Submitted:
10 August 2023
Posted:
11 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Swiss ADME
2.2. Physicochemical properties
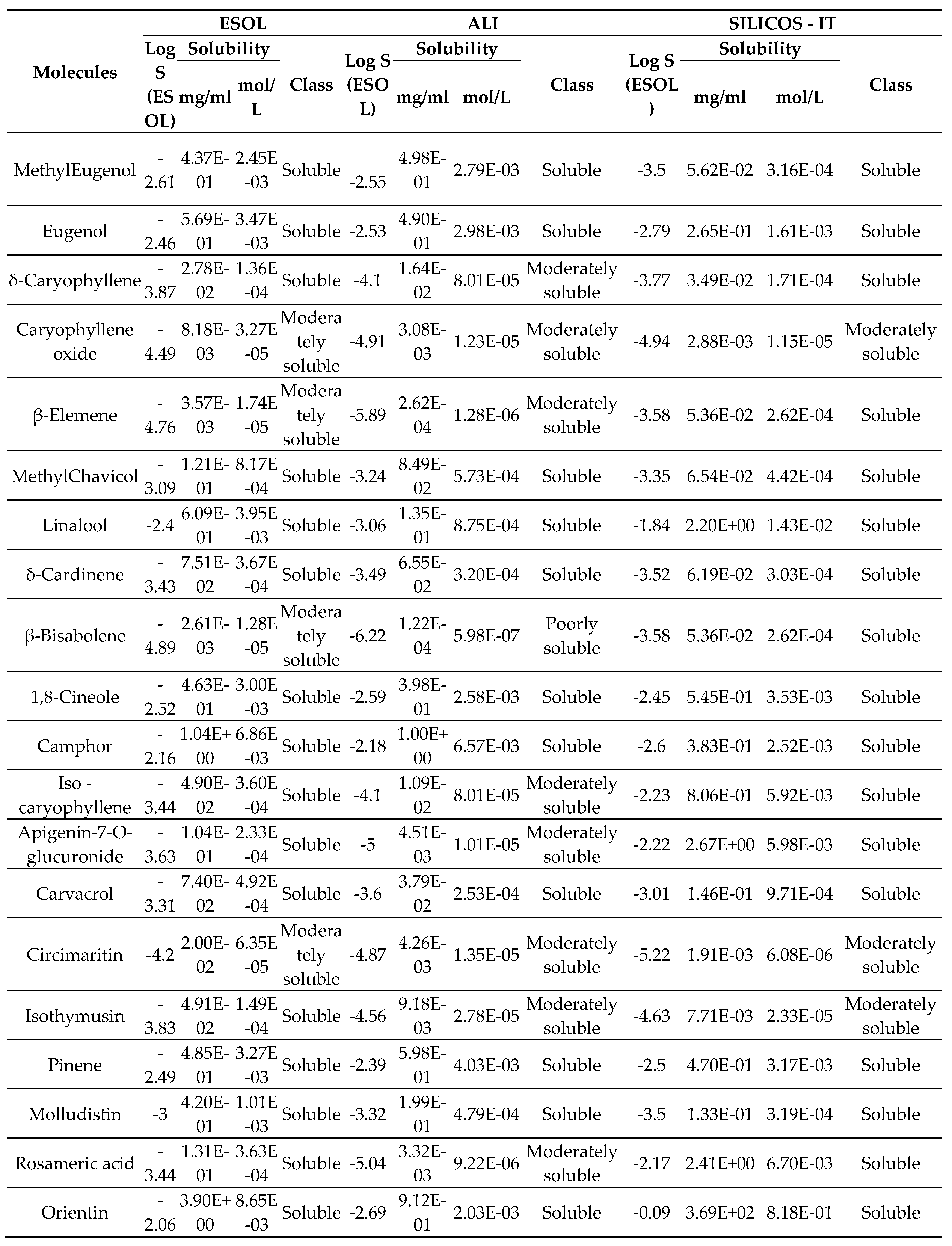
2.3. Solubility
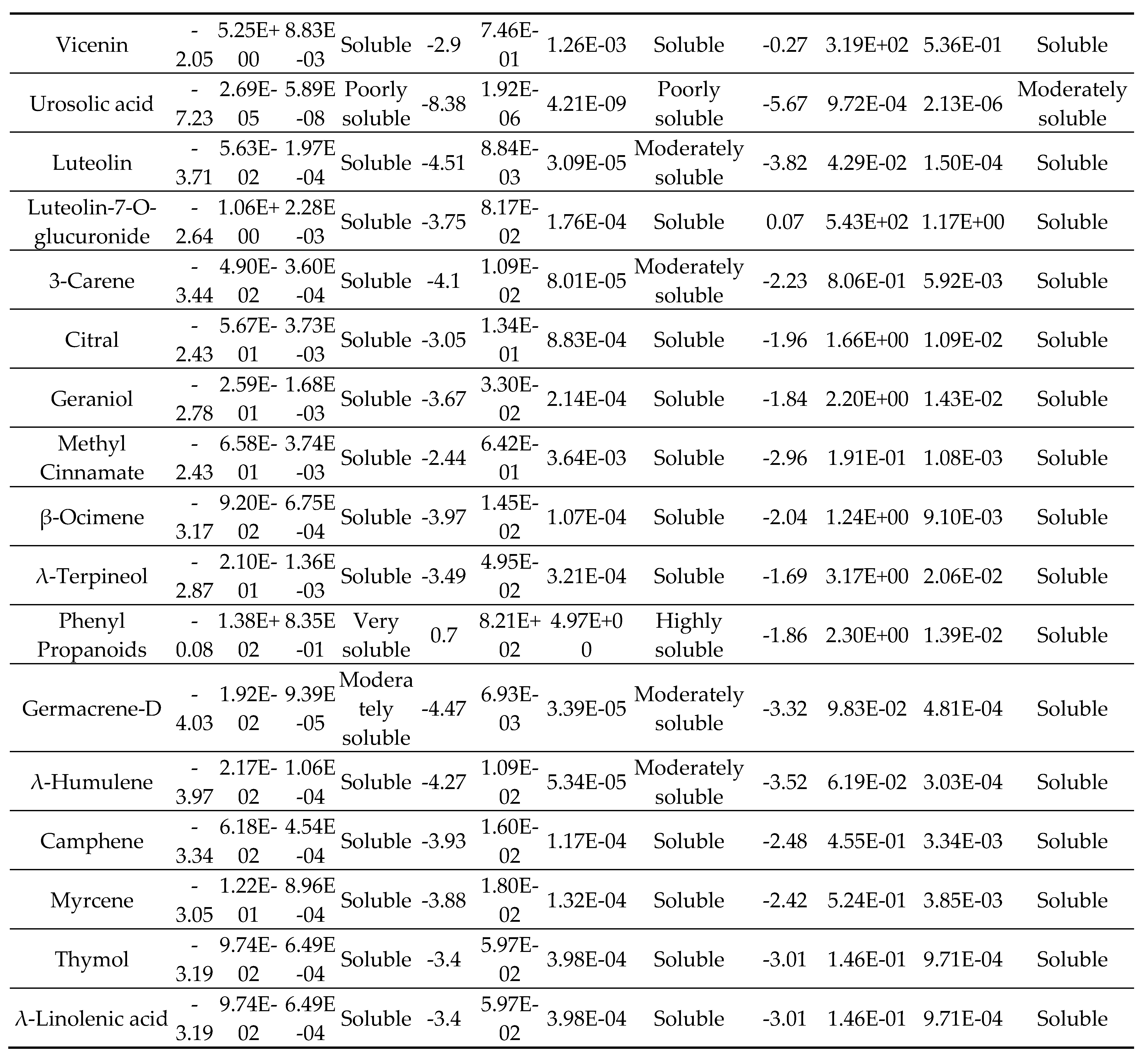
2.4. Lipophilicity
2.5. Drug likeness
2.6. Medicinal Chemistry
2.7. Pharmacokinetics
3. Results
| Sl.No | Molecules | Formula | Molecular Weight (in g/mol) |
Canonical SMILES |
|---|---|---|---|---|
| 1 | MethylEugenol | C11H14O2 | 178.23 | C=CCc1ccc(c(c1)OC)OC |
| 2 | Eugenol | C10H12O2 | 164.2 | C=CCc1ccc(c(c1)OC)O |
| 3 | δ-Caryophyllene | C15H24 | 204.35 | C/C/1=C\CCC(=C)[C@@H]2 [C@@H](CC1)C(C2)(C)C |
| 4 | Caryophyllene oxide | C17H30O | 250.42 | C[C@]12CC[C@@]3(C)O[C@]3 (CCC[C@@]2(CC1(C)C)C)C |
| 5 | β-Elemene | C15H24 | 204.35 | C=C[C@]1(C)CC[C@H] (C[C@H]1C(=C)C)C(=C)C |
| 6 | MethylChavicol | C10H12O | 148.2 | COc1ccc(cc1)CC=C |
| 7 | Linalool | C10H18O | 154.25 | C=C[C@](CCC=C(C)C)(O)C |
| 8 | δ-Cardinene | C15H24 | 204.35 | CC1=C[C@H]2C(=C(C) CC[C@@H]2C(C)C)CC1 |
| 9 | β-Bisabolene | C15H24 | 204.35 | CC(=CCCC(=C)[C@H] 1CCC(=CC1)C)C |
| 10 | 1,8-Cineole | C10H16 | 154.25 | CC12CCC(CC1)C(O2)(C)C |
| 11 | Camphor | C10H16O | 152.23 | O=C1CC2C(C1(C)CC2)(C)C |
| 12 | Isocaryophyllene | C10H16 | 136.23 | CC1=CCC2C(C1)C2(C)C |
| 13 | Apigenin-7-O-glucuronide | C21H18O11 | 446.36 | OC(=O)[C@H]1O[C@H](Oc2cc(O)c3c(c2)oc(cc3=O) c2ccc(cc2)O)[C@@H]([C@H]([C@@H]1O)O)O |
| 14 | Carvacrol | C10H14O | 150.22 | CC(c1ccc(c(c1)O)C)C |
| 15 | Circimaritin | C17H14O6 | 314.29 | COc1cc2oc(cc(=O)c2c (c1OC)O)c1ccc(cc1)O |
| 16 | Isothymusin | C17H14O7 | 330.29 | COc1c(O)c2oc(cc(=O)c 2c(c1OC)O)c1ccc(cc1)O |
| 17 | Pinene | C11H16 | 148.24 | CC1=CCC23C1C(C)(C)C2C3 |
| 18 | Molludistin | C21H20O9 | 416.38 | COc1cc(O)c2c(c1[C@@H]1OC[C@@H] ([C@@H](C1O)O)O)oc(cc2=O)c1ccc(cc1)O |
| 19 | Rosameric acid | C18H16O8 | 360.31 | O=C(O[C@@H](C(=O)O)Cc1ccc(c(c1) O)O)/C=C/c1ccc(c(c1)O)O |
| 20 | Orientin | C21H22O11 | 450.39 | OC[C@H]1O[C@@H]([C@@H]([C@H]([C@@H] 1O)O)O)c1c(O)cc(c2c1OC(=CC2O)c1ccc(c(c1)O)O)O |
| 21 | Vicenin | C27H30O15 | 594.52 | OC[C@H]1O[C@H]([C@@H]([C@H]([C@@H]1O) O)O)c1c(O)c([C@@H]2O[C@H](CO)[C@H]([C@@H] ([C@H]2O)O)O)c(c2c1oc(cc2=O)c1ccc(cc1)O)O |
| 22 | Urosolic acid | C30H48O3 | 456.7 | C[C@@H]1CC[C@]2([C@@H]([C@H]1C) C1=CC[C@H]3[C@@]([C@@]1(CC2)C)(C)CC [C@@H]1[C@]3(C)CC[C@@H](C1(C)C)O)C(=O)O |
| 23 | Luteolin | C15H10O6 | 286.24 | Oc1cc(O)c2c(c1)oc(cc2=O)c1ccc(c(c1)O)O |
