Submitted:
28 November 2023
Posted:
01 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. TDP1 knockout samples preparation
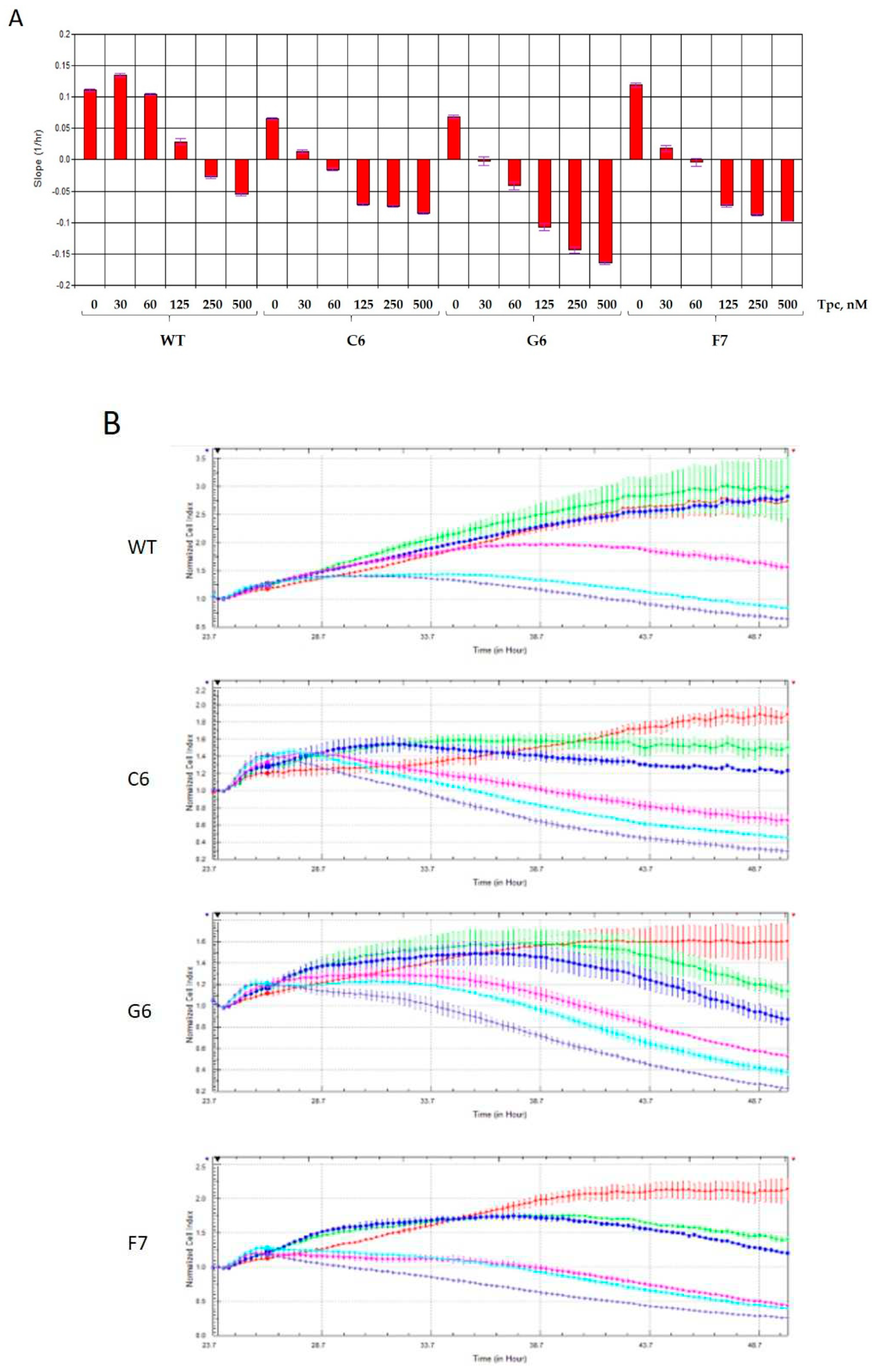
2.2. Real-time detection of cell sensitivity to topotecan for TDP1 knockout samples
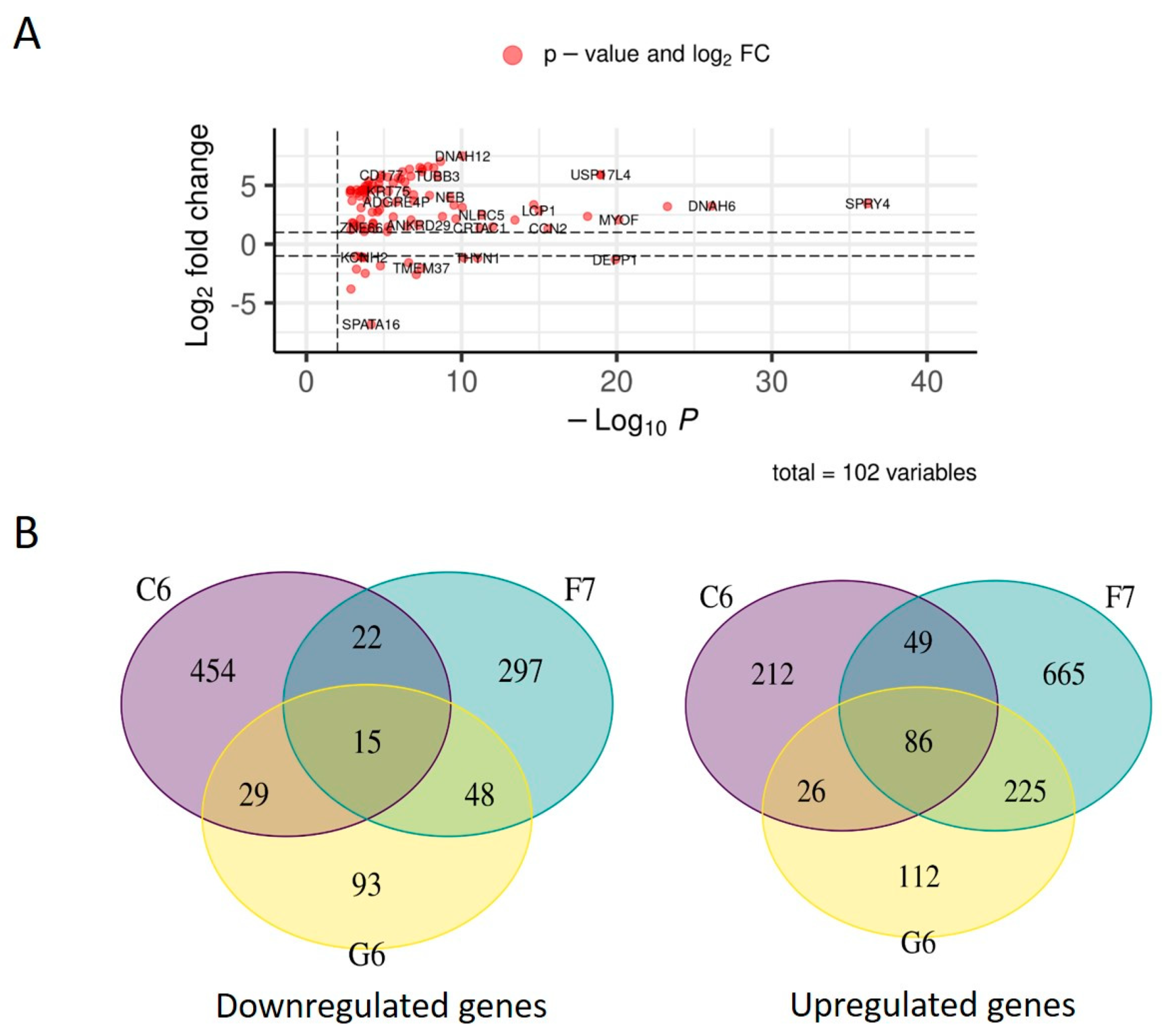
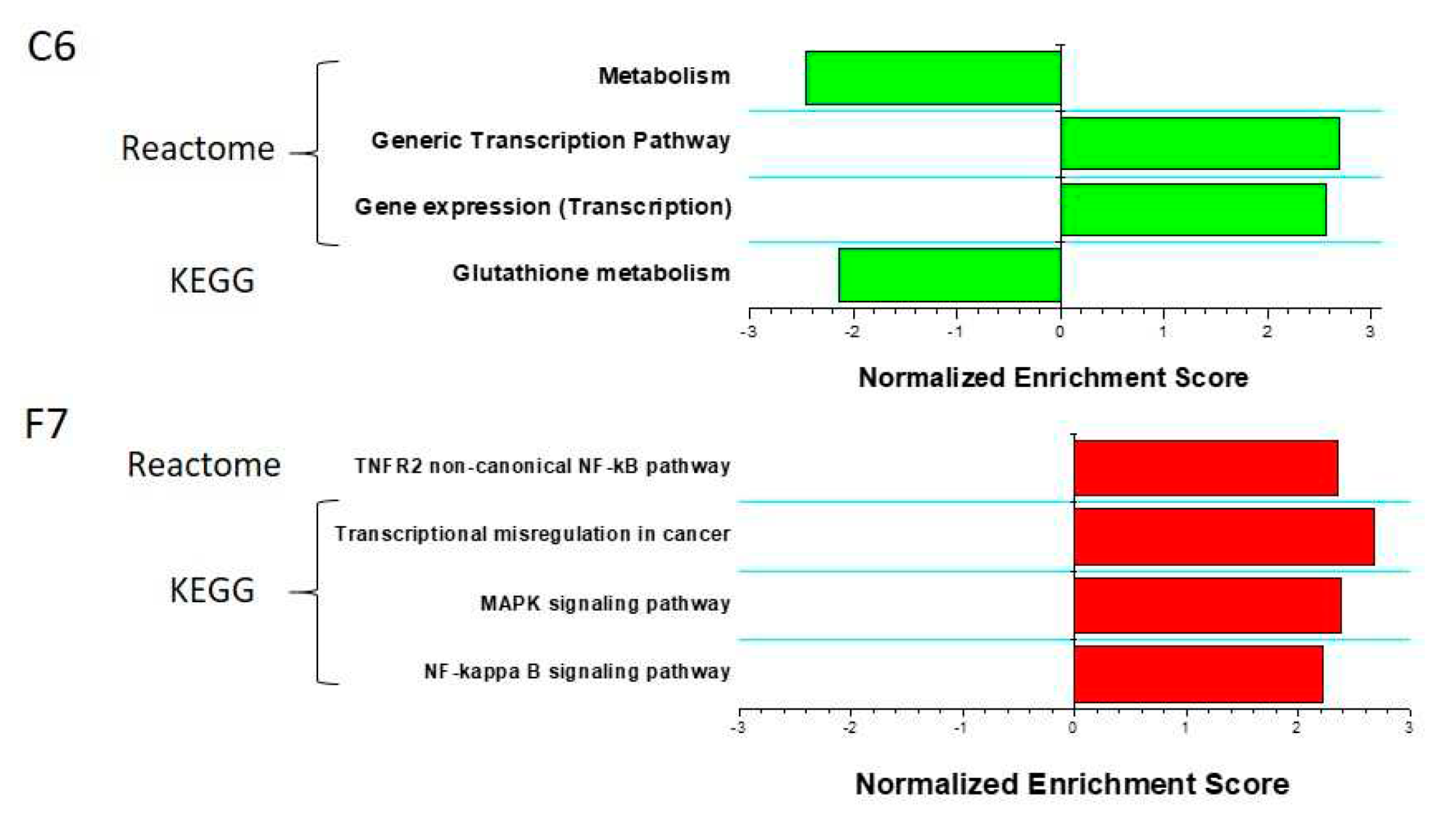
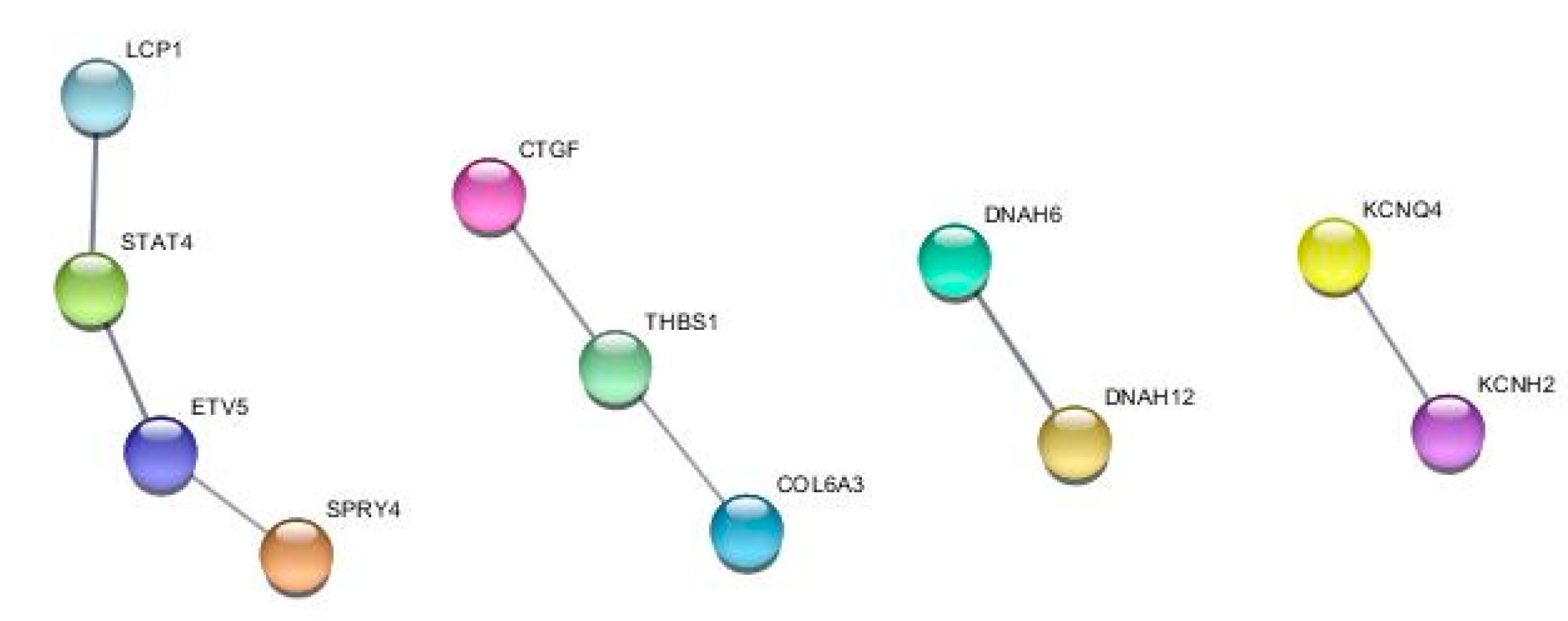
2.3. TDP1 knockout effect on transcriptome
3. Discussion
4. Materials and Methods
4.1. Tdp1 Knockout HEK293A cells generation
4.2. Cell Culture Cytotoxicity Assay
4.3. Total RNA Preparation and Transcriptome Sequencing
4.4. Bioinformatic Analysis
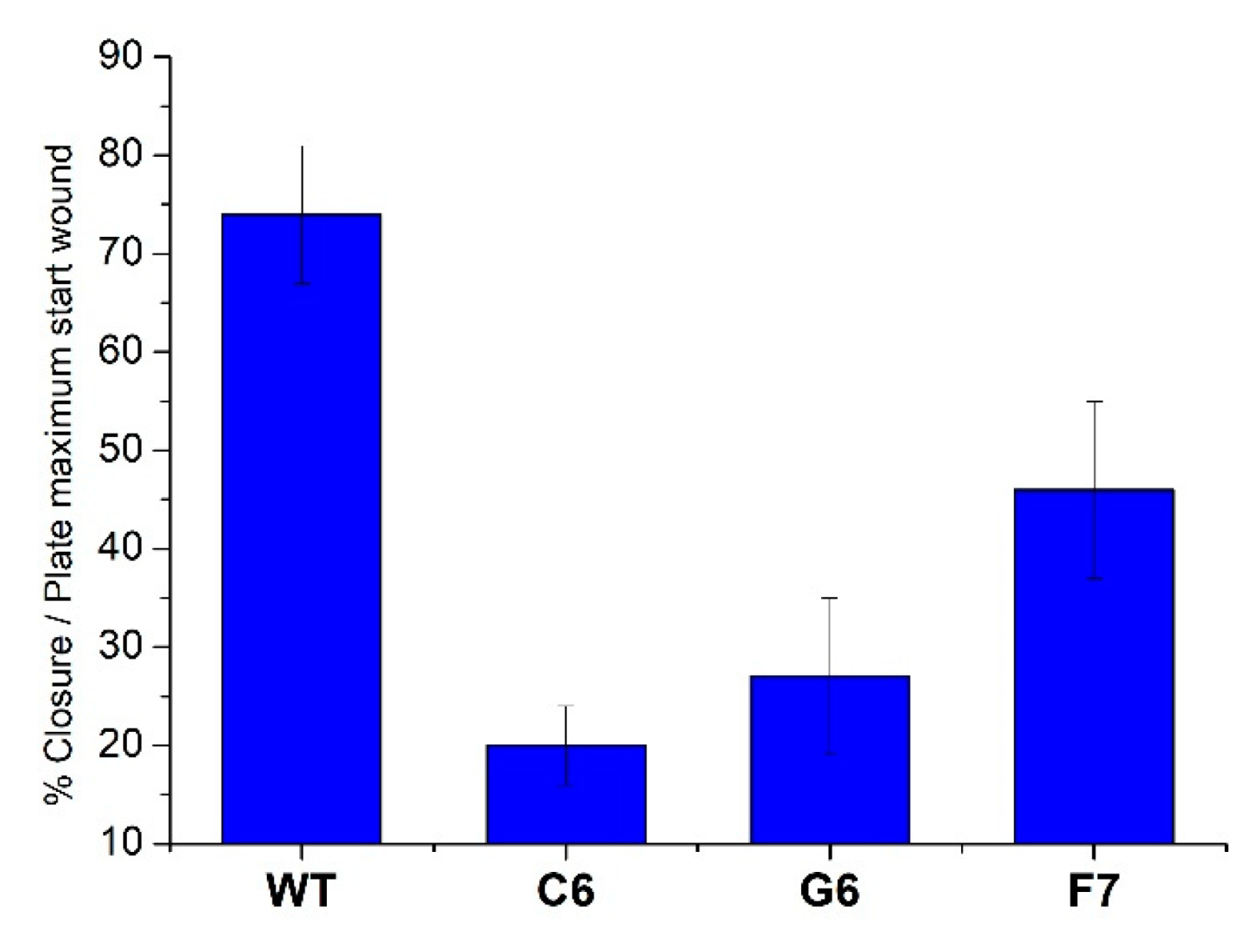
4.5. Wound healing assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Davies, D.R. , Interthal H. , Champoux J.J., Hol W.G. The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1. Structure. 2002, 10, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W. , Burgin Jr. A.B., Huizenga B.N., Robertson C.A., Yao K.C., Nash H.A. A eukaryotic enzyme that can disjoin dead-end covalentcomplexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA. 1996, 93, 11534–11539. [Google Scholar] [CrossRef] [PubMed]
- Zakharenko, A.L. , Luzina O.A., Chepanova A.A., Dyrkheeva N.S., Salakhutdinov N.F., Lavrik O.I. Natural Products and Their Derivatives as Inhibitors of the DNA Repair Enzyme Tyrosyl-DNA Phosphodiesterase 1. Int J Mol Sci. 2023, 24:5781. [CrossRef]
- Roca, J. The mechanisms of DNA topoisomerases. Trends Biochem Sci. 1995, 20, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Takashima, H. , Boerkoel C.F., John J., Saifi G.M., Salih M.A., Armstrong D., Mao Y., Quiocho F.A., Roa B.B., Nakagawa M., Stockton D.W., Lupski J.R. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002, 32, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Comeaux, E.Q. , van Waardenburg R.C. Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target. Drug Metab Rev. 2014, 46, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, N.A. , Rechkunova N.I., El-Khamisy S.F., Lavrik O.I. Tyrosyl-DNA phosphodiesterase 1 initiates repair of apurinic/apyrimidinic sites. Biochimie. 2012, 94, 1749–1753. [Google Scholar] [CrossRef] [PubMed]
- Dyrkheeva, N. , Anarbaev R., Lebedeva N., Kuprushkin M., Kuznetsova A., Kuznetsov N., Rechkunova N., Lavrik O. Human Tyrosyl-DNA phosphodiesterase 1 possesses transphosphooligonucleotidation activity with primary alcohols. Front Cell Dev Biol. 2020; 23, 604732. [Google Scholar] [CrossRef]
- Li, J. , Summerlin M., Nitiss K.C., Nitiss J.L., Hanakahi L.A. TDP1 is required for efficient non-homologous end joining in human cells. DNA Repair (Amst). 2017, 60, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.C. , Meagher M., Kassouf N., Hafezparast M., McKinnon P.J., Haywood R., El-Khamisy S.F. Mitochondrial protein-linked DNA breaks perturb mitochondrial gene transcription and trigger free radical-induced DNA damage. Sci Adv. 2017; 28, e1602506. [Google Scholar] [CrossRef]
- Chowdhuri, S.P. , Das B.B. Top1-PARP1 association and beyond: from DNA topology to break repair. NAR Cancer. 2021; 3, zcab003. [Google Scholar] [CrossRef]
- Das, B.B. , Antony S. , Gupta S., Dexheimer T.S., Redon C.E., Garfield S., Shiloh Y., Pommier Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009, 28, 3667–3680. [Google Scholar] [CrossRef] [PubMed]
- Chiang, S.C. , Carroll J., El-Khamisy S.F. TDP1 serine 81 promotes interaction with DNA ligase IIIalpha and facilitates cell survival following DNA damage. Cell Cycle. 2010, 9, 588–595. [Google Scholar] [CrossRef]
