Submitted:
29 December 2023
Posted:
30 December 2023
You are already at the latest version
Abstract
Keywords:
1. What is Cow’s Milk made of?
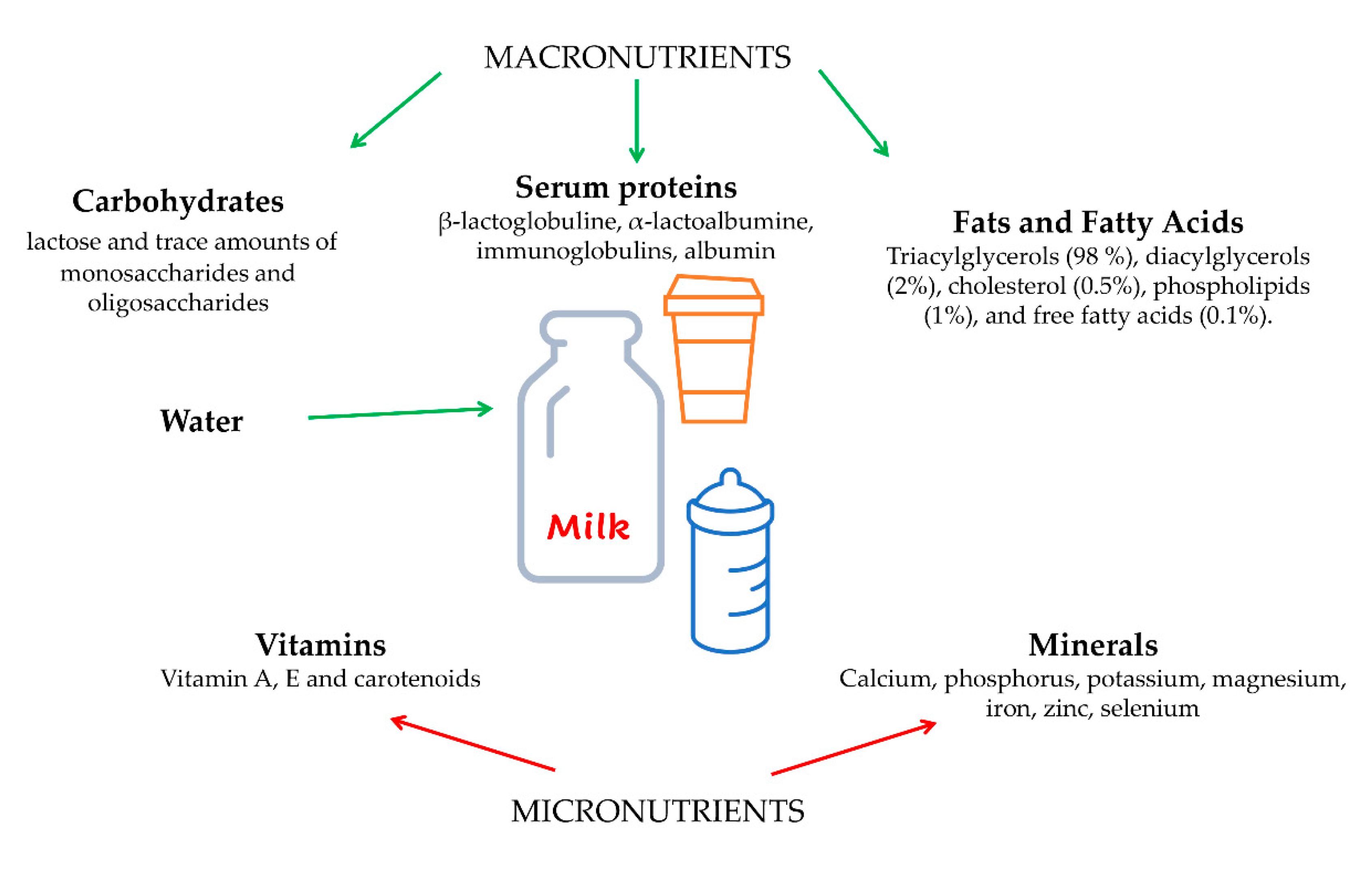
2. Cow’s Milk Proteins
3. Cow’s Milk Fats and Micronutrients
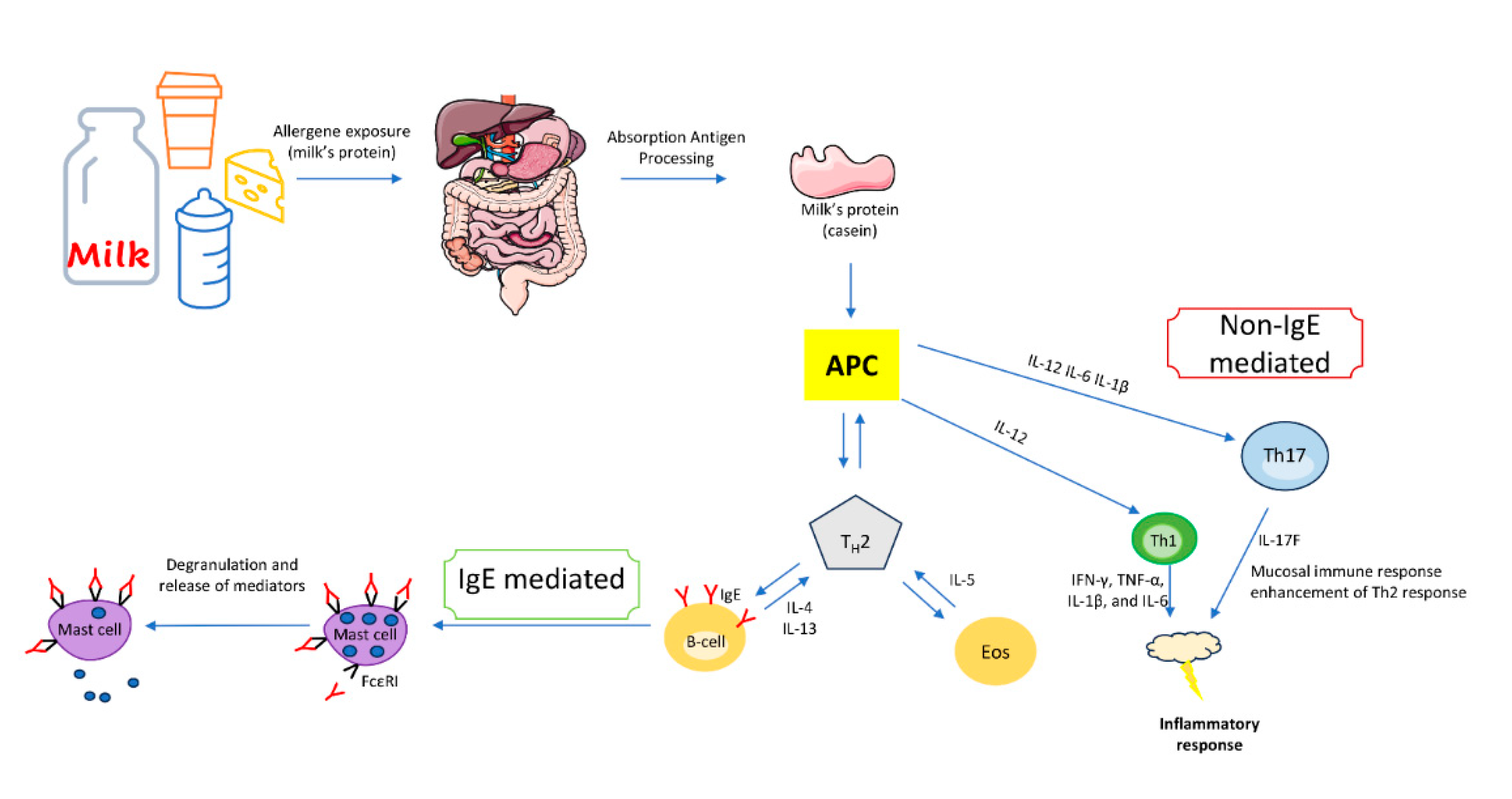
4. IgE-mediated immune response to cow’s milk proteins
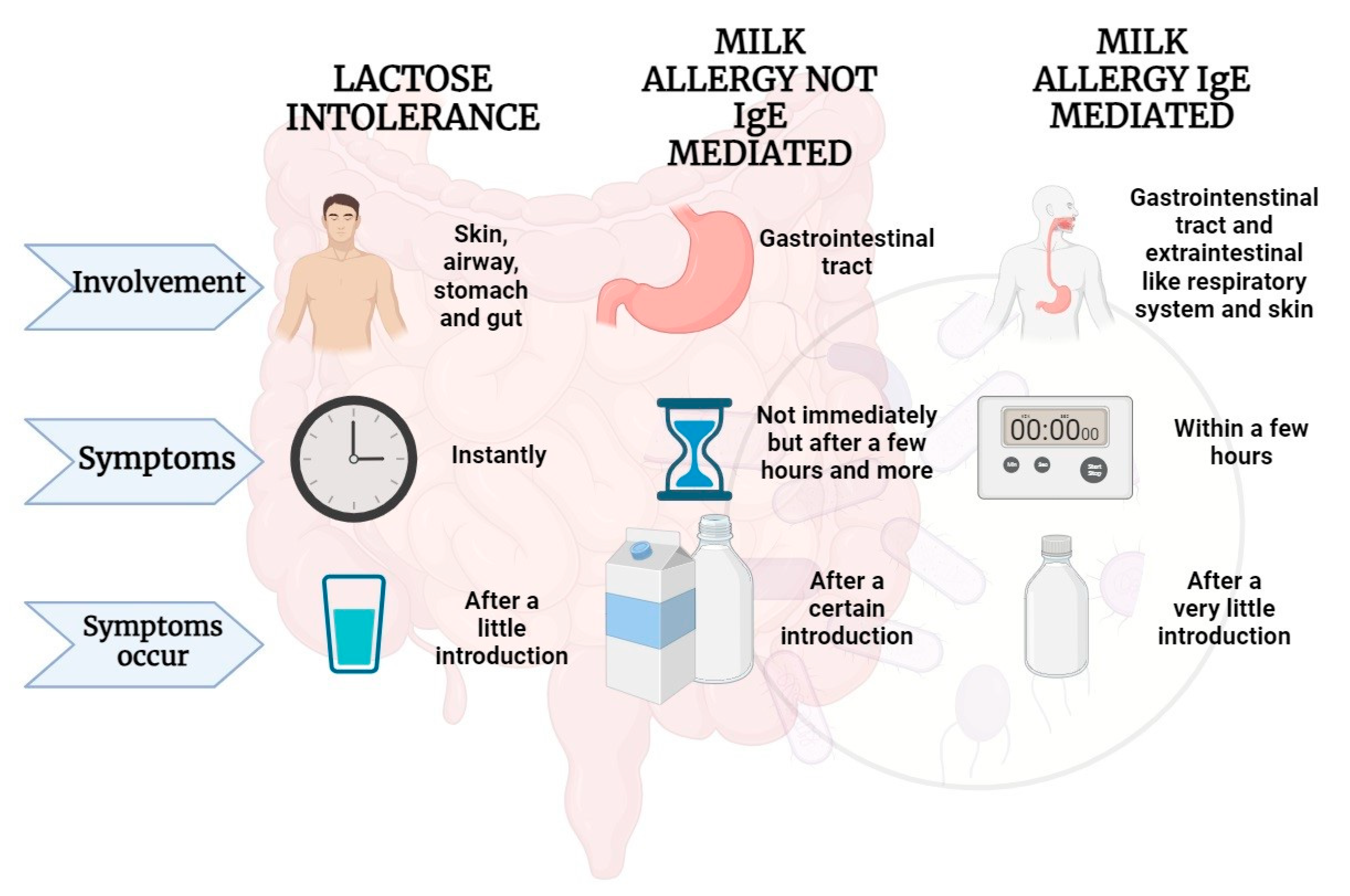
5. Non-IgE-mediated immune response to milk proteins
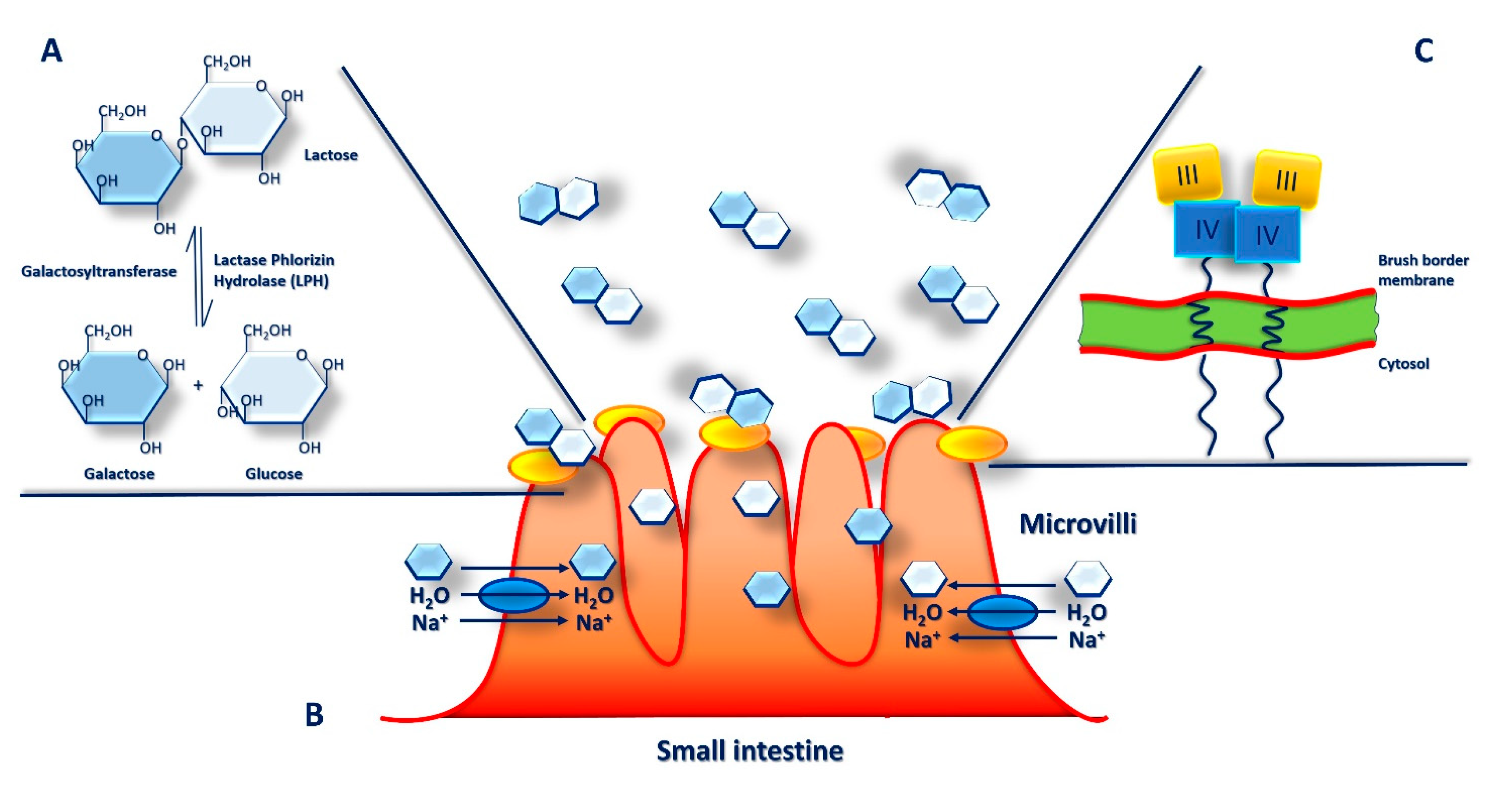
6. Microbiota and Lactose Intolerance
7. Lactase Deficiency and Lactose Intolerance
8. Omics Tools: Nutrigenetics and Epigenetics Approaches for Lactose Intolerance Management
9. Precision Nutrition through Metabolomics Approaches
10. Conclusions
- Author Contributions: Conceptualization, methodology, investigation, data curation, software, writing— original draft preparation, writing—review and editing, supervision, all authors. All authors have read and agreed to the published version of the manuscript.