| 24 | Luteolin-7-O-glucuronide | C21H20O12 | 464.38 | OC(=O)[C@H]1O[C@@H](Oc2cc3OC(=CC(c3c(c2)O) O)c2ccc(c(c2)O)O)[C@@H]([C@H]([C@@H]1O)O)O |
| 25 | 3-Carene | C10H16 | 136.23 | CC1=CC[C@@H]2[C@H](C1)C2(C)C |
| 26 | Citral | C10H16O | 152.23 | O=C/C=C(\CCC=C(C)C)/C |
| 27 | Geraniol | C10H18O | 154.25 | OC/C=C(/CCC=C(C)C)\C |
| 28 | Methyl Cinnamate | C11H12O2 | 176.21 | CC(=O)OC/C=C/c1ccccc1 |
| 29 | β-Ocimene | C10H16 | 136.23 | C=C/C(=C\CC=C(C)C)/C |
| 30 | λ-Terpineol | C10H18O | 154.25 | CC1=CCC(CC1)C(O)(C)C |
| 31 | PhenylPropanoids | C9H11NO2 | 165.19 | N[C@H](C(=O)O)Cc1ccccc1 |
| 32 | Germacrene-D | C15H24 | 204.35 | C/C/1=C/CCC(=C)/C=C\[C@@H](CC1)C(C)C |
| 33 | λ-Humulene | C15H24 | 204.35 | C/C/1=C\CC(C)(C)/C=C/C/C(=C/CC1)/C |
| 34 | Camphene | C10H16 | 136.23 | C=C1C2CCC(C1(C)C)C2 |
| 35 | Myrcene | C10H16 | 136.23 | C=CC(=C)CCC=C(C)C |
| 36 | Thymol | C10H14O | 150.22 | Cc1ccc(c(c1)O)C(C)C |
| 37 | λ-Linolenic acid | C18H30O2 | 278.43 | CC/C=C\C/C=C\C/C=C\CCCCCCCC(=O)O |
| Sl.No | Molecules | Heavy atoms | Aromatic heavy atoms | Fraction Csp3 | Rotatable bonds | H-bond acceptors | H-bond donors | Molar Refractivity | TPSA |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MethylEugenol | 13 | 6 | 0.27 | 4 | 2 | 0 | 53.53 | 18.46 |
| 2 | Eugenol | 12 | 6 | 0.2 | 3 | 2 | 1 | 49.06 | 29.46 |
| 3 | δ-Caryophyllene | 15 | 0 | 0.73 | 0 | 0 | 0 | 68.78 | 0 |
| 4 | Caryophyllene oxide | 18 | 0 | 1 | 0 | 1 | 0 | 77.87 | 12.53 |
| 5 | β-Elemene | 15 | 0 | 0.6 | 3 | 0 | 0 | 70.42 | 0 |
| 6 | MethylChavicol | 11 | 6 | 0.2 | 3 | 1 | 0 | 47.04 | 9.23 |
| 7 | Linalool | 11 | 0 | 0.6 | 4 | 1 | 1 | 50.44 | 20.23 |
| 8 | δ-Cardinene | 15 | 0 | 0.73 | 1 | 0 | 0 | 69.04 | 0 |
| 9 | β-Bisabolene | 15 | 0 | 0.6 | 4 | 0 | 0 | 70.68 | 0 |
| 10 | 1,8-Cineole | 11 | 0 | 1 | 0 | 1 | 0 | 47.12 | 9.23 |
| 11 | Camphor | 11 | 0 | 0.9 | 0 | 1 | 0 | 45.64 | 17.07 |
| 12 | Isocaryophyllene | 10 | 0 | 0.8 | 0 | 0 | 0 | 45.22 | 0 |
| 13 | Apigenin-7-O-glucuronide | 32 | 16 | 0.24 | 4 | 11 | 6 | 106.72 | 187.12 |
| 14 | Carvacrol | 11 | 6 | 0.4 | 1 | 1 | 1 | 48.01 | 20.23 |
| 15 | Circimaritin | 23 | 16 | 0.12 | 3 | 6 | 2 | 84.95 | 89.13 |
| 16 | Isothymusin | 24 | 16 | 0.12 | 3 | 7 | 3 | 86.97 | 109.36 |
| 17 | Pinene | 11 | 0 | 0.82 | 0 | 0 | 0 | 47.66 | 0 |
| 18 | Molludistin | 30 | 16 | 0.29 | 3 | 9 | 5 | 105.11 | 149.82 |
| 19 | Rosameric acid | 26 | 12 | 0.11 | 7 | 8 | 5 | 91.4 | 144.52 |
| 20 | Orientin | 32 | 12 | 0.33 | 3 | 11 | 9 | 107.27 | 200.53 |
| 21 | Vicenin | 42 | 16 | 0.44 | 5 | 15 | 11 | 139.23 | 271.2 |
