Submitted:
16 August 2023
Posted:
17 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study population
2.2. Blood culture
2.2.1. Identification of bacterial and fungal isolates
2.2.2. Antimicrobial Susceptibility Testing
2.3. Patients' COVID-19 diagnoses
2.4. Statistical Analysis
2.5. Ethical Approval
3. Results
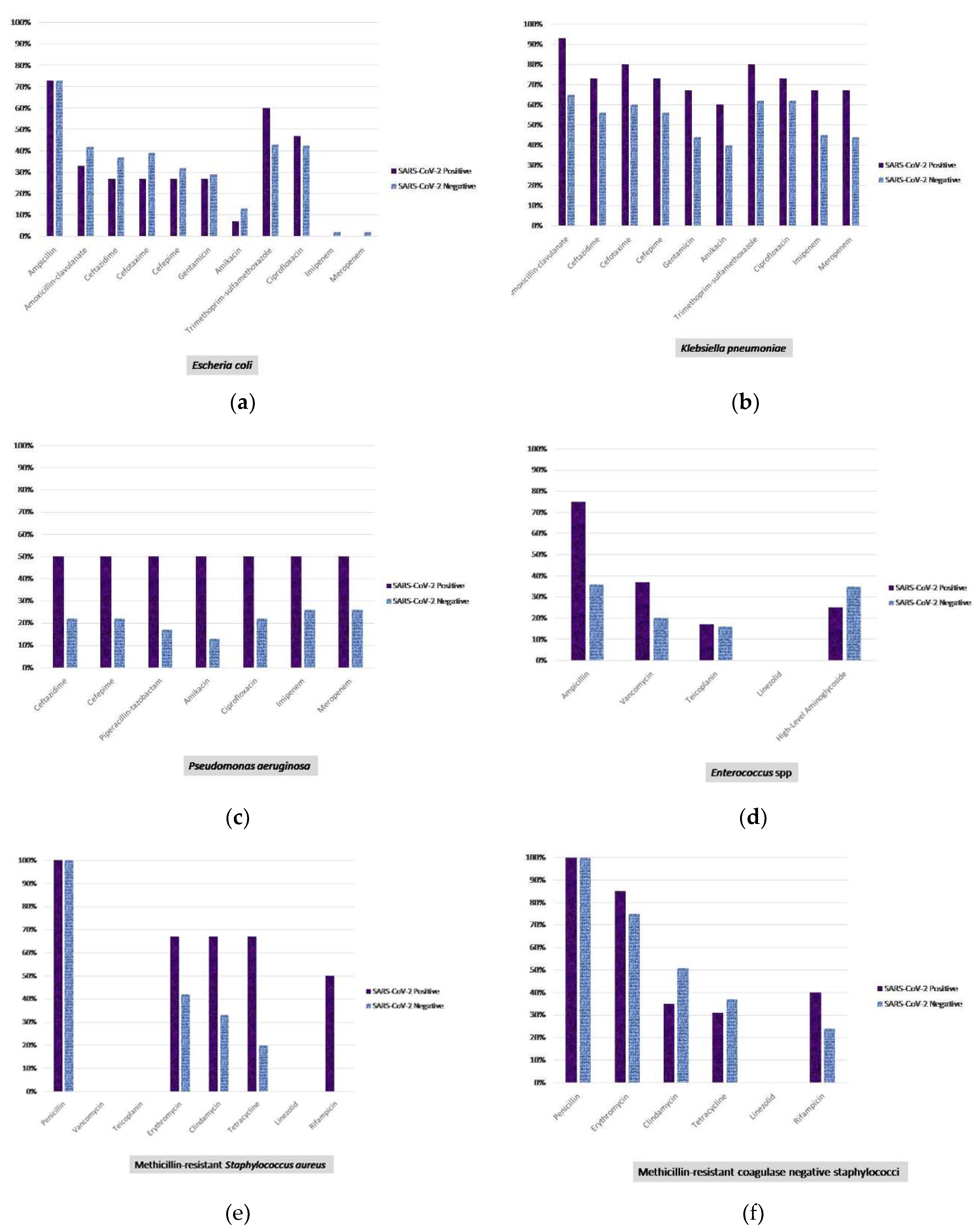
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Manohar, P.; Loh, B.; Nachimuthu, R.; Hua, X.; Welburn, S.C.; Leptihn, S. Secondary bacterial infections in patients with viral pneumonia. Front. Med. 2020, 7, 420. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Simmonds, A.; Annavajhala, M.K.; McConville, T.H.; Dietz, D.E.; Shoucri, S.M.; Laracy, J.C.; Rozenberg, F.D.; Nelson, B.; Greendyke, W.G.; Furuya, E.Y.; Whittier, S.; Uhlemann, A.C. Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J. Antimicrob. Chemother. 2021, 76, 380–384. [Google Scholar] [CrossRef]
- Bengoechea, J.A.; Bamford, C.G. SARS-CoV-2 bacterial co-infections and AMR: the deadly trio in COVID-19. EMBO. Mol. Med. 2020, 12, e12560. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Amin, A.K.; Khanna, P.; Aali, A.; McGregor, A.; Bassett, P.; Gopal Rao, G. An observational cohort study of bacterial coinfection and implications for empirical antibiotic therapy in patients presenting with COVID-19 to hospitals in North West London. J. Antimicrob. Chemother. 2021, 76, 796–803. [Google Scholar] [CrossRef]
- Yu, D.; Ininbergs, K.; Hedman, K.; Giske, C.G.; Stralin, K.; Özenci, V. Low prevalence of bloodstream infection and high blood culture contamination rates in patients with COVID-19. Plos One. 2020, 15, e0242533. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Bloodstream infection event (central line-associated bloodstream infection and non-central line-associated bloodstream infection). 2021. Available online: https://www.cdc.gov/nhsn/pdfs/pscmanual/4psc_clabscurrent.pdf (accessed on 12 December 2022).
- Gilligan, P.H.; Alby, K.; York, M.K. Blood cultures. In Clinical Microbiology Procedures Handbook, , Leber, A.L. Ed. in chief., 4th ed.; ASM Press: Washington, USA, 2016. [Google Scholar]
- CLSI. Epidemiological cutoff values for antifungal susceptibility testing. 3rd ed. CLSI supplement M59. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts; 4th informational supplement M27-S4. CLSI, Wayne, PA, 2012.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 10.0, 2020. http://www.eucast.org.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0, 2021. http://www.eucast.org.
- Centers for Disease Control and Prevention (CDC). Candida auris. Available online: https://www.cdc.gov/fungal/candida-auris/index.html (accessed on 16 February 2021).