- Hudson, J.J. , Chiang S.C., Wells O.S., Rookyard C., El-Khamisy S.F. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat Commun. 2012; 13, 733. [Google Scholar] [CrossRef]
- Das, B. B, Ghosh A., Bhattacharjee S., Bhattacharyya A. Trapped topoisomerase-DNA covalent complexes in the mitochondria and their role in human diseases. Mitochondrion. 2021, 60, 234–244. [Google Scholar] [CrossRef]
- El-Khamisy, S.F. , Caldecott K.W. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis. 2006, 21, 219–224. [Google Scholar] [CrossRef]
- Zakharenko, A. , Dyrkheeva N., Lavrik O. Dual DNA topoisomerase 1 and tyrosyl-DNA phosphodiesterase 1 inhibition for improved anticancer activity. Med Res Rev. 2019, 39, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S. , Antony S., Marchand C., Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med Chem. 2008, 8, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Meisenberg, C. , Gilbert D.C., Chalmers A., Haley V., Gollins S., Ward S.E., El-Khamisy S.F. Clinical and cellular roles for TDP1 and TOP1 in modulating colorectal cancer response to irinotecan. Mol Cancer Ther. 2015, 14, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Liu, C. , Zhou S., Begum S., Sidransky D., Westra W.H., Brock M., Califano J.A. Increased expression and activity of repair genes TDP1 and XPF in non-small cell lung cancer. Lung Cancer. 2007, 55, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Fam, H.K. , Walton C., Mitra S.A., Chowdhury M., Osborne N., Choi K., Sun G., Wong P.C., O’Sullivan M.J., Turashvili G., Aparicio S., Triche T.J., Bond M., Pallen C.J., Boerkoel C.F. TDP1 and PARP1 deficiency are cytotoxic to rhabdomyosarcoma cells. Mol Cancer Res. 2013, 11, 1179–1192. [Google Scholar] [CrossRef] [PubMed]
- Gao, R. , Das B.B., Chatterjee R., Abaan O.D., Agama K., Matuo R., Vinson C., Meltzer P.S., Pommier Y. Epigenetic and genetic inactivation of tyrosyl-DNA-phosphodiesterase 1 (TDP1) in human lung cancer cells from the NCI-60 panel. DNA Repair (Amst). 2014, 13, 1–9. [Google Scholar] [CrossRef]
- Alagoz, M. , Gilbert D.C., El-Khamisy S., Chalmers A.J. DNA repair and resistance to topoisomerase I inhibitors: mechanisms, biomarkers and therapeutic targets. Curr Med Chem. 2012, 19, 3874–3885. [Google Scholar] [CrossRef] [PubMed]
- Brettrager, E.J. , van Waardenburg R. C.A.M. Targeting Tyrosyl-DNA phosphodiesterase I to enhance toxicity of phosphodiester linked DNA-adducts. Cancer Drug Resist. 2019, 2, 1153–1163. [Google Scholar] [CrossRef]
- Leung, E. , Patel J., Hollywood J.A., Zafar A., Tomek P., Barker D., Pilkington L.I., van Rensburg M., Langley R.J., Helsby N.A., Squire C.J., Baguley B.C., Denny W.A., Reynisson J., Leung I.K.H. Validating TDP1 as an Inhibition Target for the Development of Chemosensitizers for Camptothecin-Based Chemotherapy Drugs. Oncol Ther. 2021, 9, 541–556. [Google Scholar] [CrossRef]
- Dyrkheeva, N.S. , Filimonov A.S., Luzina O.A., Zakharenko A.L., Ilina E.S., Malakhova A.A., Medvedev S.P., Reynisson J., Volcho K.P., Zakian S.M., Salakhutdinov N.F., Lavrik O.I. New Hybrid Compounds Combining Fragments of Usnic Acid and Monoterpenoids for Effective Tyrosyl-DNA Phosphodiesterase 1 Inhibition. Biomolecules. 2021; 11, 973. [Google Scholar] [CrossRef]
- Xu, M. , Xiao J. , Chen J., Li J., Yin L., Zhu H., Zhou Z., Sha J. Identification and characterization of a novel human testis-specific Golgi protein, NYD-SP12. Mol Hum Reprod. 2003, 9, 9–17. [Google Scholar] [CrossRef]