| 22 | Urosolic acid | 33 | 0 | 0.9 | 1 | 3 | 2 | 136.91 | 57.53 |
| 23 | Luteolin | 21 | 16 | 0 | 1 | 6 | 4 | 76.01 | 111.13 |
| 24 | Luteolin-7-O-glucuronide | 33 | 12 | 0.29 | 4 | 12 | 8 | 107.38 | 206.6 |
| 25 | 3-Carene | 10 | 0 | 0.8 | 0 | 0 | 0 | 45.22 | 0 |
| 26 | Citral | 11 | 0 | 0.5 | 4 | 1 | 0 | 49.44 | 17.07 |
| 27 | Geraniol | 11 | 0 | 0.6 | 4 | 1 | 1 | 50.4 | 20.23 |
| 28 | Methyl Cinnamate | 13 | 6 | 0.18 | 4 | 2 | 0 | 52.24 | 26.3 |
| 29 | β-Ocimene | 10 | 0 | 0.4 | 3 | 0 | 0 | 48.76 | 0 |
| 30 | λ-Terpineol | 11 | 0 | 0.8 | 1 | 1 | 1 | 48.8 | 20.23 |
| 31 | PhenylPropanoids | 12 | 6 | 0.22 | 3 | 3 | 2 | 45.5 | 63.32 |
| 32 | Germacrene-D | 15 | 0 | 0.6 | 1 | 0 | 0 | 70.68 | 0 |
| 33 | λ-Humulene | 15 | 0 | 0.6 | 0 | 0 | 0 | 70.42 | 0 |
| 34 | Camphene | 10 | 0 | 0.8 | 0 | 0 | 0 | 45.22 | 0 |
| 35 | Myrcene | 10 | 0 | 0.4 | 4 | 0 | 0 | 48.76 | 0 |
| 36 | Thymol | 11 | 6 | 0.4 | 1 | 1 | 1 | 48.01 | 20.23 |
| 37 | λ-Linolenic acid | 20 | 0 | 0.61 | 13 | 2 | 1 | 88.99 | 37.3 |
| Sl.No | Molecules | iLOGP | XLOGP3 | WLOGP | MLOGP | Silicos-IT Log P | Consensus Log P |
|---|---|---|---|---|---|---|---|
| 1 | MethylEugenol | 2.65 | 2.52 | 2.43 | 2.3 | 3 | 2.58 |
| 2 | Eugenol | 2.37 | 2.27 | 2.13 | 2.01 | 2.48 | 2.25 |
| 3 | δ-Caryophyllene | 3.25 | 4.38 | 4.73 | 4.63 | 4.19 | 4.24 |
| 4 | Caryophyllene oxide | 3.53 | 4.91 | 4.94 | 4.31 | 5.15 | 4.57 |
| 5 | β-Elemene | 3.37 | 6.11 | 4.75 | 4.53 | 4.5 | 4.65 |
| 6 | MethylChavicol | 2.47 | 3.37 | 2.42 | 2.67 | 2.96 | 2.78 |
| 7 | Linalool | 2.7 | 2.97 | 2.67 | 2.59 | 2.35 | 2.66 |
| 8 | δ-Cardinene | 3.41 | 3.8 | 4.73 | 4.63 | 4.12 | 4.14 |
| 9 | β-Bisabolene | 3.67 | 6.43 | 5.04 | 4.53 | 4.5 | 4.83 |
| 10 | 1,8-Cineole | 2.58 | 2.74 | 2.74 | 2.45 | 2.86 | 2.67 |
| 11 | Camphor | 2.12 | 2.19 | 2.4 | 2.3 | 2.85 | 2.37 |
| 12 | Isocaryophyllene | 2.63 | 4.38 | 3 | 4.29 | 2.79 | 3.42 |
| 13 | Apigenin-7-O-glucuronide | 1 | 1.46 | 0.14 | -1.63 | -0.1 | 0.17 |
| 14 | Carvacrol | 2.24 | 3.49 | 2.82 | 2.76 | 2.79 | 2.82 |
| 15 | Circimaritin | 2.56 | 3.32 | 2.89 | 0.47 | 3.07 | 2.46 |
| 16 | Isothymusin | 2.58 | 2.61 | 2.59 | -0.07 | 2.59 | 2.06 |
| 17 | Pinene | 2.66 | 2.74 | 3 | 4.58 | 3.06 | 3.21 |
| 18 | Molludistin | 2.21 | 0.6 | 0.71 | -1.25 | 1.37 | 0.73 |
| 19 | Rosameric acid | 1.48 | 2.36 | 1.65 | 0.9 | 1.5 | 1.58 |
| 20 | Orientin | 1.02 | -1.03 | -1.16 | -2.32 | -1.21 | -0.94 |
| 21 | Vicenin | 1.73 | -2.26 | -3.04 | -4.51 | -1.8 | -1.98 |
