Subject areas: biochemistry, structural biology, molecular biology, evolutionary biology, bioinformatics,
In a prokaryotic cell, three ribosomal RNAs (16S rRNA, 23S rRNA, and 5S rRNA) dominate the encoding of phylogenetic information. Specifically, the 16S rRNA is most commonly used to identify various microorganisms due to the rich diversity in phylogeny sequence information encoded in the gene [
1] [
2] [
3]. On the other hand, the 23S rRNA resides in the large subunit of the ribosome, and is less often used in understanding the phylogeny of microbial species [
4] [
5].
In particular, the 23S rRNA could be used to understand the evolution of the interactions between the 23S rRNA and ribosomal proteins that assemble the large ribosome subunit. Similarly, the 16S rRNA phylogeny tells the evolutionary story of interactions between the ribosomal proteins in the small ribosome subunit and 16S rRNA. This work attempts to use multiple sequence alignment and phylogenetic tree construction to understand the phylogeny of 23S rRNA and 16S rRNA in Helicobacter pylori to help tease out its future evolutionary trajectory.
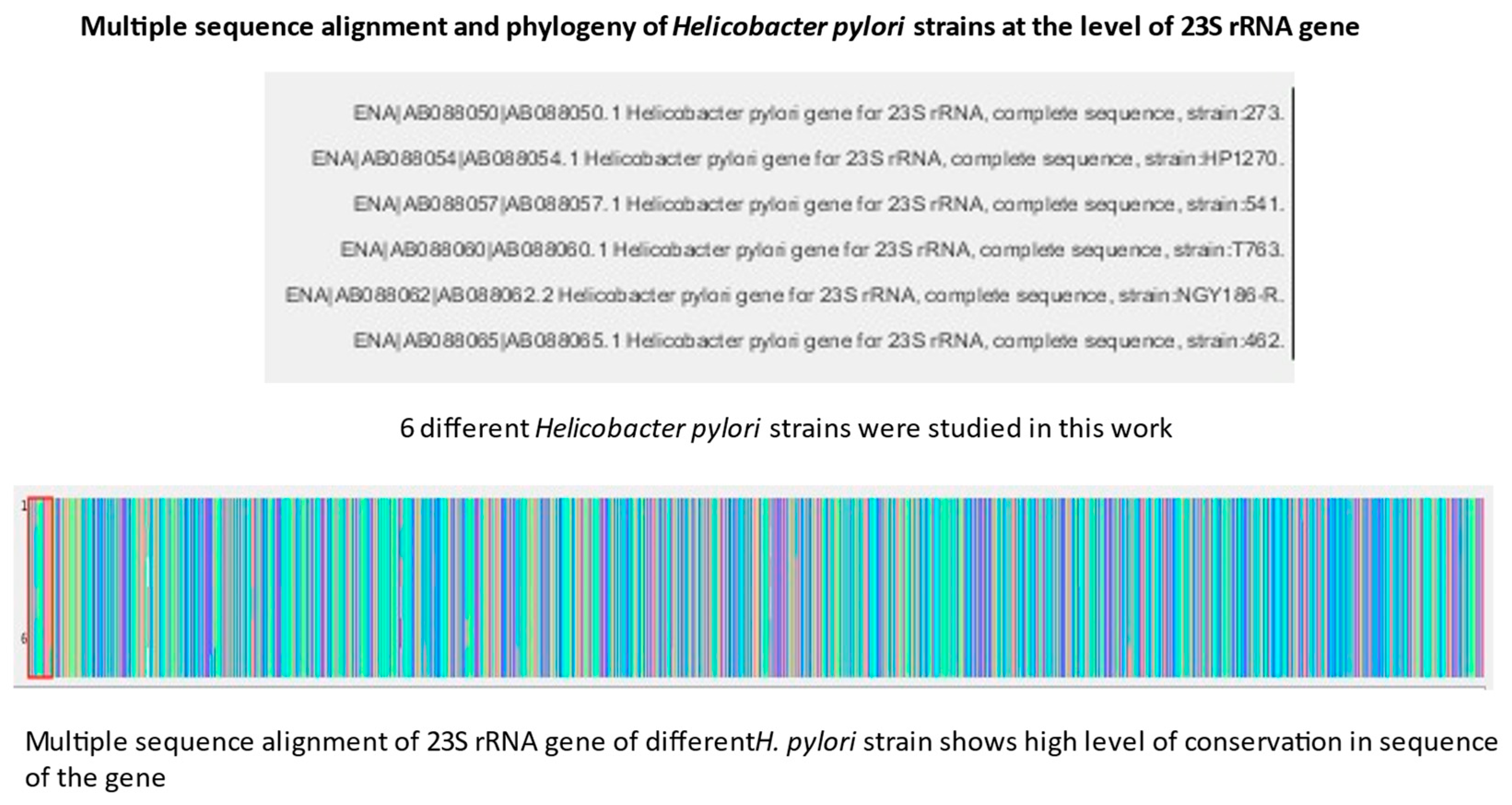
Figure 1 shows the workflow of the study where 23S rRNA gene complete sequence of six
H. pylori strains were downloaded from European Nucleotide Archive via the de Silva database portal. The sequences were processed via multiple sequence alignment in an in-house MATLAB multiple sequence alignment and phylogenetic tree construction software. Finally, the results of the multiple sequence alignment were used to construct the phylogenetic tree of 23S rRNA of
