1. Introduction
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas9 system has emerged as a flexible gene editing tool for both cell engineering and therapeutic applications [
1,
2,
3,
4,
5]. For Cas9-mediated genome editing to be effective, different components need to be delivered inside the nucleus of target cells. These include the Cas9 protein, a guide RNA that targets the Cas9 nuclease to a specific sequence in the genomic DNA and, in case of gene correction or insertion, a DNA template to enable precise gene editing via homologous recombination. The sheer size of these components, combined with the need for successful delivery of each component in optimal stoichiometric ratios are the prime reasons why effective delivery has proven to be difficult. Viral vectors can be constructed that encode the Cas9 protein along with a single guide RNA for introducing targeted double strand breaks in the genome, but the enduring expression of Cas9 in the target cell population may lead to undesired off-target effects as well as potential immunogenicity [
6,
7,
8]. More transient systems are therefore desired.
Direct delivery of the Cas9:sgRNA ribonucleoprotein complex (RNP) offers several advantages. First, it is fast as it does not depend on the transcription and translation and subsequent assembly into an active RNP inside the cell, which can often be suboptimal due to different degradation profiles of the Cas9 protein and the sgRNA. Second, its transient nature reduces the chance of off-target cutting events and potential immune responses. Third, it avoids potential risks of insertional mutagenesis which is often associated with viral vectors [
10].
To this end, different strategies have recently been explored for the delivery of Cas9 RNP into cells, including electroporation [
11], induced osmotic transduction (iTOP) [
12] as well as the use of chemical transfection reagents [
13] and non-viral vectors [
9,
14,
15,
16,
17,
18] . However, developing synthetic vectors for direct delivery of CRISPR-Cas RNP with high efficiency has been challenging. Such synthetic vectors should avidly complex or encapsulate the Cas9 RNP in order to protect it from premature proteolytic degradation, it should trigger cellular uptake, endosomal escape and vector disassembly to enable the free RNP to be transported into the nucleus through the nuclear pore complexes.
Cell-penetrating peptides (CPPs) have been explored for the delivery of Cas9 RNPs in various cell types [
19,
20,
21,
22,
23]. CPPs are short (5-30 amino acids) polycationic or amphipathic peptides that facilitate the cellular uptake of different types of cargoes, such as small molecules, proteins, plasmid DNA as well as nanoparticles [
6,
24]. The mechanism of action of CPPs is not fully understood, but is generally believed to follow two uptake routes: (1) direct translocation across the cell membrane and (2) endocytosis after binding to cell surface heparan sulphates [
6,
25]. Within the endocytic compartments the CPPs interact with the endosomal membranes, often triggered by change of pH in the endosomes, which results in endosome destabilization and partial cargo release [
6,
26].
Initial studies on CPP-mediated Cas9 RNP delivery used covalent attachment of polycationic CPPs to Cas9 to enhance its intracellular delivery, however, with moderate success. Low levels of indels were observed at relatively high doses of Cas9-CPP conjugates [
21,
27,
28]. Since covalent attachment might negatively influence RNP activity, either by modification of active site residues [
29] or by interfering with sgRNA binding, non-covalent approaches based on electrostatic complexation of the Cas9 RNP with cationic CPPs have recently been explored [
22,
23,
30]. For stable complex formation with the Cas9 RNP, cationic CPPs were modified with 1 or more lipid tails [
22,
30]. These studies showed a remarkable increase in indel efficiencies compared to covalent methods and could be further increased by the addition of PEG-PVA to the medium [
31]. Important to note is that these efficiencies were reached in HeLa or HEK293T reporter cell lines, which are known for ease of transfection, and under transfection conditions (DMEM without serum and in the presence of 5% PEG-PVA), that might be toxic to primary cells and has limited physiological relevance.
We have previously screened a large number of different CPPs, both cationic and amphipathic, for their capacity to transfect pDNA into various cell lines [
31]. From a total of >90 CPPs only 5 showed robust transfection efficiencies in the presence of serum without compromising cell viability. One of these peptides, LAH5 (KKALLALALHHLAHLAH HLALALKKA), initially described by Kichler et al (2003), showed the highest transfection efficiency of all tested. This amphipathic peptide is rich in histidine residues, which is thought to be responsible for their capability to induce endosomal escape (
Figure 1) [
32].
In this study, we have tested LAH5 for delivery of Cas9 RNP. We show for the first time that LAH5 peptides form nanocomplexes with Cas9 RNP that were efficiently internalized by a variety of different cell lines. Functional delivery of Cas9 RNP was demonstrated in HEK293T cells expressing a fluorescent Cas9 reporter construct using different ratios of RNP/LAH5 nanocomplexes. Next, CCR5 gene targeted indels was demonstrated in various cell lines including primary fibroblasts. Finally, we demonstrate that co-delivery of ssDNA as HDR template with the Cas9 RNP-LAH5 nanocomplexes results in efficient gene correction. To the best of our knowledge, this is the first research to show that CPP can deliver these three different components at the same time, which are crucial for CRISPR-mediated gene correction.
2. Materials and Methods
2.1. General Reagents
All chemicals were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands) unless otherwise specified. spCas9 and spCas9-GFP protein were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). Modified sgRNA sequences were obtained from Sigma-Aldrich (Haverhill, the United Kingdom, sequences given in
Table S1) and stored in DNAse and RNAse-free water from IDT (Integrated DNA Technologies Inc. Leuven Belgium). DNA oligonucleotides (
Table S2), HDR template (
Table S3), ATTO 550 labeled tracRNA, non-labeled crRNA (
Table S4) and Alexa fluor 647 labeled HDR template (
Table S3) were purchased from Integrated DNA Technologies Inc. Lipofectamine CRISPRMAX was obtain from Thermo Fisher Scientific (
Waltham, Massachusetts, ABD). T7 endonuclease and Q5 high fidelity 2X master mix were purchased from Bioke (Leiden, The Netherlands). The LAH5: KKALLALALHHLAHLAHHLALALKKA peptide was purchased from Synpeptide (Shanghai, China). DNA extraction and PCR purification kits were purchased from QIAGEN Benelux B.V. (Venlo, The Netherlands). TAE buffer was purchased from Biorad (Lunteren, Netherlands).
2.2. Cas9 RNP:LAH5 nanocomplex formation and particle sizing
Cas9 and sgRNA stocks were diluted in nuclease free water (Thermo Scientific). The LAH5 peptide supplied as powder was dissolved in nuclease free water with 0.1% (v/v) acetic acid at a final peptide concentration of 750μM. sgRNA (0,75µM) and Cas9 (0,75µM) dissolved in HEPES buffer pH 7.4 were mixed by pipetting and incubated for 10 minutes at room temperature to allow formation of the Cas9 ribonucleoprotein complex (RNP). Subsequently, an equal volume of LAH5 (37,5µM to 187,5µM) was added to the RNP and incubated for 10 min at RT to enable nanocomplex formation. The obtained molar ratios of Cas9:sgRNA:LAH5 varied between 1:1:50 to 1:1:250. Following the nanocomplex formation, samples were diluted in HEPES buffer pH 7.4, then 100µl of sample was transferred into a lowest sizing cell cuvette (Malvern, Malvern, UK ), and size distribution was measured with a Malvern Zetasizer Nano-ZS (Malvern Instruments, the United Kingdom). Subsequently, formulated nanocomplexes were diluted 8x times in 10mM HEPES buffer pH 7.4 then, zeta potential was measured with Zetasizer Nano Z (Malvern ALV CGS-3, United Kingdom). All measurements were done in triplicate.
2.3. Electrophoretic Mobility Shift Assay (EMSA)
Peptide mediated complexation was determined by performing a fluorescence based electrophoretic mobility shift assay by using a non-stained and non-denaturing 1.5% agarose gel in TAE buffer, pH 8.3. For this, Cas9-GFP (Merck), ATTO-550 labeled tracrRNA, unlabeled crRNA (IDT), Alexa-647 labeled HDR template (IDT), and unlabeled LAH5 peptide were used. Labeled components and unlabeled LAH5 peptide were formulated at various molar ratios as described above. All labeled components were diluted such that the final concentration reaches 1 µM when loaded on gel. All formulated nanocomplexes were adjusted to a total volume of 10 µl by adding nuclease-free water. Then, 2 µl 40% Glycerol was added after which samples were loaded into the wells of the gel. The agarose gel was run at 170 V for 15 minutes. Gel images were captured by ChemiDocTM XRS+ (Bio-Rad, Veenendaal, The Netherlands , and fluorescent images were merged and analyzed by ChemiDocTM XRS+ (Bio-Rad) with Image lab software.
2.4. Cell lines and Cell culture
HEK293T stoplight cells [
33], HEK293T HDR stoplight cells, HEK293T and primary fibroblast cells were cultured in high-glucose DMEM medium, and Hela and HEPG2 cells in Eagle’s minimum essential medium. Culture medium was supplemented with 10% (v/v) FBS (Sigma Zwijndrecht, The Netherlands) and incubated at 37⁰C and 5% CO
2. Cell culture plastics unless otherwise specified were purchased from Greiner Bio-One (Alphen aan de Rijn, The Netherlands).
2.5. Generation of HEK293T HDR Stoplight Reporter Cell line
The HDR fluorescent Stoplight Reporter construct was synthesized by Integrated DNA Technologies (Leuven, Belgium). The reporter construct was cloned into a pHAGE2-MCS-EF1a-IRES-NeoR plasmid using NotI and BamHI restriction enzymes and a NEB Quick Ligation Kit (all from New England Biolabs, MA, United States of America). The pHAGE2-EF1a-HDR Stoplight-IRES-NeoR plasmid was then used for lentiviral production and transduction. HEK293T cells were transfected with pHAGE2-EF1a-HDR Stoplight-IRES-NeoR, MD2.G plasmid, and PSPAX2 plasmid (Addgene #12259 and #12260, respectively) at a 2:1:1 ratio using 3 µg 25 kDa linearized PEI per µg of DNA. Transfection medium was removed after 24h, and replaced with DMEM with 10% FBS. After 48 h, lentiviral supernatant was harvested, and cells were cleared by a 5 minute 500 x g centrifugation step, followed by 0.45 µm syringe filter filtration. HEK293T cells were transduced with lentiviral stocks overnight with 8 µg/µl polybrene (Sigma Zwijndrecht, The Netherlands). After 24h, the transduction medium was removed, and replaced with culture medium supplemented with 1500 µg/ml G418. After 5 days, the concentration of G418 was lowered to a maintenance concentration of 1000 µg/ml G418. After two weeks of culturing in the presence of selection antibiotics, the reporter cells were sorted for mCherry+eGFP- fluorescent signals on a BD FACSAria III cell sorter.
