1. Introduction
Epigenetics is a discipline that studies the inheritance of genetic information through DNA methylation or chromatin conformation changes without altering the DNA sequence [
1]. Epigenetic phenomena include DNA methylation, genomic imprinting, maternal effects, gene silencing, RNA editing, noncoding RNA, and more [
2,
3]. Currently, there is extensive research on the modifications of DNA, RNA, and histones [
4,
5,
6]. The modification of DNA and RNA mainly includes DNA methylation, hydroxymethylation, and RNA methylation [
4,
7,
8,
9]. Research on the modification of RNA and DNA mainly focuses on the modification of nucleic acid bases and sugars [
10]. Research on histone modifications is also extensive, mainly focusing on histone phosphorylation and acetylation [
5,
11,
12,
13]. In addition to directly studying the changes in epigenetic genetic information, enzymes related to the modification of RNA, DNA, and histones are also being studied.
Abnormal DNA and RNA methylation and histone acetylation can lead to various diseases [
10,
14,
15,
16]. Abnormal methylation of RNA and DNA and histone acetylation are closely related to the occurrence of various cancers and can also cause metabolic and neurological diseases [
17,
18,
19]. Due to the close relationship between epigenetic modifications and various diseases, it has been widely studied. By detecting epigenetic biomarkers, researchers can gain a deeper understanding of the mechanisms and progression of diseases, providing more accurate and precise methods for early diagnosis and treatment. For example, detecting DNA methylation levels can improve the sensitivity and specificity of tumor detection, providing more reliable methods for early diagnosis [
20]. In addition, for some difficult-to-diagnose diseases such as autism and schizophrenia, the detection of epigenetic biomarkers can provide new diagnostic and treatment approaches [
21,
22]. Therefore, the detection of epigenetic biomarkers is of great significance for the prevention, diagnosis, and treatment of diseases. The conventional detection methods for epigenetic substances are already mature, and commonly used methods include radioanalysis [
23], chromatography [
24,
25,
26], immunological analysis [
27], and single-molecule sequencing [
28,
29,
30]. Radioactive methods require labeling of the detection substance, which may produce harmful radiation to humans and the environment. Chromatography and sequencing analysis have the disadvantage of being time-consuming and tedious. These detection methods also require expensive instruments and are not suitable for on-site testing in complex environments.
Electrochemical biosensors do not require complex equipment, and only an electrochemical workstation, electrodes, and a computer are needed to complete the detection of target analytes [
4,
31]. Compared with complex detection methods such as fluorescence [
32,
33], surface plasmon resonance [
34,
35], and surface-enhanced Raman spectroscopy [
36,
37], electrochemical biosensing detection methods have the advantages of simple operation, low equipment cost, and high sensitivity [
38]. They have been developed to detect various forms of epigenetic modifications in recent years [
39,
40,
41].
This review provides an overview of electrochemical detection methods for epigenetic modifications. It introduces the label-free method of using electrochemical detection for epigenetic modifications, the method of using signal probes based on label modification, the electrochemical detection method based on signal amplification, and the method of using nanostructure-modified electrodes. We will review the principles, characteristics, and applications of these electrochemical biosensors, and discuss challenges and future development directions in this field.
2. Label-Free Methods
The direct detection method without labeling utilizes electrochemical impedance spectroscopy (EIS) detection in electrochemistry to directly read electrical signals without any modification or amplification. The direct label-free detection methods mainly include EIS and the method using electrochemically active substances as indicators. The EIS is mainly based on the principle that the signal is generated by the increase of electrochemical impedance after the analyte binds to the capture material on the electrode, which is then used to quantify the analyte. The use of electrochemically active substances as indicators is based on the fact that these substances can bind with the analyte due to electrostatic forces. The more the analyte presents, the more electrochemically active substances will bind, and the larger the electrical signal generated, which can then reflect the content of the analyte.
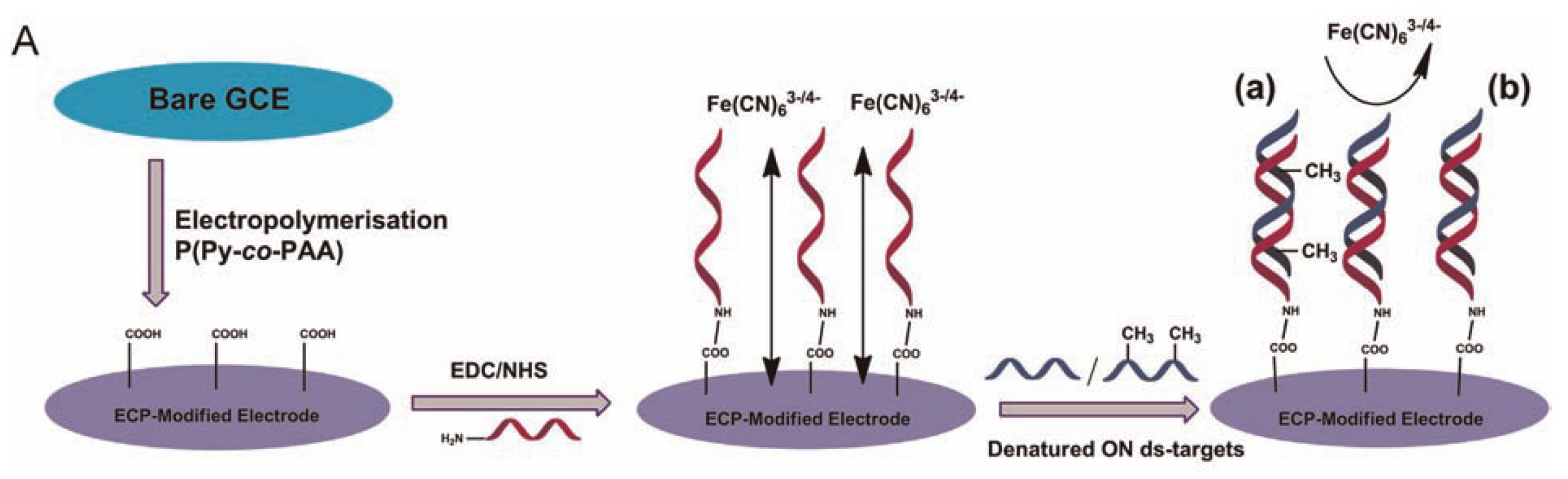
DNA methylation is the most common type of epigenetic modification. There have been many reported methods for detecting DNA methylation. Most DNA methylation detection methods rely on the principle of base complementary pairing. Sheppard’s Group developed a biological platform that takes advantage of the stability of double-stranded targets (
Figure 1) [
42]. A denaturation step is added prior to detection to take advantage of the sensitivity and selectivity of single-stranded DNA (ss-DNA) target and probe hybridization for the detection of the ss-DNA target. The authors used conductive polymer materials to modify the electrode and covalently attached on probes for bio-recognition on the modified electrode surface. Due to steric hindrances presented by methyl groups, that methylation can affect the hybridization rate. The EIS was used to detect the signal generated by the potassium ferricyanide reduction reaction to study the hybridization kinetics of double-stranded DNA to detect DNA methylation. This method detects target methylation through the kinetic changes in methylation DNA hybridization, providing a design concept for future methylation biosensors.
Currently, in contrast to the extensive research on DNA methylation, there are relatively fewer reports on RNA modification detection methods, because the content of abnormal RNA in total RNA is particularly low and difficult to detect. Since the content of RNA is relatively lower than that of DNA, more sensitive detection methods are needed to detect RNA. N6-methyladenosine (m6A) is a common RNA methylation modification in epigenetics. There have been a few reports of using electrochemical biosensors combined with competitive degradation to quantify such kind of RNA methylation.
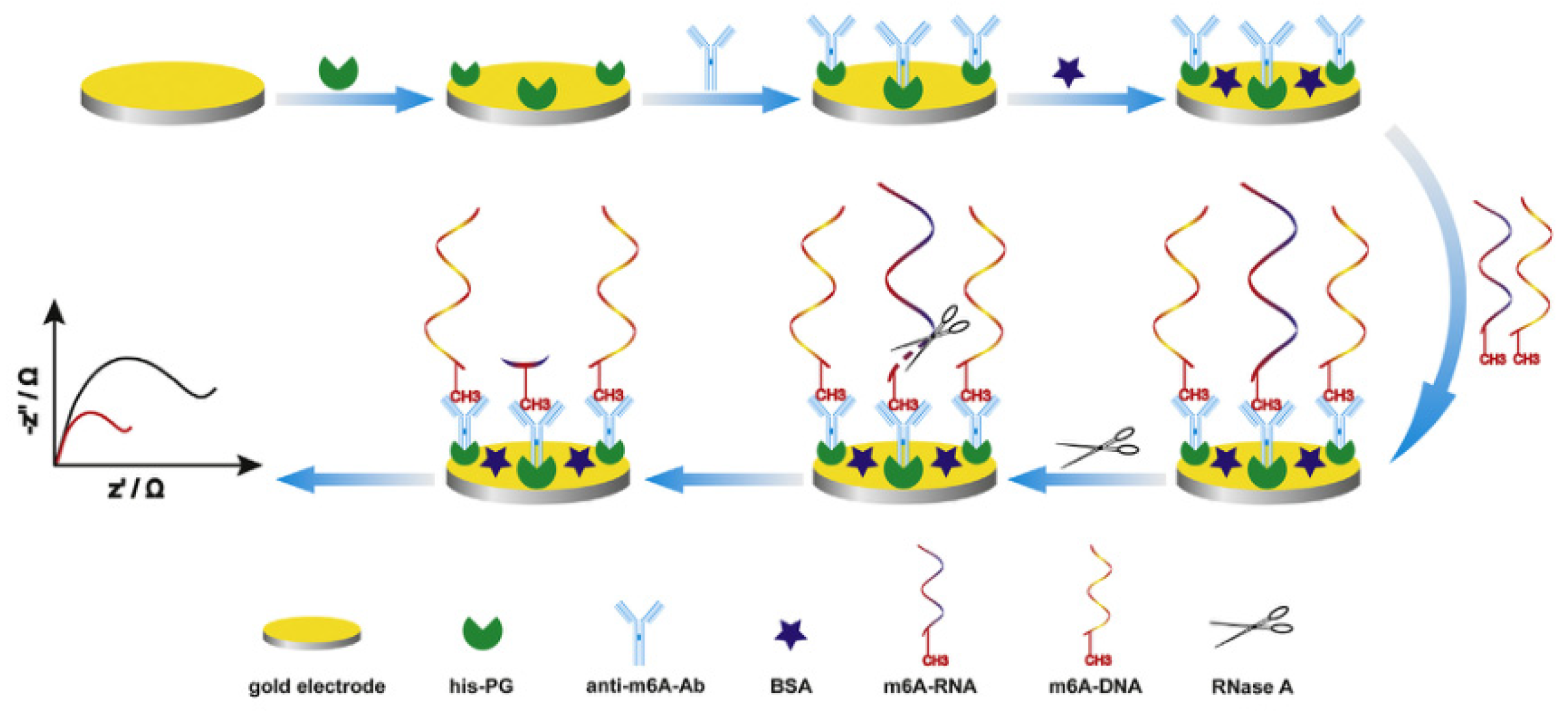
Xie’s group developed a sensitive and label-free electrochemical immunosensor for m6A RNA detection, utilizing the advantages of the sensitivity and selectivity of electrochemical biosensing technology (
Figure 2) [
43]. The key to achieving sensitivity and specificity in the developed method is the use of the specific interaction between antibody (Ab) and antigen (Ag). The authors used recombinant proteins tagged with histidine to modify the gold electrode surface. Histidine binds to the gold electrode surface, allowing the recombinant protein to be oriented. The specificity of the Ab crystal region and recombinant protein binding was then used to expose the Ab binding site, thereby improving the binding efficiency of the Ag-Ab and enhancing the detection signal, and enabling the detection of low-abundance m6A RNA. The m6A Ab used in this method can bind to both m6A RNA and m6A DNA. M6A DNA serves as a signaling molecule and participates in the reaction together with m6A RNA. After binding to the Ab, RNase A is used to hydrolyze the bound m6A RNA. The amount of m6A RNA is quantified by the decrease in EIS signal. The linear range of detection can be improved by using a method of competition reaction with m6A DNA and m6A RNA, followed by degradation. Finally, the EIS signal of the detection electrode is detected, and the decrease of signal intensity is proportional to the abundance of m6A RNA while the intensity of the signal is inversely proportional to the amount of m6A RNA in the sample. This biosensor has the advantages of simplicity and sensitivity, with a wide detection range and a sensitivity of up to 0.016 nM. This method for detecting RNA methylation is not affected by chain length or base sequence and has a certain degree of universality.
The above two methods use the principle of base complementary pairing to detect DNA methylation or RNA methylation, which can only detect methylated nucleic acids as epigenetic markers, and the methods can only detect single biomarkers. Electrochemical sensing methods can not only detect nucleic acids in epigenetics but also use Ag-Ab interactions to detect proteins. They can not only detect single biomarkers but also perform multiple detections of two different types of biomarkers. Due to their cell-type specificity, robustness, and ability to be released into body fluids, DNA methylation and histone acetylation can serve as biomarkers for cancer diagnosis in vitro.
