Submitted:
14 June 2023
Posted:
14 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plants and soil sampling
2.2. Cultural media
2.3. Isolation of endophytic and rhizosphere soil fungi
2.4. Strain identification
2.4. Data analysis
3. Results
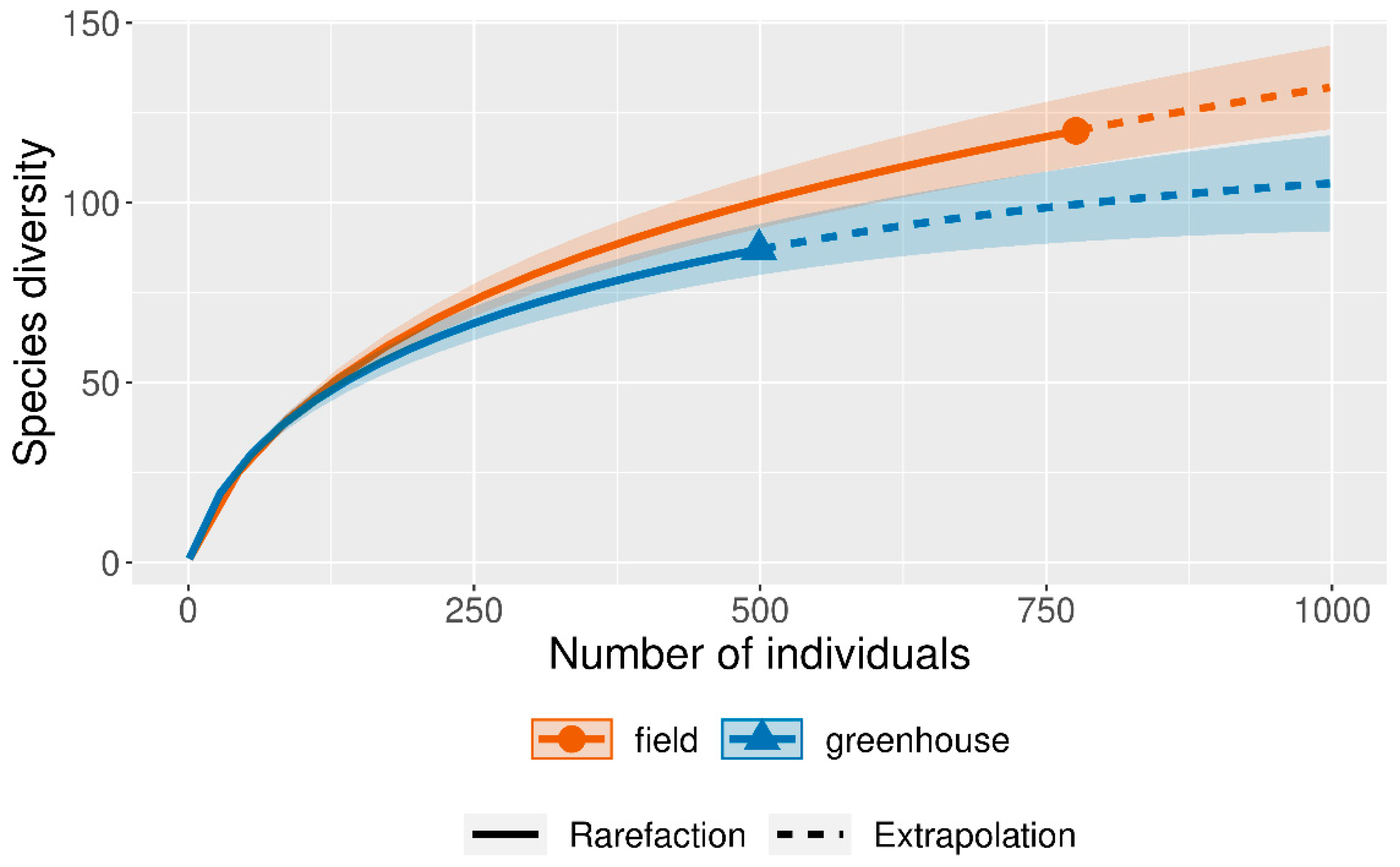
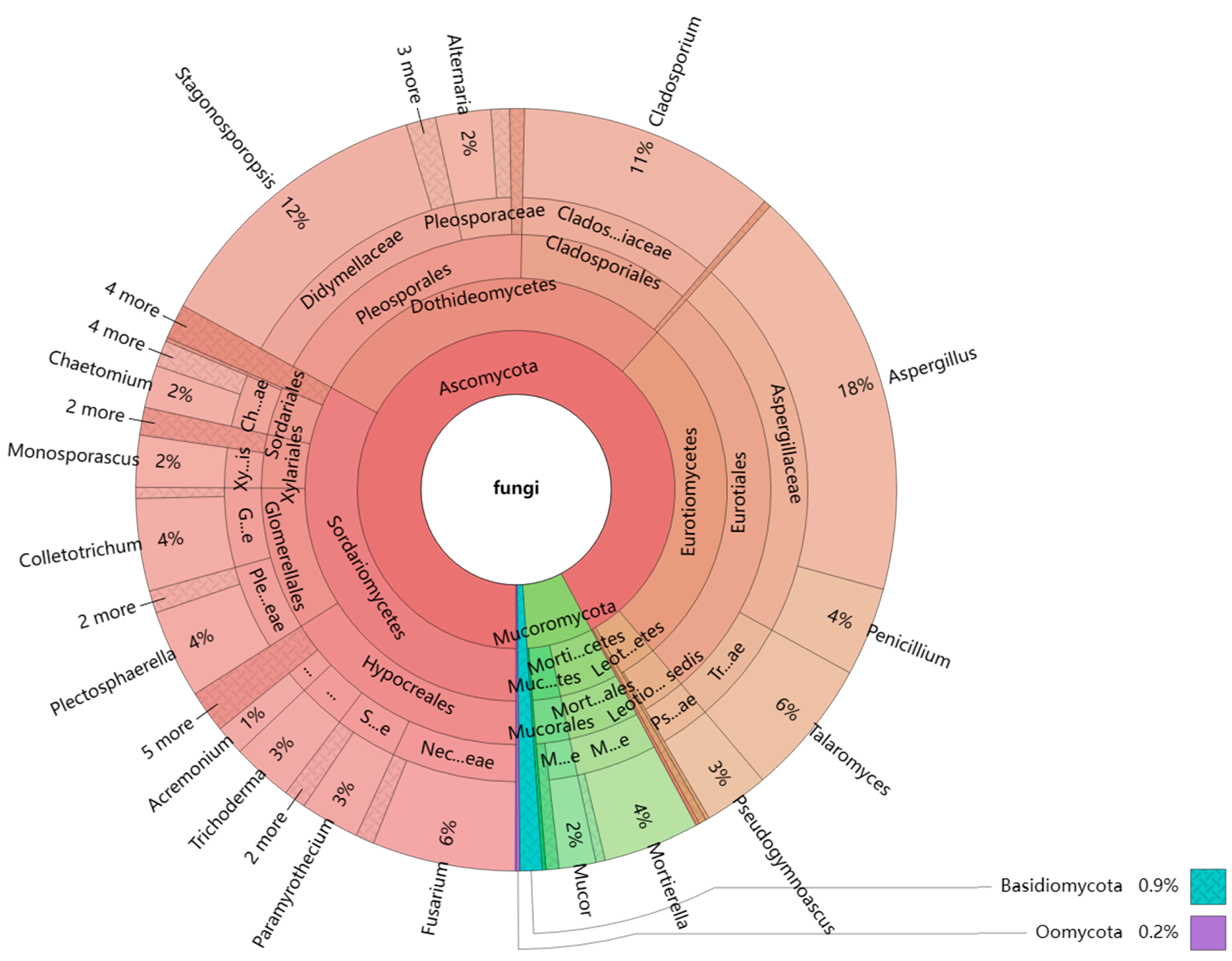
3.1. Community composition of endophytic fungi and rhizosphere soil fungi
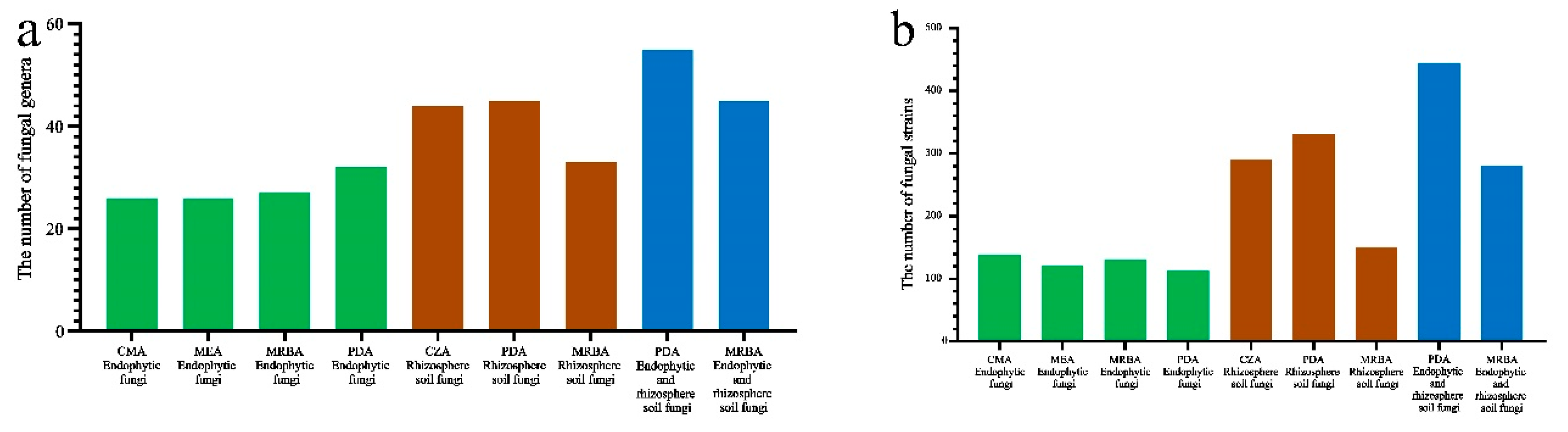
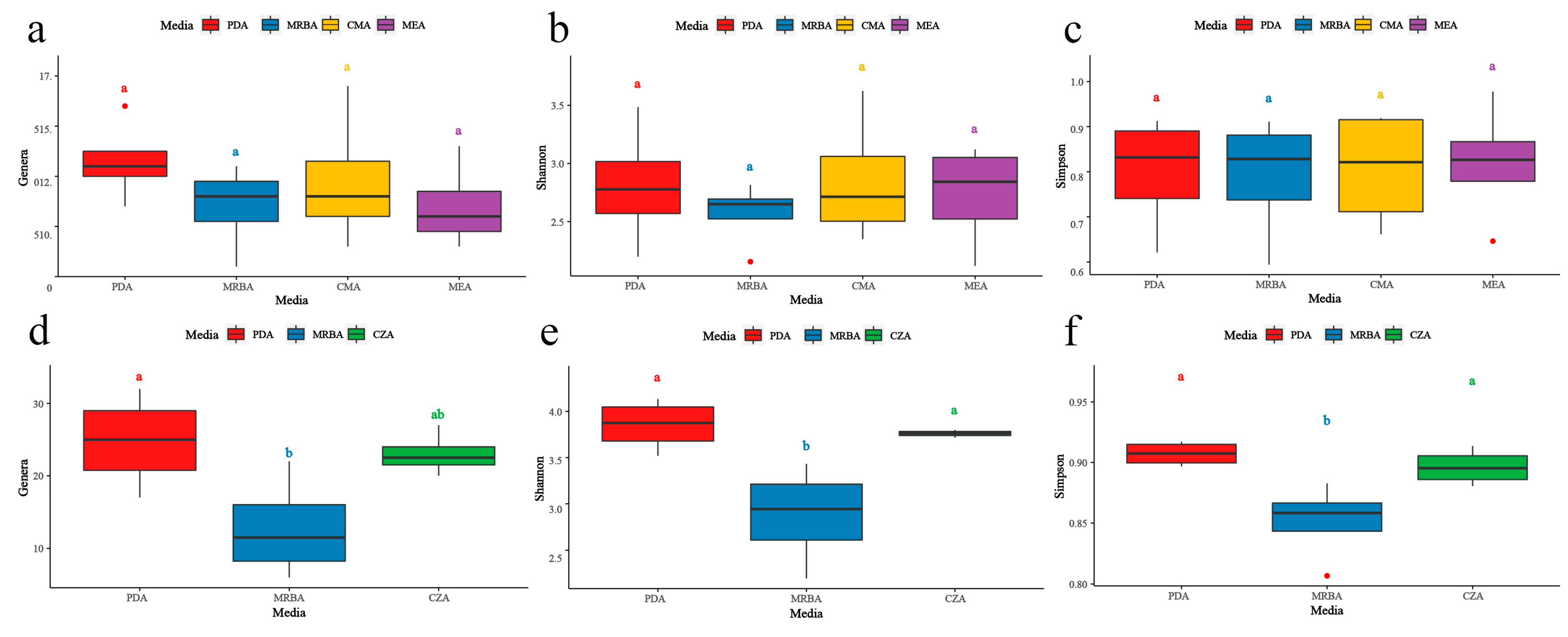
3.2. Influence of culture medium on fungal isolation
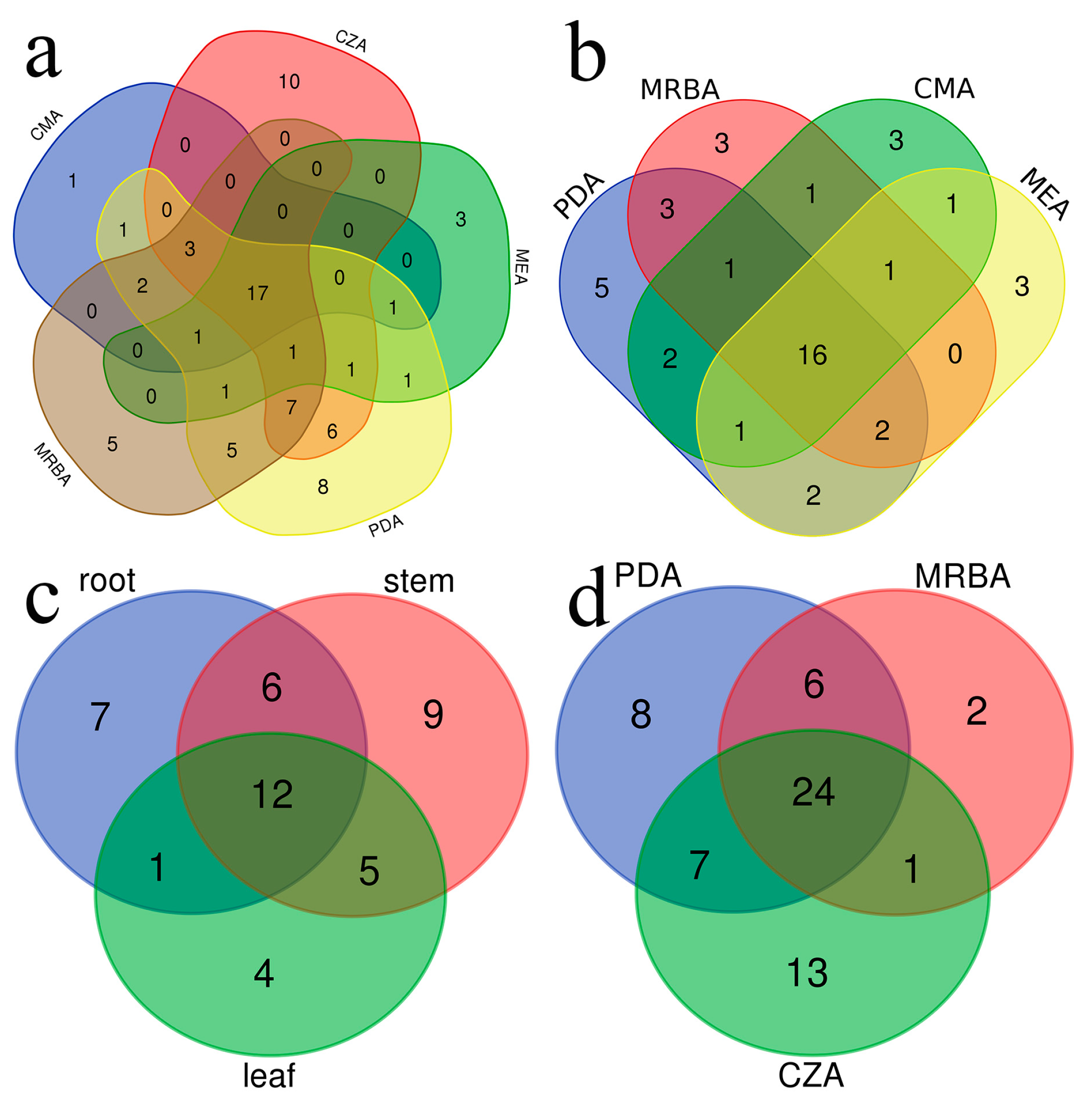
3.3. Effects of culture medium on endophytic fungal diversity
3.4. Effect of the medium type on endophytic fungal diversity in different tissues of the plant
3.5. Effects of culture medium on soil fungal diversity
3.6. Effects of culture medium on fungal genera with high isolation efficiency
3.6.1. Trichodera and Fusarium
3.6.2. Other 26 fungal genera
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgements
Conflicts of Interest
References
- Bamisile, B.S.; Dash, C.K.; Akutse, K.S.; Keppanan, R.; Afolabi, O.G.; Hussain, M.; Qasim, M.; Wang, L. Prospects of endophytic fungal entomopathogens as biocontrol and plant growth promoting agents: An insight on how artificial inoculation methods affect endophytic colonization of host plants. Microbiol. Res. 2018, 217, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma–plant–pathogen interactions. Soil Biology and Biochemistry 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Arenz, B.E.; Held, B.W.; Jurgens, J.A.; Farrell, R.L.; Blanchette, R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biology and Biochemistry 2006, 38, 3057–3064. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; Martinez-Montiel, N.; Cruz-Lopez, M.D.C.; Martinez-Contreras, R.D. Fungal diversity and community composition of culturable fungi in Stanhopea trigrina cast gibberellin producers. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Nischitha, R.; Shivanna, M.B. Antimicrobial activity and metabolite profiling of endophytic fungi in Digitaria bicornis (Lam) Roem. and Schult. and Paspalidium flavidum (Retz.) A. Camus. 3 Biotech 2021, 11. [CrossRef]
- Muggia, L.; Kopun, T.; Grube, M. Effects of growth media on the diversity of culturable fungi from lichens. Molecules 2017, 22, 824. [Google Scholar] [CrossRef]
- Prior, R.; Görges, K.; Yurkov, A.; Begerow, D. New isolation method for endophytes based on enzyme digestion. Mycol Progress 2014, 13, 849–856. [Google Scholar] [CrossRef]
- Cosoveanu, A.; Cabrera, R. Fungi as endophytes in Artemisia thuscula: Juxtaposed elements of diversity and phylogeny. Journal of Funig 2018, 4, 17. [Google Scholar] [CrossRef]
- Zeng, Z.H.; Yuan, Z.L.; Huang, X.N.; Liu, F. Isolation and diversity of mangrove endophytic fungi. Protection Forest Science and Technology 2022, 39-42, 45. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; Petrini, O. Botryosphaeria dothidea endophytic in Eucalyptus grandis and Eucalyptus nitens in South Africa. Forest Ecol Manag 1996, 89, 189–195. [Google Scholar] [CrossRef]