2.6. Transfection experiments
All cell types used in this study were plated at 30.000 cells per well in a 96 well plate (Greiner M0687-100EA) and incubated for 24h at 37 ⁰C and 5% CO2 to reach a confluency of 50%. Next, cells were transfected with Cas-CPP or Cas-HDR-CPP. As a positive control for transfection spCas9 RNP complexed with CRISPRMAX was used according to the manufacturer’s instructions (Thermo Fisher Scientific). Cas-CPP nanocomplexes were prepared following RNP formation. Increasing concentrations of LAH5 peptide were mixed with RNP by keeping the ratio between RNP:LAH5 between 1:50 and 1:250. Then, Cas-CPP mixtures were incubated for 10 min. to form nanocomplexes. For HDR experiments after the preparation of RNP’s, various concentrations of HDR template were added to the preformed spCas9 RNP to get multiple ratios 1:1, 1:2, 1:4 and incubated for 5 minutes at room temperature. Finally, LAH5 peptide was added to the spCas9 RNP and HDR template mixtures and incubated for 10 min to form Cas-HDR-CPP nanocomplexes. Opti-MEM was added to reach a total volume of 160 μl before adding the nanocomplexes to the cells at the indicated concentrations. Cas9, sgRNA and HDR template concentrations were 20nM at a 1:1:1 molar ratio in upon addition in the 96 well plate. 24h after transfection cells were washed 2 times with fresh media. Cells were incubated for another 24h at 37 ⁰C and 5% CO2. In case DNA extraction was needed for genetical experiments (T7 endonuclease and TIDE), the transfection experiments were performed in 48-well plate (Greiner M8937-100EA) to have enough DNA for post-analysis.
2.7. Stoplight gene editing and correction assays
To assess efficiency of delivery at the cellular level, upon transfection with Cas-CPP nanocomplexes two different reporter cell lines, HEK293T NHEJ Stoplight cells [
33] and HEK293T HDR Stoplight cells were used. Subsequent to transfection, gene editing, and gene correction efficiencies were assessed using flow cytometry analysis.
2.8. Flow cytometry to determine gene editing and gene correction efficiencies in HEK293T stoplight and HEK 293T HDR stoplight cells
Cells were harvested by washing twice with PBS then trypsinization step following cells were fixed in 1% paraformaldehyde, and subsequently for flow cytometry analysis transferred to a BD Falcon U-bottom 96-well plate (Becton Dickinson, Franklin Lakes, NJ, USA). Reporter fluorescence was detected by flow cytometry using the BD FACS CANTO II (Becton Dickinson, Franklin Lakes, USA). mCherry was measured using the PerCP-Cy5-5-A channel of the flow cytometer, and eGFP fluorescence was determined in the FITC channel. Flow cytometry results were analyzed by Flowlogic software (Inivai Technologies, Mentone, Australia, version 8.7). The gating strategy used for the flow cytometry analyses both for HEK293T stoplight and HEK293T HDR stoplight cells is specified in
Supplementary Figures S5 and S6. Both gene editing and gene correction efficiency depending on the cell line was identified as the number of mCherry
+ cells expressing eGFP, as described previously [
33].
2.9. Confocal Microscopy
HEK293T stoplight cells were plated at an amount of 30,000 cells per well in a 96 well imaging plate (Greiner CellStar #655090). After 24h incubation cells were treated with different concentrations of Cas-CPP and Cas-HDR-CPP nanocomplexes. After 24h incubation at 37 ⁰C and 5% CO2 cells were washed with 100 μl of high-glucose DMEM medium supplemented with 10% of FBS. Following this, cells were incubated for another 24h at 37 ⁰C and 5% CO2. Cell nuclei were stained by adding 2µg/ml Hoechst 33342 in a complete cell culture medium as the final concentration and incubated for 30 min, after which cells were imaged using the Yokogawa CV7000 Confocal Microscope. (Yokogawa Corporation, Tokyo, Japan).
2.10. Cytotoxicity assays
The cytotoxicity of the RNP/LAH5 peptide nanocomplexes was evaluated by MTS (cell viability) assay. [
34] Cells were seeded into 96-well plates with 50% confluency and incubated for 24h at 37 ⁰C and 5% CO
2. Cas-CPP nanocomplexes (prepared at molar ratios of 1:50 to 1:1000) were added at a concentration of 20nM of RNP per well in a total volume of 100 μl Opti-MEM. 24h post-transfection, cytotoxicity was determined with the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) (Promega) according to the manufacturer’s protocol. The absorbance was measured at 490nm on a Bio-Rad iMark microplate reader model 1681130. The relative cell viability was calculated by setting the aborbance value of untreated cells at 100% and those treated with 1% Triton X-100 at 0%.
2.11. Cell uptake assay
Cas9-GFP (Sigma), ATTO 550 tracrRNA (IDT), and crRNA (IDT) specific for HPRT gene and Alexa 647 labeled single stranded HDR template were used to prepare fluorescent Cas-CPP nanocomplexes for cellular uptake experiments. First ATTO 550 tracrRNA (IDT), and crRNA (IDT) were mixed at 1:1 ratio to form a ATTO 550 labeled sgRNA complex. Then, uptake of these components was tested with and without complexation with Cas9-GFP (10nM), ATTO 550 sgRNA (10nM) (RNP) with 3μM of LAH5 peptide at 1:150 ratio (m/m). A complex consisting of 10nM Cas9-GFP, ATTO 550 sgRNA (RNP), and Alexa 647 HDR template at equimolar concentrations was prepared following the previously described procedure. Additionally, the labeled RNP and labeled HDR template were mixed with 3µM LAH5 peptide at a ratio of 1:1:150 (m/m). Following the preparation of the nanocomplexes Hela cells were transfected in a 96 well- black plate. 24h after transfection nuclei of HeLa cells were stained by supplementing the complete cell medium with Hoechst 33342 dye at a final concentration of 2µg/ml, followed by incubation for 30 minutes. Microscopy images were recorded at 60x magnification using the Yokogawa CV7000s confocal microscope.
2.12. T7 Endonuclease Assay
T7E1 assay was performed in order to detect the insertion/deletion (indel) frequency after gene editing [
35]. Genomic DNA was extracted from the cells 48h after the transfection with LAH5 peptide/RNP nanocomplexes using the Qiagen DNeasy Blood & Tissue Kit Benelux B.V. (Venlo, The Netherlands) following the manufacturer’s instructions. PCR was performed using the primers designed for sgRNA target locus (
Table S2) using Q5® Hot Start High-Fidelity 2X Master Mix (Bioke). Afterwards, PCR products were purified using the QIAquick PCR Purification kit Benelux B.V. (Venlo, The Netherlands). PCR products were denatured at 95 °C for 10 min in presence of NEBuffer 2 (Bioke) and annealed by slowly lowering the temperature (95-85°C at 2 °C per second and 85-25°C at 1°C per second). Subsequently, re-annealed PCR products were incubated with 5U T7E1 enzyme (Bioke) at 37 °C for 18 min to cut heteroduplexes. DNA products were run on a 2% agarose gel in TAE buffer, pH 8.3 (Biorad). Indel frequency was calculated by determining the intensities of cleaved and uncleaved bands based on densitometry analysis using ImageJ.
2.13. TIDE analysis (Tracking of Indels by Decomposition)
Genomic DNA was isolated and the sgRNA target genomic region was amplified by PCR using the same methods as described in the section on T7E1 assay (
Table S2). The PCR products were purified using the QIAquick PCR Purification kit (Qiagen GmbH, Hilden, Germany) and submitted for unidirectional Sanger-sequencing (Macrogen Europe). Afterwards, Sanger sequence chromatograms of purified PCR products were used for TIDE analysis (
http://tide.nki.nl)[
36]. Gene modification frequencies were determined by using the sequencing chromatogram from negative control cells as a reference and comparingthe sequence chromatogram from treated samples. During the analysis parameters were set to detect maximum indel size of 15 nucleotides. The decomposition frame was set to default parameters [
36].
2.14. Statistical analysis
Statistical analysis was performed using GraphPad Prism (9.4.1). The statistical analysis methods used are specified under the specific figure legends. Values in all experiments are represented as means ± standard errors of the mean (SDs) of at least three independent experiments done in duplicate. An increase in delivery efficiency was considered significant at ****p < 0.001 using analysis of variance (ANOVA) Dunnett's multiple comparison test or ANOVA Bonferroni's multiple comparison test.
3. Results
3.1. Complexation of Cas9 RNP and HDR template with LAH5 peptides
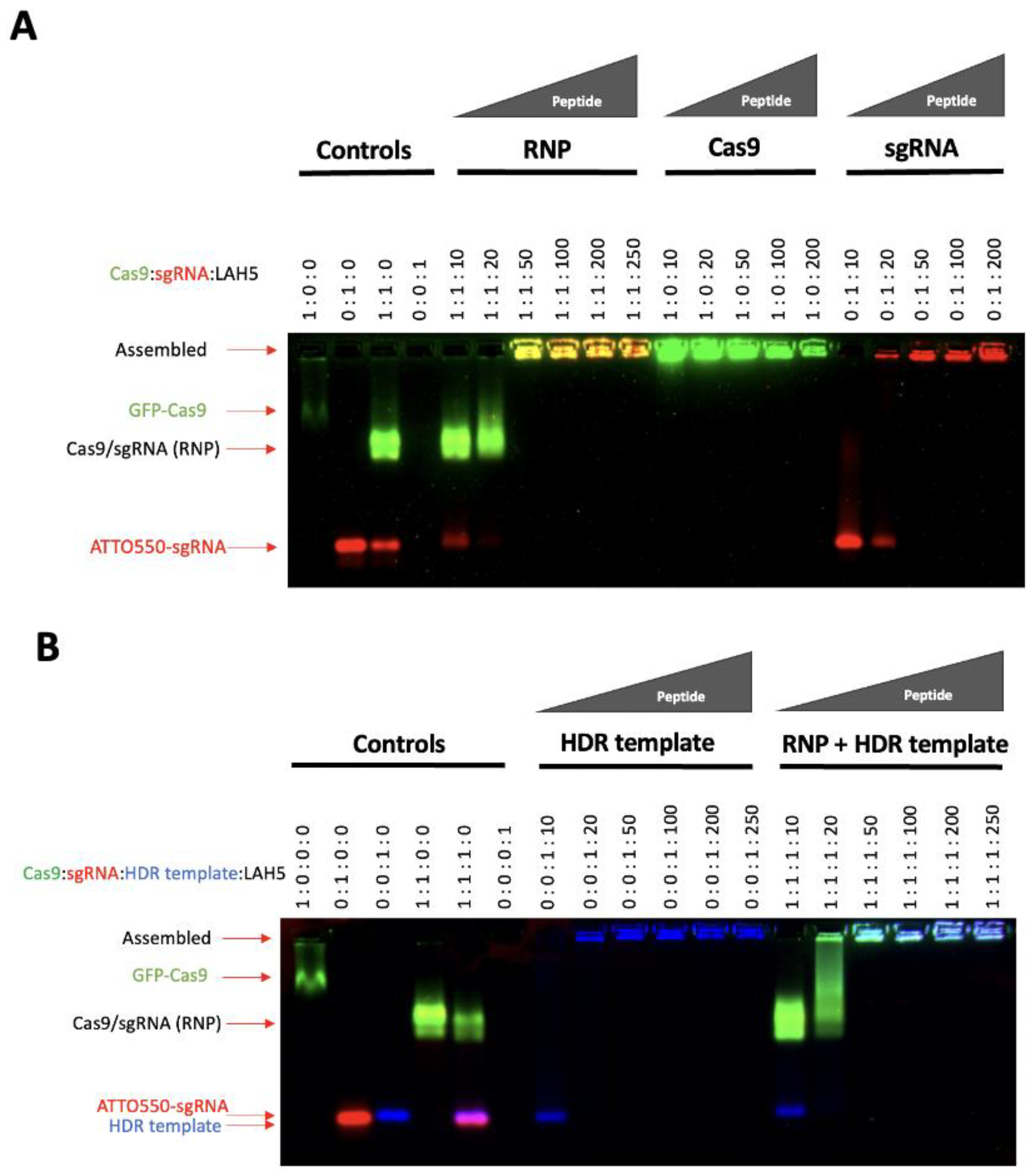
LAH5, having a net positive charge at neutral pH, is known to form stable nanocomplexes with pDNA by virtue of electrostatic complexation [
32]. Since the predicted isoelectric point of spCas9 is 9 (ExPASy) and thus positively charged at physiological pH, electrostatic complexation might not be obvious. Therefore, we first investigated the interaction of LAH5 peptide with the spCas9 protein with and without associated sgRNA and in the presence or absence of ssDNA (HDR template) using an electrophoretic mobility shift assay (EMSA). For this, fluorescently labelled spCas9 (spCas9-GFP), tracrRNA (ATTO-550) and HDR template (Alexa-647) were used. Increasing molar ratios of LAH to Cas9-GFP protein were tested for complex formation (
Figure 2). As expected, the spCas9-GFP protein (pI=8.7) itself had poor electrophoretic mobility in the agarose gel due to the presence of few negative charges under the tested conditions (TAE buffer, pH 8.3). When complexed with sgRNA, the electrophoretic mobility of spCas9-GFP increased, showing a band on gel with both GFP and ATTO-550 fluorescence in addition to free sgRNA. At molar ratios of LAH5:Cas9-GFP RNP >50 mobility of the Cas9-GFP RNP was lost, indicative of LAH5-mediated complex formation (
Figure 2A). A similar trend was seen in the presence of HDR template, which could be readily complexed together with the Cas9-GFP RNP by the LAH5 peptides at molar ratios >50 (
Figure 2B). Overall, based on the EMSA results LAH5 peptides at > 50 times molar excess formed supramolecular complexes with either RNP alone, and with RNP and HDR template together.
