Submitted:
23 June 2023
Posted:
23 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
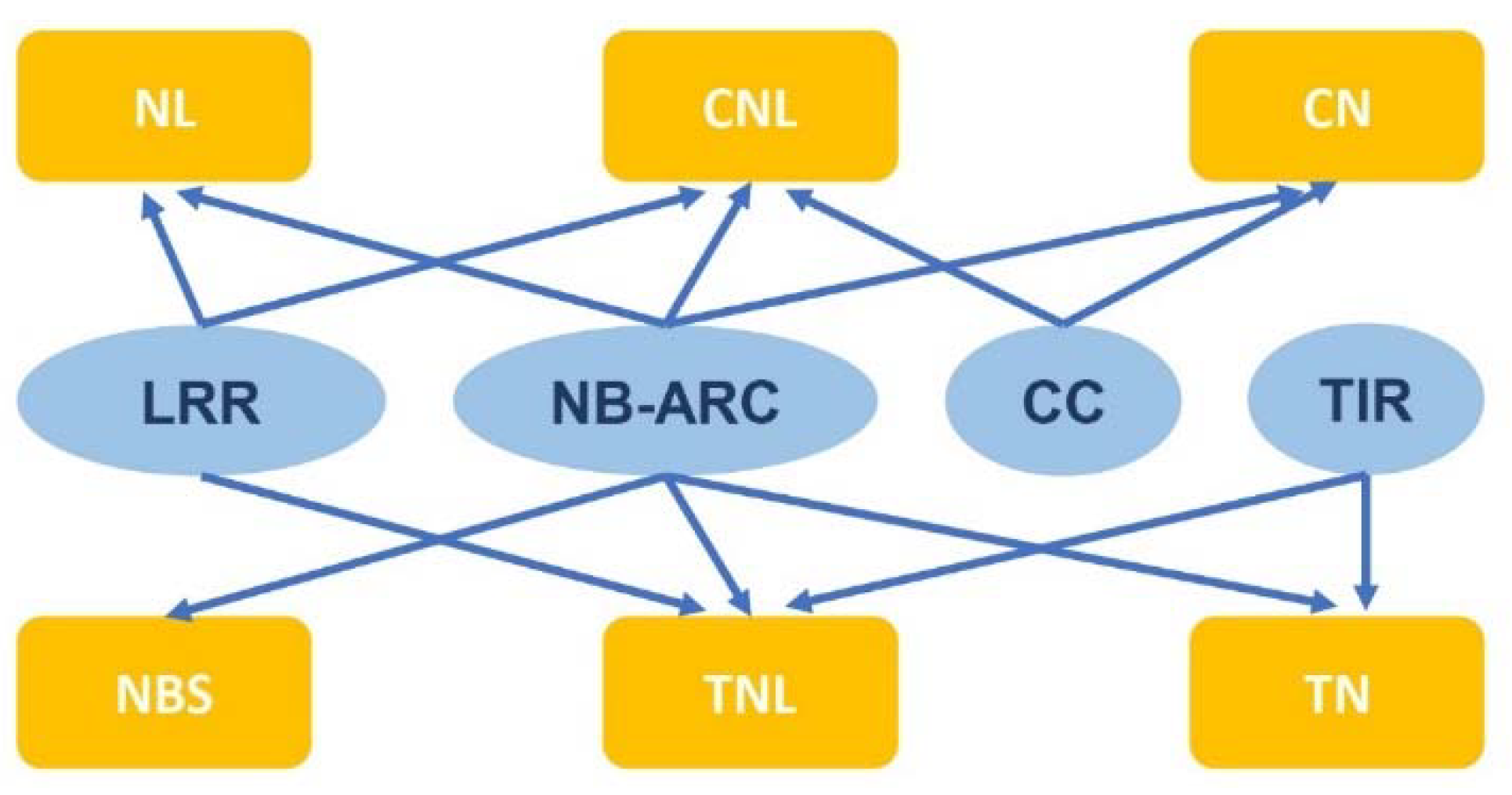
2. NLR gene structure and categories
3. Gene duplication for crop improvement
4. Diversity and divergence of NLR genes for disease resistance
5. Duplicated NLR genes are important for wheat disease resistance
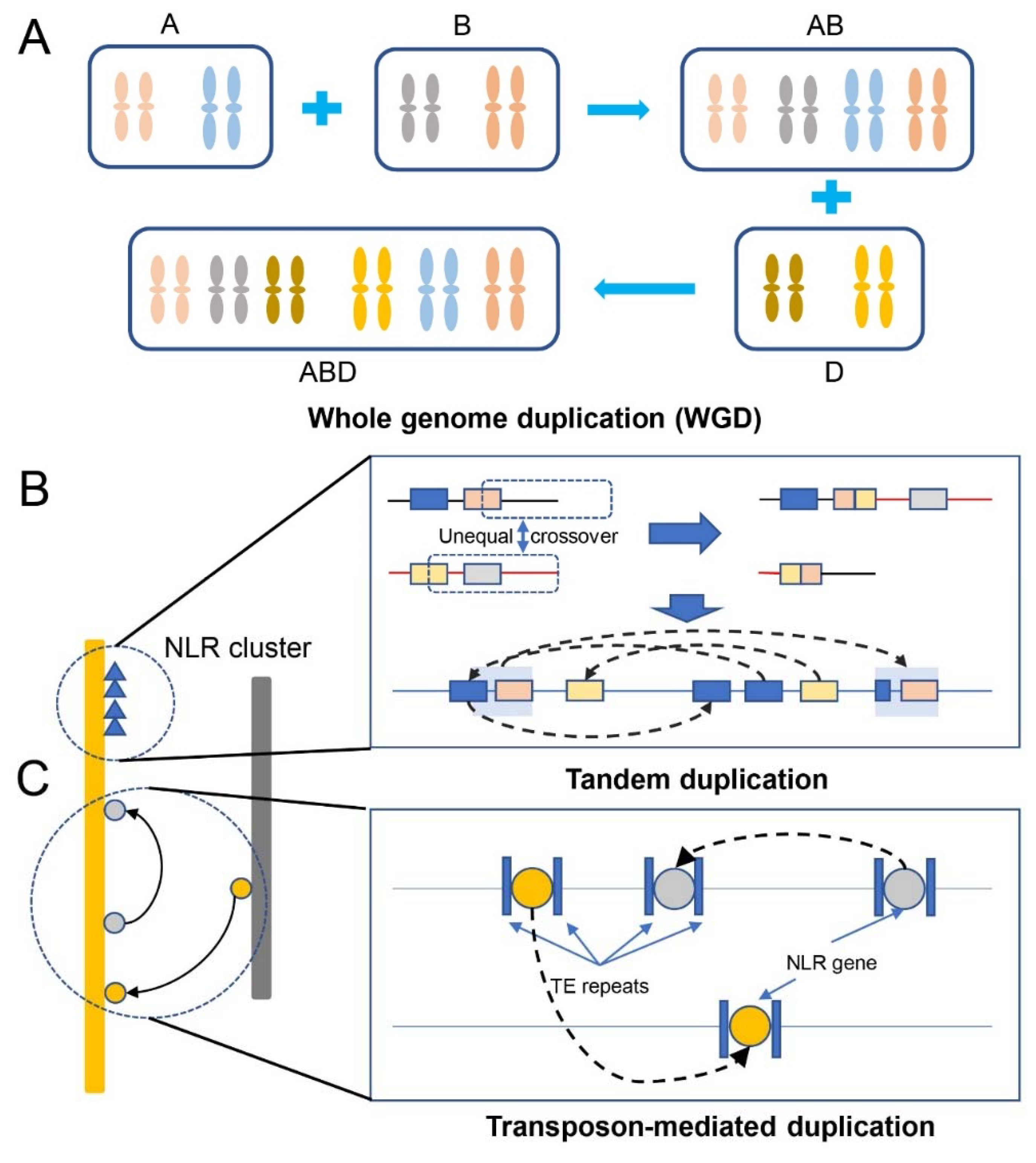
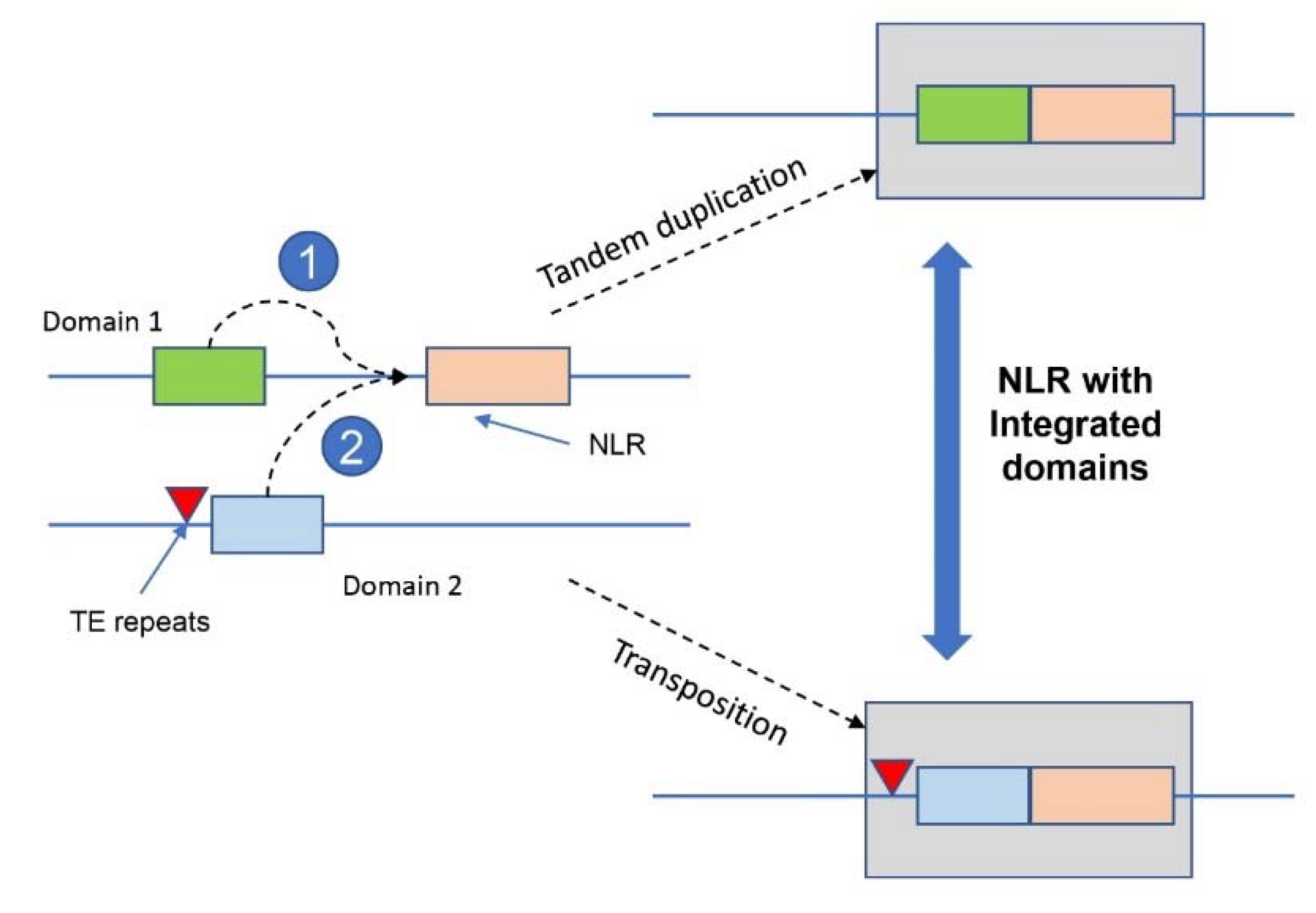
6. Mechanism of NLR gene duplication for disease resistance
7. Future perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.; Wicker, T.; Krattinger, S.G. Advances in Wheat and Pathogen Genomics: Implications for Disease Control. Annu Rev Phytopathol 2018, 56, 67–87. [Google Scholar] [CrossRef]
- Lacaze, A.; Joly, D.L. Structural specificity in plant-filamentous pathogen interactions. Mol Plant Pathol 2020, 21, 1513–1525. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef]
- Lu, Y.; Tsuda, K. Intimate Association of PRR- and NLR-Mediated Signaling in Plant Immunity. Mol Plant Microbe Interact 2021, 34, 3–14. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Brunner, S.; Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 2006, 47, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Toruno, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu Rev Phytopathol 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signaling upon Microbial Pattern Recognition. Annu Rev Phytopathol 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Plissonneau, C.; Benevenuto, J.; Mohd-Assaad, N.; Fouche, S.; Hartmann, F.E.; Croll, D. Using Population and Comparative Genomics to Understand the Genetic Basis of Effector-Driven Fungal Pathogen Evolution. Front Plant Sci 2017, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A. Fungal virulence, vertebrate endothermy, and dinosaur extinction: is there a connection? Fungal Genet Biol 2005, 42, 98–106. [Google Scholar] [CrossRef]
- Sun, Y.; Zhu, Y.X.; Balint-Kurti, P.J.; Wang, G.F. Fine-Tuning Immunity: Players and Regulators for Plant NLRs. Trends Plant Sci 2020, 25, 695–713. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Sanchez-Martin, J.; Keller, B. NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr Opin Plant Biol 2021, 62, 102053. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, X.; Hu, Y.; Qin, L.; Wang, D.; Jia, J.; Jiao, Y. A recent burst of gene duplications in Triticeae. Plant Commun 2022, 3, 100268. [Google Scholar] [CrossRef]