Sheppard’s group used graphene screen-printed electrodes to detect DNA and chromatin, because graphene screen-printed electrodes have the advantages of low cost, high signal-to-noise ratio, and no need for surface preparation [
44]. The surface of the screen-printed electrode was coated with polyaniline, a conductive polymer, to avoid the influence of defects on the graphene surface while also allowing the Ab to be surface-functionalized. The anti-5-methylcytosine Ab, which specifically binds to DNA methylation, and the anti-acetylated histone H3 Ab, which specifically binds to histone acetylation, were then directly coupled to the polyaniline-modified graphene screen-printed electrode surface. This label-free method uses EIS to detect DNA and chromatin. The authors used the developed biosensor to detect endometrial cancer cell and breast cancer cell systems and found that there was no difference in total DNA methylation, but there was a difference in histone acetylation. Unlike other methods that are time-consuming or require expensive hardware, electrochemical biosensors have the advantages of simplicity, sensitivity, and portability and can be used as important tools for DNA and histone detection. The biosensor developed by the authors can simultaneously detect two epigenetic markers. It can also be used to observe the therapeutic effect of epigenetic drugs by detecting cells in vitro. This method demonstrates the potential of using the same sample for multiple epigenetic detections.
In addition to using EIS to directly detect the binding of analytes to the electrode surface, there are also some detection methods that electrochemically active substances as indicators. Ai’s group used methylene blue as an indicator to detect DNA methylation and methyltransferase activity [
45] . Methylene blue can insert into the double-stranded DNA, providing a reliable electrochemical signal. Methylated DNA can be selectively cleaved, reducing the amount of methylene blue and producing a decrease in electrochemical signal. Li’s group chose the electroactive complex [Ru(NH
3)
6]
3+ as a signal converter [
46]. The electroactive complex can bind to double-stranded DNA through electrostatic forces. Similar to methylene blue as an indicator, it can also detect methylated DNA and has a signal amplification effect, with the ability to sensitively detect DNA methylation. The activity of DNA adenine methylation methyltransferase was detected by signal closure. Nie’s group prepared a coenzyme A silver ion-coordinated polymer with high electrocatalytic activity as a signal probe for high-sensitivity detection of coenzyme A and histone transferase activity [
47].
Direct and label-free epigenetic detection methods have the advantages of being simple and easy to operate. The methods rely on the specificity of Ag-Ab binding or DNA base complementary pairing. As the sensitivity of all methods that used EIS to detect biomarkers was often not enough, conductive material modification or interface orientation methods have been used to improve the Ab coverage and binding efficiency on the electrode interface in some degree. Direct detection of DNA methylation distinguishes between methylation patterns based on the kinetics of hybridization, while RNA methylation is quantified based on changes in electrochemical impedance caused by enzyme degradation after competition. DNA methylation and histone acetylation are detected through Ag-Ab binding. The detection methods are similar and the quantification principle is based on the signal change of the potassium ferricyanide reduction reaction. Any way, although direct detection methods are simple, their sensitivity is not enough. Even the sensitivity has been improved by interface modification, but it is still not sufficient for the detection of some low-abundance epigenetic modifications.
3. Methods with Labeled Signal Probes
Due to the low sensitivity of direct detection methods, they are not suitable for the detection of all epigenetic markers, especially in the detection of low-abundance DNA and RNA modifications. Even with the use of electroactive indicators, it is still challenging to meet the high sensitivity requirements for detection. Therefore, there is a need to develop more sensitive detection methods to improve the performance of electrochemical biosensors. Common methods to enhance sensitivity involve the use of signal probe labeling. The methods of signal probes for electrochemical biosensing detection of epigenetic markers mainly include chemically modifying specific sites, using protein interactions to bind signals, and employing signal probe methods.
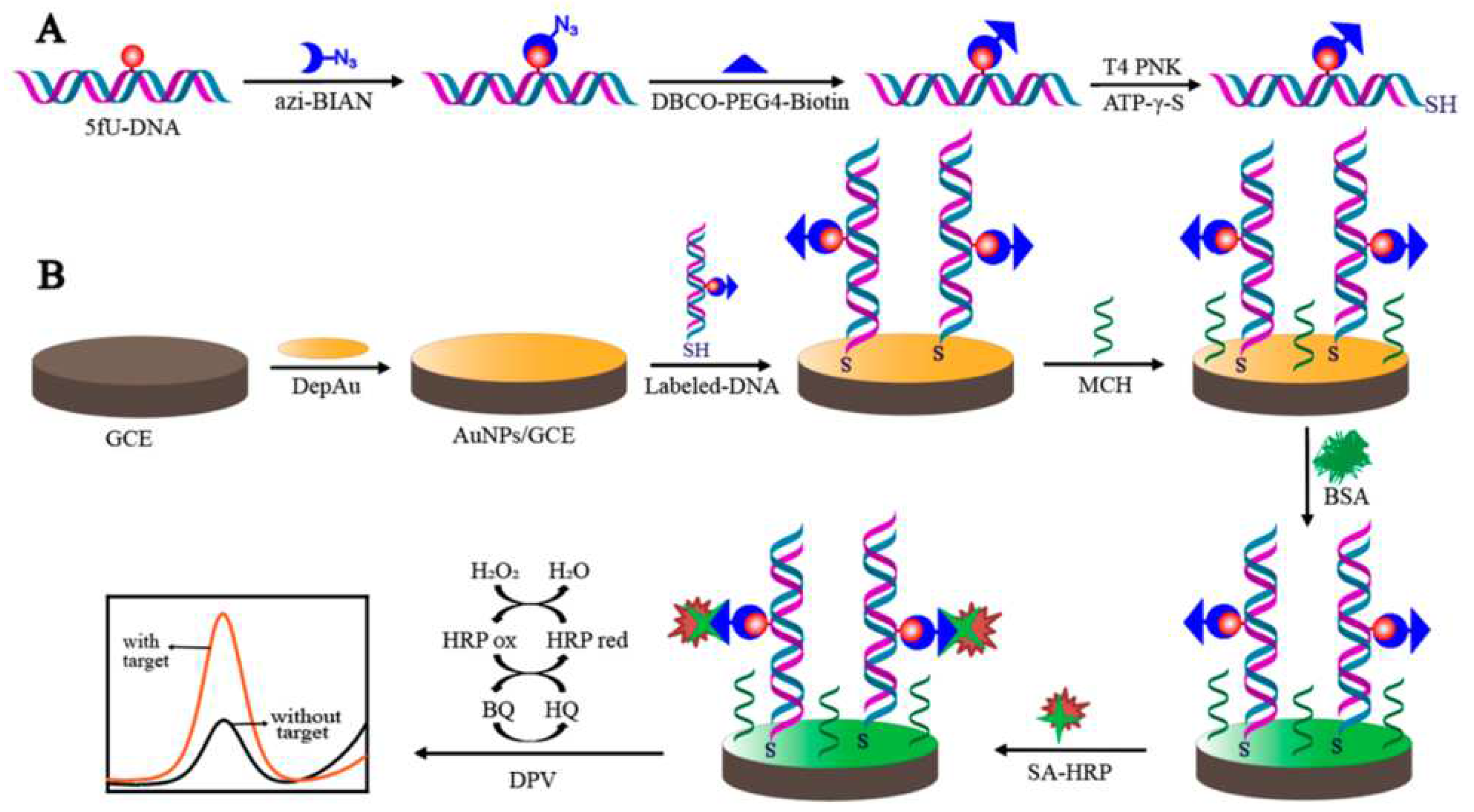
Chen’s group took advantage of the high sensitivity of electrochemistry and used a signal labeling strategy to detect low-abundance 5-formyluracil (5fU) which is closely related to the function of DNA (
Figure 3) [
48]. The most critical step of this method is to signal-modify the target site of 5fU DNA. The authors first used the azide derivative of (2-phenylimidazole) acetonitrile and the aldehyde group of 5fU to form a covalent bonding through a chemical reaction. Then, DBCO-PEG4-biotin was connected to the target DNA through a copper-free click chemistry reaction. Next, T4 polynucleotide kinase was used to catalyze the target DNA to generate a sulfhydryl group at the 5' end. The target DNA containing 5fU can be assembled on the electrode surface by the interaction of Au-S bond. Finally, specific recognition between biotin and streptavidin were used to label horseradish peroxidase onto the surface of the above electrode. Horseradish peroxidase catalyze the oxidation and reduction of hydroquinone to generate an electric current signal. Differential pulse voltammetry (DPV) was used to detect the current and enable high-sensitivity detection of 5fU. The specificity of detection comes from the specific recognition of azide to 5fU. Introducing a biotin label at the position of 5fU and then combining with streptavidin-horseradish peroxidase can significantly improve the detection sensitivity. Using T4 polynucleotide kinase to directly connect the target DNA onto the electrode through a covalent bond is more direct and does not require a capture probe. This method can avoid interference from 5-formylcytosine and apyrimidinic sites in the detection of 5-fU. In the meantime, this direct bonding method also avoids the sequence matching problem caused by the use of capture probes in traditional connection methods. This method has a good linear range and a low detection limit.
The chemical modification of signal probes requires the use of toxic chemical reagents and involves multiple steps of manipulating the target DNA during the labeling process. The labeling process may cause some loss of targets, which can introduce bias in the upstream analysis and affect the accuracy of the experimental results. Although the method uses chemical substances to recognize specific sites and achieve specific labeling, the drawbacks of the method are also evident. The complex processing steps limit the practicality of this method. Compared with the method of modifying DNA using chemical reagents, the specific binding of biological proteins is safer and more reliable. Biological proteins can selectively bind to specific groups without the need for complex processing steps, and they do not have toxic effects on the operator, making them a safer and more reliable modification labeling method.
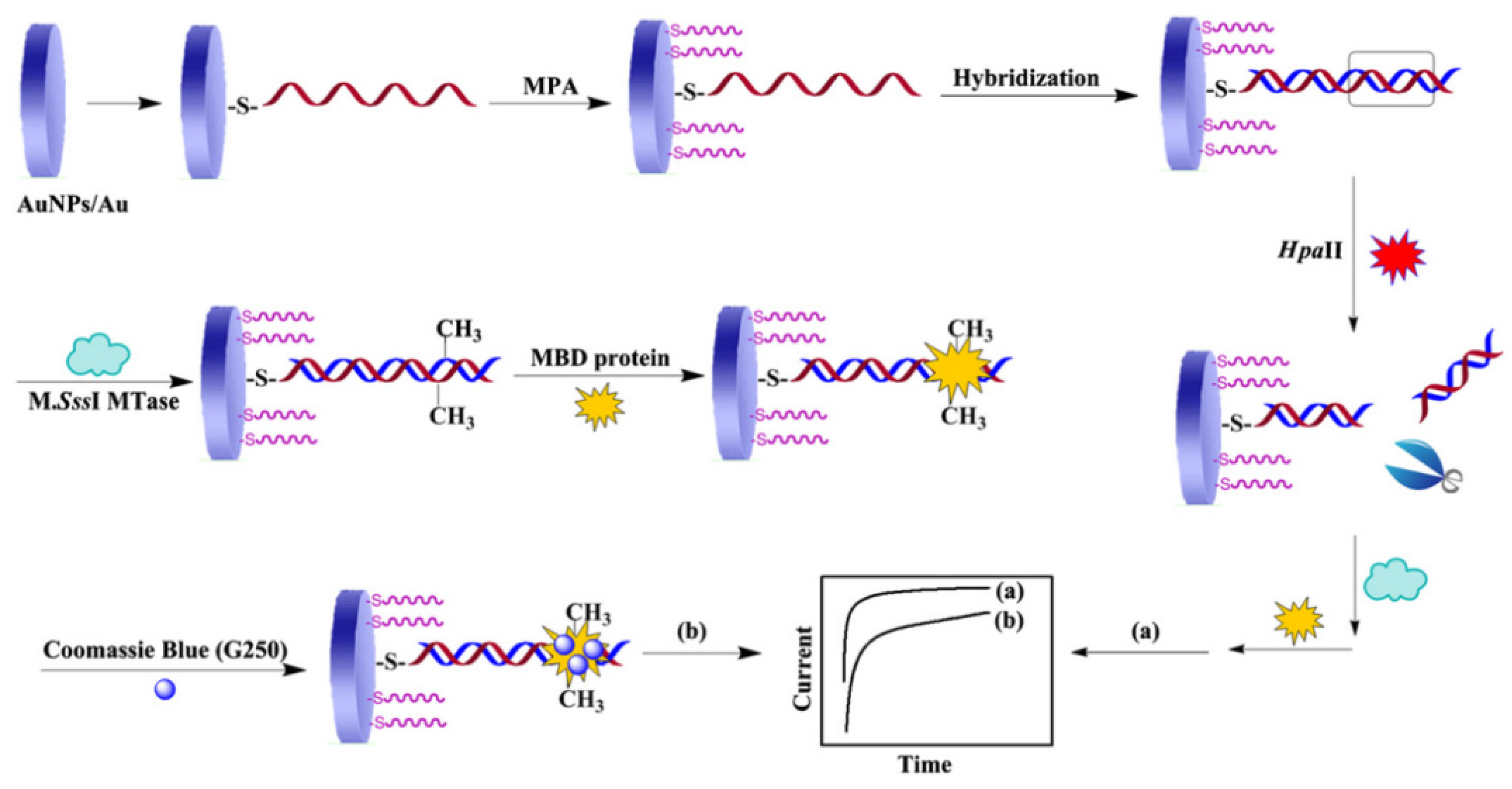
Ai’s group developed a biosensor to detect cytosine methylation of CpG dinucleotides and the activity of methyltransferase (MTase) in DNA, using a methyl-binding domain (MBD) protein that can specifically bind to CpG dinucleotides and Coomassie brilliant blue G250 (CBB-G250) as the signal label (
Figure 4) [
49]. This sample method can also be applied to screen for MTase inhibitors. The process of this method is to fix the capture DNA probe on the electrode using the Au-S bond firstly, and then hybridize the target DNA with the capture DNA probe. Treatment of the hybridized DNA with M.SssI-MTase in the presence of the methyl donor S-adenosylmethionine can methylate CpG dinucleotide specific sites. The methylated CpG region can be specifically recognized and bound by the MBD protein. CBB-G250 can bind to the MBD protein through intermolecular forces. CBB-G250 is a common electroactive molecule that can provide redox signals to methylated DNA. When the CCGG symmetrical sequence of the hybrid molecule is specifically recognized and cut by the Hpall restriction endonuclease, it cannot be methylated by MTase. The amperometric current method is used to detect the redox signal of CBB-G250, and the obtained current signal can reflect the level of DNA methylation and the activity of MTase. The authors developed a simple, portable, and sensitive biosensor that can be used to detect DNA methylation and MTase activity, and can also screen for methylation inhibitors.