H. pylori. The above procedures were repeated for 16S rRNA gene of
H. pylori.
Phylogenetic tree analysis of 23S rRNA gene of different strains of
H. pylori in
Figure 2 reveals that the sequence has diverged into two main lineages with further subdivisions. This suggests that two major selection forces are acting on the evolutionary trajectories of the 23S rRNA gene, which portends a further where, at the level of 23S rRNA gene, two different functional phenotypes in ribosomes would emerge in
H. pylori strains. Hence, one category of
H. pylori may grow faster compared to the other due to higher functional status of their 23S rRNA gene.
On the other hand, phylogenetic tree analysis in
Figure 3 showed that except for one strain, all other strains of
H. pylori show high level of similarity in their 16S rRNA gene, which indicates that this gene does not encode sufficient sequence divergence to explain the phylogeny of the different
H. pylori strains. In addition, the evolutionary distance of the outlier strain is also not too far away from other
H. pylori strains.
Combining the data and implications of the two phylogenetic tree constructions for 23S rRNA and 16S rRNA gene of H. pylori reveals that the large ribosome subunit of H. pylori is evolving fairly quickly compared to the small ribosome subunit. One possible selection force mediating this quick evolutionary trajectory of the large ribosome subunit of H. pylori is the use of antibiotics targeting the ribosome of H. pylori that engenders the adaptation of the 23S rRNA gene sequence that affords its new binding site for different ribosomal proteins in new orientations. These changes could result in changes to the shape and conformation of the large ribosome subunit that negates the binding of antibiotics used. On the other hand, lack of evolution of the 16S rRNA gene of H. pylori meant that the shape and conformation of the small ribosome subunit of H. pylori remains stable.
Funding
No funding was used in this work.
References
- H. B. Hassler et al., “Phylogenies of the 16S rRNA gene and its hypervariable regions lack concordance with core genome phylogenies,” Microbiome, vol. 10, no. 1, p. 104, Jul. 2022. [CrossRef]
- B. Idris et al., “Molecular Phylogenetic Analysis of 16S rRNA Sequences Identified Two Lineages of Helicobacter pylori Strains Detected from Different Regions in Sudan Suggestive of Differential Evolution,” Int. J. Microbiol., vol. 2020, p. e8825718, Oct. 2020. [CrossRef]
- M. Sato and K. Miyazaki, “Phylogenetic Network Analysis Revealed the Occurrence of Horizontal Gene Transfer of 16S rRNA in the Genus Enterobacter,” Front. Microbiol., vol. 8, 2017, Accessed: Jun. 30, 2023. [Online]. Available: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02225.
- F. E. Dewhirst et al., “Discordant 16S and 23S rRNA Gene Phylogenies for the Genus Helicobacter: Implications for Phylogenetic Inference and Systematics,” J. Bacteriol., vol. 187, no. 17, pp. 6106–6118, Sep. 2005. [CrossRef]
- Pei et al., “Diversity of 23S rRNA Genes within Individual Prokaryotic Genomes,” PLOS ONE, vol. 4, no. 5, p. e5437, May 2009. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).