Submitted:
06 September 2023
Posted:
08 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Samples and Pre-quantification
2.2. Mixture Sample Preparation
2.3. SpermX method for all three laboratories
2.4. Conventional Differential Extraction Method (DE)
2.5. DNA Quantification
2.6. STR Amplification and Typing
2.7. Method Comparison Studies
3. Results
3.1. Male DNA in Sperm Fractions
3.2. Total Human to Male Ratios (S/Y) in the Sperm Fractions
3.3. Male Recovery Percentage in the Sperm Fractions
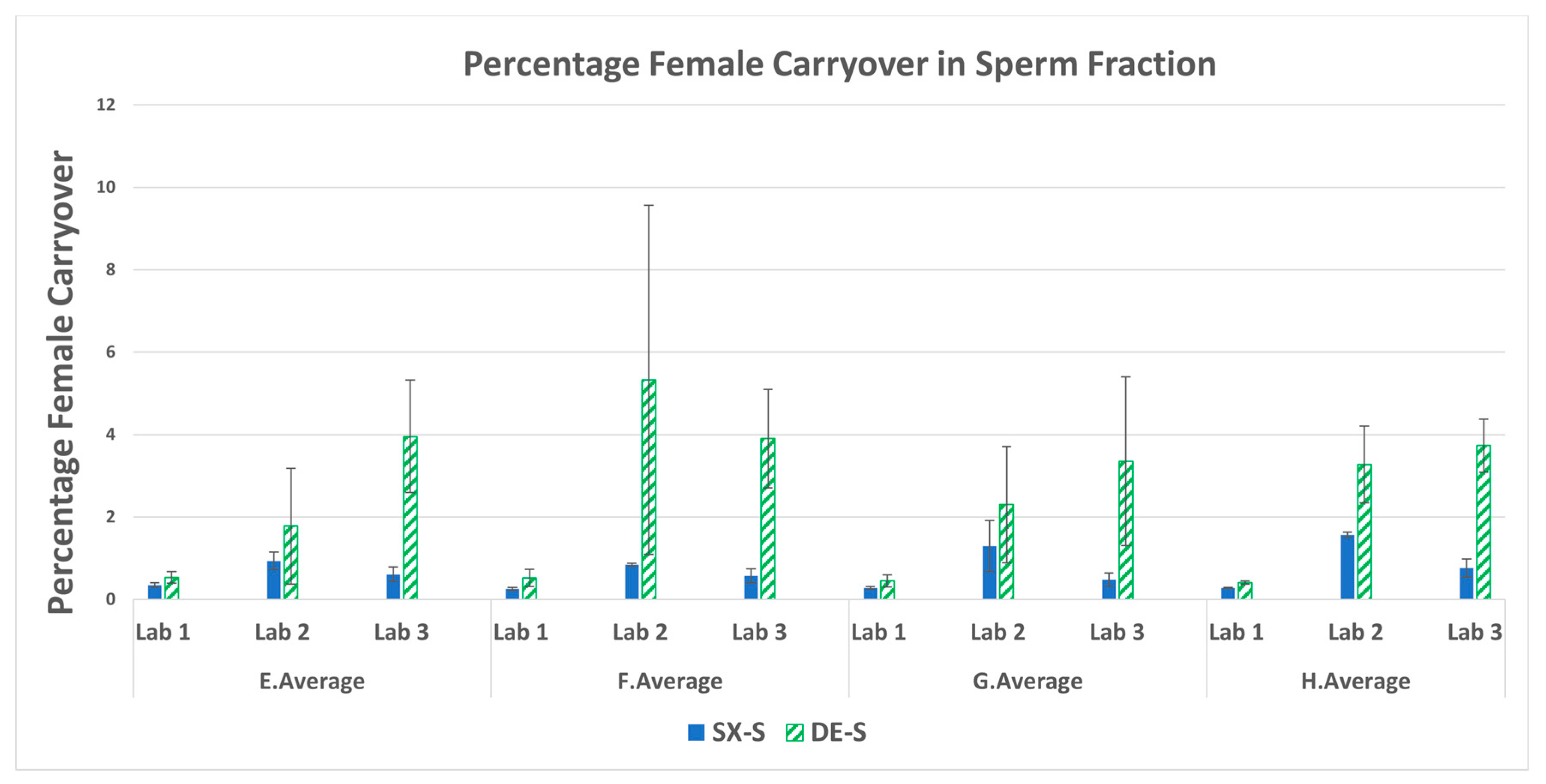
3.4. Percent Female Carryover in the Sperm Fractions
