Submitted:
26 June 2023
Posted:
26 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. RABV Genome Enrichment
2.3. Library Preparation
2.4. Nanopore Sequencing
2.5. Bioinformatics Data Processing and Analysis
3. Results
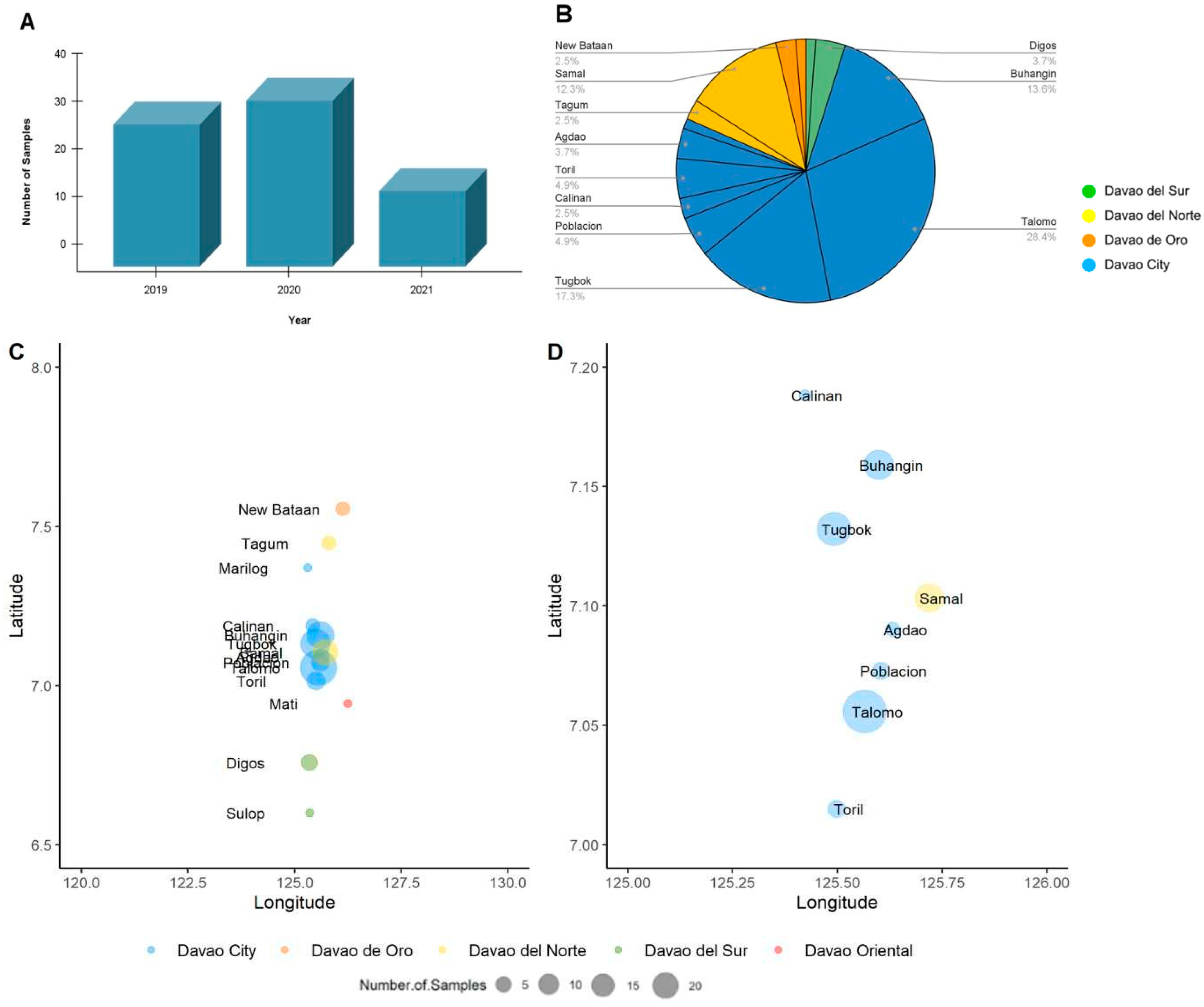
3.1. Sample Characteristics
3.2. RABV Whole Genome Sequences
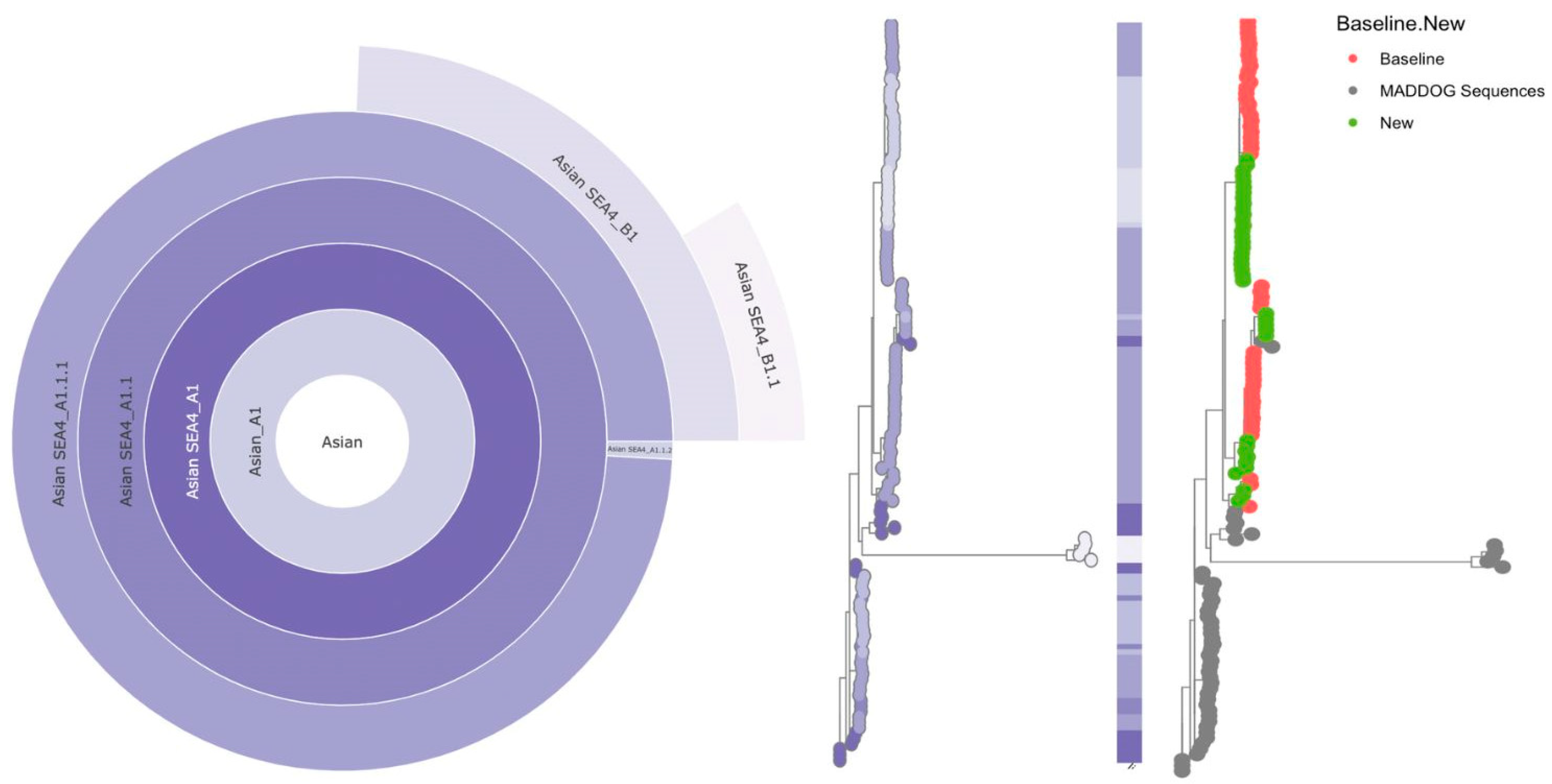
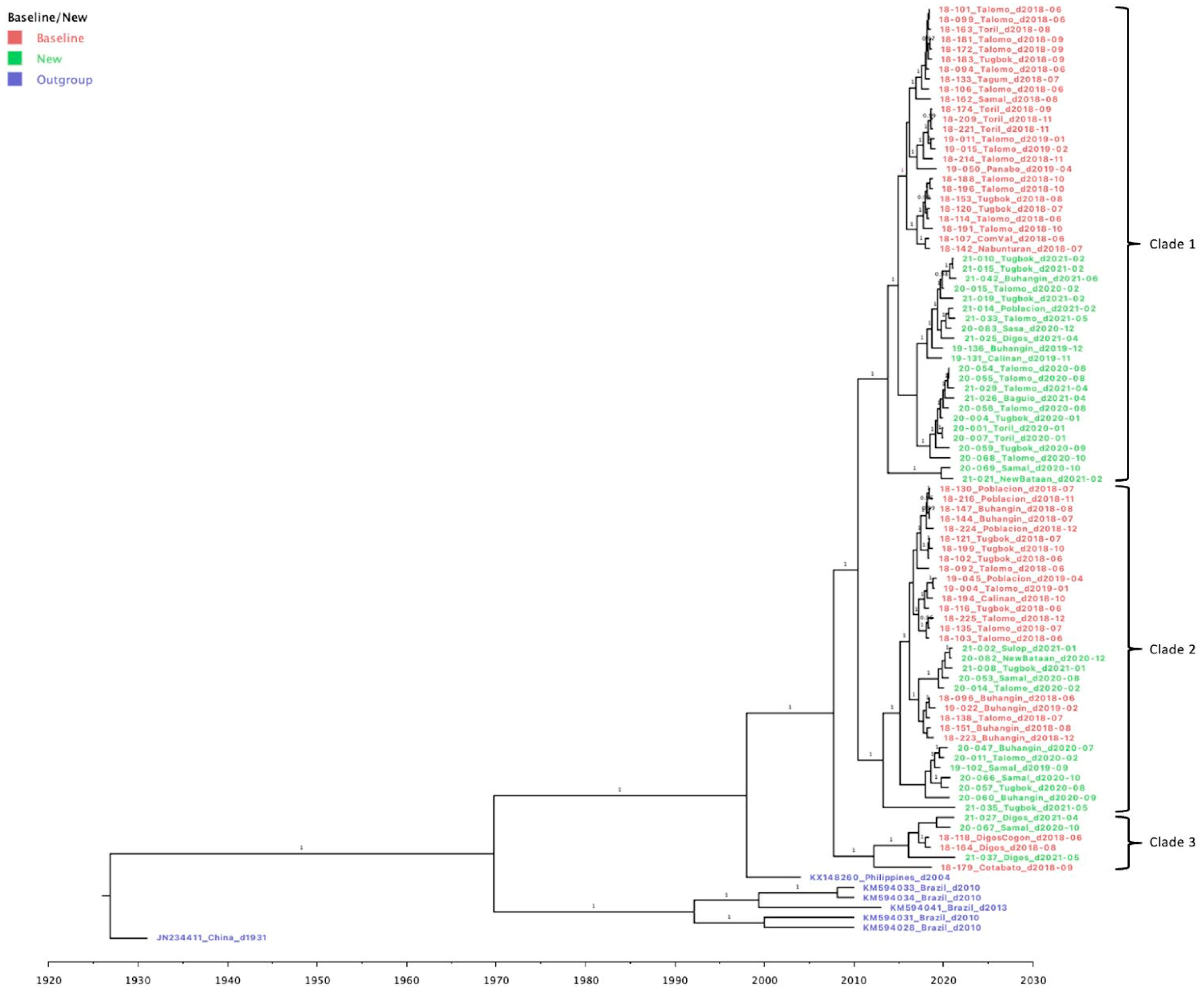
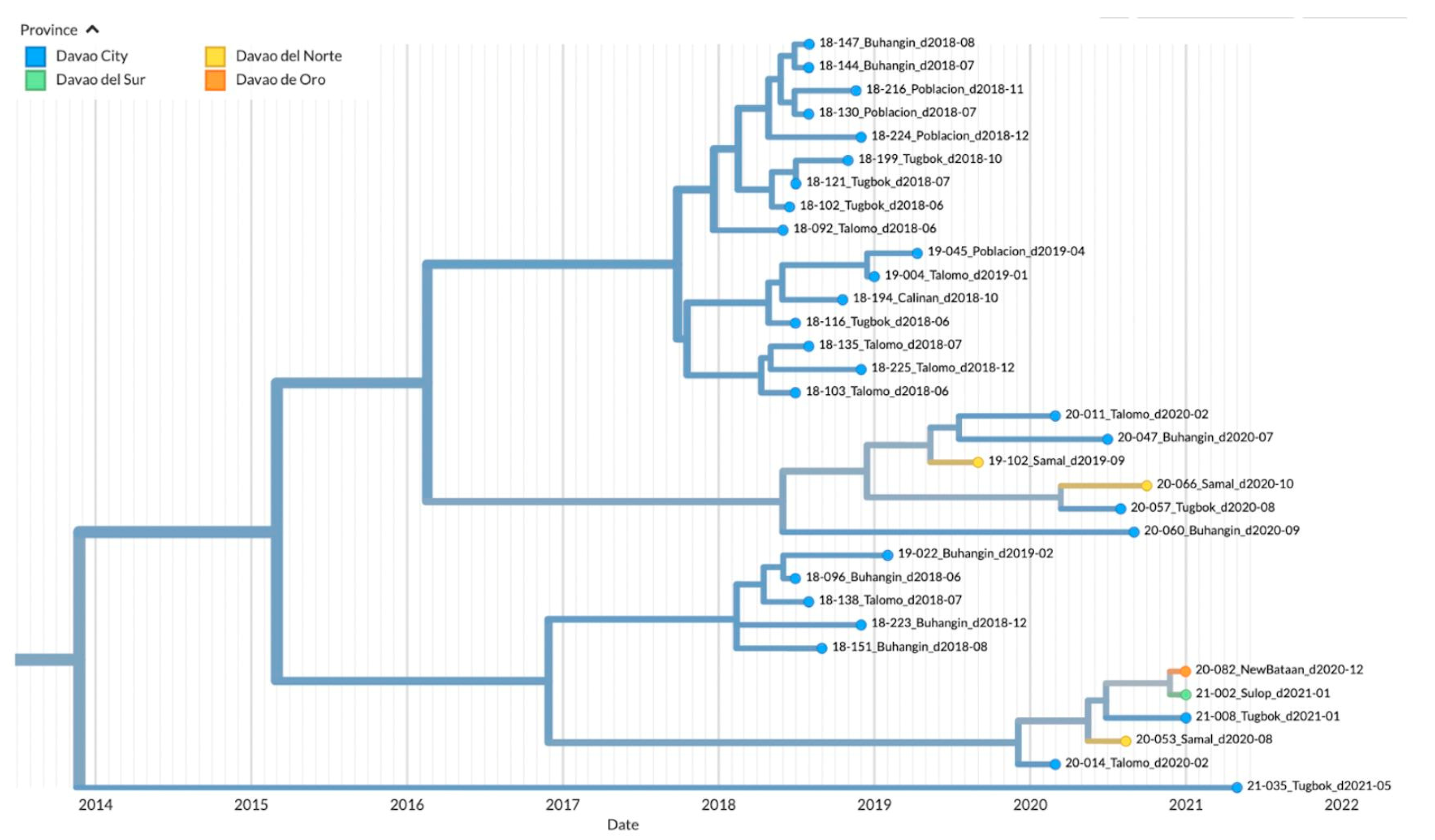
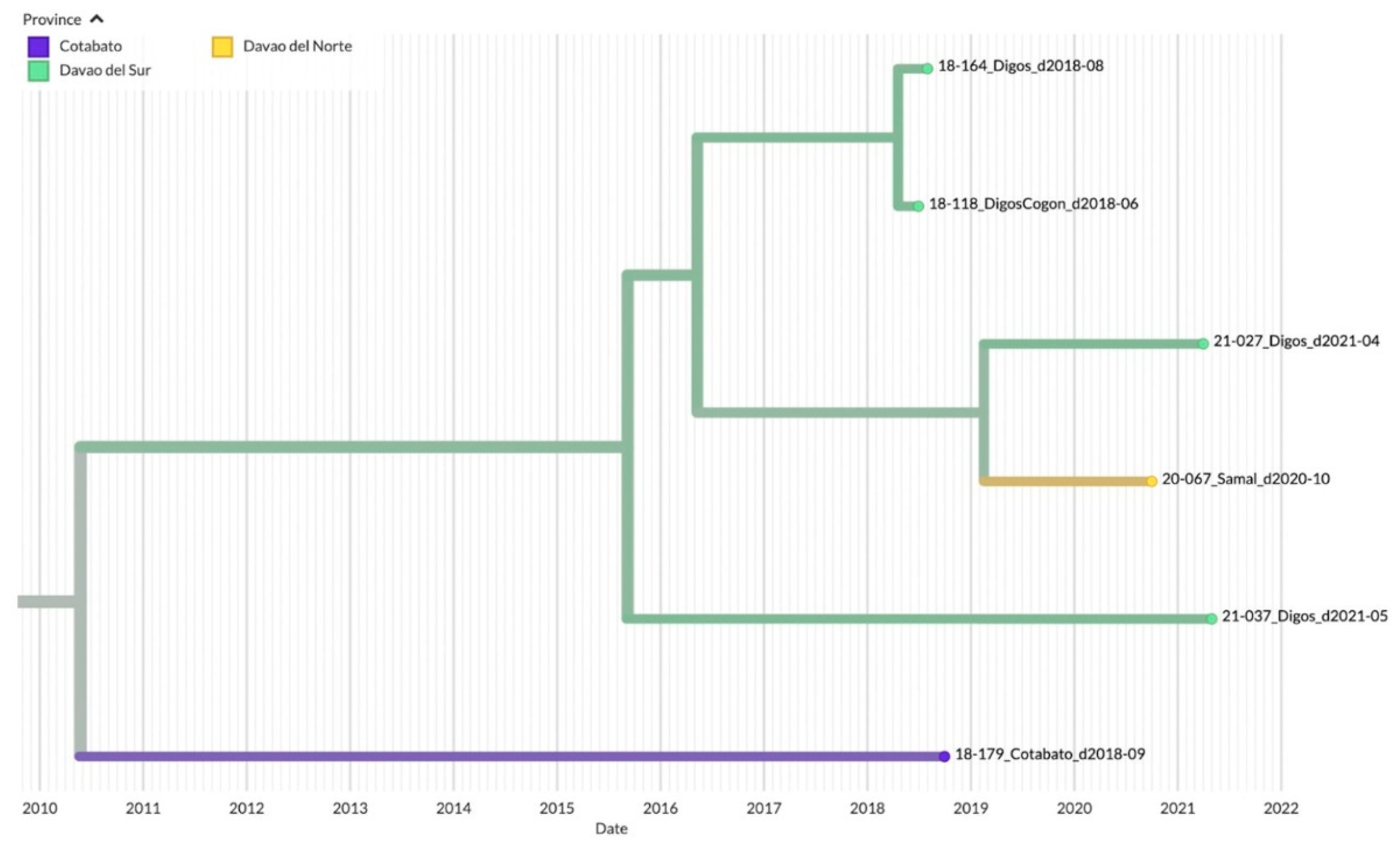
3.3. Genetic Diversity and Phylogeographic Distribution of RABV
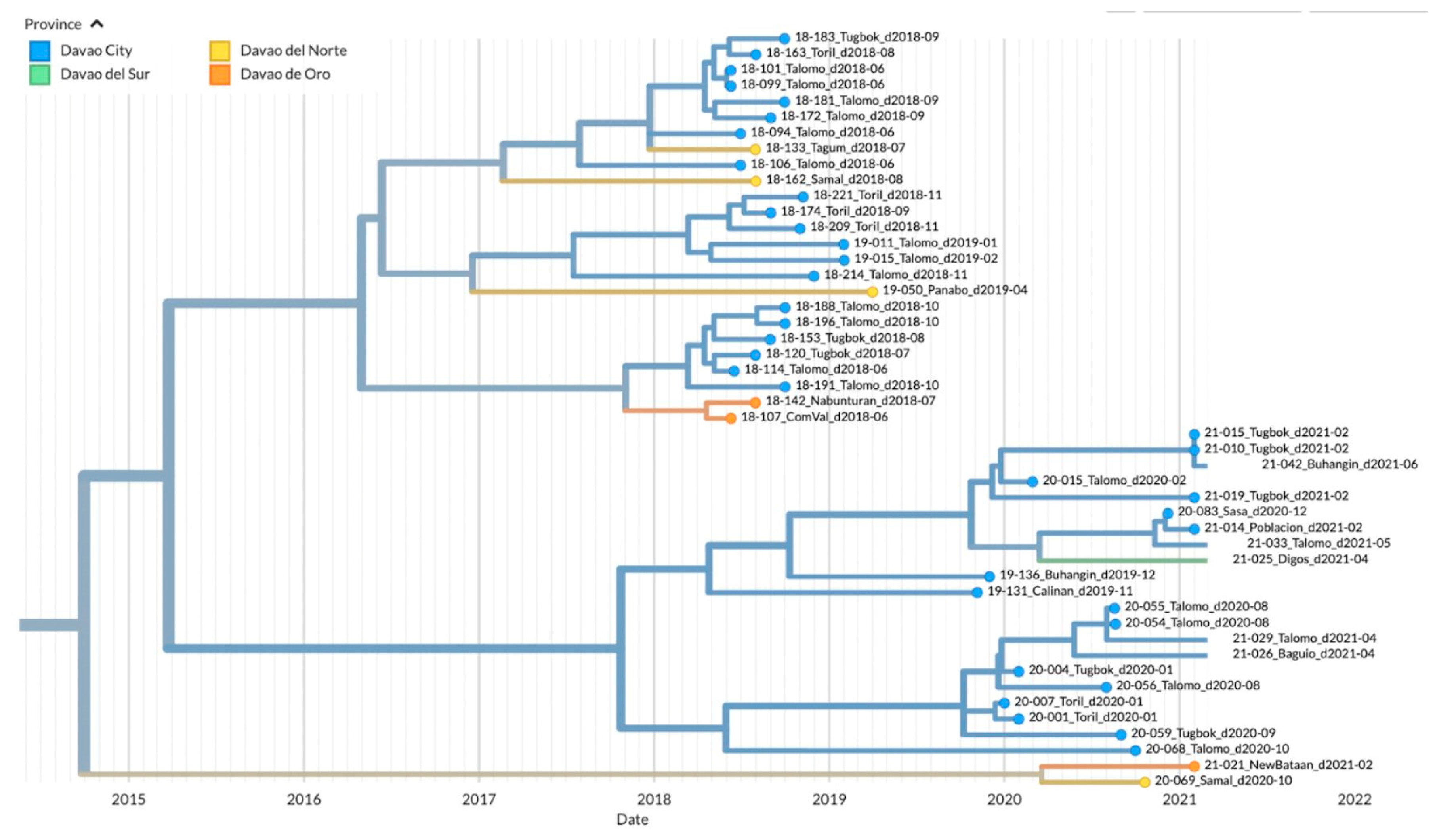
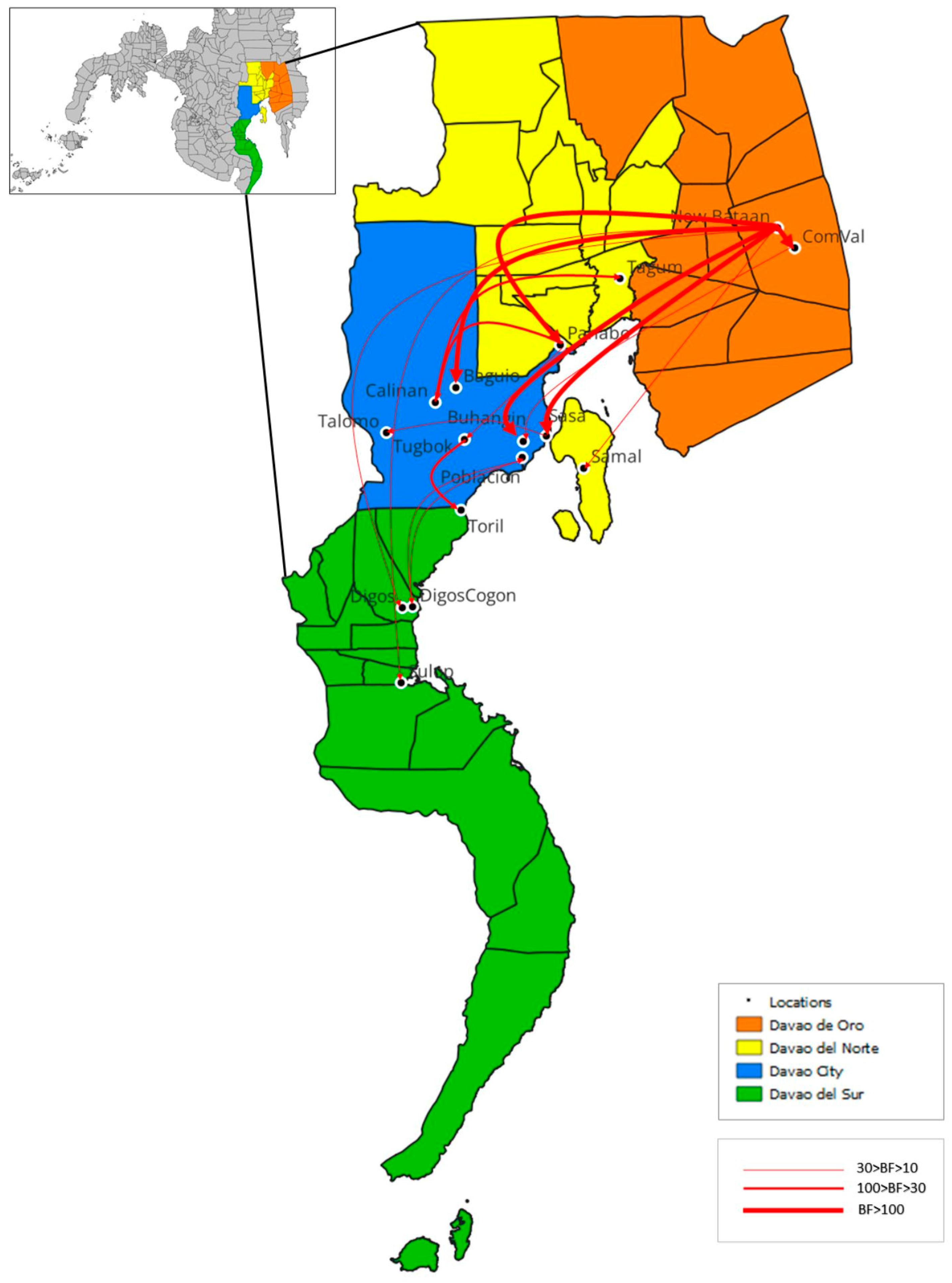
3.4. Phylodynamics of RABV in Davao Region
4. Discussion
4.1. Emerging Trends on RABV Diversity and Transmission Patterns
4.2. Genomic Surveillance in a Low Resource Setting
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample ID | Country | Year | Lineage |
|---|---|---|---|
| AB573762 | Japan | 2006 | Asian SEA4 |
| AB573763 | Japan | 2006 | Asian SEA4 |
| EU086201 | Philippines | 1994 | Asian SEA4 |
| EU086202 | Philippines | 1994 | Asian SEA4 |
| EU086203 | Philippines | 2000 | Asian SEA4 |
| EU086204 | Philippines | 2001 | Asian SEA4 |
| KF620487 | Taiwan | 2012 | Asian SEA5 |
| KF620488 | Taiwan | 2012 | Asian SEA5 |
| KF620489 | Taiwan | 2013 | Asian SEA5 |
| KP860149 | Taiwan | 2013 | Asian SEA5 |
| KP860153 | Taiwan | 2013 | Asian SEA5 |
| KX148259 | Philippines | 1994 | Asian SEA4 |
| KX148260 | Philippines | 2004 | Asian SEA4 |
| KX148261 | Philippines | 1994 | Asian SEA4 |
| KX148262 | Philippines | 1994 | Asian SEA4 |
| KX148263 | Philippines | 1994 | Asian SEA4 |
| LC571945 | Japan | 2006 | Asian SEA4 |
| LC571946 | Japan | 2020 | Asian SEA4 |
| MN726836 | Philippines | 2013 | Asian SEA4 |
| MN726837 | Philippines | 2013 | Asian SEA4 |
| MN726838 | Philippines | 2013 | Asian SEA4 |
| MN726839 | Philippines | 2015 | Asian SEA4 |
| MN726840 | Philippines | 2015 | Asian SEA4 |