- Griss, J. , Viteri G., Sidiropoulos K., Nguyen V., Fabregat A., Hermjakob H. ReactomeGSA—Efficient Multi-Omics Comparative Pathway Analysis. Mol Cell Proteomics. 2020, 19, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y. , Saha S., Wang W., Saha L.K., Huang S.N., Pommier Y. Excision repair of topoisomerase DNA-protein crosslinks (TOP-DPC). DNA Repair (Amst). 2020; 89, 102837. [Google Scholar] [CrossRef]
- Dexheimer, T.S. , Stephen A.G., Fivash M.J., Fisher R.J., Pommier Y. The DNA binding and 3’-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2010, 38, 2444–2452. [Google Scholar] [CrossRef]
- Interthal, H. , Pouliot J.J., Champoux J.J. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci USA. 2001, 98, 12009–12014. [Google Scholar] [CrossRef]
- Barthelmes, H.U. , Habermeyer M., Christensen M.O., Mielke C., Interthal H., Pouliot J.J., Boege F., Marko D. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J Biol Chem. 2004, 279, 55618–55625. [Google Scholar] [CrossRef]
- Hirano, R. , Interthal H. , Huang C., Nakamura T., Deguchi K., Choi K., Bhattacharjee M.B., Arimura K., Umehara F., Izumo S., Northrop J.L., Salih M.A., Inoue K., Armstrong D.L., Champoux J.J., Takashima H., Boerkoel C.F. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007, 26, 4732–4743. [Google Scholar] [CrossRef] [PubMed]
- Das, B.B. , Dexheimer T.S., Maddali K., Pommier Y.. Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc Natl Acad Sci USA. 2010, 107, 19790–19795. [Google Scholar] [CrossRef]
- Sjöstedt, E. , Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C., Oksvold P., Edfors F., Limiszewska A., Hikmet F., Huang J., Du Y., Lin L., Dong Z., Yang L., Liu X., Jiang H., Xu X., Wang J., Yang H., Bolund L, Mardinoglu A., Zhang C., von Feilitzen K., Lindskog C., Pontén F., Luo Y., Hökfelt T., Uhlén M., Mulder J. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020; 367, eaay5947. [Google Scholar] [CrossRef]
- Dyrkheeva, N.S. , Malakhova A.A., Zakharenko A.L., Okorokova L.S., Shtokalo D.N., Pavlova S.V., Medvedev S.P., Zakian S.M., Nushtaeva A.A., Tupikin A.E., Kabilov M.R., Khodyreva S.N., Luzina O.A., Salakhutdinov N.F., Lavrik O.I. Transcriptomic Analysis of CRISPR/Cas9-Mediated PARP1-Knockout Cells under the Influence of Topotecan and TDP1 Inhibitor. Int J Mol Sci. 2023; 24, 5148. [Google Scholar] [CrossRef]
- Zhang, Y.W. , Regairaz M., Seiler J.A., Agama K.K., Doroshow J.H., Pommier Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammalian cells. Nucleic Acids Res. 2011, 39, 3607–3620. [Google Scholar] [CrossRef]
- Das, B.B. , Huang S.Y., Murai J., Rehman I., Amé J.C., Sengupta S., Das S.K., Majumdar P., Zhang H., Biard D., Majumder H.K., Schreiber V., Pommier Y. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014, 42, 4435–4449. [Google Scholar] [CrossRef]
- Crewe, M. , Madabhushi R. Topoisomerase-Mediated DNA Damage in Neurological Disorders. Front Aging Neurosci. 2021; 13, 751742. [Google Scholar] [CrossRef]
- Szklarczyk, D. , Kirsch R., Koutrouli M., Nastou K., Mehryary F., Hachilif R., Gable A.L., Fang T., Doncheva N.T., Pyysalo S., Bork P., Jensen L.J., von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023; 51, D638–D646. [Google Scholar] [CrossRef]
- Moor, N.A. , Vasil’eva I.A., Anarbaev R.O., Antson A.A., Lavrik O.I. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015, 43, 6009–6022. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. , Huang S. N., Gao R., Das B.B., Murai J., Marchand C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst). 2014, 19, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S. , Eichler E.E., Fullerton S.M., Carrell D. SPANX gene variation in fertile and infertile males. Syst Biol Reprod Med. 2010, 55, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Leduc, F. , Maquennehan V., Nkoma G.B., Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod. 2008, 78, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, K.C. , Malik M., He X., White S.W., Nitiss J.L. Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc Natl Acad Sci USA. 2006, 103, 8953–8958. [Google Scholar] [CrossRef] [PubMed]