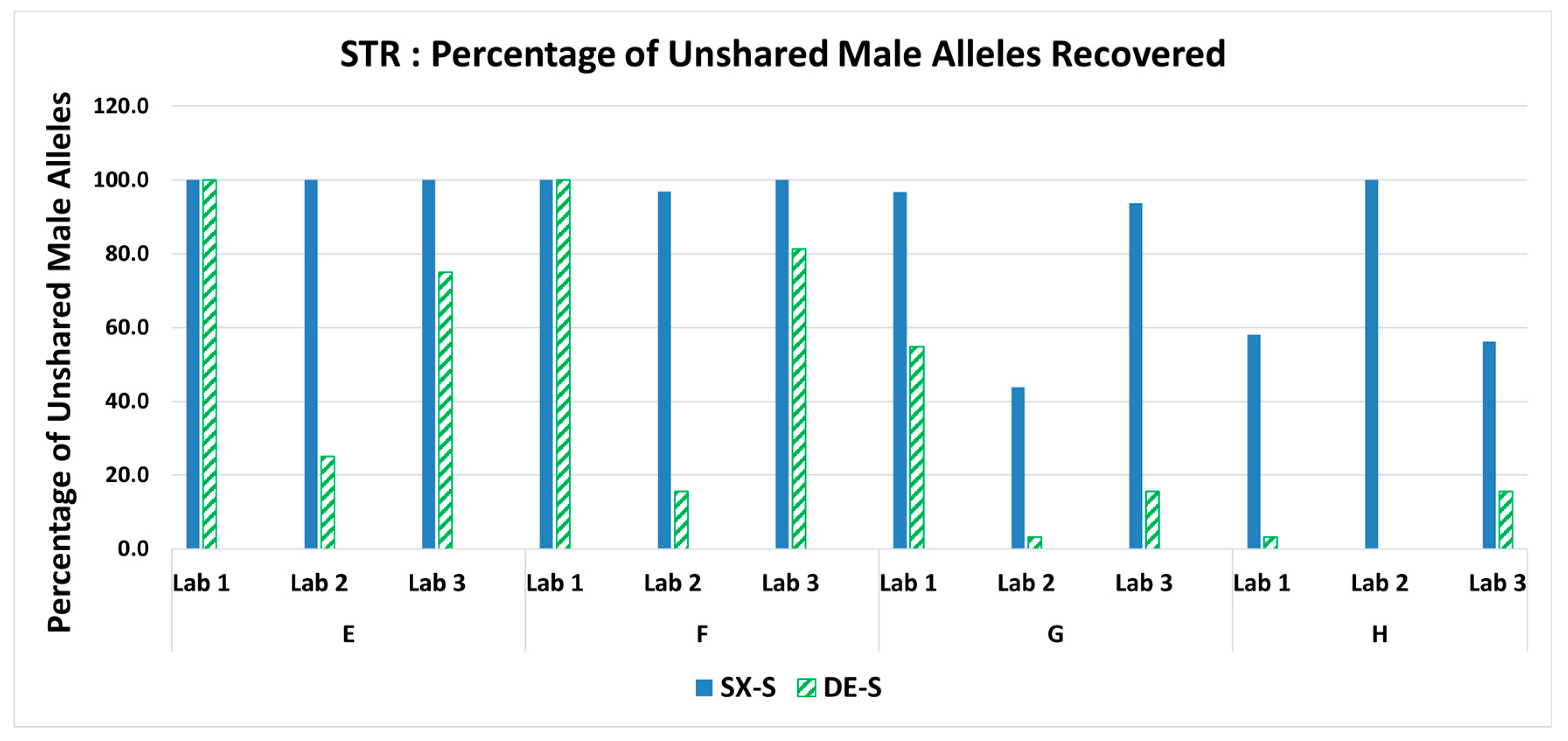
3.5. STR: Percentage of Unshared Male Alleles Recovered in the Sperm Fraction Profiles
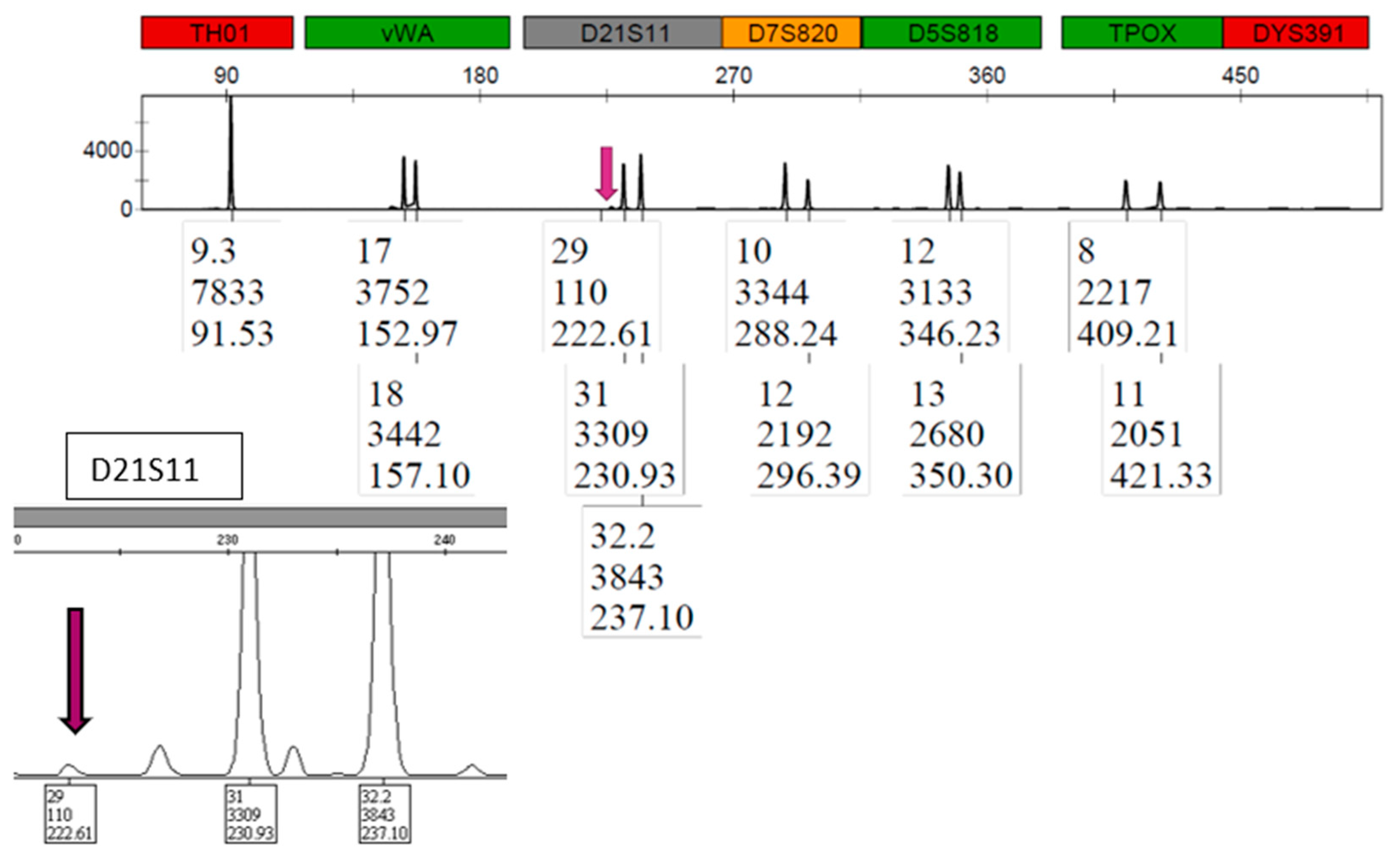
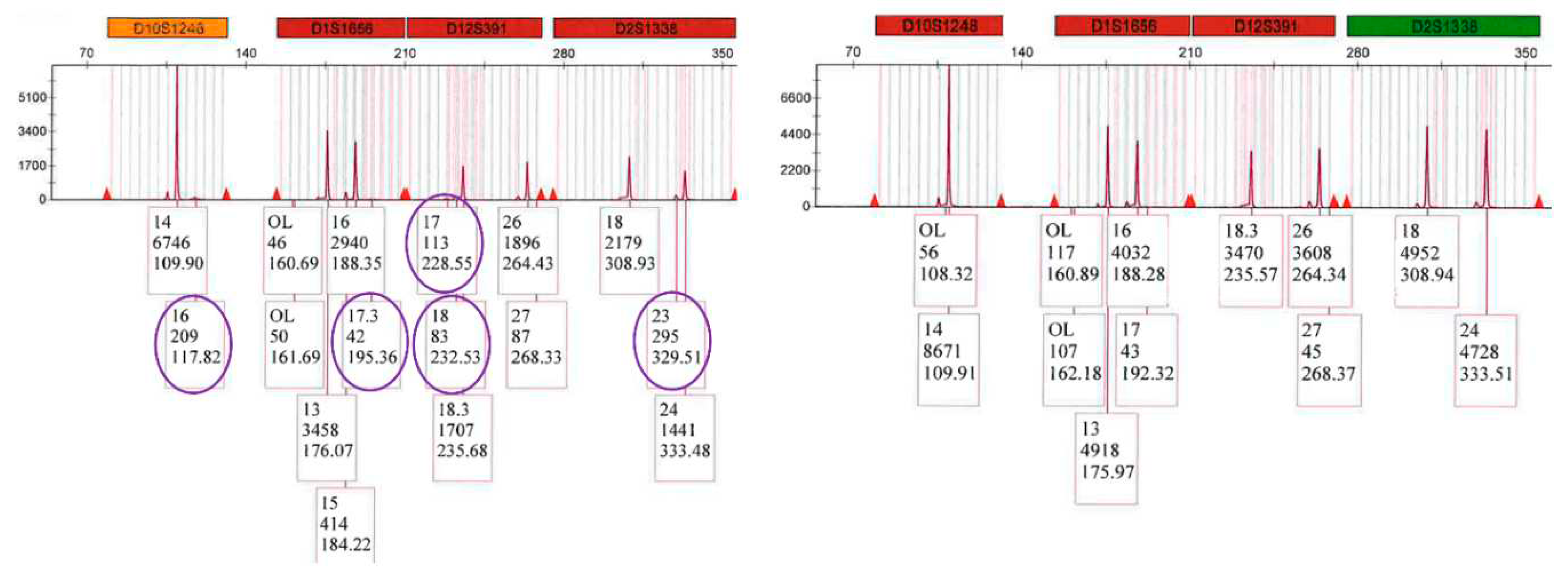
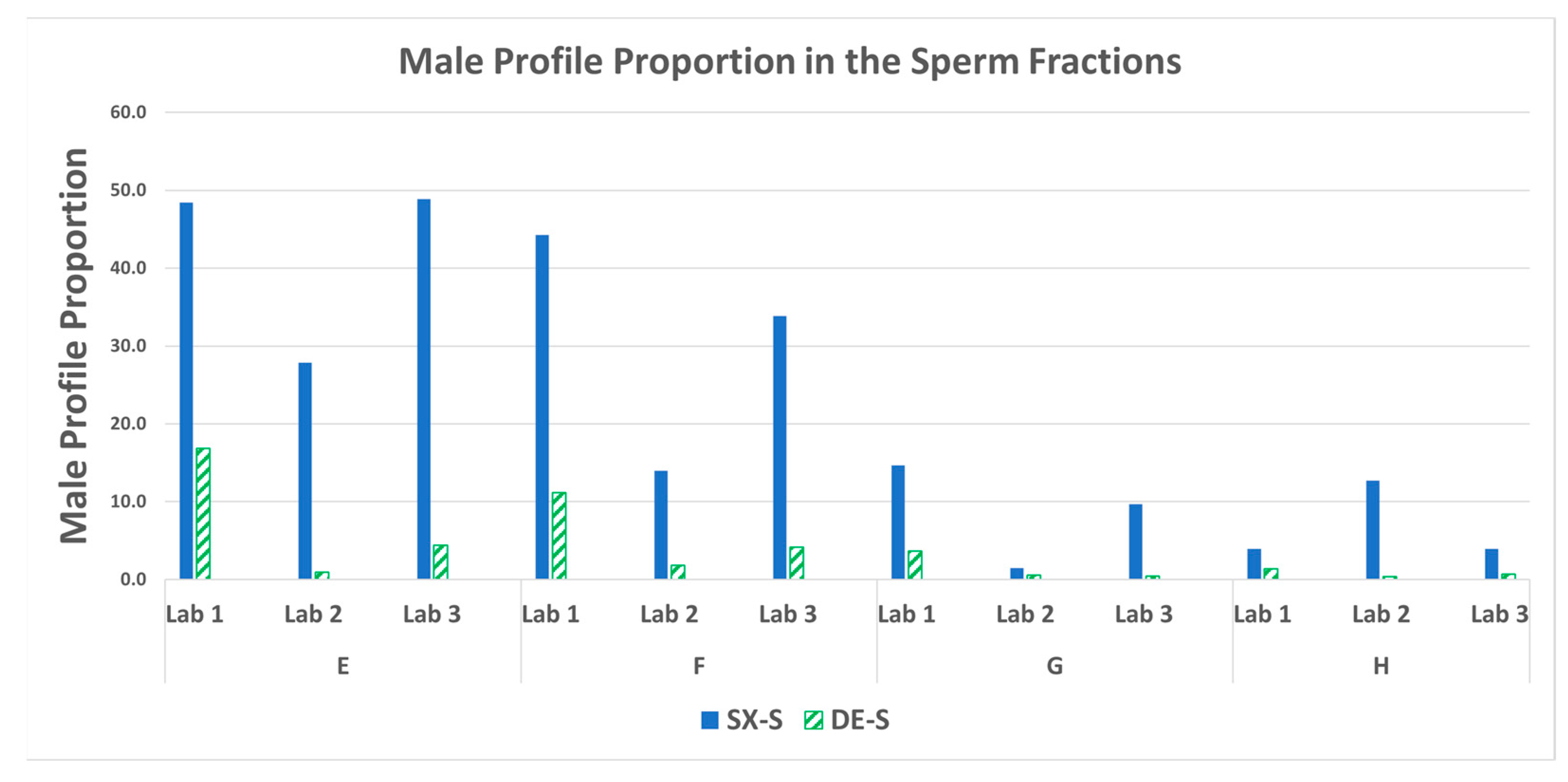
3.6. STR: Male Profile Proportion in the Sperm Fractions