| MN726841 | Philippines | 2014 | Asian SEA4 |
| MN726843 | Philippines | 2012 | Asian SEA4 |
| MN726846 | Philippines | 2015 | Asian SEA4 |
| MN726847 | Philippines | 2017 | Asian SEA4 |
| MN726848 | Philippines | 2016 | Asian SEA4 |
| MN726851 | Philippines | 2014 | Asian SEA4 |
| MN726853 | Philippines | 2012 | Asian SEA4 |
| MN726854 | Philippines | 2015 | Asian SEA4 |
| MN726855 | Philippines | 2016 | Asian SEA4 |
| MN726856 | Philippines | 2012 | Asian SEA4 |
| MN726858 | Philippines | 2016 | Asian SEA4 |
| MN726861 | Philippines | 2015 | Asian SEA4 |
| MN726862 | Philippines | 2013 | Asian SEA4 |
| MN726865 | Philippines | 2014 | Asian SEA4 |
| MN726866 | Philippines | 2014 | Asian SEA4 |
| MN726867 | Philippines | 2012 | Asian SEA4 |
| MN726868 | Philippines | 2016 | Asian SEA4 |
| MN726869 | Philippines | 2013 | Asian SEA4 |
| MN726870 | Philippines | 2014 | Asian SEA4 |
| MN726871 | Philippines | 2013 | Asian SEA4 |
| MN726872 | Philippines | 2017 | Asian SEA4 |
| MN726873 | Philippines | 2014 | Asian SEA4 |
| MN726874 | Philippines | 2012 | Asian SEA4 |
| MN726879 | Philippines | 2012 | Asian SEA4 |
| MN726881 | Philippines | 2015 | Asian SEA4 |
| MN726882 | Philippines | 2019 | Asian SEA4 |
| MN857169 | Philippines | 2019 | Asian SEA4 |
| Sample ID | New/Baseline | Length | Year | Lineage |
|---|---|---|---|---|
| 21-002_Sulop_d2021-01 | New | 10370 | 2021 | Asian SEA4_A1.1.2 |
| 21-008_Tugbok_d2021-01 | New | 10551 | 2021 | Asian SEA4_A1.1.1 |
| 21-010_Tugbok_d2021-02 | New | 11331 | 2021 | Asian SEA4_B1.1 |
| 20-047_Buhangin_d2020-07 | New | 9965 | 2020 | Asian SEA4_A1.1.1 |
| 20-053_Samal_d2020-08 | New | 9481 | 2020 | Asian SEA4_A1.1.1 |
| 20-054_Talomo_d2020-08 | New | 10851 | 2020 | Asian SEA4_A1.1.1 |
| 20-055_Talomo_d2020-08 | New | 11035 | 2020 | Asian SEA4_A1.1.1 |
| 20-056_Talomo_d2020-08 | New | 11195 | 2020 | Asian SEA4_A1.1.1 |
| 20-057_Tugbok_d2020-08 | New | 10219 | 2020 | Asian SEA4_A1.1.1 |
