1. Introduction
Causal analysis has emerged as a powerful tool for interpreting time series data across various research fields, fueled by the synergistic interplay between theory and application. A groundbreaking development in this area was a proposal of Granger Causality (GC) [
1], which tests the significance of dynamic influence between pairwise time series. Building upon this foundation, Geweke [
2] expanded GC to encompass multivariate time series, introducing conditional Granger causality (cGC). Bressler et al. [
3] and Schiatti et al. [
4] further refined cGC, enabling the estimation of causality among multivariate time series using vector autoregressive (VAR) models.
A key strength of cGC lies in its capacity to compare the magnitude of residual variance between two models. As the significance of causality estimates depends solely on residual variance magnitude, not model structure, these methods offer a flexible and versatile approach applicable to a wide array of time series models, including non-linear variants. However, GC and its derivatives have limitations in capturing dynamic characteristics of variable influence propagation. To overcome this, impulse response function (IRF) analysis has been extensively adopted for examining variable responses to shocks within an identified model.
Conceptually, the statistical significance of numerically calculated cGC and IRF should be assessed using the asymptotic method. However, real-world data analysis may yield incorrect results due to potential distribution discrepancies. In cases where the distribution function is unknown, such as with partial Granger causality [
5], the asymptotic method is inapplicable. The bootstrap strategy serves as a widely used alternative for cGC and IRF significance testing. However, with numerous bootstrap methods available, each with varying sensitivity and specificity, selecting the appropriate method remains a critical challenge when applying time series models to real data analysis.
In this study, we thoroughly evaluate the performance of four bootstrap methods designed for time series analysis. First, the uncorrelated phase-randomized bootstrap (uPRB) detects non-linearity latent in time series by transforming the original data into the spectral domain, randomizing the phase for each frequency, and transforming the result back into the time domain [
6]. We added random sequences uncorrelated for each variable to facilitate linear causality detection.
The first method is a modification of the phase randomise bootstrap (FRB) method to detect linear causality among variables. The original FRB generates surrogate time series data for hypothesis testing by randomizing the phase of each frequency component in the Fourier transform of the original time series data while preserving its amplitude spectrum. This method generates surrogate data with the same power spectrum and distribution properties as the original data but with disrupted temporal correlations. Researchers can use this method to detect non-linear dynamics in various fields by comparing the original data’s statistical properties to the surrogate data. To detect causality among variables, we introduced a modification to the method called uncorrelated Phase-randomized bootstrap (uPRB) by preparing random sequences independently for each variable.
While uFRB is a method to randomize the phase in the frequency domain, for comparison, as a second method we prepare a method to randomize the transfer in the time domain based on circular Block Bootstrap (CBB) method proposed by Politis and Romano [
7]. The CBB method is a resampling technique that addresses the issue of selecting the first and last parts of a time series data less frequently in the surrogate data generation process. To resolve this issue, circular block bootstrap combines the endpoints of the time series data, effectively creating a circular time series. By doing so, it ensures that all parts of the data are equally likely to be sampled in the surrogate data generation process. If the CBB method limits the number of blocks to 1, it can produce surrogate data in which the entire original data is randomly shifted back and forth in the time domain. The technique known as Time-shifted surrogates (TSS) was initially introduced by Quiroga el al. [
8]. Subsequent advancements in this approach were presented in [
9]. By maintaining the same state space trajectory, the TSS ensure the complete preservation of all characteristics of the initial signal.
Third, the Stationary Bootstrap (SB) calculates standard errors and constructs confidence regions for weakly dependent stationary observations [
10]. Employing random block lengths with a geometric distribution ensures weak stationarity in resampled pseudo-time series, making it an invaluable approach for stationary time series data analysis.
Finally, the AR-sieve bootstrap (ARSB) is a resampling method for time series data based on autoregressive (AR) models. This technique approximates the underlying data-generating process by fitting an AR model to capture the time series’ dependence structure [
11,
12]. The ARSB method generates resampled series by applying the bootstrap procedure to the model’s residuals, offering a simple and computationally efficient solution for handling dependent data in time series analysis.
2. Causal Analysis
2.1. Conditional Granger Causality (cGC)
Let
denote the vector variables at time
t,
, where
N denotes the length of the time series. The feedback system among the variables
can be represented by a basic vector autoregressive (VAR) model of order
p, defined as
where
denotes a set of
-dimensional coefficient matrices, and
denotes a series of shocks (disturbances), given by white noise vectors with zero mean.
Extracting the
lth variable of the VAR model gives
where
denotes the
lth row of
. Equation (
2) represents an autoregressive model with exogenous input (ARX model) that consists of an endogenous part of the
lth variable and an exogenous part of all other variables.
The ARX model, excluding the
mth variable, is called the restricted ARX (rARX) model:
where
denotes the vector
with the
mth element excluded (e.g.
,
and so on) and
denotes a set of re-estimated
-dimensional coefficient matrices. Suppose that the past values of the
mth variable
contribute to the prediction of the
lth variable
; then, the variances of the residuals should satisfy
. The significance of the difference in variances can be evaluated by a likelihood ratio test:
2.2. Impulse Response Function (IRF)
There is another representation of the VAR model of Equation (
1) using a delay operator
L, such that
:
where
denotes the
-dimensional unity matrix. The roots (eigenvalues)
of the polynomial
need to fulfill
for all
j.