- Bölükbaşı, Y.; Erköse, G.G.; Orhun, G.; Kuşkucu, M.A.; Çağatay, A.; Önel, M.; Öngen, B.; Ağaçfidan, A.; Esen, F.; Erturan, Z. First case of COVID-19 positive Candida auris fungemia in Turkey. Mikrobiyol. Bul. 2021, 55, 648–655. [Google Scholar] [CrossRef]
- Willan, J.; King, A.J.; Jeffery, K.; Bienz, N. Challenges for NHS hospitals during Covid-19 epidemic. BMJ. 2020, 368, m1117. [Google Scholar] [CrossRef]
- Sepulveda, J.; Westblade, L.F.; Whittier, S.; Satlin, M.J.; Greendyke, W.G.; Aaron, J.G.; Zucker, J.; Dietz, D.; Sobieszczyk, M.; Choi, J.J.; Liu, D.; Russell, S.; Connelly, C.; Green, D.A. Bacteremia and blood culture utilization during COVID-19 surge in New York City. J. Clin. Microbiol. 2020, 58, e00875–20. [Google Scholar] [CrossRef]
- Hughes, S.; Troise, O.; Donaldson, H.; Mughal, N.; Moore, L.S.P. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020, 26, 1395–1399. [Google Scholar] [CrossRef]
- Mahmoudi, H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS. Hyg. Infect. Control. 2020, 15, Doc35. [Google Scholar]
- Center for Disease Control and Prevention. COVID-19: US Impact on Antimicrobial Resistance, Special Report 2022; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. [Google Scholar]
- Getahun, H.; Smith, I.; Trivedi, K.; Paulin, S.; Balkhy, H.H. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull. World Health Organ. 2020, 98, 442–442A. [Google Scholar] [CrossRef]
- Bauer, K.A.; Puzniak, L.A.; Yu, K.C.; Finelli, L.; Moise, P.; Ai, C.; Watts, J.A.; Gupta, V. Epidemiology and outcomes of culture-positive bloodstream pathogens prior to and during the SARS-CoV-2 pandemic: a multicenter evaluation. BMC. Infect. Dis. 2022, 22, 841. [Google Scholar] [CrossRef] [PubMed]
- Michailides, C.; Paraskevas, T.; Karalis, I.; Koniari, I.; Pierrakos, C.; Karamouzos, V.; Marangos, M.; Velissaris, D. Impact of bacterial infections on COVID-19 patients: is timing important? Antibiotics (Basel). 2023, 12, 379. [Google Scholar] [CrossRef] [PubMed]
- Bahceci, I.; Yildiz, I.E.; Duran, O.F.; Soztanaci, U.S.; Kirdi Harbawi, Z.; Senol, F.F.; Demiral, G. Secondary bacterial infection rates among patients with COVID-19. Cureus. 2022, 14, e22363. [Google Scholar] [CrossRef] [PubMed]
- Segala, F.V.; Pafundi, P.C.; Masciocchi, C.; Fiori, B.; Taddei, E.; Antenucci, L.; De Angelis, G.; Guerriero, S.; Pastorino, R.; Damiani, A.; Posteraro, B.; Sanguinetti, M.; De Pascale, G.; Fantoni, M.; Murri, R. Incidence of bloodstream infections due to multidrug-resistant pathogens in ordinary wards and intensive care units before and during the COVID-19 pandemic: a real-life, retrospective observational study. Infection. 2023, 51, 1061–1069. [Google Scholar] [CrossRef]
- Silva, D.L.; Lima, C.M.; Magalhães, V.C.R.; Baltazar, L.M.; Peres, N.T.A.; Caligiorne, R.B.; Moura, A.S.; Fereguetti, T.; Martins, J.C.; Rabelo, L.F.; Abrahão, J.S.; Lyon, A.C.; Johann, S.; Santos, DA. Fungal and bacterial coinfections increase mortality of severely ill COVID-19 patients. J. Hosp. Infect. 2021, 113, 145–154. [Google Scholar] [CrossRef]