- Blodgett, J.T.; Swart, W.J.; Weeks, L.W.J. Species composition of endophytic fungi in Amaranthus hybridus leaves, petioles, stems, and roots. Mycologia 2000, 92, 853–859. [Google Scholar] [CrossRef]
- Broda, D.M.; Bell, R.G.; Boerema, J.A.; Musgrave, D.R. The abattoir source of culturable psychrophilic Clostridium spp. causing 'blown pack' spoilage of vacuum-packed chilled venison. J. Appl. Microbiol. 2002, 93, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Bettucci, L.; Simeto, S.; Alonso, R.; Lupo, S. Endophytic fungi of twigs and leaves of three native species of Myrtaceae in Uruguay. Sydowia 2004, 56, 8–23. [Google Scholar]
- Blodgett, J.T.; Swart, W.J.; Louw, S.V.; Weeks, W.J. Soil amendments and watering influence the incidence of endophytic fungi in Amaranthus hybridus in South Africa. Appl Soil Ecol 2007, 35, 311–318. [Google Scholar] [CrossRef]
- Xue, Q.W.; Niu, Y.C.; Deng, H. Diversity of endophytic fungi in common cucurbit plants. Mycosystema 2015, 34, 196–203. [Google Scholar] [CrossRef]
- Belfiori, B.; Rubini, A.; Riccioni, C. Diversity of endophytic and pathogenic fungi of Saffron (Crocus sativus) plants from cultivation sites in Italy. Diversity 2021, 13, 535. [Google Scholar] [CrossRef]
- Wang. Z.B.; Zhou. X.; Ma. X.B.; Wang, J.L. Isolation of endophytic fungi from medicinal plants and screening and identification of antagonism. Chinese Agricultural Science Bulletin 2022, 38, 75–81. [Google Scholar]
- Evans, H.C.; Holmes, K.A.; Thomas, S.E. Endophytes and mycoparasites associated with an indigenous forest tree, Theobroma gileri, in Ecuador and a preliminary assessment of their potential as biocontrol agents of cocoa diseases. Mycol Progress 2003, 2, 149–160. [Google Scholar] [CrossRef]
- Khan, A.R.; Waqas, M.; Ullah, I.; Khan, A.L.; Khan, M.A.; Lee, I.; Shin, J. Culturable endophytic fungal diversity in the cadmium hyperaccumulator Solanum nigrum L. and their role in enhancing phytoremediation. Environ. Exp. Bot. 2017, 135, 126–135. [Google Scholar] [CrossRef]
- Li, W.Z.; Duan, W.J.; Zhou, X.; Chen, J.; Jiang, X.Z.; Yang, Y.; Cai, L. Diversity of fungi in wheat seeds. Mycosystema 2021, 40, 487–501. [Google Scholar] [CrossRef]
- Guo, L.D.; Hyde, K.D.; Liew, E. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Xiang, L.; Gong, S.; Yang, L.; Hao, J.; Xue, M.; Zeng, F.; Zhang, X.; Shi, W.; Wang, H.; Yu, D. Biocontrol potential of endophytic fungi in medicinal plants from Wuhan Botanical Garden in China. Biol. Control. 2016, 94, 47–55. [Google Scholar] [CrossRef]
- Wang, H.X.; Yu, Y.R.; Huang, B.K. Diversity of culturable endophytic fungi from Broussonetia papyrifera. Mycosystema 2020, 39, 2399–2408. [Google Scholar] [CrossRef]