Further transformation of Equation (
5) yields
which demonstrates that a VAR model can be rewritten as
where
Equation (
8) represents the vector moving average (VMA) representation of the VAR model and can be denoted as VMA(
∞).
Partial differentiation of the VMA(
∞) model with respect to one particular shock (disturbance term)
yields a set of derivatives
The
th element of
is given by
, which represents the marginal effect (influence) from the
mth shock
to the
lth variable
. The function defining the time series of such marginal effects is called the impulse response function (IRF) and is denoted by
.
3. Bootstrap Methods
3.1. uncorrelated Phase Randomization Bootstrap (uPRB)
The Phase Randomization Bootstrap method has been proposed as a technique to generate surrogate time series data for hypothesis testing ([
6,
13]). Using the Fourier transform, the method transforms the original time series data into the frequency domain. Then, the phase of each frequency component is randomized while preserving the original amplitude spectrum. Finally, the surrogate data is obtained by applying the inverse Fourier transform to the randomized frequency components.
This process generates surrogate time series data with the same power spectrum as the original data but with disrupted temporal correlations. By comparing the original data’s non-linearity measures or other statistical properties to those of the surrogate data, researchers can test hypotheses and detect the presence of non-linear dynamics in the original time series data. This method has been applied in various fields, including the study of physiological signals ([
14]), economics and finance ([
15]), climate data ([
9]), and so on.
In order to preserve all linear auto-correlations and cross-correlations, the Phase Randomization Bootstrap adds a common random sequence to the phases of all variables. Thus, since this method cannot detect causality among variables, we prepare random sequences independently for each variable. In this paper we call this method uncorrelated Phase Randomization Bootstrap (uPRB).
The procedure for generating surrogate data sets by uPRB is as follows.
-
Step 1
Transform the original time series by applying Fourier transform to each variable as
where
l is the index of the variable, and
and
denote the amplitude and the phase, respectively.
-
Step 2
For each frequency
f, add an independent random value
following a uniform distribution throughout the interval
to
, while satisfying the symmetry property
. That is,
-
Step 3
Transform the spectral domain representation back to the time domain by applying the inverse Fourier transform to each variable as
-
Step 4
Repeat Steps 2-3 for all variables.
-
Step 5
Repeat Steps 2-4 a large number of times, thereby generating a set of surrogate data sets.
3.2. Stationary Bootstrap (SB)
The conventional Non-overlapping Block Bootstrap (NBB) has been proposed by Carlstein [
16]. By improving this conventional method, Künsch [
17] has proposed a Moving-Blocks Bootstrap (MBB). This method is useful, especially for small sample data having a wider range of blocks than the conventional method. However, in the process of random sampling, there is an edge effect of the uneven weighting of the selection at the beginning and at the end of the data. To compensate for this shortcoming, Politis and Romano [
7] have proposed a circular block bootstrap (CBB) that concatenates the start and end points of the original data. In the block bootstrap method, the stationarity of the sample is an important assumption. However, surrogate data obtained by resampling using the above mentioned method are not necessarily stationary. Therefore, the stationary bootstrap (SB) method has been proposed, in which the surrogate data are also stationary. This method is similar to the TSS method, except that the block width is not fixed but is resampled as a random variable following a geometric distribution [
10]. The procedure for generating surrogate data by SB is as follows.
-
Step 1
Set the mean block width to w. Then in the geometric distribution used in Step 3 we have .
-
Step 2
Duplicate the original data and merge it to the end of the original data such that .
-
Step 3
Generate a sequence of natural random numbers corresponding to each block width following a geometric distribution, so that the probability of the event is for Here, the value of K is determined to satisfy the condition .
-
Step 4
Generate a sequence of natural random numbers , corresponding to the index of the starting point of the block, following a uniform distribution over the interval .
-
Step 5
The blocks , constructed according to Steps 1 and 2, are arranged in the order in which they were extracted, and a pair of resamples is obtained with the first N elements as .
-
Step 6
Repeat Steps 3-5 a large number of times, thereby generating a set of surrogate data sets.
In the case of multivariate time series, there are two ways to perform Steps 3-5. One way is to use the same blocks for all variables for data shuffling, as described above, and the other is to perform Steps 3-5 for each variable independently. In this paper, we refer to the former as correlated SB (cSB) and to the latter as uncorrelated SB (ucSB).
3.3. Time Shift Surrogates (TSS)
While PRB is a method for generating surrogate data sets by randomizing the phases in the frequency domain,we evaluated the performance of TSS as a method of randomizing the phase in the time domain. This method corresponds to the special case of the CBB, where the number of blocks is limited to 1. The procedure for generating surrogate data sets by TSS is as follows.
-
Step 1
Duplicate the original data and merge it to the end of the original data such that
-
Step 2
-
Generate a natural random number s following a uniform distribution over the interval , extract a sequence of the data
, and use it as surrogate data set for the lth variable.
-
Step 3
Repeat Step 2 for all variables.
-
Step 4
Repeat Steps 2-3 a large number of times, thereby generating a set of surrogate data sets.