| 20-059_Tugbok_d2020-09 | New | 11039 | 2020 | Asian SEA4_A1.1.1 |
| 20-060_Buhangin_d2020-09 | New | 10347 | 2020 | Asian SEA4_A1.1.1 |
| 20-066_Samal_d2020-10 | New | 10737 | 2020 | Asian SEA4_A1.1.1 |
| 20-067_Samal_d2020-10 | New | 11626 | 2020 | Asian SEA4_A1.1.1 |
| 20-068_Talomo_d2020-10 | New | 11397 | 2020 | Asian SEA4_A1.1.1 |
| 20-069_Samal_d2020-10 | New | 11013 | 2020 | Asian SEA4_B1 |
| 20-082_NewBataan_d2020-12 | New | 10550 | 2020 | Asian SEA4_A1.1.1 |
| 20-083_Sasa_d2020-12 | New | 10473 | 2020 | Asian SEA4_B1.1 |
| 21-027_Digos_d2021-04 | New | 11221 | 2021 | Asian SEA4_A1.1.1 |
| 21-029_Talomo_d2021-04 | New | 11422 | 2021 | Asian SEA4_A1.1.1 |
| 21-033_Talomo_d2021-05 | New | 11619 | 2021 | Asian SEA4_B1.1 |
| 21-035_Tugbok_d2021-05 | New | 10958 | 2021 | Asian SEA4_A1.1.1 |
| 21-037_Digos_d2021-05 | New | 11570 | 2021 | Asian SEA4_A1.1.1 |
| 21-042_Buhangin_d2021-06 | New | 11587 | 2021 | Asian SEA4_B1.1 |
| 18-092_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-094_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-096_Buhangin_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-099_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-101_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-102_Tugbok_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-103_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-106_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-107_ComVal_d2018-06 | New | 11800 | 2018 | Asian SEA4_B1 |
| 18-114_Talomo_d2018-06 | New | 11800 | 2018 | Asian SEA4_B1 |
| 18-116_Tugbok_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-118_DigosCogon_d2018-06 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-120_Tugbok_d2018-07 | New | 11800 | 2018 | Asian SEA4_B1 |