- Funding: This research was funded by Finalized Research Funding (FFR 2018/2022) FRR _D15_DE BLASIO, FRR_D03_CARLISI, FRR _D03_DI LIBERTO, Università degli Studi di Palermo, Italy.
- Acknowledgments: Figures were created by using “Server Medical Art. Servier Medical Art by Servier”, licensed under a Creative Commons Attribution 3.0 Unported License, and BioRender.com.
References
- Pereira, P.C. Milk nutritional composition and its role in human health. Nutrition 2014, 30, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Heinig, M.J.; Nommsen, L.A.; Peerson, J.M.; Lonnerdal, B.; Dewey, K.G. Energy and protein intakes of breast-fed and formula-fed infants during the first year of life and their association with growth velocity: the DARLING Study. The American journal of clinical nutrition 1993, 58, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.T.; Nadeem, M.; Imran, M.; Ayaz, M.; Ajmal, M.; Ellahi, M.Y.; Khalique, A. Antioxidant capacity and fatty acids characterization of heat treated cow and buffalo milk. Lipids in health and disease 2017, 16, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Leduc, A.; Souchet, S.; Gele, M.; Le Provost, F.; Boutinaud, M.; Pascottini, O.; Carvalho, M.; Schyndel, S.; Ticiani, E.; Spricigo, J. Effect of feed restriction on dairy cow milk production. Journal of Animal Science 2021, 99, 1–1. [Google Scholar] [CrossRef] [PubMed]
- Foroutan, A.; Guo, A.C.; Vazquez-Fresno, R.; Lipfert, M.; Zhang, L.; Zheng, J.; Badran, H.; Budinski, Z.; Mandal, R.; Ametaj, B.N. Chemical composition of commercial cow’s milk. Journal of agricultural and food chemistry 2019, 67, 4897–4914. [Google Scholar] [CrossRef] [PubMed]
- Leischner, C.; Egert, S.; Burkard, M.; Venturelli, S. Potential protective protein components of cow’s milk against certain tumor entities. Nutrients 2021, 13, 1974. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Duncan, S.E.; Knowlton, K.F.; Ray, W.K.; Dietrich, A.M. Milk protein composition and stability changes affected by iron in water sources. Journal of Dairy Science 2016, 99, 4206–4219. [Google Scholar] [CrossRef]
- Dyrda-Terniuk, T.; Pryshchepa, O.; Rafińska, K.; Kolankowski, M.; Gołębiowski, A.; Gloc, M.; Dobrucka, R.; Kurzydłowski, K.; Pomastowski, P. Immobilization of silver ions onto casein. Colloids and Surfaces A: Physicochemical and Engineering Aspects 2023, 667, 131390. [Google Scholar] [CrossRef]
- Thorning, T.K.; Raben, A.; Tholstrup, T.; Soedamah-Muthu, S.S.; Givens, I.; Astrup, A. Milk and dairy products: good or bad for human health? An assessment of the totality of scientific evidence. Food & nutrition research 2016, 60, 32527. [Google Scholar]
- Carter, B.; Cheng, N.; Kapoor, R.; Meletharayil, G.; Drake, M. Invited review: Microfiltration-derived casein and whey proteins from milk. Journal of Dairy Science 2021, 104, 2465–2479. [Google Scholar] [CrossRef]
- Carter, B.; DiMarzo, L.; Pranata, J.; Barbano, D.M.; Drake, M. Efficiency of removal of whey protein from sweet whey using polymeric microfiltration membranes. Journal of Dairy Science 2021, 104, 8630–8643. [Google Scholar] [CrossRef]
- Kim, J.; Paik, H.-D.; Yoon, Y.-C.; Park, E. Whey protein inhibits iron overload-induced oxidative stress in rats. Journal of nutritional science and vitaminology 2013, 59, 198–205. [Google Scholar] [CrossRef]
- Zeng, B.; Chen, T.; Xie, M.-Y.; Luo, J.-Y.; He, J.-J.; Xi, Q.-Y.; Sun, J.-J.; Zhang, Y.-L. Exploration of long noncoding RNA in bovine milk exosomes and their stability during digestion in vitro. Journal of dairy science 2019, 102, 6726–6737. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Chen, X.; Zheng, X.; Zhu, H.; Qi, Q.; Liu, S.; Zhang, H.; Che, J. Latest trend of milk derived exosomes: Cargos, functions, and applications. Frontiers in nutrition 2021, 8, 747294. [Google Scholar] [CrossRef] [PubMed]
- Contarini, G.; Povolo, M. Phospholipids in milk fat: composition, biological and technological significance, and analytical strategies. International Journal of Molecular Sciences 2013, 14, 2808–2831. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Anaya, J.; Jiménez-Flores, R. Symposium review: The relevance of bovine milk phospholipids in human nutrition—Evidence of the effect on infant gut and brain development. Journal of dairy science 2019, 102, 2738–2748. [Google Scholar] [CrossRef] [PubMed]
- Stonehouse, W.; Klingner, B.; McJarrow, P.; Fong, B.; O’callaghan, N. Exploring in vivo dynamics of bovine milk derived gangliosides. Nutrients 2020, 12, 711. [Google Scholar] [CrossRef] [PubMed]
- Benbrook, C.M.; Butler, G.; Latif, M.A.; Leifert, C.; Davis, D.R. Organic production enhances milk nutritional quality by shifting fatty acid composition: A United States–wide, 18-month study. PloS one 2013, 8, e82429. [Google Scholar] [CrossRef] [PubMed]
- Warstedt, K.; Furuhjelm, C.; Fälth-Magnusson, K.; Fagerås, M.; Duchén, K. High levels of omega-3 fatty acids in milk from omega-3 fatty acid-supplemented mothers are related to less immunoglobulin E-associated disease in infancy. Acta Paediatrica 2016, 105, 1337–1347. [Google Scholar] [CrossRef] [PubMed]
- Kasapidou, E.; Basdagianni, Z.; Papatzimos, G.; Papadopoulos, V.; Tsiftsi, E.; Neki, I.; Nigianni, P.-A.; Mitlianga, P. Chemical composition, antioxidant profile and physicochemical properties of commercial non-cocoa-and cocoa-flavoured plant-based milk alternatives. European Food Research and Technology 2023, 1–16. [Google Scholar] [CrossRef]
- Gaucheron, F. The minerals of milk. Reproduction Nutrition Development 2005, 45, 473–483. [Google Scholar] [CrossRef]
- Sorensen, M.D. Calcium intake and urinary stone disease. Translational andrology and urology 2014, 3, 235. [Google Scholar] [PubMed]
- Heravi, A.S.; Michos, E.D. Vitamin D and calcium supplements: helpful, harmful, or neutral for cardiovascular risk? Methodist DeBakey cardiovascular journal 2019, 15, 207. [Google Scholar] [CrossRef] [PubMed]
- Gudi, S.K. Dairy consumption and risk of type-2 diabetes: the untold story. Annals of pediatric endocrinology & metabolism 2021, 26, 14. [Google Scholar]
