Submitted:
11 October 2023
Posted:
12 October 2023
You are already at the latest version
Abstract
Keywords:
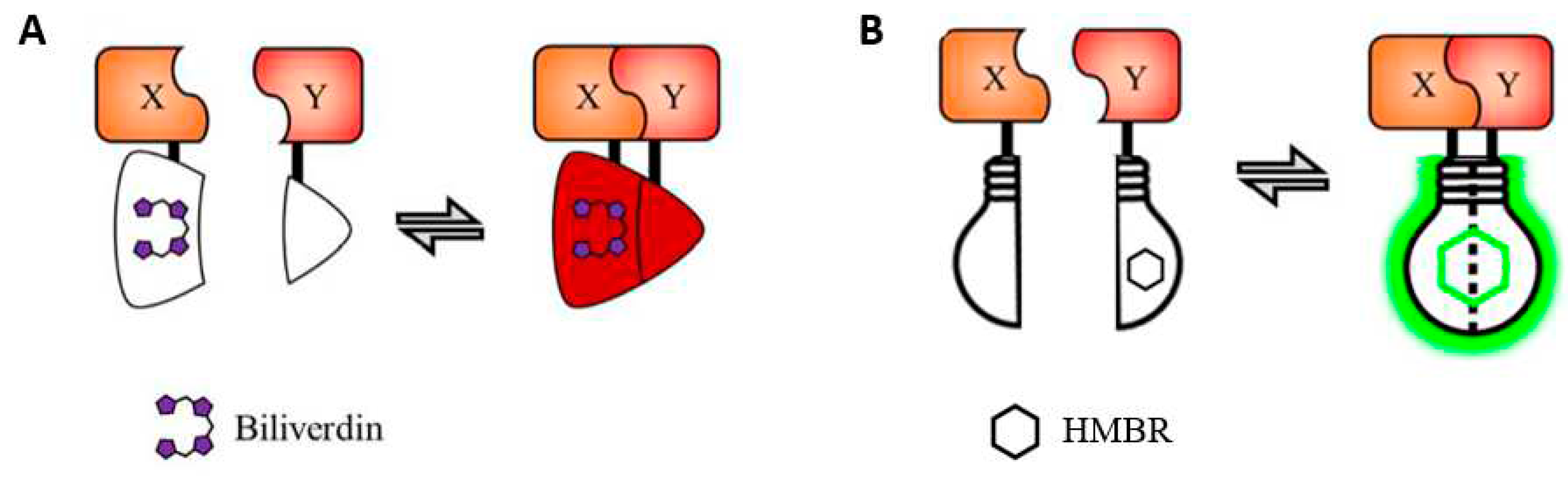
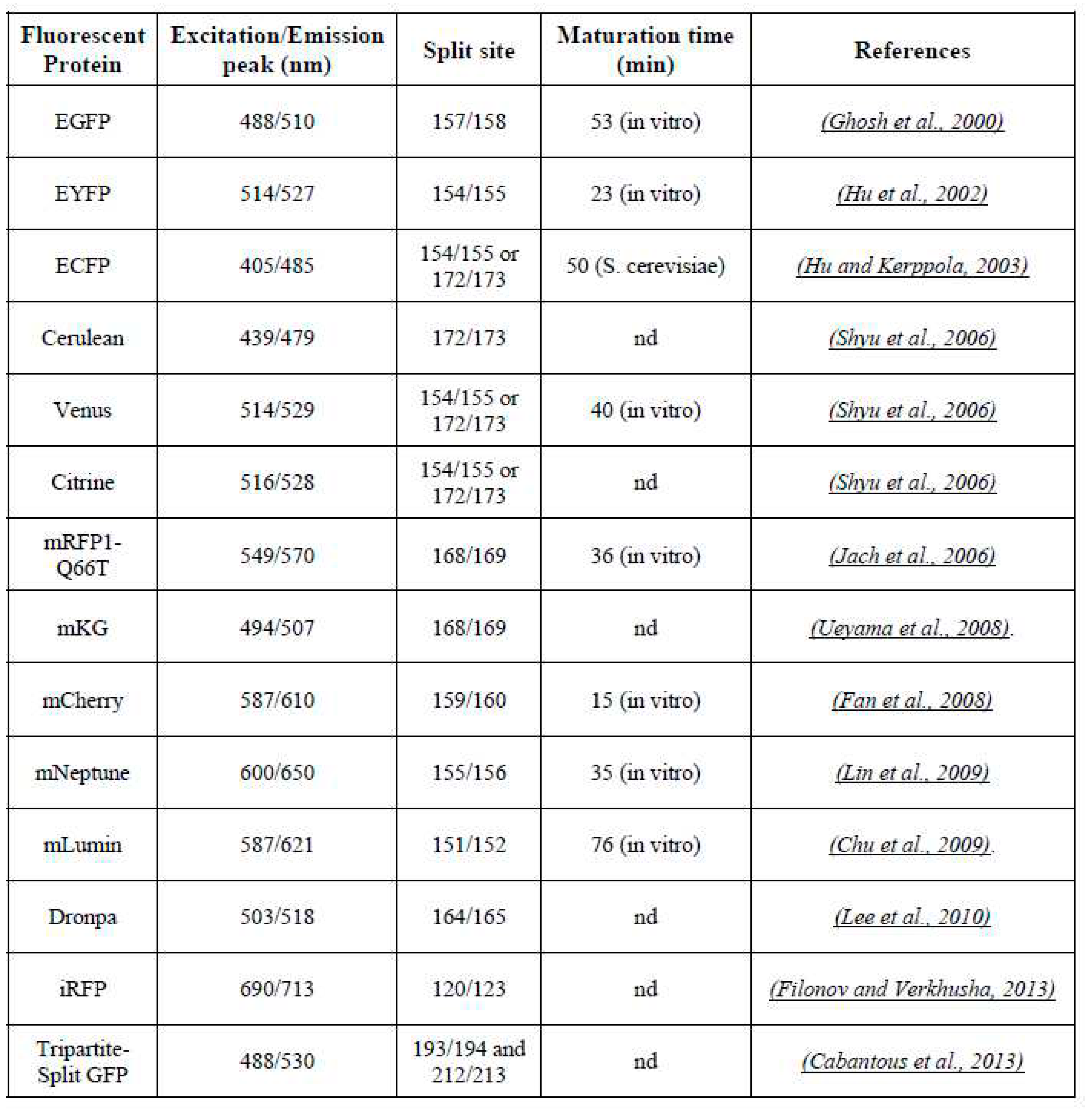
1. Fluorescent proteins for BiFC assay
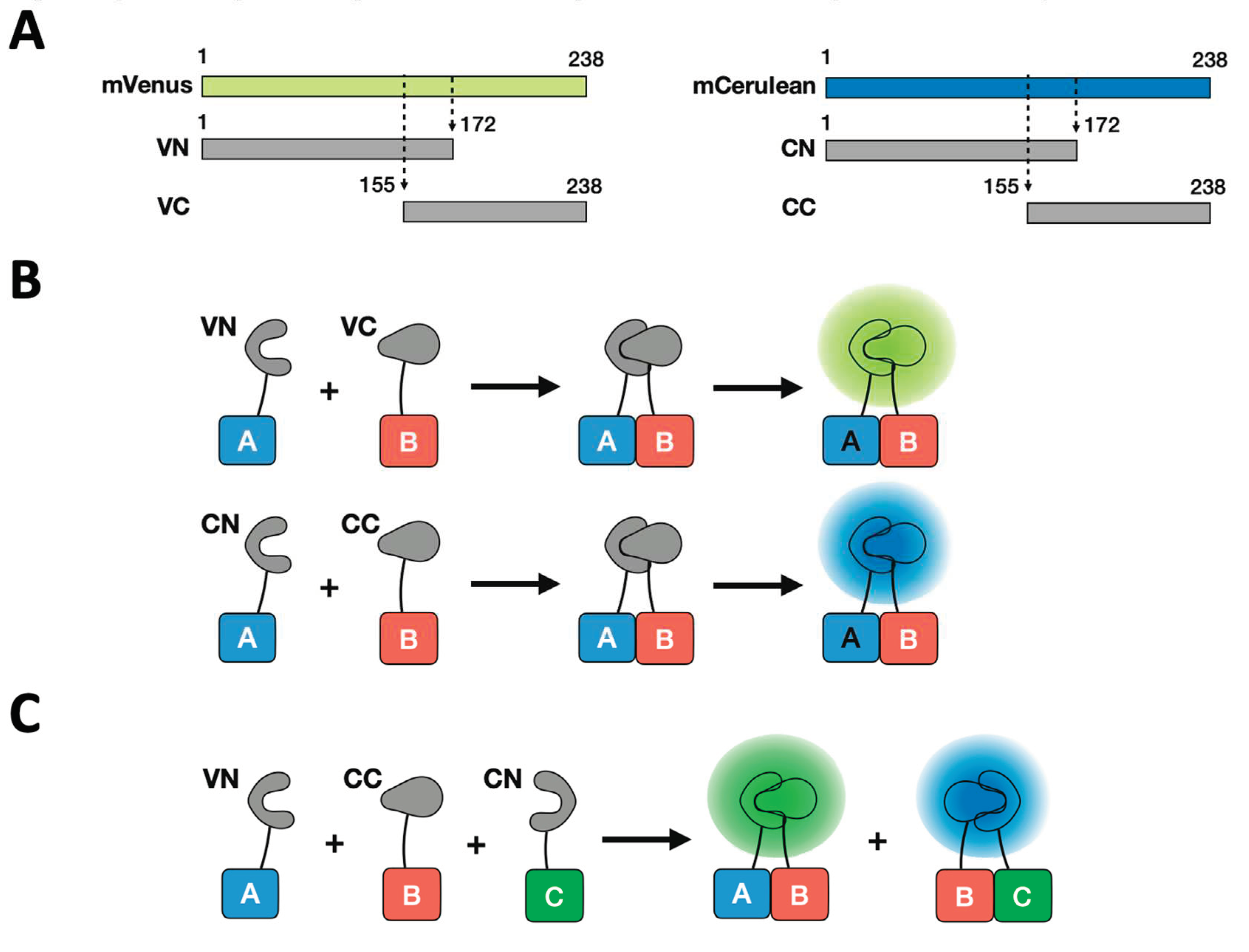
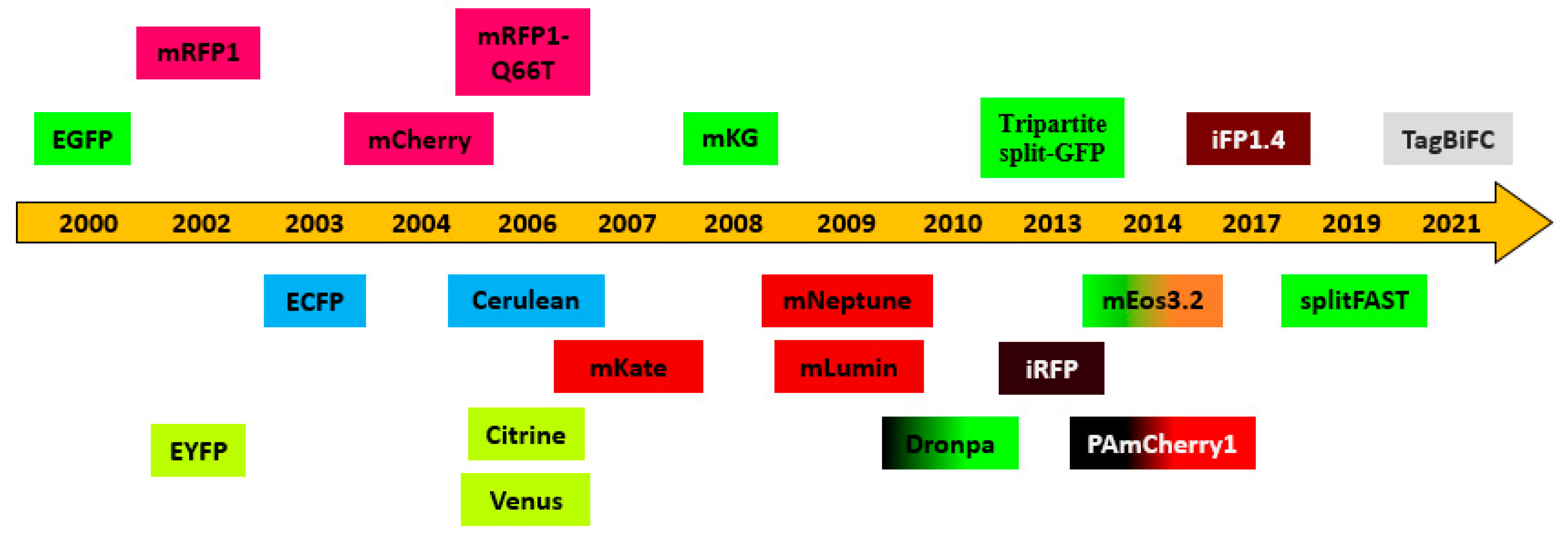
2. Advantages of BiFC assay
3. Critical considerations
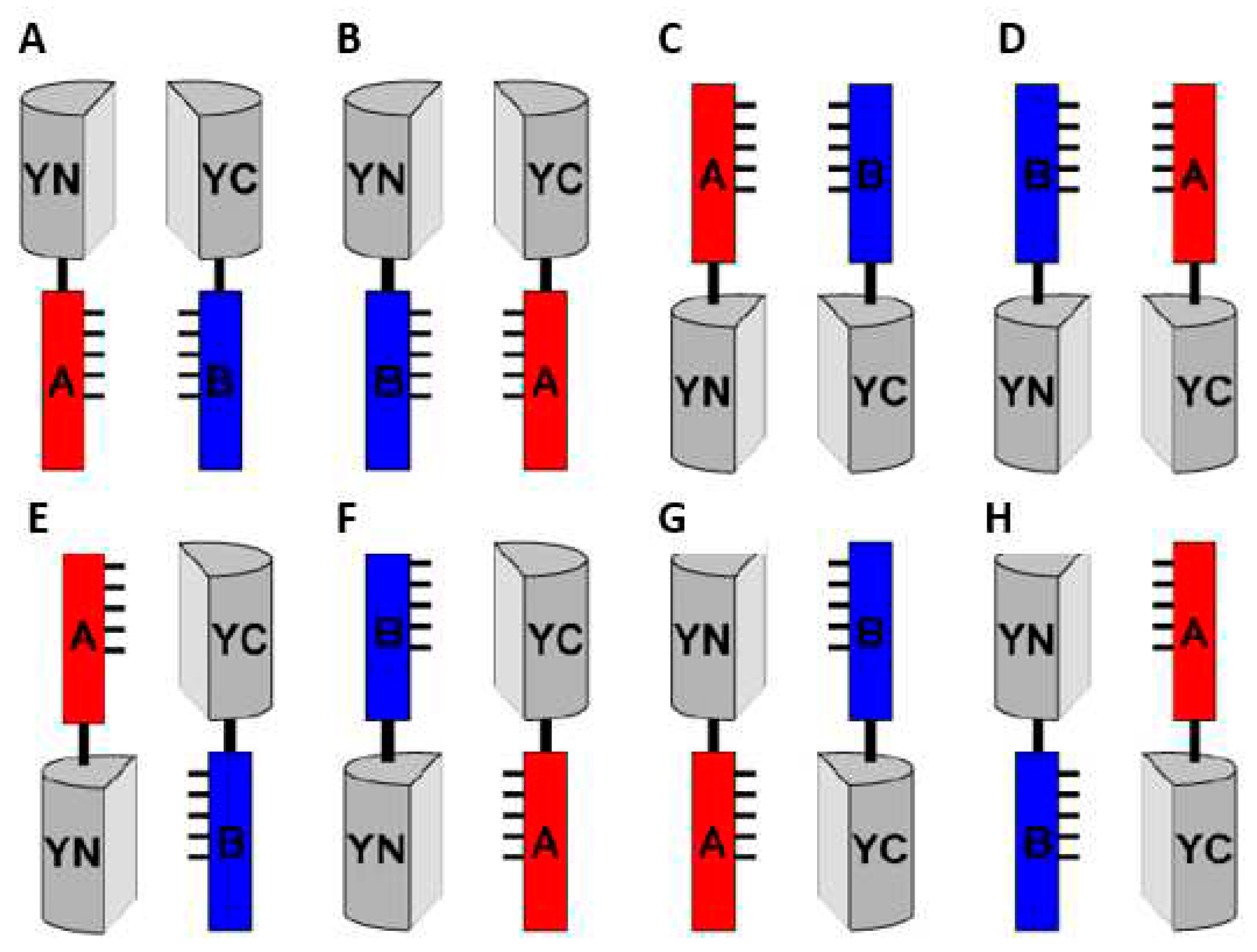
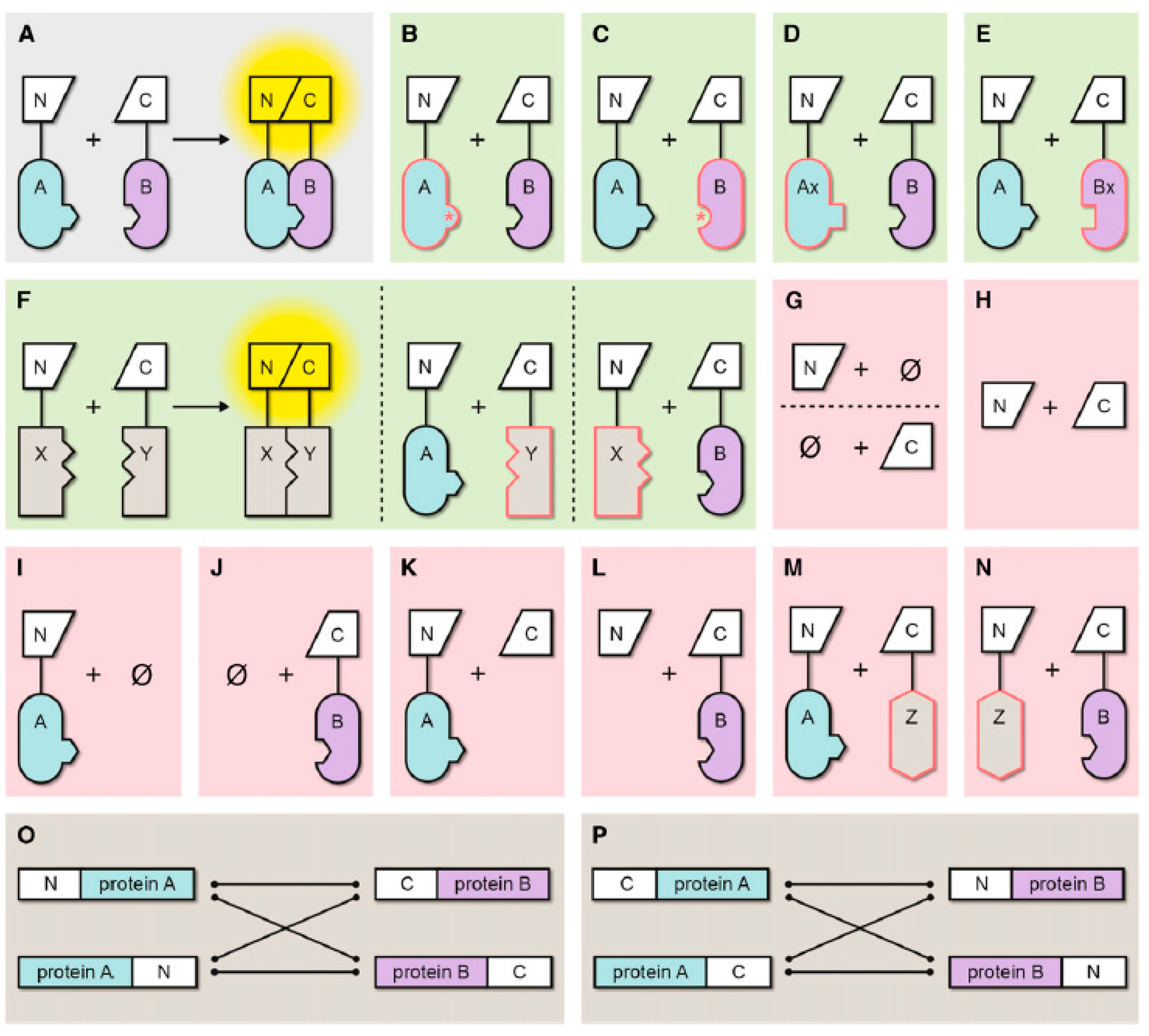
3.1. Topology of BiFC-tag fusions
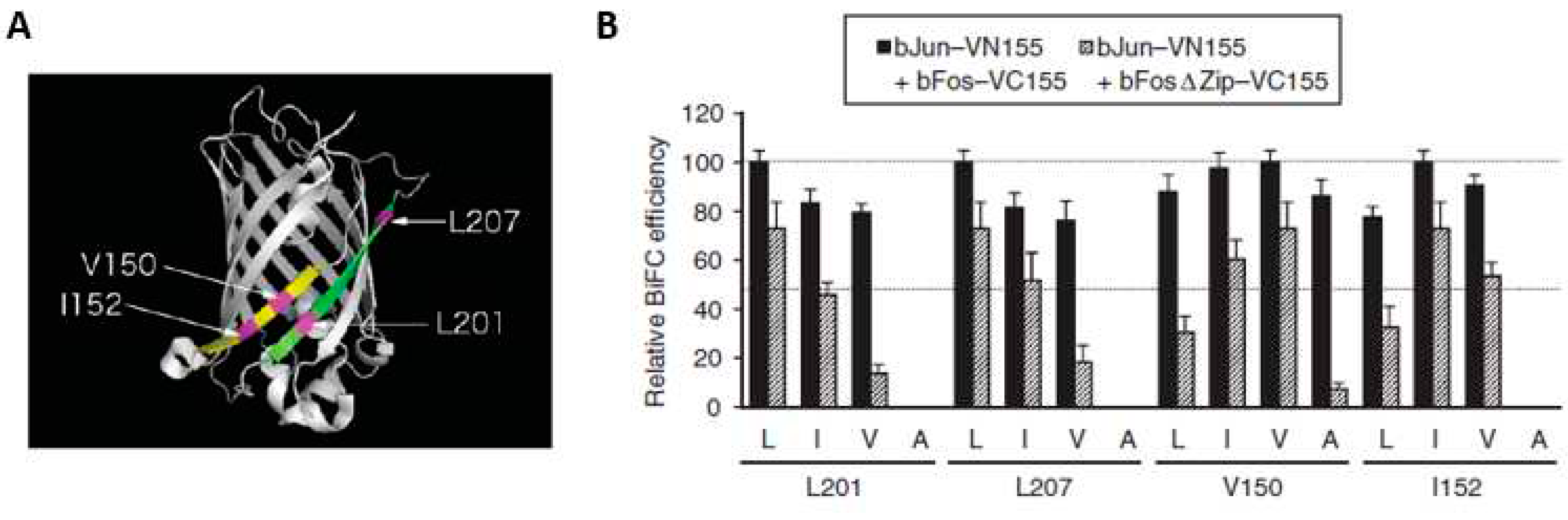
3.2. Linker selection
3.3. Improvement of signal-to-noise ratio
3.4. Controls for BiFC assay
3.5. Irreversibility of BiFC
4. Implementation of BiFC
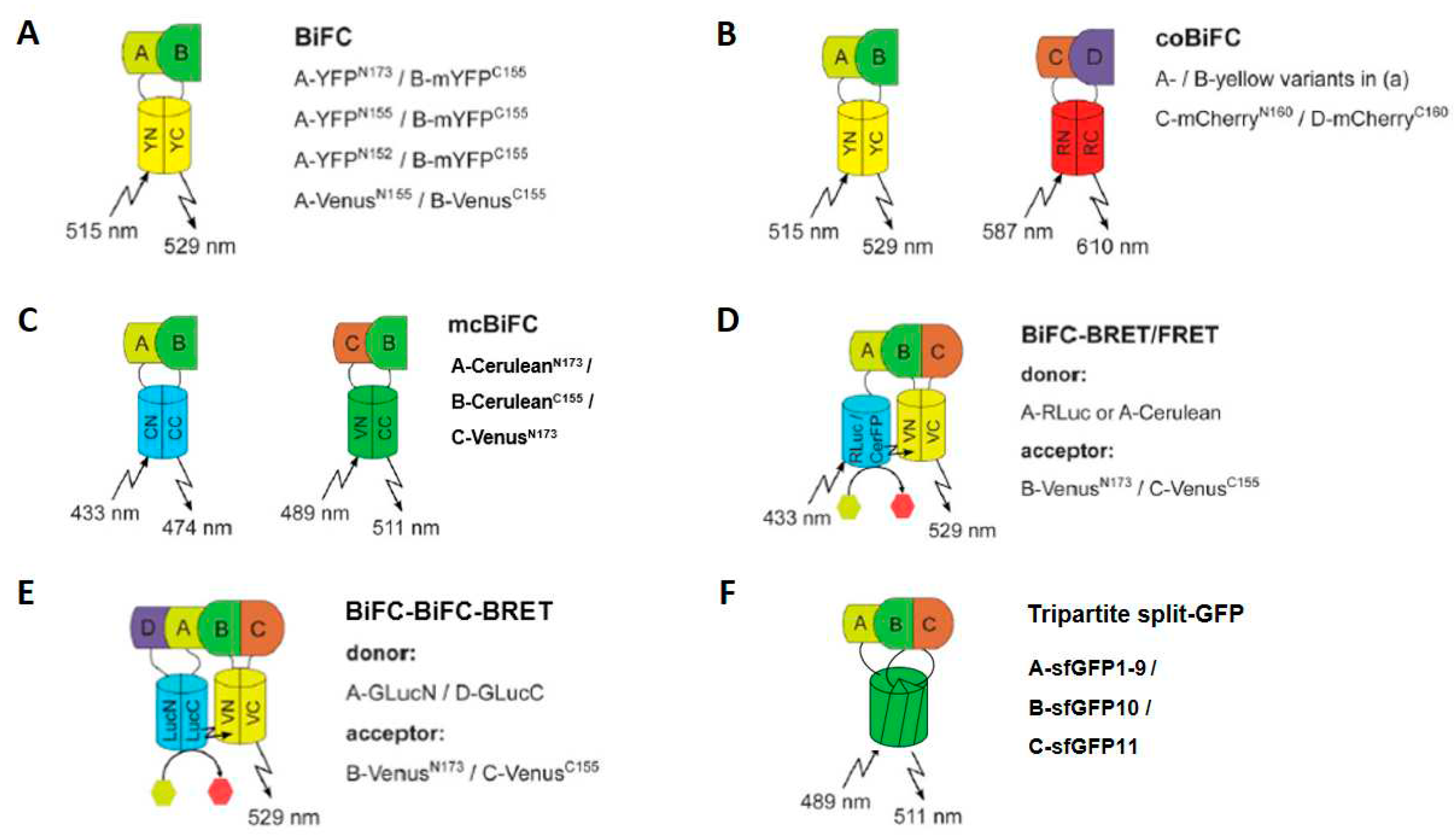
4.1. Low-throughput BiFC-based applications
4.2. Large-scale applications of BiFC
References
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Rizzo, M.A.; Springer, G.H.; Granada, B.; Piston, D.W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004, 22, 445–449. [Google Scholar] [CrossRef]
- Jia, Y.; Bleicher, F.; Reboulet, J.; Merabet, S. Bimolecular Fluorescence Complementation (BiFC) and Multiplexed Imaging of Protein-Protein Interactions in Human Living Cells. Methods Mol Biol 2021, 2350, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiological Reviews 2010, 90, 1103–1163. [Google Scholar] [CrossRef] [PubMed]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. PNAS 1994, 91, 12501–12504. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.G.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Shcherbakova, D.M.; Kuznetsova, I.M.; Turoverov, K.K.; Verkhusha, V.V. Modern fluorescent proteins: from chromophore formation to novel intracellular applications. BioTechniques 2011, 51, 313–327. [Google Scholar] [CrossRef]
- Ghosh, I.; Hamilton, A.D.; Regan, L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J. Am. Chem. Soc. 2000, 122, 5658–5659. [Google Scholar] [CrossRef]