- Figueredo, H.M.; Gonçalves, V.N.; Godinho, V.M.; Lopes, D.V.; Oliveira, F.S.; Rosa, L.H. Diversity and ecology of cultivable fungi isolated from the thermal soil gradients in Deception Island, Antarctica. Extremophiles 2020, 24, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.L.; Liu, M.; Huang, H.Q.; Zhu, J.; Bao, S.X. Diversity of soil culturable fungi in Bamen Bay Mangrove Forests. Chinese Journal of Tropical Crops 2013, 34, 181–187. [Google Scholar]
- Zhang, J.Z.; Chen, X.R.; Yang, C.D.; Xue, L. A study on the diversity of soil cultured fungi in the alpine grassland of Eastern Qilian Mountains. Acta Prataculturae Sinica 2010, 19, 124–132. [Google Scholar]
- Vasanthakumari, M.M.; Shivanna, M.B. Fungal assemblages in the rhizosphere and rhizoplane of grasses of the subfamily Panicoideae in the Lakkavalli region of Karnataka, India. Microbes Environ 2011, 26, 228–236. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Y.C.; Jiang, Y. Seasonal variation of soil fungal diversity in the rhizosphere of Phragmites australis. Journal of Liaoning Normal University (Natural Science Edition) 2017, 40, 89–94. [Google Scholar] [CrossRef]
- Ayob, Z.; Kusai, N.A.; Ali, S.R.A. Sequence-based identification and characterisation of cultivated filamentous fungi in the Alan Bunga peat ecosystems of Sarawak, Malaysia. Mires Peat 2018, 21, 1–19. [Google Scholar] [CrossRef]
- Zhou, J.; Miao, Y.F.; Fang, K.; Chen, L.; Yang, Z.P.; Dong, X.F.; Zhang, H.B. Diversity of the endophytic and rhizosphere soil fungi of Ageratina adenophora. Ecological Science 2019, 38, 1–7. [Google Scholar] [CrossRef]
- Guo, M.J.; Zhao, X.Y.; Huang, J.H.; Wei, S.Z.; Xu, X.L. Isolation and activity analysis of fungi from the rhizosphere of mangroves in Xinglin Bay, Xiamen. Microbiology 2021, 48, 1496–1503. [Google Scholar] [CrossRef]
- Vieira, F.C.S.; Nahas, E. Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 2005, 160, 197–202. [Google Scholar] [CrossRef]
- Grishkan, I.; Beharav, A.; Kirzhner, V.; Nevo, E. Adaptive spatiotemporal distribution of soil microfungi in 'Evolution Canyon' III, Nahal Shaharut, extreme southern Negev Desert, Israel. Biol. J. Linn. Soc. 2007, 90, 263–277. [Google Scholar] [CrossRef]
- Watrud, L.S.; Martin, K.; Donegan, K.K.; Stone, J.K.; Coleman, C.G. Comparison of taxonomic, colony morphotype and PCR-RFLP methods to characterize microfungal diversity. Mycologia 2006, 98, 384–392. [Google Scholar] [CrossRef]
- de Hoog, G.S.; van den Ende, A. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses 1998, 41, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evo. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Chen, T.; Liu, Y.X.; Huang, L. ImageGP: An easy-to-use data visualization web server for scientific researchers. iMeta 2022, 1, n/a-n/a. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The mathematical theory of communication. University of Illinois Press: Urbana, 1964.