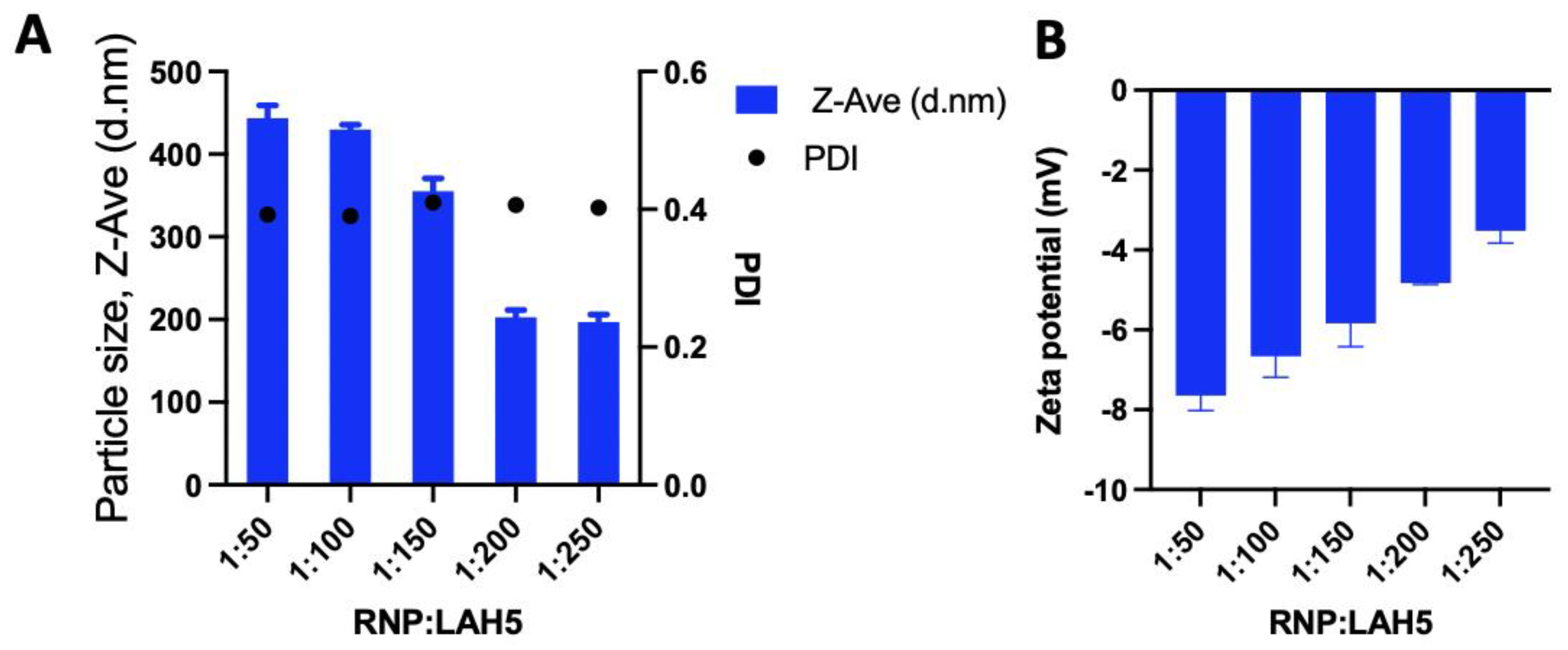
3.2. Nanocomplex size and zeta potential
The size distribution and the zeta potential of RNP/LAH5 nanocomplexes prepared at molar ratios of 1:50 to 1:500 were determined (
Figure 3). The LAH5 peptide when mixed with Cas9 RNP formed nanocomplexes with an average size ranging from 200 to 400 nm, depending on the ratios of LAH5 peptide to Cas9 RNP (
Figure 3A). The relatively high polydispersity index of the nanocomplexes ranging from 0,2 - 0,3 shows that the formed complexes are not uniform in size. Zeta potential measurements of these nanocomplexes showed an increase in zeta potential with increasing concentrations of the LAH5 peptide, but in all cases nanocomplexes were negatively charged.
3.3. Cellular uptake and functional gene editing in HeLa cells
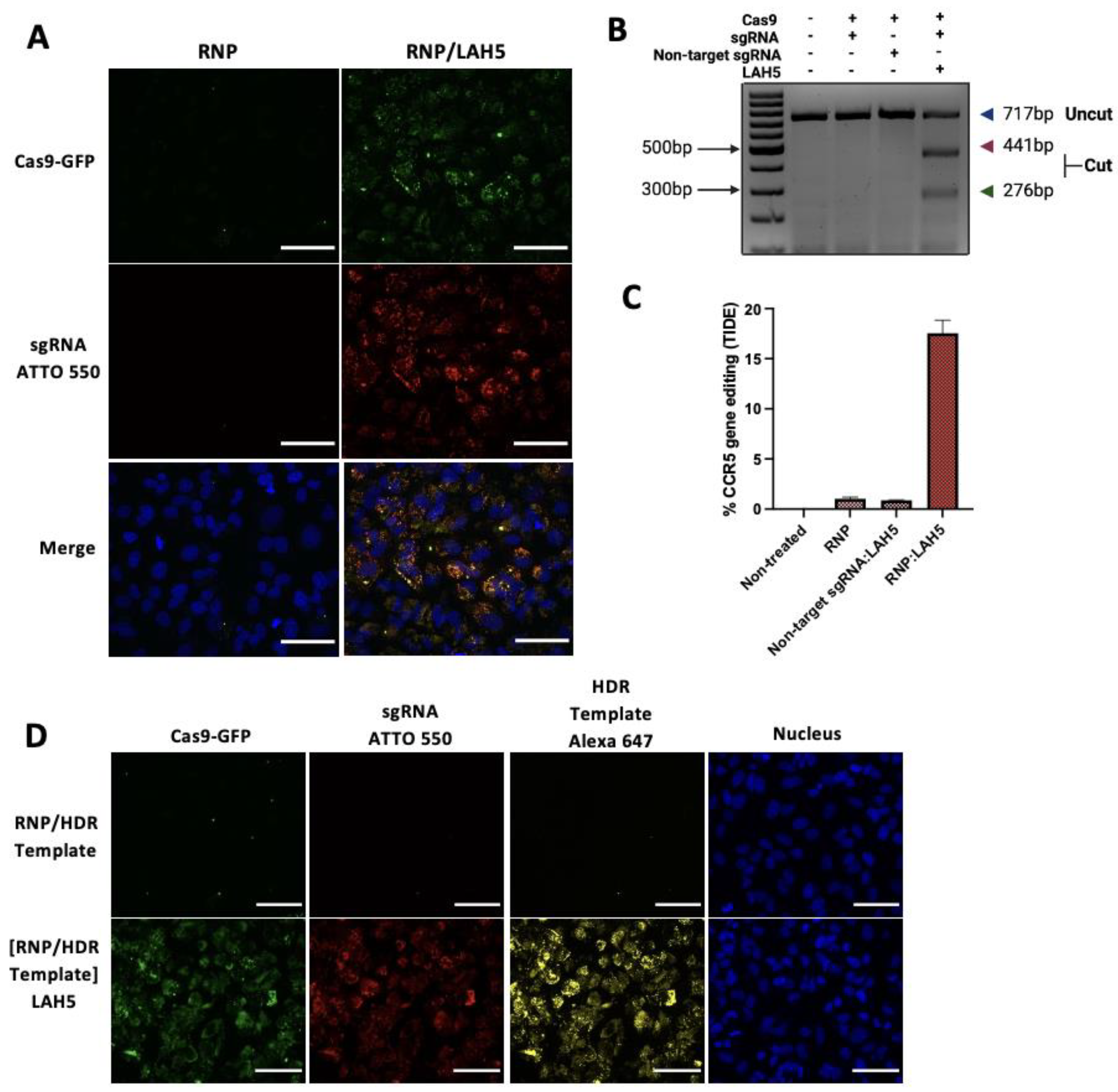
Cellular uptake of fluorescently labelled Cas9 RNP (Cas9-GFP with ATTO-550 labelled sgRNA) in free form or complexed with LAH5 peptide at a 1:150 molar ratio was monitored by live cell confocal imaging in HeLa cells (
Figure 4A). 24h following transfection, cellular uptake of Cas9 RNP without peptides was negligible, with some GFP-positive spots visible inside HeLa cells, but no ATTO-550 signal, which may suggest uptake of aggregated Cas9-GFP without sgRNA bound to it. When the Cas9 RNP were complexed with LAH5 peptide, a large increase in the cell-associated fluorescence was observed, with both Cas9-GFP and ATTO-550-sgRNA co-localizing inside the cells. The punctuate nature of the intracellular fluorescence points towards endocytic uptake.
To demonstrate functional delivery of the Cas9 RNP, which requires nuclear delivery of the RNP and hence endosomal escape, we checked the level of gene editing in HeLa cells. For this, a sgRNA was used that target the human
CCR5 gene. Cas9 RNP in free form or complexed with a 150-fold molar access of LAH5 peptide (5 μM) were added to the cells and 48 h following transfection genomic DNA was isolated and the target region of the
CCR5 gene was amplified by PCR. Gene editing was assessed semi-quantitatively by a T7 endonuclease assay (
Figure 4B) and quantitatively by the TIDE assay (
Figure 4C) [
36]. Both assays show no or low levels (<1%) of gene editing with Cas9 RNP in free form. However, when complexed with the LAH5 peptide, gene editing could be clearly observed, reaching ~17% editing as determined by the TIDE assay (
Figure 4D).