- Hu, C.-D.; Chinenov, Y.; Kerppola, T.K. Visualization of Interactions among bZIP and Rel Family Proteins in Living Cells Using Bimolecular Fluorescence Complementation. Molecular Cell 2002, 9, 789–798. [Google Scholar] [CrossRef]
- Ueyama, T.; Kusakabe, T.; Karasawa, S.; Kawasaki, T.; Shimizu, A.; Son, J.; Leto, T.L.; Miyawaki, A.; Saito, N. Sequential Binding of Cytosolic Phox Complex to Phagosomes through Regulated Adaptor Proteins: Evaluation Using the Novel Monomeric Kusabira-Green System and Live Imaging of Phagocytosis. The Journal of Immunology 2008, 181, 629–640. [Google Scholar] [CrossRef]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.-D.; Koraïchi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S. A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association. Scientific Reports 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Sawano, A.; Park, E.S.; Miyawaki, A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. PNAS 2001, 98, 3197–3202. [Google Scholar] [CrossRef] [PubMed]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.-D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 2006, 40, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-D.; Kerppola, T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nature Biotechnology 2003, 21, 539–545. [Google Scholar] [CrossRef]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nature Biotechnology 1999, 17, 969–973. [Google Scholar] [CrossRef]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. PNAS 2000, 97, 11984–11989. [Google Scholar] [CrossRef]
- Campbell, R.E.; Tour, O.; Palmer, A.E.; Steinbach, P.A.; Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. A monomeric red fluorescent protein. PNAS 2002, 99, 7877–7882. [Google Scholar] [CrossRef]
- Jach, G.; Pesch, M.; Richter, K.; Frings, S.; Uhrig, J.F. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nature Methods 2006, 3, 597–600. [Google Scholar] [CrossRef]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nature Methods 2007, 4, 741–746. [Google Scholar] [CrossRef]