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Armstrong, R.A. When to use the Bonferroni correction. Ophthalmic & Physiological Optics the Journal of the British College of Ophthalmic Opticians 2015, 34, 502–508. [Google Scholar]
- Gehan, E.A. A generalized Wilcoxon test for comparing arbitrarily singly-censored samples. Biometrika 1965, 52, 203–224. [Google Scholar] [CrossRef]
- Yurkov, A.M.; Kemler, M.; Begerow, D. Species Accumulation Curves and Incidence-Based Species Richness Estimators to Appraise the Diversity of Cultivable Yeasts from Beech Forest Soils. Plos One 2011, 6, e23671. [Google Scholar] [CrossRef]
- Wong Chin, J.M.; Puchooa, D.; Bahorun, T.; Jeewon, R. Antimicrobial properties of marine fungi from sponges and brown algae of Mauritius. Mycology 2021, 12, 231–244. [Google Scholar] [CrossRef]
- Jiang, X.L.; Ding, W.L.; Xing, X.K. Effects of different media on the isolation of symbiotic fungi from protocorm of medicinal orchid Gymnadenia conopsea. Mycosystema 2022, 41, 952–961. [Google Scholar] [CrossRef]
- Liu, S.Y.; Wang, Q.Z.; Xu, H.X.; Huang, S.Z.; Feng, J. Effects of common medium and optimized medium on fungal isolation and diversity in Fangchenggang Sea Area. Guangxi Sciences 2021, 28, 65–73. [Google Scholar] [CrossRef]
- White, J.F.; Morrow, A.C.; Morgan-Jones, G.; Chambless, D.A. Endophyte-host associations in forage grasses. XIV. Primary stromata formation and seed transmission in Epichloe typhina: Developmental and Regulatory Aspects. Mycologia 1991, 83, 72. [Google Scholar] [CrossRef]
- de Errasti, A.; Carmarán, C.C.; Novas, M.V. Diversity and significance of fungal endophytes from living stems of naturalized trees from Argentina. Fungal Divers. 2010, 41, 29–40. [Google Scholar] [CrossRef]
- Guo, L.D. Advances of researches on endophytic fungi. Mycosystema 2001, 20, 148–152. [Google Scholar] [CrossRef]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? Fems Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef]
- Abbott, M.J.; Brewer, J.S. Competition does not explain the absence of a carnivorous pitcher plant from a nutrient-rich marsh. Plant Soil 2016, 409, 495–504. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J.J.; Zhang, T.Y. Advancement on soil fungal research. Journal of Fungal Research 2005, 3, 52–58. [Google Scholar] [CrossRef]
| Collection Time | Location | Growth Stage | Plants Number | Soil Number | Treatment-Row/Pot Number |
|---|---|---|---|---|---|
| Jul. 20, 2022 | field | seedling | 16 | 16 | CC-4; CT-4; CF-4; TF-4 |
| Aug. 5, 2022 | greenhouse | seedling | 16 | 16 | |
| Aug. 14, 2022 | field | flowering | 16 | 16 | |
| Aug. 21, 2022 | greenhouse | Vine growth | 16 | 16 | |
| Jul. 20, 2022 | field | seedling | 16 | 16 | |
| total | 64 | 64 |
| Strains No. | Species Name | Per.Ident | Accession No. in GenBank of Reference Strain | Strains No. | Species Name | Per.Ident | Accession No. in GenBank of Reference Strain | ||
|---|---|---|---|---|---|---|---|---|---|