- Petrakis, V.; Panopoulou, M.; Rafailidis, P.; Lemonakis, N.; Lazaridis, G.; Terzi, I.; Papazoglou, D.; Panagopoulos, P. The impact of the Covıd-19 pandemic on antimicrobial resistance and management of bloodstream infections. Pathogens. 2023, 12, 780. [Google Scholar] [CrossRef]
- Sinto, R.; Lie, K.C.; Setiati, S.; Suwarto, S.; Nelwan, E.J.; Djumaryo, D.H.; Karyanti, M.R.; Prayitno, A.; Sumariyono, S.; Moore, C.E.; Hamers, R.L.; Day, N.P.J.; Limmathurotsakul, D. Blood culture utilization and epidemiology of antimicrobial-resistant bloodstream infections before and during the COVID-19 pandemic in the Indonesian national referral hospital. Antimicrob. Resist. Infect. Control. 2022, 11, 73. [Google Scholar] [CrossRef]
- Chowdhary, A.; Tarai, B.; Singh, A.; Sharma, A. Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April-July 2020. Emerg. Infect. Dis. 2020, 26, 2694–2696. [Google Scholar] [CrossRef]
- Arastehfar, A.; Carvalho, A.; Nguyen, M.H.; Hedayati, M.T.; Netea, M.G.; Perlin, D.S.; Hoenigl, M. COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J. Fungi (Basel). 2020, 6, 211. [Google Scholar] [CrossRef]
- Rodriguez, J.Y.; Le Pape, P.; Lopez, O.; Esquea, K.; Labiosa, A.L.; Alvarez-Moreno, C. Candida auris: a latent threat to critically ill patients with Coronavirus disease 2019. Clin. Infect. Dis. 2020, 73, e2836–7. [Google Scholar] [CrossRef] [PubMed]
- Magnasco, L.; Mikulska, M.; Giacobbe, D.R.; Taramasso, L.; Vena, A.; Dentone, C.; Dettori, S.; Tutino, S.; Labate, L.; Di Pilato, V.; Crea, F.; Coppo, E.; Codda, G.; Robba, C.; Ball, L.; Patroniti, N.; Marchese, A.; Pelosi, P.; Bassetti, M. Spread of carbapenem-resistant gram-negatives and Candida auris during the COVID-19 pandemic in critically ill patients: one step back in antimicrobial stewardship? Microorganisms. 2021, 9, 95. [Google Scholar] [CrossRef] [PubMed]
- Çaklovica-Küçükkaya, İ.; Orhun, G.; Çağatay, A.A.; Kalaycı, S.; Esen, F.; Şahin, F.; Ağaçfidan, A.; Erturan, Z. P494 Comparison of Candida colonization in intensive care unit patients with and without COVID-19: first prospective cohort study from Turkey, Medical Mycology, Volume 60, Issue Supplement_1, September 2022, myac072P494. 20 September. [CrossRef]
| Clinics | Unit | SARS CoV-2 (+) | SARS CoV-2 (-) | Not tested |
|---|---|---|---|---|
| Inpatient | Surgical | 24 | 96 | - |
| Internal | 19 | 69 | 3 | |
| Outpatient | Surgical | - | 26 | - |
| Internal | 38 | 243 | 14 | |
| Intensive Care Unit | Surgical | 5 | 43 | 1 |
| Internal | 32 | 38 | 1 |
| Microorganisms | TotalWYXWYX[n(%)] | SARS-CoV-2WYXWYXPositive [n (%)] | SARS-CoV-2WYXWYXNegative [n (%)] | SARS-CoV-2WYXWYXNon tested [n (%)] | Positive vs. Negative |