As for precise gene correction an HDR-template is required to guide the homology directed repair after CRISPR-mediated DSB, we next tested the possibility to co-deliver a ssDNA HDR template of 81bp with the Cas9 RNP/LAH5 nanocomplexes (
Figure 4D). For this, an Alexa-647 labelled HDR template and Cas9 RNP at a 1:1 molar ratio were complexed with a 250-fold molar access of LAH5 peptide. Upon delivery, it can be seen that the HDR template co-localizes with the Cas9-RNP inside HeLa cells, suggesting that the HDR template was incorporated into the LAH5 nanocomplexes, resulting in intracellular co-delivery with the Cas9 RNP complexes. No cellular uptake of the HDR template was observed when mixed with Cas9 RNP without the LAH5 peptide.
3.4. Cytotoxicity study
The cytotoxicity of RNP/LAH5 nanocomplexes on multiple cell lines was determined by performing an MTS assay (
Figure S1). In general, in all the tested cell lines no severe sign of toxicity was observed after exposure to the 1:50 to 1:500 ratios complexes, suggesting that the RNP/LAH5 complexes were not toxic to the treated cell types at concentrations used for transfection. However, when peptide concentration increased to a ratio of 1:1000 in all tested cell lines severe toxicity was observed resulting in up to 50% decline in cell viability. The results suggested that RNP/LAH5 nanocomplexes are not toxic at ratios normally used for transfection (between the range 1:50 to 1:250).
3.5. Gene editing efficiency
To further evaluate the functional delivery of Cas9 RNP using LAH5, a stoplight reporter cell for Cas9 activity [
33] was used. These HEK293T stoplight reporter cells constitutively express mCherry. eGFP expression can be induced upon CRISPR-Cas9 mediated targeted disruption of a target sequence downstream of the mCherry sequence that result in a +1 or +2 frameshift to bring the eGFP in frame with mCherry. This allows quick assessment of the indel efficiency and thereby the detection of the most optimal molar ratio between Cas9/RNP and LAH5 peptide. To assess the delivery efficiency, HEK293T stoplight cells were incubated with various combinations of Cas9 RNP/LAH5 nanocomplexes for 24h in Opti-MEM medium without serum. 24h after transfection, medium was replaced with fresh Opti-MEM supplemented with 10% FBS, and cells were incubated for an additional 24h. Cellular fluorescence was determined by both flow cytometry and confocal microscopy. Flow cytometry analysis showed that increasing the amount of peptide at a fixed Cas9 RNP concentration (20 nM), resulted in increasing gene editing. Around 70% of cells were successfully edited at the highest LAH5 peptide concentration tested (1:250 molar ratio of Cas9 RNP:LAH5 peptide)(
Figure 5A, B). The T7E1 and TIDE assays were used to corroborate these findings at the DNA level (
Figure 5C, D). In line with the flow cytometry results, these assays showed increased levels of gene editing with increasing ratios of LAH5 to Cas9 RNP.
We next varied the amount of Cas9 RNP (5-40 nM) at a fixed LAH5 peptide concentration of 5 µM (
Figure 5E,F). As expected, the lowest concentration of Cas9 RNP resulted in the lowest gene editing efficiency. Increasing the Cas9 RNP dose did not result in a linear increase in editing efficiencies, but seemed to reach a saturation point at approx. 70% of gene editing, which is close to the theoretical maximum frameshift frequency of +1nt and +2nt of 80% according to inDelphi in situ prediction [
33] for this reporter assay. No gene editing was observed when Cas9 RNP were prepared with a control sgRNA (non target sgRNA,
Table S1) to confirm sequence specificity (
Figure 5B). Gene editing was confirmed at the genomic level by the T7E1 assay (
Figure 5F). Taken together these results demonstrated that optimal gene editing could be achieved at a Cas9-RNP concentration of 20 nM complexed with 5 μM LAH5 peptide. At these concentrations, cell viability was >80% as compared to control conditions (
Figure 5E, F and
Figure S3).
Furthermore, to get insight into the kinetics of delivery of Cas9 RNP by LAH5, we measured the effect of incubation time of reporter cells with the LAH5:Cas9 RNP complexes on the overall editing efficiencies. In short, HEK293T stoplight cells were incubated from 1 minute up to 24h with the Cas9-LAH5 nanocomplexes, after which cells were washed 3x with PBS and fresh medium was added. Cells were cultured for a total of 48 h after transfection before flow cytometry analysis for eGFP expression (
Figure S3). Results demonstrated that LAH5 mediated transfection already reached 60% of gene editing when cells were exposed for only 3 h to the nanocomplexes, while similar transfection efficiencies were seen after a 24h incubation the lipid-based CRISPRMAX transfection agent. LAH5 strongly outperformed CRISPRMAX at 1 and 3h shorter incubations. This suggests that Cas9 RNP reach the cytosol much faster when delivered with LAH5 peptides than when complexed with a lipid-based transfection agent. This is most likely due to the pH-dependent membrane destabilizing effect of LAH5 that can trigger endosomal escape of its cargo [
32].
The indel efficiencies determined by flow cytometry were consistently higher than those determined by the TIDE assay (
Figure 5B, C). The higher values obtained with flow cytometry could be explained by a high copy number of the reporter construct that was introduced by lentiviral transduction. If a reporter cell line has multiple copies incorporated in its genome, editing one locus will already lead to a GFP positive signal with flow cytometry. Moreover high copy number also results in an underestimation of indels using TIDE analysis. Nevertheless, both flow cytometry and TIDE results confirmed that the optimal ratio of RNP/LAH5 nanocomplex in terms of the efficiency of gene editing is 1:250.
3.6. CCR5 editing efficiency in different cell types
We tested gene editing with Cas9-LAH5 nanocomplexes in several different human cell types, namely HEK293T, HeLa, HEPG2, ARPE-19 and primary human fibroblasts, targeting the human CCR5 gene. Gene editing outcome was determined by T7E1 and TIDE assay. The sequencing data of both negative controls and treated groups were analyzed by TIDE.
Cells were treated with Cas9/LAH5 nanocomplexes and different ratios of Cas9/LAH5 were tested ranging from 1:50 to 1:250, respectively. As a positive control CRISPRMAX was used (
Figure 6). Editing efficiencies between tested cell types varied substantially, with primary human fibroblasts showing the lowest level of gene editing and HeLa cells the highest. Despite these differences, all cell types tested showed a similar dose response, with higher amounts of LAH5 peptide leading to higher editing efficiencies. These results are clear evidence of the functioning and effective delivery potential of LAH5
in vitro.
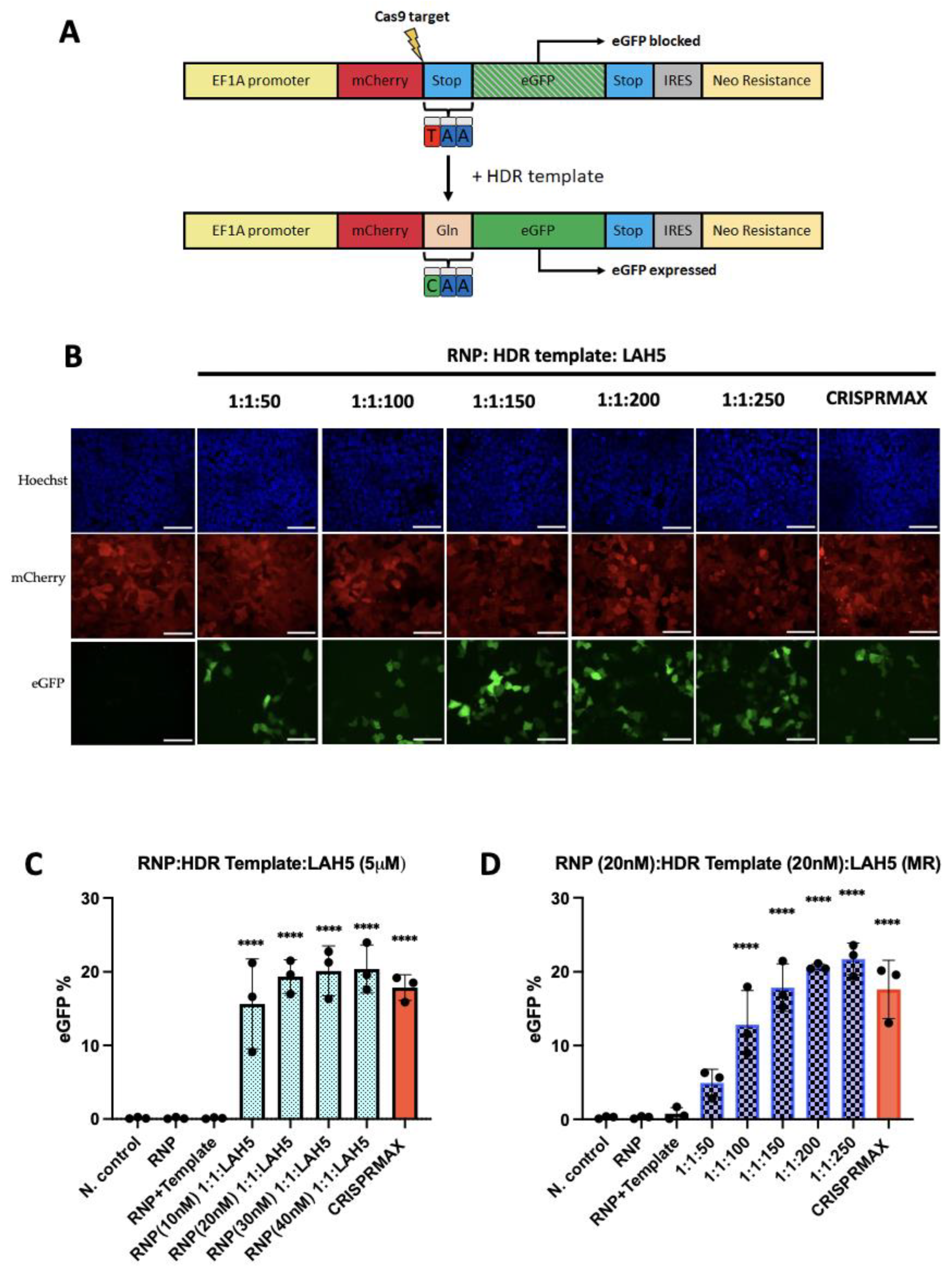
3.7. HDR-dependent gene correction
A newly developed HEK293T HDR stoplight reporter system was used as a read-out to evaluate the gene correction efficiency. HEK293T HDR Stoplight cells constitutively express mCherry, directly followed by a stop codon and an eGFP open reading frame (ORF). Including a single stranded oligonucleotide (ssODN) HDR template with CRISPR-Cas9 induces a specific replacement of the stop codon with a glutamine, resulting in the expression of a mCherry-eGFP fusion protein (
Figure 7A. HEK293T HDR stoplight cells were incubated with fixed amounts of Cas9 RNP (20 nM) and HDR ssDNA template (20 nM), which were complexed with increasing amounts of LAH5 peptides (50 to 250 fold molar excess compared to RNP) for 24h. 48 h after transfection HEK293T HDR stoplight cells were tested for mCherry and eGFP fluorescence by flow cytometry (
Figure 7 and
Figure S4). The results revealed that increasing amounts of LAH5 led to increasing gene correction efficiencies (
Figure 7C). The nanocomplex that triggered the highest gene correction efficacy was Cas9/HDR template/LAH5 peptide nanocomplexed at 1:1:250 molar ratio. This ratio of nanocomplex generated more than 20% of gene correction of the total cell population. The efficiency of HDR when the cells were treated with nanocomplexes prepared at RNP/HDR (1:2) molar ratios and complexed with LAH5 peptide concentrations at 1 μM to 5 μM is given in
Figure S4B. The level of gene correction was consistently lower as compared to nanocomplexes containing Cas9 and HDR template at a 1:1 molar ratio.