| GSSCF15Z4-6 | Acremonium camptosporum | 97.49% | MH859415.1 | FPSCC39RR-1 | Globisporangium recalcitrans | 100.00% | KJ716861.1 | ||
| GSVCC22P3-7 | Acremonium felinum | 100.00% | AB540563.1 | GPSCC05RR-1 | Glomerella lagenaria | 100.00% | AJ301965.1 | ||
| GSVCC22P3-8 | Acremonium persicinum | 99.85% | MH864511.1 | GSSCC05P3-4 | Graphium basitruncatum | 98.45% | EF165016.1 | ||
| GSSCF14Z3-9 | Acremonium sclerotigenum | 97.68% | KJ194115.1 | GPVTF34RS-3 | Harknessia ellipsoidea | 97.24% | MH865961.1 | ||
| GPVCC21CR-2 | Acrocalymma vagum | 99.55% | KJ188710.1 | FSSCF48R3-14 | Humicola fuscoatra var. fuscoatra | 98.81% | MH863738.1 | ||
| FSSCC42P3-11 | Actinomortierella ambigua | 98.76% | JX976067.1 | GPSCC07PS-1 | Humicola variabilis | 99.09% | MH863772.1 | ||
| GSVCC21Z3-4 | Albifimbria verrucaria | 98.51% | KU845891.1 | FSSCC41E4-1 | Irpex lacteus | 99.62% | OL685330.1 | ||
| GPVCC22EL-1 | Alternaria alternata | 100.00% | LC440583.1 | FSSCC39P3-4 | Lectera colletotrichoides | 98.04% | AJ301962.1 | ||
| FSFCC58Z3-7 | Alternaria atra | 99.70% | MH864090.1 | FSFCF64P3-3 | Linnemannia gamsii | 97.30% | MH859222.1 | ||
| FSFCF65Z3-8 | Antarctomyces psychrotrophicus | 97.19% | NR_164292.1 | GPVCT24PR-2 | Macrophomina phaseolina | 100.00% | EF570500.1 | ||
| GSVCC21P4-2 | Apiospora rasikravindrae | 99.58% | KF144915.1 | GSVCC20Z3-2 | Malassezia restricta | 99.87% | EU400587.1 | ||
| FSFCT60Z3-6 | Aspergillus alabamensis | 97.77% | MH863630 | GSSCC06Z3-9-2 | Marquandomyces marquandii | 97.24% | AB099511.1 | ||
| FSSCF50P3-6 | Aspergillus baeticus | 99.42% | MT990731.1 | FSFCT62P3-8-1 | Metarhizium anisopliae | 99.54% | FJ545329.1 | ||
| GSSCF14Z3-12 | Aspergillus clavatus | 97.24% | NR_121482.1 | GSSCC06Z3-2 | Meyerozyma guilliermondii | 99.88% | MH545918.1 | ||
| FSFCC58P3-1 | Aspergillus eucalypticola | 99.86% | OL711732.1 | FSFCF64Z3-4 | Microascus brevicaulis | 99.85% | MT732896.1 | ||
| FSSCC40Z4-15 | Aspergillus flavus | 100.00% | MH279408.1 | GPSCC04PS-1 | Microdochium musae | 97.14% | MH107898.1 | ||
| GPSCC04RS-1 | Aspergillus heldtiae | 98.71% | MK450656.1 | FPSCF50CS-1 | Moesziomyces aphidis | 99.03% | JQ743064.1 | ||
| FSFCC56Z3-7 | Aspergillus minisclerotigenes | 100.00% | MH279386.1 | GPVCC20PS-4 | Monosporascus cannonballus | 99.84% | JQ771932.1 | ||
| GPSCC05PS-1 | Aspergillus niger | 99.86% | MT990733.1 | FPFCT61CR-2 | Monosporascus ibericus | 97.53% | JQ973832.1 | ||
| FSSCT43Z4-7 | Aspergillus nomiae | 99.71% | MH279387.1 | GSSCC04Z3-9 | Mortierella alpina | 99.87% | AJ271630.1 | ||
| GSSCT09P3-5 | Aspergillus pallidofulvus | 98.81% | MK450639.1 | FPFCC58ER-3 | Mucor aseptatophorus | 97.89% | MZ433252.1 | ||
| FSSCC42Z3-7 | Aspergillus pseudodeflectus | 99.25% | MK450642.1 | FSSCC42P3-19 | Mucor circinelloides | 99.37% | MH854919.1 | ||
| FSSCC41Z4-1 | Aspergillus pseudonomiae | 97.29% | MH279417.1 | FSSCC41E4-2 | Mucor fragilis | 99.85% | GU566275.1 | ||