|---|---|---|---|---|---|
| All microorganisms | 671 (100) | 121 (18) | 531 (79.1) | 19 (2.8) | p value |
| FermentativeGram-negative rods | 252 (37.5) | 34 (28) | 214 (40.3) | 4 (21) | 0.012 |
| Escherichia coli | 131 (19.5) | 15 (12.4) | 113 (21.3) | 3 (15.8) | 0.026 |
| Klebsiella pneumoniae1/ Carbapenem-resistant K. pneumoniae2 | 80/38 (11.9/5.6) | 15/9 (12.4/7.4) | 64 / 28 (12.05 /5.3) | 1/1 (5.3/5.3) | 0.9171WYXWYX0.3532 |
| Klebsiella oxytoca | 5 (0.7) | 0 (0.0) | 5 (0.9) | - | 0.590 |
| Enterobacter spp. | 7 (1.05) | 1 (0.8) | 6 (1.1) | - | 0.770 |
| Serratia marcescens | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Serratia spp. | 2 (0.3) | 0 (0.0) | 2 (0.4) | - | 1.000 |
| Citrobacter spp. | 3 (0.45) | 0 (0.0) | 3 (0.6) | - | 1.000 |
| Citrobacter koseri | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Proteus mirabilis | 13 (1.9) | 1 (0.8) | 12 (2.3) | - | 0.309 |
| Morganella morganii | 3 (0.45) | 0 (0.0) | 3 (0.6) | - | 1.000 |
| Raoultella planticola | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Aeromonas spp. | 1 (0.15) | 0 (0.0) | 1 (0.2) | 1.000 | |
| Salmonella Enteritidis | 4 (0.6) | 2 (1.65) | 2 (0.4) | - | 0.105 |
| Non-FermentativeGram-negative rods | 66 (9.8) | 16 (13.2) | 48 ( 9 ) | 2 (10.5) | 0.163 |
| Pseudomonas aeruginosa | 25 (3.7) | 2 (1.65) | 23 (4.3) | - | 0.166 |
| Pseudomonas stutzeri | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Pseudomonas spp. | 5 (0.7) | 2 (1.65) | 3 (0.6) | - | 0.216 |
| Acinetobacter baumannii | 14 (2.1) | 7 (5.8) | 6 (1.1) | 1 (5.3) | 0.001 |
| Acinetobacter lwoffii | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Acinetobacter spp. | 4 (0.6) | 2 (1.65) | 2 (0.4) | - | 0.105 |
| Rhizobium radiobacter | 2 (0.3) | 2 (1.65) | 0 (0.0) | - | 0.034 |
| Achromobacter xylosoxidans | 1 (0.15) | - | - | 1 (5.3) | - |
| Ochrobactrum anthropi | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Sphingomonas paucimobilis | 2 (0.3) | 0 (0.0) | 2 (0.4) | - | 1.000 |
| Stenotrophomonas maltophilia | 7 (1.05) | 1 (0.8) | 6 (1.1) | - | 0.770 |
| Burkholderia cepacia | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Pandoraea spp. | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Non-Fermentative Gram-negative rod | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Gram-positive cocci | 300 (44.7) | 59 (48.8) | 231 (40.1) | 10 (52.6) | 0.294 |
| Methicillin resistant Staphylococcus aureus | 27 (4.0) | 3 (2.5) | 24 (4.5) | - | 0.309 |
| Methicillin sensitive Staphylococcus aureus | 53 (7.9) | 7 (5.8) | 43 (8.1) | 3 (15.8) | 0.388 |
| Methicillin resistant coagulase negative staphylococcus | 102 (15.2) | 26 (21.5) | 72 (13.5) | 4 (21.05) | 0.028 |
| Methicillin sensitive coagulase negative staphylococcus | 49 (7.3) | 12 (9.9) | 35 (6.6) | 2 (10.5) | 0.202 |