- Arafat, H.M.; Omar, J.; Shafii, N.; Naser, I.A.; Al Laham, N.A.; Muhamad, R.; Al-Astani, T.A.D.; Shaqaliah, A.J.; Shamallakh, O.M.; Shamallakh, K.M. The association between breast cancer and consumption of dairy products: a systematic review. Annals of medicine 2023, 55, 2198256. [Google Scholar] [CrossRef] [PubMed]
- Gil, H.; Chen, Q.-Y.; Khil, J.; Park, J.; Na, G.; Lee, D.; Keum, N. Milk intake in early life and later cancer risk: a meta-analysis. Nutrients 2022, 14, 1233. [Google Scholar] [CrossRef] [PubMed]
- Givens, D. MILK Symposium review: The importance of milk and dairy foods in the diets of infants, adolescents, pregnant women, adults, and the elderly. Journal of dairy science 2020, 103, 9681–9699. [Google Scholar] [CrossRef]
- Corsello, A.; Pugliese, D.; Gasbarrini, A.; Armuzzi, A. Diet and Nutrients in Gastrointestinal Chronic Diseases. Nutrients 2020, 12. [Google Scholar] [CrossRef]
- Schoemaker, A.A.; Sprikkelman, A.B.; Grimshaw, K.E.; Roberts, G.; Grabenhenrich, L.; Rosenfeld, L.; Siegert, S.; Dubakiene, R.; Rudzeviciene, O.; Reche, M. , et al. Incidence and natural history of challenge-proven cow's milk allergy in European children--EuroPrevall birth cohort. Allergy 2015, 70, 963–972. [Google Scholar] [CrossRef]
- Nwaru, B.I.; Hickstein, L.; Panesar, S.S.; Roberts, G.; Muraro, A.; Sheikh, A. Prevalence of common food allergies in Europe: a systematic review and meta-analysis. Allergy 2014, 69, 992–1007. [Google Scholar] [CrossRef]
- Katz, Y.; Rajuan, N.; Goldberg, M.R.; Eisenberg, E.; Heyman, E.; Cohen, A.; Leshno, M. Early exposure to cow's milk protein is protective against IgE-mediated cow's milk protein allergy. The Journal of allergy and clinical immunology 2010, 126, 77–82.e71. [Google Scholar] [CrossRef]
- García-Ara, M.C.; Boyano-Martínez, M.T.; Díaz-Pena, J.M.; Martín-Muñoz, M.F.; Martín-Esteban, M. Cow's milk-specific immunoglobulin E levels as predictors of clinical reactivity in the follow-up of the cow's milk allergy infants. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology 2004, 34, 866–870. [Google Scholar] [CrossRef]
- Fiocchi, A.; Terracciano, L.; Bouygue, G.R.; Veglia, F.; Sarratud, T.; Martelli, A.; Restani, P. Incremental prognostic factors associated with cow's milk allergy outcomes in infant and child referrals: the Milan Cow's Milk Allergy Cohort study. Annals of allergy, asthma & immunology : official publication of the American College of Allergy, Asthma, & Immunology 2008, 101, 166–173. [Google Scholar] [CrossRef]
- Skripak, J.M.; Matsui, E.C.; Mudd, K.; Wood, R.A. The natural history of IgE-mediated cow's milk allergy. The Journal of allergy and clinical immunology 2007, 120, 1172–1177. [Google Scholar] [CrossRef] [PubMed]
- Fiocchi, A.; Schünemann, H.J.; Brozek, J.; Restani, P.; Beyer, K.; Troncone, R.; Martelli, A.; Terracciano, L.; Bahna, S.L.; Rancé, F. , et al. Diagnosis and Rationale for Action Against Cow's Milk Allergy (DRACMA): a summary report. The Journal of allergy and clinical immunology 2010, 126, 1119–1128.e1112. [Google Scholar] [CrossRef]
- Tsabouri, S.; Douros, K.; Priftis, K.N. Cow's milk allergenicity. Endocrine, metabolic & immune disorders drug targets 2014, 14, 16–26. [Google Scholar] [CrossRef]
- Sampson, H.A. Food allergy. Part 1: immunopathogenesis and clinical disorders. The Journal of allergy and clinical immunology 1999, 103, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Vickery, B.P.; Chin, S.; Burks, A.W. Pathophysiology of food allergy. Pediatric clinics of North America 2011, 58, 363–376. [Google Scholar] [CrossRef]
- Luyt, D.; Ball, H.; Makwana, N.; Green, M.R.; Bravin, K.; Nasser, S.M.; Clark, A.T. BSACI guideline for the diagnosis and management of cow's milk allergy. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology 2014, 44, 642–672. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.M.; Paul, M.; Rothenberg, M.E. Novel immunologic mechanisms in eosinophilic esophagitis. Current opinion in immunology 2017, 48, 114–121. [Google Scholar] [CrossRef]
- Giannetti, A.; Toschi Vespasiani, G.; Ricci, G.; Miniaci, A.; di Palmo, E.; Pession, A. Cow's Milk Protein Allergy as a Model of Food Allergies. Nutrients 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. The Journal of allergy and clinical immunology 2018, 141, 41–58. [Google Scholar] [CrossRef]
- Savage, J.H.; Lee-Sarwar, K.A.; Sordillo, J.; Bunyavanich, S.; Zhou, Y.; O'Connor, G.; Sandel, M.; Bacharier, L.B.; Zeiger, R.; Sodergren, E. , et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018, 73, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Roduit, C.; Frei, R.; Depner, M.; Schaub, B.; Loss, G.; Genuneit, J.; Pfefferle, P.; Hyvärinen, A.; Karvonen, A.M.; Riedler, J. , et al. Increased food diversity in the first year of life is inversely associated with allergic diseases. The Journal of allergy and clinical immunology 2014, 133, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Perkin, M.R.; Logan, K.; Marrs, T.; Radulovic, S.; Craven, J.; Flohr, C.; Lack, G. Enquiring About Tolerance (EAT) study: Feasibility of an early allergenic food introduction regimen. The Journal of allergy and clinical immunology 2016, 137, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Venter, C.; Maslin, K.; Holloway, J.W.; Silveira, L.J.; Fleischer, D.M.; Dean, T.; Arshad, S.H. Different Measures of Diet Diversity During Infancy and the Association with Childhood Food Allergy in a UK Birth Cohort Study. The journal of allergy and clinical immunology. In practice 2020, 8, 2017–2026. [Google Scholar] [CrossRef] [PubMed]
- Koplin, J.J.; Allen, K.J.; Gurrin, L.C.; Peters, R.L.; Lowe, A.J.; Tang, M.L.; Dharmage, S.C. The impact of family history of allergy on risk of food allergy: a population-based study of infants. International journal of environmental research and public health 2013, 10, 5364–5377. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.J.; Hosking, C.S. Food allergy and atopic dermatitis in infancy: an epidemiologic study. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 2004, 15, 421–427. [Google Scholar] [CrossRef]
- Boyano-Martínez, T.; García-Ara, C.; Pedrosa, M.; Díaz-Pena, J.M.; Quirce, S. Accidental allergic reactions in children allergic to cow's milk proteins. The Journal of allergy and clinical immunology 2009, 123, 883–888. [Google Scholar] [CrossRef]