Next, we tested the effect of total concentration of Cas9 RNP and HDR template on gene correction efficiency in HEK293T HDR reporter cells. For this, the concentrations of both were varied between 10-40 nM and at molar ratios of 1:1, 1:2 and 1:4 Cas9 RNP to HDR template, respectively. The results show that gene correction increased between 10 nM to 20 nM Cas9/HDR template at 1:1 molar ratio but higher concentrations did not further increase gene correction efficiencies (
Figure 7C). We also tested the impact of increased amounts of HDR template (1:2 and 1:4 molar ratios). While the gene correction % was stable in cells treated with nanocomplexes contained RNP (20nM)/HDR template (20nM) to RNP (40nM)/HDR template (40nM) at 1:1 ratio, increased ratios 1:2 to 1:4 RNP/HDR template caused a decreasing trend of HDR percentage (
Figure S3A). These data showed that nanocomplexes prepared at a stable concentration of LAH5 peptide reached a gene correction efficacy of around 20%, however, increased concentration of RNP and HDR template did not further increase gene correction. Moreover, the increase in the ratio of the HDR template turned into a drawback and impacted gene correction adversely.
Based on the statistical analysis, the populations of the cells treated with RNP/HDR template/LAH5 mediated nanocomplexes in various conditions has significantly higher gene correction outcomes as compared to non-treated negative control. The enhancement of statistical significance is dependent on increasing ratios of LAH5, as illustrated in
Figure 7D. Moreover, HDR efficiency reached around more than 20% 1:1:200 and 1:1:250 ratios of RNP/HDR template/LAH5 peptide ratios (
Figure 7D).
4. Discussion
For efficient genome editing, intracellular delivery of the CRISPR/Cas9 components is essential. This report delves into the investigation of a promising alternative delivery strategy, wherein CPPs are employed as the chosen vehicle for effective delivery. Peptide-based delivery offer the advantage that pre-assembled Cas9 RNP can be complexed and delivered and depending on the peptide-carrier used, induce endosomal escape for cytosolic delivery of the complexes. In this report, we have repurposed the LAH5 amphipathic peptide, initially developed for transfection of pDNA (include reference), for CRISPR-Cas9 RNP delivery to a variety of human cells
in vitro. We have demonstrated that the LAH5 peptide can form stable nanocomplexes with the Cas9 RNP and with the Cas9 RNP combined with ssDNA for template-assisted homology-directed repair. We are the first to demonstrate that Cas9 RNP and ssDNA template can be stably co-entrapped inside such LAH5 peptide nanocomplexes. The electrophoretic mobility shift assay (EMSA) demonstrates that at a molar access >50 of LAH5:Cas9 RNP stable nanocomplexes are being formed by electrostatic complexation. Even though hydrophobic clustering of the LAH5 peptide itself cannot be excluded, as was demonstrated before [
37], and the formed nanocomplexes are polydisperse, we observed stable size of the nanocomplexes at 1:200 and 1:250 RNP/LAH5 ratios and confirmed this interaction via EMSA assay. This offers the opportunity to efficiently deliver multiple components needed for gene editing with a single delivery system. To show proof-of-concept for this, we have demonstrated that the co-delivery of a ssDNA template with the Cas9 RNP led to targeted gene correction in a newly developed reporter cell line. As far as we know, this is the first time that HDR-mediated targeted gene correction by delivery of all components with a peptide-based delivery system was demonstrated. A recent publication by Foss et al. elegantly showed CPP-mediated delivery of CRISPR/Cas9 RNP into human lymphocytes for gene knock-out, but it relied on co-delivery of an AAV vector carrying the HDR template for targeted insertion of a gene construct encoding a chimeric antigen receptors in those cells [
38]. However, the present study did not demonstrate peptide-mediated RNP/additional delivery of DNA templates for targeted HDR-mediated gene repair. In another recent report an amphiphlic peptide-mediated delivery system was used for the only delivery of RNP without additional DNA template to edit primary human lymphocytes [
39]. In the context of our study, it is noteworthy to emphasize the uniqueness of our findings, as successfully demonstrated the exclusive ability of a peptide-based system to co-deliver both the RNP and HDR templates. This distinctive aspect sets our report apart from existing literature in the field.
It is crucial to acknowledge that the high editing efficiencies observed in this study may not necessarily translate to scenarios requiring the co-delivery of large DNA fragments to facilitate HDR-directed repair or correction in non-dividing cells. Notably, a notable limitation of synthetic delivery systems employing cell-penetrating peptides (CPPs) likely lies in their inability to actively facilitate nuclear trafficking, despite their effectiveness in intracellular delivery. Consequently, further investigation is needed to assess the suitability of CPP-based delivery strategies for HDR in non-dividing or slow-dividing cells, similar to polyplexes and lipoplexes [
42,
43].
When comparing our results with previous publications on CPP-assisted CRISPR-Cas delivery it is important to note that our LAH5-based transfection was active at much lower concentrations of Cas9 RNP compared to recently published peptide mediated delivery studies [
38,
39] . In fact, the optimal concentrations at which we observe editing (20 nM of Cas9) are 10-1000 fold lower than earlier published results [
38,
39]. This observation may be attributed to the high histidine content present in our cell-penetrating peptide (CPP), which possesses a more favorable pKa for effective endosomal escape compared to peptides with lower histidine residue composition in their sequences [
44]. This has obvious advantages as high concentrations of Cas9 RNP may cause innate immune responses, by engaging pattern recognition receptors such as TLR3, RIG-I and PKR which in turn may lead to Cas9 specific adaptive responses [
40,
41]. Being able to lower the dose may reduce such undesired immune effects. Furthermore, robustness of our peptide-based transfection was demonstrated by showing editing in a variety of human cells. Even though editing efficiencies varied from 20-70%, in all cases sufficiently high levels of editing were achieved to be therapeutically relevant. LAH5 peptide demonstrated significantly superior performance compared to CRISPRMAX, at shorter incubation times. This observation strongly suggests that the cytosolic delivery of Cas9 RNP occurs more rapidly when facilitated by LAH5 peptide as compared to complexation with a lipid-based transfection agent.
This work demonstrated that the amphipathic LAH5 peptide can efficiently deliver Cas9 RNP with and without HDR template in a variety of cells by means of nanocomplex formation, triggering cell binding and uptake, with concomitant intracellular release of its cargo. Editing efficiencies were unprecedented showing high levels of gene editing at Cas9 RNP concentrations as low as 5-10 nM.
We do not expect that the current nanocomplex formulation is stable enough for direct in vivo applications. To make nanocomplexes, a large molar access of LAH5 peptide is needed. This points to a weak interaction with the Cas9 RNP. Intravenous injection would quickly dilute out the free peptide, leading to a change in equilibrium and potential dissociation of the nanocomplexes. Nevertheless, future research will investigate the nanocomplex stability under simulated in vivo conditions (whole serum) to get more insights into the stability. For now, LAH5 peptide-assisted delivery of Cas9 RNP could be very useful for e.g. ex vivo modification of NK or T cells as it relies on a single peptide that can be easily synthesized under good manufacturing practice (GMP).
Supplementary Materials
The following supporting information can be attached to the article. Table S1: sgRNA sequences used in this work, Table S2: PCR primers used for amplification stoplight target locus and CCR5 loci for both T7E1 and TIDE assays, Table S3: HDR template and Alexa642-HDR template sequences, Table S4: crRNA sequence used in this research, Figure S1: Cytotoxicity of RNP/LAH5 peptide nanocomplexes in multiple cell lines, Figure S2: The effectiveness of gene editing on CCR5 gene of ARPE-19 cells, Figure S3: HEK293T stoplight cells were treated with RNP/LAH5 nanocomplexes during different incubation times, Figure S4: LAH5 peptide mediated gene correction efficiency tested using the HDR stoplight reporter system, Figure S5: Gating strategy in all flow cytometry experiments using HEK293T Stoplight cells, and Figure S6: HEK293T HDR stoplight cells.
Author Contributions
Conceptualization, M. Ö., O.G.J. and E.M.; methodology, M. Ö., O.G.J. and E.M.; validation, M. Ö.; formal analysis, M. Ö.; investigation, M. Ö.; writing—original draft preparation, M. Ö.; writing—review and editing, M. Ö., O.G.J. and E.M.; visualization, M. Ö., O.G.J.; supervision, O.G.J. and E.M.; funding acquisition, E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Turkish Ministry of Education
Data Availability Statement
Supporting data to the research can be found in the supplementary materials of this manuscript.