- Borda, M.A. , Palmitelli M., Veron G., González-Cid M., de Campos Nebel M. Tyrosyl-DNA-phosphodiesterase I (TDP1) participates in the removal and repair of stabilized-Top2α cleavage complexes in human cells. Mutat Res. 2015; 781, 37–48. [Google Scholar] [CrossRef]
- Shaman, J.A. , Prisztoka R., Ward W.S. Topoisomerase IIB and an extracellular nuclease interact to digest sperm DNA in an apoptotic-like manner. Biol Reprod. 2006, 75, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G. , Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 2016; 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Huang S.-Y., N. , Pommier Y. Mammalian Tyrosyl-DNA Phosphodiesterases in the Context of Mitochondrial DNA Repair. Int J Mol Sci. 2019; 20, 3015. [Google Scholar] [CrossRef]
- Sykora, P. , Wilson D.M. 3rd, Bohr V.A. Repair of persistent strand breaks in the mitochondrial genome. Mech Ageing Dev. 2012, 133, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Haitina, T. , Lindblom J., Renström T., Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006, 88, 779–790. [Google Scholar] [CrossRef] [PubMed]
- El-Khamisy, S.F. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol Med. 2011, 3, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Rass, U. , Ahel I., West S.C. Defective DNA Repair and Neurodegenerative Disease. Cell. 2007, 130, 991–1004. [Google Scholar] [CrossRef]
- Van Waardenburg, R.C.A.M. Tyrosyl-DNA Phosphodiesterase I a critical survival factor for neuronal development and homeostasis. J Neurol Neuromedicine. 2016, 1, 25–29. [Google Scholar] [CrossRef] [PubMed]
- El-Khamisy, S.F. , Caldecott K. W. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. DNA single-strand break repair and spinocerebellar ataxia with axonal neuropathy-1. Neuroscience. 2007, 145, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Katyal, S. , El-Khamisy S.F., Russel H.R., Li Y., Ju L., Caldecott K.W., McKinnon P.J. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007, 26, 4720–4731. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-T. , Lin W.-Y., Chou I.-C., Liu H.-P., Lee C.-C., Tsai Y., Wu W.-C., Tsai F.-J. Association of Tyrosyl-DNA Phosphodiesterase 1 Polymorphism with Tourette Syndrome in Taiwanese Patients. J Clin Lab Anal. 2013, 27, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Ljungman, M. , Lane D.P. Transcription—guarding the genome by sensing DNA damage. Nat Rev Cancer. 2004, 4, 727–737. [Google Scholar] [CrossRef]
- Brou, C. , Logeat F., Gupta N., Bessia C., LeBail O., Doedens J.R., Cumano A., Roux P., Black R.A., Israël A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000, 5, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Soong, T.W. , Stea A., Hodson C.D., Dubel S.J., Vincent S.R., Snutch T.P. Structure and functional expression of a member of the low voltage-activated calcium channel family. Science. 1993, 260, 1133–1136. [Google Scholar] [CrossRef]
- Caccamo, D.V. , Herman M.M., Frankfurter A., Katsetos C.D., Collins V.P., Rubinstein L.J. An immunohistochemical study of neuropeptides and neuronal cytoskeletal proteins in the neuroepithelial component of a spontaneous murine ovarian teratoma. Primitive neuroepithelium displays immunoreactivity for neuropeptides and neuron-associated beta-tubulin isotype. Am J Pathol. 1989, 135, 801–813. [Google Scholar] [PubMed]