- Burris, A.D.; Burris, J.; Järvinen, K.M. Cow’s milk protein allergy in term and preterm infants: clinical manifestations, immunologic pathophysiology, and management strategies. NeoReviews 2020, 21, e795–e808. [Google Scholar] [CrossRef]
- Toro-Monjaraz, E.M.; Fonseca-Camarillo, G.; Zárate-Mondragón, F.; Montijo-Barrios, E.; Cadena-León, J.; Avelar-Rodríguez, D.; Ramírez-Mayans, J.; Cervantes-Bustamante, R.; Yamamoto-Furusho, J.K. Differential cytokine expression in the duodenum and rectum of children with non-immunoglobulin e-mediated cow’s milk protein allergy. Digestive Diseases and Sciences 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Vitaliti, G.; Cimino, C.; Coco, A.; Praticò, A.D.; Lionetti, E. The immunopathogenesis of cow’s milk protein allergy (CMPA). Italian journal of pediatrics 2012, 38, 1–5. [Google Scholar] [CrossRef]
- Athie-Morales, V.; Smits, H.H.; Cantrell, D.A.; Hilkens, C.M. Sustained IL-12 signaling is required for Th1 development. The Journal of Immunology 2004, 172, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Paajanen, L.; Kokkonen, J.; Karttunen, T.J.; Tuure, T.; Korpela, R.; Vaarala, O. Intestinal cytokine mRNA expression in delayed-type cow's milk allergy. Journal of pediatric gastroenterology and nutrition 2006, 43, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Veres, G.; Westerholm-Ormio, M.; Kokkonen, J.; Arato, A.; Savilahti, E. Cytokines and adhesion molecules in duodenal mucosa of children with delayed-type food allergy. Journal of pediatric gastroenterology and nutrition 2003, 37, 27–34. [Google Scholar]
- Paajanen, L.; Vaarala, O.; Karttunen, R.; Tuure, T.; Korpela, R.; Kokkonen, J. Increased IFN-γ secretion from duodenal biopsy samples in delayed-type cow's milk allergy. Pediatric allergy and immunology 2005, 16, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Monin, L.; Gaffen, S.L. Interleukin 17 family cytokines: signaling mechanisms, biological activities, and therapeutic implications. Cold Spring Harbor perspectives in biology 2018, 10, a028522. [Google Scholar] [CrossRef]
- Clayton, F.; Fang, J.C.; Gleich, G.J.; Lucendo, A.J.; Olalla, J.M.; Vinson, L.A.; Lowichik, A.; Chen, X.; Emerson, L.; Cox, K. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology 2014, 147, 602–609. [Google Scholar] [CrossRef]
- Schuyler, A.J.; Wilson, J.M.; Tripathi, A.; Commins, S.P.; Ogbogu, P.U.; Kruzsewski, P.G.; Barnes, B.H.; McGowan, E.C.; Workman, L.J.; Lidholm, J. Specific IgG4 antibodies to cow's milk proteins in pediatric patients with eosinophilic esophagitis. Journal of Allergy and Clinical Immunology 2018, 142, 139–148. [Google Scholar] [CrossRef]
- Aalberse, R.C.; Platts-Mills, T.A.; Rispens, T. The developmental history of IgE and IgG4 antibodies in relation to atopy, eosinophilic esophagitis, and the modified TH 2 response. Current allergy and asthma reports 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Pratelli, G.; Tamburini, B.; Carlisi, D.; De Blasio, A.; D’Anneo, A.; Emanuele, S.; Notaro, A.; Affranchi, F.; Giuliano, M.; Seidita, A. Foodomics-Based Approaches Shed Light on the Potential Protective Effects of Polyphenols in Inflammatory Bowel Disease. International Journal of Molecular Sciences 2023, 24, 14619. [Google Scholar] [CrossRef]
- Di Tommaso, N.; Gasbarrini, A.; Ponziani, F.R. Intestinal barrier in human health and disease. International journal of environmental research and public health 2021, 18, 12836. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N.; Hermoso, M.A. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Frontiers in immunology 2019, 277. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell host & microbe 2015, 17, 690–703. [Google Scholar]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M. Enterotypes of the human gut microbiome. nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Lugli, G.A.; Bernasconi, S.; Margolles, A.; Di Pierro, F.; Van Sinderen, D.; Ventura, M. The infant gut microbiome as a microbial organ influencing host well-being. Italian journal of pediatrics 2020, 46, 1–13. [Google Scholar] [CrossRef]
- Adak, A.; Khan, M.R. An insight into gut microbiota and its functionalities. Cellular and Molecular Life Sciences 2019, 76, 473–493. [Google Scholar] [CrossRef]
- del Carmen Tocaa, M.; Fernándezb, A.; Orsic, M.; Tabaccod, O.; Vinderolae, G. Lactose intolerance: myths and facts. An update. Arch Argent Pediatr 2022, 120, 59–66. [Google Scholar]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lecuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef]
- Corbett, A.J.; Eckle, S.B.; Birkinshaw, R.W.; Liu, L.; Patel, O.; Mahony, J.; Chen, Z.; Reantragoon, R.; Meehan, B.; Cao, H. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014, 509, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Saxena, A.; Reyes, J.-L.; McKay, D.M. Butyrate enhances antibacterial effects while suppressing other features of alternative activation in IL-4-induced macrophages. American Journal of Physiology-Gastrointestinal and Liver Physiology 2016, 310, G822–G831. [Google Scholar] [CrossRef] [PubMed]
- Albenberg, L.G.; Wu, G.D. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, B.; La Manna, M.P.; La Barbera, L.; Mohammadnezhad, L.; Badami, G.D.; Shekarkar Azgomi, M.; Dieli, F.; Caccamo, N. Immunity and nutrition: the right balance in inflammatory bowel disease. Cells 2022, 11, 455. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Zhong, Y.; Priebe, M.G.; Vonk, R.J.; Huang, C.-Y.; Antoine, J.-M.; He, T.; Harmsen, H.J.; Welling, G.W. The role of colonic microbiota in lactose intolerance. Digestive diseases and sciences 2004, 49, 78–83. [Google Scholar] [CrossRef]
- Azcarate-Peril, M.A.; Ritter, A.J.; Savaiano, D.; Monteagudo-Mera, A.; Anderson, C.; Magness, S.T.; Klaenhammer, T.R. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proceedings of the National Academy of Sciences 2017, 114, E367–E375. [Google Scholar] [CrossRef]
- Arnold, J.W.; Simpson, J.B.; Roach, J.; Bruno-Barcena, J.M.; Azcarate-Peril, M.A. Prebiotics for lactose intolerance: variability in galacto-oligosaccharide utilization by intestinal Lactobacillus rhamnosus. Nutrients 2018, 10, 1517. [Google Scholar] [CrossRef]