Acknowledgments
The authors thank Dario Fenenko and Thai Hoang Nguyen for their help with the optimization of electrophoretic mobility shift assay.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jain, P.K.; Lo, J.H.; Rananaware, S.; Downing, M.; Panda, A.; Tai, M.; Raghavan, S.; Fleming, H.E.; Bhatia, S.N. Non-viral delivery of CRISPR/Cas9 complex using CRISPR-GPS nanocomplexes. Nanoscale 2019, 11, 21317–21323. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S. E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E. H.; Doudna, J.A.; Lim, W. A.; Weissman, J. S.; . Qi, L. S. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell, 2013, 154, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung,, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol 2014, 32, 347– 355. [CrossRef]
- Shalaby, K.; Aouida, M.; El-Agnaf, O. Tissue-Specific Delivery of CRISPR Therapeutics:Strategies and Mechanisms of Non-Viral Vectors, Int. J. Mol. Sci. 2020, 21, 7353. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Mahajan, V.B.; Tsang, S.H. Gene therapy and genome surgery in the retina. J. Clin. Investig. 2018, 128, 2177–2188. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; A Glass, Z.; Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2016, 24, 144–150. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Lee, Y.-W.; Scaletti, F.; Rotello, V. In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjugate Chem. 2017, 28, 880–884. [Google Scholar] [CrossRef]
- Ghosh, Sumit.; Brown, A.M.; Jenkins, C.; Campbell, K. Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Applied Biosafety. 2020, 25, 7-18. [CrossRef]
- Laustsen, A.; Bak, R.O. Electroporation-Based CRISPR/Cas9 Gene Editing Using Cas9 Protein and Chemically Modified sgRNAs. Methods Mol Biol. 2019, 1961, 127–134. [Google Scholar] [CrossRef]
- D'Astolfo, D.S.; Pagliero, R.J.; Pras, A.; Karthaus ,W.R.; Clevers, H.; Prasad; V.; Lebbink, R.J.; Rehmann, H.; Geijsen, N. Efficient intracellular delivery of native proteins. Cell 2015, 161, 674-690. [CrossRef]
- Yu, X.; Liang, X.; Xie, H.; Kumar, S.; Ravinder, N.; Potter, J.; Jeu, X.D.M.D.;, Chesnut J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol Lett. 2016, 38, 919-29. [CrossRef]
- Rouet, R.; Thuma, B.A.; Roy, M.D.; Lintner, N.G.; Rubitski, D.M.; Finley, J.E.; Wisniewska, H.M.; Mendonsa, R.; Hirsh, A.; de Oñate, L.; et al. Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing. J. Am. Chem. Soc. 2018, 140, 6596–6603. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Cui, H.; Ying, L.; Yu, X.F. Enhanced cytosolic delivery and release of CRISPR/ Cas9 by black phosphorus nanosheets for genome editing, Angew. Chem. Int. Ed. Eng. 2018, 57, 10268–10272. [Google Scholar] [CrossRef]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Efficient Delivery of CRISPR-Cas9 for Genome Editing via Self-Assembled DNA Nanoclews. Angew. Chem. Int. Ed Engl. 2015, 54, 12029–12033. [Google Scholar] [CrossRef]
- Tan, Z.; Ganewatta, Y.J.M.S.; Kumar, R.; Keith, A.; Twaroski, K.; Pengo, T.; Tolar, J.; Lodge, T.; Reineke, T.M. Block polymer micelles enable CRISPR/Cas9 ribonucleo- protein delivery: physicochemical properties affect packaging mechanisms and gene editing efficiency. Macromolecules 2019, 52, 8197–8206. [Google Scholar] [CrossRef]
- Zhu, X.; Lv, M.-M.; Liu, J.-W.; Yu, R.-Q.; Jiang, J.-H. DNAzyme activated protein- scaffolded CRISPR–Cas9 nanoassembly for genome editing. Chem. Commun. 2019, 55, 6511–6514. [Google Scholar] [CrossRef]
- Del’Guidice, T.; Lepetit-Stoffaes, J.-P.; Bordeleau, L.-J.; Roberge, J.; Théberge, V.; Lauvaux, C.; Barbeau, X.; Trottier, J.; Dave, V.; Roy, D.-C.; et al. Membrane Permeabilizing Amphiphilic Peptide Delivers Recombinant TranscriptionFactor and CRISPR-Cas9/Cpf1 Ribonucleoproteins in Hard-to-Modify Cells. PLOS ONE 2018, 13, e0195558. [Google Scholar] [CrossRef]
- Shen, Y.; Cohen, J.L.; Nicoloro, S.M.; Kelly, M.; Yenilmez, B.; Henriques, F.; Tsagkaraki, E.; Edwards, Y.J.K.; Hu, X.; Friedline, R.H.; et al. CRISPR-Delivery Particles Targeting Nuclear Receptor–Interacting Protein 1 (Nrip1) in Adipose Cells to Enhance Energy Expenditure. J. Biol. Chem. 2018, 293, 17291–17305. [Google Scholar] [CrossRef]
- Suresh, B.; Ramakrishna, S.; Kim, H. Cell-Penetrating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA for Genome Editing. Methods. Mol. Biol. 2016, 1507, 81–94. [Google Scholar] [CrossRef]
- Gustsfsson, O., Rädler, J., Roudi, S., Lehto. T., Hällbrink, M., Lehto, T., Gupta, D., Andaloussi, S., and Nordin, J., Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP., Pharmaceutics 2021, 13, 878. [CrossRef]
- Krishnamurthy, S.; Wohlford-Lenane, C.; Kandimalla, S.; Sartre, G.; Meyerholz, D.K.; Théberge, V.; Hallée, S.; Duperré, A.-M.; Del’Guidice, T.; Lepetit-Stoffaes, J.-P.; et al. Engineered Amphiphilic Peptides Enable Delivery of Proteins and CRISPR-Associated Nucleases to Airway Epithelia. Nat. Commun. 2019, 10, 4906. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.; Frankel, A. Endocytosis and targeting of exogenous HIV-1 Tat protein. EMBO J. 1991, 10, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Madani, F.; Lindberg, S.; Langel, Ü.; Futaki, S.; Gräslund, A. Mechanisms of Cellular Uptake of Cell-Penetrating Peptides. J. Biophys. 2011, 2011, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Givens, B.E.; Naguib, Y.W.; Geary, S.M.; Devor, E.J.; Salem, A.K. Nanoparticle-Based Delivery of CRISPR/Cas9 Genome-Editing Therapeutics. AAPS J. 2018, 20, 108. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Kwaku Dad, A.-B.; Beloor, J.; Gopalappa, R.; Lee, S.-K.; Kim, H. Gene Disruption by Cell-Pene-trating Peptide-Mediated Delivery of Cas9 Protein and Guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef]
- Yin, J.; Wang, Q.; Hou, S.; Bao, L.; Yao, W.; Gao, X. Potent Protein Delivery into Mammalian Cells via a Super-charged Polypeptide. J. Am. Chem. Soc. 2018, 140, 17234–17240. [Google Scholar] [CrossRef]
- Xu, X., Wan, T., Xin, H., Li, D., Pan, P., Wu, J.,Ping, Y., Delivery of CRISPR/Cas9 for therapeutic genome editing J Gene Med. 2019, 21, e3107. [CrossRef]
- Lostalé-Seijo, I.; Louzao, I.; Juanes, M.; Montenegro, J. Peptide/Cas9 Nanostructures for Ribonucleoprotein Cell Membrane Transport and Gene Edition. Chem. Sci. 2017, 8, 7923–7931. [Google Scholar] [CrossRef]
- Raad, M. d.; Teunissen, E. A.; , Lelieveld, D.; Egan, D. A.; Mastrobattista, E. High-content screening of peptide-based non-viral gene delivery systems. Journal of Controlled Release. 2012, 158, 433–442. [CrossRef]
- Kichler, A., Leborgne, C., Marz, J., Danos, O., and Bechinger, B., Histidine-rich amphipathic peptide antibiotics promote efficient delivery of DNA into mammalian cells., PNAS. 2003, 100(4), 1564–1568. [CrossRef]
- De Jong, O.G.; Murphy, D.E.; Mäger, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; van Rheenen, J.; et al. A CRISPR-Cas9-Based Reporter System for Single-Cell Detection of Extracellular Vesicle-Mediated Functional Transfer of RNA. Nat. Commun. 2020, 11, 1113. [Google Scholar] [CrossRef]
- Segu, V.B.; Li, G.; Metz, S.A. ; Use of a soluble tetrazolium compound to assay metabolic activation of intact beta cells. Metabolism 1998, 47:824-30. [CrossRef] [PubMed]
- Mashal, R., Koontz, J., Sklar, J. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 1995, 9, 177–183. [CrossRef]
- Brinkman, E.K.; Chen, T.; Amendola, M.; Steensel, B.V. Easy quantitative assessment of genome editing by sequence trace decomposition, Nucleic Acids Research 2014, 42, e168. [CrossRef]
- Lointier, M. Structral and functional investigations of designed histidine-rich peptides. PhD thesis, Strasbourg University, Strasbourg, France, December 2020. 20 December.
- Foss, D.V.; Muldoon, J.J.; Nguyen, D.N.; et al. Peptide-mediated delivery of CRISPR enzymes for the efficient editing of primary human lymphocytes. Nat. Biomed. Eng. 2023, 7, 647–660. [Google Scholar] [CrossRef]
- Zhang, Z.; Baxter, A.E.;, Ren, D.; et al. Efficient engineering of human and mouse primary cells using peptide-assisted genome editing. Nat Biotechnol. 2023. [CrossRef]
- Wilbie, D.; Walther, J.; Mastrobattista E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Accounts of Chemical Research 2019, 52(6), 1555-1564. [CrossRef]
- Kanzler, H.; Barrat, F. J.; Hessel, E. M.; Coffman, R. L. Therapeutic Targeting of Innate Immunity with Toll-like Receptor 4 (TLR4) Antagonists. Nat. Med. 2007, 13, 552–559. [Google Scholar] [CrossRef]
- Maury, B.; Gonçalves, C.; Tresset, G.; Zeghal, M.; et al., Influence ofpDNA availability on transfection efficiency of polyplexes in non-proliferative cells. Biomaterials. 2014, 35, 5977–5985. [CrossRef]
- Belmadi, N.; Midoux, P.; Loyer, P.; Passirani, C.; Pichon, C.; Gall, T.; Jaffres, P-A.; Lehn, P.; Montier, T.; Synthetic vectors for gene delivery: An overview of their evolution depending on routes of administration. Biotechnology Journal. 2015, 10, 1370-1389. [CrossRef]
- Kichler, A.; Mason, J. A.; Bechinger, B.; Cationic amphipathic histidine-rich peptides for gene delivery. Biochimica et Biophysica Acta (BBA). 2006, 1758, 301-307. [CrossRef]
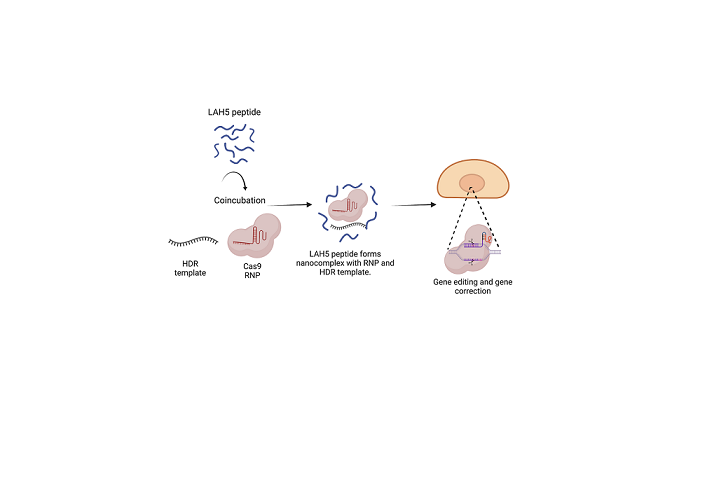
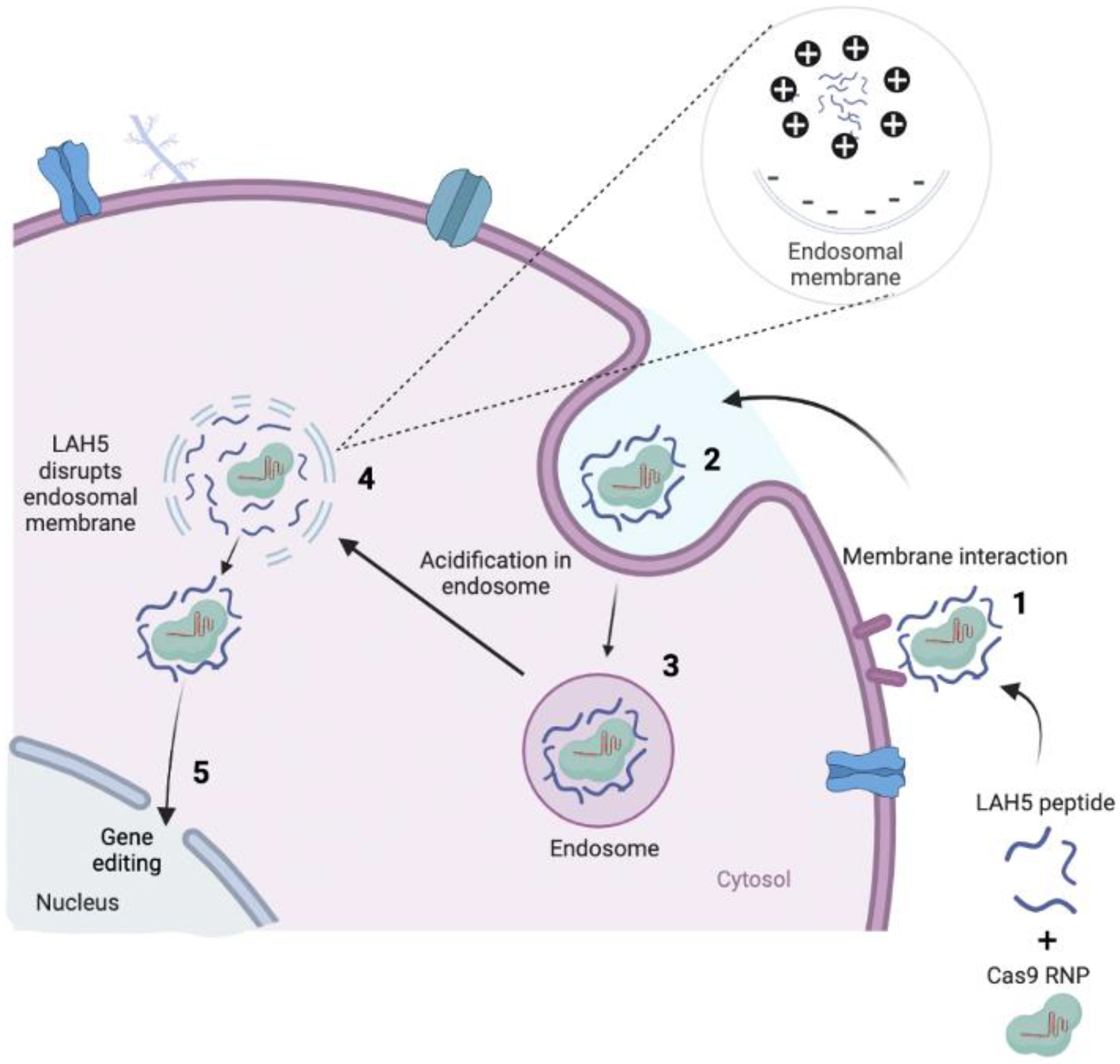
Figure 1.