| 22 | Urosolic acid | 3.95 | 7.34 | 7.09 | 5.82 | 5.46 | 5.93 |
| 23 | Luteolin | 1.86 | 2.53 | 2.28 | -0.03 | 2.03 | 1.73 |
| 24 | Luteolin-7-O-glucuronide | 1.79 | -0.13 | -0.78 | -1.94 | -1.64 | -0.54 |
| 25 | 3-Carene | 2.63 | 4.38 | 3 | 4.29 | 2.79 | 3.42 |
| 26 | Citral | 2.47 | 3.03 | 2.88 | 2.49 | 2.65 | 2.71 |
| 27 | Geraniol | 2.52 | 3.56 | 2.67 | 2.59 | 2.35 | 2.74 |
| 28 | Methyl Cinnamate | 2.17 | 2.25 | 2.15 | 2.49 | 2.57 | 2.33 |
| 29 | β-Ocimene | 2.91 | 4.26 | 3.48 | 3.56 | 2.88 | 3.42 |
| 30 | λ-Terpineol | 2.51 | 3.39 | 2.5 | 2.3 | 2.17 | 2.58 |
| 31 | PhenylPropanoids | 1.08 | -1.52 | 0.64 | -1.11 | 0.86 | -0.01 |
| 32 | Germacrene-D | 3.14 | 4.74 | 4.89 | 4.53 | 4.01 | 4.26 |
| 33 | λ-Humulene | 3.29 | 4.55 | 5.04 | 4.53 | 3.91 | 4.26 |
| 34 | Camphene | 2.58 | 4.22 | 3 | 4.29 | 3.08 | 3.43 |
| 35 | Myrcene | 2.89 | 4.17 | 3.48 | 3.56 | 3.05 | 3.43 |
| 36 | Thymol | 2.32 | 3.3 | 2.82 | 2.76 | 2.79 | 2.8 |
| 37 | λ-Linolenic acid | 2.32 | 3.3 | 5.66 | 2.76 | 2.79 | 2.8 |
| Sl.No | Molecules | Lipinski violations | Ghose violations | Veber violations | Egan violations | Muegge violations | Bioavailability Score |
|---|---|---|---|---|---|---|---|
| 1 | MethylEugenol | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 2 | Eugenol | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 3 | δ-Caryophyllene | 1 | 0 | 0 | 0 | 1 | 0.55 |
| 4 | Caryophyllene oxide | 1 | 0 | 0 | 0 | 1 | 0.55 |
| 5 | β-Elemene | 1 | 0 | 0 | 0 | 2 | 0.55 |
| 6 | MethylChavicol | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 7 | Linalool | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 8 | δ-Cardinene | 1 | 0 | 0 | 0 | 1 | 0.55 |
| 9 | β-Bisabolene | 1 | 0 | 0 | 0 | 2 | 0.55 |
| 10 | 1,8-Cineole | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 11 | Camphor | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 12 | Isocaryophyllene | 1 | 1 | 0 | 0 | 2 | 0.55 |
| 13 | Apigenin-7-O-glucuronide | 2 | 0 | 1 | 1 | 3 | 0.11 |
| 14 | Carvacrol | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 15 | Circimaritin | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 16 | Isothymusin | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 17 | Pinene | 1 | 1 | 0 | 0 | 2 | 0.55 |
| 18 | Molludistin | 0 | 0 | 1 | 1 | 0 | 0.55 |
| 19 | Rosameric acid | 0 | 0 | 1 | 1 | 0 | 0.56 |
| 20 | Orientin | 2 | 1 | 1 | 1 | 3 | 0.17 |
| 21 | Vicenin | 3 | 4 | 1 | 1 | 4 | 0.17 |
| 22 | Urosolic acid | 1 | 3 | 0 | 1 | 1 | 0.85 |
| 23 | Luteolin | 0 | 0 | 0 | 0 | 0 | 0.55 |