In addition to detecting DNA methylation, the above methods can also be used to detect DNA MTase. The detection of enzymes related to epigenetic nucleic acid markers is also essential for studying epigenetics. Detecting DNA MTase is different from directly detecting nucleic acid markers, as it requires a reactive subject. Using the specific binding ability of biological proteins and the non-specific binding method of specific groups and Coomassie blue does not directly combine with the label. The binding between the label and the electrode is not specific, which may result in insufficient accuracy in actual sample detection. Developing a method that can directly and specifically bind to the electrode can improve accuracy and sensitivity. The use of unique hairpin probes to generate capture probes and then combine them with signal probes can increase the specificity of the recognition process. It is also important to detect MTase for epigenetic research.
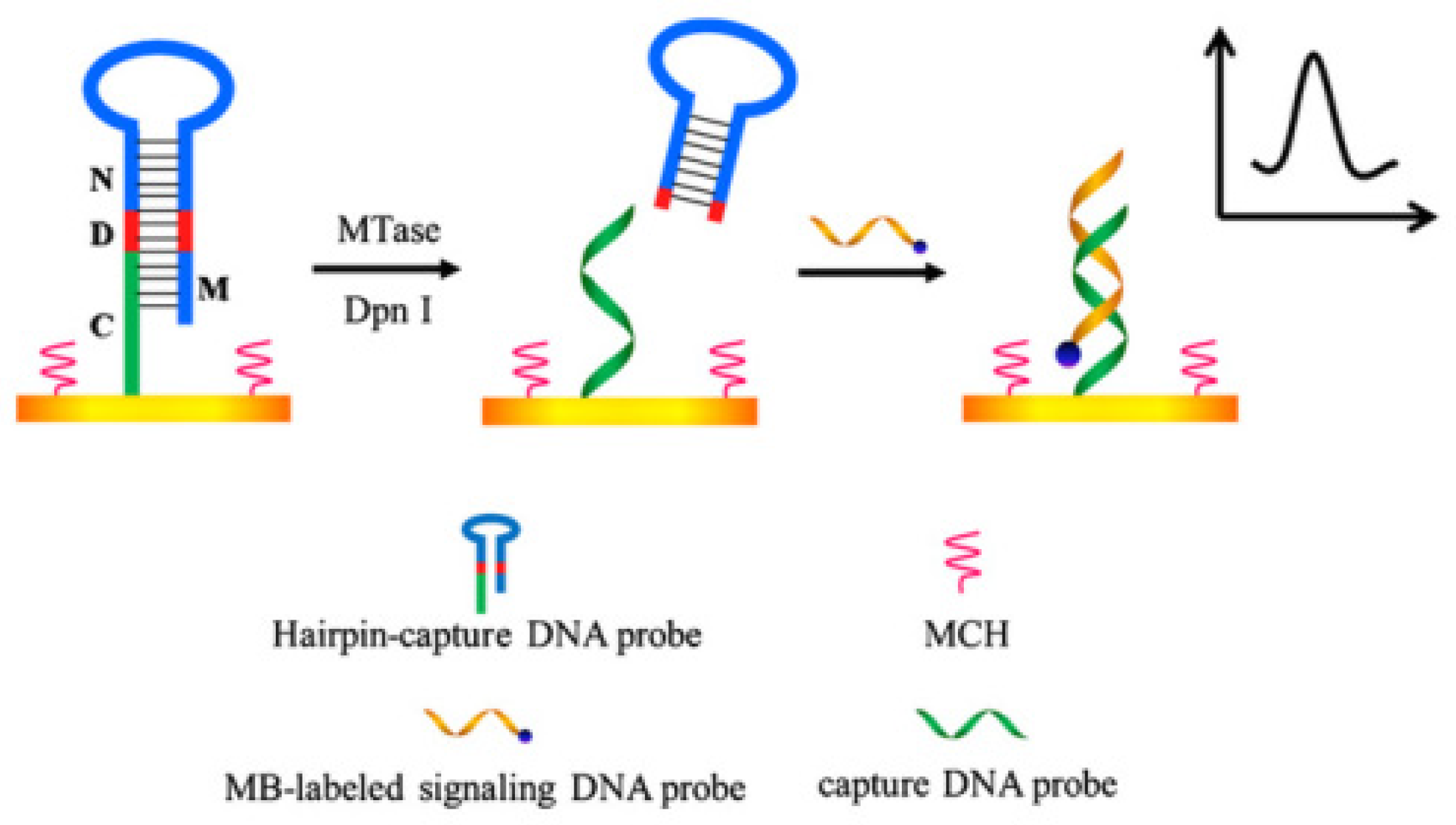
Yan’s group developed a sensitive and simple method for detecting MTase activity using hairpin DNA probes (
Figure 5) [
50]. The key design of this method is the elaborate design of the hairpin DNA probe. The 5' end of this hairpin DNA probe is modified with a sulfhydryl group, which can directly generate an Au-S covalent bond with the gold electrode, fixing the probe on the electrode surface. The hairpin DNA probe also has a methylation recognition site. After treatment with MTase or restriction endonucleases such as Dam MTase and Dpn I that can recognize methylated sites, the hairpin DNA probe is cleaved. The remained DNA fragments after cleavage are still left on the electrode as a capture probe that can hybridize with a signal probe. The signal DNA probe is modified with methylene blue, which can undergo redox reactions on the electrode surface. Unlike conventional detection methods where the signal is directly labeled on the target molecule, this method cleverly designs the capture probe to release and bind with the signal probe. When the hairpin DNA probe cannot be cleaved due to methylation, the capture probe cannot be released and cannot bind with the signal probe, avoiding false-positive results. This method is simple to prepare, easy to operate, has good selectivity, and high sensitivity. The detection limit for Dam MTase using this method is 0.07 U/mL. This method can also be applied to the screening of inhibitors and the discovery of anticancer drugs. Yuan’s group. evaluated the activity of DNA transferase using a commercial blood glucose meter [
51]. This method used biotin-avidin-peroxidase as a label to achieve sucrose catalytic hydrolysis, and used a blood glucose meter to convert the signal for detection .
Most of the label-based electrochemical detection methods for epigenetics use substances with electrochemical activity, such as CBB-G250 , methylene blue, or enzymes that catalyze reactions such as the biotin-avidin-peroxidase system. The changes in current in the electrochemical or enzyme-catalyzed redox reactions are used for detection. This method has higher sensitivity and specificity compared to direct detection methods. It can not only detect epigenetic biomarkers but also enzymes that cause abnormal changes, making the range of substances detected wider and the sensitivity higher. However, the labeling method is cumbersome and may have multiple synthesis steps. The storage time of the labeled target is also greatly limited.
4. Methods Based on Signal Amplification
Signal labeling methods can improve detection sensitivity, enabling quantitative detection of low-abundance 5-methyluracil and expanding the range of electrochemical detection. However, the signal labeling operation itself involves multiple steps, increasing the complexity of actual detection operations. Additionally, the limited storage time of the target may restrict the practical application of the method. Considering that the signal labeling method has limitations in improving sensitivity, it still lacks detection capabilities for ultra-low-abundance epigenetic biomarkers. Developing a method for ultra-sensitive detection of ultra-low-abundance markers is crucial. Signal amplification methods can effectively solve this problem. Signal amplification methods mainly include using a dual signal amplification strategy combining PCR and CRISPR/Cas12 systems, redox signal amplification combined with enzyme-catalyzed amplification, and multi-step circuit amplification design. Detection methods based on signal amplification can significantly improve detection sensitivity.
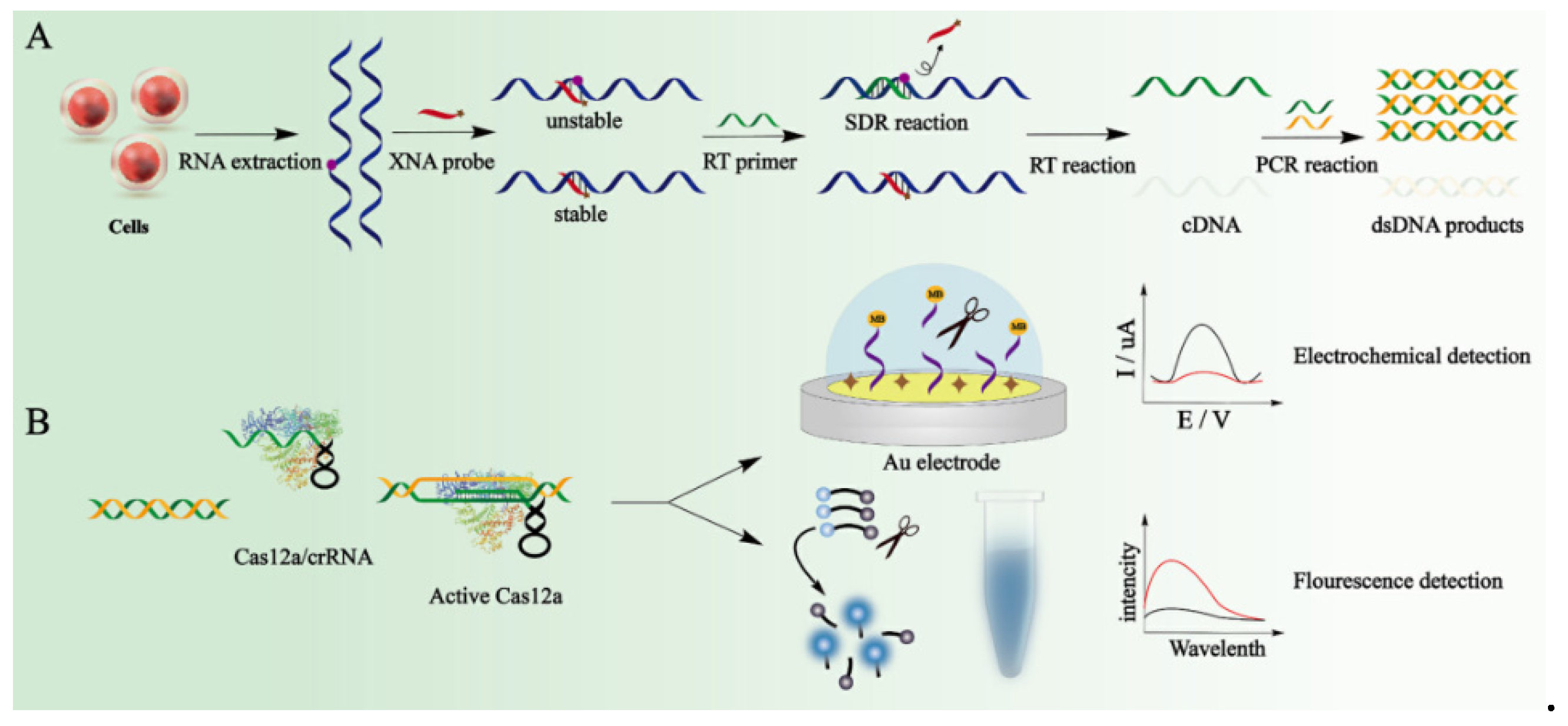
Liao’s group used the difference in the thermodynamic stability of hybridization between xeno nucleic acid (XNA) probe with m6A-RNA and A-RNA to develop a specific reverse transcription polymerase chain reaction for m6A RNA (
Figure 6) [
52]. Combined with the CRISPR/Cas12a signal amplification strategy, m6A modification can be detected with high sensitivity. In the detection process, RNA is extracted from cells to obtain RNA containing m6A-RNA and A-RNA without methylation modification. XNA probes are used to hybridize with RNA. Non-methylated RNA and XNA are more stable after hybridization, and the strand displacement reactions (SDR) with the reverse transcription primer occurs slowly. Due to the hybrid of m6A-RNA and XNA are unstable, the m6A-RNA can preferentially undergo SDR with the reverse transcription primer. The reverse transcription of non-methylated RNA is blocked by the XNA probe, directly magnify the minute differences between m6A-RNA and A-RNA. The single-stranded DNA obtained from the reverse transcription of m6A RNA is PCR amplified to generate double-stranded DNA, which were positively correlated with the m6A fraction. The authors utilized the differences in thermodynamic stability between m6A-RNA and RNA hybridization with XNA, and the difficulty of SDR to amplify the small differences in the first step after PCR amplification. Then, the CRISPR/Cas12a system is used to amplify the signal. The CRISPR-derived RNA (crRNA) in the designed CRISPR can specifically target the m6A RT-PCR amplification fragment. The CRISPR/Cas12a system is activated by the specific double-stranded DNA generated in the reverse transcription polymerase chain reaction and realized signal amplification output. A methylene blue-modified DNA probe is immobilized on the gold electrode surface, and the remaining bare area of the Au electrode is block by MCH before adding the activated Cas12 reaction system. After the DNA probe is cut away from the electrode surface, the methylene blue molecules on the electrode are released. The abundance of m6A-RNA is quantified by detecting the decrease in the square wave voltammetry signal. In this study, the authors used a dual signal amplification system. After amplifying the signal with RT-PCR, the CRISPR/Cas12a system was used to further amplify the signal. The m6A-RNA can be detected with ultra-high sensitivity and can sensitively detect 1% m6A sites.