- Chu, J.; Zhang, Z.; Zheng, Y.; Yang, J.; Qin, L.; Lu, J.; Huang, Z.-L.; Zeng, S.; Luo, Q. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosensors and Bioelectronics 2009, 25, 234–239. [Google Scholar] [CrossRef]
- Lin, M.Z.; McKeown, M.R.; Ng, H.-L.; Aguilera, T.A.; Shaner, N.C.; Campbell, R.E.; Adams, S.R.; Gross, L.A.; Ma, W.; Alber, T.; et al. Autofluorescent Proteins with Excitation in the Optical Window for Intravital Imaging in Mammals. Chemistry & Biology 2009, 16, 1169–1179. [Google Scholar] [CrossRef]
- Filonov, G.S.; Verkhusha, V.V. A Near-Infrared BiFC Reporter for In Vivo Imaging of Protein-Protein Interactions. Chemistry & Biology 2013, 20, 1078–1086. [Google Scholar] [CrossRef]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.-M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature Biotechnology 2011, 29, 757–761. [Google Scholar] [CrossRef]
- Ando, R.; Mizuno, H.; Miyawaki, A. Regulated Fast Nucleocytoplasmic Shuttling Observed by Reversible Protein Highlighting. Science 2004, 306, 1370–1373. [Google Scholar] [CrossRef]
- Lee, Y.R.; Park, J.-H.; Hahm, S.-H.; Kang, L.-W.; Chung, J.H.; Nam, K.-H.; Hwang, K.Y.; Kwon, I.C.; Han, Y.S. Development of Bimolecular Fluorescence Complementation Using Dronpa for Visualization of Protein–Protein Interactions in Cells. Mol Imaging Biol 2010, 12, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.T.; Girirajan, T.P.K.; Mason, M.D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys J 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xing, D.; Su, Q.P.; Zhu, Y.; Zhang, J.; Kong, X.; Xue, B.; Wang, S.; Sun, H.; Tao, Y.; et al. Super-resolution imaging and tracking of protein–protein interactions in sub-diffraction cellular space. Nature Communications 2014, 5, 4443. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, A.; Huang, T.; Lin, L.