| 24 | Luteolin-7-O-glucuronide | 2 | 1 | 1 | 1 | 3 | 0.11 |
| 25 | 3-Carene | 1 | 1 | 0 | 0 | 2 | 0.55 |
| 26 | Citral | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 27 | Geraniol | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 28 | Methyl Cinnamate | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 29 | β-Ocimene | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 30 | λ-Terpineol | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 31 | PhenylPropanoids | 0 | 0 | 0 | 0 | 1 | 0.55 |
| 32 | Germacrene-D | 1 | 0 | 0 | 0 | 1 | 0.55 |
| 33 | λ-Humulene | 1 | 0 | 0 | 0 | 1 | 0.55 |
| 34 | Camphene | 1 | 1 | 0 | 0 | 2 | 0.55 |
| 35 | Myrcene | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 36 | Thymol | 0 | 1 | 0 | 0 | 2 | 0.55 |
| 37 | λ-Linolenic acid | 0 | 1 | 0 | 0 | 2 | 0.55 |
| Sl.No | Molecules | PAINS | Brenk | Leadlikeness | Synthetic Accessibility |
|---|---|---|---|---|---|
| 1 | MethylEugenol | 0 | 1 | 1 | 1.71 |
| 2 | Eugenol | 0 | 1 | 1 | 1.58 |
| 3 | δ-Caryophyllene | 0 | 1 | 2 | 4.51 |
| 4 | Caryophyllene oxide | 0 | 1 | 1 | 4.46 |
| 5 | β-Elemene | 0 | 1 | 2 | 3.63 |
| 6 | MethylChavicol | 0 | 1 | 1 | 1.28 |
| 7 | Linalool | 0 | 1 | 1 | 2.74 |
| 8 | δ-Cardinene | 0 | 1 | 2 | 4.14 |
| 9 | β-Bisabolene | 0 | 1 | 2 | 3.9 |
| 10 | 1,8-Cineole | 0 | 0 | 1 | 3.65 |
| 11 | Camphor | 0 | 0 | 1 | 3.22 |
| 12 | Isocaryophyllene | 0 | 1 | 2 | 3.84 |
| 13 | Apigenin-7-O-glucuronide | 0 | 0 | 1 | 5.06 |
| 14 | Carvacrol | 0 | 0 | 1 | 1 |
| 15 | Circimaritin | 0 | 0 | 0 | 3.27 |
| 16 | Isothymusin | 0 | 1 | 0 | 3.38 |
| 17 | Pinene | 0 | 1 | 1 | 4.81 |
| 18 | Molludistin | 0 | 0 | 1 | 4.91 |
| 19 | Rosameric acid | 1 | 2 | 1 | 3.38 |
| 20 | Orientin | 1 | 1 | 1 | 5.34 |
| 21 | Vicenin | 0 | 0 | 1 | 6.4 |
| 22 | Urosolic acid | 0 | 1 | 2 | 6.21 |
| 23 | Luteolin | 1 | 1 | 0 | 3.02 |
| 24 | Luteolin-7-O-glucuronide | 1 | 1 | 1 | 5.32 |
| 25 | 3-Carene | 0 | 1 | 2 | 3.84 |
| 26 | Citral | 0 | 3 | 1 | 2.49 |
| 27 | Geraniol | 0 | 1 | 2 | 2.58 |
| 28 | Methyl Cinnamate | 0 | 0 | 1 | 1.98 |
| 29 | β-Ocimene | 0 | 2 | 2 | 3.63 |
| 30 | λ-Terpineol | 0 | 1 | 1 | 3.24 |
| 31 | PhenylPropanoids | 0 | 0 | 1 | 1.46 |
| 32 | Germacrene-D | 0 | 1 | 2 | 4.55 |
| 33 | λ-Humulene | 0 | 1 | 2 | 3.66 |
| 34 | Camphene | 0 | 1 | 2 | 3.5 |
| 35 | Myrcene | 0 | 2 | 2 | 2.85 |
| 36 | Thymol | 0 | 0 | 1 | 1 |
| 37 | λ-Linolenic acid | 0 | 0 | 1 | 1 |