3.4. AR-Sieve Bootstrap (ARSB)
The AR-Sieve Bootstrap (ARSB) method generates surrogate data sets by feeding a VAR model, which employs re-estimated parameters, with residuals from modeling [
11,
12].
The procedure for generating surrogate data sets using ARSB is as follows.
-
Step 1
Fit the VAR model of Equation (
1) to the original time series, obtaining estimates for the parameters and the residuals
.
-
Step 2
-
Compute centered residuals , where
, and generate bootstrap residuals by shuffling the indices according to a sequence of natural random numbers which was drawn randomly with replacement from a uniform distribution over the interval .
-
Step 3
Compute the surrogate time series recursively by
where
.
-
Step 4
Re-estimate the VAR parameter matrices based on the bootstrap time series.
-
Step 5
Repeat Steps 2-4 a large number of times, thereby generating a set of surrogate data sets.
Similar to the SB method, there are two ways to process Step 2 in the ARSB method. One is to use the same sequence of random values for all variables for data shuffling, as mentioned above, and the other is to perform Step 2 independently for each variable. In this paper, we refer to the former as correlated ARSB (cARSB) and to the latter as uncorrelated ARSB (ucARSB).
4. Simulation
In order to verify the performance of the methods for causal analysis and of the bootstrap methods, as discussed above, we prepare a simulation model by modifying a model proposed by [
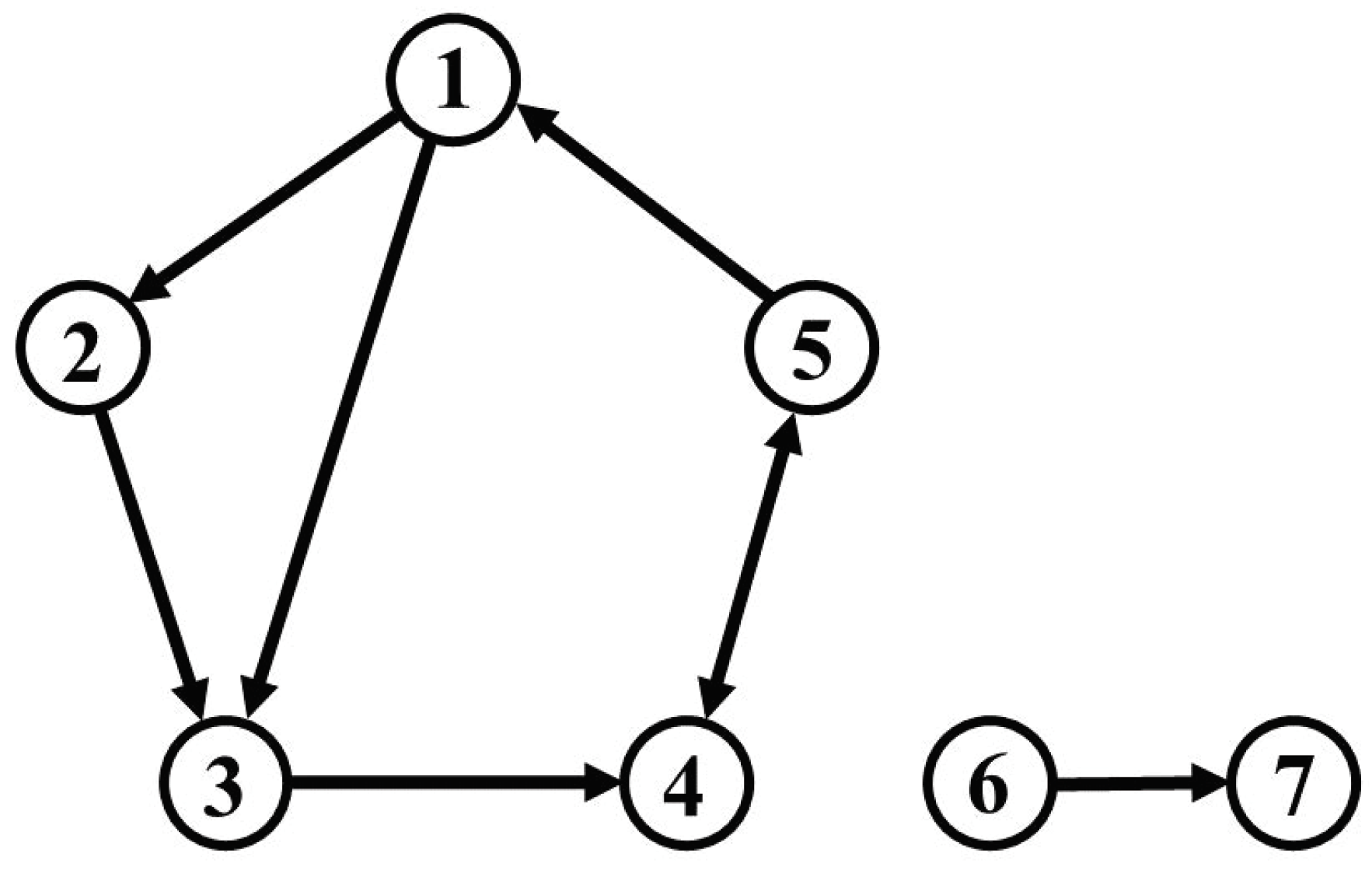
18]. We set up seven oscillatory variables, five of which are self-excited and coupled directly or indirectly by vector autoregressive (VAR) processes, constituting a dynamic feedback system. We fix the frequency of each variable to 0.1 Hz and randomly select a damping coefficient for each variable from a normal distribution. The 6th and 7th variables are isolated from the other five.