-J.; Nan, X. Photoactivated Localization Microscopy with Bimolecular Fluorescence Complementation (BiFC-PALM) for Nanoscale Imaging of Protein-Protein Interactions in Cells. PLOS ONE 2014, 9, e100589. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Zhang, H.; Zeng, Y.; Li, Y.; Sun, C.; Sun, Y. TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells. Communications Biology 2021, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. BIMOLECULAR FLUORESCENCE COMPLEMENTATION (BiFC) ANALYSIS AS A PROBE OF PROTEIN INTERACTIONS IN LIVING CELLS. Annu Rev Biophys 2008, 37, 465–487. [Google Scholar] [CrossRef]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature Protocols 2006, 1, 1278–1286. [Google Scholar] [CrossRef]
- Sekar, R.B.; Periasamy, A. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. Journal of Cell Biology 2003, 160, 629–633. [Google Scholar] [CrossRef]
- Sun, Y.; Rombola, C.; Jyothikumar, V.; Periasamy, A. Förster resonance energy transfer microscopy and spectroscopy for localizing protein–protein interactions in living cells. Cytometry Part A 2013, 83, 780–793. [Google Scholar] [CrossRef]
- Miteva, Y.V.; Budayeva, H.G.; Cristea, I.M. Proteomics-Based Methods for Discovery, Quantification, and Validation of Protein–Protein Interactions. Anal. Chem. 2013, 85, 749–768. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 2012, 196, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.-C.; Abe, K.T.; Raught, B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Current Opinion in Chemical Biology 2019, 48, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Varnaitė, R.; MacNeill, S.A. Meet the neighbors: Mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics 2016, 16, 2503–2518. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Multicolor Bimolecular Fluorescence Complementation (BiFC) Analysis of Protein Interactions with Alternative Partners. Cold Spring Harb Protoc 2013, 2013, pdb–top077164. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Schlücking, K.; Schroeder, J.I.; Kudla, J. Protein fragment bimolecular fluorescence complementation analyses for the in vivo study of protein-protein interactions and cellular protein complex localizations. Methods Mol Biol 2014, 1062, 629–658. [Google Scholar] [CrossRef] [PubMed]
