a hotspot for modular intertypic homologous and non-homologous recombination in coronavirus genomes. Mol Biol Evol 2022, msab 292. [CrossRef]
- Tsoleridis, T.; Chappell, J.G.; Onianwa, O.; Marston, D.A.; Fooks, A.R.; Monchatre-Leroy, E.; Umhang, G.; Müller, M.A.; Drexler, J.F.; Drosten, C.; Tarlinton, R.E.; McClure, C.P.; Holmes, E.C.; Ball, J.K. Shared Common Ancestry of Rodent Alphacoronaviruses Sampled Globally. Viruses 2019, 11, 125. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Research 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Research 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. "ViennaRNA Package 2.0", Algorithms for Molecular Biology 2011, 6, 26. [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
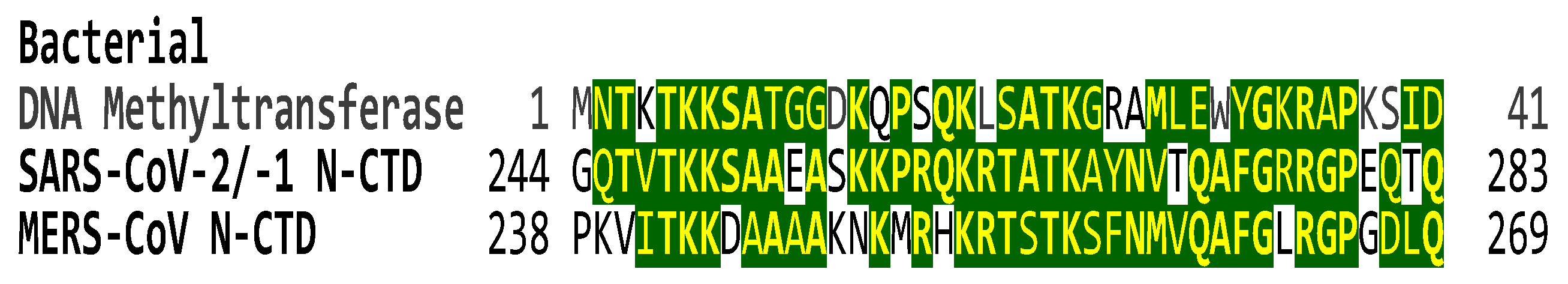

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