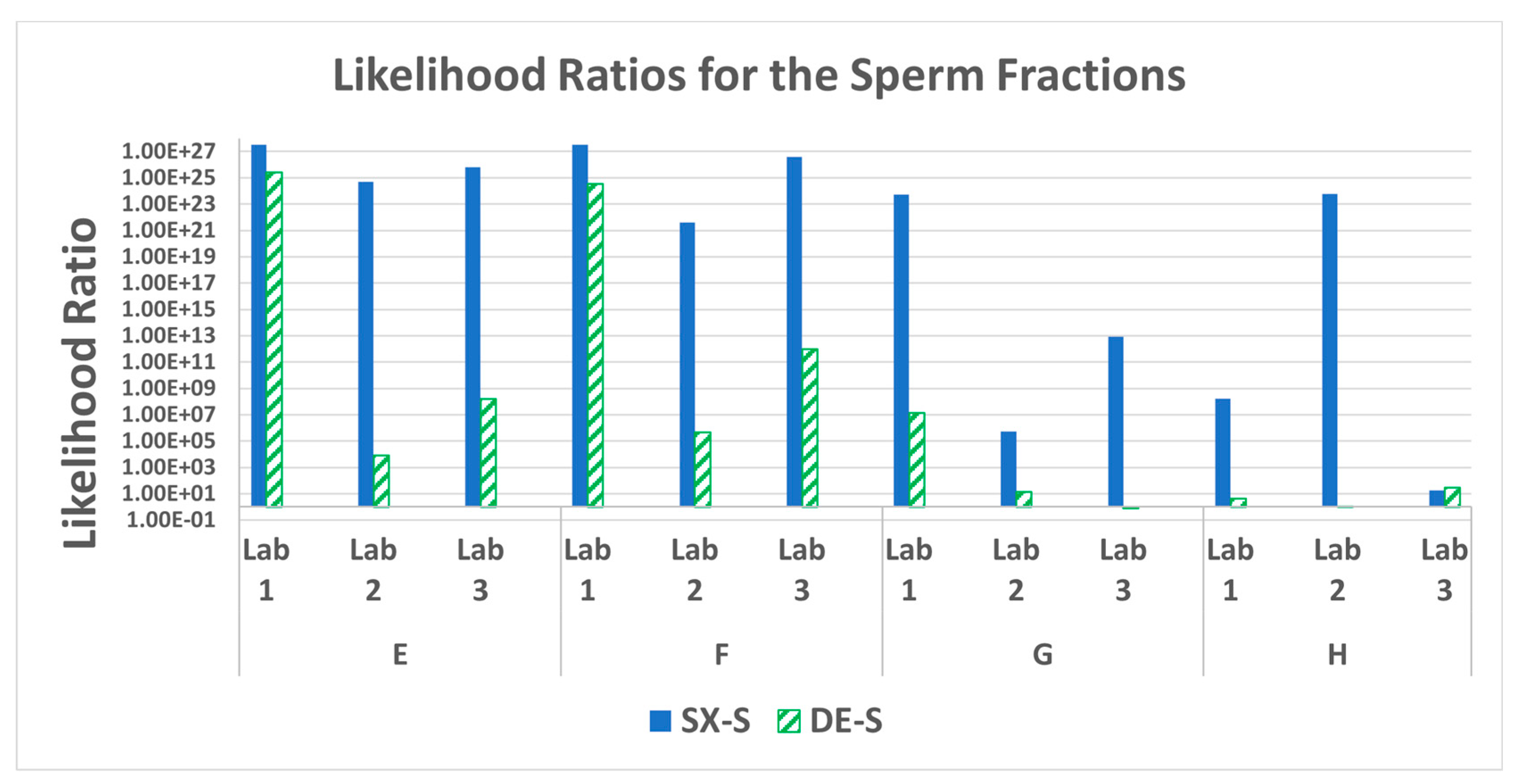
3.7. STR Statistics: Probabilistic Genotyping
4. Discussion
5. Conclusions
Supplementary Materials
References
- National Sexual Assault Kit Initiative. The National Sexual Assault Kit Initiative (SAKI). Available online: https://sakitta.org/metrics/ (accessed on 10 April 2023).