- Juery, C.; Concia, L.; De Oliveira, R.; Papon, N.; Ramirez-Gonzalez, R.; Benhamed, M.; Uauy, C.; Choulet, F.; Paux, E. New insights into homoeologous copy number variations in the hexaploid wheat genome. Plant Genome 2021, 14, e20069. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Gu, Y.Q.; Puiu, D.; Wang, H.; Twardziok, S.O.; Deal, K.R.; Huo, N.; Zhu, T.; Wang, L.; Wang, Y.; et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 2017, 551, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Danot, O.; Marquenet, E.; Vidal-Ingigliardi, D.; Richet, E. Wheel of Life, Wheel of Death: A Mechanistic Insight into Signaling by STAND Proteins. Structure 2009, 17, 172–182. [Google Scholar] [CrossRef]
- Bonardi, V.; Cherkis, K.; Nishimura, M.T.; Dangl, J.L. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol 2012, 24, 41–50. [Google Scholar] [CrossRef]
- Lukasik, E.; Takken, F.L. STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 2009, 12, 427–436. [Google Scholar] [CrossRef]
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 2002, 21, 4511–4519. [Google Scholar] [CrossRef]
- Ayliffe, M.A.; Lagudah, E.S. Molecular genetics of disease resistance in cereals. Ann Bot 2004, 94, 765–773. [Google Scholar] [CrossRef]
- Salamini, F.; Ozkan, H.; Brandolini, A.; Schafer-Pregl, R.; Martin, W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet 2002, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Zhao, G.; Zou, C.; Li, K.; Wang, K.; Li, T.; Gao, L.; Zhang, X.; Wang, H.; Yang, Z.; Liu, X.; et al. The Aegilops tauschii genome reveals multiple impacts of transposons. Nat Plants 2017, 3, 946–955. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing, C.; investigators, I.R.p.; Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; investigators, I.w.-g.a.p.; Pozniak, C.J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361. [Google Scholar] [CrossRef]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef]
- Zhou, Y.; Bai, S.; Li, H.; Sun, G.; Zhang, D.; Ma, F.; Zhao, X.; Nie, F.; Li, J.; Chen, L.; et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat Plants 2021, 7, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Bomblies, K. When everything changes at once: finding a new normal after genome duplication. Proc Biol Sci 2020, 287, 20202154. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.H. Evolution of Gene Duplication in Plants. Plant Physiol 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Matsuoka, Y. Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol 2011, 52, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Yang, Z.L.; You, F.M.; Kawahara, T.; Waines, J.G.; Dvorak, J. The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor Appl Genet 2007, 114, 947–959. [Google Scholar] [CrossRef]