The concept of how Cas9 RNP/LAH5 nanocomplexes escape from endosome. The nanocomplex firstly interacts with cellular membrane of target cells, and subsequently internalized via endocytosis. The amphiphatic character of the LAH5 peptide induces lytic membrane interactions with endosomal membrane. Following endosomal membrane disruption, nanocomplexes are released into the cytosol. Hereafter, RNP/LAH5 nanocomplexes can reach the nucleus. NLS sequences on Cas9 allow for internalization into the nucleus, where gene editing can take place. Created with BioRender.com.
Figure 1.
The concept of how Cas9 RNP/LAH5 nanocomplexes escape from endosome. The nanocomplex firstly interacts with cellular membrane of target cells, and subsequently internalized via endocytosis. The amphiphatic character of the LAH5 peptide induces lytic membrane interactions with endosomal membrane. Following endosomal membrane disruption, nanocomplexes are released into the cytosol. Hereafter, RNP/LAH5 nanocomplexes can reach the nucleus. NLS sequences on Cas9 allow for internalization into the nucleus, where gene editing can take place. Created with BioRender.com.
Figure 2.
Electrophoretic mobility shift assay (EMSA) of Cas9-GFP, Cas9-GFP RNP (A) and HDR DNA template alone or combined with Cas9-GFP RNP (B) sgRNA was labelled with ATTO550, and the HDR template was labelled with Alexa647 for visualization of each of the individual components on gel. Reduced mobility indicates charge neutralization and/or complex formation. Components were mixed at the indicated molar ratios and after a short (5 min) incubation to enable complex formation, samples were loaded onto a 1.5% agarose gel and run at 170 V for 15 minutes. Fluorescent gel images were captured with a ChemiDocTM XRS+ imager (Bio-Rad).
Figure 2.
Electrophoretic mobility shift assay (EMSA) of Cas9-GFP, Cas9-GFP RNP (A) and HDR DNA template alone or combined with Cas9-GFP RNP (B) sgRNA was labelled with ATTO550, and the HDR template was labelled with Alexa647 for visualization of each of the individual components on gel. Reduced mobility indicates charge neutralization and/or complex formation. Components were mixed at the indicated molar ratios and after a short (5 min) incubation to enable complex formation, samples were loaded onto a 1.5% agarose gel and run at 170 V for 15 minutes. Fluorescent gel images were captured with a ChemiDocTM XRS+ imager (Bio-Rad).
Figure 3.
The amphipathic LAH5 peptide forms nanocomplexes with spCas9/sgRNA (RNP). (A) Size and polydispersity index of the spCas9/sgRNA (RNP) and LAH5 peptide complexation spCas9/sgRNA RNP and increasing molar ratios of LAH5 peptide complexed together. (B) Zeta potential of the spCas9/sgRNA (RNP) spCas9/sgRNA RNP and increasing molar ratios of LAH5 peptide complexed together. Error bars indicate mean ± SD (n = 3).
Figure 3.
The amphipathic LAH5 peptide forms nanocomplexes with spCas9/sgRNA (RNP). (A) Size and polydispersity index of the spCas9/sgRNA (RNP) and LAH5 peptide complexation spCas9/sgRNA RNP and increasing molar ratios of LAH5 peptide complexed together. (B) Zeta potential of the spCas9/sgRNA (RNP) spCas9/sgRNA RNP and increasing molar ratios of LAH5 peptide complexed together. Error bars indicate mean ± SD (n = 3).
Figure 4.
Fluorescence confocal miscroscopy images of the cellular uptake of Cas9-GFP RNP Scale bar 70 μm (A) and Cas9-GFP RNP together with ssDNA HDR template (D) with and without LAH5 complexation. HeLa cells were imaged 24h (A) and (D) after addition of the indicated components. Prior to image aquisition, cells were washed 2x with fresh medium to remove excess of fluorescently-labelled compounds. The concentration of Cas9-GFP RNP was 10 nM for all experiments. The sgRNA used was fluorescently labelled with ATTO-550 and ssDNA HDR template with Alexa-647. (B) T7 endonuclease mismatch detection assay of PCR products containing the CRISPR target locus of the CCR5 gen indicate % CCR5 gene editing. (C) TIDE assay to quantify the frequency of targeted mutations in the target locus of the CCR5 gene. Data shown as mean ± SD (n=3).
Figure 4.
Fluorescence confocal miscroscopy images of the cellular uptake of Cas9-GFP RNP Scale bar 70 μm (A) and Cas9-GFP RNP together with ssDNA HDR template (D) with and without LAH5 complexation. HeLa cells were imaged 24h (A) and (D) after addition of the indicated components. Prior to image aquisition, cells were washed 2x with fresh medium to remove excess of fluorescently-labelled compounds. The concentration of Cas9-GFP RNP was 10 nM for all experiments. The sgRNA used was fluorescently labelled with ATTO-550 and ssDNA HDR template with Alexa-647. (B) T7 endonuclease mismatch detection assay of PCR products containing the CRISPR target locus of the CCR5 gen indicate % CCR5 gene editing. (C) TIDE assay to quantify the frequency of targeted mutations in the target locus of the CCR5 gene. Data shown as mean ± SD (n=3).
Figure 5.
HEK293T stoplight cells were used to determine gene editing efficiency (A) Images (60x) of Stoplight HEK293T stoplight cells from confocal microscope after treatment with different ratios of RNP:LAH5 peptide at a RNP concentration of 20 nM. Red – mCherry, green – eGFP (Cas9 gene editing), blue – Hoechst, NC: Negative control. Scale bar represents 70 μm. (B) Indel (eGFP positive) cells for increasing molar ratios of LAH5 peptide (with total 20nM RNP concentration) measured by flow cytometry. (C) TIDE and (D) T7E1 assay performed to detect gene editing of the cells transfected with same concentrations of RNP/LAH5 peptide nanocomplexes and positive control CRISPRMAX. (E) Gene editing efficiency eGFP(+) cells showed increasing concentrations of RNP and stable concentration of LAH5 (5μM) peptide, gene editing efficiency eGFP(+) cells percentage measured by flow cytometry. (F) T7E1 assay on gene editing of the cells transfected with same concentrations of RNP/LAH5 peptide nanocomplexes. Data are presented as mean ± S.E.M (n = 3).
Figure 5.
HEK293T stoplight cells were used to determine gene editing efficiency (A) Images (60x) of Stoplight HEK293T stoplight cells from confocal microscope after treatment with different ratios of RNP:LAH5 peptide at a RNP concentration of 20 nM. Red – mCherry, green – eGFP (Cas9 gene editing), blue – Hoechst, NC: Negative control. Scale bar represents 70 μm. (B) Indel (eGFP positive) cells for increasing molar ratios of LAH5 peptide (with total 20nM RNP concentration) measured by flow cytometry. (C) TIDE and (D) T7E1 assay performed to detect gene editing of the cells transfected with same concentrations of RNP/LAH5 peptide nanocomplexes and positive control CRISPRMAX. (E) Gene editing efficiency eGFP(+) cells showed increasing concentrations of RNP and stable concentration of LAH5 (5μM) peptide, gene editing efficiency eGFP(+) cells percentage measured by flow cytometry. (F) T7E1 assay on gene editing of the cells transfected with same concentrations of RNP/LAH5 peptide nanocomplexes. Data are presented as mean ± S.E.M (n = 3).
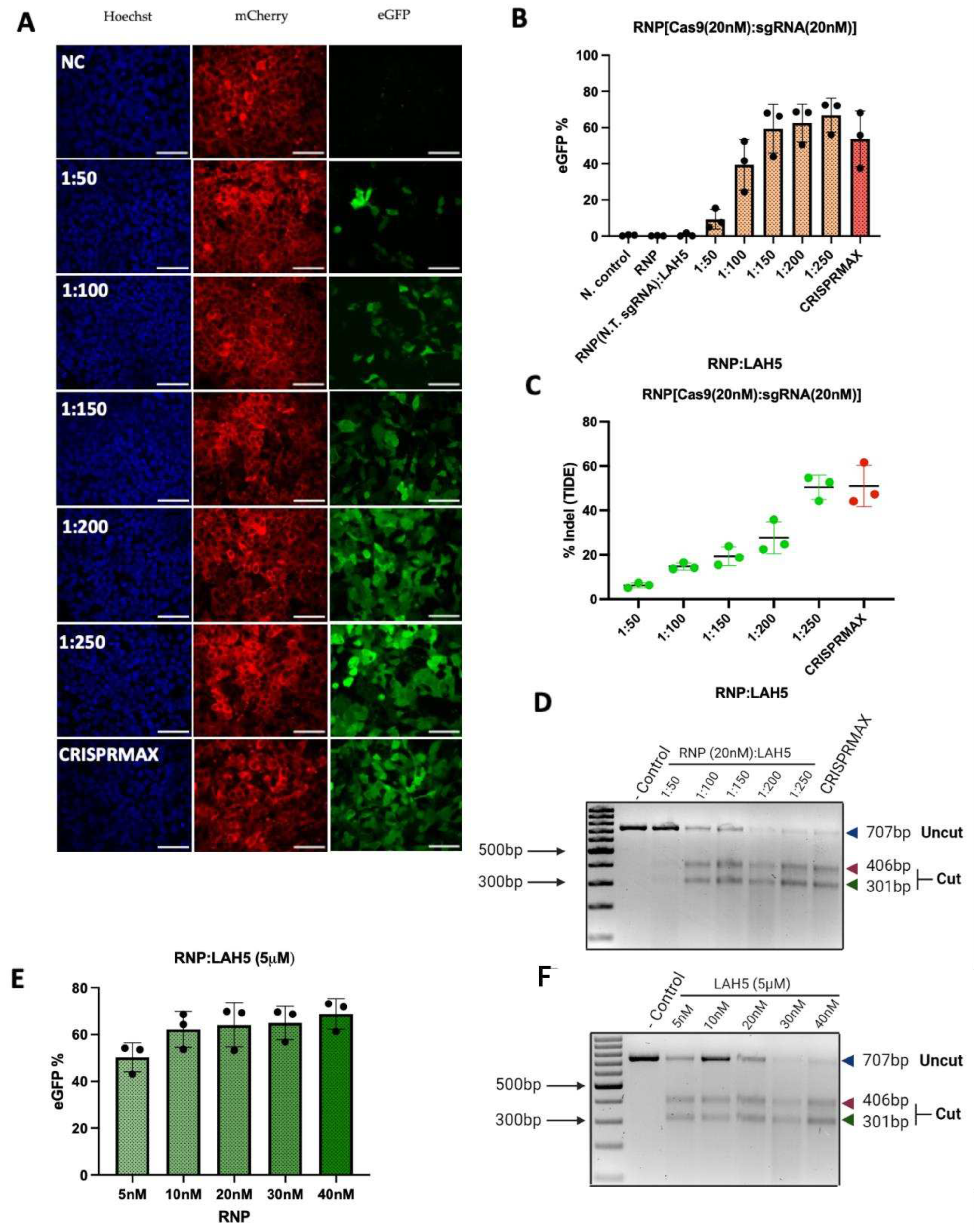
Figure 6.