| GSSCC06Z3-5 | Aspergillus seifertii | 97.37% | MK450648.1 | GSVCC23P3-7 | Mucor hiemalis | 100.00% | MF782793.1 | ||
| FSFCC57Z3-3 | Aspergillus sigurros | 97.77% | MK450650.1 | GSVCF31Z3-6 | Nalanthamala vermoesenii | 97.12% | JX456473.1 | ||
| GPVCC23PS-2 | Aspergillus sydowii | 99.70% | MT990722.1 | FSFTF68Z3-3 | Ochroconis tshawytschae | 90.06% | MH858782.1 | ||
| GSSCC04P3-2 | Aspergillus uvarum | 100.00% | OL711726.1 | FSFCF64P3-4 | Paraisaria heteropoda | 100.00% | AB084157.1 | ||
| GPSCC05PL-2 | Aureobasidium melanogenum | 99.85% | MH863678.1 | GSVCC23Z3-6 | Paramyrothecium humicola | 99.71% | MH864508.1 | ||
| FSSCC39Z3-1 | Bisifusarium delphinoides | 99.82% | KP132212.1 | GSVCF31P3-8 | Paramyrothecium roridum | 99.66% | HQ115647.1 | ||
| FSSCF48P3-1 | Boeremia exigua var. exigua | 97.48% | EU343168.1 | FSFTF70P3-2 | Paramyrothecium viridisporum | 99.69% | MH864343.1 | ||
| FPFTF68CR-1 | Boeremia exigua var. populi | 97.24% | MN973161.2 | FSSCC39Z3-11 | Penicillium aurantiocandidum | 100.00% | MH861314.1 | ||
| FSSCC42P3-6 | Calonectria ciliata | 97.82% | MH864520.1 | FSFCF65R4-2 | Penicillium cerradense | 98.53% | MT006127.1 | ||
| FPFCF64PS-3 | Cephalotheca sulfurea | 97.71% | AB278194.2 | GPVCC21RS-3 | Penicillium citrinum | 100.00% | MH858380.1 | ||
| GPVCC23ER-1 | Ceratobasidium chavesanum | 97.44% | NR_164016.1 | FSSTF54Z3-5 | Penicillium commune | 97.25% | GQ458026.1 | ||
| FSSCF50R3-2 | Chaetomidium leptoderma | 97.33% | NR_164219.1 | GSVCC21Z3-1 | Penicillium copticola | 99.84% | NR_121516.1 | ||
| FPSCC40ER-2 | Chaetomium elatum | 98.65% | MH871792.1 | FSSCF49P3-11 | Penicillium daleae | 96.56% | MH854984.1 | ||
| FPSCC41RS-4 | Chaetomium globosum | 99.85% | AB449671 | FSSCC41R3-10 | Penicillium flavigenum | 99.85% | MH862182.1 | ||
| FSSCF48Z3-3 | Chaetomium perlucidum | 99.84% | MH857726.1 | FSSCC40Z4-2 | Penicillium goetzii | 99.41% | MH859954.1 | ||
| GSSCC04P3-5 | Chaetomium piluliferum | 99.85% | MH861633.1 | FSSCC41Z3-6 | Penicillium lapidosum | 98.24% | MH860150.1 | ||
| GPSCC07RR-2 | Cladosporium acalyphae | 100.00% | MH863861.1 | FSFCC58P3-4 | Penicillium nothofagi | 99.13% | MH864386.1 | ||
| FSFCC58Z3-3 | Cladosporium anthropophilum | 99.85% | MF574171.1 | FSFCF65R3-2 | Penicillium oxalicum | 98.97% | LC386216.1 | ||
| GPSCT08PL-1 | Cladosporium dominicanum | 99.85% | MF472969.1 | GSVCC20P3-3 | Penicillium senticosum | 99.41% | MH860152.1 | ||
| GSSCC04P3-4 | Cladosporium halotolerans | 99.85% | MF473103.1 | FSSCC40Z3-7 | Penicillium sizovae | 99.70% | MH858522.1 | ||
| GSSCC06P3-11 | Cladosporium parahalotolerans | 99.85% | MF473160.1 | FSSCC41R3-13 | Phoma herbarum | 98.91% | AY337712.1 | ||
| GSSCC06Z3-7 | Cladosporium sphaerospermum | 99.70% | MF473271.1 | GPSCC04ES-1 | Plectosphaerella cucumerina | 98.58% | MH858371.1 | ||
| GPSCC04RR-2 | Cladosporium tenuissimum | 99.85% | MH864840.1 | GPVCC21ER-3 | Preussia terricola | 97.78% | MH858589.1 | ||