| Sl.No | Molecules | GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | log Kp (cm/s) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MethylEugenol | High | Yes | No | Yes | No | No | No | No | -5.6 |
| 2 | Eugenol | High | Yes | No | Yes | No | No | No | No | -5.69 |
| 3 | δ-Caryophyllene | Low | No | No | No | Yes | Yes | No | No | -4.44 |
| 4 | Caryophyllene oxide | High | Yes | No | No | No | Yes | No | No | -4.34 |
| 5 | β-Elemene | Low | No | No | No | Yes | Yes | No | No | -3.21 |
| 6 | MethylChavicol | High | Yes | No | Yes | No | No | No | No | -4.81 |
| 7 | Linalool | High | Yes | No | No | No | No | No | No | -5.13 |
| 8 | δ-Cardinene | Low | No | No | No | Yes | Yes | No | No | -4.85 |
| 9 | β-Bisabolene | Low | No | No | No | No | Yes | No | No | -2.98 |
| 10 | 1,8-Cineole | High | Yes | No | No | No | No | No | No | -5.3 |
| 11 | Camphor | High | Yes | No | No | No | No | No | No | -5.67 |
| 12 | Isocaryophyllene | Low | Yes | No | No | No | Yes | No | No | -4.02 |
| 13 | Apigenin-7-O-glucuronide | Low | No | Yes | No | No | No | No | No | -7.99 |
| 14 | Carvacrol | High | Yes | No | Yes | No | No | No | No | -4.74 |
| 15 | Circimaritin | High | No | No | Yes | No | Yes | Yes | Yes | -5.86 |
| 16 | Isothymusin | High | No | No | Yes | No | Yes | Yes | Yes | -6.46 |
| 17 | Pinene | Low | Yes | No | No | No | Yes | No | No | -5.26 |
| 18 | Molludistin | Low | No | No | No | No | No | No | No | -8.41 |
| 19 | Rosameric acid | Low | No | No | No | No | No | No | No | -6.82 |
| 20 | Orientin | Low | No | No | No | No | No | No | No | -9.78 |
| 21 | Vicenin | Low | No | Yes | No | No | No | No | No | -11.53 |
| 22 | Urosolic acid | Low | No | No | No | No | No | No | No | -3.87 |
| 23 | Luteolin | High | No | No | Yes | No | No | Yes | Yes | -6.25 |
| 24 | Luteolin-7-O-glucuronide | Low | No | Yes | No | No | No | No | No | -9.22 |
| 25 | 3-Carene | Low | Yes | No | No | No | Yes | No | No | -4.02 |
| 26 | Citral | High | Yes | No | No | No | No | No | No | -5.08 |
| 27 | Geraniol | High | Yes | No | No | No | No | No | No | -4.71 |
| 28 | Methyl Cinnamate | High | Yes | No | No | No | No | No | No | -5.78 |
| 29 | β-Ocimene | Low | Yes | No | No | No | No | No | No | -4.11 |
| 30 | λ-Terpineol | High | Yes | No | No | No | No | No | No | -4.83 |
| 31 | PhenylPropanoids | High | No | No | No | No | No | No | No | -8.39 |
| 32 | Germacrene-D | Low | No | No | No | No | Yes | No | No | -4.18 |
| 33 | λ-Humulene | Low | No | No | No | No | Yes | No | No | -4.32 |