The effectiveness of gene editing was assessed using several cell lines, and the CCR5 gene was chosen as a target. (A), (B) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HEK293T cells and % of indel calculated by (A) TIDE and (B) T7E1 assay (uncut, 717 bp; cut, 441 bp 276 bp; Cas9, 20 nM; CCR5 sgRNA, 20 nM; increasing ratios of LAH5). (C), (D) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HeLa cells and % of Indels calculated by (C) TIDE and (D) T7E1 (E), (F) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HEPG2 cells and % of Indels calculated by (E) TIDE and (F) T7E1 assay (G), (H) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated primary fibroblast cells and % of Indels calculated by (G) TIDE and (H) T7E1 assay. Difference between the dots are represented as mean ± SD (n = 3). Treated samples compared with non-treated control using analysis of variance (ANOVA) Dunnett's multiple comparison test. (*p < 0.05; * p < 0.01; *** p < 0.001; **** p < 0.0001).
Figure 6.
The effectiveness of gene editing was assessed using several cell lines, and the CCR5 gene was chosen as a target. (A), (B) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HEK293T cells and % of indel calculated by (A) TIDE and (B) T7E1 assay (uncut, 717 bp; cut, 441 bp 276 bp; Cas9, 20 nM; CCR5 sgRNA, 20 nM; increasing ratios of LAH5). (C), (D) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HeLa cells and % of Indels calculated by (C) TIDE and (D) T7E1 (E), (F) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated HEPG2 cells and % of Indels calculated by (E) TIDE and (F) T7E1 assay (G), (H) percentage indel measurements in CCR5 locus DNA isolated from RNP/LAH5 treated primary fibroblast cells and % of Indels calculated by (G) TIDE and (H) T7E1 assay. Difference between the dots are represented as mean ± SD (n = 3). Treated samples compared with non-treated control using analysis of variance (ANOVA) Dunnett's multiple comparison test. (*p < 0.05; * p < 0.01; *** p < 0.001; **** p < 0.0001).
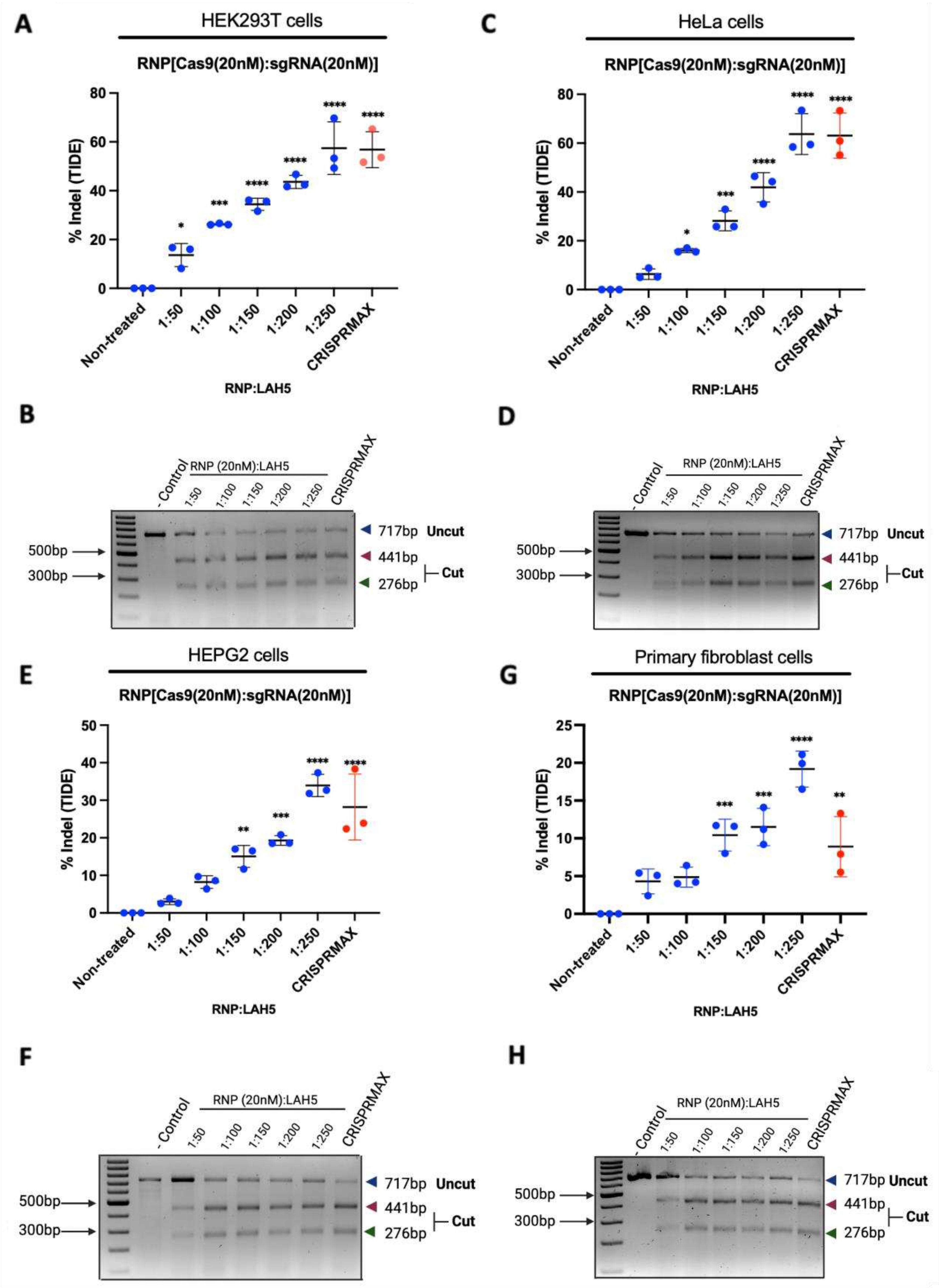
Figure 7.
LAH5 peptide-mediated gene correction efficiency tested using the HDR stoplight reporter system: To evaluate the impact of increasing concentrations of RNP/HDR template HEK293T HDR stoplight cells treated the range of 10 nM to 40 nM RNP/HDR template at 1:1 ratio and these various RNP/HDR template concentrations complexed with 5 μM of LAH5 peptide (A) The HDR Stoplight reporter system was developed as a read-out to assess HDR-mediated gene correction efficiency. A schematic depiction illustrates the mechanism of the construct. HEK293T HDR Stoplight cells exhibit continuous expression of mCherry, directly followed by a stop codon and an in-frame open reading frame (ORF) for eGFP. As translation will stop at the stop codon at the end of the mCherry ORF, eGFP expression is blocked. Introduction of a single stranded oligonucleotide (ssODN) HDR template along with CRISPR-Cas9 facilitates a targeted substitution of the stop codon with a glutamine residue, thereby inducing the synthesis of a fusion protein comprising mCherry and eGFP. (B) Images (60x) of HDR Stoplight HEK293T cells from confocal microscope after treatment with RNP (20nM)/HDR template (20 nM) and increasing ratio of LAH5 peptide range at 1:1:50 to 1:1:250, and CRISPRMAX as positive control Red – mCherry, green – eGFP (gene correction), blue – Hoechst, NC: Negative control. Scale bar 70 μm. (C) HDR efficiency quantified by flow cytometry by measuring % of eGFP positive expression (D) In order to evaluate the dose-dependent effects of the LAH5 peptide on gene correction, HEK293T HDR stoplight cells were treated with RNP/HDR template/LAH5 nanocomplexes. The nanocomplexes were prepared by combining RNP (20nM) and HDR template (20nM), along with LAH5 peptide at increasing ratios ranging from 1:1:50 to 1:1:250. Subsequently, the efficiency of gene correction was determined by calculating the percentage of eGFP positive cells. Data presented as mean ± S.E.M (n = 3). Treated samples compared with non-treated control using analysis of variance (ANOVA) Dunnett's multiple comparison test (*p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
Figure 7.
LAH5 peptide-mediated gene correction efficiency tested using the HDR stoplight reporter system: To evaluate the impact of increasing concentrations of RNP/HDR template HEK293T HDR stoplight cells treated the range of 10 nM to 40 nM RNP/HDR template at 1:1 ratio and these various RNP/HDR template concentrations complexed with 5 μM of LAH5 peptide (A) The HDR Stoplight reporter system was developed as a read-out to assess HDR-mediated gene correction efficiency. A schematic depiction illustrates the mechanism of the construct. HEK293T HDR Stoplight cells exhibit continuous expression of mCherry, directly followed by a stop codon and an in-frame open reading frame (ORF) for eGFP. As translation will stop at the stop codon at the end of the mCherry ORF, eGFP expression is blocked. Introduction of a single stranded oligonucleotide (ssODN) HDR template along with CRISPR-Cas9 facilitates a targeted substitution of the stop codon with a glutamine residue, thereby inducing the synthesis of a fusion protein comprising mCherry and eGFP. (B) Images (60x) of HDR Stoplight HEK293T cells from confocal microscope after treatment with RNP (20nM)/HDR template (20 nM) and increasing ratio of LAH5 peptide range at 1:1:50 to 1:1:250, and CRISPRMAX as positive control Red – mCherry, green – eGFP (gene correction), blue – Hoechst, NC: Negative control. Scale bar 70 μm. (C) HDR efficiency quantified by flow cytometry by measuring % of eGFP positive expression (D) In order to evaluate the dose-dependent effects of the LAH5 peptide on gene correction, HEK293T HDR stoplight cells were treated with RNP/HDR template/LAH5 nanocomplexes. The nanocomplexes were prepared by combining RNP (20nM) and HDR template (20nM), along with LAH5 peptide at increasing ratios ranging from 1:1:50 to 1:1:250. Subsequently, the efficiency of gene correction was determined by calculating the percentage of eGFP positive cells. Data presented as mean ± S.E.M (n = 3). Treated samples compared with non-treated control using analysis of variance (ANOVA) Dunnett's multiple comparison test (*p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).