The 6th variable is self-excited at 0.1 Hz, like the other five, while the 7th variable is not self-excited, but driven only by the 6th variable. If causality would be detected between either the 6th or the 7th variables, and any of the 1st to 5th variables, this would represent a false positive. The connectivity diagram is shown in
Figure 1.
We set the connectivity among variables as shown in
Figure 1 and generate a simulated time series of length
through the VAR process of Equation (
1), with
. The nominal sampling frequency is 10 Hz, and the series of shocks (disturbances)
is sampled from a 7-dimensional multivariate Gaussian noise distribution with zero mean and diagonal covariance matrix given by
.
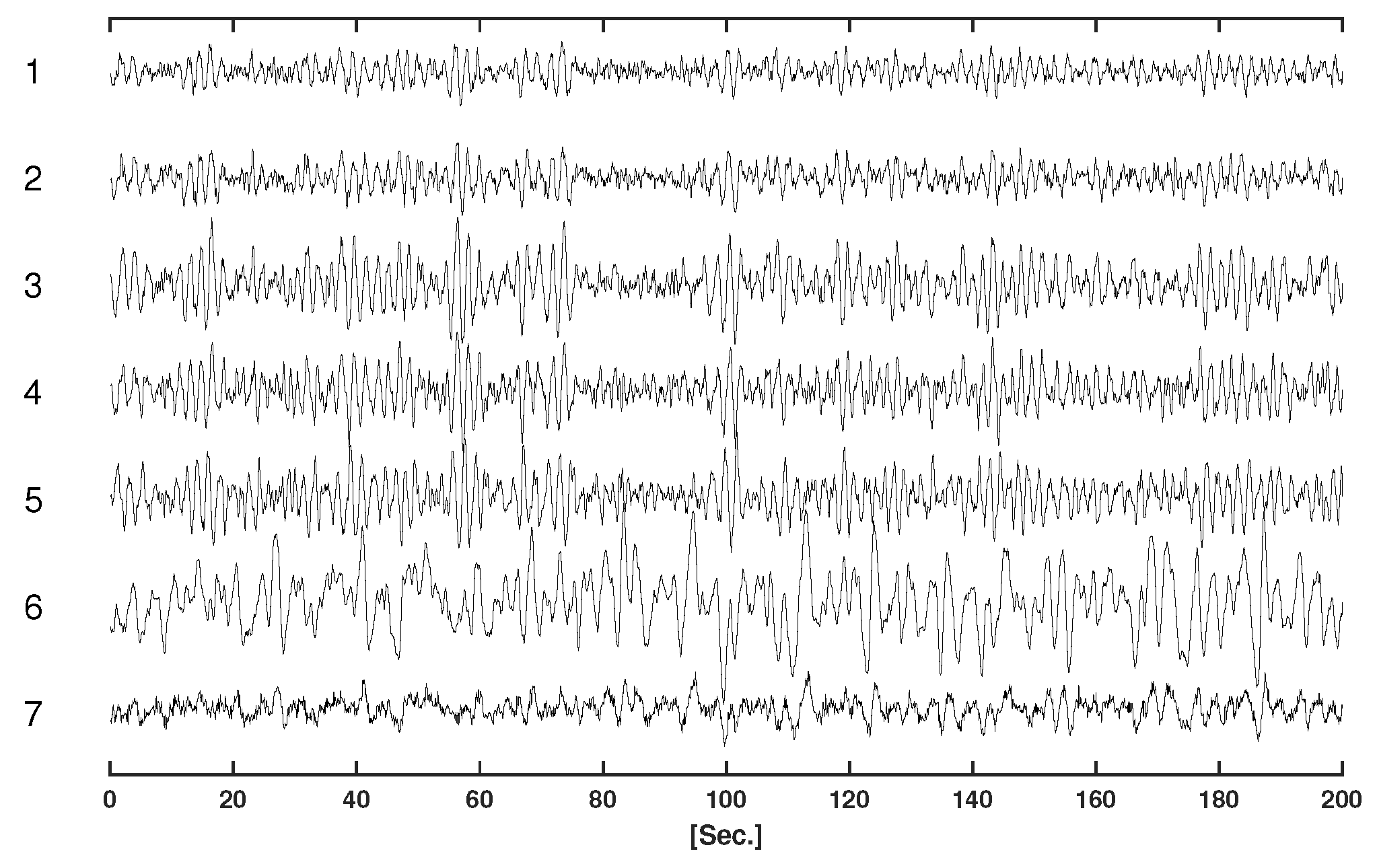
Table 1 displays the parameters of the VAR model for the simulations.
Figure 2 displays the simulated time series.