- Peterson J, Johnson D, Herz D, Graziano L, Oehler T. SEXUAL ASSAULT KIT BACKLOG STUDY, NCJ Number 238500,. https://nij.ojp.gov/library/publications/sexual-assault-kit-backlog-study. 2012.
- David Kanaris. Sexual Assault Kit Processing at the Alaska Scientific Crime Detection Laboratory-Letter- Public Information. https://dps.alaska.gov/getmedia/ac3e3f7d-6812-45d3-a730-29e0eeb9527c/April-2022-Sexual-Assault-Kit-Processing-Workflow.pdf. 1922.
- Gill P, Jeffreys AJ, Werrett DJ. Forensic application of DNA ‘fingerprints.’ Nature 1985;318(6046):577–9. [CrossRef]
- Klein SB, Buoncristiani MR. Evaluating the efficacy of DNA differential extraction methods for sexual assault evidence. Forensic Sci Int Genet 2017;29:109–17. [CrossRef]
- Bogas V, Bento AM, Serra A, Brito P, Lopes V, Sampaio L, et al. Validation of sampletype I-sep DL for differential extraction and purification with prepfiler express in the automate express DNA extraction system. Forensic Sci Int Genet Suppl Ser 2017;6:e353–4. [CrossRef]
- Paul, J. Project FORESIGHT Annual Report, 2020-2021" (2022). Faculty & Staff Scholarship. 3093. https://researchrepository.wvu.edu/faculty_publications/3093. 2022.
- Volk P, Holt A, Chen A, Hanson E, Ballantyne J. Enhancing the sexual assault workflow: Development of a rapid male screening assay incorporating molecular non-microscopic sperm identification. Forensic Sci Int Genet Suppl Ser 2019;7(1):21–2. [CrossRef]
- Timken MD, Klein SB, Kubala S, Scharnhorst G, Buoncristiani MR, Miller KWP. Automation of the standard DNA differential extraction on the Hamilton AutoLys STAR system: A proof-of-concept study. Forensic Sci Int Genet 2019;40:96–104. [CrossRef]
- Goldstein MC, Cox JO, Seman LB, Cruz TD. Improved resolution of mixed STR profiles using a fully automated differential cell lysis/DNA extraction method. Forensic Sci Res 2020;5(2):106–12. [CrossRef]
- Nittis M, Franco M, Cochrane C. New oral cut-off time limits in NSW. J Forensic Leg Med 2016;44:92–7. [CrossRef]
- Spivak, H. National Best Practices for Sexual Assault Kits: A Multidisciplinary Approach, https://nij.ojp.gov/topics/articles/national-best-practices-sexual-assault-kits-multidisciplinary-approach.
- Sinha SK, Brown H, Holt H, Khan M, Brown R, Sgueglia JB, et al. Development and validation of a novel method “SpermXTM” for high throughput differential extraction processing of sexual assault kits (SAKs) for DNA analysis. Forensic Sci Int Genet 2022;59:102690. [CrossRef]
- Casey DG, Domijan K, MacNeill S, Rizet D, O’Connell D, Ryan J. The Persistence of Sperm and the Development of Time Since Intercourse (TSI) Guidelines in Sexual Assault Cases at Forensic Science Ireland, Dublin, Ireland. J Forensic Sci 2017;62(3):585–92. [CrossRef]
- Loftus A, Murphy G, Brown H, Montgomery A, Tabak J, Baus J, et al. Development and validation of InnoQuant® HY, a system for quantitation and quality assessment of total human and male DNA using high copy targets. Forensic Sci Int Genet 2017;29:205–17. [CrossRef]
- Fedder, J. Nonsperm Cells in Human Semen: With Special Reference to Seminal Leukocytes and their Possible Influence on Fertility. Arch Androl 1996;36(1):41–65. [CrossRef]
- Shewale JG, Sikka SC, Schneida E, Sinha SK. DNA profiling of azoospermic semen samples from vasectomized males by using Y-PLEXTM6 amplification kit. J Forensic Sci 2003;48(1):127–9.
- Gillooly JF, Hein A, Damiani R. Nuclear DNA content varies with cell size across human cell types. Cold Spring Harb Perspect Biol 2015;7(7):1–27. [CrossRef]
- Qiagen. https://www.qiagen.com/us/products/human-id-and-forensics/investigator-solutions/qiaamp-dna-investigator-kit/.
- Buckleton JS, Bright JA, Gittelson S, Moretti TR, Onorato AJ, Bieber FR, et al. The Probabilistic Genotyping Software STRmix: Utility and Evidence for its Validity. J Forensic Sci 2019;64(2):393–405. [CrossRef]
- Simek E, Janssens L. Assessment of an Automated Differential Separation Utilizing a Novel Nanofiber Filter for Sexual Assault Cases. International Symposium of Human Identification, Orlando. 2021.