- Gómez-Virgilio, L. , Silva-Lucero M.D., Flores-Morelos D.S., Gallardo-Nieto J., Lopez-Toledo G., Abarca-Fernandez A.M., Zacapala-Gómez A.E., Luna-Muñoz J., Montiel-Sosa F., Soto-Rojas L.O., Pacheco-Herrero M., Cardenas-Aguayo M.D. Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators. Cells. 2022; 11, 2262. [Google Scholar] [CrossRef]
- Salcher, S. , Hermann M., Kiechl-Kohlendorfer U., Ausserlechner M.J., Obexer P. C10ORF10/DEPP-mediated ROS accumulation is a critical modulator of FOXO3-induced autophagy. Mol Cancer. 2017; 16, 95. [Google Scholar] [CrossRef]
- Stepp, M.W. , Folz R.J., Yu J., Zelko I.N. The c10orf10 gene product is a new link between oxidative stress and autophagy. Biochim Biophys Acta. 2014, 1843, 1076–1088. [Google Scholar] [CrossRef]
- Jun, J.I. , Lau L.F. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011, 10, 945–963. [Google Scholar] [CrossRef]
- Hall-Glenn, F. , Lyons K.M. Roles for CCN2 in normal physiological processes. Cell Mol Life Sci. 2011, 68, 3209–3217. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S. , Roberts D. D. Why do humans need thrombospondin-1? J Cell Commun Signal. 2023, 17, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lamandé, S.R. , Mörgelin M., Adams N.E., Selan C., Allen J.M. The C5 domain of the collagen VI alpha3(VI) chain is critical for extracellular microfibril formation and is present in the extracellular matrix of cultured cells. The Journal of Biological Chemistry. 2006, 281, 16607–16614. [Google Scholar] [CrossRef] [PubMed]
- Warmke, J.W. , Ganetsky B.A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994, 91, 3438–3442. [Google Scholar] [CrossRef] [PubMed]
- Kubisch, C. , Schroeder B.C., Friedrich T., Lutjohann B., El-Amraoui A., Marlin S., Petit C., Jentsch T.J. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999, 96, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Hu, F. , Xu K., Zhou Y., Wu C., Wang S., Xiao J., Wen M., Zhao R., Luo K., Tao M., Duan W., Liu S. Different expression patterns of sperm motility-related genes in testis of diploid and tetraploid cyprinid fish. Biol Reprod. 2017, 96, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S. , Park T., Chen Z.P., Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J BiolChem. 1993, 268, 2781–2792. [Google Scholar] [CrossRef]
- Chen, C. , Cai Q. , He W., Lam T.B., Lin J., Zhao Y., Chen X., Gu P., Huang H., Xue M., Liu H. AP4 modulated by the PI3K/AKT pathway promotes prostate cancer proliferation and metastasis of prostate cancer via upregulating L-plastin. Cell Death Dis. 2017, 8, e3060. [Google Scholar] [CrossRef]
- Miller, M.H. , Walsh S.V., Atrih A., Huang J. T.-J., Ferguson M.A. J., Dillon J.F. Serum proteome of nonalcoholic fatty liver disease: a multimodal approach to discovery of biomarkers of nonalcoholic steatohepatitis. J Gastroenterol Hepatol. 2014, 29, 1839–1847. [Google Scholar] [CrossRef]
- O’Shea, J.J. , Lahesmaa L., Vahedi G., Laurence A., Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation: new technology brings new insights and new questions. Nat Rev Immunol. 2011, 11, 239–250. [Google Scholar] [CrossRef]