5. Results
In our pursuit of accurately estimating the 99% confidence intervals (CI) for both conditional Granger causality (cGC) and impulse response function (IRF) analyses, we generated 2000 surrogate data sets using each of the previously discussed bootstrap methods. For the SB method, we determined the optimal mean block width
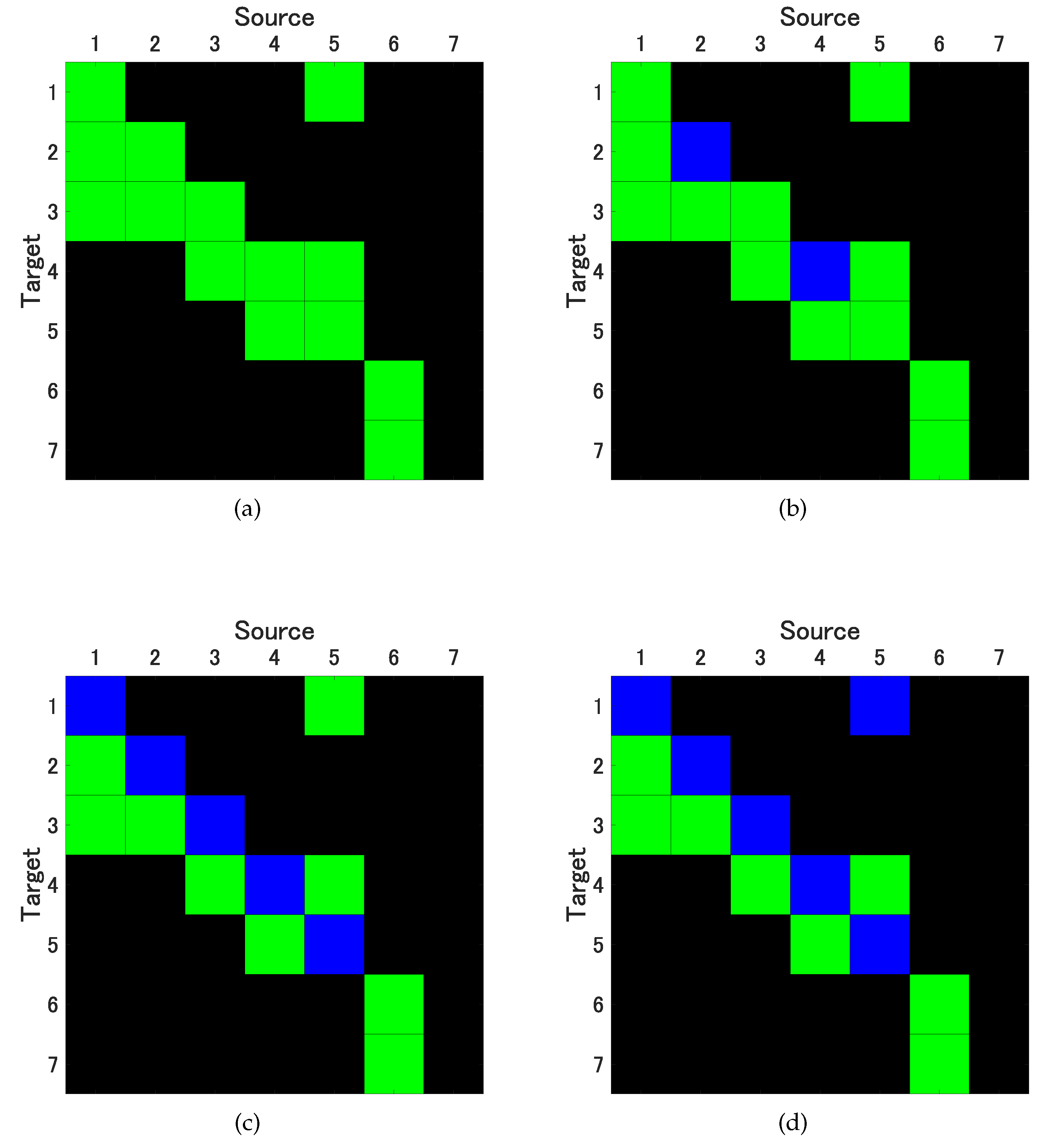
w, selecting values of 5, 10, 20, and 40. As illustrated in
Figure 3a, the cSB method consistently yields correct causality results, irrespective of the chosen mean block width. In contrast, the ucSB method produces correct causality results for
, but the detection rate of self-feedback diminishes as
w increases (see
Figure 3b,c).
While the uPRB method provides accurate causality results concerning variable interactions, it fails to detect the self-excitations of the 1st through 5th variables, yielding results akin to ucSB(20) and ucSB(40) (see
Figure 3c). If the time shift
s in the TSS method is excessively small or large, the data closely resembles the original data, consequently minimizing the phase randomization effect. We generate surrogate time series using the TSS method under the condition
.
Notably, the TSS method’s detection performance is inferior to that of the ucSB(20) and ucSB(40) methods, failing to detect causality from the 5th to the 1st variable. Importantly, our study identified no false positives within any bootstrap method.
Table 2 concisely summarizes the sensitivity and specificity for each combination of causal analysis method and bootstrap method .