| Enterococcus faecalis | 12 (1.8) | 2 (1.65) | 10 (1.9) | - | 0.865 |
| Enterococcus faecium | 12 (1.8) | 3 (2.5) | 9 (1.7) | - | 0.562 |
| Enterococcus avium | 4 (0.6) | 0 (0.0) | 4 (0.75) | - | 1.000 |
| Enterococcus gallinarum | 3 (0.45) | 1 (0.8) | 2 (0.4) | - | 0.509 |
| Enterococcus spp. | 16 (2.4) | 2 (1.65) | 14 (2.6) | - | 0.528 |
| Streptococcus pneumoniae | 4 (0.6) | 2 (1.65) | 2 (0.4) | - | 0.105 |
| Streptococcus agalactiae | 3 (0.45) | 0 (0.0) | 3 (0.6) | - | 1.000 |
| Streptococcus gallolyticus | 2 (0.3) | 0 (0.0) | 2 (0.4) | - | 1.000 |
| Streptococcus equi | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Leuconostoc pseudomesenteroides | 2 (0.3) | 0 (0.0) | 1 (0.2) | 1 (5.3) | 1.000 |
| Alpha hemolytic streptococcus | 7 (1.05) | 0 (0.0) | 7 (1.3) | - | 0.359 |
| Beta hemolytic streptococcus | 2 (0.3) | 0 (0.0) | 2 (0.4) | - | 1.000 |
| Non-hemolytic streptococcus | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Gram-positive rods | 3 (0.45) | 1 (0.8) | 2 (0.4) | 0 (0) | 0.509 |
| Corynebacterium jeikeium | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Corynebacterium striatum | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Lactobacillus casei | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Other bacteria | 5 (0.7) | 2 (1.65) | 3 (0.6) | 0 (0) | 0.216 |
| Listeria monocytogenes | 2 (0.3) | 1 (0.8) | 1 (0.2) | - | 0.251 |
| Campylobacter coli | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Campylobacter jejuni | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Moraxella nonliquefaciens | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Anaerobic bacteria | 7 (1.05) | 1 (0.8) | 6 (1.13) | 0 (0) | 0.770 |
| Bacteroides fragilis | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Bacteroides spp. | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Prevotella spp. | 2 (0.3) | 0 (0.0) | 2 (0.4) | - | 1.000 |
| Clostridium clostridioforme | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Fusobacterium nucleatum | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Anaerobic Gram-positive rod | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Fungi | 38 (5.7) | 8 (6.6) | 27 (5.1) | 3 (15.8) | 0.501 |
| Candida albicans | 15 (2.2) | 2 (1.65) | 11 (2.1) | 2 (10.5) | 0.766 |
| Candida parapsilosis complex | 7 (1.05) | 1 (0.8) | 6 (1.1) | - | 0.770 |
| Candida tropicalis | 6 (0.9) | 0 (0.0) | 5 (0.9) | 1 (5.3) | 0.590 |
| Candida kefyr | 2 (0.3) | 1 (0.8) | 1 (0.2) | - | 0.252 |
| Candida glabrata complex | 2 (0.3) | 2 (1.65) | 0 (0.0) | - | 0.034 |
| Candida metapsilosis | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Candida krusei | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Candida auris | 1 (0.15) | 1 (0.8) | 0 (0.0) | - | 0.186 |