- Morrow, C.M. , Hostetler C.E., Griswold M.D., Hofmann M.C., Murphy K.M., Cooke P.S., Hess R.A. ETV5 is required for continuous spermatogenesis in adult mice and may mediate blood testes barrier function and testicular immune privilege. Ann NY Acad Sci. 2007, 1120, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. , Dai S., Qiu C., Wang T., Zhou Y., Xue C., Yao J., Xu Y. MicroRNA-219a-5p suppress-es intestinal inflammation through inhibiting Th1/Th17-mediated immune responses in inflammatory bowel disease. Mucosal Immunol. 2020, 13, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Wagle, M.C. , Kirouac D., Klijn C., Liu B., Mahajan S., Junttila M., Moffat J., Merchant M., Huw L., Wongchenko M., Okrah K., Srinivasan S., Mounir Z., Sumiyoshi T., Haverty P.M., Yauch R.L., Yan Y., Kabbarah O., Hampton G., Amler L., Ramanujan S., Lackner M.R., Huang S.A. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis Oncol. 2018; 2, 7. [Google Scholar] [CrossRef]
- Frandsen, H.S. , Vej-Nielsen J.M., Smith L.E., Sun L., Mikkelsen K.L., Thulesen A.P., Hagensen C.E., Yang F., Rogowska-Wrzesinska A. Mapping Proteome and Lipidome Changes in Early-Onset Non-Alcoholic Fatty Liver Disease Using Hepatic 3D Spheroids. Cells. 2022, 11, 3216. [Google Scholar] [CrossRef] [PubMed]
- Chesnokov, M.S. , Yadav A., Chefetz. I. Optimized transcriptional signature for evaluation of MEK/ERK pathway baseline activity and long-term modulations in ovarian cancer. Int. J. Mol. Sci. 2022, 23, 13365. [Google Scholar] [CrossRef] [PubMed]
- Martin-Vega, A. , Earnest S., Augustyn A., Wichaidit C., Gazdar A., Girard L., Peyton M., Kollipara R.K., Minna J.D., Johnson J.E., Cobb M.H. ASCL1-ERK1/2 Axis: ASCL1 restrains ERK1/2 via the dual specificity phosphatase DUSP6 to promote survival of a subset of neuroendocrine lung cancers. bioRxiv. 2023. [Google Scholar] [CrossRef]
- Dobin, A. , Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: a quality control tool for high throughput sequence data. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Wang, L. , Wang S. , Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics (Oxford, England), 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- “Picard Toolkit.” 2019. Broad Institute, GitHub Repository. https://broadinstitute.github.
- Ewels, P. , Magnusson M., Lundin S., Käller M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 3: 1;32(19), 2016; 32, 3047–3048. [Google Scholar] [CrossRef]
- Love, M.I. , Huber W. , Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014, 15, 550. [Google Scholar] [CrossRef]
- Blighe K., Rana S., Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. 2023. R package version 1.18.0. https://github.com/kevinblighe/EnhancedVolcano.
- Korotkevich, G. , Sukhov V., Sergushichev A. Fast gene set enrichment analysis. Fast gene set enrichment analysis. bioRxiv. 2019. [CrossRef]
| log2FoldChange | p-value | p-adj | ||||||||
| C6 | G6 | F7 | C6 | G6 | F7 | C6 | G6 | F7 | ||
| 1 | KCNH2 | -1.357 | -1.111 | -1.139 | 1.6e-05 | 3.2e-04 | 1.9e-04 | 2.7e-04 | 0.004 | 0.002 |
| 2 | SYNDIG1 | -1.733 | -1.12 | -1.572 | 4.5e-08 | 2.4e-04 | 2.6e-07 | 1.9e-06 | 0.003 | 6.1e-06 |
| 3 | SERPINF1 | -1.141 | -1.698 | -1.108 | 2.2e-04 | 5.1e-08 | 2.5e-04 | 0.002 | 3.2e-06 | 0.002 |
| 4 | SPATA16 | -6.098 | -5.687 | -6.829 | 3.7e-04 | 8.6e-04 | 6.7e-05 | 0.003 | 0.008 | 7.2e-04 |
| 5 | THYN1 | -2.172 | -1.045 | -1.188 | 3.8e-30 | 2.8e-09 | 9.2e-12 | 4.2e-27 | 2.6e-07 | 5.8e-10 |
| 6 | DEPP1 | -4.185 | -1.794 | -1.31 | 1.6e-17 | 7.0e-37 | 1.2e-20 | 3.0e-16 | 3.5e-33 | 3.0e-18 |
| 7 | STX8 | -1.104 | -1.247 | -1.157 | 1.0e-09 | 4.0e-12 | 7.2e-11 | 7.4e-08 | 8.4e-10 | 3.9e-09 |
| 8 | TMEM37 | -1.735 | -2.303 | -2.042 | 4.7e-06 | 1.8e-09 | 4.0e-08 | 1.0e-04 | 1.8e-07 | 1.2e-06 |
| 9 | EMID1 | -1.112 | -1.242 | -1.078 | 2.8e-04 | 4.3e-05 | 3.0e-04 | 0.003 | 8.4e-04 | 0.002 |
| log2FoldChange | p-value | p-adj | ||||||||
| C6 | G6 | F7 | C6 | G6 | F7 | C6 | G6 | F7 | ||
| 1 | TAC1 | 1.248 | 1.678 | 2.156 | 4.2e-04 | 1.2e-06 | 2.3e-10 | 0.004 | 4.7e-05 | 1.1e-08 |