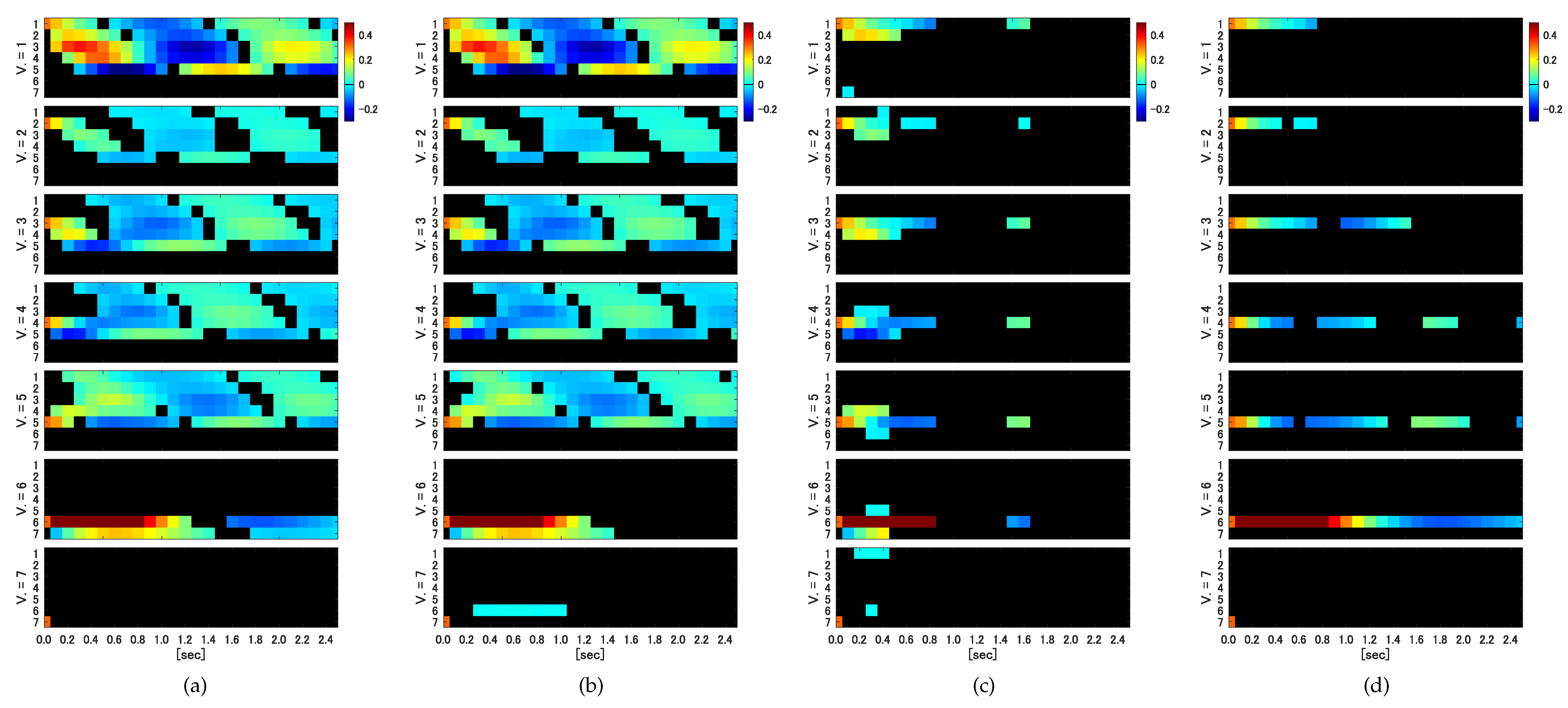
In the IRF analysis, only the ARSB method, whether correlated or uncorrelated, successfully detects both self-excitation and causality among variables in their entirety.
Figure 4a displays the IRFs for the impact assigned to each variable (
), with ucARSB evaluating the significance. This outcome aligns with the connectivity diagram depicted in
Figure 1. The cSB method demonstrates results almost identical to the ARSB method for the 1st through 5th variables. However, it detects a response from the 6th variable to the 7th variable, constituting a false positive, which was observed for all tested values of the mean block width. The uPRB method primarily detects causality between directly coupled variables (see
Figure 4c). Additionally, false positives emerge where responses occur between uncoupled variables, such as the reciprocal response between the 1st and 7th variables and between the 5th and 6th variables. For all tested values of the mean block width, the ucSB method exhibits the weakest detection performance among the evaluated bootstrap methods, detecting only self-excitation (see
Figure 4d).
To summarize the results of these IRFs like GC, we compared the true IRFs generated through the VAR parameters in
Table 1 with the IRFs detected by the respective bootstrap methods and summarized the sensitivity and specificity in
Table 2. The time interval of the IRFs used for comparison was the maximum time step before feedback occurs, i.e., the IRFs up to three time steps before the impact on the third variable propagates to the first variable via to fourth and fifth variables (see
Figure 1).
6. Discussion
In this study, we present a thorough comparison of the performance of four different bootstrap methods (encompassing twelve variations, including different values of mean block width, as well as correlated and uncorrelated derivations) to evaluate the results from causal analyses.