- Wouters, E.; Vasudevan, L.; Crans, R.A.J.; Saini, D.K.; Stove, C.P. Luminescence- and Fluorescence-Based Complementation Assays to Screen for GPCR Oligomerization: Current State of the Art. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Gokhale, R.S.; Khosla, C. Role of linkers in communication between protein modules. Current Opinion in Chemical Biology 2000, 4, 22–27. [Google Scholar] [CrossRef]
- Argos, P. An investigation of oligopeptides linking domains in protein tertiary structures and possible candidates for general gene fusion. Journal of Molecular Biology 1990, 211, 943–958. [Google Scholar] [CrossRef]
- Carayon, K.; Moulédous, L.; Combedazou, A.; Mazères, S.; Haanappel, E.; Salomé, L.; Mollereau, C. Heterologous Regulation of Mu-Opioid (MOP) Receptor Mobility in the Membrane of SH-SY5Y Cells *. Journal of Biological Chemistry 2014, 289, 28697–28706. [Google Scholar] [CrossRef]
- Armando, S.; Quoyer, J.; Lukashova, V.; Maiga, A.; Percherancier, Y.; Heveker, N.; Pin, J.-P.; Prézeau, L.; Bouvier, M. The chemokine CXC4 and CC2 receptors form homo- and heterooligomers that can engage their signaling G-protein effectors and βarrestin. The FASEB Journal 2014, 28, 4509–4523. [Google Scholar] [CrossRef] [PubMed]
- Cannaert, A.; Storme, J.; Franz, F.; Auwärter, V.; Stove, C.P. Detection and Activity Profiling of Synthetic Cannabinoids and Their Metabolites with a Newly Developed Bioassay. Anal. Chem. 2016, 88, 11476–11485. [Google Scholar] [CrossRef] [PubMed]
- Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 1999, 96, 11241–11246. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, K.; Kiuchi, T.; Shoji, K.; Sampei, K.; Mizuno, K. Visualization of cofilin-actin and Ras-Raf interactions by bimolecular fluorescence complementation assays using a new pair of split Venus fragments. BioTechniques 2012, 52, 45–50. [Google Scholar] [CrossRef]