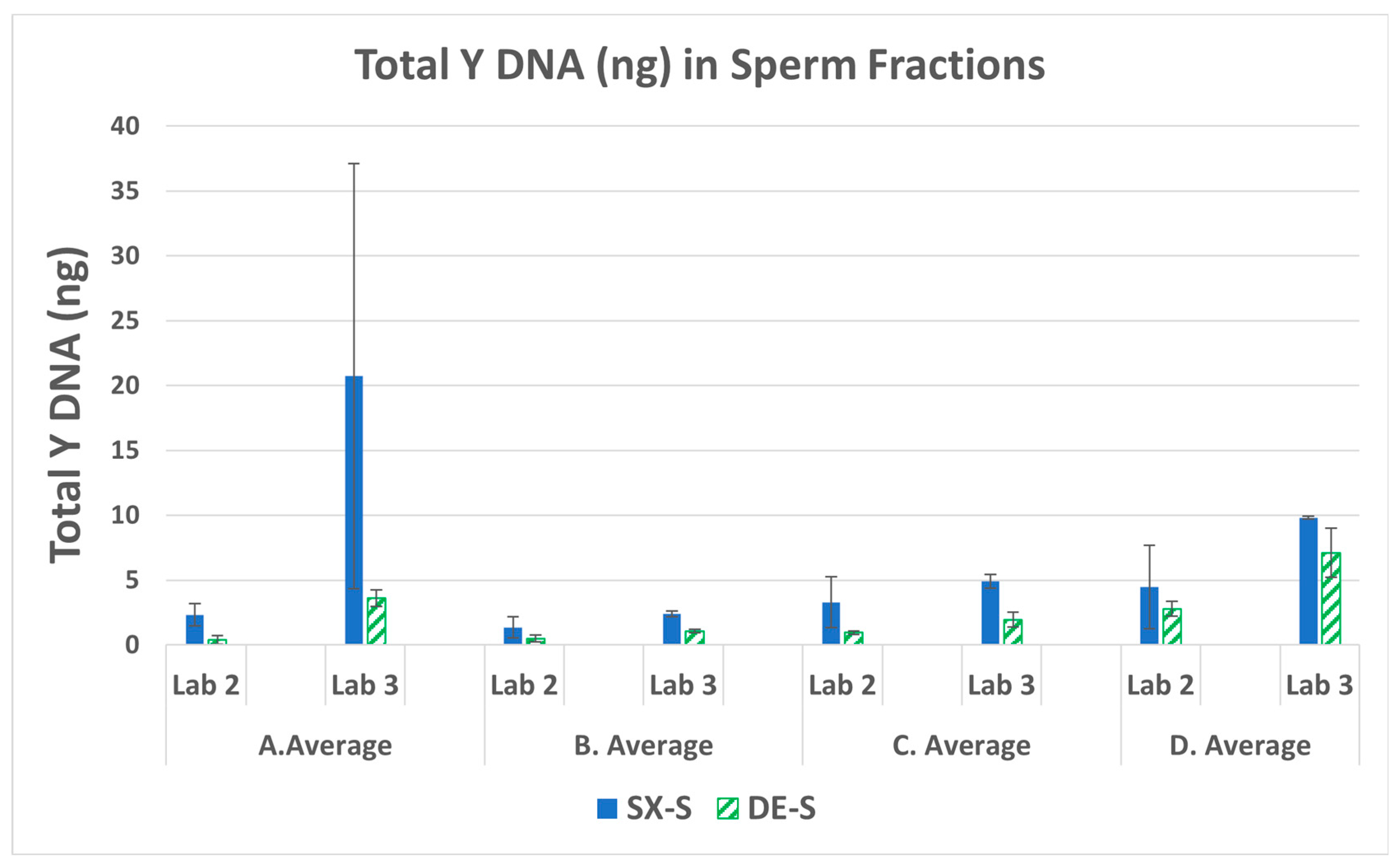
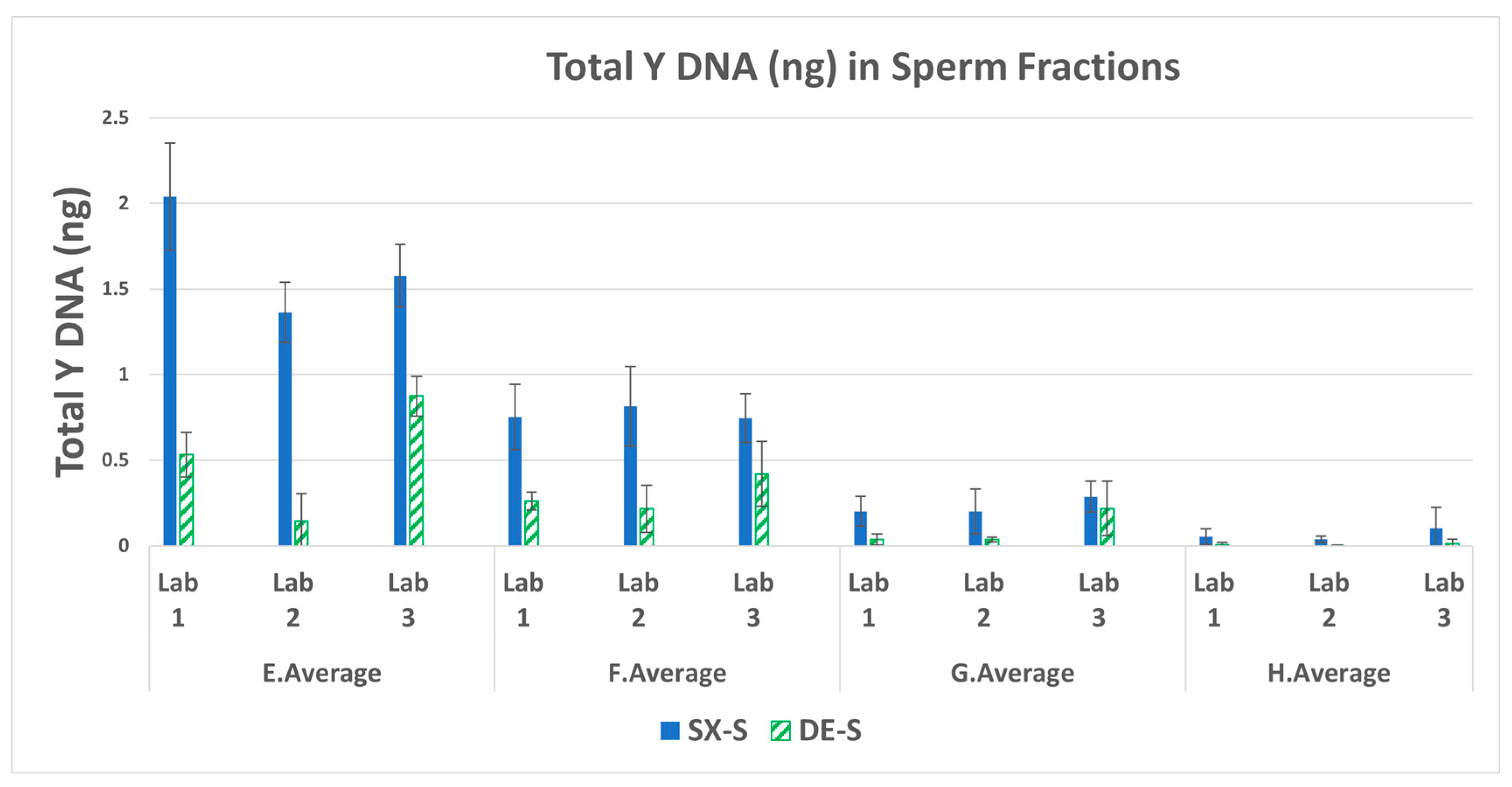
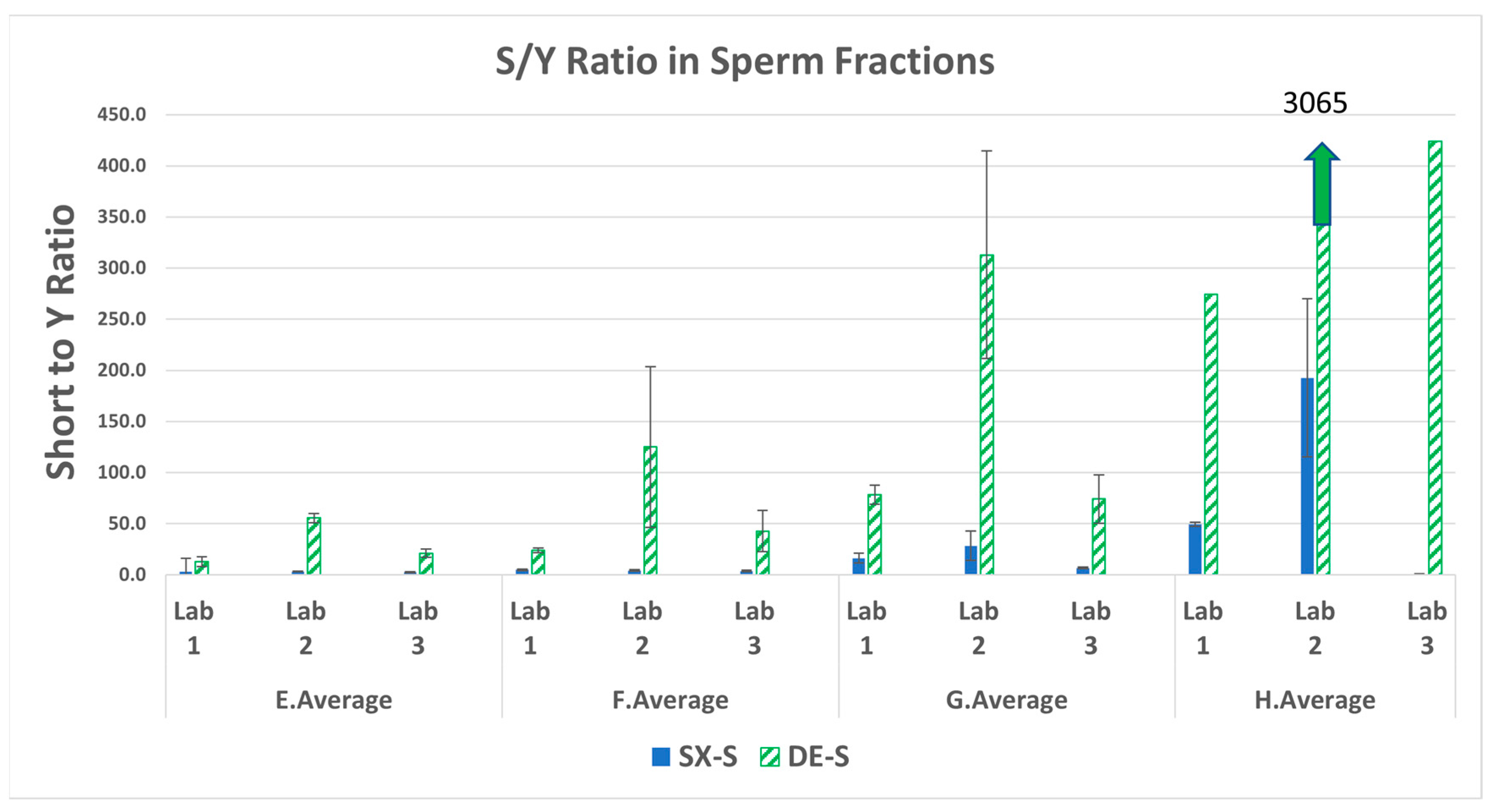
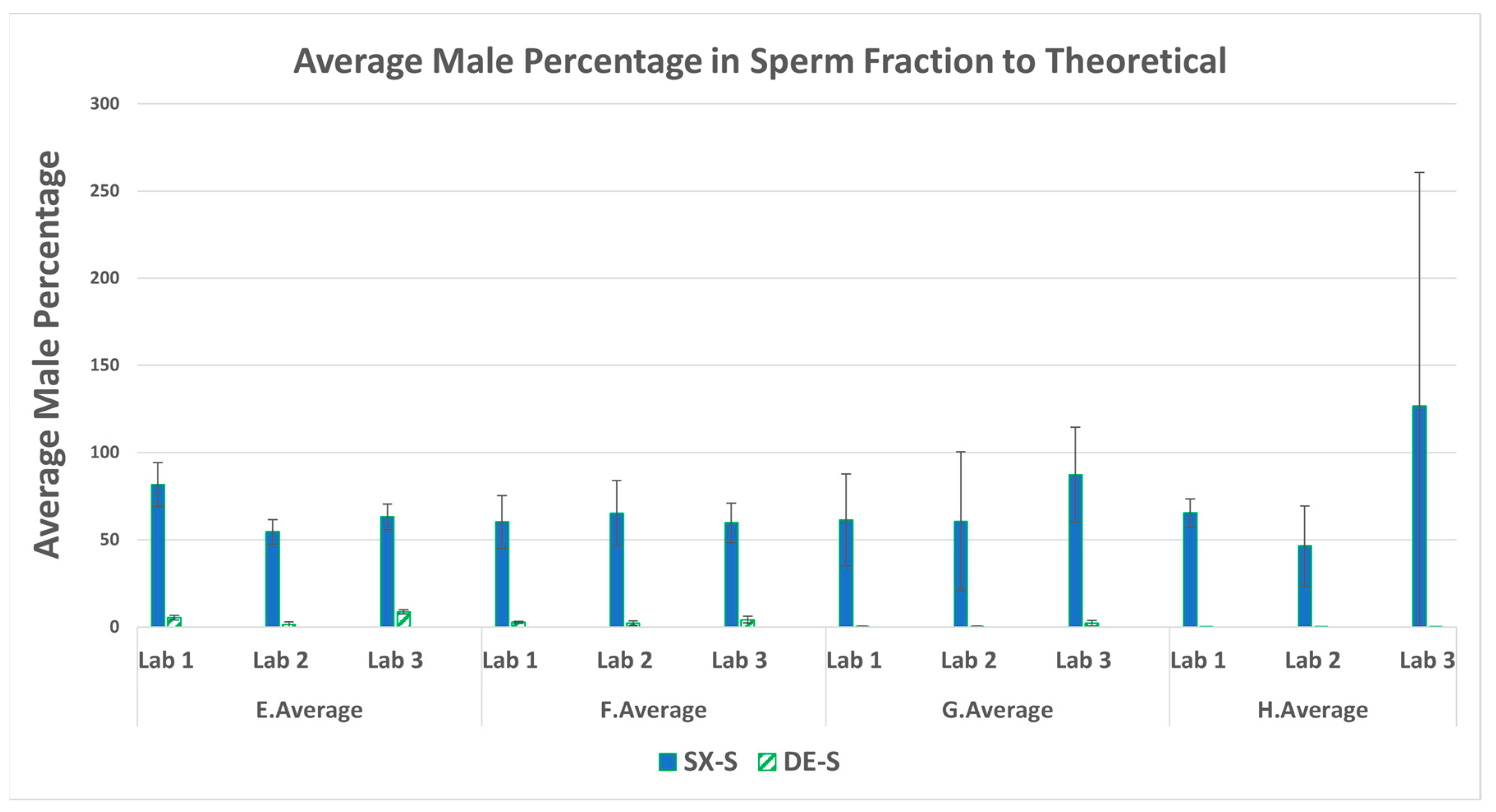

| Swab set | F:M ratio | Female (ng) | Male (ng) |
|---|---|---|---|
| A | 100:1 | 1000 | 10 |
| B | 1000:1 | 2500 | 2.5 |
| C | 1000:1 | 5000 | 5 |
| D | 750:1 | 7500 | 10 |
| E | 600:1 | 1500 | 2.5 |
| F | 1200:1 | 1500 | 1.25 |
| G | 4545:1 | 1500 | 0.33 |
| H | 18182:1 | 1500 | 0.0825 |