- Misselwitz, B.; Butter, M.; Verbeke, K.; Fox, M.R. Update on lactose malabsorption and intolerance: pathogenesis, diagnosis and clinical management. Gut 2019, 68, 2080–2091. [Google Scholar] [CrossRef]
- Leis, R.; de Castro, M.-J.; de Lamas, C.; Picáns, R.; Couce, M.L. Effects of prebiotic and probiotic supplementation on lactase deficiency and lactose intolerance: a systematic review of controlled trials. Nutrients 2020, 12, 1487. [Google Scholar] [CrossRef]
- Oak, S.J.; Jha, R. The effects of probiotics in lactose intolerance: A systematic review. Critical reviews in food science and nutrition 2019, 59, 1675–1683. [Google Scholar] [CrossRef]
- Gingold-Belfer, R.; Levy, S.; Layfer, O.; Pakanaev, L.; Niv, Y.; Dickman, R.; Perets, T.T. Use of a novel probiotic formulation to alleviate lactose intolerance symptoms—A pilot study. Probiotics and antimicrobial proteins 2020, 12, 112–118. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Venema, K.; Priebe, M.; Welling, G.; Brummer, R.J.; Vonk, R. The role of colonic metabolism in lactose intolerance. European journal of clinical investigation 2008, 38, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, M.; Canani, R.B. Lactose intolerance: common misunderstandings. Annals of Nutrition and Metabolism 2018, 73, 30–37. [Google Scholar] [CrossRef] [PubMed]
- D’Auria, E.; Venter, C. Precision medicine in cow's milk allergy. Current opinion in allergy and clinical immunology 2020, 20, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Giannetti, A.; Toschi Vespasiani, G.; Ricci, G.; Miniaci, A.; di Palmo, E.; Pession, A. Cow’s milk protein allergy as a model of food allergies. Nutrients 2021, 13, 1525. [Google Scholar] [CrossRef]
- Verduci, E.; Zuccotti, G.V.; Peroni, D.G. New Insights in Cow’s Milk and Allergy: Is the Gut Microbiota the Missing Link? MDPI: 2022; Vol. 14, p 1631.
- Koletzko, S.; Niggemann, B.; Arató, A.; Dias, J.; Heuschkel, R.; Husby, S.; Mearin, M.; Papadopoulou, A.; Ruemmele, F.; Staiano, A. Diagnostic approach and management of cow's-milk protein allergy in infants and children: ESPGHAN GI Committee practical guidelines. Journal of pediatric gastroenterology and nutrition 2012, 55, 221–229. [Google Scholar] [CrossRef]
- D’Auria, E.; Salvatore, S.; Pozzi, E.; Mantegazza, C.; Sartorio, M.U.A.; Pensabene, L.; Baldassarre, M.E.; Agosti, M.; Vandenplas, Y.; Zuccotti, G. Cow’s milk allergy: immunomodulation by dietary intervention. Nutrients 2019, 11, 1399. [Google Scholar] [CrossRef]
- Forsgård, R.A. Lactose digestion in humans: intestinal lactase appears to be constitutive whereas the colonic microbiome is adaptable. The American journal of clinical nutrition 2019, 110, 273–279. [Google Scholar] [CrossRef]
- Toca, M.; Fernández, A.; Orsi, M.; Tabacco, O.; Vinderola, G. Intolerancia a la lactosa: mitos y verdades. Actualización. Archivos argentinos de pediatría 2022, 120, 101–110. [Google Scholar]
- Facioni, M.S.; Raspini, B.; Pivari, F.; Dogliotti, E.; Cena, H. Nutritional management of lactose intolerance: the importance of diet and food labelling. Journal of translational medicine 2020, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Singh, V.; Friesen, C.A. Colonic mucosal inflammatory cells in children and adolescents with lactase deficiency. Pathology-Research and Practice 2020, 216, 152971. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Gyawali, R.; Awaisheh, S.S.; Ayivi, R.D.; Silva, R.C.; Subedi, K.; Aljaloud, S.O.; Siddiqui, S.A.; Krastanov, A. Fermented foods and probiotics: An approach to lactose intolerance. Journal of Dairy Research 2021, 88, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Vitellio, P.; Celano, G.; Bonfrate, L.; Gobbetti, M.; Portincasa, P.; De Angelis, M. Effects of Bifidobacterium longum and Lactobacillus rhamnosus on gut microbiota in patients with lactose intolerance and persisting functional gastrointestinal symptoms: A randomised, double-blind, cross-over study. Nutrients 2019, 11, 886. [Google Scholar] [CrossRef]
- Cancarevic, I.; Rehman, M.; Iskander, B.; Lalani, S.; Malik, B.H. Is there a correlation between irritable bowel syndrome and lactose intolerance? Cureus 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Alkalay, M.J. Nutrition in patients with lactose malabsorption, celiac disease, and related disorders. Nutrients 2021, 14, 2. [Google Scholar] [CrossRef]
- Jansson-Knodell, C.L.; White, M.; Lockett, C.; Xu, H.; Shin, A. Associations of food intolerance with irritable bowel syndrome, psychological symptoms, and quality of life. Clinical Gastroenterology and Hepatology 2022, 20, 2121–2131. [Google Scholar] [CrossRef]
- Usai-Satta, P.; Lai, M.; Oppia, F. Lactose malabsorption and presumed related disorders: A review of current evidence. Nutrients 2022, 14, 584. [Google Scholar] [CrossRef]
- Efremova, I.; Maslennikov, R.; Poluektova, E.; Vasilieva, E.; Zharikov, Y.; Suslov, A.; Letyagina, Y.; Kozlov, E.; Levshina, A.; Ivashkin, V. Epidemiology of small intestinal bacterial overgrowth. World Journal of Gastroenterology 2023, 29, 3400. [Google Scholar] [CrossRef] [PubMed]
- Hodges, J.K.; Cao, S.; Cladis, D.P.; Weaver, C.M. Lactose intolerance and bone health: the challenge of ensuring adequate calcium intake. Nutrients 2019, 11, 718. [Google Scholar] [CrossRef] [PubMed]
- Treister-Goltzman, Y.; Peleg, R. Primary lactase deficiency and bone mineral density in postmenopausal women. Osteoporosis International 2019, 30, 527–527. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, A.E.; Rychter, A.M.; Zawada, A.; Dobrowolska, A.; Krela-Kaźmierczak, I. Lactose intolerance in patients with inflammatory bowel diseases and dietary management in prevention of osteoporosis. Nutrition 2021, 82, 111043. [Google Scholar] [CrossRef] [PubMed]
- Zingone, F.; Bertin, L.; Maniero, D.; Palo, M.; Lorenzon, G.; Barberio, B.; Ciacci, C.; Savarino, E.V. Myths and Facts about Food Intolerance: A Narrative Review. Nutrients 2023, 15, 4969. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, A.; Ishayek, N. Lactose intolerance, dairy avoidance, and treatment options. Nutrients 2018, 10, 1994. [Google Scholar] [CrossRef]
- Malik, T.F.; Panuganti, K.K. Lactose intolerance. In StatPearls [Internet], StatPearls Publishing: 2023.