| 2 | ETV1 | 4.205 | 2.873 | 3.36 | 3.2e-23 | 2.3e-11 | 2.2e-15 | 2.4e-20 | 4.0e-09 | 2.6e-13 |
| 3 | SLAMF7 | 2.056 | 2.3 | 2.145 | 6.6e-04 | 1.2e-04 | 3.2e-04 | 0.005 | 0.002 | 0.003 |
| 4 | ADAM28 | 5.679 | 5.821 | 6.159 | 5.3e-07 | 2.3e-07 | 3.6e-08 | 1.6e-05 | 1.2e-05 | 1.1e-06 |
| 5 | PMS2P4 | 1.786 | 1.726 | 1.585 | 1.6e-04 | 2.3e-04 | 6.6e-04 | 0.002 | 0.003 | 0.005 |
| 6 | LRP2 | 2.223 | 1.749 | 1.807 | 7.3e-07 | 1.0e-04 | 5.1e-05 | 2.1e-05 | 0.002 | 5.7e-04 |
| 7 | CRTAC1 | 1.092 | 1.346 | 1.373 | 1.1e-07 | 2.8e-11 | 6.7e-12 | 4.3e-06 | 4.6e-09 | 4.3e-10 |
| 8 | RSPO4 | 5.947 | 5.69 | 6.382 | 1.8e-06 | 4.8e-06 | 2.3e-07 | 4.6e-05 | 1.5e-04 | 5.5e-06 |
| 9 | CEMIP | 2.813 | 3.226 | 3.099 | 0.001 | 1.8e-04 | 3.0e-04 | 0.009 | 0.003 | 0.003 |
| 10 | ELAVL2 | 5.263 | 6.616 | 4.564 | 1.3e-05 | 2.9e-08 | 1.5e-04 | 2.3e-04 | 1.9e-06 | 0.001 |
| 11 | DNAH6 | 1.064 | 1.722 | 3.259 | 8.0e-04 | 2.7e-08 | 7.3e-27 | 0.006 | 1.8e-06 | 4.2e-24 |
| 12 | KCNQ4 | 1.119 | 1.294 | 1.524 | 3.0e-04 | 2.0e-05 | 3.1e-07 | 0.003 | 4.5e-04 | 7.2e-06 |
| 13 | CCN2 | 1.089 | 1.108 | 1.32 | 1.7e-11 | 7.2e-12 | 2.7e-16 | 2.0e-09 | 1.4e-09 | 3.7e-14 |
| 14 | CFAP58 | 3.094 | 2.781 | 2.92 | 7.3e-06 | 5.5e-05 | 2.0e-05 | 1.4e-04 | 0.001 | 2.6e-04 |
| 15 | LRRN4 | 4.372 | 4.71 | 4.934 | 1.0e-03 | 3.4e-04 | 1.6e-04 | 0.007 | 0.004 | 0.001 |
| 16 | HOXD1 | 3.674 | 4.914 | 5.783 | 0.001 | 1.1e-05 | 1.9e-07 | 0.009 | 2.8e-04 | 4.6e-06 |
| 17 | CHAC1 | 1.582 | 1.857 | 2.057 | 9.4e-05 | 3.2e-06 | 1.8e-07 | 0.001 | 1.1e-04 | 4.4e-06 |
| 18 | CYP27A1 | 4.16 | 4.852 | 5.142 | 9.4e-04 | 7.4e-05 | 2.2e-05 | 0.007 | 0.001 | 2.8e-04 |
| 19 | LCP1 | 1.504 | 2.367 | 2.837 | 6.4e-05 | 4.7e-11 | 1.0e-15 | 8.1e-04 | 7.1e-09 | 1.2e-13 |
| 20 | THBS1 | 1.198 | 1.392 | 1.331 | 6.9e-04 | 6.9e-05 | 1.3e-04 | 0.005 | 0.001 | 0.001 |
| 21 | MYOF | 1.54 | 1.841 | 2.07 | 4.6e-12 | 9.9e-17 | 7.4e-21 | 6.1e-10 | 5.1e-14 | 1.9e-18 |
| 22 | STAT4 | 3.788 | 2.958 | 4.147 | 2.8e-07 | 7.2e-05 | 1.2e-08 | 9.4e-06 | 0.001 | 3.9e-07 |
| 23 | CYP1A1 | 1.688 | 1.873 | 3.2 | 3.1e-07 | 8.2e-09 | 5.4e-24 | 1.0e-05 | 6.5e-07 | 2.3e-21 |
| 24 | NLRC5 | 1.184 | 1.837 | 2.477 | 0.001 | 3.9e-07 | 5.1e-12 | 0.009 | 1.8e-05 | 3.4e-10 |
| 25 | FLG | 4.694 | 5.152 | 6.603 | 8.1e-05 | 1.2e-05 | 1.4e-08 | 9.8e-04 | 3.1e-04 | 4.7e-07 |
| 26 | BEND6 | 2.52 | 1.596 | 1.223 | 2.5e-11 | 3.0e-05 | 0.001 | 2.8e-09 | 6.2e-04 | 0.008 |
| 27 | ANKRD29 | 2.078 | 1.018 | 1.613 | 3.6e-12 | 7.9e-04 | 5.9e-08 | 4.8e-10 | 0.008 | 1.7e-06 |
| 28 | ADGRG4 | 7.682 | 5.224 | 4.621 | 6.0e-10 | 3.5e-05 | 2.6e-04 | 4.7e-08 | 7.1e-04 | 0.002 |
| 29 | ZNF66 | 1.866 | 1.314 | 1.413 | 2.0e-06 | 8.7e-04 | 2.9e-04 | 5.1e-05 | 0.008 | 0.002 |
| 30 | PPP1R32 | 4.354 | 4.496 | 4.625 | 1.9e-04 | 1.1e-04 | 6.1e-05 | 0.002 | 0.002 | 6.6e-04 |
| 31 | SLC25A34 | 4.231 | 4.381 | 4.656 | 6.5e-04 | 3.7e-04 | 1.3e-04 | 0.005 | 0.004 | 0.001 |
| 32 | COL6A3 | 1.836 | 1.868 | 3.135 | 2.1e-04 | 1.5e-04 | 9.1e-11 | 0.002 | 0.002 | 4.9e-09 |
| 33 | DNAJC5G | 4.164 | 4.086 | 4.386 | 4.2e-04 | 5.2e-04 | 1.7e-04 | 0.004 | 0.006 | 0.002 |
| 34 | GBX1 | 4.975 | 5.374 | 4.563 | 3.9e-05 | 7.4e-06 | 1.5e-04 | 5.4e-04 | 2.1e-04 | 0.001 |
| 35 | NOXRED1 | 4.366 | 4.502 | 4.513 | 6.4e-04 | 3.9e-04 | 3.5e-04 | 0.005 | 0.005 | 0.003 |
| 36 | KRT75 | 3.589 | 4.766 | 4.49 | 3.7e-04 | 1.4e-06 | 5.2e-06 | 0.003 | 5.1e-05 | 8.4e-05 |
| 37 | NHLH1 | 4.121 | 4.537 | 4.684 | 2.5e-04 | 4.5e-05 | 2.3e-05 | 0.002 | 8.6e-04 | 3.0e-04 |
| 38 | ABCD2 | 4.626 | 4.55 | 4.623 | 6.3e-04 | 7.4e-04 | 5.7e-04 | 0.005 | 0.008 | 0.004 |