- Zhou, J.; Lin, J.; Zhou, C.; Deng, X.; Xia, B. An improved bimolecular fluorescence complementation tool based on superfolder green fluorescent protein. Acta Biochimica et Biophysica Sinica 2011, 43, 239–244. [Google Scholar] [CrossRef]
- Nakagawa, C.; INAHATA, K.; NISHIMURA, S.; SUGIMOTO, K. Improvement of a Venus-Based Bimolecular Fluorescence Complementation Assay to Visualize bFos-bJun Interaction in Living Cells. Bioscience, Biotechnology, and Biochemistry 2011, 75, 1399–1401. [Google Scholar] [CrossRef]
- Kodama, Y.; Hu, C.-D. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. BioTechniques 2012, 53, 285–298. [Google Scholar] [CrossRef]
- Bracha-Drori, K.; Shichrur, K.; Katz, A.; Oliva, M.; Angelovici, R.; Yalovsky, S.; Ohad, N. Detection of protein–protein interactions in plants using bimolecular fluorescence complementation. The Plant Journal 2004, 40, 419–427. [Google Scholar] [CrossRef]
- Horstman, A.; Tonaco, I.A.N.; Boutilier, K.; Immink, R.G.H. A Cautionary Note on the Use of Split-YFP/BiFC in Plant Protein-Protein Interaction Studies. International Journal of Molecular Sciences 2014, 15, 9628–9643. [Google Scholar] [CrossRef]
- Kudla, J.; Bock, R. Lighting the Way to Protein-Protein Interactions: Recommendations on Best Practices for Bimolecular Fluorescence Complementation Analyses. The Plant Cell 2016, 28, 1002–1008. [Google Scholar] [CrossRef]
- Shyu, Y.J.; Hu, C.-D. Fluorescence complementation: an emerging tool for biological research. Trends in Biotechnology 2008, 26, 622–630. [Google Scholar] [CrossRef]
- Morell, M.; A, E.; Fx, A.; S, V. Detection of transient protein-protein interactions by bimolecular fluorescence complementation: the Abl-SH3 case Available online:. Available online: https://pubmed.ncbi.nlm.nih.gov/17352427/ (accessed on Sep 16, 2020).