| 18-121_Tugbok_d2018-07 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-130_Poblacion_d2018-07 | New | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-133_Tagum_d2018-07 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-135_Talomo_d2018-07 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-138_Talomo_d2018-07 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-142_Nabunturan_d2018-07 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-144_Buhangin_d2018-07 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-147_Buhangin_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-151_Buhangin_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-153_Tugbok_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-162_Samal_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-163_Toril_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-164_Digos_d2018-08 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-172_Talomo_d2018-09 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-174_Toril_d2018-09 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-179_Cotabato_d2018-09 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-181_Talomo_d2018-09 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-183_Tugbok_d2018-09 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-188_Talomo_d2018-10 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-191_Talomo_d2018-10 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-194_Calinan_d2018-10 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-196_Talomo_d2018-10 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-199_Tugbok_d2018-10 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-209_Toril_d2018-11 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-214_Talomo_d2018-11 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-216_Poblacion_d2018-11 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-221_Toril_d2018-11 | Baseline | 11800 | 2018 | Asian SEA4_B1 |
| 18-223_Buhangin_d2018-12 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-224_Poblacion_d2018-12 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 18-225_Talomo_d2018-12 | Baseline | 11800 | 2018 | Asian SEA4_A1.1.1 |
| 19-004_Talomo_d2019-01 | Baseline | 11800 | 2019 | Asian SEA4_A1.1.1 |
| 19-011_Talomo_d2019-01 | Baseline | 11800 | 2019 | Asian SEA4_B1 |
| 19-015_Talomo_d2019-02 | Baseline | 11800 | 2019 | Asian SEA4_B1 |
| 19-022_Buhangin_d2019-02 | Baseline | 11800 | 2019 | Asian SEA4_A1.1.1 |
| 19-045_Poblacion_d2019-04 | Baseline | 11800 | 2019 | Asian SEA4_A1.1.1 |
| 19-050_Panabo_d2019-04 | Baseline | 11800 | 2019 | Asian SEA4_B1 |
| 19-102_Samal_d2019-09 | Baseline | 10122 | 2019 | Asian SEA4_A1.1.1 |
| 19-131_Calinan_d2019-11 | Baseline | 9804 | 2019 | Asian SEA4_B1 |
| 19-136_Buhangin_d2019-12 | Baseline | 10697 | 2019 | Asian SEA4_B1.1 |
| 20-001_Toril_d2020-01 | Baseline | 10531 | 2020 | Asian SEA4_A1.1.1 |
| 20-004_Tugbok_d2020-01 | Baseline | 10958 | 2020 | Asian SEA4_A1.1.1 |
| 20-007_Toril_d2020-01 | Baseline | 11284 | 2020 | Asian SEA4_A1.1.1 |
| 20-011_Talomo_d2020-02 | Baseline | 9553 | 2020 | Asian SEA4_A1.1.1 |
| 20-014_Talomo_d2020-02 | Baseline | 10535 | 2020 | Asian SEA4_A1.1.1 |
| 20-015_Talomo_d2020-02 | Baseline | 11378 | 2020 | Asian SEA4_B1.1 |
| 21-014_Poblacion_d2021-02 | Baseline | 11547 | 2021 | Asian SEA4_B1.1 |
| 21-015_Tugbok_d2021-02 | Baseline | 11182 | 2021 | Asian SEA4_B1.1 |
| 21-019_Tugbok_d2021-02 | Baseline | 11404 | 2021 | Asian SEA4_B1.1 |
| 21-021_NewBataan_d2021-02 | Baseline | 11411 | 2021 | Asian SEA4_B1 |
| 21-025_Digos_d2021-04 | Baseline | 11243 | 2021 | Asian SEA4_B1.1 |
| 21-026_Baguio_d2021-04 | Baseline | 11163 | 2021 | Asian SEA4_A1.1.1 |
References
- World Health Organization. Zero by 30: the global strategic plan to end human deaths from dog-mediated rabies by 2030: United Against Rabies Collaboration: first annual progress report: Global strategic plan to end human deaths from dog-mediated rabies by 2030. No. WHO/CDS/NTD/NZD/2019.04. World Health Organization, 2019.