The method of using RT-PCR and the CRISPR/Cas12a system amplification can achieve ultra-sensitive detection of extremely low-abundance m6A RNA. However, the principle of realizing specific detection using this signal amplification method is based on the difference in complementary pairing with exogenous nucleic acid fragments. Nucleic acid-based complementary pairing detection has strong sequence specificity and can only detect nucleic acids with specific sequences. The CRISPR/Cas12a system also requires specific sequence design to complete the dual signal amplification. Through the dual specificity recognition of exogenous nucleic acid probes and crRNA, as well as the combined signal amplification of PCR and CRISPR/Cas12a, the specificity and sensitivity can be significantly improved. However, this method can only detect m6A RNA with specific sequences, which also has certain limitations.
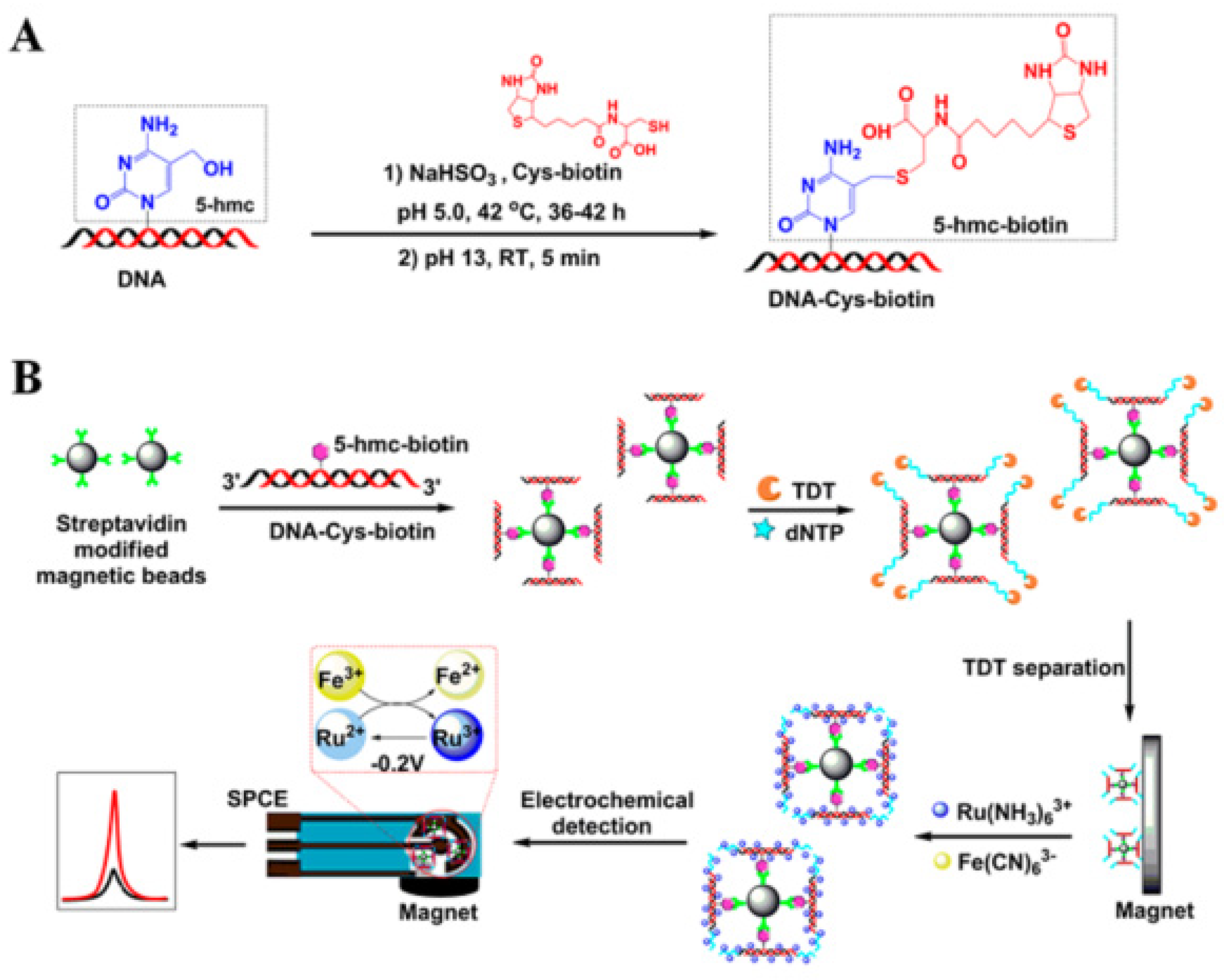
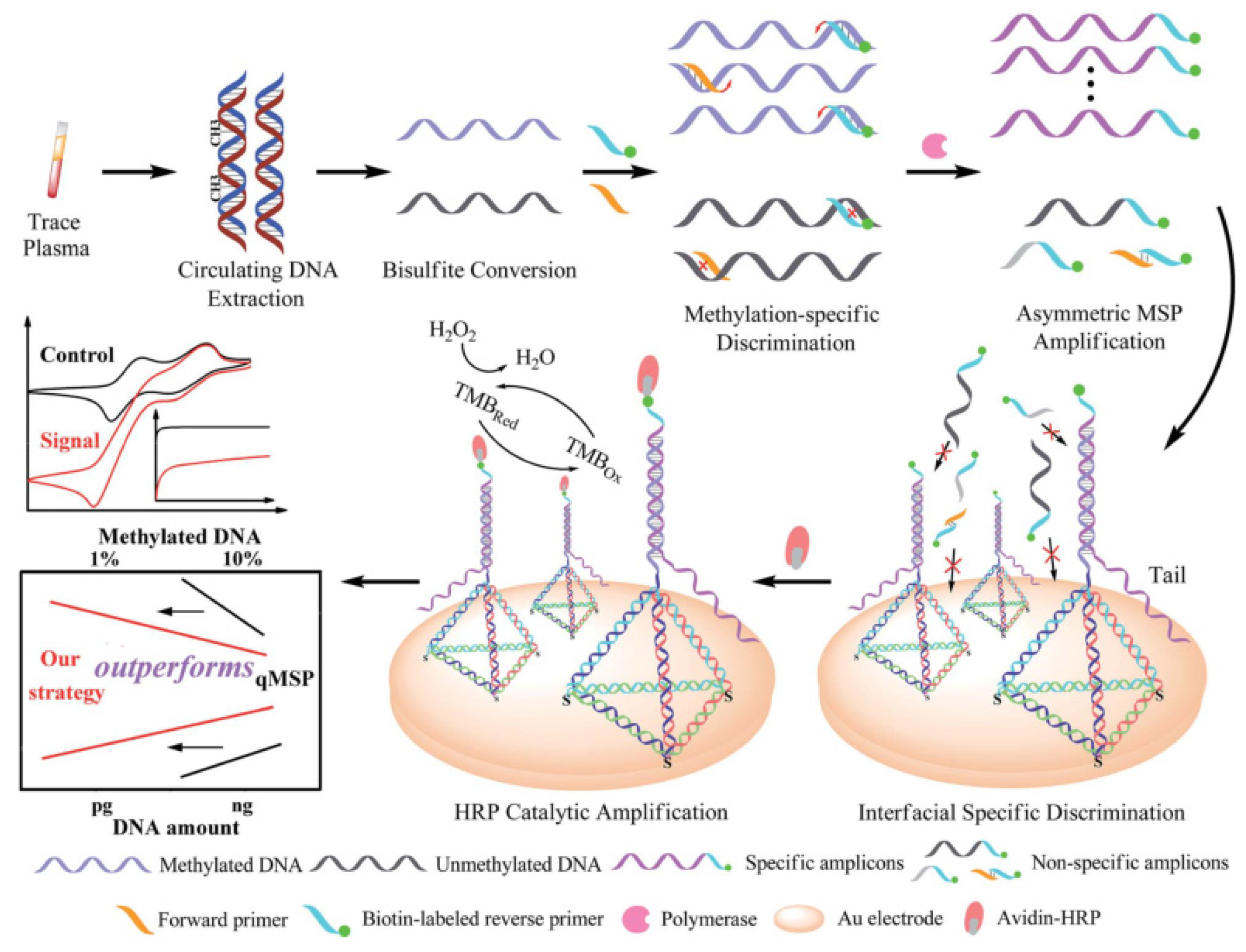
Compared with the dual signal amplification method based on nucleic acid complementary pairing, the following signal amplification method based on magnetic separation does not require specific sequence design or signal labeling and can achieve signal amplification for the detection target. Zhang’s group developed an unlabeled electrochemical magnetic biosensor for the quantitative detection of 5-hydroxymethylcytosine (5-hmC) DNA which is is closely related to cancer and is an important epigenetic biomarker for tumorigenesis (
Figure 7) [
53]. The core of this method is coupling with the terminal deoxynucleotidyl transferase (TDT) enzyme-catalyzed amplification and Ru(III) redox cycling, a dual signal amplification system, to significantly improve the sensitivity of the detection. In the detection process, 5-hmC is first modified and enriched. The 5-hmC specific site in the non-paired double-stranded DNA is modified by biotinyl-cysteine derivative. After purification using Micro-Bio-Spin P6 column, biotin-coupled 5-hmC double-stranded DNA is obtained. Because biotin has good specificity with streptavidin, the biotin-coupled double-stranded DNA is bound to magnetic beads coated with streptavidin, which links the double-stranded DNA and magnetic beads to achieve the enrichment effect. The magnetic beads linked with double-stranded DNA are elongated by terminal deoxynucleotidyl transferase treatment. Ru(NH
3)
63+ is positively charged, which can be electrostatically attracted to the negatively charged phosphate backbone of DNAs They can be combined through electrostatic interaction, allowing Ru(NH
3)
63+ to reach the vicinity of the DNA main chain. The magnetic beads linked with DNA double-strands can bind to the screen-printed electrode surface through magnetic force without immobilization. Fe(CN)
63- is negatively charged and repels the DNA main chain, making it difficult to contact. Ru(NH
3)
63+ directly participates in the electrochemical redox process. With Ru(III) being reduced to Ru(II), Ru(II) can react with Fe(CN)
63-, causing Ru(II) to be oxidized to Ru(III) again. The regenerated Ru(III) can continue to bind with DNA to participate in the redox cycle. The redox process of Ru can further amplify the electrical signal. This electrochemical biosensing detection method has good specificity and can significantly distinguish 5-hmC from 5-methylcytosine. This method does not require specific templates or special sequence design and is not limited to DNA with specific base sequences. Amplification is performed through molecular interactions without signal labeling. The dual signal amplification system enable the detection limit of as low as 9.06 fM.
Although the electrochemical magnetic biosensor method does not require signal labeling and is not limited to specific base sequences, the detection process involves column separation technology and magnetic enrichment technology, making the steps relatively complex. The above method uses terminal deoxynucleotidyl transferase treatment, which is an enzyme-dependent and magnetic-dependent complex detection technology. Although it greatly improves the detection sensitivity, the implementation of magnetic separation technology also requires specific modifications and column separation treatment for specific sites. The more complex and cumbersome the operation steps, the more potential biases may be introduced. Chemical modification of specific sites requires the use of chemical reagents and long reaction times. In contrast to the above-mentioned magnetic separation technology, the use of multi-step circuit amplification technology achieves an enzyme-free and treatment-free process. Using the multi-step circuit cycling to achieve signal amplification does not require special chemical treatment and long reaction times, making the detection process as simple as possible while improving sensitivity. In addition to direct detection and signal labeling methods, signal amplification methods have also been used for sensitive detection of DNA methylation.