| 34 | Camphene | Low | Yes | No | No | No | Yes | No | No | -4.13 |
| 35 | Myrcene | Low | Yes | No | No | No | No | No | No | -4.17 |
| 36 | Thymol | High | Yes | No | Yes | No | No | No | No | -4.87 |
| 37 | λ-Linolenic acid | High | Yes | No | Yes | No | No | No | No | -4.87 |
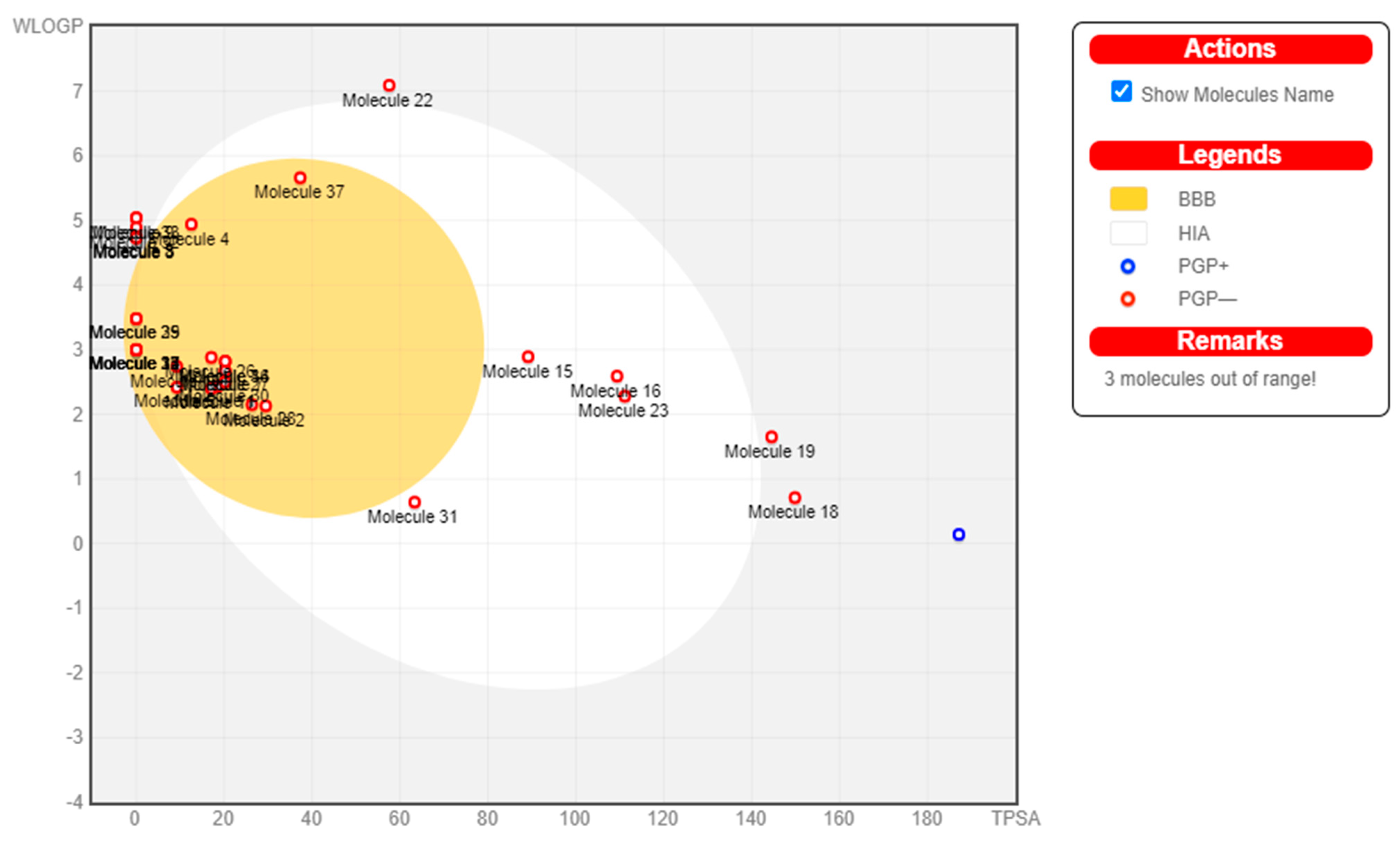
4. Discussion
5. Conclusion
References
- Pattanayak, P.; Behera, P.; Das, D.; Panda, S.K. Ocimum sanctum Linn. A reservoir plant for therapeutic applications: An overview. Pharmacogn. Rev. 2010, 4, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Bast, F.; Rani, P.; Meena, D. Chloroplast DNA Phylogeography of Holy Basil (Ocimum tenuiflorum) in Indian Subcontinent. Sci. World J. 2014, 2014, 847482. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Setzer, W.N.; da Silva, J.K. Phytoconstituents, traditional medicinal uses and bioactivities of Tulsi (Ocimum sanctum Linn. ): A review. American Journal of Essential Oils and Natural Products 2017, 5, 18–21. [Google Scholar]
- Prakash, P.; Gupta, N. Therapeutic uses of Ocimum sanctum Linn (Tulsi) with a note on eugenol and its pharmacological actions: a short review. Indian J. Physiol. Pharmacol. 2005, 49, 125–131. [Google Scholar] [PubMed]
- Viyoch, J.; Pisutthanan, N.; Faikreua, A.; Nupangta, K.; Wangtorpol, K.; Ngokkuen, J. Evaluation of in vitro antimicrobial activity of Thai basil oils and their micro-emulsion formulas against Propionibacterium acnes. Int. J. Cosmet. Sci. 2006, 28, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Magesh, V.; Lee, J.C.; Ahn, K.S.; Lee, H.J.; Lee, E.O.; Shim, B.S.; et al. Ocimum sanctum induces apoptosis in A549 lung cancer cells and suppresses the in vivo growth of Lewis lung carcinoma cells. Phytother Res 2009, 23, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Andola, H.C.; Lohani, H.; Chauhan, N. Pharmacological Review on Ocimum sanctum Linnaeus: A Queen of herbs. Journal of Pharmacy Research 2011, 4, 366–368. [Google Scholar]
- Kumar, A.; Rahal, A.; Chakraborty, S.; et al. Ocimum sanctum (Tulsi): a miracle herb and boon to medical science – A Review. International Journal of Agronomy and Plant Production 2013, 4, 1580–1589. [Google Scholar]
- Mahajan, N.; Rawal, S.; et al. A phytopharmacological overview on Ocimum species with special emphasis on Ocimum sanctum. Biomedicine & Preventive Nutrition 2013, 3, 185–192. [Google Scholar]
- Lachman LH, Lieberman, Kanig JL. The Theory and Practice of Industrial Pharmacy, Lea & Febiger, 3rd edition, 1986.