| GSVCC21Z3-3 | Cladosporium velox | 100.00% | MF473310.1 | GSSCC04P3-6 | Pseudogymnoascus pannorum | 99.85% | MH864459.1 | ||
| GPVTF35EL-4 | Cladosporium westerdijkiae | 100.00% | MF473314.1 | FSSCC41P3-5 | Purpureocillium lilacinum | 99.58% | AB103380.1 | ||
| GPVTF33PL-1 | Cladosporium xanthochromaticum | 99.54% | MF473316.1 | GSSTF18Z4-3 | Pyricularia oryzae | 97.13% | CP034204 | ||
| GPVTF33RL-2 | Cladosporium xylophilum | 100.00% | MH863875.1 | FPSCF50PS-1 | Pythium amasculinum | 99.68% | AY598671.2 | ||
| GSSCC06Z3-4 | Clonostachys rosea | 99.41% | KX958035.1 | GPVCC22ES-2 | Retroconis fusiformis | 97.66% | EU040239.1 | ||
| FSSCC41R3-14 | Collariella gracilis | 97.45% | MH864437.1 | FPSCC42CR-2 | Rhizoctonia solani | 97.01% | MH862760.1 | ||
| FPSCC40ES-2 | Colletotrichum dematium | 99.56% | AJ301954.1 | FSSCF49P4-2 | Rhizopus arrhizus | 99.05% | AB109754.1 | ||
| GPSCC05CL-1 | Colletotrichum gloeosporioides | 98.51% | AJ301908.1 | GSSCF12Z3-15 | Sarocladium kiliense | 99.66% | MF682447.1 | ||
| GSVCC22R3-2 | Coniochaeta fasciculata | 99.07% | MH855947.1 | FSSCF49P3-15 | Schizophyllum commune | 99.23% | AF350925.1 | ||
| FSSCC41P3-7 | Coniochaeta mutabilis | 99.23% | MH856122.1 | FPSTF51PS-4 | Stagonosporopsis caricae | 100.00% | MH863092.1 | ||
| GSVCC23P3-5 | Cunninghamella echinulata | 97.36% | GQ221208.1 | FSSCC39Z3-13 | Stagonosporopsis cucurbitacearum | 98.11% | EU167573.1 | ||
| GPSCF13EL-1 | Curvularia americana | 97.03% | NR_146239.1 | GSSCF13Z3-9 | Striaticonidium brachysporum | 99.84% | MH860035.1 | ||
| FSSCF50R3-10 | Didymella pomorum | 99.06% | MN983931.1 | GSSCC04P3-7 | Talaromyces amestolkiae | 100.00% | MH856395.1 | ||
| FPSCC40CS-2 | Didymella rosea | 99.63% | KT287020.1 | FPFCC55RS-3 | Talaromyces calidominioluteus | 97.93% | NR_175199.1 | ||
| GPSCC07CR-1 | Edenia gomezpompae | 97.79% | NR_156217.1 | FSFCF65P3-11 | Talaromyces liani | 100.00% | MH858781.1 | ||
| FSFCC55P4-3 | Engyodontium album | 97.93% | HM214541.1 | GSVCC20P3-5 | Talaromyces macrosporus | 97.03% | MH860495.1 | ||
| FSSCC42Z3-18 | Fusarium anthophilum | 97.41% | MH864506.1 | GSSCC06Z3-9-1 | Talaromyces pinophilus | 100.00% | CP017345.1 | ||
| GSSCC04P3-10 | Fusarium falciforme | 99.85% | MT251174.1 | FSSCF47P3-13 | Talaromyces purpureogenus | 97.59% | KM086709.1 | ||
| FSFCC57P3-5 | Fusarium fujikuroi | 99.85% | CP023090.1 | FSSCF47R3-4 | Thelonectria olida | 98.66% | MH857841.1 | ||
| FPSCF47CS-1-2 | Fusarium graminearum | 99.69% | HG970335.2 | GPSCC07RR-1 | Trametes hirsuta | 97.64% | AF516556.1 | ||
| GSVCC22R3-5 | Fusarium napiforme | 99.00% | MH862670.1 | FSSCC40Z3-11 | Trichocladium griseum | 98.79% | KU705826.1 | ||
| GPVTF34ER-1 | Fusarium oxysporum | 99.85% | LC383471.1 | GSSCC05P3-9 | Trichoderma harzianum | 97.82% | MF782824.1 | ||
| FSFCC58Z3-6 | Fusarium solani | 100.00% | MH864517.1 | FSSCC41R3-12 | Trichoderma pleuroticola | 98.49% | MH864423 | ||