- Hao, Y.; Xu, S.; lyu, Z.; Wang, H.; Kong, L.; Sun, S. Comparative Analysis of the Glutathione S-Transferase Gene Family of Four Triticeae Species and Transcriptome Analysis of GST Genes in Common Wheat Responding to Salt Stress. International Journal of Genomics 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, M.; Cui, Y.; Kong, L.; Wang, H. Genome-wide survey of the dehydrin genes in bread wheat (Triticum aestivum L.) and its relatives: identification, evolution and expression profiling under various abiotic stresses. {BMC} Genomics 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Kapitonov, V.V.; Jurka, J. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet 2007, 23, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Kim, S.; Yeom, S.I.; Choi, D. Genome-Wide Comparative Analyses Reveal the Dynamic Evolution of Nucleotide-Binding Leucine-Rich Repeat Gene Family among Solanaceae Plants. Front Plant Sci 2016, 7, 1205. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Mazzucotelli, E.; Marone, D.; Crosatti, C.; Michelotti, V.; Vale, G.; Mastrangelo, A.M. Regulation and Evolution of NLR Genes: A Close Interconnection for Plant Immunity. Int J Mol Sci 2018, 19. [Google Scholar] [CrossRef]
- Sappl, P.G.; Carroll, A.J.; Clifton, R.; Lister, R.; Whelan, J.; Harvey Millar, A.; Singh, K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J 2009, 58, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Brooks, S.; Li, W.; Fellers, J.; Nelson, J.C.; Gill, B. Evolution of new disease specificity at a simple resistance locus in a crop-weed complex: reconstitution of the Lr21 gene in wheat. Genetics 2009, 182, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wing, R.A.; Wise, R.P. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 2002, 14, 1903–1917. [Google Scholar] [CrossRef]
- van Wersch, S.; Li, X. Stronger When Together: Clustering of Plant NLR Disease resistance Genes. Trends Plant Sci 2019, 24, 688–699. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: adaptable guards. Genome Biol 2006, 7, 212. [Google Scholar] [CrossRef]