- Tarantola, Arnaud. "Four thousand years of concepts relating to rabies in animals and humans, its prevention and its cure." Tropical medicine and infectious disease 2, no. 2 (2017): 5.
- de Melo, Guilherme Dias, Florian Sonthonnax, Gabriel Lepousez, Grégory Jouvion, Andrea Minola, Fabrizia Zatta, Florence Larrous et al. "A combination of two human monoclonal antibodies cures symptomatic rabies." EMBO molecular medicine 12, no. 11 (2020): e12628. [CrossRef]
- Cantaert, Tineke, Laurence Borand, Lauriane Kergoat, Chanthy Leng, Sivlin Ung, Sotheary In, Yiksing Peng et al. "A 1-week intradermal dose-sparing regimen for rabies post-exposure prophylaxis (RESIST-2): an observational cohort study." The Lancet Infectious Diseases 19, no. 12 (2019): 1355-1362. [CrossRef]
- Department of Health. "Anti-Rabies Act (RA 9482) and Other Issuances on Rabies Control and Prevention Programs." Anti-Rabies Act (RA 9482) and Other Issuances on Rabies Control and Prevention Programs, (2007). Accessed December 16, 2022. https://rabies.doh.gov.ph/images/PDF/IRR-INSIDE-REV2_new.pdf.
- Department of Health. "National Rabies Prevention and Control Program." Strategic Plan 2020-2025, (2020). Accessed December 16, 2022. https://rr-asia.woah.org/wp-content/uploads/2020/03/final-mtp-rabies_philippines.pdf.
- Lachica, Zython Paul T., Sherelyn A. Evangelio, Eliezer O. Diamante, Abigail J. Clemente, Johanna Marie Peralta, Lyre Anni E. Murao, May Anne E. Mata, and Pedro A. Alviola IV. "Trends of canine rabies lyssavirus and impact of the intensified rabies control program in Davao city, Philippines: 2006–2017." Philipp J Sci 148 (2019): 751-763.
- Klepac, Petra, C. Jessica E. Metcalf, Angela R. McLean, and Katie Hampson. "Towards the endgame and beyond: complexities and challenges for the elimination of infectious diseases." Philosophical Transactions of the Royal Society B: Biological Sciences 368, no. 1623 (2013): 20120137. [CrossRef]
- Cotton, James A., Matthew Berriman, Love Dalén, and Ian Barnes. "Eradication genomics—lessons for parasite control." Science 361, no. 6398 (2018): 130-131.
- Balloux, Francois, Ola Brønstad Brynildsrud, Lucy Van Dorp, Liam P. Shaw, Hongbin Chen, Kathryn A. Harris, Hui Wang, and Vegard Eldholm. "From theory to practice: translating whole-genome sequencing (WGS) into the clinic." Trends in microbiology 26, no. 12 (2018): 1035-1048. [CrossRef]
- Brunker, Kirstyn, Gurdeep Jaswant, S. M. Thumbi, Kennedy Lushasi, Ahmed Lugelo, Anna M. Czupryna, Fred Ade et al. "Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes." Wellcome Open Research 5 (2020). [CrossRef]
- Suzuki, Yutaka. "Advent of a new sequencing era: long-read and on-site sequencing." Journal of Human Genetics 65, no. 1 (2020): 1-1. [CrossRef]
- Mikheyev, Alexander S., and Mandy MY Tin. "A first look at the Oxford Nanopore MinION sequencer." Molecular ecology resources 14, no. 6 (2014): 1097-1102. [CrossRef]
- Tyler, Andrea D., Laura Mataseje, Chantel J. Urfano, Lisa Schmidt, Kym S. Antonation, Michael R. Mulvey, and Cindi R. Corbett. "Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications." Scientific reports 8, no. 1 (2018): 1-12. [CrossRef]
- Bacus, Michael G., Sheryl Grace C. Buenaventura, Allan Michael C. Mamites, Hannah G. Elizagaque, Christian C. Labrador, Frederick C. Delfin, Ma Noreen J. Eng, Arlene P. Lagare, Gloria N. Marquez, and Lyre Anni E. Murao. "Genome-based local dynamics of canine rabies virus epidemiology, transmission, and evolution in Davao City, Philippines, 2018–2019." Infection, Genetics and Evolution 92 (2021): 104868. [CrossRef]
- Quick, Joshua, Nathan D. Grubaugh, Steven T. Pullan, Ingra M. Claro, Andrew D. Smith, Karthik Gangavarapu, Glenn Oliveira et al. "Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples." Nature protocols 12, no. 6 (2017): 1261-1276. [CrossRef]
- Katoh, Kazutaka, and Hiroyuki Toh. "Parallelization of the MAFFT multiple sequence alignment program." Bioinformatics 26, no. 15 (2010): 1899-1900. [CrossRef]
- Penir, Sarah. 2017. Calculating Mapping Statistics from a SAM/BAM File Using SAMtools and Awk. Retrieved from https://sarahpenir.github.io/bioinformatics/awk/calculating-mapping-stats-from-a-bam-file-using-samtools-and-awk/.
- Morobe, John Mwita, Brigitte Pool, Lina Marie, Dwayne Didon, Arnold W. Lambisia, Timothy Makori, Khadija Said Mohammed et al. "Genomic Epidemiology of SARS-CoV-2 in Seychelles, 2020–2021." Viruses 14, no. 6 (2022): 1318. [CrossRef]
- Ruis, Chris. 2022. Bammix v1.0.0. Retrieved from https://github.com/chrisruis/bammix.