| GPSCC04ES-2 | Fusarium tricinctum | 99.80% | MH931273.1 | FSFCC58Z3-2 | Trichoderma pseudokoningii | 99.72% | FJ605099.1 | ||
| FSSCC41R3-11 | Fusicolla acetilerea | 99.82% | EU860058.1 | GPSCC07RR-1 | Trichoderma simmonsii | 97.18% | CP075868.1 | ||
| FSFCF66Z3-4 | Geomyces asperulatus | 97.13% | MH861038.1 | FSSCF50P4-5 | Umbelopsis isabellina | 97.78% | MZ078794.1 | ||
| FSSCC42Z3-15 | Gibellulopsis nigrescens | 99.85% | HE972037.1 | ||||||
| Fungal Type | Media | Number of Genera | Strain Number | Specific Genera compared to PDA |
|---|---|---|---|---|
| endophytic fungi(total 44 genera 504 strains) | CMA | 26 | 138 | Acrocalymma; Boeremia; Clonostachys; Graphium; Moesziomyces1; Mucor; Paramyrothecium |
| MEA | 26 | 121 | Ceratobasidium; Curvularia; Mucor; Paramyrothecium; Preussia | |
| rhizosphere soil fungi(total 61genera 771strains) | CZA | 45 | 290 | Antarctomyces; Bisifusarium; Edenia; Geomyces; Marquandomyces; Meyerozyma; Microascus; Monosporascus; Nalanthamala; Ochroconis; Pyricularia; Sarocladium; Striaticonidium; Trichocladium |
| endophytic fungirhizosphere soil fungitotal fungi(74genera 1275strains) | PDA | 32 | 114 | - |
| 45 | 331 | |||
| 55 | 445 | |||
| endophytic fungi | MRBA | 27 | 131 | Globisporangium; Graphium; Harknessia; Mucor; Trametes |
| rhizosphere soil fungi | 33 | 150 | Chaetomidium; Irpex; Monosporascus | |
| total fungi | 45 | 281 | Chaetomidium; Globisporangium; Harknessia; Irpex;Trametes |
| Genera | Isolated with 5 types of media | Isolated with 3 types of rhizosphere soil fungi culture media | Isolated with 4 types of endophytic fungi culture media | Isolated from roots, stems and leaves of cucumber plants | |
|---|---|---|---|---|---|
| 1 | Acremonium | + | |||
| 2 | Acrocalymma | + | |||
| 3 | Albifimbria | + | |||
| 4 | Alternaria* | + | + | + | + |
| 5 | Apiospora | + | |||
| 6 | Aspergillus | + | + | + | + |
| 7 | Chaetomium | + | + | + | |
| 8 | Cladosporium | + | + | + | + |
| 9 | Clonostachys | + | |||
| 10 | Colletotrichum | + | + | + | + |
| 11 | Coniochaeta | + | + | ||
| 12 | Edenia | + | + | ||
| 13 | Fusarium | + | + | + | + |
| 14 | Humicola | + | |||
| 15 | Lectera | + | |||
| 16 | Malassezia | + | |||
| 17 | Microdochium | + | + | ||
| 18 | Monosporascus | + | + | + | |
| 19 | Mortierella | + | |||
| 20 | Mucor | + | + | ||
| 21 | Paramyrothecium | + | + | ||
| 22 | Penicillium | + | + | + | + |
| 23 | Phoma | + | |||
| 24 | Plectosphaerella | + | + | + | + |
| 25 | Pseudogymnoascus | + | + | + | |
| 26 | Stagonosporopsis | + | + | + | + |
| 27 | Talaromyces | + | + | + | + |
| 28 | Trichoderma | + | + | + | + |
| Genus | CMA | MEA | PDA | MRBA |
|---|---|---|---|---|
| root | 5.50a AB1 | 2.50b B | 3.00b AB | 6.50a A |
| stem | 7.75a AB | 6.25a AB | 8.75a B | 4.50a A |
| leaf | 4.25a A | 3.75ab A | 4.75b A | 3.25a A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