- Ellis, J.; Dodds, P.; Pryor, T. Structure, function and evolution of plant disease resistance genes. Current Opinion in Plant Biology 2000, 3, 278–284. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol 2016, 170, 2095–2109. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.Q.; Zhang, Y.M.; Hang, Y.Y.; Xue, J.Y.; Zhou, G.C.; Wu, P.; Wu, X.Y.; Wu, X.Z.; Wang, Q.; Wang, B.; et al. Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol 2014, 166, 217–234. [Google Scholar] [CrossRef]
- Glover, N.M.; Daron, J.; Pingault, L.; Vandepoele, K.; Paux, E.; Feuillet, C.; Choulet, F. Small-scale gene duplications played a major role in the recent evolution of wheat chromosome 3B. Genome Biology 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yin, H.; Jiao, C.; Fang, X.; Wang, G.; Li, G.; Ni, F.; Li, P.; Su, P.; Ge, W.; et al. Sympatric speciation of wild emmer wheat driven by ecology and chromosomal rearrangements. Proceedings of the National Academy of Sciences 2020, 117, 5955–5963. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Netherway, T.; Frioux, C.; Ferretti, P.; Coelho, L.P.; Geisen, S.; Bork, P.; Hildebrand, F. Metagenomic assessment of the global diversity and distribution of bacteria and fungi. Environ Microbiol 2021, 23, 316–326. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, Z.; Zhang, Y.M.; Li, Q.; Jiang, X.M.; Jiang, Z.; Tang, J.H.; Chen, D.; Wang, Q.; Chen, J.Q.; et al. An angiosperm NLR Atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol Plant 2021, 14, 2015–2031. [Google Scholar] [CrossRef]
- Karasov, T.L.; Kniskern, J.M.; Gao, L.; DeYoung, B.J.; Ding, J.; Dubiella, U.; Lastra, R.O.; Nallu, S.; Roux, F.; Innes, R.W.; et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 2014, 512, 436–440. [Google Scholar] [CrossRef]
- Xue, J.Y.; Zhao, T.; Liu, Y.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z.; Shao, Z.Q. Genome- Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front Genet 2019, 10, 1286. [Google Scholar] [CrossRef]
- Lin, X.; Zhang, Y.; Kuang, H.; Chen, J. Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae. BMC Genomics 2013, 14, 335. [Google Scholar] [CrossRef]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Kuromori, T.; Myouga, F.; Toyoda, T.; Li, W.H.; Shinozaki, K. Evolutionary persistence of functional compensation by duplicate genes in Arabidopsis. Genome Biol Evol 2009, 1, 409–414. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 2005, 169, 1157–1164. [Google Scholar] [CrossRef]