| Kodamaea ohmeri | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Cryptococcus neoformans | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Rhodotorula spp. | 1 (0.15) | 0 (0.0) | 1 (0.2) | - | 1.000 |
| Fungi | Antifungal MIC, µg/ml | |||||||
|---|---|---|---|---|---|---|---|---|
| Species (tested/total n) | Strain No | Fluconazole | Posaconazole | Voriconazole | İtraconazole | Amphotericin B | Caspofungin | Anidulafungin |
| Candida albicans(7/15) | 1 | 2 (S) | 0.064ᵃ (NWT) | 0.25 (I) | - | 0.5ᵃ (WT) | 0.016 (S) | 0.012 (S) |
| 2 | 2 (S) | 0.064ᵃ (NWT) | 0.047 (S) | - | - | - | - | |
| 3 | 0.75 (S) | - | - | - | - | 0.5 (I) | - | |
| 4 | 2 (S) | - | - | - | 0.25ᵃ (WT) | 0.5 (I) | 0.012 (S) | |
| 5 | 1,5 (S) | - | - | - | - | - | - | |
| 6 | - | 0.25ᵃ (NWT) | 1 (R) | - | - | 0.065 (S) | - | |
| 7 | 0.125 (S) | - | - | - | 0.047ᵃ (WT) | 0.096 (S) | 0.003 (S) | |
| Candida parapsilosiscomplex (3/7) | 1 | 0.75 (S) | - | - | - | 0.25ᵃ (WT) | 0.75 (S) | - |
| 2 | >256 (R) | 0.19ᵃ (WT) | 0.5 (I) | 2c | 0.75ᵃ (WT) | 0.38 (S) | 0.75 (S) | |
| 3 | 24 (R) | 0.25ᵃ (WT) | 0.75 (S) | |||||
| Candida tropicalis(2/6) | 1 | 0.5 (S) | - | 0.008 (S) | - | 0.25ᵃ (WT) | 0.094 (S) | 0.008 (S) |
| 2 | 0.5 (S) | - | - | - | 0.25ᵃ (WT) | - | 0.008 (S) | |
| Candida glabratacomplex (1/2) | 1.5 (SDD) | - | 0.032ᵃ (WT) | - | - | 0.25 (I) | - | |
| Candida auris(1/1)b | >256 (R) | 0.016c | 0.19c | 0,19 c | 3 (R) | 1 (S) | 0.094 (S) | |
| Cryptococcus neoformans(1/1) | 8ᵃ (WT) | - | - | - | 0.5ᵃ (WT) | - | - | |
| Unit | Microorganism | ||
|---|---|---|---|
| SARS-CoV-2 (+) [n=8] | Inpatient (n=3) | Surgical (n=1) | Candida krusei, Kodamea ohmeri |
| Internal (n=2) | Proteus mirabilis, Escherichia coli | ||
| Klebsiella pneumoniae, Candida glabrata complex | |||
| Outpatient (n=4) | Surgical (n=1) | Candida glabrata complex, Candida albicans | |
| Internal (n=3) | Streptococcus pneumoniae, Escherichia coli | ||
| Candida kefyr, Enterococcus gallinarum, Enterococcus faecium | |||
| Escherichia coli, Enterobacter spp. | |||
| Intensive Care Unit (n=1) | Surgical (n=0) | - | |
| Internal (n=1) | Enterococcus spp., Candida albicans | ||
| SARS-CoV-2 (-) [n=7] | Inpatient (n=1) | Surgical (n=2) | Candida albicans, Candida parapsilosis |
| Escherichia coli, Candida parapsilosis | |||
| Internal (n=1) | Pseudomonas aeruginosa, Acinetobacter spp. | ||
| Outpatient (n=5) | Surgical (n=1) | Citrobacter spp., Klebsiella oxytoca | |
| Internal (n=4) | Raoultella planticola, Escherichia coli | ||
| Klebsiella pneumoniae, Enterococcus spp. | |||
| Enterococcus spp., Escherichia coli, MSSA* | |||
| Enterococcus spp., Escherichia coli | |||
| Intensive Care Unit (n=1) | Surgical (n=0) | - | |
| Internal (n=1) | Proteus mirabilis, Klebsiella pneumoniae | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