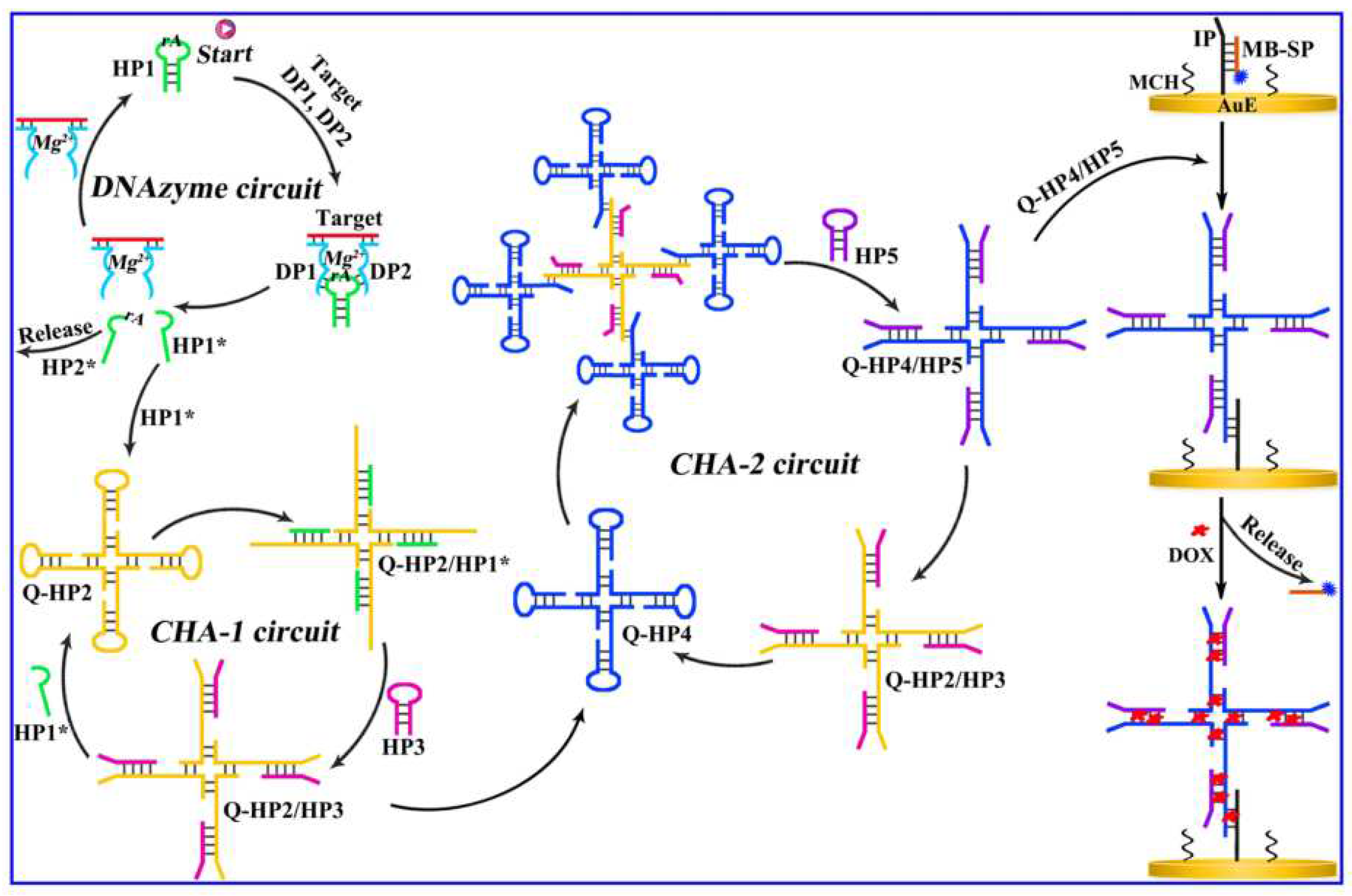
Based on a multi-step DNA amplification circuit design, Wang’s group developed a sensitive proportional electrochemical biosensor for detecting methylated DNA (
Figure 8) [
54]. The multi-step circuit amplification design which is a non-enzymatic amplification design, can use the product upstream as the initiator of the downstream circuit, effectively achieving signal amplification. The core of the non-enzymatic electrochemical biosensor designed by the authors is the design of multi-step circuit amplification, mainly including three cycles. Introduction of methylated target DNA in the first cycle triggers a Mg
2+-dependent DNA enzyme cycle, causing the hairpin structure to break and generate HP1*. The HP1* generated in the first cycle can act as an initiator for the second cycle, starting the CHA-1 cycle. QHP2/HP3 produced by the CHA-1 cycle can initiate the third cycle CHA-3, ultimately generating the four-way junction QHP4/QHP5. The multi-step amplification circuit is triggered by methylated target DNA and generates a DNA four-way junction after three cycles. The gold electrode surface is fixed with a capture probe, which binds to the signal probe of methylene blue. The DNA four-way junction can bind to the capture probe, undergo strand displacement reaction, replace the original signal probe of methylene blue, and connect the DNA four-way junction to the electrode interface. The DNA four-way junction has sufficient sites to introduce doxorubicin molecules. By detecting the ratio of doxorubicin and methylene blue signals using electrochemistry, the methylated target DNA can be sensitively detected. The ratiometric method is a way to detect biomarkers based on the ratio of two independent signals, rather than using a specific signal output value. Using the ratio of two signals to detect can significantly improve the accuracy and precision of detection. The oxidation-reduction of doxorubicin and the signal probe of methylene blue can be used as a pair of signal reporters. Although the ratiometric method produces stable and repeatable signals, amplification methods are used to increase the limited sensitivity. The multi-step circuit amplification method exhibited high sensitivity for target analysis with a detection limit of 4 aM.
Ai’s group developed an electrochemical detection method for DNA hydroxymethylation based on alkaline phosphatase-catalyzed signal amplification [
55]. This method involves specific modification of the group with glycosylation and bridging with 1,4-phenylene diboronic acid to capture alkaline phosphatase. Signal amplification is achieved through enzyme-catalyzed generation of p-nitrophenol. This method can detect 5hmC with high selectivity and sensitivity. Xie’s group developed a method combining m6A-sensitive DNA enzyme and three-way junction-mediated isothermal exponential CRISPR amplification to identify methylated RNA by using deoxyribonuclease for specific cleavage of non-methylated RNA and three-way junction. Isothermal CRISPR amplification can amplify the signal to achieve high-sensitivity detection of low-abundance m6A RNA [
56]. Zhao’s group combined sodium bisulfite conversion and PCR amplification, and developed an electrochemical biosensor with methylation specificity using PCR amplification and enzyme-catalyzed signal amplification to quantitatively detect methylation of tumor suppressor gene promoter [
57]. Our group developed a method combining glycosylation modification and enzyme-catalyzed signal amplification to detect 5hmc DNA [
58]. The developed electrochemical biosensor has high specificity and is multiplexed, capable of detecting multiple samples simultaneously. Li’s group used the peroxidase property of G-quadruplex-Cu(II) metalloenzyme to catalyze hydrogen peroxide for signal amplification to electrochemically detect histone acetyltransferase activity [
59].
Direct detection and labeled detection can detect epigenetic markers with high abundance, but there are still many biomarkers with extremely low abundance that are difficult to detect by simple signal labeling. The signal generated by a single label is not sufficient to meet the detection requirements. Signal amplification methods provide a good solution to this problem. Signal amplification methods can further amplify the signal when combined with other methods. For example, in m6A RNA detection, PCR and CRISPR/Cas12a system can be combined, or amplification circuit and ratio signal can be combined, or magnetic bead enrichment and Ru oxidation-reduction signal amplification can be combined. Signal amplification technology is no longer just using signal labeling, but combining multiple technologies to achieve electrochemical detection methods with improved sensitivity. Compared with conventional detection methods, signal amplification technology is more complex, but it provides more accurate and precise measurements, and has greater specificity, which can detect specific targets at specific sites. More importantly, signal amplification technology greatly improves the detection sensitivity.
By amplifying the signal, the sensitivity and detection limit of the sensor can be improved, enabling the detection of low-concentration target substances. In addition to using signal amplification methods, interface modification methods can also be used to enhance the stability, selectivity, and biocompatibility of biosensors, to improve signal transmission efficiency, and to reduce background noise, thereby improving the sensor's detection performance. Electrochemical biosensor interface modification and signal amplification play different roles. Signal amplification is mainly used to enhance the detection signal of the target analyte. Interface modification mainly focuses on improving the sensor surface to enhance sensor performance. They each have their advantages, but in some aspects, interface modification may be more advantageous than signal amplification. Interface modification can improve the stability of the sensor, allowing it to maintain performance over long periods of use or under different conditions, while signal amplification may introduce additional instability factors, such as enzyme inactivation or aggregation of nanomaterials. Interface modification can improve the selectivity of the sensor by modifying specific recognition elements, enabling the sensor to better distinguish between target analytes and interfering substances. In some cases, interface modification can achieve simpler operation and fewer steps, thereby improving the ease of the practical use of biosensors.
5. Methods Based on Nanostructured Modified Electrodes
Even designing various signaling methods is necessary to improve the sensitivity of epigenetic analysis, developing high-performance electrodes is extremely important for electrochemical biosensing methods, providing both high sensitivity and high signal-to-noise ratio. The development of nanostructured modified electrodes becomes a more acceptable choice because nano-bio materials and interfaces show unique performance for biosensing applications. The method of modifying nanostructures on the electrode surface is simpler and requires less sample processing. It can improve the sensitivity without complicated processing, making the detection method more concise in operation while meeting the sensitivity requirements. Nanostructure modification methods can significantly improve the signal-to-noise ratio and enhance the interference resistance of biosensors. Using nanostructure-modified electrodes can compensate for the interface defects of electrochemical biosensors, making the interface more regular, facilitating the binding of target molecules, and reducing background noise. The core of electrochemical biosensing lies in the binding of capture probes and analytes, and modifying the interface with nanomaterials is more conducive to capturing target substances. Currently, commonly used nanostructure-modified electrode methods include modifying the electrode surface with alloy nanostructures, directly generating nanostructures on the substrate surface and combining with metal nanoparticles, modifying the electrode surface with composite materials, modifying the electrode surface with gold nanoparticles, and modifying the electrode surface with DNA tetrahedra.
Recent studies have shown that non-coding RNAs play an increasingly important role in the regulation of epigenetics. Non-coding RNAs are functional RNA molecules that cannot be translated into proteins, and some common regulatory non-coding RNAs include small interfering RNAs, miRNAs, piRNAs and long-stranded non-coding RNAs [
61]. MiRNAs are short endogenous non-coding RNA that control epigenetic remodeling. Shiddiky’s group designed a gold-loaded nanoporous superparamagnetic iron oxide nanocubes as a nano-material to improve the performance of the electrochemical biosensor to detect miRNA (
Figure 9) [
60]. This nano-material is highly porous and has an exposed gold surface, which binds to DNA or RNA more efficiently through affinity interactions, thereby increasing the binding efficiency with ribonucleotides. This nanocube has catalytic activity towards Ru(III) and is an electroactive material that can amplify detection signals through catalytic redox processes. The iron oxide material is paramagnetic, which enables magnetic separation for sample enrichment and separation. In the specific detection process, RNA is first extracted from the sample, and the target RNA is extracted using the specific interaction between the streptavidin-modified magnetic beads and the biotin-modified capture probe. The extracted miRNA is then magnetically separated and released. The released miRNA is adsorbed onto the gold-loaded iron oxide nanocube. Since RNA molecules contain a negatively charged phosphate backbone, they can bind with positively charged Ru(III) through electrostatic interaction, resulting in a large number of Ru(III) molecules on the RNA molecules. The Ru(III) redox process is coupled with the [Fe(CN)
6]
3-/
4- redox system to further amplify the signal. The quantitative information of miRNA can be obtained by detect the Ru(III) bound to the electrode surface using chronoamperometry. This method solves the problem of amplifying miRNA that cannot be achieved by other methods and can achieve highly specific and sensitive detection even at low abundance and in the presence of similar RNA interference. This method uses nano-materials to modify the electrode interface and combines multiple electrochemical signal amplification systems to achieve ultra-sensitive detection of miRNA with a detection limit of 100 aM.
The method of modifying electrodes with iron oxide nanocubes loaded with gold can detect miRNA without amplification. MiRNA binds to iron oxide nanocubes loaded with gold through affinity interactions. The nanoparticles themselves do not have specific binding capabilities and do not have interference resistance to other RNAs. In the detection process, the target miRNA needs to be processed first by using magnetic separation and enrichment, and then the specific capture probe is used to extract and release the target miRNA.