- Marchal, C.; Zhang, J.; Zhang, P.; Fenwick, P.; Steuernagel, B.; Adamski, N.M.; Boyd, L.; McIntosh, R.; Wulff, B.B.H.; Berry, S.; et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants 2018, 4, 662–668. [Google Scholar] [CrossRef]
- Lin, G.; Chen, H.; Tian, B.; Sehgal, S.K.; Singh, L.; Xie, J.; Rawat, N.; Juliana, P.; Singh, N.; Shrestha, S.; et al. Cloning of the broadly effective wheat leaf rust resistance gene Lr42 transferred from Aegilops tauschii. Nat Commun 2022, 13, 3044. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, L.; Zhang, H.; Hao, Q.; Lyu, B.; Wang, M.; Epstein, L.; Liu, M.; Kou, C.; Qi, J.; et al. An ancestral NB-LRR with duplicated 3'UTRs confers stripe rust resistance in wheat and barley. Nat Commun 2019, 10, 4023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, X.; Hao, Y.; Cai, J.; Ohm, H.W.; Kong, L. A genetic map of Lophopyrum ponticum chromosome 7E, harboring resistance genes to Fusarium head blight and leaf rust. Theor Appl Genet 2011, 122, 263–270. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Yu, H.; Li, Y.X.; Li, Y.Y. Sorting out inherent features of head-to-head gene pairs by evolutionary conservation. BMC Bioinformatics 2010, 11, S16. [Google Scholar] [CrossRef]
- Liang, W.; van Wersch, S.; Tong, M.; Li, X. TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol 2019, 221, 2054–2066. [Google Scholar] [CrossRef]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol 2019, 50, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Inoue, H.; Kato, T.; Funao, T.; Shirota, M.; Shimizu, T.; Kanamori, H.; Yamane, H.; Hayano-Saito, Y.; Matsumoto, T.; et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J 2010, 64, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Marchal, C.; Michalopoulou, V.A.; Zou, Z.; Cevik, V.; Sarris, P.F. Show me your ID: NLR immune receptors with integrated domains in plants. Essays Biochem 2022, 66, 527–539. [Google Scholar] [CrossRef]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef]
- Le Roux, C.; Huet, G.; Jauneau, A.; Camborde, L.; Tremousaygue, D.; Kraut, A.; Zhou, B.; Levaillant, M.; Adachi, H.; Yoshioka, H.; et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015, 161, 1074–1088. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, M.; Jiang, L.; Zhao, W.; Li, J.; Wu, J.; Li, C.; Bai, B.; Lu, G.; Chen, H.; et al. A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol 2016, 212, 161–175. [Google Scholar] [CrossRef]