- Anguita-Ruiz, A.; Aguilera, C.M.; Gil, Á. Genetics of lactose intolerance: an updated review and online interactive world maps of phenotype and genotype frequencies. Nutrients 2020, 12, 2689. [Google Scholar] [CrossRef]
- Wanes, D.; Husein, D.M.; Naim, H.Y. Congenital lactase deficiency: mutations, functional and biochemical implications, and future perspectives. Nutrients 2019, 11, 461. [Google Scholar] [CrossRef]
- De Luca, P.; Iaconis, D.; Biffali, E.; Enza, C.; de Magistris, L.; Riegler, G.; Pappalardo, D.; Amato, M.R.; Iardino, P.; Montanino, C. Development of a novel SNP assay to detect lactase persistence associated genetic variants. Molecular Biology Reports 2021, 48, 7087–7093. [Google Scholar] [CrossRef]
- Marten, L.M.; Wanes, D.; Stellbrinck, T.; Santer, R.; Naim, H.Y. Hypomorphic variants of lactase-phlorizin hydrolase in congenital lactase deficiency are trafficking incompetent and functionally inactive. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease 2022, 1868, 166338. [Google Scholar] [CrossRef]
- Hoang, T.T.; Lei, Y.; Mitchell, L.E.; Sharma, S.V.; Swartz, M.D.; Waller, D.K.; Finnell, R.H.; Benjamin, R.H.; Browne, M.L.; Canfield, M.A. Maternal lactase polymorphism (rs4988235) is associated with neural tube defects in offspring in the National Birth Defects Prevention Study. The Journal of Nutrition 2019, 149, 295–303. [Google Scholar] [CrossRef]
- Kowalówka, M.; Kosewski, G.; Lipiński, D.; Przysławski, J. A Comprehensive Look at the-13910 C> T LCT Gene Polymorphism as a Molecular Marker for Vitamin D and Calcium Levels in Young Adults in Central and Eastern Europe: A Preliminary Study. International Journal of Molecular Sciences 2023, 24, 10191. [Google Scholar] [CrossRef]
- Domżał-Magrowska, D.; Kowalski, M.K.; Małecka-Wojciesko, E. The incidence of adult type hypolactasia in patients with irritable bowel syndrome. Gastroenterology Review/Przegląd Gastroenterologiczny 2023, 18, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Comerford, K.B.; Pasin, G. Gene–dairy food interactions and health outcomes: a review of nutrigenetic studies. Nutrients 2017, 9, 710. [Google Scholar] [CrossRef] [PubMed]
- Aruoma, O.I.; Hausman-Cohen, S.; Pizano, J.; Schmidt, M.A.; Minich, D.M.; Joffe, Y.; Brandhorst, S.; Evans, S.J.; Brady, D.M. Personalized nutrition: translating the science of nutrigenomics into practice: proceedings from the 2018 American College of Nutrition Meeting. Journal of the American College of Nutrition 2019, 38, 287–301. [Google Scholar] [CrossRef]
- Ferguson, L.R.; De Caterina, R.; Görman, U.; Allayee, H.; Kohlmeier, M.; Prasad, C.; Choi, M.S.; Curi, R.; De Luis, D.A.; Gil, Á. Guide and position of the international society of nutrigenetics/nutrigenomics on personalised nutrition: part 1-fields of precision nutrition. Journal of nutrigenetics and nutrigenomics 2016, 9, 12–27. [Google Scholar] [CrossRef]
- Robles, L.; Priefer, R. Lactose intolerance: What your breath can tell you. Diagnostics 2020, 10, 412. [Google Scholar] [CrossRef]
- Tomczonek-Moruś, J.; Wojtasik, A.; Zeman, K.; Smolarz, B.; Bąk-Romaniszyn, L. 13910C> T and 22018G> A LCT gene polymorphisms in diagnosing hypolactasia in children. United European gastroenterology journal 2019, 7, 210–216. [Google Scholar] [CrossRef]
- Mattar, R.; de Campos Mazo, D.F.; Carrilho, F.J. Lactose intolerance: diagnosis, genetic, and clinical factors. Clinical and experimental gastroenterology 2012, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Floris, M.; Cano, A.; Porru, L.; Addis, R.; Cambedda, A.; Idda, M.L.; Steri, M.; Ventura, C.; Maioli, M. Direct-to-consumer nutrigenetics testing: an overview. Nutrients 2020, 12, 566. [Google Scholar] [CrossRef] [PubMed]
- Guasch-Ferré, M.; Dashti, H.S.; Merino, J. Nutritional genomics and direct-to-consumer genetic testing: an overview. Advances in nutrition 2018, 9, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Marietta, C.; McGuire, A.L. Currents in contemporary ethics Direct-to-consumer genetic testing: is it the practice of medicine? The Journal of Law, Medicine & Ethics 2009, 37, 369–374. [Google Scholar]
- De, S.; Pietilä, A.-M.; Iso-Touru, T.; Hopia, A.; Tahvonen, R.; Vähäkangas, K. Information provided to consumers about direct-to-consumer nutrigenetic testing. Public Health Genomics 2020, 22, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Misselwitz, B.; Pohl, D.; Frühauf, H.; Fried, M.; Vavricka, S.R.; Fox, M. Lactose malabsorption and intolerance: pathogenesis, diagnosis and treatment. United European gastroenterology journal 2013, 1, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chu, H.; Cong, Y.; Deng, Y.; Long, Y.; Zhu, Y.; Pohl, D.; Fried, M.; Dai, N.; Fox, M. Self-reported lactose intolerance in clinic patients with functional gastrointestinal symptoms: prevalence, risk factors, and impact on food choices. Neurogastroenterology & Motility 2015, 27, 1138–1146. [Google Scholar]
- Tandy-Connor, S.; Guiltinan, J.; Krempely, K.; LaDuca, H.; Reineke, P.; Gutierrez, S.; Gray, P.; Davis, B.T. False-positive results released by direct-to-consumer genetic tests highlight the importance of clinical confirmation testing for appropriate patient care. Genetics in Medicine 2018, 20, 1515–1521. [Google Scholar] [CrossRef] [PubMed]
- Kuchay, R.A.H. New insights into the molecular basis of lactase non-persistence/persistence: A brief review. Drug Discoveries & Therapeutics 2020, 14, 1–7. [Google Scholar]
- Labrie, V.; Buske, O.J.; Oh, E.; Jeremian, R.; Ptak, C.; Gasiūnas, G.; Maleckas, A.; Petereit, R.; Žvirbliene, A.; Adamonis, K. Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging. Nature Structural & Molecular Biology 2016, 23, 566–573. [Google Scholar]
- Leseva, M.N.; Grand, R.J.; Klett, H.; Boerries, M.; Busch, H.; Binder, A.M.; Michels, K.B. Differences in DNA methylation and functional expression in lactase persistent and non-persistent individuals. Scientific reports 2018, 8, 5649. [Google Scholar] [CrossRef]