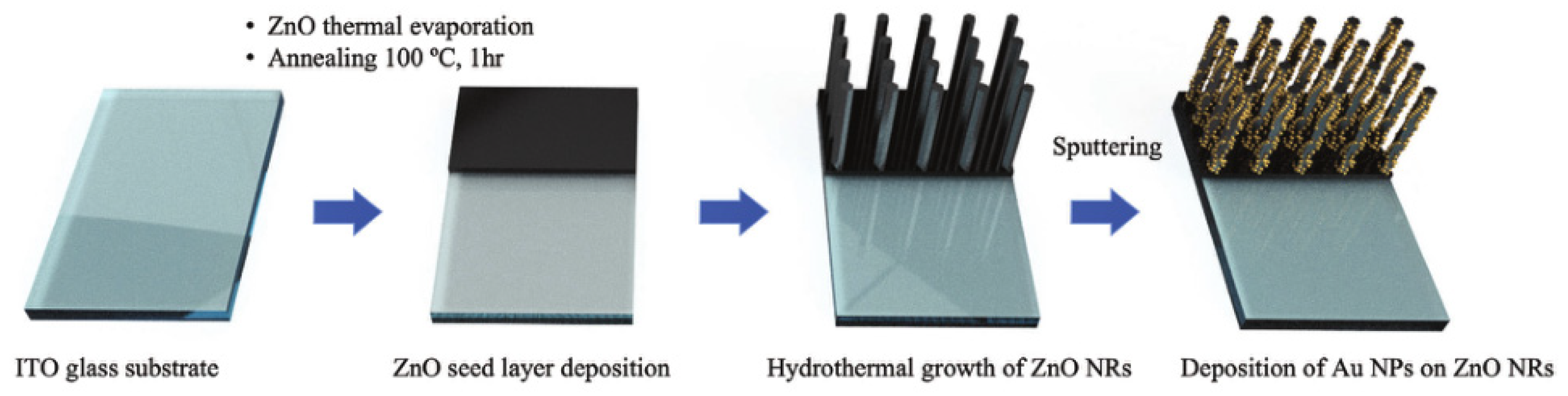
Lim’s group develops an electrochemical biosensor for the detection of 5hmc using a combination of zinc oxide nanorods (ZnO NRs) and gold nanoparticles (Au NPs) (
Figure 10) [
62]. Compared to iron oxide nanocubes loaded with gold, the generated ZnO NRs loaded with Au NPs have a certain ability to distinguish target molecules. The difference in adsorption properties can effectively distinguish between target molecules and interfering molecules without additional extraction and enrichment steps. The direct generation of ZnO NRs also makes the process between nanoparticles and electrodes interference-free, resulting in a more stable electron transfer process.
The core of the biosensor is the construction of nanostructures to detect 5hmC which is closely related to cancer epigenetics AuNPs serve as excellent platforms to detect a variety of biomolecules because of the excellent biocompatibility, conductivity, diverse surface functionalization options. The affinity of 5hmC and 5mC for Au is different, and their different adsorption and affinity abilities can be used for label-free electrochemical detection and quantification. ZnO NRs have good biocompatibility and are widely used in biochemical-related detection. ZnO NRs can be directly grown and synthesized on the electrode, which can directly contact the electrode, avoiding interference from other substances and providing a stable connection for electron transfer, thereby improving sensing performance. They also have a large surface area that can be used for modification and connection. First, they sputtered ultrathin ZnO quantum dots seed layer sputtered onto indium tin oxide glass substrate. Then, they hydrothermal growth ZnO NRs with a certain height on the zinc oxide film. Finally, they modified gold nanoparticles onto the surface of the ZnO NRs resulting in the direct generation of nano-materials on the electrode surface. Because the adsorption of 5hmC on the gold surface is lower than that of 5mC, a larger current signal is generated during current detection. The biosensor developed by the authors can also distinguish different levels of 5hmC DNA very well. This method avoids selective modification of the sample and instead constructs nanostructures on the electrode surface and modifies the surface of the biosensor in a controllable manner by directly growing nanostructures on the substrate. At the same time, the surface of the ZnO NRs can also be modified with gold nanoparticles, which can reflect the good performance and structure of the surface nanostructures. Instead of loading other capture probe on the surface, the distinction between 5hmC and interference is based on the adsorption properties of the nanostructures.
The two methods mentioned above utilize the affinity interaction between miRNA and iron oxide nanocubes loaded with gold, as well as the adsorption property of 5hmc on ZnO NRs modified with AuNPs, to successfully bind target molecules to the electrode surface. Both of these binding methods do not have high specificity, so pre-extraction and enrichment are required, or they can only distinguish between one interfering molecule and the target molecule. Compared to using the inherent affinity or adsorption of nanoparticles to directly bind to the analyte, nanoparticles can also provide a stable structural foundation for the electrode surface. The use of composite materials, such as Graphene oxide-Fe3O4-β-cyclodextrin(GO-Fe3O4-β-CD), can provide a stable cavity nano-interface for the electrode surface, increasing the surface area of the electrode and providing stable binding sites for specific capture antibodies. Subsequently, the specific binding of antigens and antibodies is used to detect DNA methylation of unknown sequences in the target. The specific interaction method that connects target molecules and electrodes significantly improves the specificity of detection.
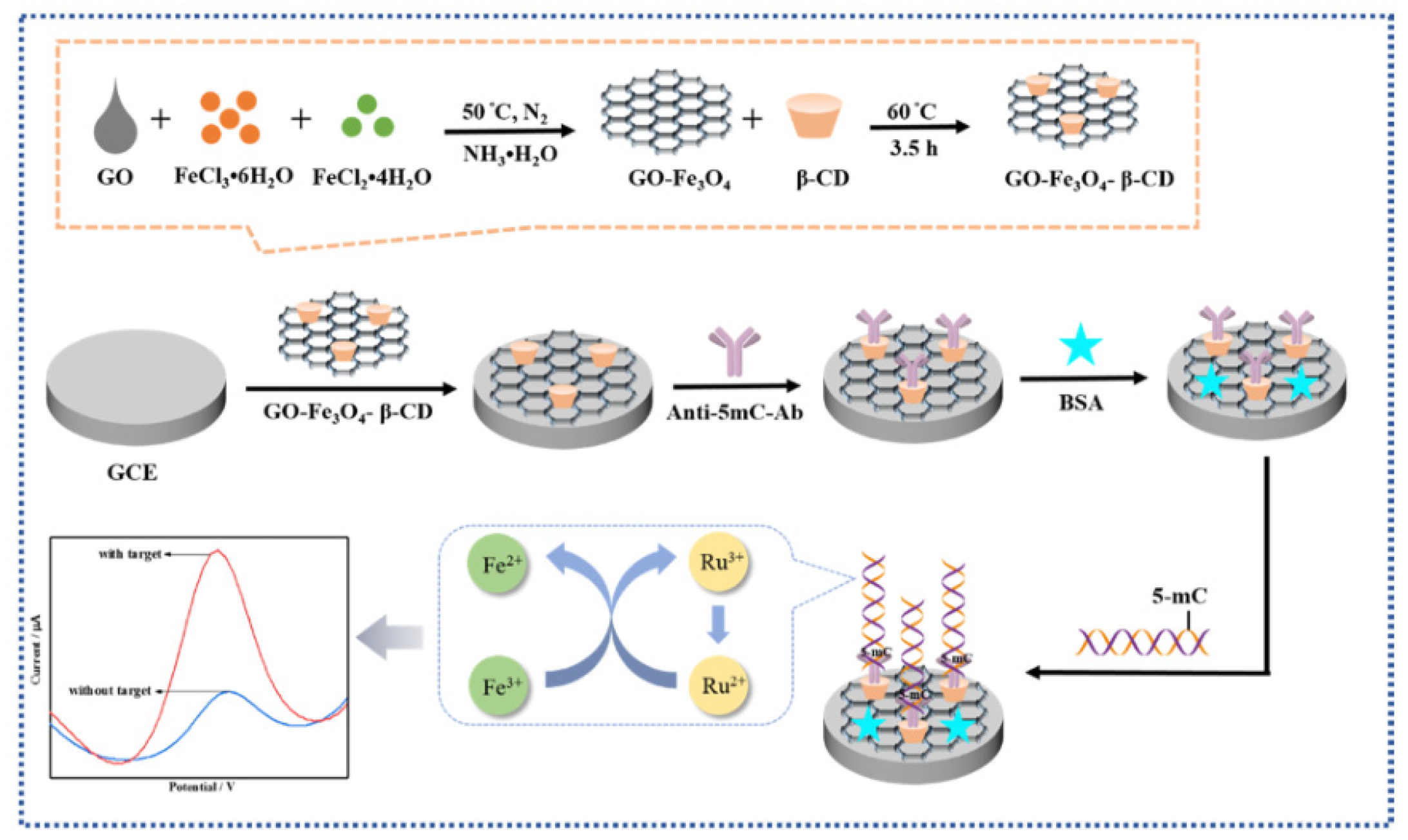
Chen’s group developed an electrochemical immunosensor that can use antibodies to recognize DNA bases to detect DNA methylation of unknown sequences(
Figure 11) [
63]. They prepared a GO-Fe
3O
4-β-CD nanocomposite material, which combines the advantages of GO-Fe
3O
4 which has good biocompatibility, a large specific surface area, and good dispersion, and β-CD which has a hydrophobic cavity that can form stable host-guest complexes. The authors dropped the synthesized GO-Fe
3O
4-β-CD composite nanomaterial on the glassy carbon electrode surface, forming a stable nanointerface with a large number of cavities, which increased the electrode's surface area and provided many antibody binding sites. Then, they immobilized the antibody for 5mc on the formed nanointerface. Through the specific interaction between the antigen and antibody, the target gene can be captured. After methylation DNA binds to the antibody, the Ru(NH
3)
63+/Fe(CN)
63- redox amplification system is used for current amplification, significantly increasing the sensitivity of detection. The current detected by the electrochemical biosensor is related to the amount of methylated DNA. The method of modifying the interface using nanomaterials can achieve a detection limit of 0.0825 pm. Using capture antibodies instead of specific sequence DNA as capture probes can detect DNA methylation of unknown sequences, but there is an issue of poor detection specificity. The combination of electrochemical detection methods and nanotechnology can achieve high sensitivity.
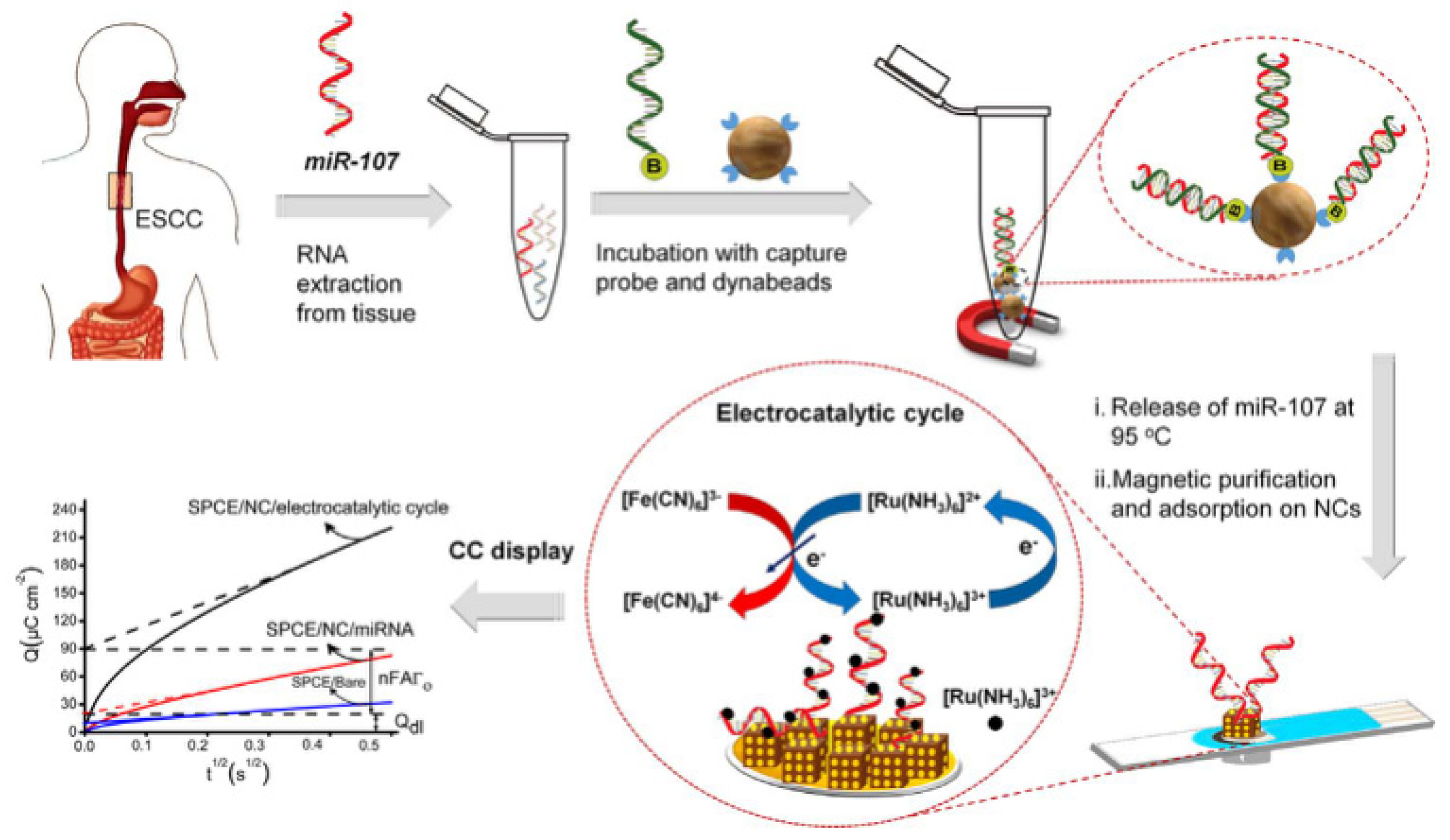
Our group used the ordered orientation, controllable spacing, good biocompatibility, and high stability of framework nucleic acids, to develop electrochemical biosensors for the detection of DNA methylation (
Figure 12) [
64,
65]. By using the nanostructure of DNA framework nucleic acids, the probe and surface can be separated from each other, making it easier to bind with the analyte in the solution, achieving high sensitivity detection of DNA methylation. We developed the method that first extracts and amplifies the circulating methylated DNA in trace plasma, with single-copy sensitivity and ultrahigh specificity. Subsequently, biotin-labeled methylation-specific primers are used for asymmetric methylation-specific PCR amplification of methylated DNA, generating a large number of biotin-labeled single-stranded amplicons. Self-assembled DNA tetrahedra modified on the gold electrode immobilize the DNA nanostructured probes on the electrode surface to capture the amplicons. Finally, horseradish peroxidase-avidin and biotin-avidin bind together, and horseradish peroxidase catalyzes the redox process to produce an electrochemical signal. The use of DNA tetrahedra modified electrodes provides stable support for the capture probe, achieving orientational order and distance control, increasing target polarizability, and reducing non-specific adsorption of byproducts, resulting in efficient and specific hybridization with a significantly reduced signal-to-noise ratio. We used the developed method to determine the DNA methylation of the p16INK4a gene promoter in trace plasma samples from lung cancer patients, and the results were in good consistency with clinical diagnosis.