- Ding, Z.; Liang, J.; Lu, Y.; Yu, Q.; Songyang, Z.; Lin, S.-Y.; Mills, G.B. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc Natl Acad Sci U S A 2006, 103, 15014–15019. [Google Scholar] [CrossRef] [PubMed]
- Remy, I.; Michnick, S.W. A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods 2004, 32, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Lönn, P.; Landegren, U. Close Encounters – Probing Proximal Proteins in Live or Fixed Cells. Trends in Biochemical Sciences 2017, 42, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Tebo, A.G.; Gautier, A. A split fluorescent reporter with rapid and reversible complementation. Nature Communications 2019, 10, 2822. [Google Scholar] [CrossRef]
- Wiens, M.D.; Campbell, R.E. Surveying the landscape of optogenetic methods for detection of protein-protein interactions. Wiley Interdiscip Rev Syst Biol Med 2018, 10, e1415. [Google Scholar] [CrossRef]
- Kodama, Y.; Wada, M. Simultaneous visualization of two protein complexes in a single plant cell using multicolor fluorescence complementation analysis. Plant Mol Biol 2009, 70, 211–217. [Google Scholar] [CrossRef]
- Dard, A.; Reboulet, J.; Jia, Y.; Bleicher, F.; Duffraisse, M.; Vanaker, J.-M.; Forcet, C.; Merabet, S. Human HOX Proteins Use Diverse and Context-Dependent Motifs to Interact with TALE Class Cofactors. Cell Reports 2018, 22, 3058–3071. [Google Scholar] [CrossRef] [PubMed]
- Vidi, P.-A.; Chemel, B.R.; Hu, C.-D.; Watts, V.J. Ligand-dependent oligomerization of dopamine D(2) and adenosine A(2A) receptors in living neuronal cells. Mol Pharmacol 2008, 74, 544–551. [Google Scholar] [CrossRef]
- Vidi, P.-A.; Przybyla, J.A.; Hu, C.-D.; Watts, V.J. Visualization of G protein-coupled receptor (GPCR) Interactions in Living Cells Using Bimolecular Fluorescence Complementation (BiFC). Curr Protoc Neurosci 2010, CHAPTER, Unit-5.29. [CrossRef]
- Kwaaitaal, M.; Keinath, N.F.; Pajonk, S.; Biskup, C.; Panstruga, R. Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiol 2010, 152, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Rebois, R.V.; Robitaille, M.; Galés, C.; Dupré, D.J.; Baragli, A.; Trieu, P.; Ethier, N.; Bouvier, M.; Hébert, T.E. Heterotrimeric G proteins form stable complexes with adenylyl cyclase and Kir3.1 channels in living cells. J Cell Sci 2006, 119, 2807–2818. [Google Scholar] [CrossRef]
- Rebois, R.V.; Robitaille, M.; Pétrin, D.; Zylbergold, P.; Trieu, P.; Hébert, T.E. Combining protein complementation assays with resonance energy transfer to detect multipartner protein complexes in living cells. Methods 2008, 45, 214–218. [Google Scholar] [CrossRef]
- Cabantous, S.; Terwilliger, T.C.; Waldo, G.S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nature Biotechnology 2005, 23, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Kellermann, S.J.; Rath, A.K.; Rentmeister, A. Tetramolecular fluorescence complementation for detection of specific RNAs in vitro. Chembiochem 2013, 14, 200–204. [Google Scholar] [CrossRef]
- Waldo, G.S.; Cabantous, S. Protein- protein interaction detection system using fluorescent protein microdomains; Los Alamos National Lab. (LANL), Los Alamos, NM (United States), 2010.
- Avilov, S.V.; Aleksandrova, N. Fluorescence protein complementation in microscopy: applications beyond detecting bi-molecular interactions. Methods Appl. Fluoresc. 2018, 7, 012001. [Google Scholar] [CrossRef]
- Lamesch, P.; Li, N.; Milstein, S.; Fan, C.; Hao, T.; Szabo, G.; Hu, Z.; Venkatesan, K.; Bethel, G.; Martin, P.; et al. hORFeome v3.1: A resource of human open reading frames representing over 10,000 human genes. Genomics 2007, 89, 307–315. [Google Scholar] [CrossRef]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 1–7. [Google Scholar] [CrossRef]
- Rual, J.-F.; Hirozane-Kishikawa, T.; Hao, T.; Bertin, N.; Li, S.; Dricot, A.; Li, N.; Rosenberg, J.; Lamesch, P.; Vidalain, P.-O.; et al. Human ORFeome Version 1.1: A Platform for Reverse Proteomics. Genome Res 2004, 14, 2128–2135. [Google Scholar] [CrossRef]
- Yang, X.; Boehm, J.S.; Yang, X.; Salehi-Ashtiani, K.; Hao, T.; Shen, Y.; Lubonja, R.; Thomas, S.R.; Alkan, O.; Bhimdi, T.; et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods 2011, 8, 659–661. [Google Scholar] [CrossRef]
- Sung, M.-K.; Huh, W.-K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 2007, 24, 767–775. [Google Scholar] [CrossRef]
- Sung, M.-K.; Lim, G.; Yi, D.-G.; Chang, Y.J.; Yang, E.B.; Lee, K.; Huh, W.-K. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res 2013, 23, 736–746. [Google Scholar] [CrossRef]
- Chang, Y.; Lim, G.; Huh, W.-K. Analysis of the TORC1 interactome reveals a spatially distinct function of TORC1 in mRNP complexes. Journal of Cell Biology 2021, 220, e201912060. [Google Scholar] [CrossRef] [PubMed]
- Snider, J.; Hanif, A.; Lee, M.E.; Jin, K.; Yu, A.R.; Graham, C.; Chuk, M.; Damjanovic, D.; Wierzbicka, M.; Tang, P.; et al. Mapping the functional yeast ABC transporter interactome. Nat Chem Biol 2013, 9, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Boruc, J.; Daele, H.V. den; Hollunder, J.; Rombauts, S.; Mylle, E.; Hilson, P.; Inzé, D.; Veylder, L.D.; Russinova, E. Functional Modules in the Arabidopsis Core Cell Cycle Binary Protein–Protein Interaction Network. The Plant Cell 2010, 22, 1264–1280. [Google Scholar] [CrossRef]