- Yalkowsky, S.H.; Valvani, S.C. Solubility and partitioning I: Solubility of nonelectrolytes in water. J. Pharm. Sci. 1980, 69, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Kah, M.; Brown, C.D. LogD: Lipophilicity for ionisable compounds. Chemosphere 2008, 72, 1401–1408. [Google Scholar] [CrossRef]
- Constantinescu, T.; Lungu, C.N.; Lung, I. Lipophilicity as a Central Component of Drug-Like Properties of Chalchones and Flavonoid Derivatives. Molecules 2019, 24, 1505. [Google Scholar] [CrossRef]
- Giaginis, C.; Tsopelas, F.; Tsantili-Kakoulidou, A. The Impact of Lipophilicity in Drug Discovery: Rapid Measurements by Means of Reversed-Phase HPLC. 2018, 1824, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.S.; Belal, A.; Aboelez, M.O.; Shokr, E.K.; Abdel-Ghany, H.; Mansour, H.S.; Shawky, A.M.; El-Remaily, M.A.E.A.A.A. Microwave-Assisted Synthesis, Biological Activity Evaluation, Molecular Docking, and ADMET Studies of Some Novel Pyrrolo [2,3-b] Pyrrole Derivatives. Molecules 2022, 27, 2061. [Google Scholar] [CrossRef] [PubMed]
- Erckes, V.; Steuer, C. A story of peptides, lipophilicity and chromatography—Back and forth in time. RSC Med. Chem. 2022, 22, 676–687. [Google Scholar] [CrossRef]
- Ginex, T.; Vazquez, J.; Gilbert, E.; Herrero, E.; Luque, F.J. Lipophilicity in drug design: an overview of lipophilicity descriptors in 3D-QSAR studies. Futur. Med. Chem. 2019, 11, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Kempińska, D.; Chmiel, T.; Kot-Wasik, A.; Mróz, A.; Mazerska, Z.; Namieśnik, J. State of the art and prospects of methods for determination of lipophilicity of chemical compounds. TrAC Trends Anal. Chem. 2019, 113, 54–73. [Google Scholar] [CrossRef]
- Dołowy, M.; Jampilek, J.; Bober-Majnusz, K. A Comparative Study of the Lipophilicity of Metformin and Phenformin. Molecules 2021, 26, 6613. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; et al. Computation of Octanol Water Partition Coefficients by Guiding an Additive Model with Knowledge. J Chem Inf. Model. 2007, 47, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Wildman, S.A.; Crippen, G.M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Moriguchi, I.; Hirono, S.; Nakagome, I.; Hirano, H. Comparison of Reliability of log P Values for Drugs Calculated by Several Methods. Chem. Pharm. Bull. 1994, 42, 976–978. [Google Scholar] [CrossRef]
- Antoine, D.; Michielin, O.; Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug likeness and medicinal chemistry friendliness of small molecules. Nature - Scientific Reports 2017, 7, 42717.1–13. [Google Scholar]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods: An Analysis of ALOGP and CLOGP Methods. J. Phys. Chem. A 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D.; et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Martin, Y.C. A Bioavailability Score. J. Med. Chem. 2005, 48, 3164–3170. [Google Scholar] [CrossRef] [PubMed]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [PubMed]
- Teague, S.; Davis, A.; Leeson, P.; Oprea, T. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed. Engl. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Hann, M.M.; Keserü, G.M. Finding the sweet spot: the role of nature and nurture in medicinal chemistry. Nat. Rev. Drug Discov. 2012, 11, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Brito Sanchez, Y.; Marrero-Ponce, Y.; Barigye, S.J.; Yaber Goenaga, I.; Morell Prez, C.; et al. Mol. Inf. 2015, 34, 308–330. [Google Scholar]
- Di, L.P.; Artursson, A.; Avdeef, G.F.; Ecker, B.; Faller, H.; Fischer, J.B.; et al. Drug Discov. Today 2012, 17, 905–912. [Google Scholar]
- Montanari, F.; Ecker, G. F. Prediction of drug-ABC-transporter interaction–Recent advances and future challenges. Adv. Drug Deliv. Rev. 2015, 86, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Testa, B.; Kraemer, S. D. The Biochemistry of Drug Metabolism – An Introduction - Testa - 2007 - Chemistry & Biodiversity - Wiley Online Library. Chem. Biodivers ( 2007.
- Di, L. The role of drug metabolizing enzymes in clearance. Expert Opin. Drug Metab. Toxicol. 2014, 10, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Mishra, N.K.; Agarwal, S.; Raghava, G.P. Prediction of cytochrome P450 isoform responsible for metabolizing a drug molecule. BMC Pharmacol. 2010, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; et al. A Systemic Review of Ocimum sanctum(Tulsi): Morphological Characteristics, Phytoconstituents and Therapeutic Applications. International Journal for Research in Applied Sciences and Biotechnology, Volume-9, Issue-2(March2022).
- Bhattacharya, A.K.; Kaul, P.N.; Rajeswara Rao, B.R. Essentialoils of Ocimum gratissimum L. and Ocimum tenuiflorum L. (Syn. Ocimum sanctum L.) grown in Andhra Pradesh. Indian Perfumer. 1996, 40, 73–75. [Google Scholar]
- Kothari SK, Bhattacharya AK, Ramesh S, Garg SN,Khanuja SPS. Volatile constituents in oil from different plant parts of methyl eugenol-rich Ocimum tenuiflorum L. f. (Syn. O. sanctum L.) grown in South India. Journal of Essential Oil Res. 2005, 17, 656–658.
- Awasthi, P.; Dixit, S. Chemical Compositions ofOcimum sanctumShyama and0cimum sanctumRama Oils from the Plains of Northern India. J. Essent. Oil Bear. Plants 2007, 10, 292–296. [Google Scholar] [CrossRef]
- Kicel, A.; Kurowska, A.; Kalemba, D. Composition of the essential oil of Ocimum sanctum L. grown in Poland during vegetation. Journal of Essential Oil Res. 2005, 17, 217–219. [Google Scholar] [CrossRef]
- Khan, A.; Ahmad, A.; Akhtar, F.; Yousuf, S.; Xess, I.; Khan, L.A.; Manzoor, N. Ocimum sanctum essential oil and its active principles exert their antifungal activity by disrupting ergosterol biosynthesis and membrane integrity. Res. Microbiol. 2010, 161, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.I.L.; Silva, M.G.V.; Matos, F.J.A.; Craveiro, A.A.; Alencar, J.W. Volatile constituents from leaves and inflorescence oil of Ocimum tenuiflorum L. f. (Syn. O.sanctum L.) grown in Northeastern Brazil. Journal of Essential Oil Res. 1999, 11, 324–326. [Google Scholar] [CrossRef]
- Pino, J.A.; Rosado, A.; Rodriguez, M.; Garcia, D. Composition of the essential oil of Ocimum tenuiflorum L. grown in Cuba. Journal of Essential Oil Res. 1988, 10, 437–438. [Google Scholar] [CrossRef]
- Brophy, J.J.; Goldsack, R.J.; Clarkson, J.R. The essential oil of Ocimum tenuiflorum L. (Lamiaceae) growing in Northern Australia. Journal of Essential Oil Res. 1993, 5, 459–461. [Google Scholar] [CrossRef]
- Kashyap, C.; Ranjeet, K.; Vikrant, A.; Vipin, K. Therapeutic Potency of Ocimum Kilimandscharicum Guerke-A Review. Global Journal of Pharmacology 2011, 5, 191–200. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