- Kumar, Sudhir, Glen Stecher, Michael Li, Christina Knyaz, and Koichiro Tamura. "MEGA X: molecular evolutionary genetics analysis across computing platforms." Molecular biology and evolution 35, no. 6 (2018): 1547. [CrossRef]
- Rambaut, Andrew, Alexei J. Drummond, Dong Xie, Guy Baele, and Marc A. Suchard. "Posterior summarization in Bayesian phylogenetics using Tracer 1.7." Systematic biology 67, no. 5 (2018): 901-904. [CrossRef]
- Hadfield, James, Colin Megill, Sidney M. Bell, John Huddleston, Barney Potter, Charlton Callender, Pavel Sagulenko, Trevor Bedford, and Richard A. Neher. "Nextstrain: real-time tracking of pathogen evolution." Bioinformatics 34, no. 23 (2018): 4121-4123. [CrossRef]
- Campbell, Kathryn, Robert J. Gifford, Joshua Singer, Verity Hill, Aine O’toole, Andrew Rambaut, Katie Hampson, and Kirstyn Brunker. "Making genomic surveillance deliver: A lineage classification and nomenclature system to inform rabies elimination." PLoS Pathogens 18, no. 5 (2022): e1010023. [CrossRef]
- Bielejec, Filip, Guy Baele, Bram Vrancken, Marc A. Suchard, Andrew Rambaut, and Philippe Lemey. "SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes." Molecular biology and evolution 33, no. 8 (2016): 2167-2169. [CrossRef]
- Rambaut, Andrew, Edward C. Holmes, Áine O’Toole, Verity Hill, John T. McCrone, Christopher Ruis, Louis du Plessis, and Oliver G. Pybus. "A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology." Nature microbiology 5, no. 11 (2020): 1403-1407. [CrossRef]
- Philippine Statistics Authority Gross Regional Domestic Product by Year Published. (2020). https://psa.gov.ph/grdp/ tables.
- Denduangboripant, Jessada, Supaporn Wacharapluesadee, Boonlert Lumlertdacha, Nipada Ruankaew, Wirongrong Hoonsuwan, Apirom Puanghat, and Thiravat Hemachudha. "Transmission dynamics of rabies virus in Thailand: implications for disease control." BMC Infectious Diseases 5, no. 1 (2005): 1-11. [CrossRef]
- Tohma, Kentaro, Mariko Saito, Taro Kamigaki, Laarni T. Tuason, Catalino S. Demetria, Jun Ryan C. Orbina, Daria L. Manalo et al. "Phylogeographic analysis of rabies viruses in the Philippines." Infection, Genetics and Evolution 23 (2014): 86-94. [CrossRef]
- Talbi, Chiraz, Philippe Lemey, Marc A. Suchard, Elbia Abdelatif, Mehdi Elharrak, Nourlil Jalal, Abdellah Faouzi et al. "Phylodynamics and human-mediated dispersal of a zoonotic virus." PLoS pathogens 6, no. 10 (2010): e1001166. [CrossRef]
- Layan, Maylis, Simon Dellicour, Guy Baele, Simon Cauchemez, and Hervé Bourhy. "Mathematical modelling and phylodynamics for the study of dog rabies dynamics and control: A scoping review." PLOS Neglected Tropical Diseases 15, no. 5 (2021): e0009449. [CrossRef]
- Zinsstag, Jakob, Monique Lechenne, Mirjam Laager, Rolande Mindekem, Service Naïssengar, Assandi Oussiguéré, Kebkiba Bidjeh et al. "Vaccination of dogs in an African city interrupts rabies transmission and reduces human exposure." Science translational medicine 9, no. 421 (2017): eaaf6984. [CrossRef]
- Bourhy, Hervé, Emmanuel Nakouné, Matthew Hall, Pierre Nouvellet, Anthony Lepelletier, Chiraz Talbi, Laurence Watier et al. "Revealing the micro-scale signature of endemic zoonotic disease transmission in an African urban setting." PLoS pathogens 12, no. 4 (2016): e1005525. [CrossRef]
- Hayman, David TS, Nicholas Johnson, Daniel L. Horton, Jessica Hedge, Philip R. Wakeley, Ashley C. Banyard, Shoufeng Zhang, Andy Alhassan, and Anthony R. Fooks. "Evolutionary history of rabies in Ghana." PLoS Neglected Tropical Diseases 5, no. 4 (2011): e1001. [CrossRef]
- De Benedictis, P., A. Sow, A. Fusaro, C. Veggiato, C. Talbi, A. Kaboré, W. G. Dundon, Hervé Bourhy, and I. Capua. "Phylogenetic analysis of rabies viruses from Burkina Faso, 2007." Zoonoses and public health 57, no. 7-8 (2010): e42-e46. [CrossRef]
- Salomao, Cristolde, Amílcar Nacima, Lutero Cuamba, Lorna Gujral, Olga Amiel, Cynthia Baltazar, Julie Cliff, and Eduardo Samo Gudo. "Epidemiology, clinical features and risk factors for human rabies and animal bites during an outbreak of rabies in Maputo and Matola cities, Mozambique, 2014: Implications for public health interventions for rabies control." PLOS Neglected Tropical Diseases 11, no. 7 (2017): e0005787. [CrossRef]
- Beran, George W. "Ecology of dogs in the Central Philippines in relation to rabies control efforts." Comparative immunology, microbiology and infectious diseases 5, no. 1-3 (1982): 265-270. [CrossRef]
- Rysava, Kristyna, Tamara Mancero, Eduardo Caldas, Mary Freire de Carvalho, André PB Castro, Veronica Gutiérrez, Daniel T. Haydon et al. "Towards the elimination of dog-mediated rabies: development and application of an evidence-based management tool." BMC infectious diseases 20, no. 1 (2020): 1-14. [CrossRef]
- Gigante, Crystal M., Gowri Yale, Rene Edgar Condori, Niceta Cunha Costa, Nguyen Van Long, Phan Quang Minh, Vo Dinh Chuong et al. "Portable rabies virus sequencing in canine rabies endemic countries using the Oxford Nanopore MinION." Viruses 12, no. 11 (2020): 1255. [CrossRef]
- Natarajan, Aravind, Alvin Han, Soumaya Zlitni, Erin F. Brooks, Summer E. Vance, Marlene Wolfe, Upinder Singh et al. "Standardized preservation, extraction and quantification techniques for detection of fecal SARS-CoV-2 RNA." Nature communications 12, no. 1 (2021): 5753. [CrossRef]
| Procedure | Challenges Encountered |
Original Protocol | Modification |
|---|---|---|---|
| RNA Extraction | Prescribed kit is unavailable | Zymo Research Quick-RNA miniprep kit, 2ml ceramic tubes (1.4mm), and Terralyzer | Biospin Viral DNA/RNA Extraction Kit, 2ml ceramic tubes (2.8mm), Bead Mill Homogenizer |