- Lee, L.-Y.; Wu, F.-H.; Hsu, C.-T.; Shen, S.-C.; Yeh, H.-Y.; Liao, D.-C.; Fang, M.-J.; Liu, N.-T.; Yen, Y.-C.; Dokladal, L.; et al. Screening a cDNA Library for Protein-Protein Interactions Directly in Planta. The Plant Cell 2012, 24, 1746–1759. [Google Scholar] [CrossRef]
- Remy, I.; Michnick, S.W. Regulation of Apoptosis by the Ft1 Protein, a New Modulator of Protein Kinase B/Akt. Mol Cell Biol 2004, 24, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.B.; Park, C.O.; Jeong, J.-Y.; Huh, W.-K. Monitoring G protein-coupled receptor activation using an adenovirus-based β-arrestin bimolecular fluorescence complementation assay. Analytical Biochemistry 2014, 449, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, J.; Watanabe, T.; Seki, T.; Karasawa, S.; Izumikawa, M.; Seki, T.; Iemura, S.-I.; Natsume, T.; Nomura, N.; Goshima, N.; et al. Novel In Vitro Protein Fragment Complementation Assay Applicable to High-Throughput Screening in a 1536-Well Format. J Biomol Screen 2009, 14, 970–979. [Google Scholar] [CrossRef]
- Berendzen, K.W.; Böhmer, M.; Wallmeroth, N.; Peter, S.; Vesić, M.; Zhou, Y.; Tiesler, F.K.E.; Schleifenbaum, F.; Harter, K. Screening for in planta protein-protein interactions combining bimolecular fluorescence complementation with flow cytometry. Plant Methods 2012, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Klopffleisch, K.; Phan, N.; Augustin, K.; Bayne, R.S.; Booker, K.S.; Botella, J.R.; Carpita, N.C.; Carr, T.; Chen, J.-G.; Cooke, T.R.; et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol 2011, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.-H.; Kim, H.; He, Q.; Baek, H.J.; Yang, D.; Chen, L.-Y.; Liang, J.; Chae, H.K.; Safari, A.; Liu, D.; et al. Genome-wide YFP Fluorescence Complementation Screen Identifies New Regulators for Telomere Signaling in Human Cells. Mol Cell Proteomics 2011, 10. [Google Scholar] [CrossRef]
- Xia, J.; Kong, L.; Zhou, L.-J.; Wu, S.-Z.; Yao, L.-J.; He, C.; He, C.Y.; Peng, H.-J. Genome-Wide Bimolecular Fluorescence Complementation-Based Proteomic Analysis of Toxoplasma gondii ROP18’s Human Interactome Shows Its Key Role in Regulation of Cell Immunity and Apoptosis. Front Immunol 2018, 9. [Google Scholar] [CrossRef]
- Lepur, A.; Kovačević, L.; Belužić, R.; Vugrek, O. Combining Unique Multiplex Gateway Cloning and Bimolecular Fluorescence Complementation (BiFC) for High-Throughput Screening of Protein–Protein Interactions. Journal of Biomolecular Screening 2016, 21, 1100–1111. [Google Scholar] [CrossRef]
| Fluorescent Protein | Bait | Prey | High-throughput Strategy | Pooled screening | Experimental cells or Organisms | References |
|---|---|---|---|---|---|---|
| Venus | VC-SUMO | VN-tagged strains (5911 ORFs) |
Individual mating (array screening) |
No | Yeast | [78] |
| Venus | 17 VC-ABC transporter genes | VN-tagged strains (209 ORFs) |
Individual mating (array screening) |
No | Yeast | [80] |
| Venus | 3 VC-Proteins from TORC1 | VN-tagged strains (5911 ORFs) |
Individual mating (array screening) | No | Yeast | [79] |
| mKG | 3 pairs of mKG_N-ProteinA/ mKG_C-ProteinB |
123599 potential PPI inhibitors | Protein solution mix (array screening) | No |
in vitro (Cell-free) |
[85] |
| GFP | 58 cGFP- core_cell_cycle proteins |
58 nGFP-core_cell_cycle proteins | Transiently coexpressed in leaf by infiltration | No | Plant (tobacco epidermal cells) | [81] |
| mYFP | YN-CPK3 | YC-cDNA plasmid library | Pooled co-transfection of YN-bait and YC-library | Yes | Plant (Arabidopsis) | [86] |
| YFP | 19 YN- or YC-proteins (G-proteins) | 33 YN- or YC-proteins (G-proteins) | individual BiFC (cotransfection) | No | Plant (Arabidopsis) | [87] |
| YFP | VirE2/VirD2/CTE-nYFP | cYFP-tagged cDNA array library (~100,000 clones) |
Cotransfection in microplate (array screening) | No | Arabidopsis leaf protoplasts | [82] |
| YFP | 6 nYFP-tagged core telomere proteins | cYFP-tagged retroviral array library (hORFeome v3.1) |
Coinfection in microplate (array screening) | No | HTC75 cells | [88] |
| Venus | VC-GPCRs | VN-β-arrestin 2 | adenovirus-based cotransduction (array screening) | No | U-2 osteosarcoma (OS) cells | [84] |
| GFP | GFP[2]-PKB | GFP[1]-tagged human brain cDNA library | Pooled co-transfection of bait and prey | Yes | COS-1 cells | [83] |
| Venus | Stable bait cell line expressing VN-AKT1 | VC-tagged prey lentivirus | Pooled infection using VN-AKT1-expressing stable cell line | Yes | Hela cells | [57] |
| YFP | Stable bait cell line expressing ROP18I/ ROP18II-nYFP | cYFP-tagged prey retrovirus (hORFeome v7.1) |
Individual infection (array screening) |
No | HTC75 cells | [89] |
| Venus | VN/VC-AHCY | VN/VC-tagged hORFs | Cotransfection (array screening) |
No | HEK293T cells | [90] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