- Fukushima, A.; Goda, T.; Motohashi, Y.; Sakuma, K. The specific expression patterns of lactase, sucrase and calbindin-D9k in weaning rats are regulated at the transcriptional level. Journal of nutritional science and vitaminology 2004, 50, 265–271. [Google Scholar] [CrossRef]
- Motohashi, Y.; Fukushima, A.; Kondo, T.; Sakuma, K. Lactase decline in weaning rats is regulated at the transcriptional level and not caused by termination of milk ingestion. The Journal of nutrition 1997, 127, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Maiuri, L.; Fusco, M.I.; Salvati, V.M.; Fuccio, A.; Auricchio, S.; Mantei, N.; Zecca, L.; Gloor, S.M.; Semenza, G. Lactase persistence versus decline in human adults: multifactorial events are involved in down-regulation after weaning. Gastroenterology 1997, 112, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-J.; Wu, X. Foodomics in microbiological investigations. Current Opinion in Food Science 2015, 4, 51–55. [Google Scholar] [CrossRef]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F. Nutrimetabolomics: an integrative action for metabolomic analyses in human nutritional studies. Molecular nutrition & food research 2019, 63, 1800384. [Google Scholar]
- Pimentel, G.; Burton, K.J.; Rosikiewicz, M.; Freiburghaus, C.; von Ah, U.; Münger, L.H.; Pralong, F.P.; Vionnet, N.; Greub, G.; Badertscher, R. Blood lactose after dairy product intake in healthy men. British Journal of Nutrition 2017, 118, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Vionnet, N.; Münger, L.H.; Freiburghaus, C.; Burton, K.J.; Pimentel, G.; Pralong, F.P.; Badertscher, R.; Vergères, G. Assessment of lactase activity in humans by measurement of galactitol and galactonate in serum and urine after milk intake. The American journal of clinical nutrition 2019, 109, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Bush, C.L.; Blumberg, J.B.; El-Sohemy, A.; Minich, D.M.; Ordovás, J.M.; Reed, D.G.; Behm, V.A.Y. Toward the definition of personalized nutrition: a proposal by the American Nutrition Association. Journal of the American College of Nutrition 2020, 39, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Toro-Martín, D.; Arsenault, B.J.; Després, J.-P.; Vohl, M.-C. Precision nutrition: a review of personalized nutritional approaches for the prevention and management of metabolic syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Sales, N.M.R.; Pelegrini, P.B.; Goersch, M. Nutrigenomics: definitions and advances of this new science. Journal of nutrition and metabolism 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Ordovas, J.M.; Ferguson, L.R.; Tai, E.S.; Mathers, J.C. Personalised nutrition and health. Bmj 2018, 361. [Google Scholar] [CrossRef]
- Authority, E.F.S. Dietary reference values for nutrients summary report; 2397-8325; Wiley Online Library: 2017.
- Dekker, P.J.; Koenders, D.; Bruins, M.J. Lactose-free dairy products: Market developments, production, nutrition and health benefits. Nutrients 2019, 11, 551. [Google Scholar] [CrossRef]
- Szilagyi, A.; Shrier, I.; Heilpern, D.; Je, J.S.; Park, S.; Chong, G.; Lalonde, C.; Cote, L.-F.; Lee, B. Differential impact of lactose/lactase phenotype on colonic microflora. Canadian Journal of Gastroenterology and Hepatology 2010, 24, 373–379. [Google Scholar] [CrossRef]
- Mutch, D.M.; Wahli, W.; Williamson, G. Nutrigenomics and nutrigenetics: the emerging faces of nutrition. The FASEB journal 2005, 19, 1602–1616. [Google Scholar] [CrossRef] [PubMed]
- Fassio, F.; Facioni, M.S.; Guagnini, F. Lactose maldigestion, malabsorption, and intolerance: a comprehensive review with a focus on current management and future perspectives. Nutrients 2018, 10, 1599. [Google Scholar] [CrossRef] [PubMed]
- RÖTTGER-WIRTZ, S.; Alie, D. Personalised Nutrition: The EU’s Fragmented Legal Landscape and the Overlooked Implications of EU Food Law. European Journal of Risk Regulation 2021, 12, 212–235. [Google Scholar] [CrossRef]
- Nordström, K.; Goossens, J. Personalized nutrition and social justice: Ethical considerations within four future scenarios applying the perspective of Nussbaum’s capabilities approach. Journal of Agricultural and Environmental Ethics 2016, 29, 5–22. [Google Scholar] [CrossRef]
- Casellas, F.; Aparici, A.; Pérez, M.; Rodríguez, P. Perception of lactose intolerance impairs health-related quality of life. European journal of clinical nutrition 2016, 70, 1068–1072. [Google Scholar] [CrossRef]
- Enko, D.; Meinitzer, A.; Brandmayr, W.; Halwachs-Baumann, G.; Schnedl, W.J.; Kriegshäuser, G. Association between increased plasma levels of homocysteine and depression observed in individuals with primary lactose malabsorption. PLoS One 2018, 13, e0202567. [Google Scholar] [CrossRef]
| TEST | APPLICATION | TEST PRINCIPLE |
| Hydrogen breath test (HBT) |
Test of choice for the diagnosis of LM/LI |
Detection of increase H2 in expiratory air after lactose intake |
| Lactose tolerance test |
LM epidemiology, low sensitivity and specificity |
Increase in blood sugar after lactose intake |
| Lactase Activity of Jejunum |
If gastroscopy is carried out for other investigations, invasive and expensive |
Evaluation of lactase enzymatic activity in duodenal biopsy sample |
| Genetic Test | LD/LNP | Test for detection of −13910:C>T polymorphism |
| Serum gaxilose or urine galactose test | Used for the diagnosis of small intestine diseases (intestinal malabsorption) |
Evaluation of D-xylose in plasma or galactose in urine after cleavage by lactase of 4-galactosylxylose oral administration |
| SNP | VARIANT | GEOGRPHIC REGION |
|---|---|---|
| rs4988235 | −13910:C>T | Europe |
| rs869051967 | −14009:T>G | Middle East |
| rs4988236 | −13908:G>A | Far East |
| rs773131166 | −13914:C>T | East Europe |
| rs41380347 | −13915:A>C | Middle East |
| rs41525747 | −13907:G>C | Middle East |
| rs820486563 | −14009:T>G | Ethiopia and Sudan |
| rs182549 | –22018:G>A | Europe |
| rs41456145 | −13913:A>G | Africa |
| rs145946881 | −14010:C>G | Kenya, Tanzania and South Africa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