Our findings reveal that, for the cSB method, the confidence interval (CI) width decreases as the mean block width increases, which is particularly evident in
Figure 5a,b. A possible explanation is that a shorter block width introduces more discontinuities in the surrogate time series, causing the dynamic properties to deviate further from those of the original time series due to the random shuffling of blocks disrupting temporal forward/backward relationships. This disruption may lead to falsely detecting causality from the 7th to 6th variables in the IRF analysis, as seen in
Figure 4b. In contrast, the CI width for the ucSB method remains wider than that of the cSB method, irrespective of the mean block width (see
Figure 5a,c), or even increases with the mean block width (
Figure 5b). This stems from each block’s starting point being determined independently for each variable, causing unnecessary destruction of causality among variables in the surrogate time series. Consequently, no causality among variables is detected in the corresponding IRF analysis (see
Figure 4d).
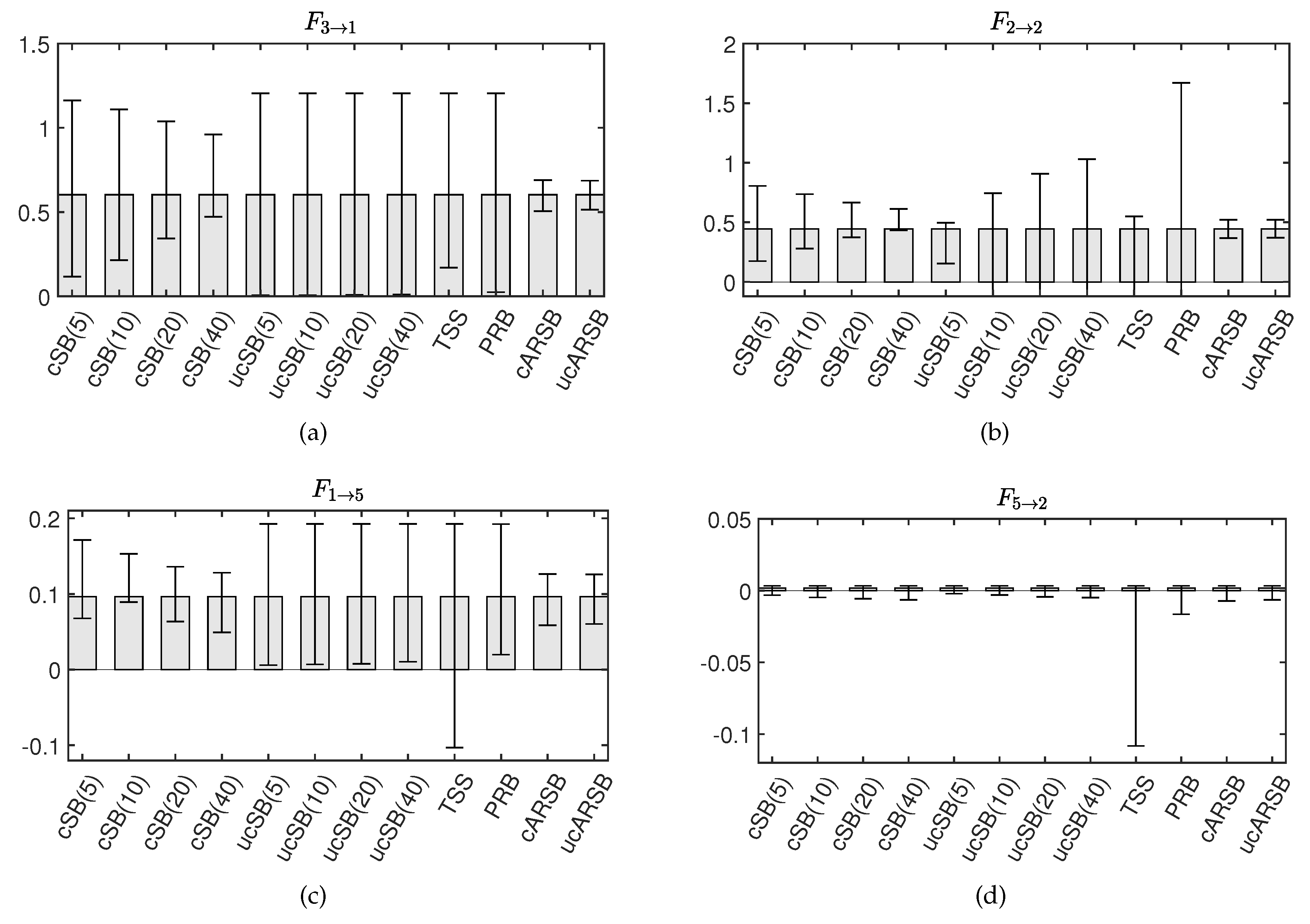
The TSS method accurately detects significance for the Granger causality
, but the CI widths for
and
are so large that they includes zero, resulting in false negatives. This could be attributed to the
th element of
having the smallest value among the VAR parameters corresponding to causality among variables, and the
th element of
having the smallest value among the VAR parameters corresponding to the attenuation rate of self-feedback. A prime example is
, which should be numerically ignorable since our simulation model does not contain causality from the 2nd to the 5th variable (see
Table 1). Although the TSS method correctly assesses non-significance, the CI width is substantially larger than that estimated by the other bootstrap methods. These results suggest that the TSS method may not provide stable estimates of bootstrap statistics, compared to other methods, when the statistic value of the original time series is small.