| 39 | DNAH12 | 4.865 | 6.219 | 7.498 | 4.7e-05 | 9.5e-08 | 8.6e-11 | 6.4e-04 | 5.3e-06 | 4.6e-09 |
| 40 | CCDC168 | 4.552 | 4.418 | 4.08 | 7.6e-05 | 1.2e-04 | 3.7e-04 | 9.4e-04 | 0.002 | 0.003 |
| 41 | KLHL11 | 1.164 | 1.35 | 1.29 | 2.6e-04 | 2.0e-05 | 4.3e-05 | 0.002 | 4.5e-04 | 5.0e-04 |
| 42 | NRIP1 | 1.391 | 1.022 | 1.071 | 4.8e-09 | 1.7e-05 | 6.1e-06 | 2.8e-07 | 4.0e-04 | 9.6e-05 |
| 43 | ANKRD30B | 4.693 | 5.804 | 5.366 | 8.2e-04 | 2.4e-05 | 9.5e-05 | 0.006 | 5.4e-04 | 9.7e-04 |
| 44 | IZUMO1 | 4.684 | 4.473 | 4.33 | 2.0e-04 | 3.8e-04 | 5.5e-04 | 0.002 | 0.005 | 0.004 |
| 45 | NEB | 3.136 | 2.901 | 3.996 | 1.8e-06 | 9.6e-06 | 5.5e-10 | 4.6e-05 | 2.5e-04 | 2.5e-08 |
| 46 | SMG1P5 | 1.843 | 2.081 | 1.824 | 0.001 | 2.1e-04 | 0.001 | 0.008 | 0.003 | 0.007 |
| 47 | CCR4 | 5.068 | 5.669 | 6.526 | 3.6e-05 | 2.7e-06 | 4.8e-08 | 5.1e-04 | 9.2e-05 | 1.4e-06 |
| 48 | TMEM232 | 5.137 | 5.778 | 6.399 | 1.5e-05 | 7.9e-07 | 3.5e-08 | 2.6e-04 | 3.3e-05 | 1.0e-06 |
| 49 | SPRY4 | 1.947 | 3.151 | 3.446 | 3.7e-12 | 8.5e-31 | 6.8e-37 | 4.9e-10 | 2.4e-27 | 6.9e-34 |
| 50 | SBSN | 3.75 | 4.183 | 5.544 | 0.001 | 2.5e-04 | 8.4e-07 | 0.008 | 0.003 | 1.7e-05 |
| 51 | UBE2Q2P1 | 1.488 | 1.806 | 1.73 | 6.2e-04 | 2.6e-05 | 5.1e-05 | 0.005 | 5.6e-04 | 5.7e-04 |
| 52 | CACNA1E | 5.203 | 5.149 | 5.616 | 9.0e-05 | 1.0e-04 | 1.8e-05 | 0.001 | 0.002 | 2.4e-04 |
| 53 | DISP3 | 2.574 | 2.956 | 3.504 | 0.001 | 2.1e-04 | 9.1e-06 | 0.009 | 0.003 | 1.4e-04 |
| 54 | CD177 | 4.994 | 5.479 | 5.877 | 3.0e-04 | 6.0e-05 | 1.5e-05 | 0.003 | 0.001 | 2.0e-04 |
| 55 | FRG2 | 3.419 | 5.366 | 5.295 | 0.002 | 3.3e-07 | 4.5e-07 | 0.01 | 1.6e-05 | 1.0e-05 |
| 56 | ZNF844 | 2.823 | 2.47 | 2.719 | 3.7e-05 | 3.1e-04 | 5.7e-05 | 5.3e-04 | 0.004 | 6.3e-04 |
| 57 | BAZ2B | 6.069 | 4.744 | 4.38 | 9.2e-06 | 6.1e-04 | 0.002 | 1.7e-04 | 0.007 | 0.009 |
| 58 | USP17L4 | 2.33 | 4.951 | 5.903 | 7.7e-04 | 3.8e-14 | 1.1e-19 | 0.006 | 1.2e-11 | 2.5e-17 |
| 59 | USP17L7 | 2.33 | 4.951 | 5.903 | 7.7e-04 | 3.8e-14 | 1.1e-19 | 0.006 | 1.2e-11 | 2.5e-17 |
| 60 | SPANXB1 | 3.724 | 4.146 | 5.179 | 9.9e-04 | 2.0e-04 | 2.5e-06 | 0.007 | 0.003 | 4.6e-05 |
| 61 | ACVR2B-AS1 | 1.743 | 2.332 | 1.776 | 0.001 | 1.3e-05 | 9.4e-04 | 0.009 | 3.3e-04 | 0.006 |
| 62 | IDI2-AS1 | 4.245 | 5.235 | 5.706 | 0.001 | 3.7e-05 | 5.5e-06 | 0.008 | 7.4e-04 | 8.8e-05 |
| 63 | KRTAP5-AS1 | 2.373 | 2.095 | 3.317 | 1.2e-05 | 1.1e-04 | 3.1e-10 | 2.1e-04 | 0.002 | 1.5e-08 |
| 64 | RNF217-AS1 | 5.109 | 5.21 | 4.564 | 3.7e-04 | 2.6e-04 | 0.001 | 0.003 | 0.003 | 0.009 |
| 65 | SMG1P1 | 2.219 | 2.956 | 2.748 | 8.5e-04 | 6.3e-06 | 2.6e-05 | 0.006 | 1.8e-04 | 3.3e-04 |
| 66 | ARHGAP31-AS1 | 4.286 | 5.36 | 5.638 | 3.3e-04 | 4.5e-06 | 1.2e-06 | 0.003 | 1.4e-04 | 2.4e-05 |
| 67 | ETV5 | 1.784 | 2.383 | 2.364 | 3.2e-11 | 4.7e-19 | 7.6e-19 | 3.4e-09 | 3.2e-16 | 1.5e-16 |
| 68 | TUBB3 | 3.8 | 4.068 | 5.783 | 1.6e-04 | 4.4e-05 | 3.7e-09 | 0.002 | 8.5e-04 | 1.4e-07 |
| 69 | LOC105372440 | 3.028 | 3.252 | 3.667 | 2.0e-05 | 3.6e-06 | 1.3e-07 | 3.2e-04 | 1.2e-04 | 3.2e-06 |
| 70 | ROCK1P1 | 2.491 | 1.522 | 2.356 | 3.2e-10 | 1.6e-04 | 1.7e-09 | 2.6e-08 | 0.002 | 7.1e-08 |
| 71 | LOC728485 | 1.26 | 1.44 | 1.484 | 2.5e-09 | 6.2e-12 | 1.0e-12 | 1.6e-07 | 1.2e-09 | 7.5e-11 |
| 72 | NAPA-AS1 | 3.621 | 4.591 | 3.81 | 3.3e-04 | 3.6e-06 | 1.3e-04 | 0.003 | 1.2e-04 | 0.001 |
| 73 | ADGRE4P | 2.481 | 3.189 | 3.587 | 0.001 | 2.4e-05 | 1.6e-06 | 0.009 | 5.4e-04 | 3.0e-05 |
| 74 | INTS4P2 | 4.105 | 4.004 | 3.689 | 3.2e-04 | 4.3e-04 | 0.001 | 0.003 | 0.005 | 0.007 |
| 75 | IQCA1L | 4.049 | 4.679 | 6.498 | 4.9e-04 | 4.0e-05 | 6.2e-09 | 0.004 | 7.8e-04 | 2.3e-07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