| Mixture | Lab | Fold Difference | Average |
|---|---|---|---|
| A | LAB 1 | Not Performed | 5.9 |
| LAB 2 | 6.1 | ||
| LAB 3 | 5.8 | ||
| B | LAB 1 | Not Performed | 9.9 |
| LAB 2 | 10.8 | ||
| LAB 3 | 8.9 | ||
| C | LAB 1 | Not Performed | 6.0 |
| LAB 2 | 6.9 | ||
| LAB 3 | 5.0 | ||
| D | LAB 1 | Not Performed | 1.5 |
| LAB 2 | 1.6 | ||
| LAB 3 | 1.4 | ||
| E | LAB 1 | 15.3 | 20.1 |
| LAB 2 | 37.6 | ||
| LAB 3 | 7.2 | ||
| F | LAB 1 | 23.0 | 22.4 |
| LAB 2 | 30.1 | ||
| LAB 3 | 14.2 | ||
| G | LAB 1 | 167.8 | 124.3 |
| LAB 2 | 165.3 | ||
| LAB 3 | 39.9 | ||
| H | LAB 1 | 878.2 | 1533.0 |
| LAB 2 | 2787.9 | ||
| LAB 3 | 932.9 |
| Mixture | Lab | SX | DE |
|---|---|---|---|
| G | LAB 1 | 30 | 17 |
| LAB 2 | 14 | 1 | |
| LAB 3 | 30 | 5 | |
| H | LAB 1 | 18 | 1 |
| LAB 2 | 32 | 0 | |
| LAB 3 | 18 | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