Huang’s group. developed a highly sensitive electrochemical gene sensor for detecting cancer long non-coding RNA using a composite interface of graphene-like tungsten disulfide/dendritic gold nanostructures [
66]. The composite nanointerface ensures the biological recognition ability of the target and capture probes. Zhang’s group developed an electrochemical immunosensor for m6A-RNA detection using a glassy carbon electrode modified with gold nanoparticles [
67]. Ai’s group used a gold electrode modified with gold nanoparticles as the substrate to develop an electrochemical immunosensor for m6A RNA detection [
68]. The developed electrochemical biosensors have high sensitivity and specificity for target detection. Raouafi’s group conducted simple and sensitive detection of miRNA-21 on the surface of electrode structures modified with gold nanoparticles [
69].
6. Other Electrochemical Biosensing Methods
Electrochemical biosensing technology can also be combined with other techniques to detect epigenetic biomarkers. When there are many interferences in the sample, pre-processing or amplification of the sample can be chosen before electrochemical detection. Matson’s group has reported using high-performance liquid chromatography (HPLC) to separate trace amounts of 7-methyl guanine and then using electrochemical detection to assess DNA methylation levels in Huntington's disease [
70]. This method takes full advantage of the powerful separation capability of HPLC and the high sensitivity of electrochemical sensors to quantify trace base changes. Lu’s group used electrochemical and linker-PCR technology coupling to detect the methylation of the 5'-CpG islands of the human p16Ink4a gene [
71]. The PCR was used to synthesize and purify target DNA fragments, which were then detected using DNA biosensors for hybridization. This electrochemical biosensor has the advantages of fast speed, high sensitivity, and low cost. Toumazou’s group applied semiconductor technology to the electrochemical detection of cancer DNA methylation, using Ion-Sensitive Field-Effect Transistors in real-time DNA methylation detection for the first time through experimental verification [
72]. The introduction of semiconductor technology in disease detection can provide new solutions for disease detection.
7. Conclusion and Prospect
Since the association between epigenetic modifications and cancers came into the spotlight in 1964, epigenetic modifications has been the subject of investigation. In this field, the great advantages of electrochemical biosensors in terms of high sensitivity, easily operated instrumentation, inexpensive, have converted them in promising tools. In this review, we have classified and summarized the methods of electrochemical biosensors for detecting epigenetic modifications, and explained the applicable scope and method characteristics of different detection methods, pointing out the advantages and disadvantages of these methods.
Simple, direct, and label-free electrochemical methods can solve some epigenetic problems, but due to the insufficient detection sensitivity and accuracy, they are not suitable for detecting low amounts of RNA. The use of signal label, which can modify the biomarkers, improve the electrochemical biosensing sensitivity of detection. However, the labeling process is complex and can introduce bias, which can affect the detection results. Moreover, the labeling signal also has certain limitations and cannot detect markers with extremely low sensitivity. Electrochemical detection methods based on signal amplification strategies can detect low-abundance RNA, and some methods can achieve sensitivity of aM. However, signal amplification strategies require high specificity and need to be designed ingeniously, and they can only detect specific epigenetic biomarkers,. The method of modifying the electrode surface with nanostructures can solve the problem of insufficient sensitivity. Nanoparticles have superior conductivity and high surface area, which can provide more binding sites to improve detection sensitivity.
It is now clear that electrochemical biosensing methods can improve the sensitivity of epigenetic analysis through signal label modification, signal amplification, or electrode surface modification with nanomaterials. However, most of the methods described in the literature were only used for proof-of-concept validation and had not yet been applied to real-life samples. Real biological samples are more complex, with excessive interference, making accurate analysis challenging. Current electrochemical biosensing methods for epigenetics mainly focus on research related to DNA methylation, with fewer reports on other epigenetic modifications and a limited research scope. In the future, it is necessary to expand the detection of other epigenetic modifications. The abundance of epigenetic biomarkers in the body is very low, and there are few detection methods that can achieve high sensitivity at low abundance. To meet the detection requirements, signal amplification and nanomaterial modification remain the main research directions for electrochemical detection of epigenetic biomarkers. For the interference problem in the detection of real-life samples, it is necessary to consider using simple and reliable specific recognition methods while also achieving signal amplification. This will be a major focus of future research. Current detection methods is mainly focused on single epigenetic molecules, with fewer reports on the combined detection of multiple related epigenetic biomarkers. It is hoped that in the future, the high-throughput advantage of electrochemical biosensors can be developed to enable rapid and multiplex detection of epigenetic biomarkers.
With the development of technology, people have higher requirements for high sensitivity and high specificity while hoping that the detection methods can meet the needs of portability, simplicity, and ease of operation. The development of nanotechnology and electrochemical technology will promote the development of electrochemical detection of epigenetics. Ultimately, the development of these detection methods needs to solve practical problems, and the future clinical application of electrochemical detection of epigenetics can be rapidly developed.
Author Contributions
Writing—original draft, Z.H. Writing—Review and Editing, S.S. Figures, Y.D. Literature retrieval, Z.H., Y.D., and J.S. Funding acquisition, S.S. Supervision, S.S. and T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Natural Science Foundation of China (21974147; 32000921).
Institutional Review Board Statement
The study did not require ethical approval.
Informed Consent Statement
This study did not involve humans.
Conflicts of Interest
The authors declare no conflict of interest..
References
- Waterland, R.A.; Michels, K.B. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr 2007, 27, 363–388. [Google Scholar] [CrossRef]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic mechanisms in mammals. Cell Mol Life Sci 2009, 66, 596–612. [Google Scholar] [CrossRef]
- Wu, Y.L.; Lin, Z.J.; Li, C.C.; Lin, X.; Shan, S.K.; Guo, B.; Zheng, M.H.; Li, F.; Yuan, L.Q.; Li, Z.H. Epigenetic regulation in metabolic diseases: mechanisms and advances in clinical study. Signal Transduct Target Ther 2023, 8, 98. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Yánez-Sedeño, P.; Pingarrón, J.M. Advances in Electrochemical (Bio)Sensing Targeting Epigenetic Modifications of Nucleic Acids. Electroanalysis 2019, 31, 1816–1832. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Y.; Yin, H. Recent advances in biosensor for histone acetyltransferase detection. Biosens Bioelectron 2021, 175, 112880. [Google Scholar] [CrossRef]
- Hulshoff, M.S.; Xu, X.; Krenning, G.; Zeisberg, E.M. Epigenetic Regulation of Endothelial-to-Mesenchymal Transition in Chronic Heart Disease. Arterioscler Thromb Vasc Biol 2018, 38, 1986–1996. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, Y.Z.; Liu, H.; Zhang, L.; Li, J.Q.; Meng, J. RNA methylation and diseases: experimental results, databases, Web servers and computational models. Brief Bioinform 2019, 20, 896–917. [Google Scholar] [CrossRef]
- Xie, S.; Chen, W.; Chen, K.; Chang, Y.; Yang, F.; Lin, A.; Shu, Q.; Zhou, T.; Yan, X. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int 2020, 20, 585. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, H.; Tian, M.; Wang, D.; He, J.; Xu, T. DNA Methylation and Hydroxymethylation in Cervical Cancer: Diagnosis, Prognosis and Treatment. Front Genet 2020, 11, 347. [Google Scholar] [CrossRef]
- Chen, Y.; Hong, T.; Wang, S.; Mo, J.; Tian, T.; Zhou, X. Epigenetic modification of nucleic acids: from basic studies to medical applications. Chem Soc Rev 2017, 46, 2844–2872. [Google Scholar] [CrossRef]
- Bhaumik, S.R.; Smith, E.; Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 2007, 14, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef]
- Shi, Y.J.; Matson, C.; Lan, F.; Iwase, S.; Baba, T.; Shi, Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 2005, 19, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, L.; Yu, C.; Yu, D.; Yu, G. Histone Acetylation Modifiers in the Pathogenesis of Alzheimer's Disease. Front Cell Neurosci 2015, 9, 226. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, Y.; Ren, J. Acetylation in cardiovascular diseases: Molecular mechanisms and clinical implications. Biochim Biophys Acta Mol Basis Dis 2020, 1866, 165836. [Google Scholar] [CrossRef]
- Chen, D.; Wu, Y.; Tilley, R.D.; Gooding, J.J. Rapid and ultrasensitive electrochemical detection of DNA methylation for ovarian cancer diagnosis. Biosens Bioelectron 2022, 206, 114126. [Google Scholar] [CrossRef]
- Campuzano, S.; Barderas, R.; Pedrero, M.; Yanez-Sedeno, P.; Pingarron, J.M. Electrochemical biosensing to move forward in cancer epigenetics and metastasis: A review. Anal Chim Acta 2020, 1109, 169–190. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J. Histone acetylation modifications: A potential targets for the diagnosis and treatment of papillary thyroid cancer. Front Oncol 2022, 12, 1053618. [Google Scholar] [CrossRef]
- Campuzano, S.; Pingarrón, J.M. Electrochemical Sensing of Cancer-related Global and Locus-specific DNA Methylation Events. Electroanalysis 2018, 30, 1201–1216. [Google Scholar] [CrossRef]
- Diwadkar, V.A.; Bustamante, A.; Rai, H.; Uddin, M. Epigenetics, stress, and their potential impact on brain network function: a focus on the schizophrenia diatheses. Front Psychiatry 2014, 5, 71. [Google Scholar] [CrossRef] [PubMed]
- Eshraghi, A.A.; Liu, G.; Kay, S.S.; Eshraghi, R.S.; Mittal, J.; Moshiree, B.; Mittal, R. Epigenetics and Autism Spectrum Disorder: Is There a Correlation? Front Cell Neurosci 2018, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, K.Y.; Eason, M.M.; Ferry, J.J.; Planck, J.L.; Walsh, C.P.; Smith, R.F.; Howitz, K.T.; Ma, H. Assay development for histone methyltransferases. Assay Drug Dev Technol 2013, 11, 227–236. [Google Scholar] [CrossRef]
- Peng, S.-Y.; Zhang, J.; Tian, M.-P.; Wang, Z.-L.; Shen, H.-Q. Determination of Global DNA Methylation in Biological Samples by Liquid Chromatography-Tandem Mass Spectrometry. Chinese Journal of Analytical Chemistry 2012, 40, 1201–1206. [Google Scholar] [CrossRef]
- Rozhon, W.; Baubec, T.; Mayerhofer, J.; Mittelsten Scheid, O.; Jonak, C. Rapid quantification of global DNA methylation by isocratic cation exchange high-performance liquid chromatography. Anal Biochem 2008, 375, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhang, L.; Chen, B.; Li, J.; Yang, Q.; Huang, Q.; Zhang, J.; Gao, Y.; Li, Z.; Cai, C. A quantitative method for detecting DNA methylation over targeted genomic regions using isotope dilution liquid chromatography tandem mass spectrometry. Talanta 2017, 169, 136–140. [Google Scholar] [CrossRef]