| Multiplex PCR | Low concentration of PCR products | Conventional multiplex PCR | Touchdown multiplex PCR |
| Library Preparation | Low concentration post barcode ligation | 5ng DNA input per sample; 70% ethanol for cleanup wash step |
50ng DNA input per sample; Freshly prepared 80% ethanol for cleanup wash step |
| Ultra II Ligation Module for Native Barcoding not available in purchased kit | Ultra II Ligation Master Mix and Ligation Enhancer | Blunt/TA Ligase Master Mix diluted with NFW | |
| Sequencing | Low read number Recycling of flow cells |
Run can finish in six hours, basecalling mode on for fast basecalling n/a |
Run extended to 16 hours or more, basecalling mode off Yes, but excluded sequences that overlapped barcodes with samples from previous run |
| Sequence Batch No. | No. of Samples a |
Concentration of Barcoded Amplicon Pool (ng/ul) |
Concentration of Final Library (ng/ul) |
|---|---|---|---|
| 1 | 12b | 3.78 | 0.956 |
| 2 | 17 | 1.39 | 1.28 |
| 3 | 17 | 1.18 | 1.21 |
| 4 | 17 | 0.84 | 0.14 |
| Sequence Batch No. | Sample ID |
Identified as RABV? |
Closest full genome ref seq |
Whole genome coverage |
Breadth of coverage | Coding region coverage | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | P | M | G | L | ||||||
| 1 | 27-21 | Yes | KX149259 | 94% | 98% | 100% | 100% | 100% | 100% | 91.2% |
| 29-21 | Yes | KX149259 | 96% | 99% | 100% | 100% | 100% | 100% | 94.4% | |
| 33-21 | Yes | KX149259 | 97% | 99% | 100% | 100% | 100% | 100% | 97.5% | |
| 35-21 | Yes | KX149259 | 92% | 99% | 100% | 100% | 100% | 100% | 90.9% | |
| 37-21 | Yes | KX149259 | 97% | 99% | 100% | 100% | 100% | 100% | 97.5% | |
| 42-21 | Yes | KX149259 | 97% | 99% | 100% | 100% | 100% | 100% | 97.5% | |
| 2 | 47-20 | Yes | KX149259 | 84% | 92% | 100% | 100% | 100% | 100% | 80.2% |
| 53-20 | Yes | KX149259 | 79% | 94% | 100% | 100% | 100% | 100% | 80.2% | |
| 54-20 | Yes | KX149259 | 91% | 97% | 100% | 100% | 100% | 100% | 90.9% | |
| 55-20 | Yes | KX149259 | 92% | 96% | 100% | 100% | 80.1% | 100% | 90.9% | |
| 56-20 | Yes | KX149259 | 94% | 96% | 100% | 100% | 100% | 100% | 94.5% | |
| 57-20 | Yes | KX149259 | 86% | 91% | 100% | 100% | 100% | 100% | 83.8% | |
| 59-20 | Yes | KX149259 | 93% | 96% | 100% | 100% | 100% | 100% | 94.5% | |
| 60-20 | Yes | KX149259 | 87% | 94% | 100% | 100% | 100% | 100% | 80.3% | |
| 66-20 | Yes | KX149259 | 90% | 97% | 100% | 100% | 100% | 100% | 83.8% | |
| 67-20 | Yes | KX149259 | 97% | 98% | 100% | 100% | 100% | 100% | 97.5% | |
| 68-20 | Yes | KX149259 | 96% | 98% | 100% | 100% | 80.1% | 87.1% | 94.3% | |
| 69-20 | Yes | KX149259 | 92% | 98% | 100% | 100% | 100% | 100% | 94.5% | |
| 82-20 | Yes | KX149259 | 88% | 99% | 100% | 100% | 100% | 100% | 83.4% | |
| 83-20 | Yes | KX149259 | 94% | 94% | 100% | 100% | 100% | 100% | 87.4% | |
| 02-21 | Yes | KX149259 | 87% | 96% | 100% | 100% | 100% | 100% | 83.4% | |
| 08-21 | Yes | KX149259 | 88% | 98% | 100% | 100% | 100% | 100% | 83.4% | |
| 10-21 | Yes | KX149259 | 95% | 99% | 100% | 100% | 100% | 100% | 94.5% | |
| 3 | 14-21 | Yes | KX149259 | 97% | 99% | 100% | 99.9% | 100% | 99.9% | 97.5% |
| 15-21 | Yes | KX149259 | 94% | 98% | 100% | 99.9% | 100% | 99.9% | 94.5% | |
| 19-21 | Yes | KX149259 | 96% | 99% | 100% | 99.9% | 100% | 99.9% | 94.5% | |
| 21-21 | Yes | KX149259 | 96% | 99% | 99.9% | 99.9% | 100% | 99.9% | 94.5% | |
| 25-21 | Yes | KX149259 | 94% | 98% | 100% | 99.9% | 100% | 99.9% | 94.5% | |
| 26-21 | Yes | KX149259 | 94% | 97% | 100% | 99.9% | 100% | 99.9% | 90.9% | |
| 4 | 98-19 | Yesb | KX149259 | 38% | 50% | 15.6% | 75.7% | 63.1% | 41.3% | 35.5% |
| 101-19 | Yesb | KX149259 | 25% | 33% | 15.6% | 75.7% | 31.2% | 17.0% | 21.6% | |
| 105-19 | Yesb | KX149259 | 25% | 32% | 15.6% | 75.7% | 31.2% | 17.0% | 21.6% | |
| 118-19 | Yesb | KX149259 | 32% | 51% | 0.0% | 71.3% | 31.2% | 41.3% | 35.5% | |
| 122-19 | Yesb | KX149259 | 47% | 56% | 15.6% | 75.7% | 63.1% | 60.8% | 49.9% | |
| 131-19 | Yes | KX149259 | 82% | 94% | 78.3% | 100% | 80.1% | 100% | 80.6% | |
| 102-19 | Yes | KX149259 | 85% | 93% | 83.8% | 100% | 100% | 100% | 83.4% | |
| 107-19 | Yesb | KX149259 | 69% | 89% | 63.3% | 100% | 80.1% | 69.8% | 63.7% | |
| 135-19 | Yesb | KX149259 | 80% | 89% | 83.9% | 100% | 100% | 100% | 72.3% | |
| 136-19 | Yes | KX149259 | 90% | 96% | 83.2% | 100% | 100% | 100% | 90.9% | |
| 01-20 | Yes | KX149259 | 88% | 94% | 83.9% | 100% | 100% | 100% | 84.9% | |
| 04-20 | Yes | KX149259 | 92% | 96% | 84.3% | 100% | 100% | 100% | 91.0% | |
| 07-20 | Yes | KX149259 | 95% | 98% | 98.9% | 100% | 100% | 100% | 94.5% | |
| 08-20a | Noa | NAa | NAa | 2% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| 11-20 | Yes | KX149259 | 80% | 90% | 83.9% | 100% | 100% | 100% | 77.7% | |
| 14-20 | Yes | KX149259 | 88% | 93% | 100% | 100% | 100% | 100% | 83.4% | |
| 15-20 | Yes | KX149259 | 95% | 96% | 98.9% | 100% | 100% | 100% | 94.5% | |
| From | To | Support |
|---|---|---|
| Intra-city transmission | ||
| Sasa1 | Talomo1 | strong |
| Tugbok1 | Toril1 | very strong |
| Inter-city transmission | ||
| Poblacion1 | DigosCogon2 | strong |
| ComVal4 | Buhangin1 | strong |
| DigosCogon2 | Poblacion1 | strong |
| Panabo3 | Tugbok1 | strong |
| Calinan1 | Tagum3 | very strong |
| Panabo3 | Calinan1 | very strong |
| New Bataan4 | Baguio1 | decisive |
| New Bataan4 | Sasa1 | decisive |
| New Bataan4 | Buhangin1 | decisive |
| Intra-province transmission | ||
| New Bataan4 | ComVal4 | decisive |
| Inter-province transmission | ||
| New Bataan4 | Digos2 | strong |
| New Bataan4 | Sulop2 | strong |
| New Bataan4 | Samal3 | strong |
| New Bataan4 | Panabo3 | decisive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