In contrast to other methods, the uPRB method’s algorithm does not require
a priori tuning of parameters, such as a mean block width for the SB method. Upon providing the original time series, surrogate time series can be generated almost instantaneously. The uPRB method is characterized by its simplicity and the ability to generate surrogate time series devoid of discontinuous time points, while preserving stationarity. However, a limitation of this method is its inability to detect self-feedback (see
Figure 5b). Although it accurately identifies causality among variables, some of the confidence intervals (CIs) have considerable width, which questions its reliability (see
Figure 5a). By constraining the interval of phase randomization in Step 2, the CI width may be reduced, thereby enhancing performance; however, adjusting the degree of restriction remains arbitrary. A key distinction between the TSS and uPRB methods is that the former shifts the phases in all frequency bands simultaneously, while the latter does so for each frequency band. As the uPRB method can selectively disturb the phases corresponding to a specific frequency band, it may prove valuable for evaluating causality in the spectral domain.
For both cGC and IRF analyses, the ARSB method outperforms the other bootstrap methods, regarding detection performance. For this method, determining the VAR model order
p is essential, which can be accomplished using the Akaike Information Criterion (AIC). As outlined in
Section 3.4, there are two derivations: correlated ARSB (cARSB), which randomly shuffles the residuals across all variables synchronously, and uncorrelated ARSB (ucARSB), which shuffles the residuals independently for each variable. Our simulation study demonstrates that both derivatives yield nearly identical results. However, observational errors (artifacts) may be concurrently superimposed onto the data when analyzing actual time series, rendering ucARSB potentially more effective for canceling noise correlation among variables.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Numbers 19K12212 and 21H04874.
References
- Granger, C.W.J. Investigating causal relations by econometric models and cross-spectral methods. Econometrica 1969, 37, 424–438. [CrossRef]
- Geweke, J. Measures of conditional linear dependence and feedback between time series. Journal of the American Statistical Association 1984, 79, 907–915. [CrossRef]
- Bressler, S.L.; Seth, A.K. Wiener-Granger causality: a well established methodology. Neuroimage 2011, 58, 323–329. [CrossRef]
- Schiatti, L.; Nollo, G.; Rossato, G.; Faes, L. Extended Granger causality: a new tool to identify the structure of physiological networks. Physiological Measurement 2015, 36, 827–843. [CrossRef]
- Guo, S.; Seth, A.K.; Kendrick, K.M.; Zhou, C.; Feng, J. Partial Granger causality – eliminating exogenous inputs and latent variables. Journal of Neuroscience Methods 2008, 172, 79–93. [CrossRef]
- Prichard, D.; Theiler, J. Generating surrogate data for time series with several simultaneously measured variables. Physical Review Letters 1994, 73, 951–954. [CrossRef]
- Politis, D.N.; Romano, J.P. A circular block resampling procedure for stationary data. In Exploring the limits of bootstrap; LePage, R.; Billard, L., Eds.; John Wiley & Sons: New York, 1992; pp. 263–270.
- Quiroga, R.Q.; Kraskov, A.; Kreuz, T.; Grassberger, P. Performance of different synchronization measures in real data: a case study on electroencephalographic signals. Physical Review E 2002, 65, 041903. [CrossRef]
- Ashkenazy, Y.; Baker, D.R.; Gildor, H.; Havlin, S. Nonlinearity and multifractality of climate change in the past 420,000 years. Geophysical Research Letters 2003, 30, 2146. [CrossRef]
- Politis, D.N.; Romano, J.P. The stationary bootstrap. Journal of the American Statistical Association 1994, 89, 1303–1313. [CrossRef]
- Bühlmann, P. Sieve bootstrap for time series. Bernoulli 1997, 3(2), 123–148. [CrossRef]
- Berg, A.; Paparoditis, E.; Politis, D. A bootstrap test for times series linearity. Journal of Statistical Planning and Inference 2010, 140, 3841–3857. [CrossRef]
- Theiler, J.; Eubank, S.; Longtin, A.; Galdrikian, B.; Farmer, J.D. Testing for nonlinearity in time series: the method of surrogate data. Physica D 1992, 58, 77–94. [CrossRef]
- Stam, C.J.; Breakspear, M.; van Walsum, A.M.v.C.; van Dijk, B.W. Nonlinear synchronization in EEG and whole-head MEG recordings of healthy subjects. Human Brain Mapping 2003, 19, 63–78. [CrossRef]
- Soofi, A.S.; Galka, A.; Li, Z.; Zhang, Y.; Hui, X. Applications of methods and algorithms of nonlinear dynamics in economics and finance. Complexity in Economics: Cutting Edge Research 2014, pp. 1–30. [CrossRef]
- Carlstein, E. The use of subseries values for estimating the variance of a general statistic from a stationary sequence. The Annals of Statistics 1986, 14, 1171–1179. [CrossRef]
- Künsch, H.R. The jackknife and the bootstrap for general stationary observations. Ann. Statist. 1989, 17, 1217–1241. [CrossRef]
- Baccalá, L.A.; Sameshima, K. Overcoming the limitations of correlation analysis for many simultaneously processed neural structures. Progress in Brain Research 2001, 130, 33–47. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).