- Wang, H.; Zou, S.; Li, Y.; Lin, F.; Tang, D. An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat Commun 2020, 11, 1353. [Google Scholar] [CrossRef]
- Wang, W.; Chen, L.; Fengler, K.; Bolar, J.; Llaca, V.; Wang, X.; Clark, C.B.; Fleury, T.J.; Myrvold, J.; Oneal, D.; et al. A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat Commun 2021, 12, 6263. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 2004, 37, 528–538. [Google Scholar] [CrossRef]
- Srichumpa, P.; Brunner, S.; Keller, B.; Yahiaoui, N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 2005, 139, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Valent, B. Wheat blast disease: danger on the move. Tropical Plant Pathology 2017, 42, 210–222. [Google Scholar] [CrossRef]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol 2011, 49, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Zhang, W.; Salcedo, A.; Rouse, M.N.; Trick, H.N.; Akhunov, E.; Dubcovsky, J. Identification of Wheat Gene Sr35 That Confers Resistance to Ug99 Stem Rust Race Group. Science 2013, 341, 783–786. [Google Scholar] [CrossRef]
- Salcedo, A.; Rutter, W.; Wang, S.; Akhunova, A.; Bolus, S.; Chao, S.; Anderson, N.; Soto, M.F.D.; Rouse, M.; Szabo, L.; et al. Variation in the AvrSr35 gene determines Sr35 resistance against wheat stem rust race Ug99. Science 2017, 358, 1604–1606. [Google Scholar] [CrossRef] [PubMed]
- Bourras, S.; McNally, K.E.; Muller, M.C.; Wicker, T.; Keller, B. Avirulence Genes in Cereal Powdery Mildews: The Gene-for-Gene Hypothesis 2.0. Front Plant Sci 2016, 7, 241. [Google Scholar] [CrossRef]
- Forderer, A.; Li, E.; Lawson, A.W.; Deng, Y.N.; Sun, Y.; Logemann, E.; Zhang, X.; Wen, J.; Han, Z.; Chang, J.; et al. A wheat resistosome defines common principles of immune receptor channels. Nature 2022, 610, 532–539. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Deng, Y.; Ning, Y.; Yang, D.L.; Zhai, K.; Wang, G.L.; He, Z. Molecular Basis of Disease Resistance and Perspectives on Breeding Strategies for Resistance Improvement in Crops. Mol Plant 2020, 13, 1402–1419. [Google Scholar] [CrossRef]
- Tanno, K.; Willcox, G. How fast was wild wheat domesticated? Science 2006, 311, 1886. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, J.; Wen, J.; Nie, X.; Xu, L.; Chen, N.; Li, Z.; Wang, Q.; Zheng, Z.; Li, M.; et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol 2019, 20, 136. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