- Ghadiali, J.E.; Lowe, S.B.; Stevens, M.M. Quantum-dot-based FRET detection of histone acetyltransferase activity. Angew Chem Int Ed Engl 2011, 50, 3417–3420. [Google Scholar] [CrossRef]
- Choy, L.Y.L.; Peng, W.; Jiang, P.; Cheng, S.H.; Yu, S.C.Y.; Shang, H.; Olivia Tse, O.Y.; Wong, J.; Wong, V.W.S.; Wong, G.L.H.; et al. Single-Molecule Sequencing Enables Long Cell-Free DNA Detection and Direct Methylation Analysis for Cancer Patients. Clin Chem 2022, 68, 1151–1163. [Google Scholar] [CrossRef]
- Han, Z.W.; Ma, F.; Zhang, C.Y. Methylation-sensitive transcription-enhanced single-molecule biosensing of DNA methylation in cancer cells and tissues. Anal Chim Acta 2023, 1251, 340996. [Google Scholar] [CrossRef]
- Shipony, Z.; Marinov, G.K.; Swaffer, M.P.; Sinnott-Armstrong, N.A.; Skotheim, J.M.; Kundaje, A.; Greenleaf, W.J. Long-range single-molecule mapping of chromatin accessibility in eukaryotes. Nat Methods 2020, 17, 319–327. [Google Scholar] [CrossRef]
- Bartosik, M.; Hrstka, R. Bioelectrochemistry of nucleic acids for early cancer diagnostics – analysis of DNA methylation and detection of microRNAs. Reviews in Analytical Chemistry 2017, 36. [Google Scholar] [CrossRef]
- Wang, G.L.; Luo, H.Q.; Li, N.B. Gold nanorods-based FRET assay for ultrasensitive detection of DNA methylation and DNA methyltransferase activity. Analyst 2014, 139, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Gao, C.; Xiong, Y.; Zhang, Y.; Liu, S.; Pu, Y. A fluorescence method for detection of DNA and DNA methylation based on graphene oxide and restriction endonuclease HpaII. Talanta 2015, 131, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Xu, J.; Shu, Y.; Wang, F.; Xia, W.; Ding, Q.; Xu, T.; Zhao, C.; Zhang, M.; Huang, P.; et al. Double recognition of oligonucleotide and protein in the detection of DNA methylation with surface plasmon resonance biosensors. Biosens Bioelectron 2010, 26, 850–853. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Song, T.; Guo, X. DNA methylation detection with end-to-end nanorod assembly-enhanced surface plasmon resonance. Analyst 2015, 140, 6230–6233. [Google Scholar] [CrossRef]
- Ishwar, D.; Venkatakrishnan, K.; Tan, B.; Haldavnekar, R. DNA Methylation Signatures of Tumor-Associated Natural Killer Cells with Self-Functionalized Nanosensor Enable Colorectal Cancer Diagnosis. Nano Lett 2023, 23, 4142–4151. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhan, D.S.; Xu, X.Y.; Zhang, Z.; Hafez, M.E.; He, Y.; Li, Y.; Li, D.W. Label-free detection of DNA methylation by surface-enhanced Raman spectroscopy using zirconium-modified silver nanoparticles. Talanta 2023, 253, 123941. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Xu, Q.; Ma, F.; Zhang, C.Y. Recent advances in biosensors for in vitro detection and in vivo imaging of DNA methylation. Biosens Bioelectron 2021, 171, 112712. [Google Scholar] [CrossRef]
- Li, C.C.; Wang, Z.Y.; Wang, L.J.; Zhang, C.Y. Biosensors for epigenetic biomarkers detection: A review. Biosens Bioelectron 2019, 144, 111695. [Google Scholar] [CrossRef]
- Krejcova, L.; Richtera, L.; Hynek, D.; Labuda, J.; Adam, V. Current trends in electrochemical sensing and biosensing of DNA methylation. Biosens Bioelectron 2017, 97, 384–399. [Google Scholar] [CrossRef]
- Hossain, T.; Mahmudunnabi, G.; Masud, M.K.; Islam, M.N.; Ooi, L.; Konstantinov, K.; Hossain, M.S.A.; Martinac, B.; Alici, G.; Nguyen, N.T.; et al. Electrochemical biosensing strategies for DNA methylation analysis. Biosens Bioelectron 2017, 94, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Booth, M.A.; Shepherd, P.; Sheppard, A.; Travas-Sejdic, J. Distinguishing cytosine methylation using electrochemical, label-free detection of DNA hybridization and ds-targets. Biosens Bioelectron 2015, 64, 74–80. [Google Scholar] [CrossRef]
- Dai, T.; Pu, Q.; Guo, Y.; Zuo, C.; Bai, S.; Yang, Y.; Yin, D.; Li, Y.; Sheng, S.; Tao, Y.; et al. Analogous modified DNA probe and immune competition method-based electrochemical biosensor for RNA modification. Biosens Bioelectron 2018, 114, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, S.R.; Abreu, C.M.; Parkes, L.; Davies, J.; Yao, S.; Sawhney, M.A.; Margarit, L.; Gonzalez, D.; Pinto, I.M.; Francis, L.W.; et al. Direct monitoring of breast and endometrial cancer cell epigenetic response to DNA methyltransferase and histone deacetylase inhibitors. Biosens Bioelectron 2019, 141, 111386. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, M.; Zhou, T.; Yin, H.; Ai, S. Electrochemical biosensing method for the detection of DNA methylation and assay of the methyltransferase activity. Sensors and Actuators B: Chemical 2013, 178, 412–417. [Google Scholar] [CrossRef]
- Wang, G.L.; Zhou, L.Y.; Luo, H.Q.; Li, N.B. Electrochemical strategy for sensing DNA methylation and DNA methyltransferase activity. Anal Chim Acta 2013, 768, 76–81. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, S.; Han, Y.; Chen, H.; Wang, Q.; Nie, Z.; Huang, Y.; Yao, S. Unique electrocatalytic activity of a nucleic acid-mimicking coordination polymer for the sensitive detection of coenzyme A and histone acetyltransferase activity. Chem Commun (Camb) 2015, 51, 17611–17614. [Google Scholar] [CrossRef]
- Tang, J.; Zou, G.; Chen, C.; Ren, J.; Wang, F.; Chen, Z. Highly Selective Electrochemical Detection of 5-Formyluracil Relying on (2-Benzimidazolyl) Acetonitrile Labeling. Anal Chem 2021, 93, 16439–16446. [Google Scholar] [CrossRef]
- Yin, H.; Zhou, Y.; Xu, Z.; Chen, L.; Zhang, D.; Ai, S. An electrochemical assay for DNA methylation, methyltransferase activity and inhibitor screening based on methyl binding domain protein. Biosens Bioelectron 2013, 41, 492–497. [Google Scholar] [CrossRef]
- Su, J.; He, X.; Wang, Y.; Wang, K.; Chen, Z.; Yan, G. A sensitive signal-on assay for MTase activity based on methylation-responsive hairpin-capture DNA probe. Biosens Bioelectron 2012, 36, 123–128. [Google Scholar] [CrossRef]
- Chen, Y.; Yi, H.; Xiang, Y.; Yuan, R. Commercial glucometer as signal transducer for simple evaluation of DNA methyltransferase activity and inhibitors screening. Anal Chim Acta 2018, 1001, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Ye, Y.; Hu, J.; Xie, C.; Zhou, X.; Yu, H.; Liao, F.; Jiang, S.; Jiang, L.; Xie, G.; et al. XNA probe and CRISPR/Cas12a-powered flexible fluorescent and electrochemical dual-mode biosensor for sensitive detection of m6A site-specific RNA modification. Talanta 2023, 252, 123754. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Hu, J.; Wang, M.; Li, C.C.; Zhang, C.Y. Label-Free and Immobilization-Free Electrochemical Magnetobiosensor for Sensitive Detection of 5-Hydroxymethylcytosine in Genomic DNA. Anal Chem 2019, 91, 1232–1236. [Google Scholar] [CrossRef]
- Zhou, H.; Shen, H.; Dou, B.; Feng, Q.; Han, X.; Wang, P. Construction of a sensitive ratiometric electrochemical sensing platform for DNA methylation detection based on the design of multistep DNA amplification circuits. Sensors and Actuators B: Chemical 2022, 370. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Li, X.; Wang, Y.; Yin, H.; Ai, S. Electrochemical biosensor for detection of DNA hydroxymethylation based on glycosylation and alkaline phosphatase catalytic signal amplification. Electrochimica Acta 2015, 174, 647–652. [Google Scholar] [CrossRef]
- Yu, H.; Pu, Q.; Weng, Z.; Zhou, X.; Li, J.; Yang, Y.; Luo, W.; Guo, Y.; Chen, H.; Wang, D.; et al. DNAzyme based three-way junction assay for antibody-free detection of locus-specific N(6)-methyladenosine modifications. Biosens Bioelectron 2021, 194, 113625. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, X.; Zhao, K. Methylation-specific electrochemical biosensing strategy for highly sensitive and quantitative analysis of promoter methylation of tumor-suppressor gene in real sample. Journal of Electroanalytical Chemistry 2016, 773, 63–68. [Google Scholar] [CrossRef]
- Chen, S.; Dou, Y.; Zhao, Z.; Li, F.; Su, J.; Fan, C.; Song, S. High-Sensitivity and High-Efficiency Detection of DNA Hydroxymethylation in Genomic DNA by Multiplexing Electrochemical Biosensing. Anal Chem 2016, 88, 3476–3480. [Google Scholar] [CrossRef]
- Cheng, W.; Ma, J.; Zhang, Y.; Xu, C.; Zhang, Z.; Hu, L.; Li, J. Bio-inspired construction of a semi-artificial enzyme complex for detecting histone acetyltransferases activity. Analyst 2020, 145, 613–618. [Google Scholar] [CrossRef]
- Islam, M.N.; Masud, M.K.; Nguyen, N.T.; Gopalan, V.; Alamri, H.R.; Alothman, Z.A.; Hossain, M.S.A.; Yamauchi, Y.; Lamd, A.K.; Shiddiky, M.J.A. Gold-loaded nanoporous ferric oxide nanocubes for electrocatalytic detection of microRNA at attomolar level. Biosens Bioelectron 2018, 101, 275–281. [Google Scholar] [CrossRef]
- Wu, G.; Jose, P.A.; Zeng, C. Noncoding RNAs in the Regulatory Network of Hypertension. Hypertension 2018, 72, 1047–1059. [Google Scholar] [CrossRef]
- Sideeq Bhat, K.; Kim, H.; Alam, A.; Ko, M.; An, J.; Lim, S. Rapid and Label-Free Detection of 5-Hydroxymethylcytosine in Genomic DNA Using an Au/ZnO Nanorods Hybrid Nanostructure-Based Electrochemical Sensor. Adv Healthc Mater 2021, 10, e2101193. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, J.; Zhao, M.; Chen, C.; Wang, F.; Chen, Z. Novel electrochemical immunosensor for O(6)-methylguanine-DNA methyltransferase gene methylation based on graphene oxide-magnetic nanoparticles-beta-cyclodextrin nanocomposite. Bioelectrochemistry 2022, 146, 108111. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, F.; Zhang, D.; Zhao, Y.; Wei, J.; Wang, L.; Song, S.; Fan, C.; Zhao, Y. Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination-amplification strategy. Chem Sci 2017, 8, 4764–4770. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Su, J.; Zhao, Z.; Shao, Y.; Dou, Y.; Li, F.; Deng, W.; Shi, J.; Li, Q.; Zuo, X.; et al. DNA Framework-Supported Electrochemical Analysis of DNA Methylation for Prostate Cancers. Nano Lett 2020, 20, 7028–7035. [Google Scholar] [CrossRef]
- Li, X.; Peng, G.; Cui, F.; Qiu, Q.; Chen, X.; Huang, H. Double determination of long noncoding RNAs from lung cancer via multi-amplified electrochemical genosensor at sub-femtomole level. Biosens Bioelectron 2018, 113, 116–123. [Google Scholar] [CrossRef]
- Yin, H.; Zhou, Y.; Yang, Z.; Guo, Y.; Wang, X.; Ai, S.; Zhang, X. Electrochemical immunosensor for N6-methyladenosine RNA modification detection. Sensors and Actuators B: Chemical 2015, 221, 1–6. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Yin, H.; Zhang, Q.; Wang, H.; Fan, H.; Ai, S. Electrochemical immunosensor based on hairpin DNA probe for specific detection of N6-methyladenosine RNA. Journal of Electroanalytical Chemistry 2017, 804, 192–198. [Google Scholar] [CrossRef]
- Zouari, M.; Campuzano, S.; Pingarron, J.M.; Raouafi, N. Amperometric Biosensing of miRNA-21 in Serum and Cancer Cells at Nanostructured Platforms Using Anti-DNA-RNA Hybrid Antibodies. ACS Omega 2018, 3, 8923–8931. [Google Scholar] [CrossRef]
- Thomas, B.; Matson, S.; Chopra, V.; Sun, L.; Sharma, S.; Hersch, S.; Rosas, H.D.; Scherzer, C.; Ferrante, R.; Matson, W. A novel method for detecting 7-methyl guanine reveals aberrant methylation levels in Huntington disease. Anal Biochem 2013, 436, 112–120. [Google Scholar] [CrossRef]
- Hou, P.; Ji, M.; Ge, C.; Shen, J.; Li, S.; He, N.; Lu, Z. Detection of methylation of human p16(Ink4a) gene 5'-CpG islands by electrochemical method coupled with linker-PCR. Nucleic Acids Res 2003, 31, e92. [Google Scholar] [CrossRef] [PubMed]
- Kalofonou, M.; Toumazou, C. Semiconductor technology for early detection of DNA methylation for cancer: From concept to practice. Sensors and Actuators B: Chemical 2013, 178, 572–580. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).